Abstract
Cereblon (CRBN), a substrate receptor of cullin 4-RING E3 ligase (CRL4) regulates the ubiquitination and degradation of c-Jun, mediating the lipopolysaccharide-induced cellular response. However, the upstream signaling pathway that regulates this process is unknown. In this study, we describe how endoplasmic reticulum (ER) stress reversely regulates sequestosome-1 (p62)and c-Jun protein levels. Furthermore, our study reveals that expression of p62 attenuates c-Jun protein levels through the ubiquitin-proteasome system. Conversely, siRNA knockdown of p62 elevates c-Jun protein levels. Immunoprecipitation and immunoblotting experiments demonstrate that p62 interacts with c-Jun and CRBN to form a ternary protein complex. Moreover, we find that CRBN knockdown completely abolishes the inhibitory effect of p62 on c-Jun. Using brefeldin A as an inducer of ER stress, we demonstrate that the p62/c-Jun axis participates in the regulation of ER stress-induced apoptosis, and that CRBN is required for this regulation. In summary, we have identified an upstream signaling pathway, which regulates p62-mediated c-Jun degradation. Our findings elucidate the underlying molecular mechanism by which p62/c-Jun axis regulates the ER stress-induced apoptosis, and provide a new molecular connection between ER stress and apoptosis.
Keywords: p62, c-Jun, CRBN, Endoplasmic reticulum stress, Apoptosis, Ubiquitination
INTRODUCTION
The ubiquitin-proteasome system (UPS) and the autophagy-lysosome pathway (ALP) are two major cellular pathways responsible for protein degradation (Ciechanover, 2005; Rubinsztein, 2006; Dikic, 2017). In the UPS, proteins are first modified by ubiquitin to form specific types of ubiquitinated proteins that are then targeted for degradation by the 26S proteasome (Grice and Nathan, 2016). In the ALP, autophagic substrates including soluble proteins, protein complexes and aggregates, are engulfed by a double-layered membrane to form autophagosomes, which are further directed to lysosomes for degradation by lysosomal hydrolases (Korolchuk et al., 2010). UPS inhibitors lead to the activation of ALP (Zhu et al., 2010; Wang et al., 2019) and ALP deficiency attenuates the UPS (Korolchuk et al., 2009; Tian et al., 2014). Therefore, there is crosstalk and interplay between these two pathways (Jian et al., 2017), and the autophagy receptor, sequestosome 1 (p62), plays important roles in both processes (Liu et al., 2016). On the one hand, p62 is a substrate of autophagy and can be degraded by the ALP by binding to microtubule-associated protein 1A/1B-light chain 3 (LC3) using its LC3-interacting region (Pankiv et al., 2007; Shvets et al., 2008). It can also be ubiquitinated and degraded through the UPS; a process that is regulated by E3 ligases including Parkin (Song et al., 2016) and X-linked inhibitor of apoptosis protein (Huang et al., 2018). On the other hand, p62 functions as a scaffold protein through its ubiquitin-associated (UBA) domain, and mediates the ubiquitination of proteins including NF-κB essential modulator (NEMO) in the presence of E3 ubiquitin ligase TRAF6 (Zotti et al., 2014). p62 is also responsible for shuttling the ubiquitinated proteins to the proteasome for degradation (Geetha et al., 2008). p62 may also inhibit the ubiquitination of proteins including NF-E2-related factor 2 (Nrf2) through binding to the E3 ligase complex component Keap1, thereby activating Nrf2 (Komatsu et al., 2010). In addition, p62 promotes the aggregation of ubiquitinated substrates for autophagic degradation (Pankiv et al., 2007; Zaffagnini et al., 2018). For example, in neurodegenerative diseases, p62 promotes the aggregation of misfolded proteins, including pathogenic ataxin-3 with expanded polyglutamine in Machado-Joseph disease (Zhou et al., 2014) and a mutant form of superoxide dismutase 1 (SOD1) in amyotrophic lateral sclerosis (Gal et al., 2007). Collectively, these studies indicate that p62 may regulate protein degradation by modulating E3 ligase-associated UPS and autophagy-associated ALP (Lippai and Lőw, 2014; Shin et al., 2020). These studies demonstrated the versatile roles played by p62 in the regulation of protein degradation in the UPS and ALP.
The UPS and ALP can regulate cell death and survival through the degradation of different substrates. However, the regulation of cell death and survival by p62 is somewhat controversial (Jin et al., 2009; Jain et al., 2010). It has been reported that p62 protects oxidative stress-induced cell death via the translocation of the transcription factor Nrf2 to the nucleus, thereby inducing the expression of its cytoprotective targets in a neuroblastoma cell line IMR-32 (Liu et al., 2007) and primary mouse hepatocytes (Ichimura et al., 2013). In contrast, p62 promotes HAMLET (a complex of oleic acids and decalcified α-lactalbumin)-induced apoptosis by activating caspase-8 in a glioma cell line U87MG (Zhang et al., 2013). These studies indicate that p62 regulates cell death and cell survival by modulating different signaling pathways.
Activator protein-1 (AP-1) transcription factor complex (Halazonetis et al., 1988; Kouzarides and Ziff, 1988) responds to a variety of cellular signals, including inflammatory stimulation (Mackman et al., 1991) and endoplasmic reticulum (ER) stress (Fuest et al., 2012), and participates in the regulation of cell death (Shaulian and Karin, 2002; Meng and Xia, 2011). It is understood that the major subunit of the AP-1 transcription factor complex, the proto-oncogene c-Jun, is responsible for its role in the regulation of cell death. c-Jun protects cells from ER stress-induced death by upregulating Down syndrome critical region 1 (DSCR1) (Zhao et al., 2008) or by downregulating tumor suppressor PTEN (Hettinger et al., 2006). It has also been reported that c-Jun promotes apoptotic cell death in NIH 3T3 fibroblasts (Bossy-Wetzel et al., 1997) and CRBN-mediated ubiquitination and degradation of c-Jun protects THP-1 cells from apoptosis induced by lipopolysaccharide (Yang et al., 2018), although the upstream signaling pathway triggering this regulation is unknown. Our previous study indicated that CRBN interacts with p62 and protects cells from death induced by pathogenic protein aggregates through its competitive binding to ubiquitinated proteins (Zhou et al., 2018). CRBN exhibits the non-enzymatic function in this regulation, while at the same time acting as a substrate receptor of the cullin 4-RING E3 ligase (CRL4), there by promoting the ubiquitination and degradation of c-Jun (Yang et al., 2018). Although both p62 and c-Jun regulate cell death and survival, it is not known whether p62 and c-Jun directly crosstalk with each other, and whether CRBN is also involved in these processes.
