Abstract
Drug addiction influences most communities directly or indirectly. Increasing studies have reported the relationship between circadian-related genes and drug addiction. Per2 disrupted mice exhibited more vulnerable behavioral responses against some drugs including methamphetamine (METH). However, its roles and mechanisms are still not clear. Transcriptional profiling analysis in Per2 knockout (KO) mice may provide a valuable tool to identify potential genetic involvement and pathways in enhanced behavioral responses against drugs. To explore the potential genetic involvement, we examined common differentially expressed genes (DEGs) in the striatum of drug naïve Per2 KO/wild-type (WT) mice, and before/after METH treatment in Per2 KO mice, but not in WT mice. We selected 9 common DEGs (Ncald, Cpa6, Pklr, Ttc29, Cbr2, Egr2, Prg4, Lcn2, and Camsap2) based on literature research. Among the common DEGs, Ncald, Cpa6, Pklr, and Ttc29 showed higher expression levels in drug naïve Per2 KO mice than in WT mice, while they were downregulated in Per2 KO mice after METH treatment. In contrast, Cbr2, Egr2, Prg4, Lcn2, and Camsap2 exhibited lower expression levels in drug naïve Per2 KO mice than in WT mice, while they were upregulated after METH treatment in Per2 KO mice. qRT-PCR analyses validated the expression patterns of 9 target genes before/after METH treatment in Per2 KO and WT mice. Although further research is required to deeply understand the relationship and roles of the 9 target genes in drug addiction, the findings from the present study indicate that the target genes might play important roles in drug addiction.
Keywords: Addiction, Methamphetamine, Per2, Gene expression
INTRODUCTION
Drug addiction is a chronic relapsing brain disease, affecting most communities directly or indirectly. Many researches and institutions have tried to identify its causes and prevent its adverse consequences, and a few drug addiction targets have been identified (Nestler and Malenka, 2004; Everitt and Robbins, 2016). Dopaminergic projections are known to be associated with the processes of drug abuse and addiction (Volkow et al., 2017; Solinas et al., 2019), and dopaminergic neurons projecting from the ventral tegmental area (VTA) release dopamine neurotransmitters to the nucleus accumbens (NAc) and the prefrontal cortex (PFC), forming mesolimbic and mesocortical pathways, respectively. In addition to the dopaminergic system, a complex interaction between exogenous (i.e., environment) and endogenous (i.e., genes and circadian rhythms) factors is also known to cause drug addiction (Abarca et al., 2002; Kim et al., 2018). Recently, increasing studies have reported the relationship between disturbed circadian rhythms and drug addiction (Abarca et al., 2002; Kovanen et al., 2010; Hasler et al., 2012). Disturbed circadian rhythms lead to increased substance use such as alcohol and cocaine indicating that circadian rhythms might play important roles in drug addiction.
Per2 is one of the key genes that generates circadian rhythms (Albrecht et al., 2001). Virtually, its mutations in mice induce different responses to drugs. For instance, mice with Per2 disruption consumed more alcohol and demonstrated increased reward effects to cocaine, morphine, or methamphetamine (METH) (Abarca et al., 2002; Spanagel et al., 2005; Perreau-Lenz et al., 2010; Kim et al., 2019). In our previous study, Per2 knockout (KO) mice (Per2tm1Drw) displayed increased METH-induced locomotor sensitization and reward responses (Kim et al., 2019). Additionally, the expression levels of dopamine-related genes and dopamine changed in the striatum depending on the expression of Per2. These findings suggest that Per2 might be one of the key genes to modulate drug addiction pathways, although mechanism of Per2 function is still not clear. Thus, further studies should focus on the mechanisms of Per2 in drug addiction.
Gene expression profiling might provide an ideal platform to elucidate the possible mechanisms of Per2 in drug addiction. The goal of this study was to further understand the role of Per2 in drug addiction using microarray analyses. In our previous study, we reported that METH-induced behavioral responses and dopamine levels in the striatum changed based on Per2 expression (Kim et al., 2019). The striatum is the brain region associated with METH addiction responses in rodents and human, and is known to increase dopamine release following psychostimulants administrations (Chang et al., 2007; Caprioli et al., 2017; Kim et al., 2019). Therefore, we analyzed the differentially expressed genes (DEGs) in the striatum of Per2 KO and wild-type (WT) mice, which showed different behavioral and molecular responses against METH.
MATERIALS AND METHODS
Drugs and materials
METH hydrochloride, purchased from Sigma-Aldrich Co (St. Louis, Mo, USA), was dissolved in physiological saline (0.9% w/v of NaCl), at a dose of 0.5 mg/kg intraperitoneally, as used in our previous study (Kim et al., 2019). TRIzol reagent was purchased from Invitrogen (Carlsbad, CA, USA), and the Hybrid-RTM kit was purchased from Geneall Biotechnology (Seoul, Korea). The AccuPower® CycleScript RT PreMix used to generate cDNA was obtained from Bioneer (Seoul, Korea). All primers for quantitative real-time PCR (qRT-PCR) were obtained from Cosmogenetech Co (Seoul, Korea), using a set of custom sequence-specific primers. SYBR® Green used to run qRT-PCR was purchased from SolGent Co (Daejeon, Korea).
Animals
Eight- to twelve-week-old male Per2tm1Drw (–/–), and C57BL/6 mice were used in this study. The heterozygous Per2tm1Drw (+/–) mutant mice (Bae et al., 2001) were obtained from the Jackson laboratory (Stock No.010492; JAX MICE®, ME, USA), and Per2tm1Drw (–/–, KO) mice were derived by intercrossing heterozygous Per2tm1Drw (+/–) and C57BL/6 mice, followed by crossing homozygous Per2tm1Drw (–/–) mutant mice. The C57BL/6 mice were used as the control group because Per2 KO were derived from C57BL/6 mice, which were obtained from Hanlim Laboratory Animals Co (Hwaseong, Korea). Each transgenic (TG) and WT mouse group was housed in an animal room under controlled conditions (12 h/12 h light/dark cycle, 7 AM-7 PM, and 22 ± 2°C) and fed ad libitum. All animal treatment and maintenance were performed in accordance with the Principles of Laboratory Animal Care (NIH Publication No. 85-23 revised 1985), and the Animal Care and Use Guidelines of Sahmyook University, Seoul, Korea (SYUIACUC2018-004).
Experiment time
METH treatment and collection of brain tissue samples were conducted at ZT 4-5, wherein ZT 0 indicates lights on (e.g., 7 AM) and ZT 12 indicates lights off (e.g., 7 PM), as previously described (Abarca et al., 2002; Hood et al., 2010; Kim et al., 2019). ZT 4-5 was selected based on previous reports (Abarca et al., 2002; Hood et al., 2010; Kim et al., 2019).
mRNA-Seq Data Analyses (Microarray)
Drug naïve animals and animals treated with METH for 7 days (n=3/group) were sacrificed by decapitation at ZT 4-5, and the extracted brain samples were separated into the target area, striatum, using the mouse brain matrix, and stored at –80°C. Total RNA was isolated, purified, and checked for quality. Total RNA was used to construct cDNA libraries with the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA). The protocol involved polyA-selected RNA extraction, RNA fragmentation, random hexamer primed reverse transcription and 100nt paired-end sequencing by Illumina NovaSeq 6000 (Illumina). The libraries were quantified using qPCR according to the qPCR Quantification Protocol Guide and qualified using an Agilent Technologies 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA).
