Abstract
The skin microbiota of atopic dermatitis (AD) patients is characterized by increased Staphylococcus aureus colonization, which exacerbates disease symptoms and has been linked to reduced bacterial diversity. Skin bacterial communities in AD patients have mostly been described at family and genus levels, while species-level characterization has been limited. In this study, we investigated the role of the bacteria belonging to the Staphylococcus genus using targeted sequencing of the tuf gene with genus-specific primers. We compared staphylococcal communities on lesional and non-lesional skin of AD patients, as well as AD patients with healthy controls, and determined the absolute abundance of bacteria present at each site. We observed that the staphylococcal community, bacterial alpha diversity, and bacterial densities were similar on lesional and non-lesional skin, whereas AD severity was associated with significant changes in staphylococcal composition. Increased S. aureus, Staphylococcus capitis, and Staphylococcus lugdunensis abundances were correlated with increased severity. Conversely, Staphylococcus hominis abundance was negatively correlated with severity. Furthermore, S. hominis relative abundance was reduced on AD skin compared to healthy skin. In conclusion, various staphylococcal species appear to be important for skin health.
Keywords: atopic dermatitis, skin microbiome, skin microbiota, Staphylococcus, S. aureus, coagulase-negative staphylococci, tuf amplicon sequencing, 16S rRNA amplicon sequencing, 16S rRNA qPCR
1. Introduction
Atopic dermatitis (AD) is a common inflammatory skin disease characterized by dry and itchy skin and recurring flares. The disease etiology is complex and involves interplays between immunological dysregulation and impaired skin barrier function, the most well-known genetic risk factor being loss-of-function mutations in the gene encoding the skin barrier protein filaggrin (FLG) [1,2]. Furthermore, AD patients are more often colonized with Staphylococcus aureus on their skin than healthy individuals, with a metanalysis showing a 70% colonization prevalence on lesional skin and 39% on non-lesional skin [3].
A reduction in the bacterial alpha diversity on skin, sometimes referred to as bacterial dysbiosis, has been considered to be a hallmark of the AD skin phenotype [4,5]. Yet, reduced alpha diversity may not be universal, but depend on S. aureus skin colonization [4,6] and the microenvironmental skin conditions, such as moisture and sebum content [7,8]. Some studies have found that alpha diversity is significantly reduced on lesional compared to non-lesional skin [9,10] whereas other have found no difference in alpha diversity between the two skin sites [5,11]. These contradictory results may suggest a continuous transition between lesional and non-lesional skin within AD patients.
The immune dysregulation and skin barrier impairment of AD patients are believed to promote skin colonization with S. aureus. Thus, filaggrin deficiency and reduced levels of free fatty acids in AD skin lead to increased skin pH, which, in turn, has been shown to enhance S. aureus growth and adherence to keratinocytes [12,13,14,15,16,17], and FLG loss-of-function mutations have previously been associated with increased S. aureus skin colonization among AD patients [18]. On the other hand, S. aureus skin colonization has been shown to exacerbate AD [4,19]. The elimination of S. aureus from clinically infected skin by the topical application of antibiotics is, therefore, one strategy used to treat AD [20]. However, this treatment approach does not have a long-lasting effect and has been shown to select for antibiotic-resistant S. aureus [21,22]; hence, new strategies for controlling S. aureus skin colonization in AD are needed. One approach has been to identify natural compounds secreted from skin commensal bacteria with bacteriocidal or bacteriostatic activity against S. aureus. Bacterial antagonism is more likely to occur between closely related organisms [23]; thus, other staphylococcal species may be a likely source of new antimicrobial compounds that target S. aureus. In accordance, some skin commensal coagulase-negative staphylococci (CoNS) are able to inhibit S. aureus growth, through the production of selective bacteriocins [6,24,25,26], or by inhibiting S. aureus virulence through the interference of the accessory gene regulator (agr) quorum-sensing system [27,28,29]. Interestingly, the application of viable bacteriocin-producing Staphylococcus epidermidis or Staphylococcus hominis on the skin of AD patients has been shown to significantly decrease the cutaneous S. aureus abundance 24 h after treatment when compared to non-treated patients [6], emphasizing the therapeutic potential of using skin commensal staphylococci to limit S. aureus colonization in AD.
Despite their predominantly commensal relationship to their host, many CoNS are opportunistic pathogens that may infect wounds and eczematous lesions, and their role in AD may, therefore, be different to their role in healthy skin. Whereas many studies have focused on the role of S. aureus in AD, only a few, and often smaller, studies focusing on CoNS at species level have been published [5,19,30,31]. One of the reasons for this may be that many studies investigating the skin microbiota are based on targeted amplicon sequencing of 16S rRNA gene regions (V1–V3 or V3–V4 regions) that suffer from limited staphylococcal species discrimination of [32,33].
Here, our primary aim was to study the diversity of the staphylococcal communities on the lesional and non-lesional skin of AD patients and skin of healthy individuals using an amplicon sequencing-based method targeting the tuf gene. The tuf gene is conserved within the Staphylococcus genus but contains highly variable regions that can be used to discriminate between different species [32]. We also investigated staphylococcal communities on AD skin in correlation to AD severity and skin barrier dysfunction, including transepidermal water loss (TEWL) and skin pH. Secondly, we compared the overall bacterial abundance and alpha diversity on AD lesional, AD non-lesional and healthy skin using 16S rRNA (V3-V4) amplicon sequencing and qPCR.
2. Materials and Methods
2.1. Participants
Adult AD patients from the Dermatology Outpatient Clinic at Bispebjerg Hospital (Copenhagen, Denmark) were enrolled at two time periods: November 2016–January 2017 [34] and March 2018–September 2019. Inclusion criteria were age ≥ 18 years and presence of AD according to U.K. criteria [35]. Patients were invited to participate in the study regardless of topical of systemic treatment of AD; however, any treatment within the past three months was registered (Table S1). Disease severity was assessed by scoring AD (SCORAD), which is based on an evaluation of the skin (i.e., erythema, oedema, oozing, excoriation, lichenification, dryness and eczema extent) and a subjective assessment of symptoms (itch and sleeplessness) [36]. Common loss-of-function mutations in FLG (R501X, 2282del4, and R2447X) were determined using a blood test and the skin barrier function assessed by measurements of TEWL and skin pH on the volar forearm on non-lesional skin (DermaLab, Cortex Technology, Denmark) (Table 1), as described previously [34].
Table 1.
Description of study populations.
| Covariates | AD Patients (n = 94) |
Sub-Group of AD Patients (n = 36) | Healthy Individuals (n = 92) | |
|---|---|---|---|---|
| Demographic data | ||||
| Sex | Female:Male ratio | 44:50 | 18:18 | 47:45 |
| Age (years) | Median (range) | 38 (18–71) | 36 (21–71) | 40 (18–66) |
| Clinical data | ||||
| Other atopic diseases | Asthma | 47 (50%) | 19 (53%) | 1 (1%) |
| Allergic rhinitis | 67 (71%) | 26 (72%) | 13 (14%) | |
| Type 1 allergy | 79 (83%) | 29 (81%) | 14 (15%) | |
| AD severity (SCORAD) 1 | Median (range) | 38 (5–88) | 34.0 (4.7–88) | NR |
| Mild:Moderate:Severe | 25:48:21 | 14:17:5 | NR | |
| FLG loss-of-function mutations 2 | Mutations | 34 (36%) | 12 (33%) | NE |
| WT | 50 (53%) | 22 (61%) | NE | |
| Skin pH 3 | Median (range) | 5.7 (4.5–7.0) | 5.7 (4.7–6.8) | NE |
| TEWL 3 (g/m2/h) | Median (range) | 10 (3–50) | 10 (4–34) | NE |
| S. aureus culture data | ||||
| Colonization of LS | 58 (62%) | 20 (56%) | NR | |
| Colonization of NLS | 32 (34%) | 10 (28%) | 5 (6%) | |
| Colonization of Nares | 61 (66%) | 20 (56%) | 31 (34%) | |
| Sample location | ||||
| LS sample location 4 | Vf:Ac:Other | 50:17:27 | 0:17:0 | NR |
| NLS sample location 4 | Vf:Ac:Other | 73:19:2 | 0:19:0 | 0:92:0 |
| Microenvironment of LS | Dry:Moist:Sebaceous | 64:18:12 | 0:17:0 | NR |
| Microenvironment of NLS | Dry:Moist:Sebaceous | 75:19:0 | 0:19:0 | 0:92:0 |
1 Mild disease: SCORAD < 25; Moderate disease: SCORAD 25–50; Severe disease: SCORAD > 50. 2 Mutations: R501X, 2282del4, and/or R2447X. Wild type (WT) represents absence of any of the three mutations. Ten patients were not tested for FLG mutations. 3 skin pH and TEWL were measured on non-lesional skin of the volar forearm. 4 Vf: volar forearm. Ac: antecubital crease. “Other” includes the back, cheeks, chest, hand, leg, neck, popliteal crease, and thigh. Abbreviations: AD: atopic dermatitis; LS: lesional skin; NLS: non-lesional skin; FLG: filaggrin gene; TEWL: transepidermal water loss; NR: Not relevant; NE: Not examined.
