Abstract
Objective
Autophagy is a physiological self-eating process that can promote cell survival or activate cell death in eukaryotic cells. In skeletal muscle, it is important for maintaining muscle mass and function that is critical to sustain mobility and regulate metabolism. The UV radiation resistance-associated gene (UVRAG) regulates the early stages of autophagy and autophagosome maturation and plays a key role in endosomal trafficking. This study investigated the essential in vivo role of UVRAG in skeletal muscle biology.
Methods
To determine the role of UVRAG in skeletal muscle in vivo, we generated muscle-specific UVRAG knockout mice using the Cre-loxP system driven by Myf6 promoter that is exclusively expressed in skeletal muscle. Myf6-Cre+ UVRAGfl/fl (M-UVRAG−/−) mice were compared to littermate Myf6-Cre+ UVRAG+/+ (M-UVRAG+/+) controls under basal conditions on a normal chow diet. Body composition, muscle function, and mitochondria morphology were assessed in muscles of the WT and KO mice at 24 weeks of age.
Results
M-UVRAG−/− mice developed accelerated sarcopenia and impaired muscle function compared to M-UVRAG+/+ littermates at 24 weeks of age. Interestingly, these mice displayed improved glucose tolerance and increased energy expenditure likely related to upregulated Fgf21, a marker of muscle dysfunction. Skeletal muscle of the M-UVRAG−/− mice showed altered mitochondrial morphology with increased mitochondrial fission and EGFR accumulation reflecting defects in endosomal trafficking. To determine whether increased EGFR signaling had a causal role in muscle dysfunction, the mice were treated with an EGFR inhibitor, gefitinib, which partially restored markers of muscle and mitochondrial deregulation. Conversely, constitutively active EGFR transgenic expression in UVRAG-deficient muscle led to further detrimental effects with non-overlapping distinct defects in muscle function, with EGFR activation affecting the muscle fiber type whereas UVRAG deficiency impaired mitochondrial homeostasis.
Conclusions
Our results show that both UVRAG and EGFR signaling are critical for maintaining muscle mass and function with distinct mechanisms in the differentiation pathway.
Keywords: UVRAG, Skeletal muscle, Fgf21, Mitochondrial dynamics, EGFR
Highlights
-
•
Deletion of UVRAG in skeletal muscle accelerates muscle wasting with aging.
-
•
UVRAG in skeletal muscle regulates mitochondrial dynamics and function.
-
•
UVRAG deletion leads to EGFR accumulation in skeletal muscle.
-
•
Constitutively active EGFR contributes to muscle fiber type determination.
1. Introduction
Sarcopenia, or progressive loss of skeletal muscle mass and strength, is one of the hallmarks of aging that can directly affect health and lifespan. Several cellular processes are implicated in the development of age-related sarcopenia. These include inefficient protein turnover with accumulation of abnormal and dysfunctional proteins within skeletal muscle [1]. Autophagy and the ubiquitin-proteasome system are the main catabolic processes that facilitate degradation of skeletal muscle proteins, thereby playing a critical role in maintaining muscle homeostasis [2,3]. Autophagy is an essential physiological process for cellular homeostasis and a key regulator of several physiological processes including metabolism, development, and differentiation. As such, dysregulation in autophagy leads to many diseases such as neuro- and muscular-degenerative diseases, metabolic diseases, or cancer [[4], [5], [6], [7], [8]].
Autophagy is also critical for mitochondrial homeostasis, known as mitophagy. Defects in mitophagy have been associated with impaired mitochondrial function that is observed with age-related decline in muscle health [9,10]. Mitochondria are dynamic organelles that undergo continuous fusion and fission that play an important role in regulating cell survival and proliferation [11]. Mitochondrial fission occurs early in apoptosis, whereas autophagy and fusion are associated with increased cell survival [[12], [13], [14]]. The dynamic balance of fusion and fission is disturbed in many pathologies as well as with aging [[15], [16], [17], [18], [19]].
The ultraviolet radiation resistance-associated gene (UVRAG) is a novel gene that has been shown to promote autophagy through interaction with Beclin1-PI3KC3 complex [20]. UVRAG inactivation, through either null or frameshift mutations, is associated with cancer development [21,22]. In addition to its role in autophagosome maturation, UVRAG is also shown to critically regulate the endocytic pathway through its interaction with Beclin1-Vps34 complex [23]. However, whether these mechanistic processes are essential for UVRAG in skeletal muscle homeostasis and function in vivo is unknown.
In the current study, we assessed the essential in vivo role of UVRAG in skeletal muscle using tissue-specific UVRAG knockout (M-UVRAG−/−) mice. We show that M-UVRAG−/− mice exhibit an accelerated decline of muscle mass and strength with aging. Using genetic, pharmacologic, and proteomic approaches, our study reveals that UVRAG and EGFR signaling together are critical for maintaining skeletal muscle mass and function with aging, with distinct mechanisms in the muscle differentiation pathway.
2. Materials and methods
2.1. Animal model and diet
UVRAGfl/fl mice were kindly provided by T.W. Mak, and their generation and genotyping were previously described [24]. Skeletal muscle-specific UVRAG knockout mice were generated by breeding Myf6-Cre+ mice (Jackson Laboratories) with UVRAGfl/fl mice to generate Myf6-Cre+UVRAGfl/+ mice, which were then intercrossed to generate Myf6-Cre+UVRAGfl/fl mice, referred to herein as M-UVRAG−/− mice. Their wild-type littermate Myf6-Cre+UVRAG+/+ mice, referred to herein as M-UVRAG+/+ mice, were used as controls. Genotypes were identified by PCR of ear clip DNA [24]. Skeletal muscle-specific double transgenic mice were generated by breeding M-UVRAG+/+ and M-UVRAG−/− mice with EGFR+/L858R mice to generate Myf6-Cre+UVRAG+/+EGFR+/L858R and Myf6-Cre+UVRAG−/−EGFR+/L858R mice. Mouse lines were then intercrossed to generate Myf6-Cre+UVRAG+/+EGFR+/+, Myf6-Cre+UVRAG+/+EGFRL858R/L858R, Myf6-Cre+UVRAG−/−EGFR+/+, and Myf6-Cre+UVRAG−/−EGFRL858R/L858R mice. Rosa26mTmGMyf6-Cre+ mice were generated by breeding Rosa26mTmG mice (Jackson Laboratories) with Myf6-Cre+ mice. All of the animals were housed in a specific pathogen-free facility with a 12:12-h light/dark cycle with free access to water and standard irradiated rodent chow (5% fat, Harlan Teklad). All of the mice were maintained on a C57BL/6 background. All of the animal experiments were approved by the Toronto General Hospital Research Institute's Animal Care Committee.
2.2. In vivo metabolic studies
Glucose tolerance tests (GTTs) were performed on 14-16 h-fasted mice using a glucose dose of 1 g/kg body weight injected intraperitoneally. Insulin tolerance tests (ITTs) were performed on animals fasted for 4 h using insulin lispro (Lilly) 1 U/kg body weight injected intraperitoneally. Blood glucose measurements were obtained at 0, 15, 30, 45, 60, and 120 min after injection using an Accu-Chek blood glucose-monitoring system.
2.3. Treadmill exercise and grip strength test
The mice were familiarized to a treadmill (Columbus Instruments) for 5 days for acclimatization, after which they were exercised with the following protocol: 5 min at 5 m/min, 15 min at 10 m/min, 5 min at 12.5 m/min, 5 min at 15 m/min, 5 min at 17.5 m/min, 5 min at 20 m/min, and 5 min at 22.5 m/min and exercised until exhaustion at 25 m/min. A Grip Strength Meter (BIO-GS3, Bioseb) was used to measure their forelimb grip strength. The test was performed on one mouse at a time, with at least 1 min of rest between each of the five tests per animal. Measurements were normalized to body weight.
2.4. Analysis of serum Fgf21
Blood was collected from the tail vein and centrifuged at 5,000 g for 10 min at 4 °C. Serum Fgf21 levels were determined by a FGF-21 Quantikine ELISA kit (R&D Systems).
2.5. Cell culture
C2C12 myoblasts (CRL 1772, ATCC) were maintained in a growth medium consisting of DMEM (Dulbecco's modified Eagle's medium, 4.5 g/l glucose) supplemented with 10% FBS (Gibco) and 100 U/ml of penicillin and streptomycin (Gibco). Differentiation was induced by switching to differentiation medium consisting of DMEM supplemented with 2% horse serum (Gibco) when the cells reached confluence. The cells were maintained under a humidified condition of 95% air and 5% CO2 at 37 °C. All of the experiments were performed after 4 days of differentiation.
2.6. SiRNA transfection
For siRNA transfection, C2C12 cells cultured in growth medium were transfected with 50 nM of scramble or UVRAG siRNA (Ambion) using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's protocol. The cells were switched to differentiation medium after 48 h of transfection and cultured for 4 days. The cells were collected for subsequent analyses.
2.7. Oxygen consumption rate (OCR) measured using Seahorse XF analyzer
C2C12 cells were seeded at 1 × 104 cells/well in 24-well XF plates and transfected with scramble or UVRAG siRNA. After 4 days of differentiation, myotubes were switched to Assay Medium (Seahorse Biosciences) and incubated for 1 h at 37 °C in a non-CO2 incubator. OCR was measured following a cycle that included 3 min of mixing, 2 min of waiting, and measurement for 3 min. Three measurements were obtained at baseline and following injection of oligomycin (1 μM, Sigma), FCCP (0.5 μM, Sigma), rotenone, and antimycin A (1 μM, Sigma). The measurements were normalized to the total protein content per well.
2.8. Histology
Muscle tissues were harvested after an overnight fast and fixed in 4% paraformaldehyde (PFA) in 0.1 M of PBS (pH 7.4). Sections were stained with hematoxylin and eosin (H&E). Immunostaining for succinate dehydrogenase (SDH) was obtained on frozen sections of tibialis anterior (TA) muscle that were air dried for 30 min at room temperature (RT) and incubated in 0.2 M of phosphate buffer (pH 7.4), 0.1 M of succinic acid, and 1.2 mM of nitroblue tetrazolium for 1 h at 37 °C in a humidified chamber. Immunostaining for cytochrome c oxidase (COX) was incubated in 0.2 M of phosphate buffer (pH 7.4), 1.1 mM of cytochrome c, 342 mM of sucrose, 4.3 mM of catalase, and DAB (Dako) for 1 h at 37 °C in a humidified chamber. Following incubation, slides were washed with distilled water 3 times, dehydrated using a graded series of ethanol (70%, 80%, 90%, and 100%), and then cover-slipped using an aqueous mounting medium. The immunostained density of the target protein was analyzed on the mean gray value using ImageJ software. For immunofluorescent staining of phospho-Drp1, C2C12 cells were seeded on cover slips and siRNA transfected for 48 h and then differentiated for 4 days. Mitochondria were stained in live cells using 100 nM of MitoTracker (red, Thermo Fisher Scientific) for 15 min in the dark. Following live cell staining, they were washed with PBS 3 times and fixed with 4% PFA (Sigma) for 15 min at RT. Primary antibody anti-phospho-Drp1 (S616) was incubated (1:500) overnight at 4 °C, and HRP-conjugated secondary antibody (rabbit green, 1:300) was incubated for 1 h at RT. After immunostaining, the cells were washed 3 times for 10 min in PBS. Nuclei were stained with 4’,6-diamino-2-phenylindole (DAPI, blue) for 5 min and mounted with Immuno-Mount (Thermo Fisher Scientific) for 2 h at RT. For immunofluorescent staining for mitophagy, the previously described protocols were followed for C2C12 transfection and mitochondrial staining. Following MitoTracker incubation, the media was removed and lysosomes were stained in live cells using 50 nM of LysoTracker (green, Thermo Fisher Scientific) for 5 min in the dark.
