Abstract
Functional toll-like receptors (TLRs) could modulate anti-tumor effects by activating inflammatory cytokines and the cytotoxic T-cells response. However, excessive TLR expression could promote tumor progression, since TLR-induced inflammation might stimulate cancer cells expansion into the microenvironment. Myd88 is involved in activation NF-κB through TLRs downstream signaling, hence in the current study we provided, for the first time, a complex characterization of expression of TLR2, TLR4, TLR7, TLR9, and MYD88 as well as their splicing forms in two distinct compartments of the microenvironment of chronic lymphocytic leukemia (CLL): peripheral blood and bone marrow. We found correlations between MYD88 and TLRs expressions in both compartments, indicating their relevant cooperation in CLL. The MYD88 expression was higher in CLL patients compared to healthy volunteers (HVs) (0.1780 vs. 0.128, p < 0.0001). The TLRs expression was aberrant in CLL compared to HVs. Analysis of survival curves revealed a shorter time to first treatment in the group of patients with low level of TLR4(3) expression compared to high level of TLR4(3) expression in bone marrow (13 months vs. 48 months, p = 0.0207). We suggest that TLRs expression is differentially regulated in CLL but is similarly shared between two distinct compartments of the microenvironment.
Keywords: CLL (chronic lymphocytic leukemia), TLRs (toll-like receptors), Myd88 (myeloid differentiation primary response protein 88)
1. Introduction
Chronic lymphocytic leukemia (CLL) is a disease with the accumulation of aberrant B cells in peripheral blood as well as proliferation and accumulation of CLL cells in the bone marrow and peripheral lymphoid organs. CLL patients are characterized by different prognoses, as well as profound molecular and immune defects [1,2,3]. Immune deregulations result in high susceptibility to infections as well as a failure to improve effective antitumor immune responses [4,5,6]. The nonspecific immune response could be continuously stimulated in CLL patients, although it has been suggested that the stimulation of toll-like receptors (TLRs) expressed on CLL cells could increase the immunogenicity of tumor cells and thus potentially contribute to the induction of tumor-specific immune response [7,8].
TLRs represent a family of transmembrane receptors that recognize a broad spectrum of pathogen-associated molecular patterns (PAMPs), such as highly conserved structures of viral (TLR7 or TLR9), bacterial (TLR2 or TLR4), and endogenous molecules released by injured tissues [8]. TLRs regulate innate immunity and determine the polarization and function of adaptive immunity mediated by B and T cells. TLRs expression is highly up-regulated through B-cell receptor (BCR) triggering of naive B-cells, indicating synergism between TLR and BCR leading to B-cell proliferation as well as differentiation [9]. All TLRs downstream signaling pathways except TLR3 are conducted by adaptor molecule Myd88 (myeloid differentiation primary response protein 88). It has been proven that TLRs types including 5, 7, 8, 9, 11 initiate the Myd88-dependent pathway directly. However, Myd88-dependent signaling pathway through TLRs types 1, 2, 4, 6 involves TIR—domain-containing adaptor protein (TIRAP). TLR3 and TLR4 initiate an alternative pathway which is MyD88-independent by recruiting TIR-domain-containing adaptor protein, inducing IFN-β (TRIF) [10]. Activation of MyD88 plays a crucial role in inflammatory cytokine secretion. The MyD88-dependent signaling pathway leads to the early phase of nuclear factor-κB (NF-κB) activation, whereas the MyD88-independent signaling pathway initiates the late phase of NF-κB activation. TLR-dependent signals could be also involved in the regulation of B lymphocytes function by inducing TLR tolerance or auto-reactivity promotion [11]. TLR7 and TLR9 engaged together with BCR also participate in the response to auto-antigens [12]. Recent studies indicated that both auto- and exogenous antigens might be engaged in the initiation and progression of CLL. The majority of data emphasized the role of adaptive immune receptors, including BCR, in the pathogenesis and progression of the disease [13,14,15].
The consequence of the occurrence of the MYD88 mutation includes chronic activation of TLRs, thus the constitutive activation of the NF-κB promotes cell proliferation and survival [16]. Recent studies confirmed the prognostic value of MYD88 mutation in CLL [17]. Since the MYD88 mutation is harboring in 1–10% CLL patients, the current study aimed to characterize the association between expression and mutational status of MYD88 and TLRs expression in the bone marrow and peripheral blood in CLL.
Splicing variants of TLRs might have different abilities to induce signal transduction since alternative splicing produces various transcripts and, as a result, various proteins from a single gene [18]. The current study aimed to present an expression pattern of TLR2, TLR7, TLR9, and splicing variants expression of TLR4 (TLR4(1), TLR4(3), TLR4(4)) on the mRNA level as well as perform a comparison in two different microenvironmental compartments, peripheral blood and bone marrow, referring to recognized prognostic markers as well as clinical outcome.
2. Materials and Methods
2.1. Study Subjects
The material was obtained from 94 untreated CLL patients diagnosed at the Department of Hematooncology and Bone Marrow Transplantation, the Medical University of Lublin (60 males, 34 females, mean age 65). In the current study, we included patients with CLL diagnosed by aberrant immunophenotype, including CD5+ CD19+. The median value of CD19+ CD5+ expression analyzed on peripheral blood mononuclear cells (PBMC) was always >90%.
Thirty-three patients were classified in stage A, thirty-four in stages B, and eleven in stages C, according to the Binet classification. Detailed characteristics of CLL patients are shown in Table 1. This material was also obtained from 27 healthy volunteers. This study was approved by the Local Ethics Committee (KE-0254/7/2019), and the patients were informed about the use of their blood for scientific purposes.
Table 1.
Clinical characteristics of chronic lymphocytic leukemia (CLL) patients.
| Characteristic | Number of Patients |
|---|---|
| Sex | |
| Female | 34 (36.2%) |
| Male | 60 (63.8%) |
| Binet’s classification | |
| A | 38 (40.43%) |
| B | 39 (41.49%) |
| C | 14 (14.89%) |
| ZAP-70 (cut off 20%) | |
| Positive | 28 (29.8%) |
| Negative | 58 (61.7%) |
| NA (not available) | 8 (8.5%) |
| CD38 (cut off 30%) | |
| Positive | 18 (19.2%) |
| Negative | 68 (8.5%) |
| NA (not available) | 8 (72.3%) |
| Mutational status of the immunoglobulin heavy-chain variable-region (IGHV) | |
| Mutated | 28(29.8%) |
| Unmutated | 41 (43.6%) |
| NA (not available) | 25 (26.6%) |
2.2. Isolation of Mononuclear Cells
Mononuclear cells from peripheral blood (PBMC) and bone marrow (BMMC) were isolated by Ficoll (Biochrom AG, Berlin, Germany) density gradient. Next, cells were washed twice in phosphate-buffered saline (Biochrom AG, Berlin, Germany) and counted. The viability of obtained PBMC and BMMC was always >95%, as determined by trypan blue exclusion (Sigma-Aldrich, Schnelldorf, Germany). The viable cells were quantified in a Neubauer chamber (Zeiss, Jena, Germany) and stored for RNA preparation at −192 °C in liquid nitrogen.
2.3. RNA Isolation and Reverse Transcription
For the isolation of mRNA from PBMC, the QIAamp RNA Blood Mini Kit (Qiagen, Venlo, The Netherlands) was used according to the manufacturer’s instructions. One µg of mRNA was reverse transcribed into 20 μL of cDNA using a QuantiTect Reverse Transcription Kit (Qiagen, Venlo, The Netherlands). For each RT-PCR, 1 μL of the cDNA preparation was used.
2.4. Quantitative Polymerase Chain Reaction (q-PCR)
For the quantitative measurement of the mRNA expression of TLR2, TLR4, TLR7 and TLR9, real-time q-PCR was performed using the Light Cycler SYBR Green I technology according to the manufacturer’s protocol (Roche Diagnostics, Rotkreuz, Switzerland). The sequences of the primers for q-PCR are shown in Table 2 (Roche Diagnostics, Rotkreuz, Switzerland).
Table 2.