Blood donors ≥ 18 years (blood donor clinic, Hvidovre Hospital, Copenhagen, Denmark) were enrolled as healthy controls in the months of May 2019 and January 2020. Study exclusion criteria were history of skin diseases, including AD.
2.2. Samples
Primary samples were collected from skin using eSwabs (Copan, Brescia, Italy), and kept on dry ice for up to five hours before storing at −80 °C until further processing. For patients, lesional and non-lesional skin samples were collected depending on the site of eczema, primarily from the volar forearm and the antecubital crease (Table 1). Skin samples from healthy individuals were collected from the antecubital crease.
S. aureus skin colonization was determined by culturing, by plating sample aliquots on S. aureus-selective plates (SaSelect, BioRad, Marnes-la-Coquette, France; or chromID, bioMérieux, France). S. aureus classification of colonies was validated using a spa-specific PCR, as previously described [37].
2.3. DNA Extraction, Amplicon Sequencing, and Quantitative PCR
Bacterial cells were enzymatically pre-lysed and DNA extracted using a MagNA Pure 96 purification instrument (Roche, Mannheim, Germany). The V3–V4 region of the 16S rRNA gene was selected for the analysis of complete bacterial communities [38], and the tuf gene was selected for analysis of staphylococcal species [32]. The selected gene regions were amplified in a two-step PCR and sequenced on a MiSeq instrument using 600-cycle MiSeq Reagent Kit v3 (Illumina Inc., San Diego, CA, USA) kits. Sequences are available at the European Nucleotide Archive under the project identification number: PRJEB42898. The absolute abundance of bacteria was estimated by qPCR using primers identical to the 16S rRNA V3–V4 primers used for amplicon sequencing. See Supplemental Methodology for details on primers and protocols.
2.4. Bioinformatics and Statistics
DADA2 v.1.12.1 [39] was used for the inference of amplicon sequence variants (ASVs) and taxonomic assignments using the Silva reference database v.132 [40] for 16S rRNA-derived ASVs and an inhouse reference database was used for tuf-derived ASVs [32]. The tuf reference database can be accessed here: https://github.com/ssi-dk/staphylome/tree/master/database, accessed on 18 February 2021. All statistical analyses were performed in R v.4.0.2 [41], using the R package phyloseq v.1.33.0 [42] for data processing, and ggplot2 v.3.3.2. [43] for the visual presentation of results. Putative contaminants were removed prior to downstream analysis using the R package Decontam v.3.6.1 [44].
The absolute abundance of bacteria was quantified as the total number of 16S rRNA gene copies within 1 µl of DNA eluate. For graphical visualizations, log10-transformed counts were used. The absolute abundance of Staphylococcus was estimated by combining 16S rRNA qPCR and amplicon data (relative abundance of Staphylococcus (amplicon data) x absolute abundance of bacteria (qPCR data)), as previously suggested in [45,46]. Absolute abundances of staphylococcal species were similarly calculated (see Supplemental Methodology for an example).
Taxonomic counts resulting from 16S rRNA V3–V4 amplicon sequencing were agglomerated at genus level for all analyses except for alpha diversity measures, which was estimated at ASV level using the Shannon index. Differences in Shannon index, absolute abundance of bacteria, and absolute abundance of Staphylococcus between skin sites were tested using Wilcoxon signed-rank test and Mann–Whitney U test for paired (lesional versus non-lesional AD skin) and un-paired (AD skin versus healthy skin) samples, respectively. In addition, differences between AD lesional and non-lesional skin were tested using a linear mixed model including the skin microenvironment of the sampling area (i.e., dry, moist or sebaceous skin) as a fixed effect (Supplemental methodology). Bacterial compositional differences between skin sites (beta diversity) were measured using Bray–Curtis dissimilarities on Hellinger-transformed counts and tested with permutational multivariable analysis of variance (PERMANOVA) (function adonis in R package vegan v.2.5.6. [47]). The determination of differentially abundant bacterial genera between skin sites was performed using analysis of composition of microbiomes with bias correction (ANCOM-BC) modelling, which estimates a change between test groups for each taxon using log-transformed values of absolute sequence counts [48]. The 30 most abundant genera were included in the analysis and results were corrected for multiple testing using the Benjamini-Hochberg method (i.e., controlling the false discovery rate (fdr)). Both PERMANOVA and ANCOM-BC methods allow for the adjustment of covariates and a variable containing information on the skin microenvironment of the sampling area was, therefore, included in the comparative analyses of lesional and non-lesional skin. R package Metacoder v.0.3.4 [49] was used for graphical presentation of the 30 most abundant bacterial genera on skin.
The tuf-based analysis of Staphylococcus was performed at species level. Staphylococcal species composition was compared between skin sites using a PERMANOVA test as described above and visualized using principal coordinates analysis (PCoA). Partitioning around medoid (PAM) clustering (R package cluster v.2.1.0. [50]), based on the Jensen-Shannon distance on Hellinger transformed counts, was used to group AD lesional and non-lesional skin samples into staphylococcal community clusters. Differences in bacterial Shannon diversity, absolute abundance of bacteria, SCORAD values, and TEWL measures between the four identified community clusters were tested using a Kruskal–Wallis test. If significant, a Dunn’s post hoc test was performed with correction for multiple testing using the Benjamini–Hochberg method. Differences in pH across the four community clusters were tested with an analysis of variance (ANOVA) test. The identification of differentially abundant species between skin sites and between AD severity groups was performed using ANCOM-BC as described for the 16S rRNA analysis. Spearman’s correlations were calculated between the absolute abundance of selected species and the following variables: Bacterial Shannon diversity, SCORAD values, skin pH measures, and TEWL measures. Results were corrected for multiple testing (Benjamini–Hochberg method) in cases where several species were included in a test. See Supplemental methodology for an extended description.
2.5. Ethical Approval
Sample and data collection was approved by the local Science Ethics Committee (Videnskabsetisk Komite, Region-Hovedstaden) (Inclusion of AD patients: H-1-2014-039 (approved 8 July 2014). Inclusion of healthy controls: H-16023435 (approved on 6 June 2016)) and by the Danish Data protection Agency. All participants provided signed informed consent.
3. Results
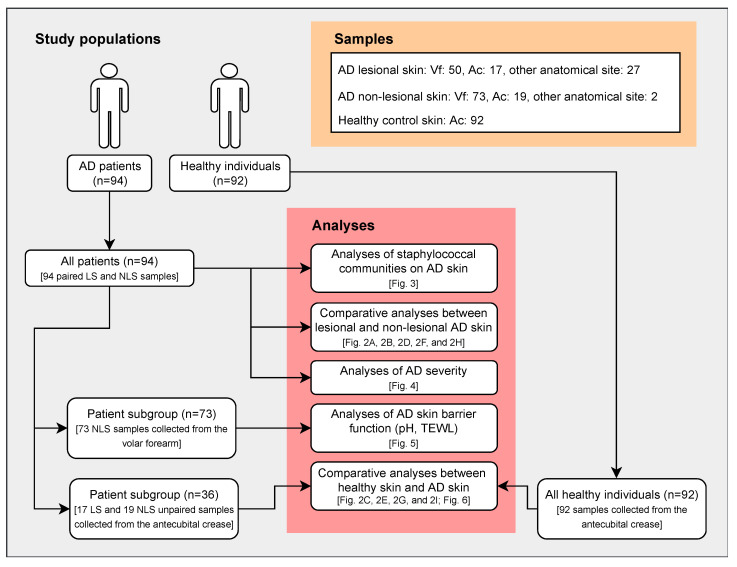
The study included 94 AD patients with active disease and 92 healthy individuals, with similar sex and age distributions as the patient cohort (Table 1). Samples were collected from both lesional and non-lesional skin areas of AD patients, primarily from the volar forearm (dry skin) and the antecubital crease (moist skin), and from the antecubital crease of healthy individuals. Since previous studies have shown that the bacterial diversity and composition can differ between anatomical skin sites [4,7,8,51], only AD skin samples collected from the antecubital crease were used in the comparative analyses with the healthy individuals. This subgroup of AD patients (n = 36) reflected the complete patient cohort, according to demographic and clinical characteristics (Table 1). An outline of the study design is shown in Figure 1.
