Abstract
The RV144 trial represents the only vaccine trial to demonstrate any protective effect against HIV-1 infection. While the reason(s) for this protection are still being evaluated, it serves as justification for widespread efforts aimed at developing new, more effective HIV-1 vaccines. Advances in our knowledge of HIV-1 immunogens and host antibody responses to these immunogens are crucial to informing vaccine design. While the envelope (Env) protein is the only viral protein present on the surface of virions, it exists in a complex trimeric conformation and is decorated with an array of variable N-linked glycans, making it an important but difficult target for vaccine design. Thus far, efforts to elicit a protective humoral immune response using structural mimics of native Env trimers have been unsuccessful. Notably, the aforementioned N-linked glycans serve as a component of many of the epitopes crucial for the induction of potentially protective broadly neutralizing antibodies (bnAbs). Thus, a greater understanding of Env structural determinants, most critically Env glycosylation, will no doubt be of importance in generating effective immunogens. Recent studies have identified the HIV-1 Env signal peptide (SP) as an important contributor to Env glycosylation. Further investigation into the mechanisms by which the SP directs glycosylation will be important, both in the context of understanding HIV-1 biology and in order to inform HIV-1 vaccine design.
Keywords: HIV-1, HIV envelope, glycosylation, signal peptide, PNGS, broadly neutralizing antibodies, vaccine
1. Introduction
Infection of CD4+ T cells by the human immunodeficiency virus type 1 (HIV-1) leads to a drastic decrease in their number and causes acquired immune deficiency syndrome (AIDS), which can eventually lead to death. Treatment of individuals infected with HIV-1 consists of the administration of highly effective antiretroviral medications, which allow patients to live relatively normal lives assuming that treatment regimens are adhered to [1,2,3]. However, resistance to these medications is of some concern, and therefore additional treatment approaches are of importance [4]. In the absence of a cure, prevention of new infections via prophylactic vaccination is likely to have a more widespread and lasting effect and is therefore the main focus of HIV-1 research [5].
Due to the high selective pressure induced by the immune system and the high mutation rate of HIV-1, chronic infection often consists of multiple viral quasispecies [6,7]. The immune response of an HIV-1 infected individual therefore needs to effectively prevent multiple viral quasispecies from infecting CD4+ T cells. This can be achieved via the induction of a broad milieu of antibodies (Abs) with varying specificities that act in concert to prevent infection but is ideally mediated via antibodies capable of binding epitopes that are conserved between many HIV-1 strains [8]. These antibodies are known as broadly neutralizing antibodies (bnAbs) and attempts to induce their production via vaccination have been a focal point of HIV-1 vaccine design.
The trimeric envelope (Env) that is responsible for initiating HIV-1 infection is the sole target for the induction of neutralizing antibodies (nAbs). Much effort has been expended to create recombinant Env trimers that share great structural similarity to native Env trimers (termed SOSIP, uncleaved prefusion optimized (UFO), single-chain (SC), and native-flexible-linker (NFL), among others), with the hope of inducing bnAbs. However, none have thus far succeeded in this aim. These efforts are no doubt a crucial step in immunogen design and have been reviewed elsewhere [9,10]. Complicating these efforts is the existence of a wide array of N-linked glycan structures on the Env surface that modulate its interaction with the host immune response and whose composition is dependent on a number of variable viral and host factors. One such viral factor, the Env signal peptide (SP), has recently been shown to greatly influence the composition of N-glycans on Env [11]. Notably, glycosylation of the HIV-1 Env can modulate the antibody response. This review highlights challenges in HIV-1 vaccine design related to Env glycosylation, with emphasis on the contribution of the Env SP in directing said glycosylation.
2. HIV-1 Envelope Structure and Immunogenicity
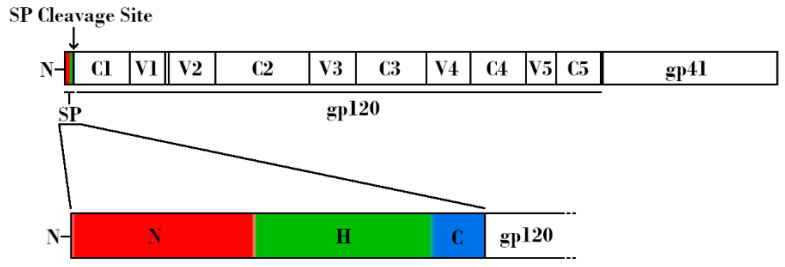
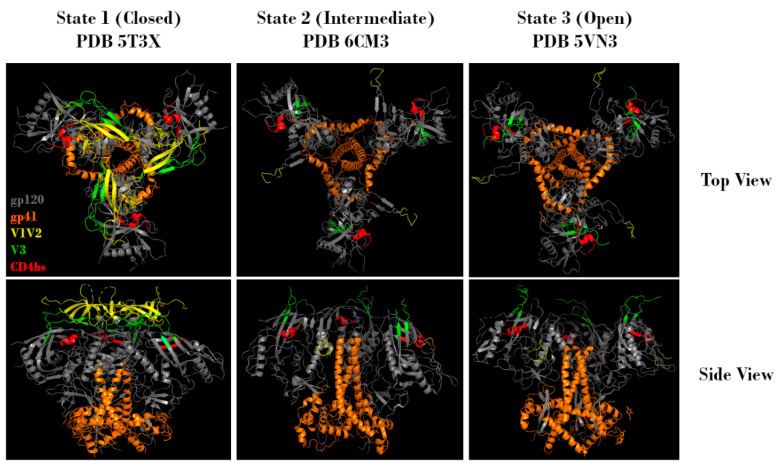
In order to understand how Env glycosylation complicates efforts to create effective Env-based immunogens, it is important to understand the structure, function, and inherent immune evasion properties of Env. HIV-1 Env exists as a trimeric spike on the viral surface, consisting of three heterodimers. These heterodimers arise from furin-mediated cleavage of glycoprotein 160 (gp160) proteins, resulting in a glycoprotein 120 (gp120) non-covalently linked to a glycoprotein 41 (gp41) (Figure 1, top). The gp120 subunit can be further subdivided into five conserved regions (C1-C5) and five hypervariable regions (V1-V5) [12,13,14,15]. Binding to host cells is facilitated by the gp120 subunit, which contains both the CD4 and co-receptor (either CCR5 or CXCR4) binding sites. Upon binding, Env undergoes conformational changes that allow gp41 to mediate fusion of the viral membrane with the host cell membrane, leading to infection [6,13,16,17]. Env is a dynamic molecule that exists in one of three conformations at any given time: a metastable closed conformation (state 1), a partially open intermediate conformation (state 2), and an open conformation (state 3) (Figure 2). Env transitions between these three conformations, but the majority of known bnAbs, isolated from HIV-1 infected individuals, target Env in state 1. Of note, the current recombinant Env trimers present Env in a stabilized version of state 2 [18]. In state 2 or 3, the variable loops V1V2 and V3 are more available for immune recognition. This functionally means that the initial antibody response is often specific to the infecting strain, and that escape mutations are common. Uncleaved gp160 monomers, as well as gp41 stumps lacking an associated gp120, also exist on the viral surface. These nonfunctional proteins display epitopes that may lead to the generation of non-neutralizing antibodies (nnAbs). Combined with the relatively low number of functional Env trimers, these mechanisms serve to effectively divert the host immune response [19,20,21,22,23,24,25].