Primer sequences for the examined toll-like receptors (TLRs).
| Type of TLR (Splicing Variants) |
Sequences of Primers |
|---|---|
| TLR2 | F: 5′ CAA GCA GGA TCC AAA GGA GA 3′ R: 5′ ACC CAC ACC ATC CAC AAA GT 3′ |
| TLR4 (1) | F: 5′GGC TCG AGG AAG AGA AGA CA 3′ R: 5′ATT AGG AAC CAC CTC CAC GC 3′ |
| TLR4 (3) | F: 5′CTG TGG GGC GGC TCG AGG AA 3′ R: 5′GCC AAG TCT CCA CGC AGG GCT 3′ |
| TLR4 (4) | F: 5′CGG TGA TAG CGA GCC ACG CA 3′ R: 5′GGA TTT CAC ACC TCC ACG CAG GG 3′ |
| TLR7 | F: 5′ ATC ACT CCA TGC CAT CAA GA 3′ R: 5′ CCC CAA GGA GTT TGG AAA TTA 3′ |
| TLR9 | F: 5′ GCC AGA CCC TCT GGA GAA 3′ R: 5′ AGA CTT CAG GAA CAG CCA GTT G 3′ |
F, forward, R, reverse.
The glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) was used as a housekeeping gene. An initial denaturation step at 95 °C for 10 min was followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. The qPCR reactions were carried out using 7300 Real Time PCR System (Applied Biosystems, Foster City, CA, USA).
The TLRs mRNA expression was calculated as an inverse ratio of the difference in cycle threshold (ΔCt), where ΔCt is the Ct value of the target gene minus Ct value of GAPDH.
For the quantitative measurements of the mRNA expression of MYD88, qPCR was performed using TaqMan Universal PCR Master Mix and TaqMan Gene Expression Assay primer/probe mixes (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. GAPDH was used as a reference gene. Thermocycling program was set for 40 cycles of 15 s at 95 °C and 1 min at 60 °C on the 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The MYD88 mRNA expression was calculated as an inverse ratio of the difference in cycle threshold (ΔCt), where ΔCt is the Ct value of the target gene minus Ct value of GAPDH.
2.5. Detection of the MYD88 L265P Mutation by the Allele Specific-Polymerase Chain Reaction (AS-PCR)
The MYD88 L265P mutation was analyzed in 61 CLL patients in blood samples using the three primers: the mutant-specific reverse primer—5′-CCT TGT ACT TGA TGG GGA ACG-3′, the wild-type-specific reverse primer—5′-GCC TTG TAC TTG ATG GGG AAC A-3′, and the common forward primer—5′-AAT GTG TGC CAG GGG TAC TTA G-3′. Two reverse primers were designed to differentiate the mutant and wild-type allele of MYD88 L265P before the PCR reaction DNA isolation was performed using QIAamp DNA Blood Mini Kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s instructions. PCR was performed in a total reaction volume of 25 μL with 10 pmol of each primer and 100 ng DNA using QIAGEN Multiplex PCR Kit (Qiagen, Hilden, Germany). Thermal cycling conditions were: 2 min of preheating at 94 °C followed by 40 cycles of 94 °C for 30 s, 57 °C for 30 s, and 68 °C for 30 s, with a final extension at 68 °C for 5 min. The PCR products (159 bp) were separated on 2% agarose gel electrophoresis and visualized under UV light.
2.6. Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5. All results are presented as median values with rage. The U Mann–Whitney test and Kruskal–Wallis test was used to evaluate the difference between subgroups of patients. The correlations of variables were computed with Spearman’s rank correlation coefficient. Survival curves were calculated for time to first treatment (TTFT) of CLL patients according to the Kaplan–Meier method using a log-rank test. TTFT was calculated in months since the date of initial diagnosis until the date of initial treatment. For the subgroup of patients who had never received any treatment, we characterized TTFT as the date of the last follow up.
3. Results
3.1. Aberrant Expression of TLR2, TLR7, TLR9 and Splicing Variants of TLR4 in CLL Patients Compared to Healthy Volunteers
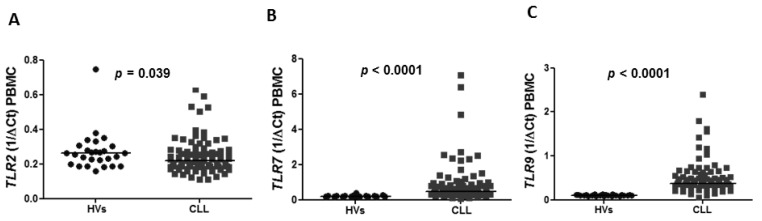
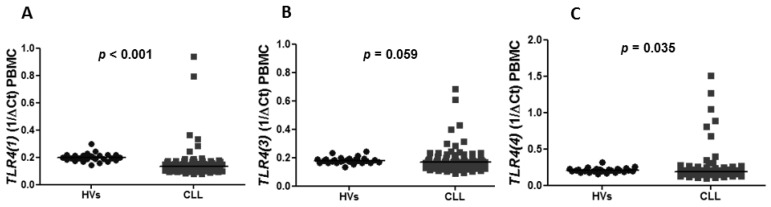
The expression of TLR2, TLR7, TLR9 and splicing variants of TLR4 was confirmed in PBMC in CLL patients as well as in healthy volunteers (HVs). The expression of TLR2 in peripheral blood was found to be lower in CLL patients compared to HVs with a median 0.2185 vs. 0.2632 (p = 0.039) (Figure 1A). Similarly, the expression of splicing variants of TLR(4), including TLR4(1) and TLR4(4), was significantly lower in CLL patients than in HVs (median of TLR4(1): 0.1330 vs. 0.1970, p < 0.0001 and median of TLR4(4): 0.1840 vs. 0.2066, p = 0.0353) (Figure 2A,C). There was no difference in the expression of TLR4(3) in CLL patients and HVs (0.1680 vs. 0.1775, p = 0.0592) (Figure 2B). The expression of TLR7 and TLR9 was significantly higher in CLL patients compared to HVs (0.4790 vs. 0.1877, p < 0.0001), (0.3735 vs. 0.1066, p < 0.0001), respectively (Figure 1B,C).
Figure 1.
Expression of TLR2, TLR7, TLR9 in CLL patients compared to healthy volunteers (HVs) in peripheral blood mononuclear cells (PBMC). (A) Lower expression of TLR2 in PBMC in CLL patients compared to healthy volunteers (HVs) (0.2185 vs. 0.2632 (p = 0.039). (B) Higher expression of TLR7 in PBMC in CLL patients compared to HVs (0.4790 vs. 0.1877, p < 0.0001). (C) Expression of TLR9 in blood samples of CLL patients was significantly higher compared to HVs (0.3735 vs. 0.1066, p < 0.0001). Each dot (HV) or square (CLL) represents one individual case.
Figure 2.
TLR4 splice variants expression in CLL patients compared to healthy volunteers (HVs) in peripheral blood mononuclear cells (PBMC). (A) Lower expression of TLR4(1) in CLL patients than in HVs in PBMC (0.1330 vs. 0.1970, p < 0.0001). (B) No difference in expression of TLR4(3) in HV and CLL patients (0.1775 vs. 0.1680, p = 0.0592). (C) Lower expression of TLR4(4) in CLL patients compared to HVs (0.1840 vs. 0.2066, p = 0.0353). Each dot (HV) or square (CLL) represents one individual case.
3.2. Expression of TLR2, TLR7, TLR9 and Splicing Variants of TLR4 in Peripheral Blood and Bone Marrow Compartments in CLL
To analyze a difference in the regulation of TLRs expression in biologically various compartments, we performed a comparison of the levels of expression of TLRs in peripheral blood and bone marrow mononuclear cells. There was no difference in TLR2, TLR7, and TLR9 expression in PBMC and BMMC with the median expression 0.2185 vs. 0.2 (p = 0.178); 0.479 vs. 0.4665 (p = 0.7215); 0.3735 vs. 0.368 (p = 0.8333), respectively. There was also no difference in expression of splicing variants of TLR4 including TLR4(1), TLR4(3), TLR4(4) in PBMC and BMMC (0.1330 vs. 0.1270, p = 0.8117), (0.1680 vs. 0.1520, p = 0.0952), (0.184 vs. 0.167, p = 0.0952).