In this study, we explore the possibility of the interplay between p62 and c-Jun and their role in regulating cell death under ER stress. We first measured the expression of p62 and c-Jun under ER stress induced by different stimuli. We then examined the potential interaction between p62, c-Jun, and CRBN using immunoprecipitation and immunoblotting techniques. We further investigated the regulation of p62 on c-Jun protein levels, and the role of CRBN in this regulation. Finally, we comment on the potential function of CRBN in ER stress-induced apoptosis and the underlying molecular mechanism.
MATERIALS AND METHODS
Materials
The primary antibodies used in this work were obtained from the following sources: anti-HA (sc-7392) and anti-ubiquitin (sc-8017) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, USA); anti-c-Jun (CPA1634) antibody was from Cohesion Biosciences (London, UK); anti-FLAG (0912-1) and anti-GFP (EM30501) antibodies were from HuaAn Biotechnology (Hangzhou, China); mouse anti-CRBN antibody was a gift from Dr. Xiu-Bao Chang (Mayo Clinic College of Medicine, Scottsdale, AZ, USA); rabbit anti-CRBN antibody (11435-1-AP) and anti-GAPDH (60004-1) antibody were from ProteinTech Group (Rosemont, IL, USA); anti-cleaved caspase 3 (9661S) and anti-PARP1 (9532S) antibodies were from Cell Signaling Technology (Danvers, MA, USA); anti-p62 (P0067) antibody was from Sigma (Saint Louis, MO, USA); anti-LC3 (NB100-2220) antibody was from Novus Biologicals (Centennial, CO, USA); anti-FLAG affinity gel (B23102) and anti-HA magnetic beads (B26301) were from Bimake (Houston,TX, USA); rabbit IgG (A7016) for control immunoprecipitation was from Beyotime Biotechnology (Haimen, Jiangsu, China); and HRP-labeled secondary antibodies were from Beyotime Biotechnology and Thermo Fisher (Waltham, MA, USA).
Chemicals were from the following companies: MG132 (CC2775) was from ChemCatch (Suwanee, GA, USA); brefeldin A (s7046) was from Selleck (Houston, TX, USA); puromycin (P8230) was from Solarbio (Beijing, China); Hoechst (C1022) was from Beyotime Biotechnology; and propidium iodide (PI, KGA214-50) was from Nanjing KeyGen BioTech (Nanjing, Jiangsu, China).
Strep-FLAG (SF)-c-Jun, FLAG-p62, HA-CRBN, and GFP-p62 plasmids were from our previous work (Yang et al., 2018; Zhou et al., 2018). The control siRNAs (ID: 35164) and sip62 were synthesized by GenePharma (Shanghai, China) or RiboBio Co (Guangzhou, Guangdong, China). The siRNA sequences for human sip62 are sense #1: CAUGUCCUACGUGAAGGAUGATT, antisense #1: UCAUCCUUCACGUAGGACAUGTT; sense #2: GCAUUGAAGUUGAUAUCGAUTT; antisense #2: AUCGAUAUCAACUUCAAUGCTT.
Cell culture
Human embryonic kidney (HEK) 293T and HeLa cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (PAN Biotech, Aidenbach, Germany and Lonsera, Ciudad de la Costa, Uruguay) and 100 units/mL penicillin and 100 µg/mL streptomycin (Gibco, Waltham, MA, USA). Cells were passaged every two or three days.
Transfection
Plasmids were transfected with lipofectamine 3000 (Life Technologies, Carlsbad, CA, USA) or polyethyleneimine (PEI, Sigma) transfection reagent and siRNAs were transfected with lipofectamine 2000 transfection reagent (Life Technologies) or riboFECTTM CP reagent (RiboBio Co) according to the manufacturer’s instructions. Culture medium was changed 6 h following transfection for subsequent experiments.
Construction of plasmids and stable cell lines
The 21-nucleotide sequence for shRNA targeting human CRBN was CCCAGACACTGAAGATGAAAT and the corresponding sequence was inserted into the pLKO.1-TRC lentiviral vector to construct the pLKO.1-shCRBN plasmid. In order to produce lentiviral particles, pLKO.1-TRC or pLKO.1-shCRBN plasmid was transfected into HEK293T cells together with the packaging plasmids psPAX2 and pMD2G at a ratio of 3:2:1. At 48 and 72 h following transfection, the culture medium containing the lentiviral particles was collected and filtered by a 0.45 μm filter (Merck Millipore, Billerica, MA, USA). The lentiviral particles were then used to infect HEK293T cells in a six-well plate. In order to obtain stable transduced cell lines, the infected cells were selected using puromycin (2 μg/mL) for 2 weeks.
Preparation of cell lysate
Following transfection or drug treatment, cells were lysed in modified RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, and 1 mM EDTA) on ice with brief sonication. Cell lysates were obtained after centrifugation (13, 000 g) for 15 min at 4°C.
Immunoprecipitation
FLAG-tagged proteins were purified with anti-FLAG affinity gel according to a previously reported method (Zhu et al., 2018). The affinity gel was first prewashed three times with TBST (TBS with 0.1% Tween 20) and incubated with cell lysates at 4°C overnight. The gel was centrifuged at 500 g for 1 min, washed three times with the modified RIPA buffer and three times with RIPA buffer containing 0.3 M NaCl. SF-c-Jun and its interacting proteins were eluted twice with RIPA buffer containing 200 μg/mL FLAG peptide (DYKDDDDK, ChinaPeptides, Shanghai, China). The eluate was combined and used for Western blotting analyses.
HA-tagged CRBN was purified with anti-HA-magnetic beads according to a method described previously (Zhu et al., 2018). Briefly, anti-HA-magnetic beads were prewashed three times with TBS. Beads were further blocked with 5% BSA in TBST and then incubated with cell lysates at 4°C overnight. The magnetic beads were separated from cell lysates using a magnetic separation stand, and then washed ten times with the modified RIPA buffer containing 0.6 M NaCl to remove the nonspecific binding proteins. Finally, the beads were boiled twice with 50 μL SDS sample loading buffer and the eluate was combined for immunoblotting analysis.
Immunoprecipitation of endogenous c-Jun was carried out using a procedure described previously (Yang et al., 2018). Briefly, cell lysates were pre-incubated with protein A/G agarose beads to remove nonspecific binding proteins, incubated with 1 μg of control IgG or c-Jun antibody at 4°C overnight, and further incubated with prewashed protein A/G agarose beads (40 μL) at 4°C for 5 h. The protein A/G agarose beads were washed four times with PBS and proteins were eluted by heating with 60 μL of 2× SDS sample loading buffer.