2.9. Immunofluorescent images from tissues of Rosa26mTmG;Myf6-Cre+ mice
Rosa26mTmG/+;Myf6-Cre+ mice were anesthetized with isoflurane and trans-cardially perfused with 1% PFA. Their tissues were harvested and fixed with 4% PFA overnight. The tissues were then washed with PBS, equilibrated in 30% sucrose overnight, embedded in optimal cutting temperature (OCT) medium, sectioned into 20 μm slices using cryotome (Leica), and mounted on slides with mounting medium (Vector Laboratories). Fluorescent signals were then visualized using a Nikon A1R confocal microscope system.
2.10. Mitochondrial morphology analysis via transmission electron microscopy (TEM)
The extensor digitorum longus (EDL) was fixed in 2% glutaraldehyde in 0.1 M of cacodylate buffer for 2 h at RT. Muscle tissue was cut into thin sections and subsequently mounted on cover slips containing Quetol resin. Embedded samples were cut into 90 nm sections, placed on copper grids, and stained with uranyl acetate and lead citrate. Once the samples were prepared, they were imaged on a FEI Technai 20 TEM. TEM micrographs of muscle tissue of M-UVRAG−/− and controls were obtained at 25,000 x magnification (n = 3, each, 16 images per n). Only clear discernible mitochondria were traced on ImageJ software and assessed by area (μm2), perimeter (distance surrounding the actual shape, μm), aspect ratio (major to minor axis ratio), roundness (4 × area/π × major axis2), circularity (4 π × area/perimeter2), and Feret's diameter (the longest distance between any two points).
2.11. Mitochondrial DNA (mtDNA) analysis
Total DNA was extracted from muscle tissue using GenElute Mammalian Genomic DNA Miniprep kits (Sigma) according to the manufacturer's protocol. The MtDNA content was calculated using real-time PCR by measuring the expression levels of a mitochondrial-encoded gene (Cox2) and normalized to the expression levels of a nuclear-encoded gene (cyclophilin A: Ppia).
2.12. Proteomic analysis
The sample preparation was carried out at the SickKids Proteomics, Analytics, Robotics, and Chemical Biology Center (SPARC BioCenter). The resulting raw mass spectra from each fraction were processed simultaneously using MaxQuant (version 1.6.2.10). To assess the false-positive rate, a reverse hit database was created using MaxQuant. A false-discovery rate (FDR) of 1% was specified. Carbamidomethylation of cysteines was set as fixed modifications and oxidation of methionine residues, and N-terminal acetylation was set as variable modifications and up to two missed cleavages were allowed. Trypsin/Lys-C was selected as the digestion enzyme and a fragment tolerance of 0.5 Da and a parent tolerance of 20 Da were used. Protein was identified with a minimum of one unique peptide. Quantification was performed using unmodified unique and razor peptides and a minimum of one counted ratio. Re-quantification and matching between runs were selected. Tandem mass tag isobaric labels (126, 127N, 127C, 128N, 128C, 129N, 129C, and 131) and quantification at the MS3 level were selected. The data were further analyzed using Perseus software (version 1.6.10.45). The data sets were filtered to remove false positives and common contaminants. Reporter ion intensities from each TMT channel were normalized to adjust the average intensity of each to the average total intensity across the 8 channels and then assigned to each experimental group (2 channels per group). Imputation of missing values was accomplished by replacing them with random numbers representing low-abundance measurements drawn from a normal distribution. The analysis was then restricted to the proteins quantified in both samples of at least one experimental group (M-UVRAG+/+, M-UVRAG−/−, and M-UVRAG−/−EGFRL858R/L858R). To test for statistical difference between M-UVRAG+/+ and M-UVRAG−/−, and M-UVRAG+/+ and M-UVRAG−/−EGFRL858R/L858R, the log2 transformed values of the normalized reporter ion intensities were compared using Student's two-tailed t test (p < 0.05) and corrected for multiple comparisons using the Benjamini-Hochberg method (q < 0.1).
2.13. Total RNA isolation and quantitative RT-PCR
Total RNA from muscle and liver tissues was isolated using TRIzol Reagent (Invitrogen). mRNA was reverse transcribed with random primers using the M-MLV enzyme (Invitrogen), and real-time PCR was performed using specific primers and SYBR Green Master Mix (Applied Biosystems) on a 7900HT Fast Real-Time PCR System (Applied Biosystems). Primer sequences are listed in the Supporting Information, Table S1. The relative mRNA expression of each gene was normalized to expression levels of the housekeeping gene L32.
2.14. Western blotting
Total protein from muscle tissues was lysed using RIPA buffer with protease inhibitor and protein lysates were separated by SDS-PAGE and immunoblotted for LC3 (#4108), EGFR (#4267), UVRAG (#13115), Stat3 (#4904), p-Stat3 (#9145), Akt (#4691), p-Akt (#9271), p-Drp1 (#3455), tubulin (#2148), GAPDH (# 2118, Cell Signaling Technology) and SQSTM1 (sc-28359), Drp1 (sc-271583), VDAC1 (sc-390996), Fgf21 (R &D, AF3057), and total OXPHOS (ab110413). Primary antibodies were diluted to 1:1,000 and secondary antibodies to 1:5,000. Band intensities were quantified using ImageJ software.
2.15. Statistics
The data are presented as mean ± S.E.M. The data were analyzed using Student's two-tailed unpaired t test for comparisons between two groups and by one-way ANOVA with Tukey's post hoc test for comparisons between multiple groups using GraphPad Prism. P values < 0.05 were considered statistically significant.
3. Results
3.1. Expression of muscle UVRAG decreased with aging
To assess for changes in autophagy gene expression that occurs with aging, we measured Beclin1, Atg5, Atg7, and MuRF1 in the skeletal muscle of young and aged mice at 8 and 24 weeks, respectively. As shown by others, the gene expression of MuRF1 increased, whereas Beclin1, Atg5, and Atg7 decreased in muscle at 24 weeks of age compared to 8 weeks (Figure S1A). We next examined the gene expression of UVRAG, a novel autophagy gene, in the skeletal muscle at these ages. Similar to other autophagy genes, the mRNA expression of UVRAG also decreased with aging (Figure S1B) as well as its protein levels (Figure S1C). The LC3-II/I ratio was downregulated with the accumulation of p62 in muscle at 24 weeks of age (Figure S1C), suggesting that UVRAG plays a regulatory role in autophagy in skeletal muscle with aging.
3.2. Generation of skeletal muscle-specific UVRAG knockout mice
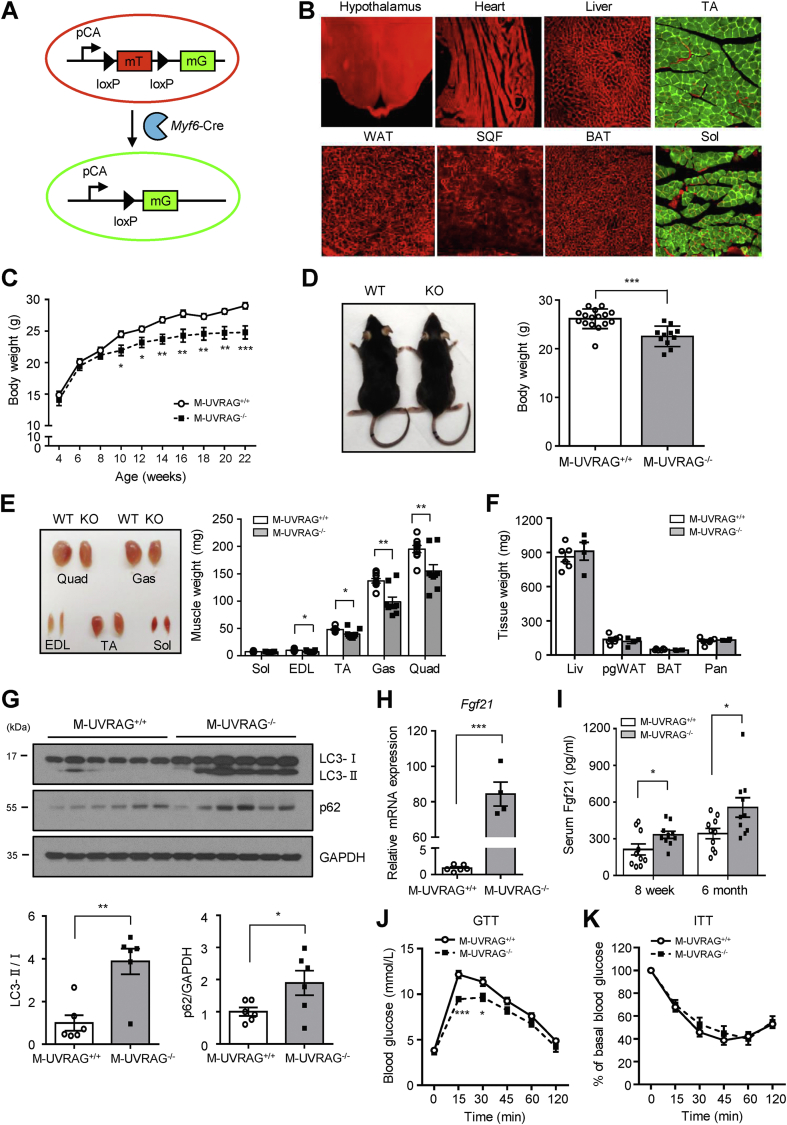
To investigate the essential in vivo role of UVRAG, we generated skeletal muscle-specific UVRAG knockout (M-UVRAG−/−) mice using the Cre-loxP system driven by Myf6 promoter. A fluorescent reporter mouse in which the transgene expression of mouse red fluorescent protein (mTomato) converted to green fluorescent protein (mGFP) following Cre recombinase-mediated excision (mT/mG) [25] validated that Cre transgene was activated exclusively in the skeletal muscle tissue, and no other tissues, including the cardiac muscle (Figure 1A,B). The M-UVRAG−/− mice had significantly decreased expression of UVRAG gene in all of their muscle groups, including the soleus, extensor digitorum longus (EDL), tibialis anterior (TA), gastrocnemius, and quadriceps (Figure S1D), and decreased protein expression was also observed in their quadriceps (Figure S1E). UVRAG deletion specifically in skeletal muscle was confirmed as its levels in multiple other tissues, including the brain, liver, pancreas, spleen, heart, and brown adipose tissue (BAT) were similar between the two groups (Figure S1F).