3.3. Prognostic Value of the TLR2 Expression in Peripheral Blood and Bone Marrow Compartments in CLL
To characterize the significance of TLRs in CLL in a prognostic context we characterized the impact of TLRs expression on the clinical outcome and association with the recognized prognostic markers in both compartments, including PBMC and BMMC.
We observed no difference in TTFT in subgroups of patients with a high and low level of the TLR2 expression in PBMC (p = 0.7229) as well as in BMMC (p = 0.4523).
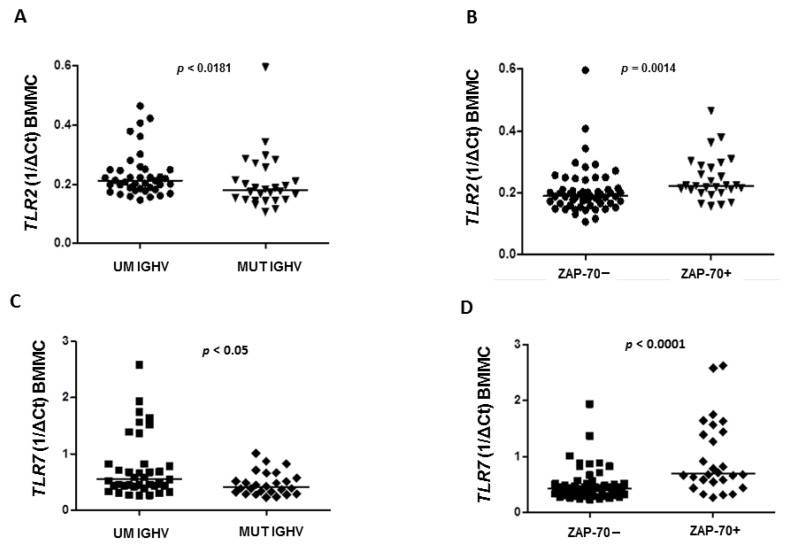
We found that the expression of TLR2 in BMMC was significantly higher in CLL patients with the unmutated status of the immunoglobulin heavy chain variable (UM IGHV) genes than in patients with IGHV mutation (MUT) with a median 0.212 vs. 0.1795 (p = 0.0181), respectively (Figure 3A).
Figure 3.
TLR2 and TLR7 expression in prognostically different CLL subgroups in refers to expression of ZAP-70 and mutational status of immunoglobulin heavy chain variable (IGHV) in bone marrow mononuclear cells (BMMC). (A) Higher expression of TLR2 in CLL patients with unmutated (UM) IGHV than in patients with IGHV mutation (MUT) (0.212 vs. 0.1795, p < 0.05). Each dot (UM IGHV) and triangle (MUT IGHV) represents an individual case. (B) Higher expression of TLR2 in ZAP-70+ patients compared to ZAP-70− (0.2225 vs. 0.1885, p = 0.0014). Each dot (ZAP-70+) and triangle (ZAP-70-) represents an individual case. (C) Higher expression of TLR7 in IGHV UM patients compared to IGHV MUT (0.549 vs. 0.407 p < 0.0361). Each square (UM IGHV) and rhombus (MUT IGHV) represents an individual case. (D) Higher expression of TLR7 in ZAP-70+ patients compared to ZAP-70− (0.694 vs. 0.424, p < 0.0001). Each square (ZAP-70-) and rhombus (ZAP-70+) represents an individual case.
The expression of TLR2 in BMMC was confirmed to be higher in ZAP-70+ (defined as cytoplasmatic expression >20% CLL cells) patients compared to ZAP-70−, with a median 0.2225 vs. 0.1885 (p = 0.0014), respectively (Figure 3B). However, in PBMC there were no differences in the expression of TLR2 depending on ZAP-70 expression (median expression 0.215 vs. 0.222, p = 0.8538) as well as the mutational status of IGHV genes (median expression 0.225 vs. 0.204, p = 0.8690).
We found that there was no difference in TLR2 expression in PBMC (median expression 0.212 vs. 0.2185, p = 0.5737) and BMMC (median expression 0.2055 vs. 0.2, p = 0.9788) in the groups of patients defined by CD38 expression (cut-off value for the positive expression = 30%).
Additionally, there were no statistical differences in TLR2 expression in stages A, B, and C according to Binet’s classification (Table S1). We did not find any correlation between expression of TLR2 in PBMC and BMMC and CLL patient’s age (r = −0.04755, p = 0.6582), (r = 0.05174, p = 0.6301), respectively.
3.4. Prognostic Value of the TLR7 Expression in Peripheral Blood and Bone Marrow Compartments in CLL
Analysis of survival curves found no difference in TTFT in subgroups of patients with a high and low level of the TLR7 expression in PBMC (p = 0.6819) as well as in BBMC (p = 0.5472).
In the groups of CLL patients categorized by the mutational status of IGHV genes and ZAP-70 expression, we found that in PBMC there were no differences in TLR7 expression in IGHV MUT and IGHV UM CLL patients (0.4805 vs. 0.583, p > 0.05) as well as in ZAP-70− and ZAP-70+ CLL patients (0.4935 vs. 0.532, p = 0.3738). Interestingly, in BMMC we found that expression of TLR7 was significantly higher in IGHV UM patients compared to IGHV MUT (0.549 vs. 0.407 vs., p < 0.0361) (Figure 3C) as well as in ZAP-70+ patients compared to ZAP-70− (0.694 vs. 0.424 vs., p < 0.0001) (Figure 3D).
There was no difference in TLR7 expression in CD-38+ and CD-38− patients in both peripheral blood and bone marrow compartments (0.668 vs. 0.4805, p = 4515 and 0.6895 vs. 0.4435, p = 0.0647), respectively. No association was observed in TLR7 expression in peripheral blood and bone marrow with the clinical stage of disease according to Binet’s classification (Table S1).
We did not find a correlation between the expression of TLR7 in peripheral blood and bone marrow samples and CLL patient’s age (r = 0.1755, p = 0.1, r = 0.03711, p = 0.7299), respectively.
3.5. Prognostic Value of the TLR9 Expression in Peripheral Blood and Bone Marrow Compartments in CLL
We showed the tendency of shorter TTFT in groups of CLL patients with high TLR9 expression in comparison to low TLR9 expression in BBMC (12 vs. 45, p = 0.0655). In PBMC there was no difference in TTFT, referring to the level of TLR9 expression (p = 0.3210).
There was no difference in the expression of TLR9 in peripheral blood as well as in bone marrow in IGHV MUT CLL patients mutated compared to IGHV UM (0.4345 vs. 0.367, p = 0.2104; median expression 0.3375 vs. 0.353, p = 0.6380, respectively).
There was no difference in TLR9 expression between groups of patients characterized by the expression of ZAP-70 both in peripheral blood (0.3524 vs. 0.4190, p = 0.2327) and bone marrow samples (0.3375 vs. 0.407, p = 0.1038), respectively. In PBMC we found that expression of TLR9 was significantly higher in CD-38− compared to CD-38+ patients (0.4005 vs. 0.295, p = 0.0234). In contrast, in bone marrow samples we did not find a statistically significant difference in TLR9 expression between CD-38+ and CD-38− patients (0.4025 vs. 0.3583, p = 0.6519).
No differences were observed in TLR9 expression in peripheral blood between stages A, B, and C according to Binet’s classification in PBMC and BMMC (Table S1). We did not find any correlation between the expression of TLR9 in peripheral blood and bone marrow samples and CLL patient‘s age (r = −0.01817, p = 0.8658: r = 0.07727, p = 0.4717), respectively.