Western blotting
Western blotting analysis was carried out based on a previously used method (Yang et al., 2018). Briefly, cell lysates or affinity-purified samples were separated by SDS-PAGE electrophoresis and proteins were transferred to PVDF membranes. Membranes were further blocked with 5% non-fat milk in TBST at room temperature for 1 h, incubated with the indicated primary antibodies at 4°C for overnight, and washed three times with TBST at room temperature (each for 10 min). After the washing steps, membranes were incubated with secondary antibodies at room temperature for 1 h, and washed three times with TBST at room temperature (each for 10 min). Proteins were visualized on membranes using Western blotting chemiluminescent horseradish peroxidase substrate (NCM Biotech, Suzhou, China), and signals were recorded using a Tanon 5200 imaging system (Tanon, Shanghai, China).
Propidium iodide (PI) staining
PI staining was described in a previous study (Hou et al., 2015). Briefly, HEK293T cells stably expressing shNC or shCRBN in 24-well plates were transfected with FLAG or FLAG-p62 plasmids for 24 h. Cells were stained with PI for 5 min, and then washed once with PBS. Cells were further stained with Hoechst for 7 min, and images were captured using an inverted microscope (IX71, Olympus, Tokyo, Japan).
TdT-mediated dUTP Nick-End Labeling (TUNEL) staining
The colorimetric TUNEL apoptosis assay kit was purchased from Beyotime Biotechnology. HEK293T cells stably expressing shNC or shCRBN were used to perform the TUNEL assay according to the manufacturer’s instructions. The cells were washed once with PBS, fixed with 4% paraformaldhyde for 30 min, permeabilized with 0.3% Triton X-100 for 5 min, washed three times with PBS, and then stained with the TUNEL solution at room temperature for 1 h. The cells were further incubated with Hoechst for 5 min to visualize the cellular nuclei. The images were captured using an inverted microscope (IX71, Olympus).
Statistics
Statistical analysis was performed using two-tailed Student’s t-test for the number of biological replicates indicated in the figure legends. *p<0.05; **p<0.01; ***p<0.001.
RESULTS
Autophagic degradation of p62 leads to c-Jun upregulation
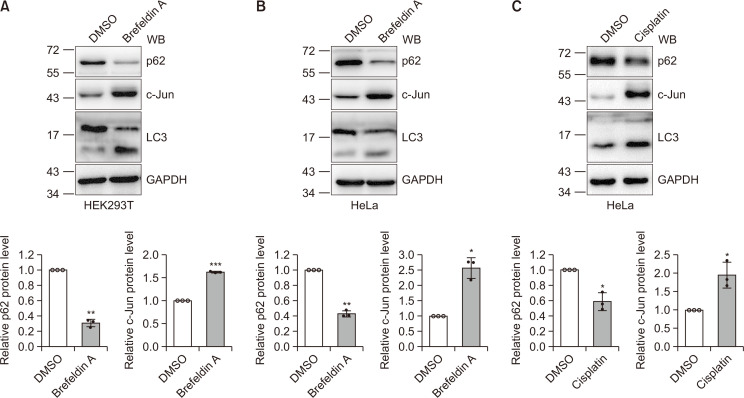
Activation of autophagy may result in a decrease of p62 protein levels (Wu et al., 2018). Since c-Jun responds to a variety of cellular signals, we examined whether c-Jun is affected by the activation of autophagy. Consistent with previous studies (Ding et al., 2007; Lin et al., 2017), our results showed an increase in LC3II and the LC3II/LC3I ratio upon brefeldin A and cisplatin treatment, indicating the activation of autophagy in HEK293T and HeLa cells (Fig. 1). Furthermore, we found that c-Jun protein levels were increased after cells were treated with brefeldin A, HBSS, and cisplatin (Fig. 1A, Supplementary Fig. 1). These results suggest that p62 may participate in the regulation of c-Jun in ER stress-induced autophagy.
Fig. 1.
Autophagic degradation of p62 leads to c-Jun upregulation. (A, B) HEK293T (A) or HeLa (B) cells were treated with brefeldin A (5 µg/mL) for 24 h and then the cell lysates were immunoblotted with the antibodies indicated. Detection of LC3 I/II and p62 indicated autophagic activity. The means and standard deviations (mean ± SDs) from three biological replicates are depicted in the bar graph. Student’s t-test,*p<0.05; **p<0.01; ***p<0.001. (C) HeLa cells were treated with cisplatin (12 μg/mL) for 24 h and then the cell lysates were subjected to immunoblotting with the antibodies indicated. Detection of LC3 I/II and p62 indicated autophagic activity. Quantitative data were obtained from three independent experiments. Student’s t-test, *p< 0.05.
p62 downregulates c-Jun protein levels through the UPS
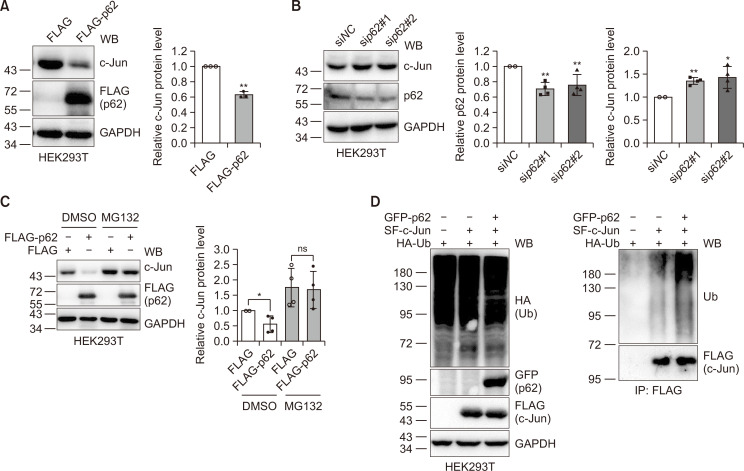
It has been reported that p62 may regulate the cellular degradation pathway (Pankiv et al., 2007; Geetha et al., 2008; Zotti et al., 2014; Zaffagnini et al., 2018). Since ER stress-induced autophagy reversely regulates p62 and c-Jun (Fig. 1), we investigated the possible regulation of c-Jun protein levels by p62. Ectopic expression of p62 in HEK293T cells resulted in a decrease in c-Jun protein levels (Fig. 2A), while siRNA knockdown of p62 increased c-Jun protein levels (Fig. 2B). These experiments demonstrate that p62 may indeed regulate c-Jun protein levels. Given the crosstalk between the ALP and the UPS, the fact that blockade of the UPS activates the ALP (Wang and Wang, 2015), and deficiency in the ALP leads to the accumulation of p62 (which in turn binds to ubiquitinated proteins, delaying their degradation by the UPS) (Korolchuk et al., 2009), we examined the degradation of c-Jun following overexpression of p62. Since the UPS may degrade c-Jun, we considered the UPS. Therefore, we transfected FLAG or FLAG-p62 plasmids into HEK293T cells and then divided the transfected cells into two plates for further treatment with DMSO, or with the proteasome inhibitor MG132. The data clearly show that MG132 inhibits the reduction in c-Jun protein following p62 expression (Fig. 2C). This suggests that p62 downregulates c-Jun protein levels, most probably through the UPS. Furthermore, we found that p62 could promote the ubiquitination of c-Jun (Fig. 2D). Collectively, these data demonstrate that p62 may promote the ubiquitination and degradation of c-Jun.