Figure 1.
M-UVRAG−/−mice had accelerated sarcopenia with aging. (A) A schematic of ROSA mT/mG reporter mouse. (B) Immunofluorescent images of mouse tissue expressing fluorescent mT (tomato red) and mG (GFP green) from the ROSA mT/mG; Myf6-Cre+ mice (n = 3). (C) Body weights of male mice fed a standard chow diet (n = 9–17). (D) Left, a representative image of the M-UVRAG+/+ (WT) and M-UVRAG−/− (KO) mice at 6 months of age. Right, body weights at 6 months of age (n = 11–16). (E) Gross images (left) and weights (right) of the various muscle groups from the M-UVRAG+/+ (WT) and M-UVRAG−/− (KO) mice at 6 months of age (n = 9). (F) Weights of other metabolic tissues at 6 months of age; Liv: liver, pgWAT; perigonadal white adipose tissue (visceral fat), BAT: brown adipose tissue, Pan: pancreas (n = 4–5). (G) Immunoblots (upper) and quantification (lower) of LC3 and p62 from quadriceps at 6 months of age (n = 6). GAPDH was used as a loading control. (H) Relative Fgf21 expression in quadriceps from the M-UVRAG−/− mice relative to the M-UVRAG+/+ mice at 6 months of age (n = 4–6). (I) Random serum Fgf21 from 8-week-old and 6-month-old mice (n = 10, each). (J and K) Glucose tolerance test (GTT, n = 12–17) and insulin tolerance test (ITT, n = 9–15) at 6 months of age. Data shown are mean ± S.E.M. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 using Student's t test.
3.3. M-UVRAG−/− mice had accelerated sarcopenia with aging
M-UVRAG−/− mice developed normally and showed no difference in total body weight and metabolic tissue weight compared to the controls at 8 weeks of age (Figure S2A). We next assessed for the LC3-II/I ratio and p62 in muscle, which showed no change in the M-UVRAG−/− compared with control mice, suggesting no defect in autophagy at this age (Figure S2B).
With aging however, the M-UVRAG−/− mice had attenuated body weight gain compared with the controls on a standard chow diet (Figure 1C,D). At 6 months of age, the M-UVRAG−/− mice showed decreased weight of all of their muscle groups with no difference in weights of other tissues, except the soleus, which was comprised primarily of slow-twitch type I fibers compared to the control mice (Figure 1E,F). These results supported that the decreased weight in the M-UVRAG−/− mice was likely attributed predominantly to loss in their muscle mass. We next assessed for autophagy genes in the muscle tissue of these aged mice. The gene expression of Beclin1 decreased, whereas Atg5 and Atg7 were unchanged in the muscle of the M-UVRAG−/− mice (data not shown). The LC3-II/I ratio increased with the accumulation of p62 in muscle in the M-UVRAG−/− mice compared with the control mice (Figure 1G), indicating impaired lysosomal degradation of the autophagosome and a defect in autophagy completion in the aged M-UVRAG−/− mice.
Biomarkers for muscle dysfunction including fibroblast growth factor 21 (Fgf21) and growth/differentiation factor 15 (Gdf15) were dramatically increased in the muscle of the 6-month-old M-UVRAG−/− mice compared to the controls (Figures 1H, S2C, and S2D). This upregulation of Fgf21 and Gdf15 was present as early as 8 weeks of age in the M-UVRAG−/− mice compared to the controls (Figure S2E). Serum Fgf21 also increased as early as 8 weeks of age, becoming progressively more significant at 6 months of age in the M-UVRAG−/− mice compared to the controls (Figure 1I). To determine whether the increase in Fgf21 was a direct result of UVRAG deficiency or a secondary consequence and a marker of muscle dysfunction, we assessed the expression of Fgf21 in the muscle of the M-UVRAG−/− mice at 4 weeks of age. At this early age, the expression of muscle Fgf21 was not different between the M-UVRAG−/− and control mice (Figure S2F). Importantly, Fgf21 from the liver, which is normally the predominant source in the circulation, was not different between the two groups at 8 weeks and 6 months of age (Figure S2G). These results indicated that the increased circulating Fgf21 likely contributed by increased production from the skeletal muscle in the M-UVRAG−/− mice. To assess the metabolic effects of progressive increases in muscle Fgf21, we performed GTT and ITT. Supporting the beneficial role of Fgf21 in metabolism, the M-UVRAG−/− mice had improved glucose homeostasis as assessed by GTT with similar ITT at 6 months of age compared to the controls (Figure 1J,K).
3.4. Muscle dysfunction in M-UVRAG−/− mice with aging
In association with decreased muscle mass in the M-UVRAG−/− mice at 6 months of age, they had dramatically compromised muscle function as assessed by lower grip strength and decreased inverted holding impulse (body mass x hanging time) compared to the control mice (Figure 2A,B). Furthermore, these mice showed premature exhaustion in response to acute exercise on the treadmill, with significantly shorter running distances compared to the control mice (Figure 2C). The male and female M-UVRAG−/− mice were analyzed separately and showed similar defects in body weight, muscle mass, and strength compared to the controls at 6 months (Figure S3A-S3E). Muscle loss was more accelerated with evident kyphosis in the female M-UVRAG−/− mice at 1 year of age (Figure S3F and S3G). To assess for bone mass, we measured the bone mineral density and bone mineral content using dual-energy X-ray absorptiometry (DEXA) in the female mice at 1 year of age. These mice showed decreased total body length, bone mineral density, and bone mineral content, indicating that muscle loss in the M-UVRAG−/− mice was associated with bone loss (Figure S3H-S3J). In contrast to the females, the male M-UVRAG−/− mice at 1 year and 5 months of age showed similar muscle weakness at 6 months of age (Figure S3K and S3L).
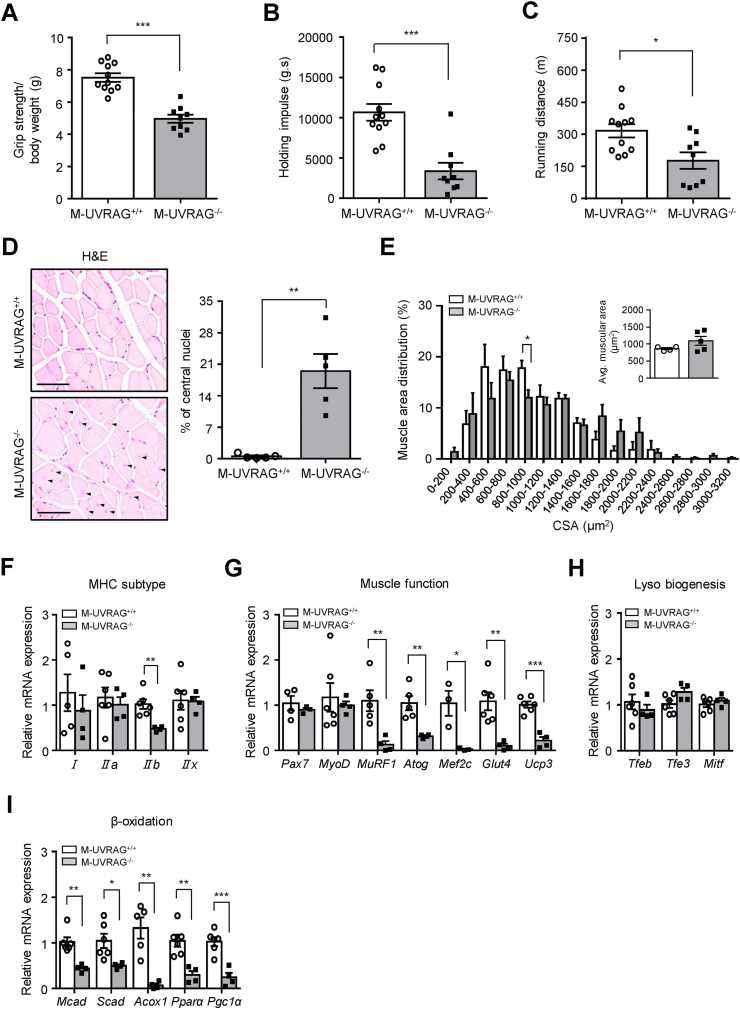
Figure 2.
Muscle dysfunction in the M-UVRAG−/−mice with aging. (A) Grip strength and (B) holding impulse (inverted hanging time) measurements from the 6-month-old male mice (n = 9–11). (C) Total running distance on the treadmill at 6 months of age (n = 9–11). (D) H&E staining (left) and quantification of central nuclei (right) from the tibialis anterior at 6 months of age (n = 5). Central nuclei are indicated by arrows. Scale bar, 50 μm. (E) Quantified distribution and average muscle fiber cross-sectional area (CSA) from the tibialis anterior at 6 months of age (n = 5). (F–I) Relative gene expression of myosin heavy-chain (MHC) subtypes, markers of muscle function, lysosomal biogenesis, and β-oxidation in quadriceps from the M-UVRAG−/− mice relative to the M-UVRAG+/+ mice at 6 months of age (n = 4–6). Data shown are mean ± S.E.M. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 using Student's t test.
We next performed a histological analysis of the muscle tissue. The presence of central nuclei, a hallmark of muscle pathology, significantly increased with no difference in the muscle fiber cross-sectional area (CSA) in the M-UVRAG−/− mice compared to the control muscle at 6 months of age (Figure 2D,E). To assess whether a defect in a specific fiber type was associated with muscle weakness in the M-UVRAG−/− mice, we analyzed for myosin heavy-chain (MHC) isoforms. Interestingly, the only major defect was the downregulation in gene expression of MHC IIb from the quadriceps of the M-UVRAG−/− mice compared to the control mice (Figure 2F).