3.6. Prognostic Value of the Expression of Splicing Variants of TLR4 (TLR4(1), TLR4(3), TLR4(4)) in Peripheral Blood and Bone Marrow Compartments in CLL
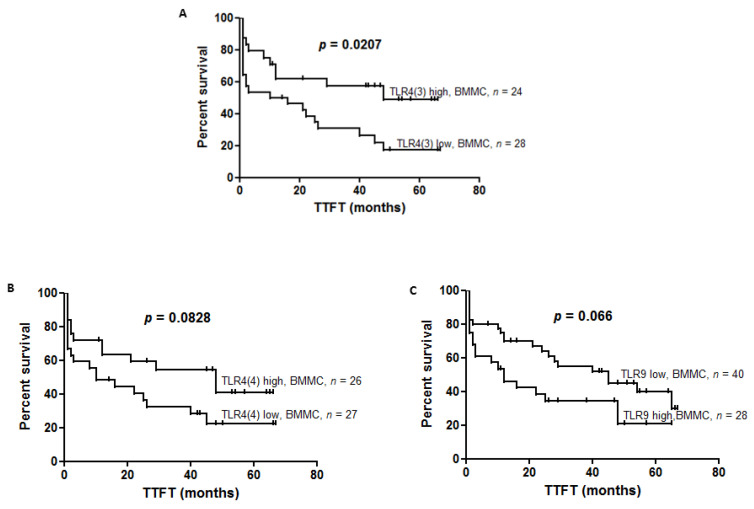
To define the prognostic value of TLR(4) splicing variants, we divided TTFT according to the level of the TLR4(1), TLR4(3), TLR4(4) expression in PBMC and BMMC. Analysis of survival curves found that TTFT was not different when referring to the level of TLR4(1) expression in PBMC (p = 0.5674) as well as BMMC (p = 0.3008). We found shorter TTFT in patients with low expression of TLR4(3) compared to patients with high expression of TLR4(3) in BBMC (13 vs. 48, p = 0.0207) (Figure 4A). A tendency to have shorter TTFT was found in patients with low expression of TLR4(4) compared to high expression in BBMC (10 vs. 48, p = 0.0828) (Figure 4B). However, in PBMC we did not observe changes in TTFT regarding TLR4(3) (p = 0.01761) and TLR4(4) expression.
Figure 4.
Time to first treatment (FFTF) in CLL patients divided according to the level of the TLRs expression in peripheral blood mononuclear cells (PBMC) and bone marrow mononuclear cells (BMMC). (A) TTFT was significantly shorter in the group of patients with a low level of TLR4(3) expression compared to the group of patients with a high level of TLR4(3) expression in BMMC (13 vs. 48, p = 0.0207). (B) TTFT tended to be shorter in the group of patients with a low level of TLR4(4) expression compared to the group of patients with a high level of TLR4(4) expression in BMMC (10 vs. 48, p = 0.0828). (C) TTFT tended to be shorter in the group of patients with a high level of TLR9 expression compared to the group of patients with a low level of TLR9 expression in BMMC (12 vs. 45, p = 0.0655).
We analyzed the expression of splicing variants of TLR4 (TLR4(1), TLR4(3), TLR4(4)) with respect to the mutational status of IGHV genes. There were no differences in expression of TLR4(1) in IGHV UM CLL cases compared to IGHV MUT (0.124 in vs. 0.141, p = 0.1316) in PBMC as well as in BMMC (0.1265 vs. 0.131, p = 0.493). We found that expression of TLR4(3) and TLR(4) in PBMC were lower in patients with IGHV UM compared to IGHV MUT (0.158 vs. 0.1815, p = 0.0233) (0.1695 vs. 0.2015 vs, p = 0.00642), respectively. The expression was found to be no different in BBMC in patients with IGHV MUT compared to IGHV UM (0.1585 vs. 0.137, p = 0.3188).
The expressions of TLR4(1), TLR4(3) and TLR4(4) were analyzed in two groups of CLL patients: ZAP-70+ and ZAP-70−. The expression of TLR4(1) in PBMC was found to be lower in ZAP-70+ patients than in ZAP-70+ (median expression 0.122 vs. 0.139, p = 0.0305). In bone marrow samples we also did not find differences in expression of TLR4(1) in ZAP-70+ and ZAP-70− CLL patients (0.157 vs. 0.152, p = 0.7583). We evaluated the expression of TLR4(3) in those groups of CLL patients. We observed that there was no difference in expression of TLR4(3) in PBMC and BMMC in ZAP-70− and ZAP-70+ patients (0.172 vs. 0.159, p = 0.0541), (0.152 vs. 0.157, p = 0.7583), respectively. The expression of TLR4(4) was similar in ZAP-70− and ZAP-70+ patients (0.1965 vs. 0.173, p = 0.2039) in PBMC as well as in BMMC (0.17 vs. 0.168, p = 0.804).
We determined the expression of splicing variants of TLR4 (TLR4(1), TLR4(3), TLR4(4)) in CD38+ and CD38− CLL patients. There were no differences in expression of TLR4(1) (0.126 vs. 0.137, p = 0.3911), TLR4(3) (0.16 vs. 0.171, p = 0.3079) and TLR4(4) (0.192 vs. 0.184, p = 0.5018) in CD-38+ and CD-38− groups in PBMC. In bone marrow, differences in expression of TLR4(1) (0.121 vs.0.13 p = 0.4011), TLR4(3) (0.156 vs. 0.152, p = 0.6144), TLR4(4) (0.176 vs. 0.168, p = 0.679) in CD38+ and CD38− CLL patients were not also observed.
We did not find any difference in expression of splicing variants of TLR4 in stages A, B, and C according to Binet’s classification in PBMC and BMMC (Table S1). We did not find correlation between expression of splicing variants of TLR4 in peripheral blood (TLR4(1): r = −0.0705, p = 0.5188, TLR4(3): r = −0.1203, p = 0.2698, TLR4(4): r = 0.8704) and bone marrow samples TLR4(1):r = −0.1613, p = 0.1378, TLR4(3): r = −0.04505, p = 0.6804, TLR4(4): r = −0.02286, p = 0.8345) and CLL patient’s age.
3.7. Prognostic Value of the MYD88 Expression and the Association with TLRs Expression in CLL
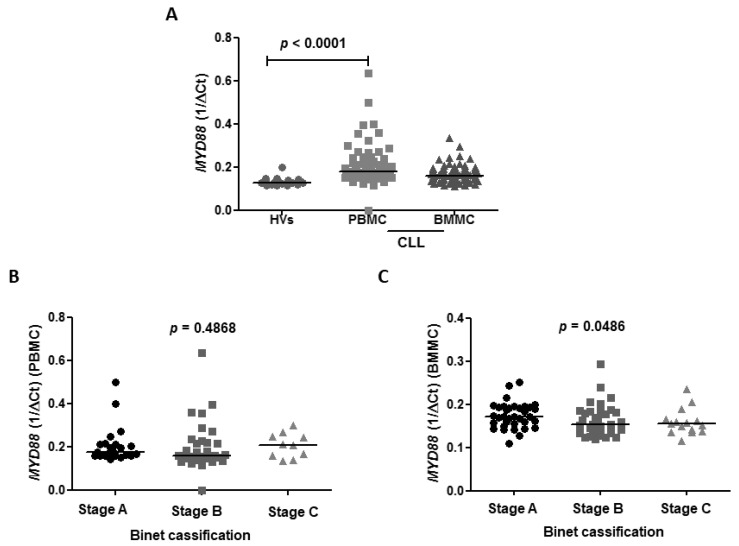
The MYD88 expression was higher in CLL patients compared to HVs with a median of 0.1780 vs. 0.128 (p < 0.0001), respectively. The median expression of MYD88 in BMMC was 0.1600 (Figure 5A). We revealed no differences in TTFT in subgroups of patients with high and low expression of MYD88 in bone marrow (10 vs. 26, p = 0.92) as well as in peripheral blood (11 vs. 21, p = 0.59).
Figure 5.