Figure 1.
Schematic linear representation of the nascent HIV-1 Envelope (Env) protein attached to the HIV-1 signal peptide (SP). (Top) Regions corresponding to SP, glycoprotein 120 (gp120), and glycoprotein 41 (gp41) are indicated. Variable and constant regions of gp120 are indicated by V1-V5 and C1-C5, respectively. SP cleavage site is indicated by an arrow. (Bottom) Expanded schematic representation of the HIV-1 SP. The N-terminal hydrophilic positively charged region is shown in red, the central hydrophobic region is shown in green, and the slightly polar C-terminal region is shown in blue.
Figure 2.
Representative top and side views of Env trimers in state 1 (closed), state 2 (intermediate), and state 3 (open) conformations. Regions corresponding to gp120 (grey), gp41 (orange), V1V2 (yellow), V3 (green), and the CD4 binding site (red) are indicated. Protein Data Bank identifier (PDB ID) numbers are also indicated. Structures adapted from Gristick et al., 2016 [26], Bjorkman et al., 2018 [27], and Ozorowski et al., 2017 [28].
3. HIV-1 Env N-linked Glycosylation
The existence of a wide array of N-glycans on the Env surface adds an additional layer of complexity to the already highly variable Env. Each Env of the trimer contains between 18 and 33 potential N-linked glycosylation sites (PNGSs), whose occupancy by glycans can account for up to half of the trimer’s total mass and cover up to 50–70% of the Env surface [7,29,30,31,32]. In this way, these poorly immunogenic glycans shield underlying protein residues from immune recognition. The occupancy of any particular PNGS is variable, with most being less than 90% conserved between HIV-1 strains [33,34]. Most PNGSs exist on gp120, but ~4–5 can be found on gp41 as well [33].
The mechanism by which Env is processed prior to deposition on the viral surface is a key determinant in the makeup of the Env “glycan shield”. Shortly after SP-mediated delivery of the nascent Env polypeptide to the endoplasmic reticulum (ER) for processing, PNGS occupancy is initiated via the placement of a Glc3Man9GlcNAc2 residue at each site. During subsequent transit through the ER and Golgi, this precursor glycan is modified by a milieu of host enzymes to yield its final glycoform (Figure 3A). However, the accessibility of these enzymes is greatly affected by steric constraints related to PNGS occupancy, which is in turn dependent on viral strain, host cell type, and acquired mutations [33,35,36,37,38,39]. Due to the high level of overall PNGS occupancy, these steric constraints result in a glycan landscape that consists predominantly of high-mannose (immature/unprocessed Man5-9GlcNAc2) glycans, with a smaller proportion of more mature/highly-processed complex and hybrid glycans [39,40,41,42,43,44,45,46,47]. Even with such constraints, the total number of variables involved in this process leads to incredibly diverse glycan landscapes from the outset [41,46,48].
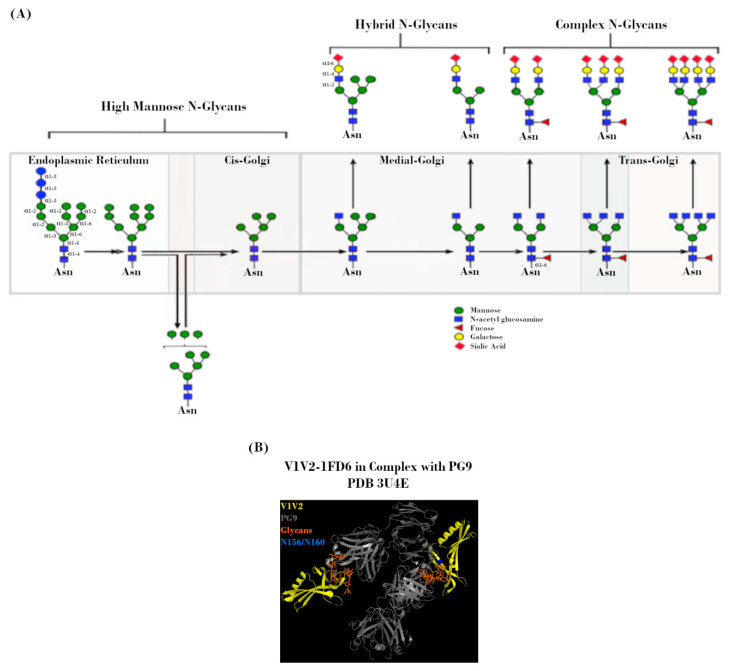
Figure 3.
N-linked glycans on the HIV-1 envelope (Env). (A) Potential N-linked glycosylation site (PNGS) occupancy is initiated via the placement of a Glc3Man9GlcNAc2 residue, which is subsequently modified during transit through the endoplasmic reticulum (ER) and Golgi by a milieu of host enzymes to yield its final glycoform. Figure adapted from Jan et al., 2019 [49]. (B) Crystal structure (2.19 Å) of PG9 mAb in complex with V1V2 region from HIV-1. Regions corresponding to V1V2 (yellow), PG9 (grey), PNGS N156 and N160 (blue), and N-glycans (orange) are indicated. Protein Data Bank identifier (PDB ID) number is also indicated. Adapted from McLellan et al., 2011 [50].
In much the same way that these glycans sterically hinder host glycosidases, they also confer resistance to nAbs [30,51,52]. Interactions between antibodies and these host-derived glycans are often low affinity, due to both the nature of such interactions as well as the fact that said glycans are immunologically “self” [53,54,55]. That is not to say that glycan-targeting bnAbs cannot exist, merely that glycans are often only one component of larger, non-linear epitopes [56]. Occupancy of specific PNGSs, such as N156 and N160, has been shown to be crucial for the formation of conformational epitopes recognized by bnAbs, such as PG9 and PG16 (Figure 3B) [57]. Additionally, one of the most common bnAb targets is a high-mannose patch centered around either N332 or N334, dependent on viral strain and antibody lineage [58,59,60,61]. This so-called glycan supersite is also the target of 2G12, which binds an epitope derived solely of glycans. Due to their dependence on higher-order conformation, attempts to elicit these bnAbs in the context of vaccination with recombinant Env constructs have not been successful. There is also evidence that glycans can modulate the composition of immune responses via interactions with antigen presenting cells. Glycans of the C2V3 region of Env have been shown in a mouse model to be capable of inducing a bias for unfavorable IgG1 antibody subtype and TH2 T cell responses [62].