The raw reads from the sequencer were preprocessed to remove low quality and adapter sequences before analysis and aligned the processed reads to the Mus musculus (mm10) using HISAT v2.1.0 (The HISAT-genotype source code is available in a public GitHub repository). HISAT utilizes two types of indexes for alignment (a global, whole-genome index and tens of thousands of small local indexes). These indexes are constructed using the same BWT (Burrows–Wheeler transform)/ graph FM index (GFM) as Bowtie2 (Johns Hopkins University, MD, USA). Owing to its use of these efficient data structures and algorithms, HISAT generates spliced alignments several times faster than Bowtie and BWA. The reference genome sequence of Mus musculus (mm10) and annotation data were downloaded from the NCBI (https://www.ncbi.nlm.nih.gov/genome/). The transcript assembly of known transcripts was then processed using StringTie v1.3.4d (Johns Hopkins University, MD, USA). Base on the results obtained, the expression abundance of the transcript and gene were calculated as read count or FPKM value (fragments per kilobase of exon per million fragments mapped) per sample. The expression profiles were used to perform additional analysis such as those of DEGs. In groups with different conditions, DEGs or transcripts can be filtered through statistical hypothesis testing.
Validation of microarray data by quantitative real-time PCR (qRT-PCR)
Drug naïve animals and animals treated with METH for 7 days (n=6-8/group) were sacrificed at ZT 4-5, and each brain sample was kept at –80°C immediately after extraction. Total RNA from the striatum was isolated and purified using TRIzol reagent and the Hybrid-RTM kit. The AccuPower® CycleScript RT PreMix was used to prepare cDNA, and all procedures were conducted in compliance with the manufacturer’s instructions. Table 1 shows the sequences of each target gene’s primer. A total volume of 20 μL (SYBR® Green 10 μL+forward primer 1 μL+reverse primer 1 μL+cDNA 2 μL+distilled water 6 μL) was used to run qRT-PCR. A housekeeping gene (GAPDH) was used to normalize each value. All data were analyzed using the ΔΔCt method (Ct is defined as the threshold cycle).
Table 1.
Selected 9 genes primers for Real-time PCR
| Forward (5`→3`) | Reverse (5`→3`) | |
|---|---|---|
| Ncald | AGC ATG GAC AGA CTT CGT GG | CAG GAC TGG ATG GGT TTC CC |
| Cpa6 | GTC TGG ATA GAC TGC GGC ATT | GTC TCT TGA CCG GGT CTT TC |
| Pklr | TAG GAG CAC CAG CAT CAT TG | CAT CCC TGC CTT GAT CAT CT |
| Ttc29 | CTT CCC ATG ACT CAT ACA AGG C | CCC TTT GAA ATT CTG TTC CAG GT |
| Cbr2 | GGG CAG GGA AAG GGA TTG G | CCA CAC ACA CGG GCT CTA TTC |
| Egr2 | GCC AAG GCC GTA GAC AAA ATC | CCA CTC CGT TCA TCT GGT CA |
| Prg4 | CGC CTT TTC CAA AGA TCA ATA CTA | GTG GTA ATT GCT CTT GCT GTT |
| Lcn2 | ATG TAT GGC CGG TAC ACT CAG | AAC AAA TGC GAC ATC TGG CAC |
| Camsap2 | CGA CCT CAG GCG GCT AAA | GAG GGC CTT TGA TCG GTG T |
Statistical analysis of gene expression level
The relative abundances of gene were measured in read count using StringTie. We performed statistical analysis to find DEGs using the estimates of abundances for each gene in the samples. Genes with more than one ‘zero’ read count values in the samples were excluded. Filtered data were log2-transformed and subjected to RLE normalization. Statistical significance of the differential expression data was determined using the nbinomWaldTest using DESeq2 and fold change (FC); the null hypothesis was that no difference exists among groups. False discovery rate (FDR) was controlled by adjusting the p value using the Benjamini-Hochberg algorithm. For the DEG set, hierarchical clustering analysis was performed using complete linkage and Euclidean distance as a measure of similarity. Gene-enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses for all DEGs were also performed based on Gene Ontology (http://geneontology.org/), and KEGG pathway (https://www.genome.jp/kegg/) database, respectively. We performed functional analysis using STRING database online (https://string-db.org/) which aims to collect, score, and integrate all publicly available sources of protein-protein interaction (PPI) information, and to complement these with computational predictions.