Expression of MYD88 in peripheral blood mononuclear cells (PBMC) and bone marrow mononuclear cells (BMMC) of CLL compared to healthy volunteers (HVs) and association of MYD88 expression with clinical stage of CLL according to Binet’s scale. (A) Higher expression of MYD88 in CLL vs. HVs in PBMC (0.1780 vs. 0.1280, p < 0.0001). The median expression of MYD88 in BMMC, p = 0.1600. Each dot (HV), square (PBMC) and triangle (BMMC) represent an individual case. (B) No association of expression of MYD88 with clinical stage of CLL according to Binet’s scale in PBMC with the median expression in A, B, C Binet stages: 0.1765 vs. 0.1605 vs. 0.2070, p = 0.4868, respectively. Each dot (Stage A), square (Stage B) and triangle (Stage C) represents an individual case. (C) Higher expression of MYD88 in A Binet stage compared to B and C Binet stage in BMMC (0.1720 vs. 0.1535 vs. 0.1550, p < 0.0001). Each dot (Stage A), square (Stage B) and triangle (Stage C) represents an individual case.
To identify association of MYD88 expression and TLRs expression, we assessed correlations in blood and bone marrow samples (Table 3).
Table 3.
Correlations between TLR and MYD88 expression in peripheral blood mononuclear cells (PBMC) and bone marrow mononuclear cells (BMMC) of CLL patients.
| PBMC | BMMC | |||||
|---|---|---|---|---|---|---|
| 95% CI | r | p | 95% CI | r | p | |
|
TLR2
MYD88 |
0.112–4.919 0.000–0.634 |
0.722 | <0.0001 | 0.106–0.612 0.110–0.335 |
0.388 | 0.0001 |
|
TLR4(1)
MYD88 |
0.078–0.332 0.115–0.634 |
0.599 | <0.001 | 0.080–0.341 0.110–0.335 |
0.284 | 0.007 |
|
TLR4(3)
MYD88 |
0.095–0.685 0.115–0.634 |
0.558 | <0.001 | 0.078–0.471 0.110–0.335 |
0.299 | 0.005 |
|
TLR4(4)
MYD88 |
0.104–1.271 0.115–0.634 |
0.560 | <0.001 | 0.063–0.654 0.110–0.335 |
0.440 | <0.001 |
|
TLR7
MYD88 |
0.066–6.382 0.000–0.398 |
0.580 | <0.001 | 0.137–11.641 0.110–0.335 |
0.358 | <0.001 |
|
TLR9
MYD88 |
0.096–2.392 0.000–0.497 |
0.492 | <0.001 | 0.166–7.692 0.110–0.294 |
0.541 | <0.001 |
We showed a strong correlation between MYD88 expression and TLR2 in PBMC (r = 0.722, p ≤ 0.0001) and medium correlation in BMMC (r = 0.389, p = 0.0001) (Figure S1A,B). There were medium correlations between expression of MYD88 and expression of TLR4 splicing variants including TLR4(1) (r = 0.559, p < 0.001), TLR(3) (r = 0.558, p < 0.001), and TLR4(4) (r = 0.56, p < 0.001) in PBMC (Figure S2A,C,E). In BMMC there were weak associations between expression of MYD88 and expression of TLR4(1) (r = 0.284, p < 0.007), TLR(3) (r = 0.294, p < 0.005), TLR4(4) (r = 0.44, p < 0.001) (Figure S2B,D,F). We observed medium correlation between MYD88 expression and TLR7 expression in PBMC (r = 0.580, p < 0.001) and medium correlation in BMMC (r = 0.358, p < 0.001) (Figure S3A,B) There were medium correlations between MYD88 expression and TLR9 expression in PBMC (r = 0.492, p < 0.001) as well as in BMMC (r = 0.541, p < 0.001) (Figure S4A,B).
To determine the significance of MYD88 in a different compartment we performed statistical analyses referring to CLL subgroups of patients with different prognoses in blood as well as bone marrow samples. We observed no association of expression of MYD88 with clinical stage of CLL according to Binet’s scale in PBMC with the median expression in A, B, C Binet’s stage: 0.1765 vs. 0.1605 vs. 0.2070, p = 0.4868, respectively (Figure 5B). However, BMMC expression of MYD88 was found to be higher in A Binet stage compared to B and C Binet stages in BMMC (0.1720 vs. 0.1535 vs. 0.1550, p < 0.0001) (Figure 5C).
No correlation between MYD88 expression in blood and bone marrow samples in CLL patients (r = 0.04, p = 0.715) was found.
We analyzed if the expression of MYD88 in blood and bone marrow samples in CLL patients depends on the prognostic markers including the mutational status of IGHV genes, ZAP-70 expression, CD38 expression, and Binet stage. We observed no differences in the expression of MYD88 in IGHV MUT patients compared to IGHV UM in PBMC with a median 0.1831 vs. 0.1926 (p = 0.4934), respectively, as well as in BMMC with a median 0.1603 vs. 0.1605 (p = 0.8730), respectively. There were no differences in the expression of MYD88 in ZAP-70+ CLL patients compared to ZAP-70− with a median 0.1734 vs. 0.1816 (p = 0.6426) in PBMC and BMMC with a median 0.1646 vs. 0.1601 (p = 0.9589). There were also no differences in the expression of MYD88 in CD38+ CLL patients compared to CD38− in PBMC with a median 0.2112 vs. 0.1778 (p = 0.867), respectively, as well as in BMMC with a median 0.1553 vs.0.1636 (p = 0.1855), respectively.
There were no correlations between MYD88 expression and the age of CLL patients in peripheral blood (p = 0.7874) and bone marrow (p = 0.3189) samples. We did not find any associations between MYD88 expression and morphological parameters, including the level of white blood cells (WBC) (p = 0.7873), red blood cells (RBC) (p = 0.537), platelets (PLT) (p = 0.2014), neutrophils (p = 0.981), and the level of hemoglobin (Hgb) (p = 0.570) and hematocrit (Hct) (p = 0.5017) in blood samples. There were no correlations between MYD88 expression and morphological parameters, including the level of white blood cells (WBC) (p = 0.9314), red blood cells (RBC) (p = 0.1792), platelets (PLT) (p = 0.2209), neutrophils (0.1007), and the level of hemoglobin (Hgb) (p = 0.3833) and hematocrit (Hct) (p = 0.3389) in bone marrow.
3.8. MYD88 L265P Mutation in CLL Patients
MYD88 L265P mutation occurred in 2/61 (3.28%) CLL patients in PBMC. Since a small number of the cohort had a MYD88 mutation, we did not aim to obtain median and statistical data. We obtained mean values. In patients with MYD88 L265P mutation, the mean expression of splicing variants of TLR4 (1/ΔCt) was as follows: TLR4(1)—0.165, TLR4(3)—0.153, TLR4(4)—0.138. In the group of patients without MYD88 L265P mutation, the median expression of splicing variants of TLR4 (1/ΔCt) was 0.138 for TLR4(1), 0.171 for TLR4(3), and 0.184 for TLR4(4). In the population of MYD88-mutated patients, the mean expression of TLR2 (1/ΔCt) was 0.203, meanwhile patients without MYD88 mutation had a median of 0.225. The mean expression of TLR7 (1/ΔCt) in patients with MYD88 L265P mutation was 0.866, and in the case of patients without MYD88 L265P mutation, the median expression of TLR7 (1/ΔCt) was 0.573. In patients harboring the MYD88 L265P mutation, the mean expression of TLR9 (1/ΔCt) was 0.517, meanwhile patients without MYD88 L265P mutation had a median of 0.405.
4. Discussion
In tumors, functional TLRs expression could influence preferable anti-tumor effects by activating inflammatory cytokine expression and cytotoxic T-cells response. However, an excessive TLRs activation could promote tumor progression, since TLR-induced inflammation stimulates cancer cells boost in the microenvironment [16]. Several reports demonstrated changes of immunological parameters between accumulation and proliferation compartments of the microenvironment in CLL [19,20]. There are some studies that suggest differential signaling trough BCR in an accumulative and proliferative compartment in CLL microenvironment and provide their differential involvement in biology and pathogenesis in this disease [19,21]. It was found that especially in the lymph node microenvironment of CLL, both BCR and TLR signaling could contribute to NF-κB activation, which is essential in immune response regulation, oncogenesis, and tumor progression [21]. So far there are many questions about the expression pattern of TLRs and their prognostic significance in CLL, especially in the bone marrow microenvironment [12,22,23].