Over the course of disease progression, selective pressure drives mutations that further aid in immune escape. These may manifest as altered PNGS locations, such as the N332 to N334 mutation that has been shown to mutate in both directions to avoid immune recognition [63]. They may also manifest as alterations in glycoforms, due to genetic or steric determinants [33,36,48,63,64,65,66,67,68,69,70]. Furthermore, single mutations that alter the occupancy or glycoform at any particular PNGS may have cascading effects on the processing of glycans at other PNGSs. Taken together, these factors contribute to a complex and constantly moving target for the immune system [71].
Despite this variation, a number of generalized structural and glycan motifs have been identified in HIV-1 isolates from varying stages of disease progression. In the vast majority of heterosexual infection events (>80%), a single transmitted/founder (T/F) strain is responsible for the establishment of infection [72,73,74,75,76]. For this reason, T/F strains are a prime target of interest for vaccines, microbicides, and pre- and post-exposure prophylactic measures. Cases involving multiple infecting strains are often tied to alternative transmission routes (such as intravenous drug use), or breakdown of mucosal barriers [75,77,78]. These T/F strains have been shown to possess a number of characteristics that separate them from chronic isolates and may partially explain why they are effective at establishing infection. For instance, T/F viruses from clades A, C, and D have been shown to possess shorter V1V2 regions and a reduced number of occupied PNGSs compared to chronic isolates [65,77,79,80,81,82,83,84]. Additionally, some clade B isolates have been shown to possess a shorter V5 region, as well as a V3 region that contains fewer occupied PNGSs and less positively charged residues [77,81,85,86,87,88,89,90]. As these variable regions are commonly targeted by nAbs, their shortening presents fewer potential targets for the immune system [91,92,93]. Due to their important roles in infection, many conserved sites (such as the CD4 and coreceptor binding sites) within these variable regions are rarely, if ever, directly occluded from immunological access via glycans. The lower PNGS occupancy observed in T/F viruses may reflect an adaptation to divert the immune response away from these crucial sites and onto residues in the vicinity of unoccupied PNGSs. Indeed, the earliest nAbs targeting T/F viruses are often specific for non-conserved residues within the variable regions of Env.
Wagh et al. recently utilized a computational model in order to characterize the relationship between the completeness of the glycan shield at transmission and the development of nAbs [51]. They found that a more complete glycan shield at transmission correlated with more rapid development of a broad nAb response. One possibility for this observation is that an increased initial PNGS occupancy represents fewer opportunities for escape. While not all escape mutations result in increased PNGS occupancy, we know that chronic isolates have more complete glycan shielding and a higher proportion of complex/hybrid glycans, suggesting a directionality to the process [6,68,77,94,95,96,97]. Additionally, as PNGS become occupied by primarily high-mannose glycans, the virus becomes more susceptible to capture and degradation by the C-type lectin DC-SIGN on dendritic cells and macrophages [49,98,99,100,101,102]. This leads to increased antigen presentation and therefore may increase the rate at which these rounds of escape and adaptation occur. Eventual increases in the proportions of complex/hybrid glycans decrease viral capture by DC-SIGN but are also temporally associated with the development of bnAbs [49,103]. However, virus that does end up captured via this route is protected from degradation by these more mature glycans, resulting in increased levels of CD4+ T cell transinfection and thus representing an additional viral route of infection free of bnAb interference.
It is clear that the contribution of glycosylation at all stages of HIV-1 infection is great, and therefore, understanding the mechanisms governing it is going to be crucial to the development of an effective vaccine. Recently, the Env SP has been implicated as an important determining factor in this process.
4. Role of the Signal Peptide
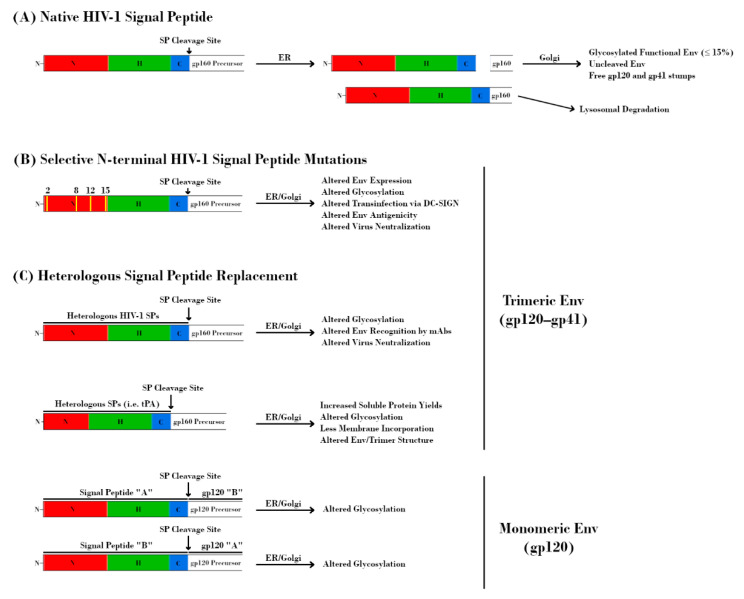
As with all membrane-bound and secreted proteins, the HIV-1 Env contains an N-terminal SP that is responsible for targeting the nascent polypeptide to the ER for processing. In general, SPs consist of an N-terminal hydrophilic positively charged region, a central hydrophobic region responsible for translocation to the ER membrane, and a slightly polar C-terminal region containing a cleavage site (Figure 1, bottom) [104]. The N-terminal region of the HIV-1 SP is relatively unique in that it is both longer and contains more positively charged residues than the average SP [105]. Additionally, the SP remains attached to gp160 until right before it reaches the Golgi, which is also unusual among SPs, most of which are cleaved co-translocationally (Figure 4A) [105,106,107,108].
Figure 4.