Fig. 2.
p62 downregulates c-Jun through the ubiquitin-proteasome system. (A) HEK293T cells were transfected with FLAG or FLAG-p62 plasmid for 48 h and the resulting cell lysates were immunoblotted with the antibodies indicated. Mean ± SDs from three biological replicates were depicted. Student’s t-test, **p<0.01. (B) HEK293T cells were transfected with siNC or sip62 using lipofectamine 2000 (Life Technologies, Carlsbad, USA) for 48 h. Cell lysates were immunoblotted with the antibodies indicated. Quantification was carried out as described in (A) for four biological replicates. Student’s t-test, *p<0.05; **p<0.01. (C) HEK293T cells were transfected with FLAG or FLAG-p62 plasmid for 36 h and then treated with DMSO or MG132 (10 µM) for 12 h. Cell lysates were immunoblotted with the antibodies indicated. Experiments were repeated four times for quantification. Student’s t-test, *p<0.05; ns: not significant. (D) HEK293T cells were transfected with HA-Ub, SF-c-Jun, and GFP-p62 for 48 h and the cell lysates were subjected to anti-FLAG immunoprecipitation. The immunoprecipitates and cell lysates were subjected to immunoblotting with the antibodies indicated.
p62 interacts with c-Jun
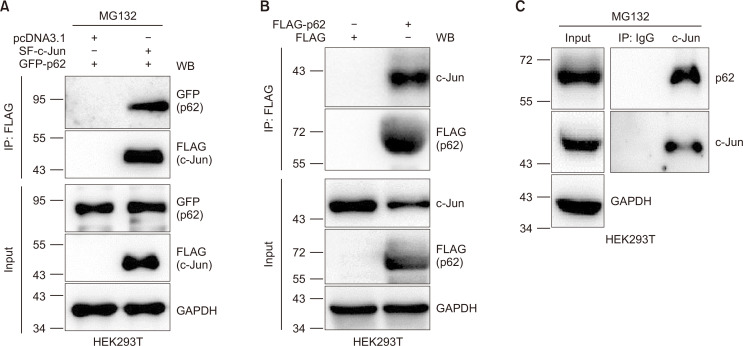
In our previous work, we discovered that CRBN interacts with p62 (also shown in Supplementary Fig. 2) and prevents the formation of pathogenic protein aggregates in cell lines and primary neuronal cells (Zhou et al., 2018). We have also demonstrated that CRBN interacts with c-Jun and reduces c-Jun protein levels through the UPS, leading to the attenuation of the transcription activity of the associated transcription factor complex AP-1 and suppression of the inflammatory response (Yang et al., 2018). Since p62 downregulates c-Jun (Fig. 2), we sought to examine whether p62 also interacts with c-Jun. First, we co-expressed Strep-FLAG-(SF)-c-Jun with GFP or GFP-p62 in HEK293T cells and immunoprecipitated SF-c-Jun and its interacting proteins. Immunoblotting images unambiguously indicated the presence of p62 in the c-Jun immunoprecipitate (Fig. 3A). We also expressed FLAG or FLAG-p62 in HEK293T cells and immunoprecipitated FLAG-p62 and its interacting proteins. The results demonstrate that FLAG-p62 binds to c-Jun (Fig. 3B). We further used an anti-c-Jun antibody to immunoprecipitate the endogenous c-Jun for immunoblotting analysis, and confirmed the endogenous interaction between p62 and c-Jun (Fig. 3C).
Fig. 3.
p62 interacts with c-Jun. (A) HEK293T cells were transfected with GFP-p62 and pcDNA3.1 or Strep-FLAG (SF)-c-Jun plasmids using PEI transfection reagent. At 36 h post-transfection, cells were treated with MG132 (10 µM) for 12 h and lysed in RIPA buffer. SF-c-Jun was purified with anti-FLAG affinity gel. Cell lysates and purified samples were immunoblotted using the antibodies indicated. Note: MG132 was added to prevent the degradation of c-Jun upon p62 expression, which resulted in approximately equal c-Jun protein levels in two cell lysates. (B) HEK293T cells were transfected with FLAG or FLAG-p62 for 48 h. The resulting cell lysates were subjected to anti-FLAG immunoprecipitation. The immunoprecipitates and cell lysates were subjected to immunoblot with the antibodies indicated. (C) UntransfectedHEK293T cells were treated with MG132 (10 µM) for 12 h and lysed in RIPA buffer. IgG or anti-c-Jun antibodies were used for immunoprecipitation of endogenous proteins. Endogenous proteins in cell lysates and the immunoprecipitates were immunoblotted with the antibodies indicated.
p62, CRBN, and c-Jun form a ternary protein complex
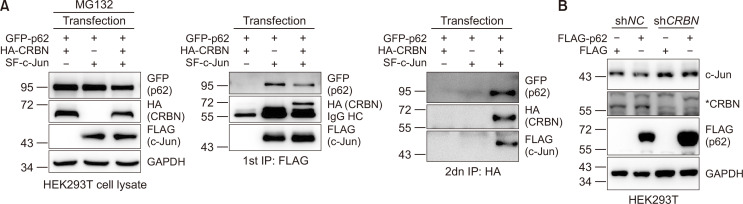
Our previous work (Yang et al., 2018) and the data described above indicates that c-Jun interacts with both CRBN and p62. We then asked whether p62, CRBN, and c-Jun form a ternary complex. Therefore, we co-expressed GFP-p62 with HA-CRBN and SF-c-Jun in HEK293T cells and performed two rounds of immunoprecipitation. We slightly adjusted the amount of transfected plasmids to obtain similar levels of protein expression in these samples (Fig. 4A, left panel). In the first round of immunoprecipitation, we purified SF-c-Jun with anti-FLAG affinity gel. Immunoblotting of the purified samples shows that both CRBN and p62 are detected in the c-Jun immunoprecipitate but not in the mock immunoprecipitate (Fig. 4A, middle panel). Although this demonstrates that c-Jun interacts with CRBN and p62, it does not establish if they form a ternary protein complex, or alternatively, if c-Jun interacts with CRBN and p62 in two different complexes. In order to distinguish between these two possibilities, we performed a second round of immunoprecipitation using anti-HA magnetic beads to purify HA-CRBN and its interacting proteins from the first round immunoprecipitate. The resulting Western blotting images show that both c-Jun and p62 are detected in the CRBN immunoprecipitate but not in the mock immunoprecipitate (Fig. 4A, right panel).