All qRT-PCR data are expressed as mean ± SEM and analyzed using two-way analysis of variance (ANOVA). When significant group differences were observed, the Bonferroni post-hoc tests were performed. Statistical analyses were performed using GraphPad Prism v7 (GraphPad Software Inc., San Diego, CA, USA), and p<0.05 was considered statistically significant.
RESULTS
Striatal gene expression changes in drug naïve Per2 KO/WT mice
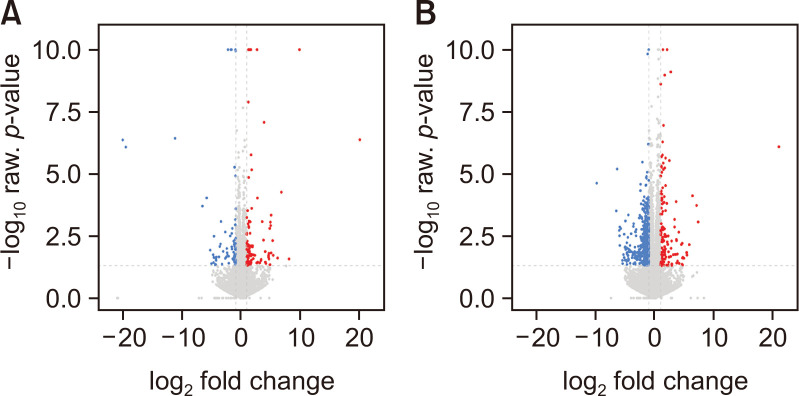
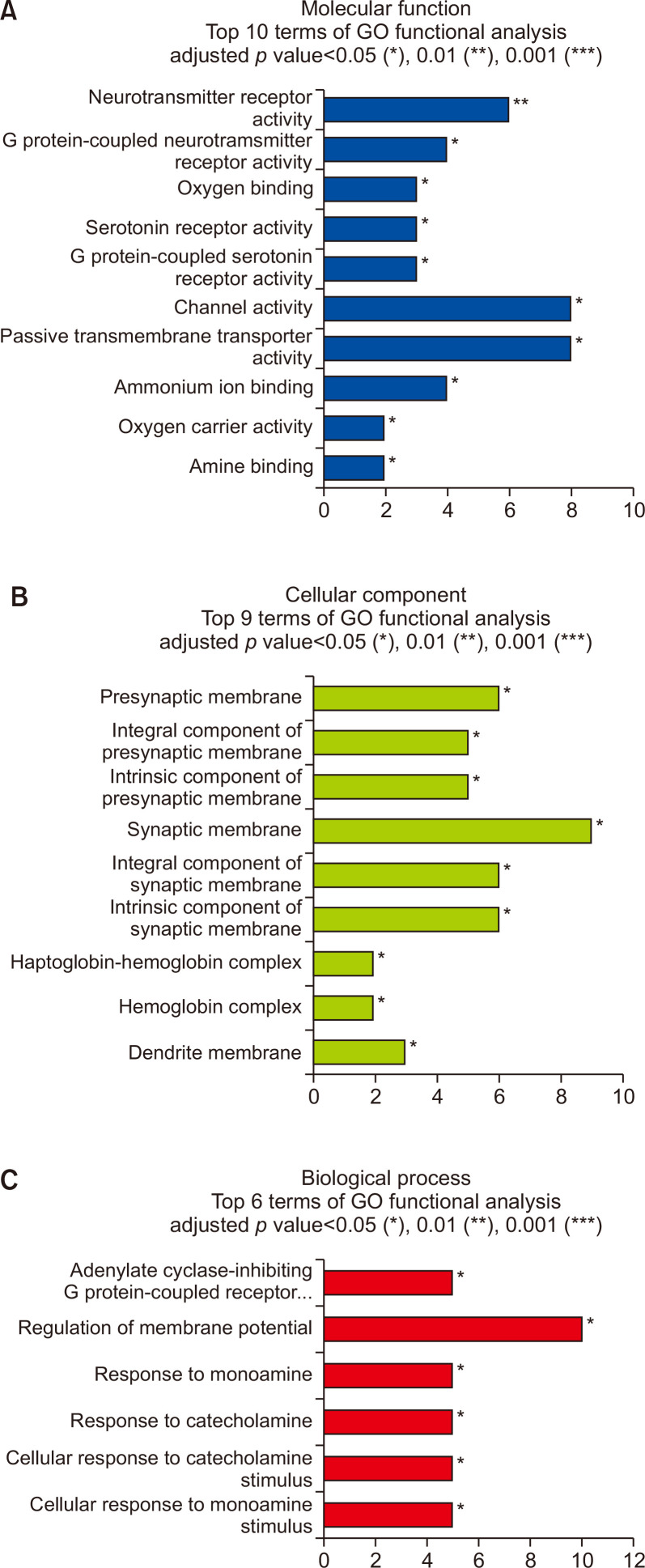
Microarray analyses indicated that 122 genes were differentially expressed in the striatum of drug naïve Per2 KO/WT mice with |FC|≥2, p≤0.05 (Fig. 1). Among them, 64 genes were upregulated, and 58 genes were downregulated in Per2 KO mice compared to WT mice (Supplementary Table 1, 2). Total DEGs, 122 genes in drug naïve Per2 KO/WT mice were analyzed using GO analysis (Fig. 2). Results showed that DEGs in drug naïve groups were strongly enriched in response to channel activity, neurotransmitter receptor activity, G-protein coupled receptor (GPCR) activity, and serotonin receptor activity in molecular functions (Fig. 2A). These genes were also enriched in response to synaptic membrane, presynaptic membrane, integral component of synaptic membrane, intrinsic component of synaptic membrane, and dendritic membrane in cellular compartment (Fig. 2B). These results indicated that DEGs in Per2 KO mice might be associated with synaptic structures and functions. All DEGs in drug naïve Per2 KO/WT mice are also associated with membrane potential regulation, GPCR regulation, and responses to monoamine/catecholamine in biological process (Fig. 2C) indicating that DEGs in Per2 KO would play important roles in GPCR-related neurotransmitter. In addition, KEGG analysis suggests that DEGs in drug naïve Per2 KO/WT mice are also related with neuroactive ligand-receptor interaction, cAMP signaling, chemokine signaling, and cholinergic synapse pathways (Supplementary Fig. 1). These pathways are similar to the results of GO analysis results.
Fig. 1.
Hierarchical clustering analysis of DEGs. Volcano plot showing the transcriptional changes in the striatum of drug naïve Per2 KO/WT mice (A), and before/after METH treatments in Per2 KO mice (B). Each circle represents one gene. The log fold change (FC) in the indicated genotype versus wild type is represented on the x-axis. The y-axis shows the log10 of the p value. A p value of 0.05 and a FC of 2 are indicated by grey lines. In the plot, the area above the genes is colored if they pass the thresholds for FDR and Log FC, red if they are upregulated and blue if they are downregulated.
Fig. 2.
Gene ontology (GO) enrichment analysis of DEGs between drug naïve Per2 KO and WT mice. (A) Molecular process, (B) cellular component, and (C) biological process. *p<0.05, **p<0.01, ***p<0.001.
Striatal gene expression changes in drug naïve Per2 KO/Per2 KO mice treated with METH
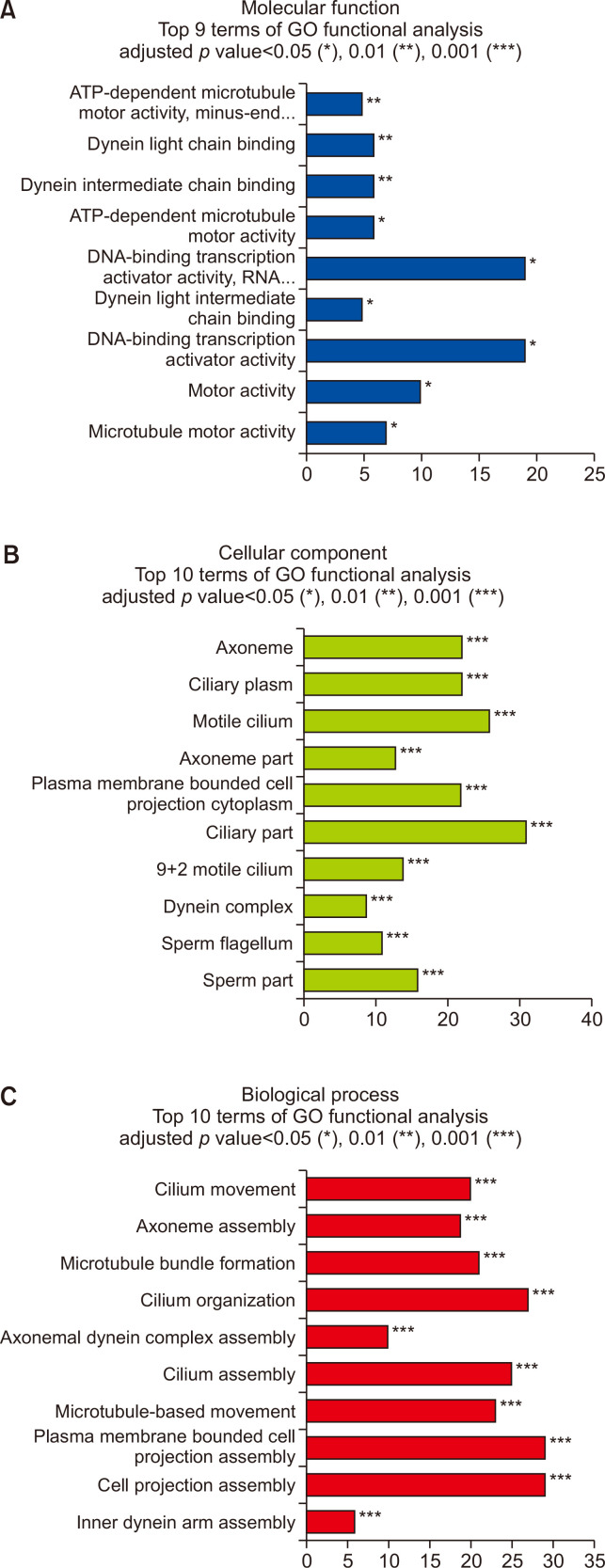
When the animals were treated with METH for 7 days, 90 genes were upregulated, and 347 genes were downregulated in Per2 KO mice after treatment (Supplementary Table 3, 4). In GO analysis, 437 DEGs in Per2 KO mice before and after METH treatment were strongly enriched in response to DNA-binding transcription activator activity, motor activity, and ATP-dependent microtubule motor activity in molecular functions (Fig. 3A). In the cellular compartment, the DEGs were strongly enriched in ciliary part, motile cilium, axoneme, and ciliary plasm (Fig. 3B). Additionally, 437 DEGs were also associated with membrane bound cell projection assembly, cell projection assembly, cilium organization, microtubule-based movement, and cilium movement in biological processes (Fig. 3C). These findings suggest that Per2 roles in METH-induced responses might be associated with intracellular cilia or microtubule activities. In KEGG analysis, 437 DEGs were significantly enriched in neuroactive ligand-receptor interaction, cAMP signaling, PI3K-Akt signaling, actin cytoskeleton, and calcium signaling pathways (Supplementary Fig. 2). Results of the KEGG analysis in Per2 KO before/after treatment with METH were similar to the results from drug naïve Per2 KO/WT mice.