Effect of signal peptide (SP) modification/replacement on envelope (Env). (A) The HIV-1 SP is responsible for targeting the nascent glycoprotein 160 (gp160) precursor to the endoplasmic reticulum (ER) for processing. Uniquely, the SP remains attached to this protein throughout the ER and is cleaved prior to delivery of gp160 to the Golgi apparatus. Further retention of the SP results in lysosomal degradation of Env and serves as a quality control mechanism. (B) Mutations at specific residues within the N-terminal region of the HIV-1 SP result in alterations to Env expression, glycosylation, DC-SIGN-mediated transinfection, antigenicity, and neutralization. (C, top) Replacement of the native HIV-1 SP with heterologous HIV-1 SPs alters glycosylation, Env recognition by monoclonal antibodies (mAbs), and virus neutralization. Replacement with non-HIV-1 SPs is a common strategy for increasing immunogen yield. However, this approach may also have detrimental effects on Env glycosylation, structure, and antigenicity. The perceived advantage of increased soluble protein yields may be offset by a higher proportion of Env immunogens with under-processed glycans due to the absence of HIV-1 SP-mediated effects on processing. (C, bottom) The glycosylation profiles of HIV-1 glycoprotein 120 (gp120) proteins are dependent on their SPs. Transposition of SPs from HIV-1 species with different proportions of glycan types is sufficient to alter glycosylation profiles.
The effects on Env processing due to these unique features may be explained by examining SPs from other systems. For instance, an increased positive charge in the N-terminal region is commonly seen in bacteria, especially Gram-positives [109]. Of note, many of these bacterial SPs are never cleaved from their mature proteins and perform additional functions related to membrane incorporation, secretion, and anchoring [110]. The HIV-1 SP is of course cleaved, as lack of SP cleavage prevents maturation of the protein, but it is quite possible that some of these functions overlap [111,112]. The N-terminal positive charge has also been shown in multiple systems to be crucial for SP orientation, which in turn affects additional SP functions such as facilitating membrane incorporation and regulating the timing of SP cleavage. Indeed, there is evidence that post-translocational cleavage of the HIV-1 SP is enabled via occlusion of the C-terminal cleavage site by SP itself [105,108,111,113]. The effect of increased SP length has also been studied extensively in other systems. The absolute length of SPs in each system varies greatly, but increased SP length is often associated with additional functions [114]. Of particular note are the SPs from human interleukin-15 and murine C4b-binding protein, which provide evidence for SP involvement in glycosylation efficiency and increased protein processing time, respectively [114]. These are but two examples of many but provide a precedent for the possibility that these functions, and potentially others, are being performed by the HIV-1 SP as well. Therefore, it is reasonable to speculate that the increased positive charge of the HIV-1 SP functions to facilitate membrane association of both the SP and nascent gp160. In the case of the SP, this helps to orient the SP in such a way that, in tandem with its increased length, occludes its cleavage site and prevents premature cleavage. Both the increased SP length and delayed cleavage provide more time for the processing of gp160, in the form of protein folding and enzymatic glycan modifications. The close membrane association also sterically influences glycan accessibility by glycosidases and increases the likelihood that functional Env trimers are incorporated into virions. A number of recent studies support these assertions and are discussed below [112,115,116,117].
The aforementioned differences in glycosylation patterns exhibited by T/F and chronic HIV-1 isolates were found to be correlated with mutations at specific sites within the N-terminal region of the SP. Broadly speaking, T/F strains were found to have a higher proportion of neutral and basic residues, which were lost in chronic isolates [82,115,118,119,120]. Chronic isolates were also shown to have increased basic residues within their hydrophobic region. This argues that the HIV-1 SP is under selective pressure during infection, and that the charge of these regions may affect membrane association and therefore alter glycosylation [82,118,121,122]. Several groups have utilized varying approaches in order to elucidate the role of these SP differences as they pertain to glycosylation and Env function. These include making complete SP swaps as well as more targeted changes, and utilize recombinant proteins, pseudoviruses, and infectious molecular clones (IMC).
The basic histidine residue at position 12 (H12) within acute/early viruses has been proposed to provide higher overall Env expression, which correlates with higher Env incorporation into virions and increased infectivity [82,115,123]. Using a targeted approach, Upadhyay et al. mutated basic residues within the N-terminal region of the Env SP and assessed their effects on Env functions and virus phenotype (Figure 4B) [121]. Three separate mutations at position 12 (H12R/Q/Y) resulted in decreased Env incorporation. Additionally, mutations at positions 8, 12, and 15 increased resistance to monoclonal antibodies (mAbs) targeting the V1V2 region of Env. Capture and transmission of virus via DC-SIGN was also affected by these mutations, with H12Q/Y exhibiting decreased capture and K2G, R15G, and H12/R/Q/Y exhibiting decreased transmission [121]. Importantly, these mutations were shown to alter the levels of α1-3/α1-2 mannose- and fucose-containing glycans on Env, as determined via lectin binding experiments and liquid chromatography–mass spectrometry (LC-MS/MS). These findings were observed in two separate HIV-1 strains (REJO and JRFL), demonstrating that they may be broadly applicable [121]. Further studies by this group assessed in more detail the contribution of glycans, and SP mutations affecting glycans, on DC-SIGN-mediated transinfection and interaction with antiviral lectins [49]. While the use of pseudoviruses (as opposed to IMCs) resulted in a few areas of divergence, these experiments further underscored the contribution of the SP in directing Env glycan composition. Of particular note was the observation that only SP-related mutations, not mutations directly affecting PNGS occupancy or overall gp120 conformation, had observable effects on DC-SIGN-dependent virus capture/transmission or lectin-mediated transmission inhibition [124,125]. Therefore, these findings argue that even modest changes in the SP at key residues can have widespread effects on the way that HIV-1 Env interacts with host cells and processes. Most recently, Upadhyay et al. observed that replacement of HIV-1 SPs with those from other HIV-1 isolates altered the proportions of oligomannose and complex glycans at the Env trimer apex (V1V2 and V3) and base (gp41 membrane proximal external region, MPER) (Figure 4C) [11]. These changes resulted in altered Env binding and virus neutralization by known mAbs, most prominently those targeting V2i and V2q epitopes. Furthermore, they demonstrated that these effects varied based on the Env backbone (CMU06, REJO, or SF162), as well as the host cell type (293T or PBMC) used for virus production. Utilizing recombinant gp120 proteins, Yolitz et al. also demonstrated that swapping SPs between HIV-1 strains affected the relative proportions of glycans such that they more closely resembled the strain from which the SP was derived (Figure 4C). The mass, glycosylation, interaction with DC-SIGN, and susceptibility to mAbs were all altered in these proteins [103]. Taken together, these studies underscore the important contribution of the HIV-1 SP with regards to glycosylation.