Fig. 4.
c-Jun, p62, and CRBN form a ternary complex and CRBN is required for p62-mediated c-Jun degradation. (A) HEK293T cells were first transfected with GFP-p62 and HA-CRBN, GFP-p62 and SF-c-Jun, or GFP-p62, HA-CRBN, and SF-c-Jun plasmids using PEI transfection reagent. The amount of plasmids was slightly adjusted to express proteins at similar levels in different samples and pcDNA3.1 was used to balance the total amount of plasmids used. At 36 h post-transfection, cells were treated with MG132 (10 µM) for 12 h and lysed. In the first round of immunoprecipitation, SF-c-Jun and its interacting proteins were purified with anti-FLAG affinity gel and eluted with FLAG peptides. The eluate was further purified with HA magnetic beads in the second round of immunoprecipitation. The cell lysates and the first and second round affinity-purified samples were immunoblotted with the antibodies indicated. (B) HEK293T cells stably expressing shNC or shCRBN were transiently transfected with FLAG or FLAG-p62 for 48 h. The cell lysates were subjected to immunoblotting with the antibodies indicated. The CRBN antibody was used to examine the knockdown efficiency.
These results demonstrate that p62 interacts with, and downregulates, c-Jun through the UPS and that a ternary protein complex is formed between p62, c-Jun, and CRBN. Our previous studies also showed that CRBN interacts with c-Jun and that c-Jun functions as a substrate receptor of the CRL4 E3 ligase for the ubiquitination and degradation of c-Jun (Yang et al., 2018). Furthermore, we have also found that CRBN binds to p62 but does not affect p62 protein levels (Zhou et al., 2018). Therefore, we decided to test whether the regulation of c-Jun by p62 is dependent on CRBN. Our results show that although p62 expression reduces c-Jun, CRBN knockdown almost completely abolishes this reduction (Fig. 4B), indicating the essential role of CRBN in the regulation of c-Jun by p62. Collectively, these data suggest that p62 recruits CRBN and c-Jun to form a ternary complex, which then attenuates c-Jun protein through the UPS.
p62/c-Jun axis regulates ER stress-induced apoptosis
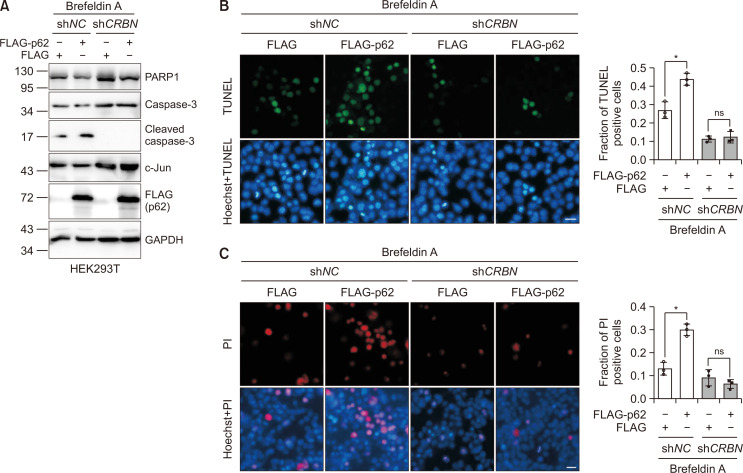
The experiments described above indicate that p62 may participate in the regulation of c-Jun under ER stress. Since c-Jun protected cells against ER stress-induced apoptosis (Supplementary Fig. 3), we examined the role of p62, CRBN, and c-Jun in ER stress. We first constructed the stable control or CRBN knockdown HEK293T cell lines using shRNA. We then used these cell lines to express FLAG or FLAG-p62 to investigate the effect of p62 on apoptosis under ER stress. Using brefeldin A as an ER stress inducer (Moon et al., 2012; Zhu et al., 2017), we found that overexpression of p62 reduced PARP1 protein levels and increased cleaved caspase-3 protein levels, indicating the role of p62 in brefeldin A-induced apoptosis activation in HEK293T cells. Furthermore, CRBN knockdown eliminated this regulation, indicating the essential role of CRBN in this process (Fig. 5A). We also used propidium iodide (PI) and a TUNEL apoptosis assay kit (Beyotime Biotechnology) to stain the apoptotic cells induced by ER stress. The immunofluorescence measurement of TUNEL and PI stained cells show that expression of p62 increased the apoptosis induced by brefeldin A, which was abolished in CRBN-deficient cells (Fig. 5B, 5C). Collectively, these data demonstrate that the p62/c-Jun axis participates in the regulation of ER stress-induced apoptosis (Fig. 6).
Fig. 5.
p62/c-Jun axis regulates ER stress-induced apoptosis. (A) HEK293T cells stably expressing shNC or shCRBN were transfected with FLAG or FLAG-p62 plasmid using PEI transfection reagent for 48 h and then treated with brefeldin A (5 µg/mL) for 24 h to activate ER stress-induced apoptosis. Cell lysates were immunoblotted with the antibodies indicated. The caspase 3 antibody was used to indicate the activation of apoptosis. (B) Cells transfected as in (A) were treated with brefeldin A (5 µg/mL) for 12 h to activate ER stress-induced apoptosis. Cells were subjected to immunofluorescence analysis after staining with TUNEL and Hoechst. TUNEL was used to label early apoptosis. Scale bar: 20 μm. Mean ± SDs were obtained for data from three biological replicates. Student’s t-test, *p<0.05; ns: not significant. (C) Cells were treated as in (A) and were subjected to immunofluorescence analysis after staining with PI and Hoechst. The PI was used to label the late apoptosis. Scale bar: 20 μm. Mean ± SDs were obtained for data from three biological replicates. Student’s t-test, *p<0.05; ns: not significant.
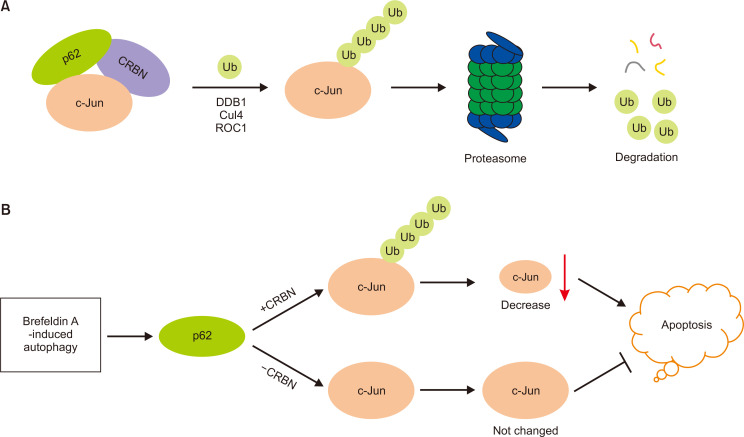
Fig. 6.