Results of our work showed aberrant expression of TLRs in CLL patients compared to HVs, proving that TLRs expression is differentially regulated in CLL. We showed higher expression of TLR7 and TLR9 in CLL compared to HVs, which was confirmed in earlier reports [24,25]. Arvaniti et al. [11] demonstrated high expression of TLR7, while the expression of TLR9 was showed to be low on a mRNA level. Other studies showed variable levels of mRNA and low protein expression of TLR9 [26]. Chatzouli et al. [27] reported that TLR7 and TLR9 stimulation with agonists results in apoptosis of CLL cells but only in IGHV mutated patients. Recent studies provided by Zhao et al. [28] demonstrated that phenotypically identical cells nevertheless express very different levels of each receptor. The authors suggested that the engagement of multiple receptors, such as TLR7/8/9, may be necessary for improving the host anti-tumor response. Activation of TLR7/8/9 leads to antigen-specific humoral responses through B-lymphocyte activation, but also the inhibition of B-cell apoptosis. In contrast to higher TLR7 and TLR9 expression in CLL, expression of TLR2 and TLR4 was confirmed to be lower in CLL compared to HVs. Our results are in line with previous studies [7,23,24,25]. The expression of TLR2 and TLR7 was correlated with negative known prognostic markers, but only in the bone marrow compartment. Expanded expression analysis of splicing variants of TLR4 was assessed in CLL patients for the first time. Here, we showed that the expression of splicing variants of TLR4 (TLR4(1) and TLR4(4)) were significantly lower in PBMC in CLL compared to HV. The decreased expression of splicing variants of TLR4 observed in this study might then be a result of an impaired host response in CLL patients. Several TLR agonists have been used in clinical trials of CLL patients as adjuvants to improve the efficacy of chemotherapy, for example, the agonist of TLR4. Different studies demonstrate positive as well as negative effects of TLR4 stimulation on cancer development or treatment. Hwang et al. [29] indicated that stimulation of TLR4 results in IL-6, IL-8, IL-12, TNF, INF-γ, and CCL5 secretion. Stimulation of TLR4 that can enhance the anti-tumor response and the beneficial effects of TLR4 stimulation while eliminating the negative effects remains a challenge for cancer researchers.
Signal transduction through TLRs is involved in B cell biology, including activation of naïve B cells, differentiation, and induction proliferation of memory B cells. Expression of TLR is associated with their ability to respond to TLR agonists. Naïve B cells are characterized by low expression of TLR7 and TLR9, as well as TLR1, TLR6, TLR10, while memory B cells express a high level of TLR7 and TLR9, accompanied by a low level of TLR2, TLR4, TLR8 [11,30]. Comparing our defined TLR profile of CLL cells to normal B cells, we can indicate that it is parallel to memory B cells. This analogy is in line with the previous study [30].
No differences in TLRs expression between peripheral blood and bone marrow indicate similarities shared between these two distinct compartments of the CLL microenvironment. However, we suggest that differential regulation of TLRs accompanies its variable prognostic value, depending on the microenvironment of the specific compartment of CLL. Interestingly, our results for the first time showed the impact of splicing variants of TLR4(3) on clinical outcomes in CLL. Analysis of survival curves revealed that TTFT was significantly shorter in the group of patients with a low level of TLR4(3) expression compared to the group of patients with a high level of TLR4(3) expression in BMMC, thereby we could indicate the negative prognostic value of low TLR4(3) expression assessed in the bone marrow compartment in CLL. Moreover, our results might indicate also a potential negative prognostic value of the low level of TLR4(4) in bone marrow since expression of TTFT tended to be shorter in the group of patients with a low level of TLR4(4) expression compared to the group of patients with a high level of TLR4(4) expression in BMMC. Analogous results were not obtained in peripheral blood, as well as in terms of TLR4(1). Thereby, we might indicate that deregulation of TLR4 signaling via TLR4(3) and TLR(4) is more relevant in bone marrow as a proliferative than accumulative compartment. These differences might be explained by differential posttranscriptional regulation of TLR4(1), TLR4(3) and TLR(4) and diverse functional and cellular interactions in those two compartments of CLL. Although TLR4 splicing variants differ in terms of the number of exons and length of the extracellular domain, their functional importance in a cell has not been analyzed [31,32]. The significance of TLR4 in disease progression was shown also in human lung cancers [33], as well as ovarian cancer [34]. In mantle cell lymphoma (MCL), Wang et al. [35] showed that signaling through TLR4 triggers a cascade leading to the growth of cells and evasion from immune surveillance, contributing to disease progression. However, Nunez et al. [36] showed that TLR4-activated tumor cells are engaged in an antitumoral immune response. This discrepancy might be associated with the type of cytokines that are secreted upon stimulation in a specific tumor microenvironment.
Results of our study revealed a tendency for shorter TTFT in groups of CLL patients with high TLR9 expression in comparison to low TLR9 expression in BBMC, indicating the potential prognostic significance of high mRNA TLR9 expression in bone marrow in CLL. In PBMC, there was no difference in TTFT when referring to the level of TLR9 expression, while the results of our previous study [22] revealed that high expression of TLR9 on the protein level in CLL patients is correlated with longer TTFT. Interestingly, significant discrepancies were identified between mRNA and protein levels for certain TLR expression, and a high expression on a mRNA level did not always correspond to strong protein expression. Several factors could be responsible for these discrepancies, such as cellular intraclonal heterogeneity, differential activation status of malignant cells, and different cell viability in different samples. The expression of TLR2 and TLR7 was correlated with known negative prognostic markers but only in the bone marrow compartment.
Since downstream signaling pathways through TLRs involve Myd88 as an adaptor molecule, we provided MYD88 expression patterns in peripheral blood and bone marrow, referring to CLL prognostic factors as well as an association between MYD88 and TLRs expression in both compartments. In the literature, there is only data about the prognostic significance of MYD88 mutation [17] or MYD88 expression. We revealed higher MYD88 expression in CLL patients compared to HVs, although Antosz et al. [37] revealed lower expression of MYD88 on a mRNA level in CLL compared to control. They found that TLR agonist stimulation did not result in changes in Myd88 protein expression and suggested some defects in Myd88 proteins in CLL. Myd88 involvement by acting downstream of TLRs in carcinogenesis was shown in many reports concerning cancer of the skin, pancreas, liver, colon, sarcoma [38], whereas the data about the mRNA role of MYD88 expression in tumors are limited. Chen et al. [39] showed that MYD88 as well as TLR4 mRNA expression was higher in breast cancer tissue compared to adjacent normal tissue. Moreover, subsequent research showed a higher protein level of Myd88 and TLR4 in breast carcinoma than paracarcinoma tissue, as well as a correlation between their expression and axillary lymph node metastasis that providing the metastatic potential role of TLR4/Myd88 signaling in breast cancer [40]. However, in ovarian cancer it was also revealed that Myd88 expression strongly correlated with TRL4 expression and provided a favorable prognosis [41].
All TLRs expressions that were analyzed (TLR2, TLR7, TLR9, and TLR4 isoforms) represent components of downstream cell signaling pathways by Myd88 that finally activate NF-κB, which is essential for CLL survival. It has been proven that TLRs, including 7 and 9, initiate the Myd88-dependent pathway directly, while TLRs types 2 and 4 indirectly impact the involvement of TIRAP. Specifically, TLR4 initiates an alternative pathway which is MyD88-independent by recruiting TRIF that eventually activates NF-κB.
These mutual associations indicate their relevant cooperation which points to the utilization of Myd88 by TLRs signaling in CLL cells. Since the MyD88-dependent signaling pathway leads to the early phase of NF-κB activation whereas the MyD88-independent signaling pathway initiates the late phase of NF-κB activation, this suggests that both mechanisms might be utilized in CLL.
To sum up, our results proved correlations between MYD88 and analyzed TLRs expressions in both compartments, indicating their relevant cooperation in signal transduction in CLL cells. The MYD88 expression was higher in CLL patients compared to HVs. The TLRs expression was aberrant in CLL patients compared to HVs. Differences in TTFT indicate a negative prognostic value of low TLR4(3) expression in the bone marrow, which might suggest an importance of deregulation of the signaling pathway typical for TLR(4) (Myd88-independent/TRIF-dependent NF-κB activation) in this compartment.