The crucial role of the HIV-1 SP in directing Env glycosylation no doubt has interesting implications for vaccine development as well. A major obstacle in the production of Env immunogens is the inherently low level of expression of these proteins under the control of the native HIV-1 SP. Heterologous SPs have long been used in lieu of HIV-1 SPs in recombinant vaccine constructs, as they have been shown to potentially increase immunogen yields in insect and mammalian expression systems (Figure 4C) [29,105,126,127,128,129]. Various heterologous SPs have been utilized in this way, with tissue plasminogen activator (tPA) being one of the most common, especially among the native-like trimers mentioned previously. Notably, this SP does not share the increased length and increased number of positively charged residues of the HIV-1 Env SP [122]. It is no doubt of importance to express usable quantities of immunogen, but the similarity of the immunogen to native HIV-1 Env is arguably more important. If modest changes to the SP can have such widespread effects on glycan composition and occupancy, then replacing it entirely is sure to complicate efforts to produce effective immunogens. Indeed, there is evidence of similar glycan composition on SOSIP.664 trimers despite being derived from early and late stages of infection, presumably due to the use of the same heterologous SP and in contrast to what is observed in HIV-1 strains [130]. The role of the HIV-1 Env SP in ensuring proper protein processing via its retention is also worth considering, as levels of functional Env are low (≤15%) even in the context of wild-type virus [131]. It is important to remember that the SP and Env protein have co-evolved. In these systems, there exists a balance between the amount of protein and the secretory machinery such that excess protein, even if it is functional, can be degraded if the system is overwhelmed. Heterologous SPs that increase total protein yield may be artificially selecting for products that mature more quickly due to co-translocational SP cleavage but do not present epitopes important for the generation of productive bnAbs [109]. Recent analyses of the PNGS occupancy of native-like trimers have indicated that, while there are generally high levels of occupancy, regions with many closely-spaced PNGSs (such as the V1V2 region) can sometimes have lower occupancy levels in these constructs [36,37,38,130]. This is likely due to a combination of an increased rate of processing due to codon optimization and a lack of the aforementioned quality control mechanisms. There has been a recent interest in creating membrane-bound native-like trimers, the thought being that the close proximity to the membrane may influence enzyme accessibility and therefore glycosylation. Interestingly, replacement of the SP has also been detrimental to these efforts, as it appears to decrease the amount of membrane incorporation compared to that of the native SP [122]. Therefore, it is possible that the benefits of using heterologous SPs may come at the cost of immunogen quality. Proponents of utilizing heterologous SPs may point to the replacement of the HIV-1 SP with that of the herpes simplex virus 1 (HSV-1) glycoprotein D (gD) SP in the RV144 trial as evidence that this practice is well tolerated. However, this replacement was done alongside an 11 amino acid deletion from the N-terminus of the gp120 immunogens, which was made to reduce errant immunogen dimerization and promote proper folding. Post-trial studies utilizing RV144 patient serum suggest that this deletion was also sufficient for enhanced antigenicity to C1, V2, and V1V2 epitopes [132,133]. It is then possible that utilizing native HIV-1 SPs may further increase the effectiveness of such an immunogen. However, it is worth noting that removal of these 11 amino acids also removes a region of Env shown to be important for SP retention and may therefore affect Env processing and glycosylation in much the same way its complete replacement does [111]. Further investigation focused on retaining native-like glycosylation and processing while also increasing protein yield is no doubt warranted.
5. RV144 and Beyond
Of the four HIV-1 vaccine approaches evaluated in clinical trials, only one—RV144—has demonstrated modest efficacy in preventing HIV-1 acquisition [133,134,135]. In this trial, two previously investigated but non-protective vaccines were combined (ALVAC-HIV and AIDSVAX B/E), and a prime-boost strategy was used. ALVAC-HIV (prime) was administered at 0, 4, 12, and 24 weeks and AIDSVAX B/E (boost) was administered at 12 and 24 weeks only. ALVAC-HIV utilizes a recombinant canarypox vector to express HIV-1 LAI Gag and Pol alongside monomeric 92TH023 gp120 linked to the transmembrane anchoring portion of LAI gp41. AIDSVAX B/E is a formulation of recombinant Env gp120 proteins from MN and A244 strains wherein the first 11 amino acids of Env and the native SPs are replaced with a 27 amino acid sequence from HSV-1 gD [134,136]. While the findings of this study are the subject of some controversy, it remains the best evidence that vaccine-mediated protection is a realistic and achievable goal [137,138,139,140,141]. For obvious reasons, the exact mechanism(s) by which this protective effect occurred continue to be a topic of much interest. The vaccine regimen induced HIV-specific humoral and cellular immune responses, and much emphasis has subsequently been placed on characterizing the contribution of antibodies targeting HIV-1 with regards to protection [135,142]. Of note, nnAbs containing Fc effector function and directed against conserved epitopes within the variable regions (V1V2 and V3) of Env were identified in the serum of RV144 patients [143,144]. Furthermore, the existence of these antibodies was correlated with decreased infection risk [145]. This humoral immune response was found to be robust, both immediately after the initial vaccine regimen and upon boost 6–8 years later, but in both cases, the response was short-lived [134,141]. Nonetheless, this trial highlighted the importance of antibody responses targeting V2 epitopes in vaccine-mediated protection. Based on the success of RV144, the HVTN100 vaccine trial was designed and carried out in South Africa [136,146]. This trial utilized a similar prime-boost strategy as RV144 but utilized immunogens based on clade C HIV, as this better reflected the HIV-1 strains circulating in that part of the world. These immunogens were also formulated with a more potent adjuvant than that utilized in RV144, and an additional boost at 12 months was included to test durability. Increased antibody titers and immune response durability were observed compared to RV144, and interim results were favorable enough to justify an additional trial intended to assess the effect of additional boosts on durability. Unfortunately, this trial—HVTN702—was discontinued in early 2020 due to lack of evidence that it had any efficacy in preventing HIV-1 infection. More specifically, an independent data and safety monitoring board (DSMB) examined HIV-1 infection rates after 60% of study participants had been participating for longer than 18 months and found that similar numbers of infections occurred in both the vaccine and placebo groups. While no additional vaccinations are being performed, the study participants are still being followed.
6. Broadly Neutralizing Antibodies and Current Vaccine Approaches
While not induced in the RV144 or HVTN100 vaccinees, bnAbs have been found in some chronically infected individuals. As of now, no bnAb has been identified that is capable of clearing infection in humans. The protective capacity of such antibodies has however been shown via passive and active immunization studies in mice and non-human primates [147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168]. Unfortunately, as in so many cases, promising results in animals do not necessarily translate to clinical efficacy in humans. Nonetheless, a number of bnAbs targeting the CD4 binding site and/or V3 glycan epitopes (such as VRC01 and PGT121) have been or are currently being evaluated as therapeutics in clinical trials [169]. The administration of heterologous bnAbs does, however, come with its own set of challenges, such as limited therapeutic duration due to relatively short half-lives. As such, a major focus of vaccines has been to induce the production of bnAbs within vaccinated individuals.