Figure 1.
Study design. Illustration showing study subjects, samples, and performed analyses. Skin samples from healthy individuals were all collected at the antecubital crease. Consequently, only AD skin samples collected from the antecubital crease were included in comparative analyses between AD patients and healthy individuals. TEWL and pH were measured on the volar forearm on non-lesional AD skin, and only samples collected from this site were, therefore, included in analyses of skin barrier function. Abbreviations: AD: atopic dermatitis; LS: lesional skin; NLS: non-lesional skin; TEWL: transepidermal water loss; Vf: volar forearm; Ac: antecubital crease.
3.1. Skin Bacterial Community Profiling
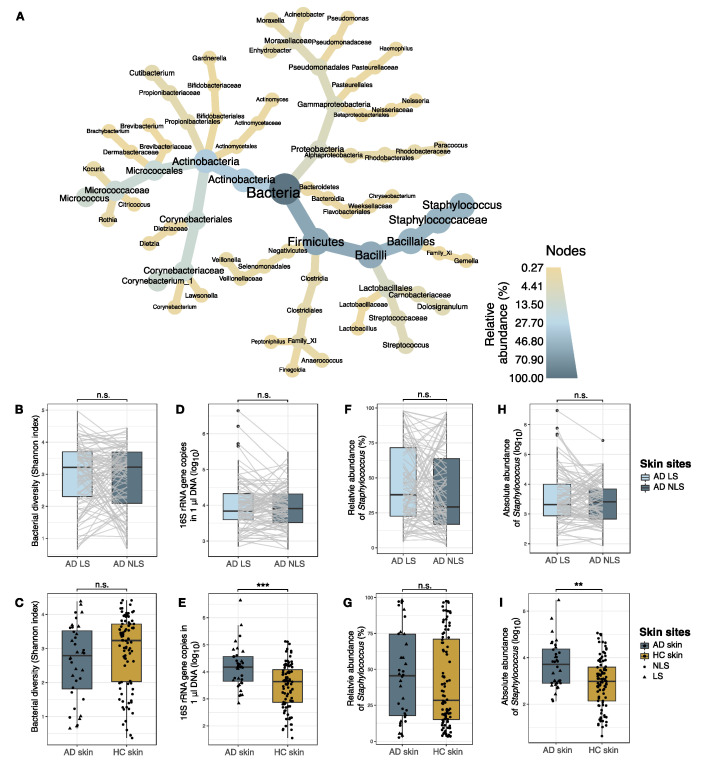
Staphylococcus (Firmicutes), Corynebacterium, and Micrococcus (Actinobacteria) were the three most prevalent and abundant bacterial genera identified on the skin of both AD patients and healthy individuals (Figure 2A and Figure S1).
Figure 2.
Skin bacterial communities. (A) Taxonomic illustration of the 30 most abundant genera on AD skin. Color and size of nodes and leaves correspond to the average relative abundance of taxa. AD lesional and non-lesional skin samples were grouped together. (B,C). Bacterial alpha diversity measured by Shannon index. (D,E) Absolute abundance of bacteria, measured as 16S rRNA gene copies within 1 µl DNA sample eluate. (F,G) Relative abundance (%) of Staphylococcus genus. Differences between skin sites were tested using ANCOM-BC modelling on log transformed sequence counts. (H,I) Absolute abundance of Staphylococcus, calculated by combining 16S rRNA qPCR and amplicon sequence data. In B, D, F, and H, lesional and non-lesional skin of AD patients (n = 94) were compared. Samples collected from the same patient are indicated by grey lines. In C, E, G, and I, skin samples collected from AD patients (n = 36) and healthy individuals (n = 92) were compared. Here, AD lesional (triangles) and non-lesional (dots) skin samples were grouped together as AD skin. Boxplots represent the median and interquartile range (IQR) with whiskers extending to the minimum/maximum value, but no longer than 1.5xIQR. Asterisks indicate statistical significance: ** p < 0.01, *** p < 0.001, n.s.: not significant. Abbreviations: AD: atopic dermatitis; LS: lesional skin; NLS: non-lesional skin; HC: healthy control skin.
3.1.1. Comparison of Bacterial Communities in AD Lesional and Non-lesional Skin
When comparing the overall bacterial population on lesional and non-lesional skin from AD patients we found no differences with regards to alpha diversity (Figure 2B), or absolute abundance of bacteria (Figure 2D). Furthermore, the bacterial community structures were similar on lesional and non-lesional AD skin (R2 = 0.005), with none of the 30 most abundant bacterial genera, including Staphylococcus, being differentially abundant between the two skin sites. The absolute abundance of Staphylococcus was also similar on lesional and non-lesional skin (Figure 2H).
3.1.2. Comparison of Bacterial Communities in AD and Skin
Since we observed similar bacterial populations of lesional and non-lesional AD skin, the samples were grouped together for the comparison of AD with healthy individuals. The absolute abundance of bacteria on the antecubital crease was significantly higher on the skin of AD patients when compared to healthy skin (median fold-change = 3.4, p < 0.001) (Figure 2E). Alpha diversity was reduced on AD skin (median: 2.8) compared with healthy skin (median: 3.2), although not statistically significant (p = 0.1) (Figure 2C). A significant difference in the composition of bacterial genera on the skin was identified between AD and healthy individuals (R2 = 0.02, p = 0.004). Differential abundance analysis indicated that Streptococcus was enriched and Bacillus and Kocuria abundances were reduced on AD skin compared to healthy skin (Figure S2), whereas there was no significant difference in the abundance of Staphylococcus on AD and healthy skin. However, the absolute abundance of Staphylococcus was significantly higher on AD skin (median fold change: 5.6, p = 0.002) (Figure 2I).
3.2. Staphylococcal Communities on AD Skin
To further investigate the staphylococcal species composition on skin, we performed Staphylococcus-specific amplicon sequencing of the tuf gene.
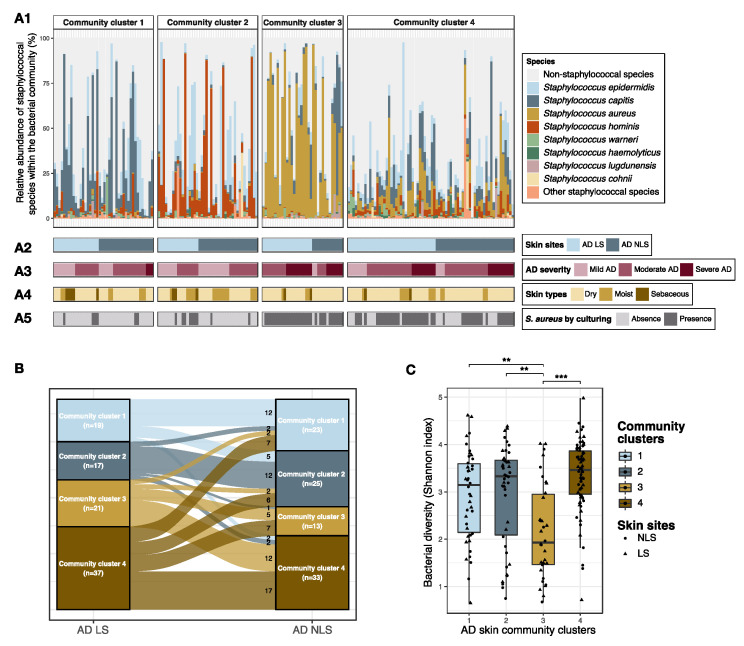
Four distinct staphylococcal community clusters were identified based on species compositional (dis)similarities (Figure 3A, Figure S3). Community cluster 1 was characterized by high proportions of Staphylococcus capitis and S. epidermidis, community cluster 2 by S. hominis and S. epidermidis and community cluster 3 by S. aureus, whereas community cluster 4 was more diverse. AD skin samples belonging to community cluster 3 were further characterized by a reduced bacterial alpha diversity as compared to skin samples assigned to the other three clusters (adj. p < 0.001) (Figure 3C), indicating an association between S. aureus proportional abundance and bacterial alpha diversity. To examine this further, we performed a correlation analysis, which showed that an increase in the absolute abundance of S. aureus was significantly correlated with a reduced Shannon diversity within both lesional (ρ = −0.4, p < 0.0001) and non-lesional skin (ρ = −0.2, p =0.02) (Figure S4). The absolute abundance of bacteria on skin was similar in the four defined community clusters (Figure S5).