Fig. 3.
Gene ontology (GO) enrichment analysis of DEGs in the Per2 KO mice before and after METH treatment. (A) molecular process, (B) cellular component, and (C) biological process. *p<0.05, **p<0.01, ***p<0.001.
Selected DEGs in METH-induced dependent responses
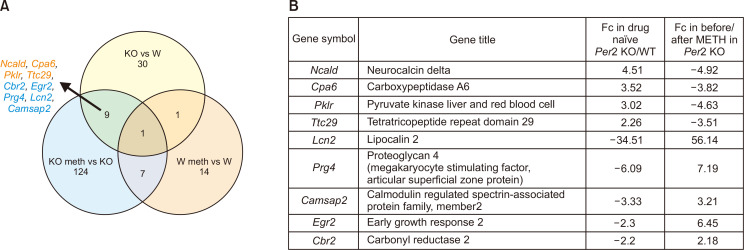
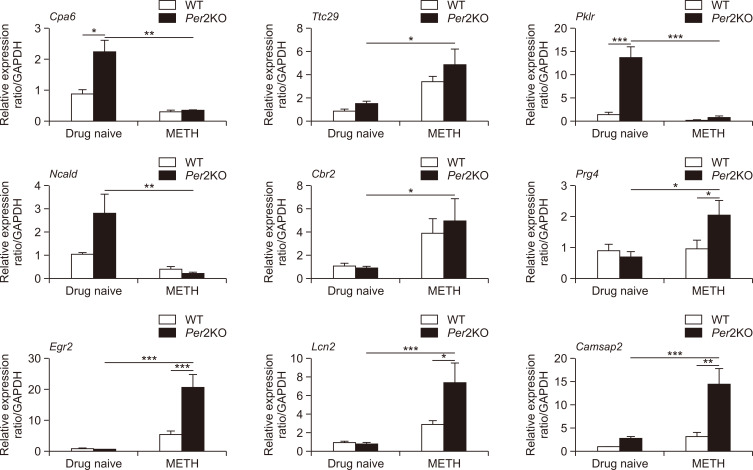
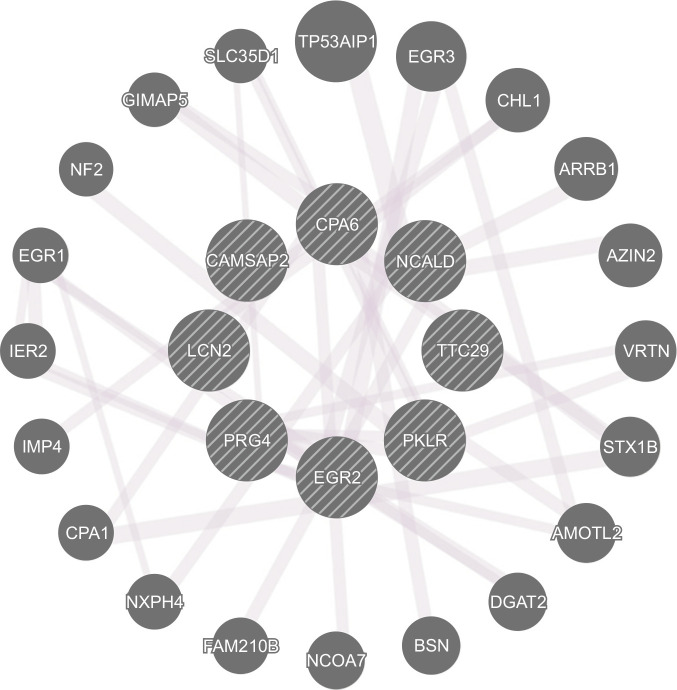
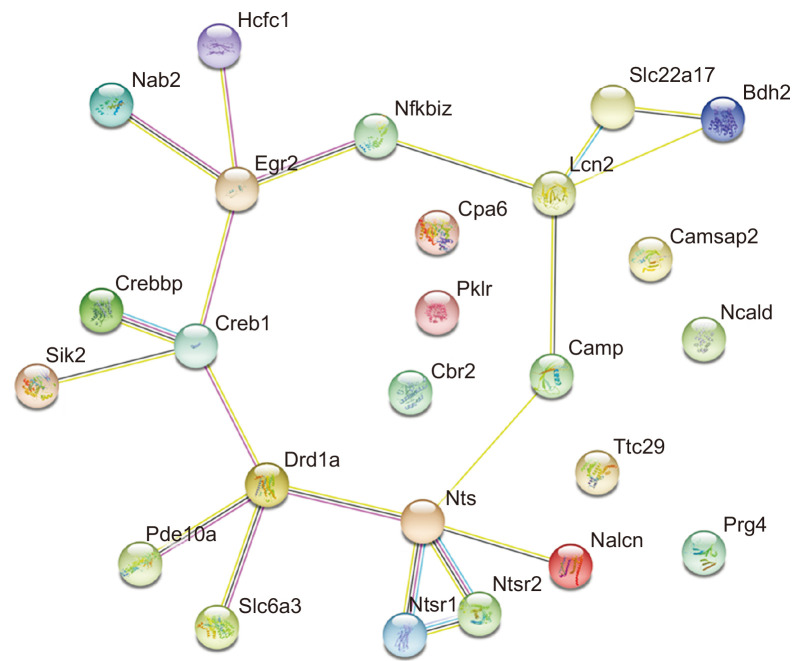
We analyzed the common DEGs in drug naïve Per2 KO/WT mice and before/after METH treatment in Per2 KO mice to explore some of the potential genetic underpinnings of the relationship between Per2 and METH-induced dependent responses. Firstly, we selected some DEGs associated with brain and drug addiction through literature searches. Among the total DEGs, 41 genes were chosen after literature searches in drug naïve Per2 KO/WT mice, while 141 genes were chosen in Per2 KO mice before and after METH treatment (Fig. 4). In WT mice before/after treatment with METH, only 23 genes were differentially expressed. We selected 9 genes that changed commonly in drug naïve Per2 KO/WT mice and in Per2 KO mice before/after METH treatment, but not in WT mice before/after METH. Interestingly, Ncald, Cpa6, Pklr, and Ttc29 were upregulated in drug naïve Per2 KO mice compared to drug naïve WT mice, while the genes were downregulated in Per2 KO mice after METH treatment. Cbr2, Egr2, Prg4, Lcn2, and Camsap2 were more downregulated in drug naïve Per2 KO mice than in drug naïve WT mice, while the genes were more upregulated in Per2 KO mice after METH treatment. qRT-PCR was used to confirm the selected 9 genes identified in microarray. All qRT-PCR results confirmed with the microarray results except Ttc29 in Per2 KO mice after METH treatment and Camsap2 in drug naïve Per2 KO mice (Fig. 5). In addition, GeneMANIA (http://www.genemania.org/) was used to predict the functions of the selected 9 genes (Fig. 6). Results of GeneMANIA suggested that the selected 8 genes except Cbr2 might be associated with response to gonadotropin stimulus, response to cAMP, response to organophosphorus, and regulation of neuronal synaptic plasticity. To obtain the PPI network, the DEGs of the 9 target genes were entered into the STRING database. Fig. 7 shows the PPI network of target genes with 25 nodes and 20 edges (average node degree: 1.6, PPI enrichment p value: 7.15e-11).