The methods utilized to elicit bnAbs via vaccination are extremely varied, both in their design and efficacy. However, they generally involve some combination of expression vectors, recombinant proteins, and genetic material. As mentioned above, both RV144 and HVTN100 utilized canarypox vectors to express monomeric Env immunogens alongside the administration of recombinant Env proteins [134,136]. Synthetic immunogens containing known bnAb epitopes coupled to immune-stimulating carriers have also been generated but have only resulted in nnAbs [170]. Another approach that is currently being investigated is the use of so-called mosaic immunogens which contain bioinformatically selected epitopes from multiple viral strains and proteins [171]. An advantage to this approach is that immunogens from many disparate geographical isolates can all be represented in one vaccine. This approach was initially designed to improve the breadth of cellular immune responses but has been shown to also induce cross-clade binding antibodies. A formulation utilizing an adenovirus serotype 26 (Ad26) vector expressing various Env/Gag/Pol immunogens in combination with recombinant glycoprotein 140 (gp140) has been shown to induce comparable immune responses in humans and rhesus macaques. A clinical trial utilizing this vaccine regimen, HV705, is currently underway in sub-Saharan Africa [171].
While elicitation of bnAbs is a rare event, even in the context of chronically infected individuals, attempts to induce protective bnAbs must take into account the possibility that the immunogen(s) being utilized are not accurate enough mimics of their native counterparts. The aforementioned native-like Env trimers represent efforts to address this possibility [9,10]. However, designing these trimers is likewise complicated. Iterative modifications intended to address various issues with stability, solubility, expression levels, and purification may very well compromise their effectiveness as immunogens. Furthermore, there has been recent interest in developing methods to mimic bnAb development during the course of infection, via engagement of naïve B cells producing germline bnAbs. These constructs have shown promise in proof-of-principle experiments utilizing transgenic animals [58,172,173]. However, these transgenic animal models represent optimal conditions for bnAb lineage development and therefore lack much of the complexity found in humans. The affinities of native-like trimers for the naïve B cell populations of interest will likely need to be specifically increased in order to overcome obstacles such as a lower frequency of B cell targets as well as the existence of other naïve B cell populations that may interfere. Regardless of the exact approach utilized, the contribution of steric and conformational determinants on epitope structure is increasingly being recognized as crucial to immunogen design, and therefore to the induction of bnAbs. This review highlights the importance of such determinants, especially those dictated by the unique features of the HIV-1 SP. Future attempts to elicit bnAbs would no doubt benefit from the use of native-like Env trimers generated using native HIV-1 SPs or SPs engineered to mimic them, given the important biosynthetic mechanisms (and, by extension, structural determinants such as glycosylation and higher-order protein structure) that they govern. The use of HIV-1 SPs will also likely aid in attempts to generate membrane-bound versions of these immunogens via increased membrane incorporation, which should further increase their similarity to their native counterparts. Finally, expressing these immunogens in cells that produce more similar glycosylation profiles to HIV-1 target cells could help as well.
7. Conclusions
Taken together, it is clear that the mechanisms by which HIV-1 evades the immune response—both in order to establish infection and in chronically infected patients—are extremely complex. A major contributor to this complexity is the existence of an array of N-linked glycans decorating the surface of the HIV-1 Env. The mechanisms governing the location, heterogeneity, and immunogenicity of these glycans are likewise complex, incorporating genetic, structural, and host-derived determinants. Regardless, evidence suggests that the HIV-1 SP plays an integral role in determining the composition of its glycan shield and must therefore be taken into consideration when designing immunogens for use in vaccination. Further investigation into the mechanisms by which this occurs will be important, both in the context of understanding HIV-1 biology and in order to inform HIV-1 vaccine design.
Author Contributions
G.S.L. wrote the manuscript, helped in the conception of the paper, and contributed to reference collection. C.U. supervised the overall conceptualization, draft writing, and finalization of the paper. All authors have read and agreed to the published version of the manuscript.
Funding
The authors were supported by NIH grant AI140909 (CU).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Harrigan P.R., Whaley M., Montaner J.S. Rate of HIV-1 RNA rebound upon stopping antiretroviral therapy. AIDS. 1999;13:F59–F62. doi: 10.1097/00002030-199905280-00001. [DOI] [PubMed] [Google Scholar]
- 2.Antiretroviral Therapy Cohort Collaboration Survival of HIV-positive patients starting antiretroviral therapy between 1996 and 2013: A collaborative analysis of cohort studies. Lancet HIV. 2017;4:e349–e356. doi: 10.1016/S2352-3018(17)30066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson L.F., Mossong J., Dorrington R.E., Schomaker M., Hoffmann C.J., Keiser O., Fox M.P., Wood R., Prozesky H., Giddy J., et al. Life expectancies of South African adults starting antiretroviral treatment: Collaborative analysis of cohort studies. PLoS Med. 2013;10:e1001418. doi: 10.1371/journal.pmed.1001418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyidogan P., Anderson K.S. Current perspectives on HIV-1 antiretroviral drug resistance. Viruses. 2014;6:4095–4139. doi: 10.3390/v6104095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pham H.T., Mesplede T. The latest evidence for possible HIV-1 curative strategies. Drugs Context. 2018;7:212522. doi: 10.7573/dic.212522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richman D.D., Wrin T., Little S.J., Petropoulos C.J. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korber B., Gaschen B., Yusim K., Thakallapally R., Kesmir C., Detours V. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 2001;58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- 8.Forthal D.N., Landucci G., Gorny M.K., Zolla-Pazner S., Robinson W.E., Jr. Functional activities of 20 human immunodeficiency virus type 1 (HIV-1)-specific human monoclonal antibodies. AIDS Res. Hum. Retrovir. 1995;11:1095–1099. doi: 10.1089/aid.1995.11.1095. [DOI] [PubMed] [Google Scholar]