We proved that TLRs expression is differentially regulated in CLL but is similarly shared between two distinct compartments of the CLL microenvironment. We indicate that differential regulation of TLRs might accompany the various role of TLRs signaling in peripheral blood and bone marrow, representing two different microenvironmental compartments of CLL.
Acknowledgments
The authors would like to thank to Magdalena Osiak, Natalia Pajak, Malgorzata Zajac, Joanna Zaleska, Marta Karp, Maciej Grzywnowicz for laboratory support and Waldemar Tomczak for patient recruitment and providing clinical data.
Supplementary Materials
The following are available online at https://www.mdpi.com/2077-0383/10/4/867/s1, Figure S1: 637 Correlations between TLR2 expression and MYD88 expression in PBMC (A) and BMMC (B) in CLL., Figure S2: 638 Correlations between TLR4 splice variants expression and MYD88 expression in PBMC (A, C, E), and BMMC (B,D,F) in CLL., Figure S3: 639 Correlations between TLR7 expression and MYD88 expression in PBMC (A) and BMMC (B) in CLL., Figure S4: 640 Correlations between TLR9 expression and MYD88 expression in PBMC (A) and BMMC (B) in CLL., Table S1. Expression of TLRs in A, B, C clinical stage of disease according to Binet scale.
Author Contributions
Conceptualization, K.G.; methodology, K.S., A.K.; software, K.S., A.K.; validation, K.S., P.W. and K.G.; formal analysis, K.S.; investigation, K.S., A.K.; resources, K.G.; data curation, K.S., A.K., P.W.; writing—original draft preparation, K.S., P.W.; writing—review and editing, K.G., K.S.; visualization, K.S.; supervision, K.G.; project administration, K.G.; funding acquisition, K.G. K.G. designed the research. K.S., A.K. performed the research. K.S., P.W. and A.K. analyzed data. K.S. and P.W. wrote the paper. K.G., K.S. and P.W. discussed results. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by educational grants of the Medical University of Lublin (DS462) and National Science Centre UMO: 2018/29/B/NZ5/02706. The APC was funded by educational grants of the Medical University of Lublin and National Science Centre UMO: 2018/29/B/NZ5/02706.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Local Ethics Committee of the Medical University of Lublin (KE-0254/7/2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mosquera Orgueira A., Antelo Rodríguez B., Alonso Vence N., Bendaña López Á., Díaz Arias J.Á., Díaz Varela N., González Pérez M.S., Pérez Encinas M.M., Bello López J.L. Time to Treatment Prediction in Chronic Lymphocytic Leukemia Based on New Transcriptional Patterns. Front. Oncol. 2019;9:79. doi: 10.3389/fonc.2019.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kittai A.S.A.S., Lunning M., Danilov A.V., Lunning M. Relevance of Prognostic Factors in the Era of Targeted Therapies in CLL. Volume 14. Springer; Berlin/Heidelberg, Germany: 2019. pp. 302–309. [DOI] [PubMed] [Google Scholar]
- 3.Skórka K., Kot M., Knap J., Giannopoulos K. Deregulation of the immune system in patients with chronic lymphocytic leukemia. Postepy Hig. Med. Dosw. 2019;73:117–132. doi: 10.5604/01.3001.0013.0876. [DOI] [Google Scholar]
- 4.Hallek M., Cheson B.D., Catovsky D., Caligaris-Cappio F., Dighiero G., Döhner H., Hillmen P., Keating M., Montserrat E., Chiorazzi N., et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood. 2018;131:2745–2760. doi: 10.1182/blood-2017-09-806398. [DOI] [PubMed] [Google Scholar]
- 5.García-Muñoz R., Galiacho V.R., Llorente L. Immunological aspects in chronic lymphocytic leukemia (CLL) development. Ann. Hematol. 2012;91:981–996. doi: 10.1007/s00277-012-1460-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forconi F., Moss P. Perturbation of the normal immune system in patients with CLL. Blood. 2015;126:573–581. doi: 10.1182/blood-2015-03-567388. [DOI] [PubMed] [Google Scholar]
- 7.Grandjenette C., Kennel A., Faure G.C., Béné M.C., Feugier P. Expression of functional toll-like receptors by B-chronic lymphocytic leukemia cells. Haematologica. 2007;92:1279–1281. doi: 10.3324/haematol.10975. [DOI] [PubMed] [Google Scholar]
- 8.Montero Vega M.T., de Andrés Martín A. The significance of toll-like receptors in human diseases. Allergol. Immunopathol. 2009;37:252–263. doi: 10.1016/j.aller.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Bernasconi N.L., Onai N., Lanzavecchia A. A role for Toll-like receptors in acquired immunity: Up-regulation of TLR9 by BCR triggering in naive B cells and constitutive expression in memory B cells. Blood. 2003;101:4500–4504. doi: 10.1182/blood-2002-11-3569. [DOI] [PubMed] [Google Scholar]
- 10.Harsini S., Beigy M., Akhavan-Sabbagh M., Rezaei N. Toll-like receptors in lymphoid malignancies: Double-edged sword. Crit. Rev. Oncol. Hematol. 2014;89:262–283. doi: 10.1016/j.critrevonc.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Arvaniti E., Ntoufa S., Papakonstantinou N., Touloumenidou T., Laoutaris N., Anagnostopoulos A., Lamnissou K., Caligaris-Cappio F., Stamatopoulos K., Ghia P., et al. Toll-like receptor signaling pathway in chronic lymphocytic leukemia: Distinct gene expression profiles of potential pathogenic significance in specific subsets of patients. Haematologica. 2011;96:1644–1652. doi: 10.3324/haematol.2011.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muzio M., Scielzo C., Bertilaccio M.T.S., Frenquelli M., Ghia P., Caligaris-Cappio F. Expression and function of toll like receptors in chronic lymphocytic leukaemia cells. Br. J. Haematol. 2009;144:507–516. doi: 10.1111/j.1365-2141.2008.07475.x. [DOI] [PubMed] [Google Scholar]
- 13.Lanemo Myhrinder A., Hellqvist E., Sidorova E., Söderberg A., Baxendale H., Dahle C., Willander K., Tobin G., Bäckman E., Söderberg O., et al. A new perspective: Molecular motifs on oxidized LDL, apoptotic cells, and bacteria are targets for chronic lymphocytic leukemia antibodies. Blood. 2008;111:3838–3848. doi: 10.1182/blood-2007-11-125450. [DOI] [PubMed] [Google Scholar]
- 14.Zaleska J., Skorka K., Zajac M., Karczmarczyk A., Karp M., Tomczak W., Hus M., Wlasiuk P., Giannopoulos K. Specific cytotoxic T-cell immune responses against autoantigens recognized by chronic lymphocytic leukaemia cells. Br. J. Haematol. 2016;174:582–590. doi: 10.1111/bjh.14098. [DOI] [PubMed] [Google Scholar]