Fig. 4.
Venn diagrams and target genes. (A) Venn diagrams showing the number of DEGs in the striatum of drug naïve Per2 KO/WT mice, before/after METH in Per2 KO mice, and before/after METH in WT mice (B) target genes that commonly changed in drug naïve Per2 KO/WT mice, and before/after METH in Per2 KO mice that showed more vulnerable responses against METH.
Fig. 5.
Confirmed changes in the striatal gene expression of Per2 KO and WT mice. qRT-PCR validated expressions of target genes (Ncald, Cpa6, Pklr, Ttc29, Cbr2, Egr2, Prg4, Lcn2, and Camsap2) in the striatum of Per2 KO and WT mice before and after METH chronic treatments. *p<0.05, **p<0.01, ***p<0.001.
Fig. 6.
Network analysis of target genes by GeneMANIA.
Fig. 7.
Network analysis of target genes by STRING. Results of protein-protein interaction (PPI) network for the target genes from STRING database. PPI network of target genes shows 25 nodes and 20 edges (average node degree: 1.6, PPI enrichment p value: 7.15e-11).
DISCUSSION
The findings from our previous behavioral studies indicate that the dependence responses of chronic METH treatment are associated with the expression level of Per2 as evidenced by more developed METH-induced locomotor sensitization and reward effects in Per2 KO mice, while Per2 overexpressed mice exhibited lesser responses against METH compared to WT mice (Kim et al., 2019). In addition to our previous study, increasing studies have reported that disrupted Per2 expression can increase the vulnerability to drugs such as cocaine, alcohol, and morphine (Abarca et al., 2002; Spanagel et al., 2005; Perreau-Lenz et al., 2010; Brager et al., 2013). Several studies have attempted to reveal the mechanism of Per2 in drug addiction, however it remains unclear. To elucidate the possible mechanisms of Per2 in METH-induced dependence responses, we analyzed DEGs in the striatum of Per2 KO and WT mice before and after METH treatment. Disruption of Per2 might regulate alterations in the expression of multiple genes that are associated with biological functions.
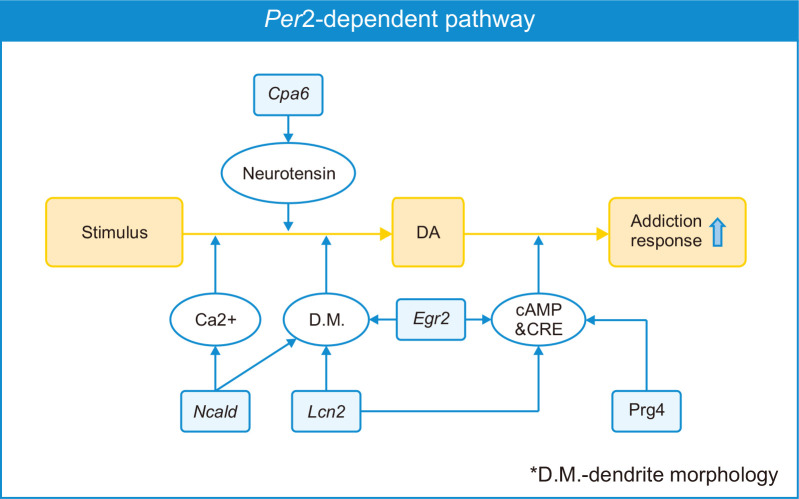
Among the common DEGs in drug naïve Per2 KO/WT mice and before/after METH treatment in Per2 KO mice, Cpa6 is the gene encoding Carboxypeptidase A6 (CPA6), a metallocarboxypeptidase enzyme. CPA6 converts enkephalins and neurotensin into inactive forms (Lyons et al., 2008). Neurotensin is an endogenous neuropeptide known to modulate dopaminergic pathways (Kasckow and Nemeroff, 1991; Binder et al., 2001). Enhanced neurotensin decreased dopamine release in the NAc and blocked stimulant-induced hyper-locomotor activity (Binder et al., 2001; Servonnet et al., 2017). Previous studies using Per2 transgenic (TG) mice have reported that disruption or overexpression of Per2 might lead to changes in dopaminergic systems and stimulant-induced behavior responses (Abarca et al., 2002; Hampp et al., 2008; Kim et al., 2018, 2019). In contrast, alteration of the dopamine expression level influenced Per2 expression (Hood et al., 2010; Gravotta et al., 2011). All these results suggest that Per2 may influence stimulant-induced dependence responses through the interaction of Cpa6, neurotensin, and dopamine. However, PPI networks (Fig. 7) did not exhibit any network between Cpa6 and neurotensin-related proteins, although some studies have reported the relationship between them. Thus, further studies are required to deepen our insights into the relationship between Per2 and Cpa6 on drug dependence.
Egr2 (also termed Krox20) belongs to the family of early growth response (Egr) genes associated with neuronal development (Gabet et al., 2010; dela Peña et al., 2015). Egr2 is also associated with long-term potentiation (Williams et al., 1995; Belluardo et al., 2005). Belluardo and colleagues (2005) reported that Egr2 may play key roles as a cognitive enhancer in nicotine dependence (Belluardo et al., 2005). Growing studies have reported the changes of Egr2 expression by drug administration. Rats treated with heroin demonstrated an increase in Egr2 expression and exhibited reinforcement effects and conditioned place preference (CPP) against heroin (Kuntz et al., 2008; Xia et al., 2018). Chronic morphine treatment increased Krox20 (Egr2), Per2, and cAMP responsive element binding protein (CREB) in the frontal cortex of Wistar rats (Ammon et al., 2003). The spontaneously hypertensive rat (SHR) treated with amphetamine also showed increased expression level of Egr2 and Per2 in the striatum and exhibited reinforcement effects and CPP (dela Peña et al., 2015). METH administration induced changes in Egr2 expression in the striatum. Acute METH treatment increased Egr2 expression in the striatum of rats, while chronic METH administration decreased Egr2 and Creb expressions (McCoy et al., 2011). Based on these results, we can assume that Per2 may influence drug addiction through Egr2 and Creb activities. Our PPI networks also show the relationship between EGR2 and CREB (Fig. 7).