Proposed model for the regulation of c-Jun degradation and ER stress-induced apoptosis by p62. (A) p62 forms a ternary complex with c-Jun and CRBN. CRBN forms a CRL4 E3 ligase and promotes the ubiquitination and proteasomal degradation of c-Jun. (B) In the presence of CRBN, p62 promotes c-Jun ubiquitination and degradation, decreases c-Jun protein levels, and promotes ER stress-induced apoptosis. In the absence of CRBN, p62-mediated c-Jun degradation is attenuated, leading to high c-Jun protein levels and the protection of ER stress-induced cell apoptosis.
DISCUSSION
Activation of autophagy causes the autophagic degradation of p62, leading to reduced apoptosis (Levine and Kroemer, 2008; Lamark et al., 2009). The molecular mechanisms underlying autophagy-regulated apoptosis are largely unknown (Tang et al., 2019; Tilija Pun et al., 2020). In the present study, we found that activation of autophagy leads to the downregulation of p62 and upregulation of c-Jun (Fig. 1, 4), and that p62 promotes the proteasomal degradation of c-Jun in a CRBN-dependent manner (Fig. 2, 4). This suggests crosstalk between the UPS and ALP cellular degradation pathways (Hewitt et al., 2016; Bustamante et al., 2018). Indeed, it has been shown that p62 functions as a scaffolding protein and binds to ubiquitinated proteins through its UBA domain (Seibenhener et al., 2004) and LC3 through its LC3-interacting region (Pankiv et al., 2007).
In this study, we confirmed that p62-mediated apoptosis under brefeldin treatment is due to the proteasomal degradation of c-Jun, which may occur following the recruitment of the substrate receptor CRBN of the CRL4 E3 ligase leading to the ubiquitination of c-Jun and its degradation. This finding is consistent with a previous study which found that autophagic degradation of p62 protected cells from ER-stress induced apoptosis (Ogata et al., 2006). Furthermore, we identified a novel molecular mechanism by which p62 upregulates c-Jun, and thereby protects cells from apoptosis. The mitochondrial pathway may be activated upon induction of ER stress, which causes the release of cytochrome c, and eventually activates the executioner caspases including caspase-3, leading to PARP1 cleavage (Iurlaro and Muñoz-Pinedo, 2016). ER stress also activates the extrinsic or death receptor pathway, which may activate caspase-8, and thus caspase-3 (Yamaguchi and Wang, 2004; Sano and Reed, 2013). Since the activation of caspase-3 is important in ER stress-induced apoptosis (Masud et al., 2007), we confirmed caspase-3-associated apoptosis in the p62/c-Jun axis (Fig. 5, 6). However, it has also been reported that p62 could enhance bortezomib resistance in multiple myeloma cells, and then promote their survival (Milan et al., 2015; Marino et al., 2017), indicating that p62 may regulate survival or death in different cell lines.
Our previous studies showed that CRBN suppresses the formation of p62 bodies (Zhou et al., 2018) and promotes the ubiquitination and proteasomal degradation of c-Jun (Yang et al., 2018). In the present study, we found that p62, c-Jun, and CRBN form a ternary protein complex (Fig. 3), suggesting that p62 promotes the ubiquitination and degradation of c-Jun in the p62 bodies in the presence of CRBN. The present study also indicates that p62 may act as a scaffold protein to promote the function of the CRL4CRBN E3 ligase. Indeed, it has been reported that p62 may enhance the TRAF6-mediated ubiquitination of NEMO (Zotti et al., 2014) and increase Parkin-induced mitochondrial clustering (Narendra et al., 2010). This is in accordance with the discovery that the amyloid precursor protein may act as a substrate recognition unit of CRL4CRBN E3 ligase to regulate the ubiquitination of cytosolic and membrane-bound proteins leading to their subsequent degradation (Del Prete et al., 2016).
In summary, we have identified that p62 forms a ternary complex with c-Jun and CRBN and thus promotes the ubiquitin-mediated c-Jun degradation. Downregulation of p62 induced by ER stress exhibits the protective role against apoptosis by reducing the c-Jun degradation. This work revealed the essential role of CRBN in the regulation of c-Jun by p62 and elucidated a new molecular mechanism by which the p62/c-Jun axis regulates the ER stress-induced apoptosis.
Supplementary Materials
ACKNOWLEDGMENTS
We are grateful to Dr. Xiu-Bao Chang from Mayo Clinic College of Medicine (USA) for generously providing the mouse CRBN antibody and to Dr. Guanghui Wang for kindly providing p62 and HA-Ub plasmids. This work was supported by the National Key R&D Program of China (2018YFC1705505), National Natural Science Foundation of China (31670833 & 81703535), Jiangsu Key Laboratory of Neuropsychiatric Diseases (BM2013003), National Center for International Research (2017B01012), a project funded by the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
Footnotes
CONFLICT OF INTEREST
None.