Figure 3.
Staphylococcal communities on AD skin. Lesional and non-lesional skin from AD patients (n = 94) were grouped into four distinct community clusters based on compositional similarities of staphylococcal species. (A1) Bar plot showing the relative abundances of the eight most abundant staphylococcal species within the bacterial community. (A2) Tile plot showing whether samples were collected from lesional or non-lesional skin. (A3) Tile plot showing the AD severity of patients from whom the samples were collected. Mild AD: SCORAD < 25; Moderate AD: SCORAD 25–50; and Severe AD: SCORAD > 50. (A4) Tile plot showing whether samples were collected from dry, moist, or sebaceous skin areas. (A5) Tile plot showing S. aureus colonization status, defined by culturing. (B) Sankey diagram for intra-individual comparison of staphylococcal community cluster assignment of lesional and non-lesional skin. (C) Comparison of skin bacterial alpha diversity, measured by Shannon index, across the four defined staphylococcal community clusters. Lesional and non-lesional skin samples were visualized as triangles and dots, respectively, and boxplots represent the median and interquartile range (IQR) with whiskers extending to the minimum/maximum value, but no longer than 1.5 × IQR. Asterisks indicate statistical significance: ** adj. p < 0.01, *** adj. p < 0.001. Abbreviations: AD: atopic dermatitis; LS: lesional skin; NLS: non-lesional skin.
3.2.1. Comparison of Staphylococcal Communities on AD Lesional and Non-Lesional Skin
There was no difference in the composition of staphylococcal species on lesional and non-lesional AD skin (R2 = 0.009) (Figure S6). In alignment, no significant difference in the proportion of lesional and non-lesional skin samples assigned to each community cluster was found (Figure 3A,B). Approximately half of the patients had lesional and non-lesional skin assigned to the same cluster (n = 46, 49%) (Figure 3B).
Differential abundance analysis showed that S. epidermidis was enriched in the staphylococcal community within non-lesional skin compared to lesional skin (mean fold change: 1.9, adj. p = 0.007), whereas the abundance of S. aureus was similar at the two sites (Figure S7). It may be of more clinical relevance to examine changes in absolute abundance of colonizing bacteria rather than compositional abundances within the staphylococcal community. Therefore, the absolute abundance of staphylococcal species was calculated by combining the proportional species abundance within the bacterial community with the total bacterial abundance quantified by qPCR. These data revealed that the absolute abundance of S. aureus was significantly increased on lesional compared to non-lesional skin (median fold change: 1.8, adj. p = 0.002), whereas there was no difference in the absolute abundance of S. epidermidis (Figure S7).
3.2.2. Staphylococcal Communities and AD Severity
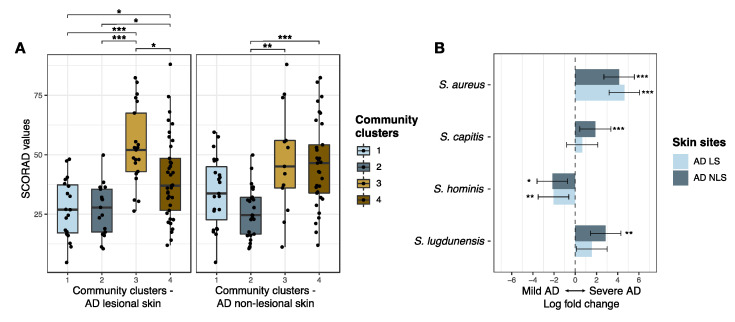
AD patients with skin staphylococcal communities assigned to cluster 3 and cluster 4 had more severe AD as compared to patients with skin staphylococcal communities belonging to cluster 1 or cluster 2 (Figure 3A, Figure 4A). Differential abundance analysis showed that S. aureus was enriched and S. hominis reduced within the staphylococcal community on lesional and non-lesional skin among patients with severe AD (SCORAD > 50, n = 21) compared to patients with mild AD (SCORAD < 25, n = 25) (Figure 4B). S. capitis and S. lugdunensis were also found to be enriched on the non-lesional skin of patients with severe AD (Figure 4B). In accordance, SCORAD was positively correlated with the absolute abundance of S. aureus on both lesional and non-lesional skin (ρ = 0.6 and ρ = 0.5 for lesional and non-lesional skin, respectively, adj. p-values < 0.0001). On non-lesional skin, absolute abundances of S. capitis (ρ = 0.3, adj. p = 0.03) and S. lugdunensis (ρ = 0.2, adj. p = 0.04), were positively correlated with SCORAD, whereas S. hominis absolute abundance was negatively correlated (ρ = −0.3, adj. p = 0.02) (Figure S8).
Figure 4.
AD severity and cutaneous staphylococcal communities. (A) AD severity, assessed by SCORAD, and its association to staphylococcal community cluster assignments of lesional and non-lesional skin of AD patients (n = 94). Boxplots represent the median and interquartile range (IQR) with whiskers extending to the minimum/maximum value, but no longer than 1.5xIQR. (B) Differential abundant staphylococcal species on lesional and non-lesional skin from patients with mild AD (SCORAD < 25, n = 25) as compared to patients with severe AD (SCORAD > 50, n = 21). Bars represent the ANCOM-BC estimated log fold change in species abundance between compared groups (effect size) and error bars, with the 95% confidence interval. Log fold change refers to the natural logarithm. Species with a fdr-adjusted p-value < 0.05 were considered significant and are shown in the plot. Asterisks indicate statistical significance: * adj. p < 0.05, ** adj. p < 0.01, *** adj. p < 0.001. Abbreviations: AD: atopic dermatitis; LS: lesional skin; NLS: non-lesional skin; SCORAD: Scoring Atopic Dermatitis.
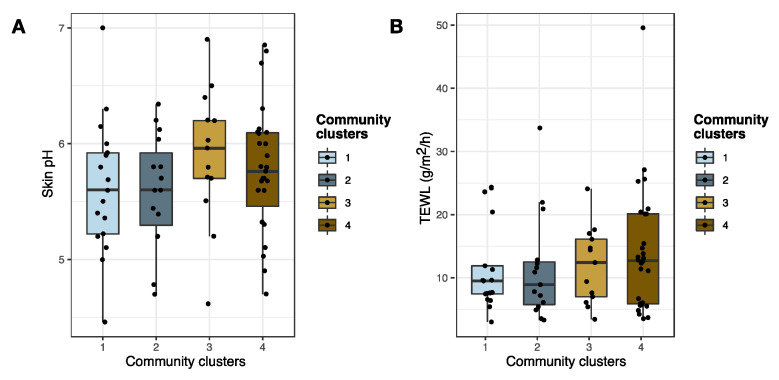
3.2.3. Staphylococcal Communities and AD Skin Barrier Function
Changes in the skin barrier function may have an impact on Staphylococcus skin colonization [52], and we therefore investigated if skin pH and TEWL were associated with changes in the staphylococcal community composition. TEWL and pH were measured on non-lesional skin of the volar forearm, and thus analyses were only performed on samples collected at this skin site (n = 73 patients) (Figure 1). The average skin pH was higher for skin assigned to staphylococcal community cluster 3 compared to the pH of skin assigned to the three other community clusters (Figure 5A), though this was not of statistical significance. TEWL measurements were found to be similar across all four community clusters (Figure 5B). Previous studies have shown that pH affects S. aureus growth in vitro [16,17,53], and we therefore tested for a possible correlation between the absolute abundance of S. aureus and skin pH. A tendency towards increasing quantities of S. aureus with increasing skin pH was observed, though this was not of statistical significance (ρ = 0.2, p = 0.07) (Figure S9). The absolute abundance of S. aureus was found to be positively correlated with TEWL (ρ = 0.3, p = 0.01) (Figure S9).
Figure 5.
Skin barrier function (pH and TEWL) and cutaneous staphylococcal communities. Skin pH (A) and TEWL (B) and their association to staphylococcal community cluster assignment of non-lesional AD skin (n = 73). Boxplots represent the median and interquartile range (IQR) with whiskers extending to the minimum/maximum value, but no longer than 1.5xIQR. Abbreviations: AD: atopic dermatitis; TEWL: transepidermal water loss.
No significant associations were found between carriage of FLG loss-of-function mutations and assignment of lesional or non-lesional skin samples to the four staphylococcal community clusters.