To investigate the role of UVRAG in muscle function, we performed qRT-PCR analysis in muscle of M-UVRAG−/− mice. Many of the genes related to muscle function including MuRF1, Atrogin1, myocyte-specific enhancer factor 2c (Mef2c), glucose transporter type 4 (Glut4), and uncoupling protein 3 (Ucp3) were significantly downregulated in the muscle of the M-UVRAG−/− mice compared to the controls at 6 months of age (Figure 2G). The expression of genes related to lysosomal biogenesis, including Tfeb, Tfe3, and Mitf, was not different between the two groups (Figure 2H). Interestingly, the expression of genes related to β-oxidation, including medium-chain acyl-CoA dehydrogenase (Mcad), short-chain acyl-CoA dehydrogenase (Scad), peroxisomal acyl-coenzyme A oxidase 1 (Acox1), peroxisome proliferator-activated receptors alpha (Pparα), and peroxisome proliferator-activated receptor gamma coactivator-1α (Pgc1α) were significantly downregulated in the muscle of the M-UVRAG−/− mice compared to the controls (Figure 2I).
3.5. Impaired mitochondrial function in UVRAG-deficient muscle
We next sought to determine, in an unbiased fashion, proteins that were dysregulated in the muscles of the M-UVRAG−/− mice at 6 months of age. To this end, we performed mass spectrometry and identified a total of 3,160 proteins. Following the removal of false positives and contaminants, we restricted our analysis to the proteins quantified in both replicates of at least one experimental group (3,034 proteins). Comparison of the reporter ion intensities between the 2 groups revealed 1,063 differentially expressed proteins. Pathway enrichment analysis revealed that the differentially expressed proteins were involved in aerobic respiration, TCA cycle, and NADH pathways (Figure S7B). In keeping with these data, proteins involved in the mitochondrial function including TFAM and CYC were significantly downregulated in the M-UVRAG−/− mice (Figure 3A).
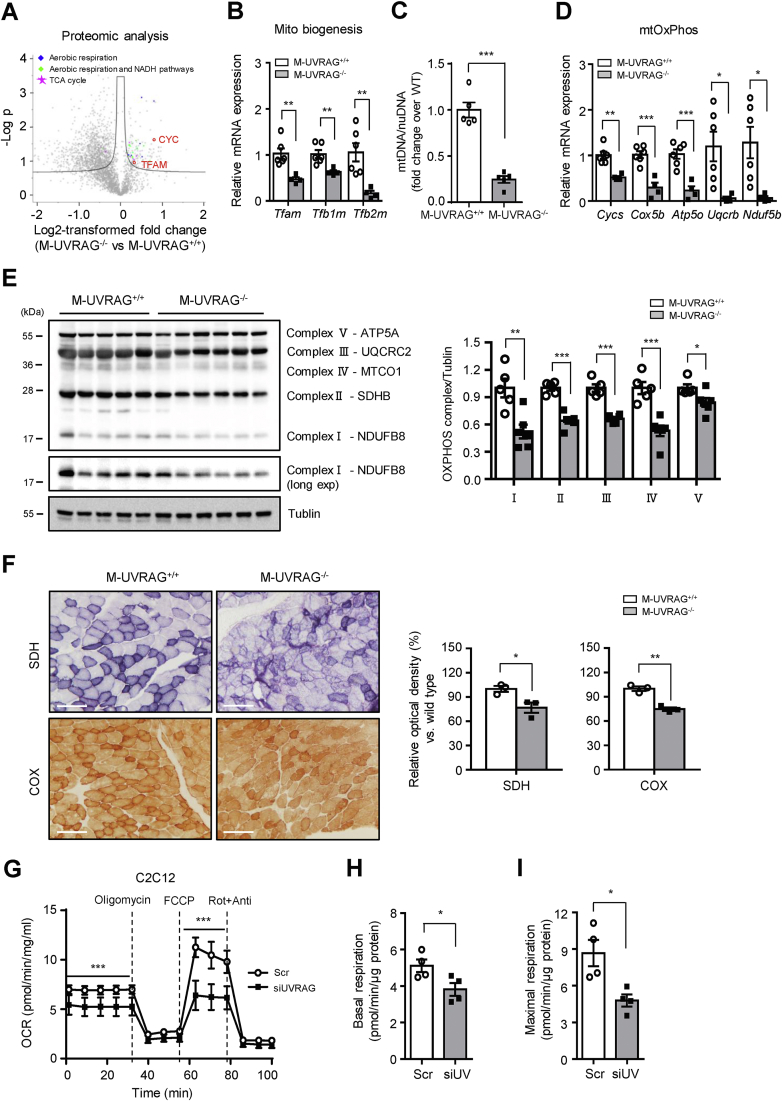
Figure 3.
Impaired mitochondrial function in UVRAG-deficient muscle. (A) Volcano plot illustrating statistically significant protein abundance differences in the tibialis anterior muscle between the M-UVRAG−/− and M-UVRAG+/+ mice. Proteins significantly downregulated in the M-UVRAG−/− mice are shown in red. Data points shown are differentially expressed proteins using Student's t test (p < 0.05) with the Benjamini-Hochberg adjustment (q < 0.1). (B) Relative expression of genes related to mitochondrial biogenesis from quadriceps of the M-UVRAG−/− mice relative to the M-UVRAG+/+ mice at 6 months of age (n = 4–6). (C) Mitochondrial DNA (mtDNA) copy number calculated as the ratio of Cox2 to cyclophilin A (Ppia) levels measured by real-time quantitative PCR from quadriceps of the 6-month-old mice (n = 5). nuDNA: nuclear DNA. (D–E) mRNA and protein expression of mitochondrial oxidative phosphorylation (mtOxPhos) from quadriceps of the M-UVRAG−/− and M-UVRAG+/+ mice at 6 months of age (n = 4–6). (F) Representative SDH and COX staining (left) on frozen muscle (tibialis anterior) sections from the 6-month-old mice (n = 3). Relative optical density (right) is expressed as a percentage of the SDH and COX immunoreactivity detected in muscle of the M-UVRAG−/− mice compared to the M-UVRAG+/+mice. Scale bar, 100 μm. (G) Oxygen consumption rate (OCR) measured from C2C12 myotubes using Seahorse XF24 analyzer in response to 1 μM of oligomycin, 0.5 μM of FCCP, 1 μM of rotenone (Rot), and antimycin A (Anti) (n = 3 per group). Basal (H) and maximal (I) respiration calculated from (G). Data shown are mean ± S.E.M. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 using Student's t test.
Because the expression of genes related to β-oxidation was significantly downregulated in the M-UVRAG−/− mice and Pgc1α serves critical roles in regulating cellular energy metabolism and mitochondrial function [26,27], we next assessed the role of UVRAG in mitochondrial homeostasis. Consistent with decreased protein levels of TFAM and CYC from the proteomic analysis, the muscles of the M-UVRAG−/− mice showed a reduction in the expression of genes related to mitochondrial biogenesis, including mitochondrial transcription factor A (Tfam) and mitochondrial transcription factor B1 (Tfb1m) and B2 (Tfb2m) compared to the controls (Figure 3B). In addition, we observed a significantly decreased mitochondrial DNA (mtDNA) copy number in the M-UVRAG−/− muscle (Figure 3C). Furthermore, the expression of genes related to mitochondrial oxidative phosphorylation (mtOxPhos) including cytochrome c (Cycs), cytochrome c oxidase subunit 5B (Cox5b), ATP synthase subunit O (Atp5o), complex III subunit 7 (Uqcrb), protein levels (Figure 3E), and complex I subunit (Ndufb5) were significantly downregulated in the muscle of the M-UVRAG−/− mice compared to the controls at 6 months of age (Figure 3D). These defects in gene expression involved in β-oxidation, mtOxPhos, and mitochondrial biogenesis were present as early as 8 weeks of age in the M-UVRAG−/− mice to a lesser extent, while the mtDNA content was similar to the controls at this age (data not shown). Consistent with the decreased mitochondrial biogenesis genes and mtDNA content, we observed significantly less staining for succinate dehydrogenase (SDH) and cytochrome c oxidase (COX) in the muscle of the M-UVRAG−/− mice compared to the controls, indicating impaired mitochondrial enzyme activity (Figure 3F).
We next assessed whether these findings in the muscle of the M-UVRAG−/− mice were indeed due to cell autonomous function of UVRAG without systemic confounders by examining UVRAG-deficient C2C12 myotubes in vitro. To assess for mitochondrial function, we measured the oxygen consumption rate (OCR) using a Seahorse XF24 Analyzer. UVRAG-deficient cells had a decrease in the basal OCR (Figure 3G,H). Addition of an uncoupler, carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP), to maximize mitochondrial respiration led to a further increase in the OCR in scramble siRNA transfected cells, whereas an additional increase in the OCR was absent in UVRAG knockdown cells (Figure 3G,I). Overall, UVRAG-deficient myotubes had a significant compromise in both basal and maximal respiration compared to the controls, further supporting the essential role of UVRAG in the mitochondrial capacity.
3.6. Defects in mitochondrial morphology in muscle in the M-UVRAG−/− mice
Mitochondria are dynamic organelles that continuously undergo homeostatic fission and fusion that are critical for maintaining optimal skeletal muscle function [28]. We first examined for mitochondrial morphology from the muscle of the M-UVRAG−/− mice at 6 months of age using electron microscopy, which showed altered mitochondrial structures with defects in the inner and outer membranes, with some appearing swollen with paucity or even the complete absence of cristae (Figure 4A and S4). We also analyzed mitochondrial shapes in intermyofibrillar (IMF) regions of muscle from the M-UVRAG−/− mice [28], which showed significantly smaller areas and perimeters, which corresponds to their lower Feret's diameter (defined as the longest distance between any two points within the selected mitochondrion) compared to the controls (Figure 4B). Furthermore, the mitochondria in the M-UVRAG−/− muscle were rounder with more circular shapes (0 referring to a straight line and 1 as a perfect circle) with lower aspect ratios (defined as the major to minor axis ratio), less elongated, and more fragmented (Figure 4B), which were associated with dysfunction.
