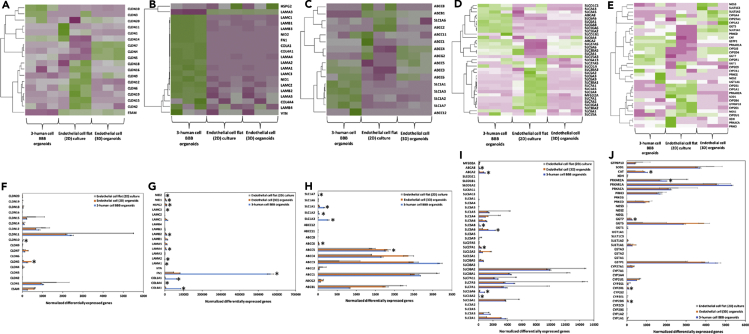
Figure 6.
Transcriptomic (RNA-Seq) analysis
Heatmap of RNA-Seq and differentially expressed genes (DEGs) upregulated analysis of 3-human cell spheroids and 2D and 3D endothelial cell monocultures (n = 3 for each culture condition). Green and pink indicate up-regulation and down-regulation, respectively. Average of hierarchical clustering indicates the interclass correlation between all three groups. Selected differential expression of genes encoding for (A and F) tight junction proteins, (B and G) extracellular matrix (ECM) proteins, (C, D, H, and I) ABC efflux transporters, solute carriers (SLCs) and other nutrient transporters, and (E and J) metabolic enzymes. ∗Significantly differentially expressed genes (DEG) (padj < 0.05, | fold change | >2, base mean ≥ 20). To provide optional filtering criteria in addition to the padj, additional criteria of |fold change| >2 (|log2 fold change| >1) and average expression level higher than 20 (base Mean > 20) were used.