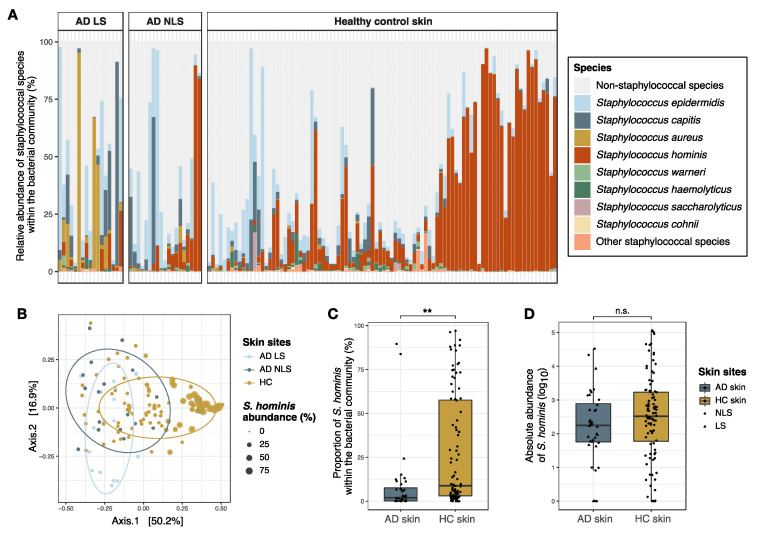
3.3. Comparison of Staphylococcal Communities on AD Skin and Healthy Skin
The staphylococcal community on the skin of the antecubital crease was significantly different between AD patients and healthy individuals, with a greater difference observed between lesional and healthy skin (R2 = 0.13, p < 0.001) compared to AD non-lesional and healthy skin (R2 = 0.06, p < 0.001) (Figure 6A,B). Differential abundance analysis indicated that S. aureus and S. capitis were enriched within cutaneous staphylococcal communities of AD patients, whereas S. hominis and Staphylococcus cohnii were more abundant on the skin of healthy individuals (Figure S10). The relative abundance of S. cohnii within the bacterial community was, on average, below 1% and its clinical significance might, therefore, be questioned. The absolute abundance of S. hominis was calculated in order to adjust for the observed differences in bacterial densities on the skin of AD patients and healthy individuals (Figure 2E). Though the proportions of S. hominis within the bacterial community were four times higher on healthy skin compared to AD skin (p < 0.001) (Figure 6C), no significant difference in the absolute abundance was found between the two skin sites (Figure 6D).
Figure 6.
Staphylococcal communities on the antecubital crease of AD patients and healthy individuals. (A) Bar plot showing the relative abundances of the eight most abundant staphylococcal species within the bacterial community of AD lesional skin (n = 17), AD non-lesional skin (n = 19), and healthy control skin (n = 92). (B) PCoA plot showing the compositional differences in staphylococcal communities on AD skin and healthy control skin. Point sizes are scaled based on the relative abundance of S. hominis within the bacterial community. Of note, two AD non-lesional skin samples with high S. hominis abundance cluster together with healthy control skin on the right side of the plot. (C) The relative abundance of S. hominis within the cutaneous bacterial community. (D) The absolute abundance of S. hominis on skin, which was calculated by combining proportional amplicon sequence data with 16S rRNA qPCR data. Thus, the absolute abundance is equivalent to the number of S. hominis 16S rRNA gene copies within 1µl DNA eluate (log10 transformed counts). Box plots represent the median and interquartile range (IQR) with whiskers extending to the minimum/maximum value, but no longer than 1.5 × IQR, and lesional and non-lesional skin samples are visualized as triangles and dots, respectively. Asterisks indicate a significant difference: ** p < 0.01, n.s.: not significant. Abbreviations: AD: atopic dermatitis; LS; lesional skin; NLS: non-lesional skin; HC skin: healthy control skin.
4. Discussion
This is the first study to perform an in-depth assessment of staphylococcal species in AD patients and relate this to disease severity. The study underlines that species-level characterization of Staphylococcus is important and contributes significantly to the already existing knowledge on S. aureus and AD.
Our results indicate that the absolute abundance of bacteria, as well as the bacterial alpha diversity and composition are similar on lesional and non-lesional skin of AD patients. Furthermore, compositional analysis suggests that the staphylococcal community is generally similar between lesional and non-lesional AD skin, whereas we found significant differences between AD and healthy skin as well as between AD severity groups. These findings indicate that AD pathogenesis has an impact on the overall skin microbiota, comprising staphylococcal species, independent of the presence of eczema. The presence of a universal bacterial community within AD skin sites has also previously been suggested [8,11], and Brandwein et al. found that increasing AD severity, rather than the presence or absence of lesions, was most important regarding changes in bacterial community within AD skin compared to healthy skin [11]. The present study of species-level characterization of Staphylococcus further supports this theory.
In the present study, four significantly distinct staphylococcal community clusters were identified with community cluster 1 characterized by a high proportional abundance of S. capitis and S. epidermidis, community cluster 2 by S. hominis and S. epidermidis, and community cluster 3 by S. aureus. Community cluster 4 was more diverse, with both S. epidermidis, S. aureus, S. capitis, and S. hominis. Bacterial alpha diversity was significantly reduced in AD skin samples belonging to community cluster 3. In accordance, an increase in the absolute abundance of S. aureus was correlated with a decreased bacterial alpha diversity, which supports previous reports [4,6]. Patients with lesional and non-lesional skin samples assigned to community clusters 3 and 4 had more severe AD compared to patients within community cluster 1 or 2. In accordance, increases in the absolute S. aureus abundance on lesional and non-lesional skin were correlated with increasing disease severity. These findings support the well-documented association between S. aureus skin colonization and AD severity [3,18]. The absolute abundances of S. capitis and S. lugdunensis on non-lesional skin were also associated with severe AD, whereas S. hominis absolute abundance was associated with a milder AD presentation. In a previous study, S. hominis skin colonization was found to be negatively associated with AD development during the first two years of life, whereas cutaneous S. aureus was predictive of AD development [54]. Although a causative relationship is uncertain, this could indicate that S. hominis is able to inhibit S. aureus skin colonization, leading to a reduced risk of AD development. In accordance, Nakatsuji et al. showed that some S. hominis strains produce bacteriocins acting against S. aureus, and that topical application of these S. hominis strains onto AD skin led to a significant reduction in the absolute abundance of S. aureus [6]. S. lugdunensis is also able to produce a bacteriocin that has a bactericidal effect on S. aureus, and it has been shown that individuals colonized with S. lugdunensis in the anterior nares are unlikely to be S. aureus nasal carriers [24]. Thus, it is surprising that we here observed that S. aureus and S. lugdunensis co-colonized the skin of patients with severe AD. A possible explanation could be nutritional differences between skin and nasal epithelium that could impact the growth and colonization properties of these species.
The composition of the staphylococcal communities on the antecubital crease of healthy individuals was significantly different from that on AD lesional and non-lesional skin, with reduced proportional abundances of S. hominis and S. cohnii, and increased abundance of S. aureus and S. capitis on AD skin. Baurecht et al. also found S. hominis abundance to be reduced on the antecubital crease of adult AD patients compared to healthy individuals, whereas no difference in abundance was found on dry skin of the arms [30]. S. hominis and S. capitis proportional abundances were not associated with either AD disease or disease exacerbation in children [4,19], however, these studies included less than 12 volunteers, which limits their ability to detect such associations. The importance of S. cohnii for human skin health is not clear, but one study has shown a negative correlation between S. aureus and S. cohnii colonization of the nasal epithelium in pigs [55]. This may indicate an antagonistic interaction between S. aureus and S. cohnii. Differences in the proportional abundance of staphylococcal species between AD patients and healthy individuals likely reflect pathophysiological abnormalities in AD skin with immune activation and decreased expression of skin barrier proteins, creating niches that facilitate different staphylococcal communities. Furthermore, one study has indicated strain-level differences in the CoNS colonizing skin of AD patients and healthy individuals [6], suggesting that it could be of relevance to investigate and compare virulence gene expression in CoNS from AD and healthy skin in future studies. Such studies could provide insight into whether S. capitis and S. lugdunensis could contribute to disease exacerbation in AD. Furthermore, staphylococcal species communities likely differ between skin anatomical areas [56], and thus it will be relevant to perform additional comparative studies of AD skin and healthy skin at other skin sites than the antecubital crease.