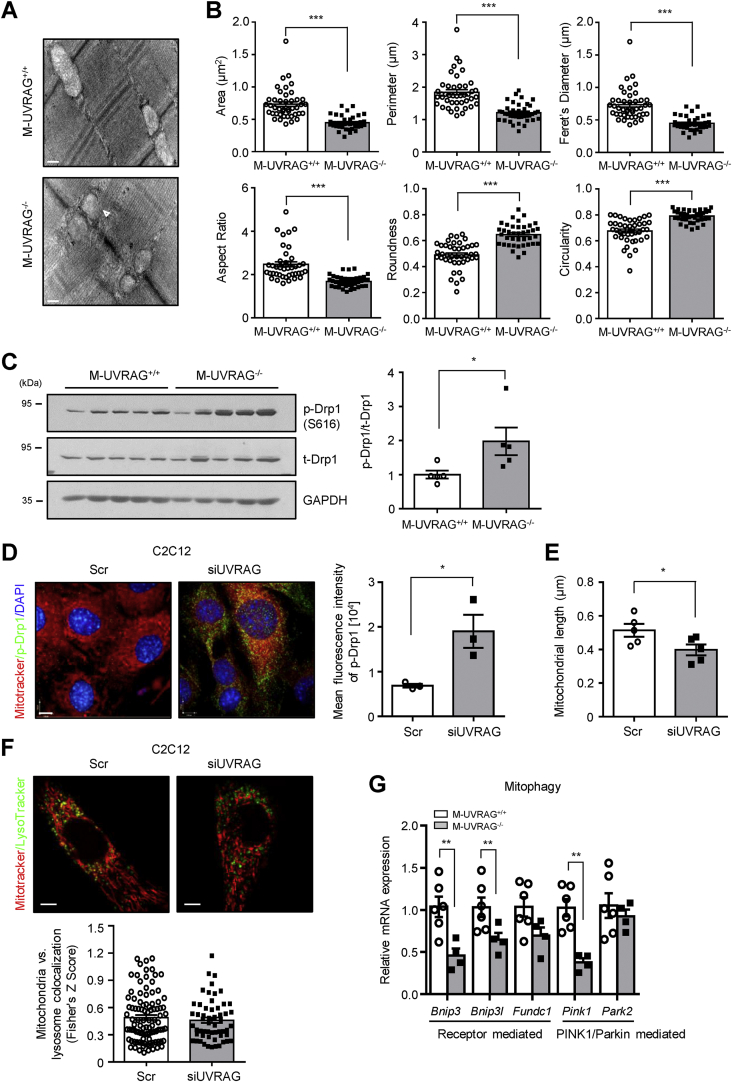
Figure 4.
Mitochondrial morphology in UVRAG-deficient muscle. (A) Representative TEM images of intermyofibrillar (IMF) mitochondria and quantification (B) of mitochondrial morphology from the extensor digitorum longus (EDL) at 6 months of age (n = 3). Swollen mitochondrion is indicated by arrows. Scale bar, 200 nm. (C) Immunoblots (left) and quantification (right) of total- and phospho-Drp1 from quadriceps of thee 6-month-old mice (n = 5). GAPDH was used as a loading control. (D) Representative immunofluorescent images (left) of MitoTracker (red), phospho-Drp1 (green), DAPI (blue), and quantification of green fluorescence intensity (right) from C2C12 myotubes transfected with siUVRAG or scramble (Scr) knockdown (n = 3). Scale bar, 7 μm. (E) ImageJ quantitation of mitochondrial length from MitoTracker-stained C2C12 myotubes transfected with siUVRAG or scramble (Scr) knockdown (n = 5). (F) Representative immunofluorescent images (left) of MitoTracker (red) and LysoTracker (green) and quantification of colocalization (right) from C2C12 myotubes transfected with siUVRAG or scramble (Scr) knockdown (n = 3). Scale bar, 5 μm. (G) Relative expression of genes related to mitophagy from quadriceps of the M-UVRAG−/− mice relative to the M-UVRAG+/+ mice at 6 months of age (n = 4–6). Data shown are mean ± S.E.M. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 using Student's t test.
Dynamin-related protein 1 (Drp1) is a key factor that facilitates mitochondrial fission, and phosphorylation of Drp1 at S616 is required for this process. To determine whether the activation of Drp1 was affected by UVRAG deletion, we assessed the expression of Drp1 and its phosphorylation status and observed increased phosphorylation at S616 of Drp1 in the muscle of the M-UVRAG−/− mice compared to the control mice (Figure 4C). In addition, increased phospho-Drp1 was observed in UVRAG knockdown myotubes as assessed by immunofluorescence (Figure 4D) with decreased mitochondrial length (Figure 4E). We next assessed whether these mitochondrial defects were associated with changes in mitophagy by examining colocalization of mitochondria and lysosomes using MitoTracker and LysoTracker staining and observed no differences under basal conditions in C2C12 myotubes (Figure 4F). Moreover, the skeletal muscle of the M-UVRAG−/− mice had generally decreased gene expression of markers of mitophagy, including mitophagy-receptors BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3), BNIP3-like (BNIP3L), and FUN14 domain-containing protein 1 (FUNDC1) and markers related to ubiquitin-mediated mitophagy, PTEN-induced kinase 1 (PINK1), at 24 weeks of age (Figure 4G).
3.7. EGFR accumulation in UVRAG-deficient muscle
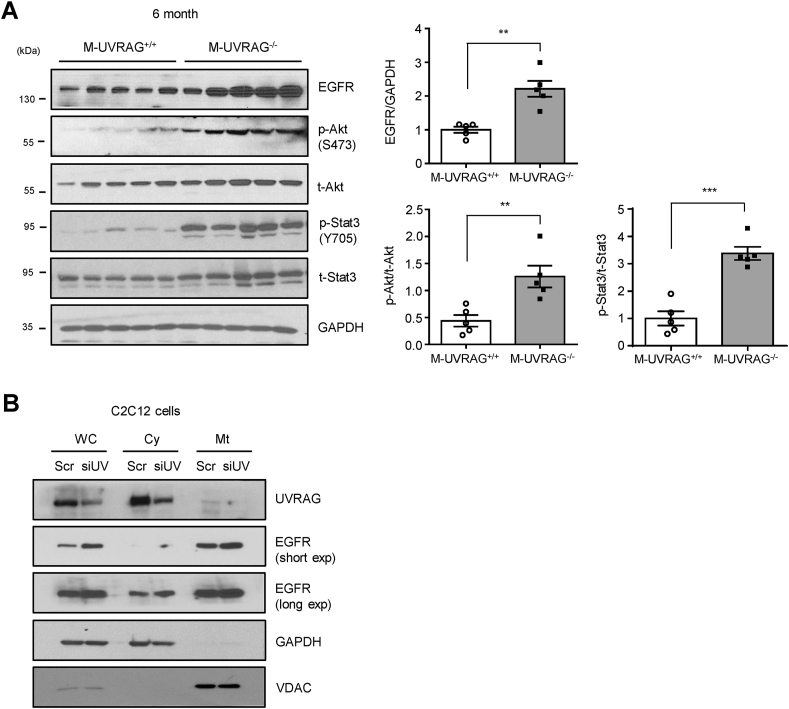
Autophagic processes have distinct and overlapping pathways with endosomal trafficking. UVRAG has been shown to be involved in both of these processes in molecular studies using 293T cells [23]; however, its role in skeletal muscle was unclear. To investigate whether UVRAG played a role in endosomal trafficking in muscle, we assessed for the accumulation of epidermal growth factor receptor (EGFR), which classically occurs with a defect in endocytic machinery. Indeed, EGFR accumulation was markedly increased in the muscle of the M-UVRAG−/− mice compared with the control mice at 6 months of age (Figure 5A). We next analyzed for the phosphorylation status of major downstream targets of EGFR, Akt, and Stat3. In line with the enhanced EGFR accumulation in the M-UVRAG−/− muscle, phosphorylation of Akt and Stat3 significantly increased compared to the controls (Figure 5A).
Figure 5.
EGFR accumulation in UVRAG-deficient muscle. (A) Immunoblots (left) and quantification (right) of epidermal growth factor receptor (EGFR), phospho- and total-Akt, and phospho- and total-Stst3 from quadriceps of the 6-month-old mice (n = 5). (B) Immunoblots of UVRAG and EGFR in the cytosolic (Cy) and mitochondrial (Mt) fractions of C2C12 myotubes transfected with scramble (Scr) or UVRAG siRNA (siUV) (n = 3). GAPDH and VDAC are representative cytosolic and mitochondrial control proteins respectively. WC: whole cell lysate. Data shown are mean ± S.E.M. ∗∗p < 0.01 and ∗∗∗p < 0.001 using Student's t test.
To determine whether the accumulation of EGFR was in the cytosolic or mitochondrial fractions, we separated subcellular fractions in C2C12 myotubes with UVRAG or scramble knockdown. Consistent with accumulation of EGFR in the muscle tissue of the M-UVRAG−/− mice, we observed increased EGFR in the UVRAG knockdown C2C12 myotubes, and its accumulation was more predominant in the mitochondrial fractions (Figure 5B). In contrast, UVRAG was more prominently present in the cytosolic fractions (Figure 5B).
3.8. EGFR inhibitor restored markers of muscle dysfunction in the M-UVRAG−/− mice
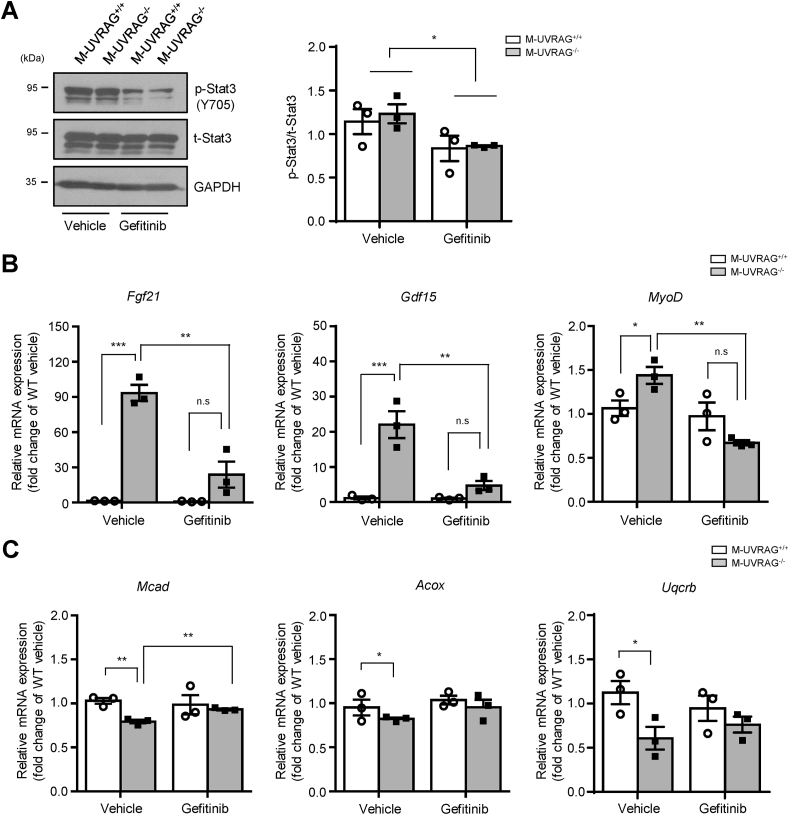
To assess whether the increased EGFR signaling in the UVRAG deficient muscle played a causal role in skeletal muscle function, we disrupted EGFR signaling pharmacologically using gefitinib, an EGFR tyrosine kinase inhibitor. The M-UVRAG−/− and control mice were given gefitinib by oral gavage 5 days per week for 21 days starting at 4 weeks of age, when the muscle Fgf21 levels were unaltered (Figure S2F). After 21 days of oral gavage, body and muscle weight as well as grip strength were assessed and no differences between the two groups were found with vehicle or gefitinib treatment (Figure S5). To confirm the efficacy of gefitinib in the skeletal muscle, we examined the phosphorylation status of Stat3 in the muscle after 21 days of gefitinib treatment. We observed a significant decrease in phosphorylation of Stat3 in both the M-UVRAG+/+ and M-UVRAG−/− mice compared to the vehicle treatment groups, supporting the efficacy of the drug treatment (Figure 6A).