LCN2 is an iron-related protein encoded by lipocalin 2 (Lcn2) that enhances the activation of nuclear factor kappa B subunit 2 (NF-κB) pathway (Ye et al., 2014). LCN2 also establishes a network with Egr2 through NF-κB in our PPI networks. Some previous studies have reported that NF-κB plays key roles in drug rewarding effects and drug-related memory (Zhang et al., 2011; Ferreira et al., 2013). Inhibition of NF-κB blocked CPP against morphine. In contrast to these studies, Lcn2 expression reduced after chronic morphine treatment (Ye et al., 2014). Lcn2 also participates in the regulation of spine morphology and neuronal excitability (Mucha et al., 2011; Ferreira et al., 2013). Upregulated Lcn2 induced a reduction in dendritic spine actin’s mobility in the hippocampus of mice (Mucha et al., 2011). Recently, several studies have suggested that increased Lcn2 expression might be associated with neurotoxicity and neuroinflammation of dopamine neurons (Kim et al., 2016; Xu et al., 2018). In contrast, LCN2 null mice exhibited decreased LTP in the hippocampus and impaired cognitive abilities in special learning tests (Ferreira et al., 2013). Although some studies reviewed various functions of Lcn2 in the central nervous system (Ferreira et al., 2015; Jha et al., 2015), evidences to support the roles of Lcn2 in drug addiction are not sufficient. Thus, further studies are required to confirm the relationship between Per2 and Lcn2 in drug addiction.
It was also reported that neurocalcin delta (Ncald), a member of the neuronal calcium sensor (NCS) family, was associated with neuronal morphology. Overexpressed Ncald induced a reduction in axon outgrowth and branching of cultured hippocampal neurons (Yamatani et al., 2010), and Ncald KO restored axonal growth in zebrafish and mice (Riessland et al., 2017). In contrast, Upadhyay et al. (2019) reported that Ncald KO impaired neurogenesis in adult mice, while Ncald heterozygous mice did not exhibit significant changes. Additionally, Ncald was downregulated in the obesity-induced memory impairment animal model (Ma et al., 2017). Thus, the study suggested that Ncald might play important roles in memory. Consistently, Ncald expression was lower in patients with Alzheimer’s disease (Shimohama et al., 1996; Miller et al., 2013). Ncald is also associated with coffee and caffeine consumption (Yamamoto et al., 2015; Lee, 2018). Ncald is one of the SNPs associated with coffee consumption. However, evidences showing the relationship between Ncald and substance dependence are insufficient. In addition, NCALD encoded by Ncald is one of the key proteins that contribute to the release of neurotransmitters (Ivings et al., 2002; Vercauteren et al., 2007). Thus, Ncald might be a good target to better understand drug addiction.
PRG4 (proteoglycan4), also known as lubricin, encoded by Prg4, acts as a joint lubricant. Interestingly, Prg4 expression significantly changed in the striatum of drug naïve Per2 KO and WT mice, and before and after METH treatment in Per2 KO mice, but not before and after METH treatment in WT mice. Some previous studies reported that Prg4 expression was associated with cAMP activities in chondrocyte or synoviocyte (Wu et al., 2017; Qadri et al., 2018). Wu et al. (2017) reported that the kappa opioid receptor agonist induced changes of Prg4 expression through the cAMP/CREB pathway. Taken together, Prg4 could be one of the new candidates to elucidate the drug addiction pathway.
We found that the roles of selected common DEGs reported by other previous studies are associated with the GO and KEGG analyses of total DEGs such as intrinsic component of synaptic membrane, dendritic membrane in cellular components, calcium signaling pathways, GPCR regulation, cAMP signaling, etc., which have relevance to drug addiction pathways. This fact indicates that the selected common DEGs including Cpa6, Ncald, Egr2, Lcn2, and Prg4, might play key roles in drug addiction, and their pathways may be associated with Per2 regulation. Per2 KO mice exhibited more vulnerable METH addiction responses in our previous study, and in the current study, we found the expression levels of some genes such as Cpa6, Ncald, Egr2, Lcn2, and Prg4 changed in Per2 KO mice after METH treatments. Taken together, the selected genes might play key roles in vulnerable responses of Per2 KO mice against METH. Furthermore, the potential pathway of the selected 5 genes (Cpa6, Ncald, Egr2, Lcn2, and Prg4) under Per2 regulation is illustrated in Fig. 8. Thus, further studies should be performed to investigate the link between these genes and the effects on drug dependence responses. These target genes would be new candidates to reveal the mechanisms of drug addiction.
Fig. 8.
Schematic diagram (Per2-mediated addiction behavior hypothesis). Diagram illustrates that Per2 expression might influence drug addiction through the pathways of target genes.
Supplementary Materials
ACKNOWLEDGMENTS
This study was supported by a grant from the National Research Foundation of Korea (NRF) (2020R1F1A1075633) funded by the Korean government (MSIT), and a grant from Ministry of Food and Drug Safety in 2019 (19182MFDS410).