We found that the absolute S. aureus abundance on non-lesional AD skin was positively correlated with TEWL. A tendency for an association of increasing S. aureus abundance with increasing skin pH was also observed. A culture-based study has found that S. aureus skin colonization was associated with an increase in TEWL, whereas no difference in skin pH was seen between patients with S. aureus culture-positive or culture-negative skin swabs [52]. In vitro studies have shown that acidification of bacterial growth media, corresponding to the pH of healthy skin, was associated with impaired S. aureus growth [16,17,53] and decreased expression of S. aureus genes, which are important for the adherence to keratinocytes [16]. Thus, increasing skin pH might be an important mediator of enhanced S. aureus growth and persistent colonization in AD. Furthermore, increased TEWL reflecting impaired skin barrier function may cause enhanced penetration of S. aureus into the dermis, where it can trigger an immunological response [57]. At the same time, S. aureus may also induce skin barrier disruption by secreting proteases and cytotoxic toxins [27,58]. Since pH and TEWL were only assessed on non-lesional skin, it is unknown if the same associations would be present on lesional skin.
Some studies have shown reduced bacterial alpha diversity on AD skin compared to healthy skin [4,5,59], often denoted as microbial dysbiosis. In the present study, Shannon diversity varied between individuals, both among patients and healthy individuals, with no significant difference between AD lesional, AD non-lesional, and healthy skin. The majority of the included patients were treated with topical corticosteroids, which may have influenced the bacterial diversity of their skin [59,60]. In accordance, Kong et al. found that Shannon diversity on the antecubital crease was similar between AD patients with intermittent treatment and healthy individuals [4]. Furthermore, changes in bacterial diversity seem to be dependent on S. aureus colonization and abundance [4,6], which is supported by our results.
Many studies have investigated cutaneous bacterial communities in AD using compositional-based analysis, but only a few studies have examined the absolute abundance of bacteria on AD skin [9,57,59]. These studies found a significantly higher bacterial load on lesional compared to non-lesional skin, which is in contrast to the results presented in this study, where we found similar bacterial quantities on lesional and non-lesional skin. In two of these studies [9,57], lesional and non-lesional skin samples were collected from different anatomic sites, i.e., lesional samples from the antecubital crease (moist skin) and non-lesional samples from the upper arm (dry skin), which potentially could have influenced their results. In line with this, we observed a tendency towards higher bacterial density on AD non-lesional skin of moist compared to dry skin areas (p = 0.06, Figure S11). We also found, in agreement with previous findings [57,59] that the absolute bacterial abundance was significantly higher on AD skin compared to healthy skin, which indicates that an increase in total bacterial density is related to AD.
A major advantage of amplicon-based sequencing of bacterial target genes is the possibility of identifying fastidious bacterial species, that are difficult to culture, and thus the approach is essential when examining species diversity and microbial community composition. However, amplicon sequence data are compositional, meaning that an increase in the proportional abundance of one taxon will result in a decrease in other taxa, which is not necessarily synonymous with a change in density, as indicated by the results presented here. Thus, whereas the proportional abundance of S. hominis within the bacterial community was significantly reduced on AD skin compared to healthy skin, no difference in the absolute abundance of S. hominis was found between the two skin sites. This indicates that the reduced proportional abundance of S. hominis on AD skin was caused by a relative increase in other staphylococcal species, mainly S. aureus, and not a decrease in S. hominis density. Species competition within microbial communities is likely dependent on the absolute abundance of species [61]. Furthermore, the absolute abundance of a species can, in some cases, also be important for regulation of bacterial virulence, as was observed with the population density-modulated agr quorum sensing pathway, which regulates the expression of important S. aureus virulence genes [62,63]. Thus, investigating changes in absolute rather than relative abundance is likely of clinical relevance.
Our study has some limitations. One relates to the inconsistency of skin anatomical sites being sampled from AD patients and healthy individuals. Consequently, only a minor subset of AD skin samples was included in the comparative analyses with healthy skin, limiting the statistical power. Furthermore, the use of topical and systemic treatments among some of the patients may have influenced the results. However, this reflects the situation in a real-life clinical setting. Another limitation is that the absolute abundance of species was estimated using 16S rRNA qPCR, without correcting for copy number variations in the gene across species. Thus, our findings should be confirmed in future studies quantifying staphylococcal species abundance by qPCR targeting Staphylococcus-specific single-copy genes. However, the quantification of absolute abundance, and not solely proportional abundances, constitutes a significant strength of the study. Another major strength of the study is the use of tuf-based amplicon sequencing leading to reliable staphylococci classification at species level and, finally, the high number of included patients compared to previous studies in this field of research, which allows for stronger statistical power.
In conclusion, we found that increased abundance of S. aureus, S. capitis, and S. lugdunensis are positively correlated with increasing severity, whereas S. hominis abundance showed a negative correlation, and that S. hominis proportional abundance was reduced on AD skin compared to healthy skin. This indicates that multiple staphylococcal species, and not just S. aureus, have an important role in AD regarding chronic disease and exacerbations. Furthermore, our data suggest that the pathophysiological abnormalities in AD skin have a universal impact on the skin microbiota, that is independent of the presence of current eczema status.
Acknowledgments
We would like to thank technicians Elvira Chapka, Ditte Marie Brix, Emine Yüksel Coskun, and Christian Høiby Aumayr at Statens Serum Institut for laboratory assistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/2/432/s1, Supplemental Methodology, Table S1: Information on treatments, Figure S1: Taxonomic illustration of the 30 most abundant genera on healthy skin, Figure S2: Differential abundance analysis of bacteria; comparison between AD skin and healthy skin, Figure S3: Comparison of species composition within defined staphylococcal community clusters on AD skin, Figure S4: Correlations between absolute abundance of Staphylococcus aureus and bacterial alpha diversity, Figure S5: Bacterial absolute abundances on skin in relation to staphylococcal community cluster assignment, Figure S6: Comparison of staphylococcal species composition within lesional and non-lesional AD skin, Figure S7: Comparation of S. aureus and S. epidermis abundances between lesional and non-lesional skin, Figure S8: Correlations between SCORAD and absolute abundances of staphylococcal species, Figure S9: Correlations between the absolute abundance of S. aureus with pH and TEWL, Figure S10: Differential abundance analysis of staphylococcal species; comparisons between AD skin and healthy skin, Figure S11: Comparison of bacterial diversity and density on non-lesional AD skin collected at dry and moist skin areas.