Figure 6.
Pharmacologic inhibition of EGFR signaling partially restored markers of muscle dysfunction in the M-UVRAG−/−mice. (A) Representative immunoblots (left) and quantification (right) of phospho- and total-Stat3 in quadriceps following oral gavage with vehicle or EGFR inhibitor gefitinib for 21 days (n = 3, each group). (B and C) Relative expression of genes related to muscle dysfunction including Fgf21, Gdf15, and MyoD and mitochondrial function including Mcad, Acox, and Uqcrb in tibialis anterior from the M-UVRAG−/− mice relative to the M-UVRAG+/+ mice after treatment with vehicle or gefitinib for 21 days (n = 3, each group). Data shown are mean ± S.E.M. ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001 using Student's t test.
Importantly, gene expression of Fgf21, Gdf15, and MyoD, markers of muscle dysfunction or damage, which were significantly upregulated in the M-UVRAG−/− vehicle group, were significantly attenuated by gefitinib (Figure 6B). Furthermore, downregulation of genes involved in mitochondrial function including Mcad, Acox1 and Uqcrb in the muscle of the M-UVRAG−/− mice were also attenuated by gefitinib (Figure 6C). These results supported a potential causal role of EGFR accumulation and signaling in mitochondrial deregulation and muscle dysfunction in the M-UVRAG−/− mice that was at least partially attenuated with pharmacologic inhibition of EGFR signaling.
3.9. Distinct effects of EGFR activation and UVRAG deficiency on skeletal muscle differentiation and function
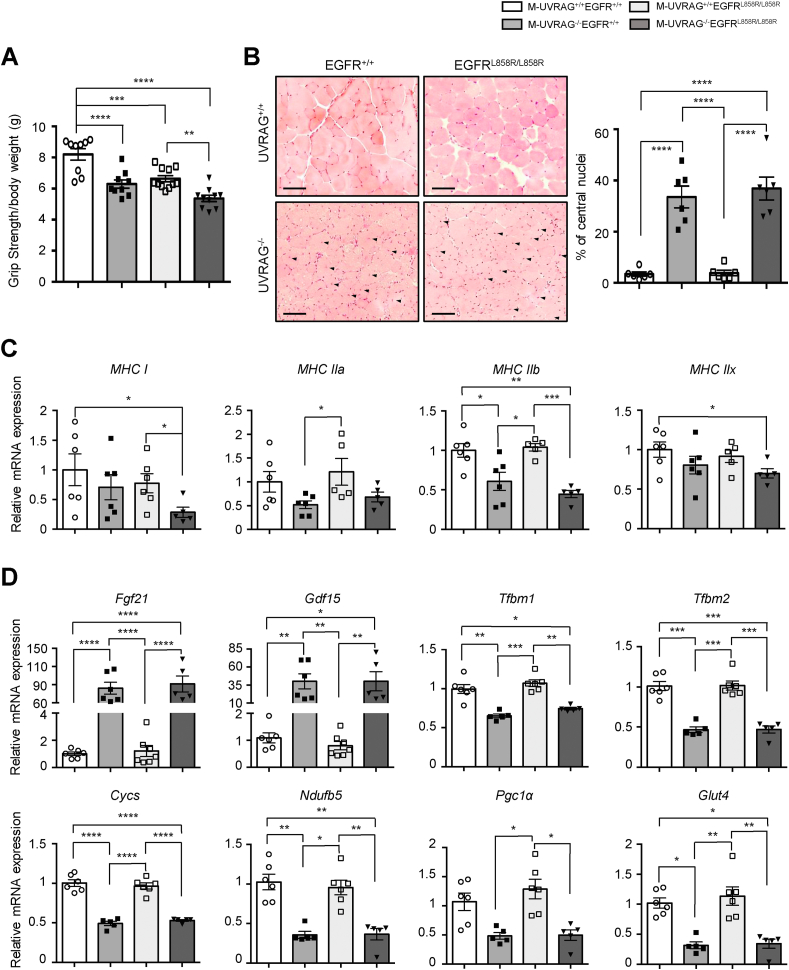
To further elucidate the mechanisms of how EGFR signaling affects skeletal muscle function, we generated a knock-in mouse in which EGFR was constitutively active with a point mutation (L858R) in EGFR specifically in the skeletal muscle. This mutation enhances the downstream EGFR signaling pathways due to a conformational change in the receptor [29,30]. Similar to the M-UVRAG−/− mice, the skeletal muscle-specific EGFR mutant (EGFRL858R/L858R) mice developed normally and displayed weak grip strengths of similar degrees to that of the M-UVRAG−/− mice (Figure 7A). These mice also showed decreased muscle weights, with no differences in other tissue weights at 6 months of age (Figure S6A and S6B). Furthermore, concomitant disruption of UVRAG and activation of EGFR with the generation of double-mutant M-UVRAG−/−EGFRL858R/L858R mice showed an additive weakness in grip strength (Figure 7A).
Figure 7.
Distinct effects of EGFR activation and UVRAG deletion in skeletal muscle function. (A) Grip strength measurement of 6-month-old male mice (n = 9–11). (B) Representative H&E staining (left) and quantification of central nuclei (right) from tibialis anterior muscle at 6 months of age (n = 6). Central nuclei are indicated by arrows. Scale bar, 50 μm. (C) Relative expression of myosin heavy-chain (MHC) subtypes in tibialis anterior muscle of the 6-month-old male mice (n = 5–6). (D) Relative expression of genes related to mitochondrial deregulation (Fgf21 and Gdf15), mitochondrial biogenesis (Tfb1m and Tfb2m), mitochondrial oxidative phosphorylation (Cycs and Ndufb5), and muscle function (Pgc1α and Glut4) from tibialis anterior of the 6-month-old male mice (n = 5–7). Data shown are mean ± S.E.M. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 using one-way ANOVA with Tukey's post hoc test.
We next assessed for histologic abnormalities in the skeletal muscle of the EGFRL858R/L858R mice. Interestingly, central nuclei that were frequently present in the M-UVRAG−/− mice were absent in the EGFRL858R/L858R mice, similar to the WT littermate controls, while the double-mutant M-UVRAG−/−EGFRL858R/L858R mice showed equivalent levels of central nuclei compared to the M-UVRAG−/− littermate controls at 6 months of age (Figure 7B).
To assess for potential mechanisms to account for these differences, we again took an unbiased proteomics approach to compare the skeletal muscle tissue from the M-UVRAG−/−EGFRL858R/L858R and M-UVRAG+/+ mice. We identified 1,403 proteins significantly different between the M-UVRAG−/−EGFRL858R/L858R and M-UVRAG+/+ mice. Some of the most significant pathways enriched among these proteins included integrin signaling, cytoskeletal regulation, and amino acid metabolism (Figure S7A and S7B). The MYH family genes including MYH3, MYH4, MYH7, MYH9, and MYH14 were significantly upregulated in the M-UVRAG−/−EGFRL858R/L858R mice (Figure S7A). However, examination of the gene expression of skeletal muscle fiber types by MHC subtype demonstrated a decrease in type I muscle fibers (MHC I) with both UVRAG deletion and EGFR activation showing an additive decline in the double-mutant M-UVRAG−/−EGFRL858R/L858R mice compared to the WT controls (Figure 7C). These data corroborate with the recently published finding that EGFR activation negatively correlates with slow-twitch muscle (type I) fiber expression [31].
Interestingly, markers of muscle dysfunction, Fgf21 and Gdf15, as well as markers of mitochondrial biogenesis (Tfbm1 and Tfbm2), mitochondrial oxidative phosphorylation (Cycs and Ndufb5), and muscle function (Pgc1α and Glut4) that were abnormal in the muscle of the M-UVRAG−/− mice were not affected in the EGFRL858R/L858R mice and showed a similar degree of abnormality in the M-UVRAG−/−EGFRL858R/L858R mice as that of the M-UVRAG−/−EGFR+/+ mice (Figure 7D). Similarly, the gene expression of mitochondrial proteins such as TFAM, COX5B, and CYC remained significantly lower in the M-UVRAG−/−EGFRL858R/L858R mice to a similar degree as the muscle of the M-UVRAG−/− mice (Figure S7A and 3A). Other genes involved in muscle development (Lbx1, Mox2, myogenin, and myostatin) and potentially relevant in EGFR signaling such as Wnt signaling (β-catenin, LRP6, and GSK3β), Notch signaling (Notch1 and Jagged1), and fibrosis markers (Col1a1 and TGFβ1) were also only affected in the muscle of the M-UVRAG−/− mice but unchanged by concomitant EGFRL858R/L858R mutation in the skeletal muscle (Figure S6C-S6F).
4. Discussion
Several cellular processes are implicated in the development of age-related sarcopenia, including denervation, mitochondrial dysfunction, or inflammatory or hormonal changes. Autophagy can impact many of these processes, and indeed, defective autophagy is tightly associated with age-related declines in muscle mass and function [1,32,33]. For example, skeletal muscle-specific Atg5 deletion in mice shows loss of muscle mass with the accumulation of protein aggregates [34]. Similarly, ablation of skeletal muscle Atg7 results in the accumulation of p62, mitochondrial dysfunction, and muscle atrophy [3]. UVRAG is a novel autophagy gene with other non-autophagy-related functions; however, its essential in vivo role in skeletal muscle was unclear. In this study, we showed that UVRAG is critical for maintaining skeletal muscle function and mitochondrial homeostasis with aging.
It was recently shown that mice with disruption of the UVRAG gene by piggyBac transposon insertion in multiple tissues showed impaired autophagic flux in the heart and developed age-related cardiomyopathy with increased inflammatory response [35]. Furthermore, UVRAG deficiency aggravates DOX-induced cardiotoxicity [36], while iron overload has been shown to impair autophagic flux in skeletal muscle by disrupting mTORC1-UVRAG signaling [37].
In this study, we showed that UVRAG was not essential during homeostasis at a young age or development of skeletal muscle at least during Myf6 expression. Thus, our M-UVRAG−/− mice appeared normal at 8 weeks of age. However, with aging, UVRAG is clearly essential for maintaining skeletal muscle homeostasis, such that UVRAG deficiency leads to defects in autophagy and decreased muscle mass in groups containing predominantly myosin heavy chain (MHC) isoform type IIb. Interestingly, age-related muscle weakness is associated with these type IIb fibers, which are more susceptible to mitochondrial dysfunction and atrophy. In contrast, type I fibers have high mitochondrial content and are more resistant to age-dependent loss of muscle mass [38,39]. Of note, type I fiber-predominant soleus muscle from the M-UVRAG−/− mice was spared from a decline in muscle mass with aging.