REFERENCES
- Bossy-Wetzel E., Bakiri L., Yaniv M. Induction of apoptosis by the transcription factor c-Jun. EMBO J. 1997;16:1695–1709. doi: 10.1093/emboj/16.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante H. A., González A. E., Cerda-Troncoso C., Shaughnessy R., Otth C., Soza A., Burgos P. V. Interplay between the autophagy-lysosomal pathway and the ubiquitin-proteasome system: a target for therapeutic development in Alzheimer's disease. Front. Cell. Neurosci. 2018;12:126. doi: 10.3389/fncel.2018.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- Del Prete D., Rice R. C., Rajadhyaksha A. M., D'Adamio L. Amyloid precursor protein (APP) may act as a substrate and a recognition unit for CRL4CRBN and Stub1 E3 ligases facilitating ubiquitination of proteins involved in presynaptic functions and neurodegeneration. J. Biol. Chem. 2016;291:17209–17227. doi: 10.1074/jbc.M116.733626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I. Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 2017;86:193–224. doi: 10.1146/annurev-biochem-061516-044908. [DOI] [PubMed] [Google Scholar]
- Ding W. X., Ni H. M., Gao W., Hou Y. F., Melan M. A., Chen X., Stolz D. B., Shao Z. M., Yin X. M. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J. Biol. Chem. 2007;282:4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- Fuest M., Willim K., MacNelly S., Fellner N., Resch G. P., Blum H. E., Hasselblatt P. The transcription factor c-Jun protects against sustained hepatic endoplasmic reticulum stress thereby promoting hepatocyte survival. Hepatology. 2012;55:408–418. doi: 10.1002/hep.24699. [DOI] [PubMed] [Google Scholar]
- Gal J., Ström A. L., Kilty R., Zhang F., Zhu H. p62 accumulates and enhances aggregate formation in model systems of familial amyotrophic lateral sclerosis. J. Biol. Chem. 2007;282:11068–11077. doi: 10.1074/jbc.M608787200. [DOI] [PubMed] [Google Scholar]
- Geetha T., Seibenhener M. L., Chen L., Madura K., Wooten M. W. p62 serves as a shuttling factor for TrkA interaction with the proteasome. Biochem. Biophys. Res. Commun. 2008;374:33–37. doi: 10.1016/j.bbrc.2008.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice G. L., Nathan J. A. The recognition of ubiquitinated proteins by the proteasome. Cell. Mol. Life Sci. 2016;73:3497–3506. doi: 10.1007/s00018-016-2255-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis T. D., Georgopoulos K., Greenberg M. E., Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988;55:917–924. doi: 10.1016/0092-8674(88)90147-X. [DOI] [PubMed] [Google Scholar]
- Hettinger K., Vikhanskaya F., Poh M. K., Lee M. K., de Belle I., Zhang J. T., Reddy S. A. G., Sabapathy K. c-Jun promotes cellular survival by suppression of PTEN. Cell Death Differ. 2006;14:218–229. doi: 10.1038/sj.cdd.4401946. [DOI] [PubMed] [Google Scholar]
- Hewitt G., Carroll B., Sarallah R., Correia-Melo C., Ogrodnik M., Nelson G., Otten E. G., Manni D., Antrobus R., Morgan B. A., von Zglinicki T., Jurk D., Seluanov A., Gorbunova V., Johansen T., Passos J. F., Korolchuk V. I. SQSTM1/p62 mediates crosstalk between autophagy and the UPS in DNA repair. Autophagy. 2016;12:1917–1930. doi: 10.1080/15548627.2016.1210368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X. O., Si J. M., Ren H. G., Chen D., Wang H. F., Ying Z., Hu Q. S., Gao F., Wang G. H. Parkin represses 6-hydroxydopamine-induced apoptosis via stabilizing scaffold protein p62 in PC12 cells. Acta Pharmacol. Sin. 2015;36:1300–1307. doi: 10.1038/aps.2015.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Wang X. N., Yuan X. D., Wu W. Y., Lobie P. E., Wu Z. XIAP facilitates breast and colon carcinoma growth via promotion of p62 depletion through ubiquitination-dependent proteasomal degradation. Oncogene. 2018;38:1448–1460. doi: 10.1038/s41388-018-0513-8. [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Waguri S., Sou Y. S., Kageyama S., Hasegawa J., Ishimura R., Saito T., Yang Y., Kouno T., Fukutomi T., Hoshii T., Hirao A., Takagi K., Mizushima T., Motohashi H., Lee M. S., Yoshimori T., Tanaka K., Yamamoto M., Komatsu M. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Iurlaro R., Muñoz-Pinedo C. Cell death induced by endoplasmic reticulum stress. FEBS J. 2016;283:2640–2652. doi: 10.1111/febs.13598. [DOI] [PubMed] [Google Scholar]
- Jain A., Lamark T., Sjøttem E., Larsen K. B., Awuh J. A., Øvervatn A., McMahon M., Hayes J. D., Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian Y., Gao W., Geng C., Zhou H., Leng Y., Li Y., Chen W. Arsenic trioxide potentiates sensitivity of multiple myeloma cells to lenalidomide by upregulating cereblon expression levels. Oncol. Lett. 2017;14:3243–3248. doi: 10.3892/ol.2017.6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Li Y., Pitti R., Lawrence D., Pham V. C., Lill J. R., Ashkenazi A. Cullin 3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y. S., Ueno I., Sakamoto A., Tong K. I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Korolchuk V. I., Mansilla A., Menzies F. M., Rubinsztein D. C. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol. Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk V. I., Menzies F. M., Rubinsztein D. C. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–8139. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- Kouzarides T., Ziff E. The role of the leucine zipper in the fos-jun interaction. Nature. 1988;336:646–651. doi: 10.1038/336646a0. [DOI] [PubMed] [Google Scholar]
- Lamark T., Kirkin V., Dikic I., Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. F., Lin Y. C., Tsai T. F., Chen H. E., Chou K. Y., Hwang T. I. S. Cisplatin induces protective autophagy through activation of BECN1 in human bladder cancer cells. Drug Des. Devel. Ther. 2017;11:1517–1533. doi: 10.2147/DDDT.S126464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippai M., Lőw P. The role of the selective adaptor p62 and ubiquitin-like proteins in autophagy. Biomed Res. Int. 2014;2014:832704. doi: 10.1155/2014/832704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. J., Ye L., Huang W. F., Guo L. J., Xu Z. G., Wu H. L., Yang C., Liu H. F. p62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation. Cell. Mol. Biol. Lett. 2016;21:29. doi: 10.1186/s11658-016-0031-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Kern J. T., Walker J. R., Johnson J. A., Schultz P. G., Luesch H. A genomic screen for activators of the antioxidant response element. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5205–5210. doi: 10.1073/pnas.0700898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackman N., Brand K., Edgington T. S. Lipopolysaccharide-mediated transcriptional activation of the human tissue factor gene in THP-1 monocytic cells requires both activator protein 1 and nuclear factor kappa B binding sites. J. Exp. Med. 1991;174:1517–1526. doi: 10.1084/jem.174.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S., Petrusca D. N., Silberman R., Toscani D., Anderson J. L., Giuliani N., Xie X. Q., Kurihara N., Roodman G. D. Inhibition of p62-ZZ domain-mediated signaling overcomes bortezomib resistance in multiple myeloma cells independent of their p53 status. Blood. 2017;130:4421. [Google Scholar]