Author Contributions
Conceptualization, S.M.E., M.S., and P.S.A.; funding acquisition, S.M.E.; investigation, T.A., L.B.N., C.M.O., M.-L.C., and S.M.E.; data curation, S.M.E., M.-L.C., L.B.N., C.M.O., and A.C.I.; project administration, S.M.E., M.-L.C., and C.M.O.; methodology, S.M.E., S.I., J.S.J., A.C.I., and B.L.; formal analysis, S.M.E.; validation, S.M.E.; writing—original draft preparation, S.M.E.; writing—review and editing, S.M.E., P.S.A., M.S., T.A., A.C.I., B.L., M.-L.C., L.B.N., C.M.O., J.S.J., and S.I.; visualization, S.M.E. and M.S.; supervision, P.S.A., T.A., M.S., M.-L.C., and S.M.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Augustinus Fonden, grant number 19-3201.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the local Ethics Committee (Videnskabsetisk Komite, Region-Hovedstaden, protocol code H-1-2014-039 (approval date: 8 July 2014) and H-16023435 (approval date: 6 June 2016)).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All sequences are available through the European Nucleotide Archive (project number: PRJEB42898). R scripts and phyloseq objects are available at: https://github.com/ssi-dk/AD_staphylome_project, accessed on 18 February 2021. Due to national data protection regulations regarding personally identifiable information, only a limited number of variables are included in the sample data file.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Leung D.Y., Guttman-Yassky E. Deciphering the complexities of atopic dermatitis: Shifting paradigms in treatment approaches. J. Allergy Clin. Immunol. 2014;134:769–779. doi: 10.1016/j.jaci.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabanillas B., Novak N. Atopic dermatitis and filaggrin. Curr. Opin. Immunol. 2016;42:1–8. doi: 10.1016/j.coi.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Totte J.E., van der Feltz W.T., Hennekam M., van Belkum A., van Zuuren E.J., Pasmans S.G. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: A systematic review and meta-analysis. Br. J. Dermatol. 2016;175:687–695. doi: 10.1111/bjd.14566. [DOI] [PubMed] [Google Scholar]
- 4.Kong H.H., Oh J., Deming C., Conlan S., Grice E.A., Beatson M.A., Nomicos E., Polley E.C., Komarow H.D., Murray P.R., et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clausen M.L., Agner T., Lilje B., Edslev S.M., Johannesen T.B., Andersen P.S. Association of Disease Severity with Skin Microbiome and Filaggrin Gene Mutations in Adult Atopic Dermatitis. JAMA Dermatol. 2018;154:293–300. doi: 10.1001/jamadermatol.2017.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakatsuji T., Chen T.H., Narala S., Chun K.A., Two A.M., Yun T., Shafiq F., Kotol P.F., Bouslimani A., Melnik A.V., et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grice E.A., Kong H.H., Conlan S., Deming C.B., Davis J., Young A.C., Bouffard G.G., Blakesley R.W., Murray P.R., Green E.D., et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ottman N., Barrientos-Somarribas M., Fyhrquist N., Alexander H., Wisgrill L., Olah P., Tsoka S., Greco D., Levi-Schaffer F., Soumelis V., et al. Microbial and transcriptional differences elucidate atopic dermatitis heterogeneity across skin sites. Allergy. 2020 doi: 10.1111/all.14606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callewaert C., Nakatsuji T., Knight R., Kosciolek T., Vrbanac A., Kotol P., Ardeleanu M., Hultsch T., Guttman-Yassky E., Bissonnette R., et al. IL-4Ralpha Blockade by Dupilumab Decreases Staphylococcus aureus Colonization and Increases Microbial Diversity in Atopic Dermatitis. J. Investig. Dermatol. 2019 doi: 10.1016/j.jid.2019.07.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi B., Bangayan N.J., Curd E., Taylor P.A., Gallo R.L., Leung D.Y.M., Li H. The skin microbiome is different in pediatric versus adult atopic dermatitis. J. Allergy Clin. Immunol. 2016;138:1233–1236. doi: 10.1016/j.jaci.2016.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandwein M., Fuks G., Israel A., Sabbah F., Hodak E., Szitenberg A., Harari M., Steinberg D., Bentwich Z., Shental N., et al. Skin Microbiome Compositional Changes in Atopic Dermatitis Patients Accompany Dead Sea Climatotherapy. Photochem. Photobiol. 2019 doi: 10.1111/php.13119. [DOI] [PubMed] [Google Scholar]
- 12.Jakasa I., Koster E.S., Calkoen F., McLean W.H., Campbell L.E., Bos J.D., Verberk M.M., Kezic S. Skin barrier function in healthy subjects and patients with atopic dermatitis in relation to filaggrin loss-of-function mutations. J. Investig. Dermatol. 2011;131:540–542. doi: 10.1038/jid.2010.307. [DOI] [PubMed] [Google Scholar]
- 13.Seidenari S., Giusti G. Objective assessment of the skin of children affected by atopic dermatitis: A study of pH, capacitance and TEWL in eczematous and clinically uninvolved skin. Acta Derm. Venereol. 1995;75:429–433. doi: 10.2340/0001555575429433. [DOI] [PubMed] [Google Scholar]
- 14.Kezic S., Kemperman P.M., Koster E.S., de Jongh C.M., Thio H.B., Campbell L.E., Irvine A.D., McLean W.H., Puppels G.J., Caspers P.J. Loss-of-function mutations in the filaggrin gene lead to reduced level of natural moisturizing factor in the stratum corneum. J. Investig. Dermatol. 2008;128:2117–2119. doi: 10.1038/jid.2008.29. [DOI] [PubMed] [Google Scholar]
- 15.van Smeden J., Bouwstra J.A. Stratum Corneum Lipids: Their Role for the Skin Barrier Function in Healthy Subjects and Atopic Dermatitis Patients. Curr. Probl. Dermatol. 2016;49:8–26. doi: 10.1159/000441540. [DOI] [PubMed] [Google Scholar]
- 16.Miajlovic H., Fallon P.G., Irvine A.D., Foster T.J. Effect of filaggrin breakdown products on growth of and protein expression by Staphylococcus aureus. J. Allergy Clin. Immunol. 2010;126:1184–1190. doi: 10.1016/j.jaci.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mempel M., Schmidt T., Weidinger S., Schnopp C., Foster T., Ring J., Abeck D. Role of Staphylococcus aureus surface-associated proteins in the attachment to cultured HaCaT keratinocytes in a new adhesion assay. J. Investig. Dermatol. 1998;111:452–456. doi: 10.1046/j.1523-1747.1998.00293.x. [DOI] [PubMed] [Google Scholar]
- 18.Clausen M.L., Edslev S.M., Andersen P.S., Clemmensen K., Krogfelt K.A., Agner T. Staphylococcus aureus colonization in atopic eczema and its association with filaggrin gene mutations. Br. J. Dermatol. 2017;177:1394–1400. doi: 10.1111/bjd.15470. [DOI] [PubMed] [Google Scholar]
- 19.Byrd A.L., Deming C., Cassidy S.K.B., Harrison O.J., Ng W.I., Conlan S., Belkaid Y., Segre J.A., Kong H.H. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen F.S., Simonsen L., Melgaard A., Wendicke K., Henriksen A.S. An efficient new formulation of fusidic acid and betamethasone 17-valerate (fucicort lipid cream) for treatment of clinically infected atopic dermatitis. Acta Derm. Venereol. 2007;87:62–68. doi: 10.2340/00015555-0174. [DOI] [PubMed] [Google Scholar]
- 21.Edslev S.M., Clausen M.L., Agner T., Stegger M., Andersen P.S. Genomic analysis reveals different mechanisms of fusidic acid resistance in Staphylococcus aureus from Danish atopic dermatitis patients. J. Antimicrob. Chemother. 2018;73:856–861. doi: 10.1093/jac/dkx481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harkins C.P., McAleer M.A., Bennett D., McHugh M., Fleury O.M., Pettigrew K.A., Oravcova K., Parkhill J., Proby C.M., Dawe R.S., et al. The widespread use of topical antimicrobials enriches for resistance in Staphylococcus aureus isolated from patients with atopic dermatitis. Br. J. Dermatol. 2018;179:951–958. doi: 10.1111/bjd.16722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russel J., Røder H.L., Madsen J.S., Burmølle M., Sørensen S.J. Antagonism correlates with metabolic similarity in diverse bacteria. Proc. Natl. Acad. Sci. USA. 2017;114:10684–10688. doi: 10.1073/pnas.1706016114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zipperer A., Konnerth M.C., Laux C., Berscheid A., Janek D., Weidenmaier C., Burian M., Schilling N.A., Slavetinsky C., Marschal M., et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016;535:511–516. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 25.O’Sullivan J.N., Rea M.C., O’Connor P.M., Hill C., Ross R.P. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 2019;95 doi: 10.1093/femsec/fiy241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Freire Bastos M.D.C., Miceli de Farias F., Carlin Fagundes P., Varella Coelho M.L. Staphylococcins: An update on antimicrobial peptides produced by staphylococci and their diverse potential applications. Appl. Microbiol. Biotechnol. 2020;104:10339–10368. doi: 10.1007/s00253-020-10946-9. [DOI] [PubMed] [Google Scholar]