- 9.Seabright G.E., Doores K.J., Burton D.R., Crispin M. Protein and Glycan Mimicry in HIV Vaccine Design. J. Mol. Biol. 2019;431:2223–2247. doi: 10.1016/j.jmb.2019.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders R.W., Moore J.P. Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol. Rev. 2017;275:161–182. doi: 10.1111/imr.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Upadhyay C., Feyznezhad R., Cao L., Chan K.W., Liu K., Yang W., Zhang H., Yolitz J., Arthos J., Nadas A., et al. Signal peptide of HIV-1 envelope modulates glycosylation impacting exposure of V1V2 and other epitopes. PLoS Pathog. 2020;16:e1009185. doi: 10.1371/journal.ppat.1009185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starcich B.R., Hahn B.H., Shaw G.M., McNeely P.D., Modrow S., Wolf H., Parks E.S., Parks W.P., Josephs S.F., Gallo R.C., et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 13.Checkley M.A., Luttge B.G., Freed E.O. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J. Mol. Biol. 2011;410:582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore J.P., Willey R.L., Lewis G.K., Robinson J., Sodroski J. Immunological evidence for interactions between the first, second, and fifth conserved domains of the gp120 surface glycoprotein of human immunodeficiency virus type 1. J. Virol. 1994;68:6836–6847. doi: 10.1128/JVI.68.11.6836-6847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed E.O. HIV-1 replication. Somat. Cell Mol. Genet. 2001;26:13–33. doi: 10.1023/A:1021070512287. [DOI] [PubMed] [Google Scholar]
- 16.Rong R., Bibollet-Ruche F., Mulenga J., Allen S., Blackwell J.L., Derdeyn C.A. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J. Virol. 2007;81:1350–1359. doi: 10.1128/JVI.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan D.C., Kim P.S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/S0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q., Finzi A., Sodroski J. The Conformational States of the HIV-1 Envelope Glycoproteins. Trends Microbiol. 2020;28:655–667. doi: 10.1016/j.tim.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachmann M.F., Rohrer U.H., Kundig T.M., Burki K., Hengartner H., Zinkernagel R.M. The influence of antigen organization on B cell responsiveness. Science. 1993;262:1448–1451. doi: 10.1126/science.8248784. [DOI] [PubMed] [Google Scholar]
- 20.Zhu P., Liu J., Bess J., Jr., Chertova E., Lifson J.D., Grise H., Ofek G.A., Taylor K.A., Roux K.H. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 21.Pancera M., Wyatt R. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology. 2005;332:145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 22.Parren P.W., Burton D.R., Sattentau Q.J. HIV-1 antibody--debris or virion? Nat. Med. 1997;3:366–367. doi: 10.1038/nm0497-366d. [DOI] [PubMed] [Google Scholar]
- 23.Moore P.L., Crooks E.T., Porter L., Zhu P., Cayanan C.S., Grise H., Corcoran P., Zwick M.B., Franti M., Morris L., et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sattentau Q.J., Moore J.P. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 1995;182:185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders R.W., Derking R., Cupo A., Julien J.P., Yasmeen A., de Val N., Kim H.J., Blattner C., de la Pena A.T., Korzun J., et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gristick H.B., von Boehmer L., West A.P., Jr., Schamber M., Gazumyan A., Golijanin J., Seaman M.S., Fatkenheuer G., Klein F., Nussenzweig M.C., et al. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat. Struct. Mol. Biol. 2016;23:906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H., Barnes C.O., Yang Z., Nussenzweig M.C., Bjorkman P.J. Partially Open HIV-1 Envelope Structures Exhibit Conformational Changes Relevant for Coreceptor Binding and Fusion. Cell Host Microbe. 2018;24:579–592. doi: 10.1016/j.chom.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozorowski G., Pallesen J., de Val N., Lyumkis D., Cottrell C.A., Torres J.L., Copps J., Stanfield R.L., Cupo A., Pugach P., et al. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature. 2017;547:360–363. doi: 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lasky L.A., Groopman J.E., Fennie C.W., Benz P.M., Capon D.J., Dowbenko D.J., Nakamura G.R., Nunes W.M., Renz M.E., Berman P.W. Neutralization of the AIDS retrovirus by antibodies to a recombinant envelope glycoprotein. Science. 1986;233:209–212. doi: 10.1126/science.3014647. [DOI] [PubMed] [Google Scholar]
- 30.Pancera M., Zhou T., Druz A., Georgiev I.S., Soto C., Gorman J., Huang J., Acharya P., Chuang G.Y., Ofek G., et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang M., Gaschen B., Blay W., Foley B., Haigwood N., Kuiken C., Korber B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- 32.Leonard C.K., Spellman M.W., Riddle L., Harris R.J., Thomas J.N., Gregory T.J. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 1990;265:10373–10382. doi: 10.1016/S0021-9258(18)86956-3. [DOI] [PubMed] [Google Scholar]
- 33.Go E.P., Ding H., Zhang S., Ringe R.P., Nicely N., Hua D., Steinbock R.T., Golabek M., Alin J., Alam S.M., et al. Glycosylation Benchmark Profile for HIV-1 Envelope Glycoprotein Production Based on Eleven Env Trimers. J. Virol. 2017;91 doi: 10.1128/JVI.02428-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pritchard L.K., Spencer D.I., Royle L., Bonomelli C., Seabright G.E., Behrens A.J., Kulp D.W., Menis S., Krumm S.A., Dunlop D.C., et al. Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies. Nat. Commun. 2015;6:7479. doi: 10.1038/ncomms8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torrents de la Pena A., Rantalainen K., Cottrell C.A., Allen J.D., van Gils M.J., Torres J.L., Crispin M., Sanders R.W., Ward A.B. Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics. PLoS Pathog. 2019;15:e1007920. doi: 10.1371/journal.ppat.1007920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao L., Diedrich J.K., Kulp D.W., Pauthner M., He L., Park S.R., Sok D., Su C.Y., Delahunty C.M., Menis S., et al. Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat. Commun. 2017;8:14954. doi: 10.1038/ncomms14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao L., Pauthner M., Andrabi R., Rantalainen K., Berndsen Z., Diedrich J.K., Menis S., Sok D., Bastidas R., Park S.R., et al. Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer. Nat. Commun. 2018;9:3693. doi: 10.1038/s41467-018-06121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struwe W.B., Chertova E., Allen J.D., Seabright G.E., Watanabe Y., Harvey D.J., Medina-Ramirez M., Roser J.D., Smith R., Westcott D., et al. Site-Specific Glycosylation of Virion-Derived HIV-1 Env Is Mimicked by a Soluble Trimeric Immunogen. Cell Rep. 2018;24:1958–1966. doi: 10.1016/j.celrep.2018.