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
REFERENCES
- Abarca C., Albrecht U., Spanagel R. J. Cocaine sensitization and reward are under the influence of circadian genes and rhythm. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9026–9030. doi: 10.1073/pnas.142039099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U., Zheng B., Larkin D., Sun Z. S., Lee C. C. mPer1 and mPer2 are essential for normal resetting of the circadian clock. J. Biol. Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- Ammon S., Mayer P., Riechert U., Tischmeyer H., Höllt V. J. Microarray analysis of genes expressed in the frontal cortex of rats chronically treated with morphine and after naloxone precipitated withdrawal. Mol. Brain Res. 2003;112:113–125. doi: 10.1016/S0169-328X(03)00057-3. [DOI] [PubMed] [Google Scholar]
- Bae K., Jin X., Maywood E. S., Hastings M. H., Reppert S. M., Weaver D. R. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/S0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- Belluardo N., Olsson P., Mudo G., Sommer W., Amato G., Fuxe K. Transcription factor gene expression profiling after acute intermittent nicotine treatment in the rat cerebral cortex. Neuroscience. 2005;133:787–796. doi: 10.1016/j.neuroscience.2005.01.061. [DOI] [PubMed] [Google Scholar]
- Binder E. B., Kinkead B., Owens M. J., Nemeroff C. B. Neurotensin and dopamine interactions. Pharmacol. Rev. 2001;53:453–486. [PubMed] [Google Scholar]
- Brager A. J., Stowie A. C., Prosser R. A., Glass J. D. The mPer2 clock gene modulates cocaine actions in the mouse circadian system. Behav. Brain Res. 2013;243:255–260. doi: 10.1016/j.bbr.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli D., Venniro M., Zhang M., Bossert J. M., Warren B. L., Hope B. T., Shaham Y. Role of dorsomedial striatum neuronal ensembles in incubation of methamphetamine craving after voluntary abstinence. J. Neurosci. 2017;37:1014–1027. doi: 10.1523/JNEUROSCI.3091-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Alicata D., Ernst T., Volkow N. Structural and metabolic brain changes in the striatum assoiciated with methamphetamine abuse. Addiction. 2007;102:16–32. doi: 10.1111/j.1360-0443.2006.01782.x. [DOI] [PubMed] [Google Scholar]
- dela Peña I., de la Peña J. B., Kim B. N., Han D. H., Noh M., Cheong J. H. Gene expression profiling in the striatum of amphetamine-treated spontaneously hypertensive rats which showed amphetamine conditioned place preference and self-administration. Arch. Pharm. Res. 2015;38:865–875. doi: 10.1007/s12272-014-0470-x. [DOI] [PubMed] [Google Scholar]
- Everitt B. J., Robbins T. W. Drug addiction: updating actions to habits to compulsions ten years on. Annu. Rev. Psychol. 2016;67:23–50. doi: 10.1146/annurev-psych-122414-033457. [DOI] [PubMed] [Google Scholar]
- Ferreira A. C., Da Mesquita S., Sousa J. C., Correia-Neves M., Sousa N., Palha J. A., Marques F. From the periphery to the brain: Lipocalin-2, a friend or foe? Prog. Neurobiol. 2015;131:120–136. doi: 10.1016/j.pneurobio.2015.06.005. [DOI] [PubMed] [Google Scholar]
- Ferreira A. C., Pinto V., Mesquita S. D., Novais A., Sousa J. C., Correia-Neves M., Sousa N., Palha J. A., Marques F. Lipocalin-2 is involved in emotional behaviors and cognitive function. Front. Cell. Neurosci. 2013;7:122. doi: 10.3389/fncel.2013.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabet Y., Baniwal S. K., Leclerc N., Shi Y., Kohn-Gabet A. E., Cogan J., Dixon A., Bachar M., Guo L., Turman J. E., Jr, Frenkel B. Krox20/EGR2 deficiency accelerates cell growth and differentiation in the monocytic lineage and decreases bone mass. Am. J. Hematol. 2010;116:3964–3971. doi: 10.1182/blood-2010-01-263830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravotta L., Gavrila A. M., Hood S., Amir S. J. Global depletion of dopamine using intracerebroventricular 6-hydroxydopamine injection disrupts normal circadian wheel-running patterns and PERIOD2 expression in the rat forebrain. J. Mol. Neurosci. 2011;45:162–171. doi: 10.1007/s12031-011-9520-8. [DOI] [PubMed] [Google Scholar]
- Hampp G., Ripperger J. A., Houben T., Schmutz I., Blex C., Perreau-Lenz S., Brunk I., Spanagel R., Ahnert-Hilger G., Meijer J. H., Albrecht U. Regulation of monoamine oxidase A by circadian-clock components implies clock influence on mood. Curr. Biol. 2008;18:678–683. doi: 10.1016/j.cub.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Hasler B., Smith L., Cousins J., Bootzin R. J. Circadian rhythms, sleep, and substance abuse. Sleep Med. Rev. 2012;16:67–81. doi: 10.1016/j.smrv.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood S., Cassidy P., Cossette M. P., Weigl Y., Verwey M., Robinson B., Stewart J., Amir S. Endogenous dopamine regulates the rhythm of expression of the clock protein PER2 in the rat dorsal striatum via daily activation of D2 dopamine receptors. J. Neurosci. 2010;30:14046–14058. doi: 10.1523/JNEUROSCI.2128-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivings L., Pennington S. R., Jenkins R., Weiss J. L., Burgoyne R. D. Identification of Ca2+-dependent binding partners for the neuronal calcium sensor protein neurocalcin δ: interaction with actin, clathrin and tubulin. Biochem. J. 2002;363:599–608. doi: 10.1042/bj3630599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha M. K., Lee S., Park D. H., Kook H., Park K. G., Lee I. K., Suk K. Diverse functional roles of lipocalin-2 in the central nervous system. Neurosci. Biobehav. Rev. 2015;49:135–156. doi: 10.1016/j.neubiorev.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Kasckow J., Nemeroff C. B. The neurobiology of neurotensin: focus on neurotensin-dopamine interactions. Regul. Pept. 1991;36:153–164. doi: 10.1016/0167-0115(91)90053-J. [DOI] [PubMed] [Google Scholar]
- Kim B. W., Jeong K. H., Kim J. H., Jin M., Kim J. H., Lee M. G., Choi D. K., Won S. Y., McLean C., Jeon M. T., Lee H. W., Kim S. R., Suk K. Pathogenic upregulation of glial lipocalin-2 in the parkinsonian dopaminergic system. J. Neurosci. 2016;36:5608–5622. doi: 10.1523/JNEUROSCI.4261-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Custodio R. J., Botanas C. J., de la Peña J. B., Sayson L. V., Abiero A., Ryoo Z. Y., Cheong J. H., Kim H. J. The circadian gene, Per2, influences methamphetamine sensitization and reward through the dopaminergic system in the striatum of mice. Addict. Biol. 2019;24:946–957. doi: 10.1111/adb.12663. [DOI] [PubMed] [Google Scholar]
- Kim M., De La Pena J. B., Cheong J. H., Kim H. J. J. Neurobiological functions of the period circadian clock 2 gene, Per2. Biomol. Ther. (Seoul) 2018;26:358–367. doi: 10.4062/biomolther.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovanen L., Saarikoski S. T., Haukka J., Pirkola S., Aromaa A., Lönnqvist J., Partonen T. Circadian clock gene polymorphisms in alcohol use disorders and alcohol consumption. Alcohol Alcohol. 2010;45:303–311. doi: 10.1093/alcalc/agq035. [DOI] [PubMed] [Google Scholar]
- Kuntz K. L., Patel K. M., Grigson P. S., Freeman W. M., Vrana K. E. Heroin self-administration: II. CNS gene expression following withdrawal and cue-induced drug-seeking behavior. Pharmacol. Biochem. Behav. 2008;90:349–356. doi: 10.1016/j.pbb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. H. Investigating the possible causal association of coffee consumption with osteoarthritis risk using a Mendelian randomization analysis. Clin. Rheumatol. 2018;37:3133–3139. doi: 10.1007/s10067-018-4252-6. [DOI] [PubMed] [Google Scholar]