- Masud A., Mohapatra A., Lakhani S. A., Ferrandino A., Hakem R., Flavell R. A. Endoplasmic reticulum stress-induced death of mouse embryonic fibroblasts requires the intrinsic pathway of apoptosis. J. Biol. Chem. 2007;282:14132–14139. doi: 10.1074/jbc.M700077200. [DOI] [PubMed] [Google Scholar]
- Meng Q., Xia Y. c-Jun, at the crossroad of the signaling network. Protein Cell. 2011;2:889–898. doi: 10.1007/s13238-011-1113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan E., Perini T., Resnati M., Orfanelli U., Oliva L., Raimondi A., Cascio P., Bachi A., Marcatti M., Ciceri F., Cenci S. A plastic SQSTM1/p62-dependent autophagic reserve maintains proteostasis and determines proteasome inhibitor susceptibility in multiple myeloma cells. Autophagy. 2015;11:1161–1178. doi: 10.1080/15548627.2015.1052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J. L., Kim S. Y., Shin S. W., Park J. W. Regulation of brefeldin A-induced ER stress and apoptosis by mitochondrial NADP+-dependent isocitrate dehydrogenase. Biochem. Biophys. Res. Commun. 2012;417:760–764. doi: 10.1016/j.bbrc.2011.12.030. [DOI] [PubMed] [Google Scholar]
- Narendra D., Kane L. A., Hauser D. N., Fearnley I. M., Youle R. J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K., Shiosaka S., Hammarback J. A., Urano F., Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Rubinsztein D. C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Sano R., Reed J. C. ER stress-induced cell death mechanisms. Biochim. Biophys. Acta. 2013;1833:3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener M. L., Babu J. R., Geetha T., Wong H. C., Krishna N. R., Wooten M. W. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell. Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E., Karin M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- Shin W. H., Park J. H., Chung K. C. The central regulator p62 between ubiquitin proteasome system and autophagy and its role in the mitophagy and Parkinson's disease. BMB Rep. 2020;53:56–63. doi: 10.5483/BMBRep.2020.53.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvets E., Fass E., Scherz-Shouval R., Elazar Z. The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J. Cell Sci. 2008;121:2685–2695. doi: 10.1242/jcs.026005. [DOI] [PubMed] [Google Scholar]
- Song P., Li S., Wu H., Gao R., Rao G., Wang D., Chen Z., Ma B., Wang H., Sui N., Deng H., Zhang Z., Tang T., Tan Z., Han Z., Lu T., Zhu Y., Chen Q. Parkin promotes proteasomal degradation of p62: implication of selective vulnerability of neuronal cells in the pathogenesis of Parkinson's disease. Protein Cell. 2016;7:114–129. doi: 10.1007/s13238-015-0230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Kang R., Berghe T. V., Vandenabeele P., Kroemer G. The molecular machinery of regulated cell death. Cell Res. 2019;29:347–364. doi: 10.1038/s41422-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z., Wang C., Hu C., Tian Y., Liu J., Wang X. Autophagic-lysosomal inhibition compromises ubiquitin-proteasome system performance in a p62 dependent manner in cardiomyocytes. PLoS ONE. 2014;9:e100715. doi: 10.1371/journal.pone.0100715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilija Pun N., Jang W. J., Jeong C. H. Role of autophagy in regulation of cancer cell death/apoptosis during anti-cancer therapy: focus on autophagy flux blockade. Arch. Pharm. Res. 2020;43:475–488. doi: 10.1007/s12272-020-01239-w. [DOI] [PubMed] [Google Scholar]
- Wang C., Wang X. The interplay between autophagy and the ubiquitin-proteasome system in cardiac proteotoxicity. Biochim. Biophys. Acta. 2015;1852:188–194. doi: 10.1016/j.bbadis.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Xu Q., Yuan Q., Jia M., Niu H., Liu X., Zhang J., Young C. Y., Yuan H. Proteasome inhibition boosts autophagic degradation of ubiquitinated-AGR2 and enhances the antitumor efficiency of bevacizumab. Oncogene. 2019;38:3458–3474. doi: 10.1038/s41388-019-0675-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Hao Z., Ren H., Wang G. Loss of VAPB regulates autophagy in a Beclin 1-dependent manner. Neurosci. Bull. 2018;34:1037–1046. doi: 10.1007/s12264-018-0276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Wang H. G. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J. Biol. Chem. 2004;279:45495–45502. doi: 10.1074/jbc.M406933200. [DOI] [PubMed] [Google Scholar]
- Yang J., Huang M., Zhou L., He X., Jiang X., Zhang Y., Xu G. Cereblon suppresses lipopolysaccharide-induced inflammatory response through promoting the ubiquitination and degradation of c-Jun. J. Biol. Chem. 2018;293:10141–10157. doi: 10.1074/jbc.RA118.002246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini G., Savova A., Danieli A., Romanov J., Tremel S., Ebner M., Peterbauer T., Sztacho M., Trapannone R., Tarafder A. K., Sachse C., Martens S. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018;37:e98308. doi: 10.15252/embj.201798308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. B., Gong J. L., Xing T. Y., Zheng S. P., Ding W. Autophagy protein p62/SQSTM1 is involved in HAMLET-induced cell death by modulating apotosis in U87MG cells. Cell Death Dis. 2013;4:e550. doi: 10.1038/cddis.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Xiao X., Kim A. S., Leite M. F., Xu J., Zhu X., Ren J., Li J. c-Jun inhibits thapsigargin-induced ER stress through up-regulation of DSCR1/Adapt78. Exp. Biol. Med. (Maywood) 2008;233:1289–1300. doi: 10.3181/0803-RM-84. [DOI] [PubMed] [Google Scholar]
- Zhou L., Hao Z., Wang G., Xu G. Cereblon suppresses the formation of pathogenic protein aggregates in a p62-dependent manner. Hum. Mol. Genet. 2018;27:667–678. doi: 10.1093/hmg/ddx433. [DOI] [PubMed] [Google Scholar]
- Zhou L., Wang H., Chen D., Gao F., Ying Z., Wang G. p62/Sequestosome 1 regulates aggresome formation of pathogenic ataxin-3 with expanded polyglutamine. Int. J. Mol. Sci. 2014;15:14997–15010. doi: 10.3390/ijms150914997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K., Dunner K., Jr, McConkey D. J. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2010;29:451–462. doi: 10.1038/onc.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Huang L., Gong J., Shi C., Wang Z., Ye B., Xuan A., He X., Long D., Zhu X., Ma N., Leng S. NF-κB pathway link with ER stress-induced autophagy and apoptosis in cervical tumor cells. Cell Death Dis. 2017;3:17059. doi: 10.1038/cddiscovery.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Lei Q., Li D., Zhang Y., Jiang X., Hu Z., Xu G. Proteomic and biochemical analyses reveal a novel mechanism for promoting protein ubiquitination and degradation by UFBP1, a key component of ufmylation. J. Proteome Res. 2018;17:1509–1520. doi: 10.1021/acs.jproteome.7b00843. [DOI] [PubMed] [Google Scholar]
- Zotti T., Scudiero I., Settembre P., Ferravante A., Mazzone P., D'Andrea L., Reale C., Vito P., Stilo R. TRAF6-mediated ubiquitination of NEMO requires p62/sequestosome-1. Mol. Immunol. 2014;58:27–31. doi: 10.1016/j.molimm.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.