- 27.Williams M.R., Costa S.K., Zaramela L.S., Khalil S., Todd D.A., Winter H.L., Sanford J.A., O’Neill A.M., Liggins M.C., Nakatsuji T., et al. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aat8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paharik A.E., Parlet C.P., Chung N., Todd D.A., Rodriguez E.I., Van Dyke M.J., Cech N.B., Horswill A.R. Coagulase-Negative Staphylococcal Strain Prevents Staphylococcus aureus Colonization and Skin Infection by Blocking Quorum Sensing. Cell Host Microbe. 2017;22:746–756. doi: 10.1016/j.chom.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canovas J., Baldry M., Bojer M.S., Andersen P.S., Grzeskowiak P.K., Stegger M., Damborg P., Olsen C.A., Ingmer H. Cross-Talk between Staphylococcus aureus and Other Staphylococcal Species via the agr Quorum Sensing System. Front. Microbiol. 2016;7:1733. doi: 10.3389/fmicb.2016.01733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baurecht H., Ruhlemann M.C., Rodriguez E., Thielking F., Harder I., Erkens A.S., Stolzl D., Ellinghaus E., Hotze M., Lieb W., et al. Epidermal lipid composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration. J. Allergy Clin. Immunol. 2018;141:1668–1676. doi: 10.1016/j.jaci.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 31.Soares J., Lopes C., Tavaria F., Delgado L., Pintado M. A diversity profile from the staphylococcal community on atopic dermatitis skin: A molecular approach. J. Appl. Microbiol. 2013;115:1411–1419. doi: 10.1111/jam.12296. [DOI] [PubMed] [Google Scholar]
- 32.Iversen S., Johannesen T.B., Ingham A.C., Edslev S.M., Tevell S., Månsson E., Nilsdotter-Augustinsson Å., Söderquist B., Stegger M., Andersen P.S. Alteration of Bacterial Communities in Anterior Nares and Skin Sites of Patients Undergoing Arthroplasty Surgery: Analysis by 16S rRNA and Staphylococcal-Specific tuf Gene Sequencing. Microorganisms. 2020;8:1977. doi: 10.3390/microorganisms8121977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meisel J.S., Hannigan G.D., Tyldsley A.S., SanMiguel A.J., Hodkinson B.P., Zheng Q., Grice E.A. Skin Microbiome Surveys Are Strongly Influenced by Experimental Design. J. Investig. Dermatol. 2016;136:947–956. doi: 10.1016/j.jid.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clausen M.L., Edslev S.M., Nørreslet L.B., Sørensen J.A., Andersen P.S., Agner T. Temporal variation of Staphylococcus aureus clonal complexes in atopic dermatitis: A follow-up study. Br. J. Dermatol. 2019;180:181–186. doi: 10.1111/bjd.17033. [DOI] [PubMed] [Google Scholar]
- 35.Williams H.C., Burney P.G., Hay R.J., Archer C.B., Shipley M.J., Hunter J.J., Bingham E.A., Finlay A.Y., Pembroke A.C., Graham-Brown R.A., et al. The, U.K. Working Party’s Diagnostic Criteria for Atopic Dermatitis. I. Derivation of a minimum set of discriminators for atopic dermatitis. Br. J. Dermatol. 1994;131:383–396. doi: 10.1111/j.1365-2133.1994.tb08530.x. [DOI] [PubMed] [Google Scholar]
- 36.Kunz B., Oranje A.P., Labreze L., Stalder J.F., Ring J., Taieb A. Clinical validation and guidelines for the SCORAD index: Consensus report of the European Task Force on Atopic Dermatitis. Dermatology. 1997;195:10–19. doi: 10.1159/000245677. [DOI] [PubMed] [Google Scholar]
- 37.Larsen A.R., Stegger M., Sorum M. spa typing directly from a mecA, spa and pvl multiplex PCR assay-a cost-effective improvement for methicillin-resistant Staphylococcus aureus surveillance. Clin. Microbiol. Infect. 2008;14:611–614. doi: 10.1111/j.1469-0691.2008.01995.x. [DOI] [PubMed] [Google Scholar]
- 38.Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glöckner F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Callahan B.J. Silva taxonomic training data formatted for DADA2 (Silva version 132) Zenodo. 2018 doi: 10.5281/zenodo.1172783. [DOI] [Google Scholar]
- 41.R_Core_Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2020. [(accessed on 18 February 2021)]. Available online: https://www.R-project.org/ [Google Scholar]
- 42.McMurdie P.J., Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; New York, NY, USA: 2016. [Google Scholar]
- 44.Davis N.M., Proctor D.M., Holmes S.P., Relman D.A., Callahan B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu C.M., Price L.B., Hungate B.A., Abraham A.G., Larsen L.A., Christensen K., Stegger M., Skov R., Andersen P.S. Staphylococcus aureus and the ecology of the nasal microbiome. Sci. Adv. 2015;1:e1400216. doi: 10.1126/sciadv.1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jian C., Luukkonen P., Yki-Järvinen H., Salonen A., Korpela K. Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PLoS ONE. 2020;15:e0227285. doi: 10.1371/journal.pone.0227285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O’Hara R.B., Simpson G.L., Solymos P., et al. Vegan: Community Ecology Package. R Package Version 2.5-6. [(accessed on 18 February 2021)];2019 Available online: https://CRAN.R-project.org/package=vegan.
- 48.Lin H., Peddada S.D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020;11:3514. doi: 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foster Z.S., Sharpton T.J., Grünwald N.J. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput. Biol. 2017;13:e1005404. doi: 10.1371/journal.pcbi.1005404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maechler M., Rousseeuw P., Struyf A., Hubert M., Hornik K. Cluster: Cluster Analysis Basics and Extensions. R Package Version 2.1.0. [(accessed on 18 February 2021)];2019 Available online: https://cran.r-project.org/web/packages/cluster/news.html.
- 51.Oh J., Byrd A.L., Deming C., Conlan S., Kong H.H., Segre J.A. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simpson E.L., Villarreal M., Jepson B., Rafaels N., David G., Hanifin J., Taylor P., Boguniewicz M., Yoshida T., De Benedetto A., et al. Patients with Atopic Dermatitis Colonized with Staphylococcus aureus Have a Distinct Phenotype and Endotype. J. Investig. Dermatol. 2018;138:2224–2233. doi: 10.1016/j.jid.2018.03.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lambers H., Piessens S., Bloem A., Pronk H., Finkel P. Natural skin surface pH is on average below 5, which is beneficial for its resident flora. Int. J. Cosmet. Sci. 2006;28:359–370. doi: 10.1111/j.1467-2494.2006.00344.x. [DOI] [PubMed] [Google Scholar]
- 54.Meylan P., Lang C., Mermoud S., Johannsen A., Norrenberg S., Hohl D., Vial Y., Prod’hom G., Greub G., Kypriotou M., et al. Skin Colonization by Staphylococcus aureus Precedes the Clinical Diagnosis of Atopic Dermatitis in Infancy. J. Investig. Dermatol. 2017;137:2497–2504. doi: 10.1016/j.jid.2017.07.834. [DOI] [PubMed] [Google Scholar]
- 55.Verstappen K.M., Willems E., Fluit A.C., Duim B., Martens M., Wagenaar J.A. Staphylococcus aureus Nasal Colonization Differs among Pig Lineages and Is Associated with the Presence of Other Staphylococcal Species. Front. Vet. Sci. 2017;4:97. doi: 10.3389/fvets.2017.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahle C.M., Stødkilde K., Afshar M., Poehlein A., Ogilvie L.A., Söderquist B., Hüpeden J., Brüggemann H. Staphylococcus saccharolyticus: An Overlooked Human Skin Colonizer. Microorganisms. 2020;8:1105. doi: 10.3390/microorganisms8081105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakatsuji T., Chen T.H., Two A.M., Chun K.A., Narala S., Geha R.S., Hata T.R., Gallo R.L. Staphylococcus aureus Exploits Epidermal Barrier Defects in Atopic Dermatitis to Trigger Cytokine Expression. J. Investig. Dermatol. 2016;136:2192–2200. doi: 10.1016/j.jid.2016.05.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong S.W., Choi E.B., Min T.K., Kim J.H., Kim M.H., Jeon S.G., Lee B.J., Gho Y.S., Jee Y.K., Pyun B.Y., et al. An important role of alpha-hemolysin in extracellular vesicles on the development of atopic dermatitis induced by Staphylococcus aureus. PLoS ONE. 2014;9:e100499. doi: 10.1371/journal.pone.0100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gonzalez M.E., Schaffer J.V., Orlow S.J., Gao Z., Li H., Alekseyenko A.V., Blaser M.J. Cutaneous microbiome effects of fluticasone propionate cream and adjunctive bleach baths in childhood atopic dermatitis. J. Am. Acad. Dermatol. 2016;75:481–493.e488. doi: 10.1016/j.jaad.2016.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwon S., Choi J.Y., Shin J.W., Huh C.H., Park K.C., Du M.H., Yoon S., Na J.I. Changes in Lesional and Non-lesional Skin Microbiome During Treatment of Atopic Dermatitis. Acta Derm. Venereol. 2019;99:284–290. doi: 10.2340/00015555-3089. [DOI] [PubMed] [Google Scholar]
- 61.Hibbing M.E., Fuqua C., Parsek M.R., Peterson S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji G., Beavis R.C., Novick R.P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jenul C., Horswill A.R. Regulation of Staphylococcus aureus Virulence. Microbiol. Spectr. 2019;7 doi: 10.1128/microbiolspec.GPP3-0031-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences are available through the European Nucleotide Archive (project number: PRJEB42898). R scripts and phyloseq objects are available at: https://github.com/ssi-dk/AD_staphylome_project, accessed on 18 February 2021. Due to national data protection regulations regarding personally identifiable information, only a limited number of variables are included in the sample data file.