In addition to the well-appreciated role of skeletal muscle in its contractile function, myocytes have also been shown to be capable of producing and secreting peptides into the circulation, known as myokines. Physical exercise and muscle dysfunction can affect the release of various myokines that can regulate distant metabolic tissues [40]. Fgf21 and Gdf15 are well-known myokines that can serve as biomarkers of muscle dysfunction [41,42]. Fgf21 is classically derived from the liver as a critical regulator of energy homeostasis and metabolism by stimulating fatty acid oxidation and inhibiting gluconeogenesis and lipogenesis [43,44]. However, Fgf21 can also be derived from dysfunctional muscle and may contribute to sarcopenia as evidenced by skeletal muscle specific-Fgf21 knockout mice that were shown to be protected from muscle loss during fasting [45]. Gdf15 has been suggested as a marker of muscle wasting after cardiovascular surgery [46,47], is secreted by skeletal muscle during exercise, and promotes lipolysis [48]. In addition, circulating Gdf15 levels have been shown to increase with age and may reflect mitochondrial dysfunction that occurs with age-related diseases [49,50]. Our data showed that upregulation of Fgf21 and Gdf15 was associated with muscle dysfunction and imbalance of mitochondrial homeostasis in the M-UVRAG−/− mice. These increased myokines likely played a causal role in the paradoxical improvement in glucose homeostasis in these mice despite decreased muscle mass and function.
Our unbiased proteomic analysis demonstrated decreased expression of proteins involved in aerobic respiration, the TCA cycle, and NADH pathways in the muscle of the M-UVRAG−/− mice. We showed that the skeletal muscle of the M-UVRAG−/− mice had impaired mitochondrial function with downregulated expression of genes related to β-oxidation, mitochondrial biogenesis, and mtOxPhos and decreased mtDNA content. Pgc1α is a major regulator mitochondrial biogenesis, and Akt activation has been shown to phosphorylate and inhibit Pgc1α [51]. While the molecular mechanism of how UVRAG regulates mitochondrial function is not entirely clear, we can postulate that activated Akt signaling by EGFR accumulation that we observed in the M-UVRAG−/− mice can potentially inhibit transcription of mitochondrial biogenesis and function through decreased Pgc1α expression. Furthermore, the UVRAG-deficient myotubes showed a decreased oxygen consumption rate (OCR) in vitro, indicating that UVRAG was critical for mitochondrial function in muscle.
Autophagic and endosomal trafficking processes are tightly associated, and UVRAG is shown to critically regulate the endocytic pathway in addition to autophagosome maturation [23]. Endocytic trafficking of growth factor receptors is one of the vital cellular mechanisms, and EGFR is classically investigated for this endosomal recycling. We found that in the muscle of the M-UVRAG−/− mice, the accumulation of EGFR increased, and phosphorylation of Akt and Stat3, downstream targets of EGFR, was also upregulated. Mitochondrial EGFR has been shown to regulate mitochondrial dynamics by increasing the expression of fusion proteins, PHB2, and OPA1 [52], and interacting with Mfn1, disrupting its activity [53]. Our data showed that deletion of UVRAG in the myotubes led to the accumulation of EGFR in both cytosolic and mitochondrial fractions. Gefitinib, an EGFR inhibitor, selectively binds to its tyrosine kinase domain, preventing ATP from binding and blocking receptor autophosphorylation, which results in the inhibition of signal transduction. It has been shown that prolonged exposure to gefitinib induces drug resistance and mitochondrial dysfunction [54,55], but short-term treatment (72 h) with gefitinib enhances mitochondrial function by improving respiratory enzyme succinate-tetrazolium reductase activity in lung cancer cells [56]. In addition, inhibition or knockdown of EGFR has been shown to increase slow-twitch fiber genes (type I) with mitochondrial activity in C2C12 myotubes [31]. To assess whether the accumulation of EGFR in UVRAG-deficient muscle had a causal role in its dysfunction, we inhibited EGFR signaling pharmacologically in vivo and showed that this partially attenuated the increase in markers of muscle dysfunction in the M-UVRAG−/− mice. Measurements of muscle strength were not changed by gefitinib likely because the drug treatment was given at an early age (4 weeks of age) for a short term of 21 days.
It has previously been shown that silencing EGFR in C2C12 myoblasts triggers skeletal muscle differentiation [57]. Furthermore, EGFR inhibition both in vitro and in vivo promotes a slow-twitch skeletal muscle fiber determination [31]. However, the relationship between UVRAG and EGFR in skeletal muscle has yet to be reported. We determined that UVRAG and EGFR did not interact directly in protein–protein interaction analysis using a mammalian-membrane two-hybrid assay (data not shown). To further investigate the role of EGFR signaling in vivo, we generated EGFR mutant mice with constitutive activation of EGFR in skeletal muscle. The EGFR L858R mutant was a point mutation in exon 21 located in the EGFR activation loop that disrupted interactions that stabilize the inactive conformation of EGFR, causing the receptor to become constitutively active [30,58]. Proteomic analysis demonstrated differential expression mainly in pathways involved in integrin signaling, cytoskeleton regulation, and amino acid metabolism in the M-UVRAG−/−EGFRL858R/L858R mice. Furthermore, our data showed that mutant EGFR did not affect markers of muscle and mitochondrial deregulation. The expression of proteins involved in the MYH family including MYH3, MYH4, MYH7, MYH9, and MYH14 increased in the M-UVRAG−/−EGFRL858R/L858R mice, but the mRNA expression of genes related to slow-twitch muscle fiber (type I) decreased in both UVRAG deletion and EGFR mutation. The differential results between our gene expression and proteomic analysis likely reflected the complexity in post-transcriptional and translational modifications that regulate the stability of mRNA processing and the protein degradation pathway, which may have been differentially regulated in the M-UVRAG−/− and M-UVRAG−/−EGFRL858R/L858R mice. Our data suggested that EGFR L858R mutation affected the skeletal muscle fiber type, with a decrease in MHC type I expression, indicating a negative regulatory role in slow-twitch fibers to further impact in an additive manner the effects of UVRAG deletion. This corresponded to additive defects in skeletal muscle function as demonstrated by the weak grip strength in the double-mutant mice.
The proportion of type I to type II muscle fibers in skeletal muscle has a significant clinical impact, as a decline in type I skeletal muscle fibers increases susceptibility to loss of muscle mass. Indeed, markers of muscle dysfunction with UVRAG deficiency were partially attenuated by pharmacologic inhibition of EGFR and aggravated by constitutively active EGFR signaling in UVRAG-deficient muscle.
In conclusion, we showed an essential role of UVRAG in maintaining muscle mass and function with aging, with a distinct role of EGFR signaling that is at least partially required for defects observed in UVRAG-deficient muscle. We also showed the non-overlapping role of constitutive EGFR activation and UVRAG deficiency in muscle dysfunction and sarcopenia in an additive manner. Overall, we identified UVRAG and EGFR as new regulatory factors for mitochondrial and muscle homeostasis that occurs with aging.
Author contributions
MJK and DF performed the experiments, analyzed the data, and wrote the manuscript. SF and AK analyzed the proteomics data and generated corresponding volcano plots. TG and IC conducted the experiments and analyzed the data for mitochondrial morphology and Drp1 phosphorylation. JES and CCH obtained immunofluorescent images from the tissue of the Rosa26mTmG mice. EPT, YJP, TS, and SJO stained and analyzed the SDH, COX, and H&E data. JY performed the experiments and analyzed the qRT-PCR data. RR, HHC, and JVR stained cells and obtained images of MitoTracker and LysoTracker colocalization. PS and IS used the mammalian-membrane two-hybrid assay to assess UVRAG and EGFR protein interactions. ZH and TWM produced UVRAG floxed mice. MW designed and supervised the study, secured funding, and wrote the manuscript.
Acknowledgments
This work study supported by operating grants from the Canadian Institutes of Health Research (CIHR) MOP 142193 and CIHR PJT 159505 to MW. MW holds the Canada Research Chair in Signal Transduction in Diabetes Pathogenesis and the Soham and Shaila Ajmera Family Chair in Molecular Diabetes Research. MJK was supported by a National Research Foundation of Korea grant funded by the Korea government (2020R1C1C1014670) and a Banting and Best Diabetes Centre, University of Toronto (BBDC)-Kangbuk Samsung Hospital postdoctoral fellowship. DF is a recipient of the BBDC-University Health Network Graduate Award, Ontario Graduate Scholarship, and Queen Elizabeth II/Dr. Arnie Aberman Graduate Scholarship in Science and Technology.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101185.