- 15.Chu C.C., Catera R., Zhang L., Didier S., Agagnina B.M., Damle R.N., Kaufman M.S., Kolitz J.E., Allen S.L., Rai K.R., et al. Many chronic lymphocytic leukemia antibodies recognize apoptotic cells with exposed nonmuscle myosin heavy chain IIA: Implications for patient outcome and cell of origin. Blood. 2010;115:3907–3915. doi: 10.1182/blood-2009-09-244251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J.Q., Jeelall Y.S., Ferguson L.L., Horikawa K. Toll-Like Receptors and Cancer: MYD88 Mutation and Inflammation. Front. Immunol. 2014;5:367. doi: 10.3389/fimmu.2014.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin S.-C., Xia Y., Miao Y., Zhu H.-Y., Wu J.-Z., Fan L., Li J.-Y., Xu W., Qiao C. MYD88 mutations predict unfavorable prognosis in Chronic Lymphocytic Leukemia patients with mutated IGHV gene. Blood Cancer J. 2017;7:651. doi: 10.1038/s41408-017-0014-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adamia S., Haibe-Kains B., Pilarski P.M., Bar-Natan M., Pevzner S., Avet-Loiseau H., Lode L., Verselis S., Fox E.A., Burke J., et al. A Genome-Wide Aberrant RNA Splicing in Patients with Acute Myeloid Leukemia Identifies Novel Potential Disease Markers and Therapeutic Targets. Clin. Cancer Res. 2014;20:1135–1145. doi: 10.1158/1078-0432.CCR-13-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caligaris-Cappio F., Bertilaccio M.T.S., Scielzo C. How the microenvironment wires the natural history of chronic lymphocytic leukemia. Semin. Cancer Biol. 2014;24:43–48. doi: 10.1016/j.semcancer.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Grzywnowicz M., Karczmarczyk A., Skorka K., Zajac M., Zaleska J., Chocholska S., Tomczak W., Giannopoulos K. Expression of Programmed Death 1 Ligand in Different Compartments of Chronic Lymphocytic Leukemia. Acta Haematol. 2015;134:255–262. doi: 10.1159/000430980. [DOI] [PubMed] [Google Scholar]
- 21.Herishanu Y., Pérez-Galán P., Liu D., Biancotto A., Pittaluga S., Vire B., Gibellini F., Njuguna N., Lee E., Stennett L., et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117:563–574. doi: 10.1182/blood-2010-05-284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Własiuk P., Tomczak W., Zając M., Dmoszyńska A., Giannopoulos K. Total expression of HLA-G and TLR-9 in chronic lymphocytic leukemia patients. Hum. Immunol. 2013;74:1592–1597. doi: 10.1016/j.humimm.2013.08.277. [DOI] [PubMed] [Google Scholar]
- 23.Antosz H., Sajewicz J., Marzec-Kotarska B., Dmoszyńska A., Baszak J., Jargiełło-Baszak M. TLR2 may influence the behavior of the malignant clone in B-CLL. Blood Cells. Mol. Dis. 2012;49:32–40. doi: 10.1016/j.bcmd.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Rybka J., Butrym A., Wróbel T., Jaźwiec B., Bogucka-Fedorczuk A., Poręba R., Kuliczkowski K. The Expression of Toll-Like Receptors in Patients with B-Cell Chronic Lymphocytic Leukemia. Arch. Immunol. Ther. Exp. 2016;64:147–150. doi: 10.1007/s00005-016-0433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barcellini W., Imperiali F.G., Zaninoni A., Reda G., Consonni D., Fattizzo B., Lonati S., Nobili L., Zanella A., Cortelezzi A. Toll-like receptor 4 and 9 expression in B-chronic lymphocytic leukemia: Relationship with infections, autoimmunity and disease progression. Leuk. Lymphoma. 2014;55:1768–1773. doi: 10.3109/10428194.2013.856426. [DOI] [PubMed] [Google Scholar]
- 26.Longo P.G., Laurenti L., Gobessi S., Petlickovski A., Pelosi M., Chiusolo P., Sica S., Leone G., Efremov D.G. The Akt signaling pathway determines the different proliferative capacity of chronic lymphocytic leukemia B-cells from patients with progressive and stable disease. Leukemia. 2007;21:110–120. doi: 10.1038/sj.leu.2404417. [DOI] [PubMed] [Google Scholar]
- 27.Chatzouli M., Ntoufa S., Papakonstantinou N., Chartomatsidou E., Anagnostopoulos A., Kollia P., Ghia P., Muzio M., Stamatopoulos K., Belessi C. Heterogeneous functional effects of concomitant B cell receptor and TLR stimulation in chronic lymphocytic leukemia with mutated versus unmutated Ig genes. J. Immunol. 2014;192:4518–4524. doi: 10.4049/jimmunol.1302102. [DOI] [PubMed] [Google Scholar]
- 28.Zhao B.G., Vasilakos J.P., Tross D., Smirnov D., Klinman D.M. Combination therapy targeting toll like receptors 7, 8 and 9 eliminates large established tumors. J. Immunother. cancer. 2014;2:12. doi: 10.1186/2051-1426-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang S.H., Cho H.K., Park S.H., Lee W., Lee H.J., Lee D.C., Oh J.H., Park S.H., Kim T.-G., Sohn H.-J., et al. Toll like receptor 3 & 4 responses of human turbinate derived mesenchymal stem cells: Stimulation by double stranded RNA and lipopolysaccharide. PLoS ONE. 2014;9:e101558. doi: 10.1371/journal.pone.0101558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rozková D., Novotná L., Pytlík R., Hochová I., Kozák T., Bartůnková J., Spísek R. Toll-like receptors on B-CLL cells: Expression and functional consequences of their stimulation. Int. J. cancer. 2010;126:1132–1143. doi: 10.1002/ijc.24832. [DOI] [PubMed] [Google Scholar]
- 31.Hoang T.X., Duong C.N., Kim J.Y. Identification and Characterization of a Splicing Variant in the 5′ UTR of the Human TLR5 Gene. Biomed Res. Int. 2017;2017:1–7. doi: 10.1155/2017/8727434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wells C., Chalk A., Forrest A. Alternate transcription of the Toll-like receptor sigaling cascade. Genome Biol. 2006;7:R10. doi: 10.1186/gb-2006-7-2-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He W., Liu Q., Wang L., Chen W., Li N., Cao X. Corrigendum to “TLR4 signaling promotes immune escape of human lung cancer cells by inducing immunosuppressive cytokines and apoptosis resistance” [Mol. Immunol. 44 (2007) 2850–2859] Mol. Immunol. 2020;122:232–234. doi: 10.1016/j.molimm.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Szajnik M., Szczepanski M.J., Czystowska M., Elishaev E., Mandapathil M., Nowak-Markwitz E., Spaczynski M., Whiteside T.L. TLR4 signaling induced by lipopolysaccharide or paclitaxel regulates tumor survival and chemoresistance in ovarian cancer. Oncogene. 2009;28:4353–4363. doi: 10.1038/onc.2009.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L., Zhao Y., Qian J., Sun L., Lu Y., Li H., Li Y., Yang J., Cai Z., Yi Q. Toll-like receptor-4 signaling in mantle cell lymphoma. Cancer. 2013;119:782–791. doi: 10.1002/cncr.27792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nunez N.G., Andreani V., Crespo M.I., Nocera D.A., Breser M.L., Moron G., Dejager L., Libert C., Rivero V., Maccioni M. IFN Produced by TLR4-Activated Tumor Cells Is Involved in Improving the Antitumoral Immune Response. Cancer Res. 2012;72:592–603. doi: 10.1158/0008-5472.CAN-11-0534. [DOI] [PubMed] [Google Scholar]
- 37.Antosz H., Sajewicz J., Marzec-Kotarska B., Dmoszyńska A., Baszak J., Jargiełło-Baszak M. Aberrant TIRAP and MyD88 expression in B-cell chronic lymphocytic leukemia. Blood Cells. Mol. Dis. 2013;51:48–55. doi: 10.1016/j.bcmd.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Salcedo R., Cataisson C., Hasan U., Yuspa S.H., Trinchieri G. MyD88 and its divergent toll in carcinogenesis. Trends Immunol. 2013;34:379–389. doi: 10.1016/j.it.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X., Zhao F., Zhang H., Zhu Y., Wu K., Tan G. Significance of TLR4/MyD88 expression in breast cancer. Int. J. Clin. Exp. Pathol. 2015;8:7034–7039. [PMC free article] [PubMed] [Google Scholar]
- 40.Wu K., Zhang H., Fu Y., Zhu Y., Kong L., Chen L., Zhao F., Yu L., Chen X. TLR4/MyD88 signaling determines the metastatic potential of breast cancer cells. Mol. Med. Rep. 2018;18:3411–3420. doi: 10.3892/mmr.2018.9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang A.-C., Ma Y.-B., Wu F.-X., Ma Z.-F., Liu N.-F., Gao R., Gao Y.-S., Sheng X.-G. TLR4 induces tumor growth and inhibits paclitaxel activity in MyD88-positive human ovarian carcinoma in vitro. Oncol. Lett. 2014;7:871–877. doi: 10.3892/ol.2013.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.