- Lyons P. J., Callaway M. B., Fricker L. D. Characterization of carboxypeptidase A6, an extracellular matrix peptidase. J. Biol. Chem. 2008;283:7054–7063. doi: 10.1074/jbc.M707680200. [DOI] [PubMed] [Google Scholar]
- Ma W. W., Ding B. J., Yuan L. H., Zhao L., Yu H. L., Xiao R. Neurocalcin-delta: a potential memory-related factor in hippocampus of obese rats induced by high-fat diet. Afr. Health Sci. 2017;17:1211–1221. doi: 10.4314/ahs.v17i4.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy M. T., Jayanthi S., Wulu J. A., Beauvais G., Ladenheim B., Martin T. A., Krasnova I. N., Hodges A. B., Cadet J. L. Chronic methamphetamine exposure suppresses the striatal expression of members of multiple families of immediate early genes (IEGs) in the rat: normalization by an acute methamphetamine injection. Psychopharmacology (Berl.) 2011;215:353–365. doi: 10.1007/s00213-010-2146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. A., Woltjer R. L., Goodenbour J. M., Horvath S., Geschwind D. H. Genes and pathways underlying regional and cell type changes in Alzheimer's disease. Genome Med. 2013;5:48. doi: 10.1186/gm452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucha M., Skrzypiec A. E., Schiavon E., Attwood B. K., Kucerova E., Pawlak R. Lipocalin-2 controls neuronal excitability and anxiety by regulating dendritic spine formation and maturation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:18436–18441. doi: 10.1073/pnas.1107936108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler E. J., Malenka R. C. The addicted brain. Sci. Am. 2004;290:78–85. doi: 10.1038/scientificamerican0304-78. [DOI] [PubMed] [Google Scholar]
- Perreau-Lenz S., Sanchis-Segura C., Leonardi-Essmann F., Schneider M., Spanagel R. Development of morphine-induced tolerance and withdrawal: involvement of the clock gene mPer2. Eur. Neuropsychopharmacol. 2010;20:509–517. doi: 10.1016/j.euroneuro.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Qadri M. M., Jay G. D., Ostrom R. S., Zhang L. X., Elsaid K. A. cAMP attenuates TGF-β's profibrotic responses in osteoarthritic synoviocytes: involvement of hyaluronan and PRG4. Am. J. Physiol. Cell Physiol. 2018;315:C432–C443. doi: 10.1152/ajpcell.00041.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riessland M., Kaczmarek A., Schneider S., Swoboda K. J., Löhr H., Bradler C., Grysko V., Dimitriadi M., Hosseinibarkooie S., Torres-Benito L., Peters M., Upadhyay A., Biglari N., Kröber S., Hölker I., Garbes L., Gilissen C., Hoischen A., Nürnberg G., Nürnberg P., Walter M., Rigo F., Bennett C. F., Kye M. J., Hart A. C., Hammerschmidt M., Kloppenburg P., Wirth B. Neurocalcin delta suppression protects against spinal muscular atrophy in humans and across species by restoring impaired endocytosis. Am. J. Hum. Genet. 2017;100:297–315. doi: 10.1016/j.ajhg.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servonnet A., Minogianis E. A., Bouchard C., Bédard A. M., Lévesque D., Rompré P. P., Samaha A. N. Neurotensin in the nucleus accumbens reverses dopamine supersensitivity evoked by antipsychotic treatment. Neuropharmacology. 2017;123:10–21. doi: 10.1016/j.neuropharm.2017.05.015. [DOI] [PubMed] [Google Scholar]
- Shimohama S., Chachin M., Taniguchi T., Hidaka H., Kimura J. Changes of neurocalcin, a calcium-binding protein, in the brain of patients with Alzheimer's disease. Brain Res. 1996;716:233–236. doi: 10.1016/0006-8993(96)00070-4. [DOI] [PubMed] [Google Scholar]
- Solinas M., Belujon P., Fernagut P. O., Jaber M., Thiriet N. J. Dopamine and addiction: what have we learned from 40 years of research. J. Neural Transm. (Vienna) 2019;126:481–516. doi: 10.1007/s00702-018-1957-2. [DOI] [PubMed] [Google Scholar]
- Spanagel R., Pendyala G., Abarca C., Zghoul T., Sanchis-Segura C., Magnone M. C., Lascorz J., Depner M., Holzberg D., Soyka M., Schreiber S., Matsuda F., Lathrop M., Schumann G., Albrecht U. The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nat. Med. 2005;11:35–42. doi: 10.1038/nm1163. [DOI] [PubMed] [Google Scholar]
- Upadhyay A., Hosseinibarkooie S., Schneider S., Kaczmarek A., Torres-Benito L., Mendoza-Ferreira N., Overhoff M., Rombo R., Grysko V., Kye M. J., Kononenko N. L., Wirth B. Neurocalcin delta knockout impairs adult neurogenesis whereas half reduction is not pathological. Front. Mol. Neurosci. 2019;12:19. doi: 10.3389/fnmol.2019.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercauteren F. G., Flores G., Ma W., Chabot J. G., Geenen L., Clerens S., Fazel A., Bergeron J. J., Srivastava L. K., Arckens L., Quirion R. An organelle proteomic method to study neurotransmission-related proteins, applied to a neurodevelopmental model of schizophrenia. Proteomics. 2007;7:3569–3579. doi: 10.1002/pmic.200700379. [DOI] [PubMed] [Google Scholar]
- Volkow N. D., Wise R. A., Baler R. J. The dopamine motive system: implications for drug and food addiction. Nat. Rev. Neurosci. 2017;18:741–752. doi: 10.1038/nrn.2017.130. [DOI] [PubMed] [Google Scholar]
- Williams J., Dragunow M., Lawlor P., Mason S., Abraham W., Leah J., Bravo R., Demmer J., Tate W. Krox20 may play a key role in the stabilization of long-term potentiation. Mol. Brain Res. 1995;28:87–93. doi: 10.1016/0169-328X(94)00187-J. [DOI] [PubMed] [Google Scholar]
- Wu L., Zhang S., Shkhyan R., Lee S., Gullo F., Eliasberg C. D., Petrigliano F. A., Ba K., Wang J., Lin Y. J., Evseenko D. Kappa opioid receptor signaling protects cartilage tissue against posttraumatic degeneration. JCI Insight. 2017;2:e88553. doi: 10.1172/jci.insight.88553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B., Li Y., Li R., Yin D., Chen X., Li J., Liang W. Effect of sirtuin-1 on synaptic plasticity in nucleus accumbens in a rat model of heroin addiction. Med. Sci. Monit. 2018;24:3789–3803. doi: 10.12659/MSM.910550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Wang Y., Song N., Wang J., Jiang H., Xie J. New progress on the role of glia in iron metabolism and iron-induced degeneration of dopamine neurons in Parkinson's disease. Front. Mol. Neurosci. 2018;10:455. doi: 10.3389/fnmol.2017.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto V. J., Paula V. d. J. R. d., Forlenza O. V., Santos B. D., Kerr D. S. Association study in Alzheimer's disease of single nucleotide polymorphisms implicated with coffee consumption. Arch. Clin. 2015;42:69–73. doi: 10.1590/0101-60830000000050. [DOI] [Google Scholar]
- Yamatani H., Kawasaki T., Mita S., Inagaki N., Hirata T. Proteomics analysis of the temporal changes in axonal proteins during maturation. Dev. Neurobiol. 2010;70:523–537. doi: 10.1002/dneu.20794. [DOI] [PubMed] [Google Scholar]
- Ye J., Yang Z., Li C., Cai M., Zhou D., Zhang Q., Wei Y., Wang T., Liu Y. NF-κB signaling and vesicle transport are correlated with the reactivation of the memory trace of morphine dependence. Diagn. Pathol. 2014;9:142. doi: 10.1186/1746-1596-9-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Cui Y., Jing J., Cui Y., Xin W., Liu X. Involvement of p38/NF-κB signaling pathway in the nucleus accumbens in the rewarding effects of morphine in rats. Behav. Brain Res. 2011;218:184–189. doi: 10.1016/j.bbr.2010.11.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.