Conflicts of interest
The authors declare no competing financial interests.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Rubinsztein D.C., Marino G., Kroemer G. Autophagy and aging. Cell. 2011;146(5):682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 2.Grumati P., Bonaldo P. Autophagy in skeletal muscle homeostasis and in muscular dystrophies. Cells. 2012;1(3):325–345. doi: 10.3390/cells1030325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masiero E., Agatea L., Mammucari C., Blaauw B., Loro E., Komatsu M. Autophagy is required to maintain muscle mass. Cell Metabolism. 2009;10(6):507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Aredia F., Guaman Ortiz L.M., Giansanti V., Scovassi A.I. Autophagy and cancer. Cells. 2012;1(3):520–534. doi: 10.3390/cells1030520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quan W., Jung H.S., Lee M.S. Role of autophagy in the progression from obesity to diabetes and in the control of energy balance. Archives of Pharmacal Research. 2013;36(2):223–229. doi: 10.1007/s12272-013-0024-7. [DOI] [PubMed] [Google Scholar]
- 6.Papackova Z., Cahova M. Important role of autophagy in regulation of metabolic processes in health, disease and aging. Physiological Research. 2014;63(4):409–420. doi: 10.33549/physiolres.932684. [DOI] [PubMed] [Google Scholar]
- 7.Wang F., Jia J., Rodrigues B. Autophagy, metabolic disease, and Pathogenesis of heart dysfunction. Canadian Journal of Cardiology. 2017;33(7):850–859. doi: 10.1016/j.cjca.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Rocchi A., He C. Regulation of exercise-induced autophagy in skeletal muscle. Curr Pathobiol Rep. 2017;5(2):177–186. doi: 10.1007/s40139-017-0135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson C.M., Johannsen D.L., Ravussin E. Skeletal muscle mitochondria and aging: a review. J Aging Res. 2012;2012:194821. doi: 10.1155/2012/194821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvani R., Joseph A.M., Adhihetty P.J., Miccheli A., Bossola M., Leeuwenburgh C. Mitochondrial pathways in sarcopenia of aging and disuse muscle atrophy. Biological Chemistry. 2013;394(3):393–414. doi: 10.1515/hsz-2012-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westermann B. Mitochondrial fusion and fission in cell life and death. Nature Reviews Molecular Cell Biology. 2010;11(12):872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y.J., Jeong S.Y., Karbowski M., Smith C.L., Youle R.J. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Molecular Biology of the Cell. 2004;15(11):5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. The EMBO Journal. 2008;27(2):433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomes L.C., Scorrano L. Mitochondrial elongation during autophagy: a stereotypical response to survive in difficult times. Autophagy. 2011;7(10):1251–1253. doi: 10.4161/auto.7.10.16771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bossy-Wetzel E., Barsoum M.J., Godzik A., Schwarzenbacher R., Lipton S.A. Mitochondrial fission in apoptosis, neurodegeneration and aging. Current Opinion in Cell Biology. 2003;15(6):706–716. doi: 10.1016/j.ceb.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 16.Chan D.C. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125(7):1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chauhan A., Vera J., Wolkenhauer O. The systems biology of mitochondrial fission and fusion and implications for disease and aging. Biogerontology. 2014;15(1):1–12. doi: 10.1007/s10522-013-9474-z. [DOI] [PubMed] [Google Scholar]
- 19.Seo A.Y., Joseph A.M., Dutta D., Hwang J.C., Aris J.P., Leeuwenburgh C. New insights into the role of mitochondria in aging: mitochondrial dynamics and more. Journal of Cell Science. 2010;123(Pt 15):2533–2542. doi: 10.1242/jcs.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang C., Feng P., Ku B., Dotan I., Canaani D., Oh B.H. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nature Cell Biology. 2006;8(7):688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 21.He S., Zhao Z., Yang Y., O'Connell D., Zhang X., Oh S. Truncating mutation in the autophagy gene UVRAG confers oncogenic properties and chemosensitivity in colorectal cancers. Nature Communications. 2015;6:7839. doi: 10.1038/ncomms8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He S., Liang C. Frameshift mutation of UVRAG: switching a tumor suppressor to an oncogene in colorectal cancer. Autophagy. 2015;11(10):1939–1940. doi: 10.1080/15548627.2015.1086523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang C., Lee J.S., Inn K.S., Gack M.U., Li Q., Roberts E.A. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nature Cell Biology. 2008;10(7):776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Afzal S., Hao Z., Itsumi M., Abouelkheer Y., Brenner D., Gao Y. Autophagy-independent functions of UVRAG are essential for peripheral naive T-cell homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(4):1119–1124. doi: 10.1073/pnas.1423588112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 26.Scarpulla R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochimica et Biophysica Acta. 2011;1813(7):1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 28.Kalkhoran S.B., Munro P., Qiao F., Ong S.B., Hall A.R., Cabrera-Fuentes H. Unique morphological characteristics of mitochondrial subtypes in the heart: the effect of ischemia and ischemic preconditioning. Discoveries. 2017;5(1) doi: 10.15190/d.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carey K.D., Garton A.J., Romero M.S., Kahler J., Thomson S., Ross S. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Research. 2006;66(16):8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- 30.Yun C.H., Boggon T.J., Li Y., Woo M.S., Greulich H., Meyerson M. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11(3):217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciano M., Mantellato G., Connolly M., Paul-Clark M., Willis-Owen S., Moffatt M.F. EGF receptor (EGFR) inhibition promotes a slow-twitch oxidative, over a fast-twitch, muscle phenotype. Scientific Reports. 2019;9(1):9218. doi: 10.1038/s41598-019-45567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madeo F., Tavernarakis N., Kroemer G. Can autophagy promote longevity? Nature Cell Biology. 2010;12(9):842–846. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- 33.Jiao J., Demontis F. Skeletal muscle autophagy and its role in sarcopenia and organismal aging. Current Opinion in Pharmacology. 2017;34:1–6. doi: 10.1016/j.coph.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 34.Raben N., Hill V., Shea L., Takikita S., Baum R., Mizushima N. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Human Molecular Genetics. 2008;17(24):3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song Z., An L., Ye Y., Wu J., Zou Y., He L. Essential role for UVRAG in autophagy and maintenance of cardiac function. Cardiovascular Research. 2014;101(1):48–56. doi: 10.1093/cvr/cvt223. [DOI] [PubMed] [Google Scholar]
- 36.An L., Hu X.W., Zhang S., Hu X., Song Z., Naz A. UVRAG deficiency exacerbates doxorubicin-induced cardiotoxicity. Scientific Reports. 2017;7:43251. doi: 10.1038/srep43251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jahng J.W.S., Alsaadi R.M., Palanivel R., Song E., Hipolito V.E.B., Sung H.K. Iron overload inhibits late stage autophagic flux leading to insulin resistance. EMBO Reports. 2019;20(10) doi: 10.15252/embr.201947911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wanagat J., Cao Z., Pathare P., Aiken J.M. Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting, and oxidative damage in sarcopenia. The FASEB Journal. 2001;15(2):322–332. doi: 10.1096/fj.00-0320com. [DOI] [PubMed] [Google Scholar]
- 39.Trappe S., Gallagher P., Harber M., Carrithers J., Fluckey J., Trappe T. Single muscle fibre contractile properties in young and old men and women. Jornal of Physiol. 2003;552(Pt 1):47–58. doi: 10.1113/jphysiol.2003.044966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leal L.G., Lopes M.A., Batista M.L., Jr. Physical exercise-induced myokines and muscle-adipose tissue crosstalk: a review of current knowledge and the implications for health and metabolic diseases. Frontiers in Physiology. 2018;9:1307. doi: 10.3389/fphys.2018.01307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boenzi S., Diodato D. Biomarkers for mitochondrial energy metabolism diseases. Essays in Biochemistry. 2018;62(3):443–454. doi: 10.1042/EBC20170111. [DOI] [PubMed] [Google Scholar]
- 42.Chung H.K., Ryu D., Kim K.S., Chang J.Y., Kim Y.K., Yi H.S. Growth differentiation factor 15 is a myomitokine governing systemic energy homeostasis. The Journal of Cell Biology. 2017;216(1):149–165. doi: 10.1083/jcb.201607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kharitonenkov A., Shiyanova T.L., Koester A., Ford A.M., Micanovic R., Galbreath E.J. FGF-21 as a novel metabolic regulator. Journal of Clinical Investigation. 2005;115(6):1627–1635. doi: 10.1172/JCI23606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inagaki T., Dutchak P., Zhao G., Ding X., Gautron L., Parameswara V. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metabolism. 2007;5(6):415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Oost L.J., Kustermann M., Armani A., Blaauw B., Romanello V. Fibroblast growth factor 21 controls mitophagy and muscle mass. J Cachexia Sarcopenia Muscle. 2019;10(3):630–642. doi: 10.1002/jcsm.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bloch S.A., Lee J.Y., Wort S.J., Polkey M.I., Kemp P.R., Griffiths M.J. Sustained elevation of circulating growth and differentiation factor-15 and a dynamic imbalance in mediators of muscle homeostasis are associated with the development of acute muscle wasting following cardiac surgery. Critical Care Medicine. 2013;41(4):982–989. doi: 10.1097/CCM.0b013e318274671b. [DOI] [PubMed] [Google Scholar]
- 47.Nakajima T., Shibasaki I., Sawaguchi T., Haruyama A., Kaneda H., Nakajima T. Growth differentiation factor-15 (GDF-15) is a biomarker of muscle wasting and renal dysfunction in preoperative cardiovascular surgery patients. Journal of Clinical Medicine. 2019;8(10) doi: 10.3390/jcm8101576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laurens C., Parmar A., Murphy E., Carper D., Lair B., Maes P. Growth and differentiation factor 15 is secreted by skeletal muscle during exercise and promotes lipolysis in humans. JCI Insight. 2020;5(6) doi: 10.1172/jci.insight.131870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujita Y., Taniguchi Y., Shinkai S., Tanaka M., Ito M. Secreted growth differentiation factor 15 as a potential biomarker for mitochondrial dysfunctions in aging and age-related disorders. Geriatrics and Gerontology International. 2016;16(Suppl 1):17–29. doi: 10.1111/ggi.12724. [DOI] [PubMed] [Google Scholar]
- 50.Kim H., Kim K.M., Kang M.J., Lim S. Growth differentiation factor-15 as a biomarker for sarcopenia in aging humans and mice. Experimental Gerontology. 2020;142:111115. doi: 10.1016/j.exger.2020.111115. [DOI] [PubMed] [Google Scholar]
- 51.Li X., Monks B., Ge Q., Birnbaum M.J. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447(7147):1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 52.Bollu L.R., Ren J., Blessing A.M., Katreddy R.R., Gao G., Xu L. Involvement of de novo synthesized palmitate and mitochondrial EGFR in EGF induced mitochondrial fusion of cancer cells. Cell Cycle. 2014;13(15):2415–2430. doi: 10.4161/cc.29338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Che T.F., Lin C.W., Wu Y.Y., Chen Y.J., Han C.L., Chang Y.L. Mitochondrial translocation of EGFR regulates mitochondria dynamics and promotes metastasis in NSCLC. Oncotarget. 2015;6(35):37349–37366. doi: 10.18632/oncotarget.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhosle J., Kiakos K., Porter A.C., Wu J., Makris A., Hartley J.A. Treatment with gefitinib or lapatinib induces drug resistance through downregulation of topoisomerase IIalpha expression. Molecular Cancer Therapeutics. 2013;12(12):2897–2908. doi: 10.1158/1535-7163.MCT-12-1049. [DOI] [PubMed] [Google Scholar]
- 55.Okon I.S., Coughlan K.A., Zhang M., Wang Q., Zou M.H. Gefitinib-mediated reactive oxygen specie (ROS) instigates mitochondrial dysfunction and drug resistance in lung cancer cells. Journal of Biological Chemistry. 2015;290(14):9101–9110. doi: 10.1074/jbc.M114.631580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takenaka T., Katayama M., Sugiyama A., Hagiwara M., Fujii I., Takatani-Nakase T. Gefitinib enhances mitochondrial biological functions in NSCLCs with EGFR mutations at a high cell density. Anticancer Research. 2017;37(9):4779–4788. doi: 10.21873/anticanres.11884. [DOI] [PubMed] [Google Scholar]
- 57.Leroy M.C., Perroud J., Darbellay B., Bernheim L., Konig S. Epidermal growth factor receptor down-regulation triggers human myoblast differentiation. PloS One. 2013;8(8) doi: 10.1371/journal.pone.0071770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kosaka T., Yatabe Y., Endoh H., Kuwano H., Takahashi T., Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Research. 2004;64(24):8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.