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonomelli C., Doores K.J., Dunlop D.C., Thaney V., Dwek R.A., Burton D.R., Crispin M., Scanlan C.N. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS ONE. 2011;6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Behrens A.J., Struwe W.B., Crispin M. Glycosylation profiling to evaluate glycoprotein immunogens against HIV-1. Expert Rev. Proteom. 2017;14:881–890. doi: 10.1080/14789450.2017.1376658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong L., Sheppard N.C., Stewart-Jones G.B.E., Robson C.L., Chen H., Xu X., Krashias G., Bonomelli C., Scanlan C.N., Kwong P.D., et al. Expression-system-dependent modulation of HIV-1 envelope glycoprotein antigenicity and immunogenicity. J. Mol. Biol. 2010;403:131–147. doi: 10.1016/j.jmb.2010.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raska M., Takahashi K., Czernekova L., Zachova K., Hall S., Moldoveanu Z., Elliott M.C., Wilson L., Brown R., Jancova D., et al. Glycosylation patterns of HIV-1 gp120 depend on the type of expressing cells and affect antibody recognition. J. Biol. Chem. 2010;285:20860–20869. doi: 10.1074/jbc.M109.085472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doores K.J., Bonomelli C., Harvey D.J., Vasiljevic S., Dwek R.A., Burton D.R., Crispin M., Scanlan C.N. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc. Natl. Acad. Sci. USA. 2010;107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Binley J.M., Ban Y.E., Crooks E.T., Eggink D., Osawa K., Schief W.R., Sanders R.W. Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. J. Virol. 2010;84:5637–5655. doi: 10.1128/JVI.00105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jan M., Upadhyay C., Sharma A., Hioe C.E., Arora S.K. Short Communication: Manalpha1-2Man-Binding Anti-HIV Lectins Enhance the Exposure of V2i and V3 Crown Neutralization Epitopes on the V1/V2 and V3 Hypervariable Loops of HIV-1 Envelope. AIDS Res. Hum. Retrovir. 2017;33:941–945. doi: 10.1089/aid.2016.0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Go E.P., Herschhorn A., Gu C., Castillo-Menendez L., Zhang S., Mao Y., Chen H., Ding H., Wakefield J.K., Hua D., et al. Comparative Analysis of the Glycosylation Profiles of Membrane-Anchored HIV-1 Envelope Glycoprotein Trimers and Soluble gp140. J. Virol. 2015;89:8245–8257. doi: 10.1128/JVI.00628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Go E.P., Liao H.X., Alam S.M., Hua D., Haynes B.F., Desaire H. Characterization of host-cell line specific glycosylation profiles of early transmitted/founder HIV-1 gp120 envelope proteins. J. Proteome Res. 2013;12:1223–1234. doi: 10.1021/pr300870t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Behrens A.J., Vasiljevic S., Pritchard L.K., Harvey D.J., Andev R.S., Krumm S.A., Struwe W.B., Cupo A., Kumar A., Zitzmann N., et al. Composition and Antigenic Effects of Individual Glycan Sites of a Trimeric HIV-1 Envelope Glycoprotein. Cell Rep. 2016;14:2695–2706. doi: 10.1016/j.celrep.2016.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jan M., Upadhyay C., Hioe C.E. HIV-1 Envelope Glycan Composition as a Key Determinant of Efficient Virus Transmission via DC-SIGN and Resistance to Inhibitory Lectins. iScience. 2019;21:413–427. doi: 10.1016/j.isci.2019.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McLellan J.S., Pancera M., Carrico C., Gorman J., Julien J.P., Khayat R., Louder R., Pejchal R., Sastry M., Dai K., et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagh K., Kreider E.F., Li Y., Barbian H.J., Learn G.H., Giorgi E., Hraber P.T., Decker T.G., Smith A.G., Gondim M.V., et al. Completeness of HIV-1 Envelope Glycan Shield at Transmission Determines Neutralization Breadth. Cell Rep. 2018;25:893–908.e7. doi: 10.1016/j.celrep.2018.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang Y., Guttman M., Williams J.A., Verkerke H., Alvarado D., Hu S.L., Lee K.K. Changes in Structure and Antigenicity of HIV-1 Env Trimers Resulting from Removal of a Conserved CD4 Binding Site-Proximal Glycan. J. Virol. 2016;90:9224–9236. doi: 10.1128/JVI.01116-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wardemann H., Yurasov S., Schaefer A., Young J.W., Meffre E., Nussenzweig M.C. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 54.Haynes B.F., Verkoczy L. AIDS/HIV. Host controls of HIV neutralizing antibodies. Science. 2014;344:588–589. doi: 10.1126/science.1254990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cohen M. Notable Aspects of Glycan-Protein Interactions. Biomolecules. 2015;5:2056–2072. doi: 10.3390/biom5032056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crispin M., Bowden T.A. Antibodies expose multiple weaknesses in the glycan shield of HIV. Nat. Struct. Mol. Biol. 2013;20:771–772. doi: 10.1038/nsmb.2627. [DOI] [PubMed] [Google Scholar]
- 57.Doores K.J., Burton D.R. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J. Virol. 2010;84:10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steichen J.M., Kulp D.W., Tokatlian T., Escolano A., Dosenovic P., Stanfield R.L., McCoy L.E., Ozorowski G., Hu X., Kalyuzhniy O., et al. HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies. Immunity. 2016;45:483–496. doi: 10.1016/j.immuni.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doores K.J., Kong L., Krumm S.A., Le K.M., Sok D., Laserson U., Garces F., Poignard P., Wilson I.A., Burton D.R. Two classes of broadly neutralizing antibodies within a single lineage directed to the high-mannose patch of HIV envelope. J. Virol. 2015;89:1105–1118. doi: 10.1128/JVI.02905-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gray E.S., Madiga M.C., Hermanus T., Moore P.L., Wibmer C.K., Tumba N.L., Werner L., Mlisana K., Sibeko S., Williamson C., et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blazkova J., Refsland E.W., Clarridge K.E., Shi V., Justement J.S., Huiting E.D., Gittens K.R., Chen X., Schmidt S.D., Liu C., et al. Glycan-dependent HIV-specific neutralizing antibodies bind to cells of uninfected individuals. J. Clin. Investig. 2019;129:4832–4837. doi: 10.1172/JCI125955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hess R., Storcksdieck Genannt Bonsmann M., Lapuente D., Maaske A., Kirschning C., Ruland J., Lepenies B., Hannaman D., Tenbusch M., Uberla K. Glycosylation of HIV Env Impacts IgG Subtype Responses to Vaccination. Viruses. 2019;11:153. doi: 10.3390/v11020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moore P.L., Gray E.S., Wibmer C.K., Bhiman J.N., Nonyane M., Sheward D.J., Hermanus T., Bajimaya S., Tumba N.L., Abrahams M.R., et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat. Med. 2012;18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Freund N.T., Wang H., Scharf L., Nogueira L., Horwitz J.A., Bar-On Y., Golijanin J., Sievers S.A., Sok D., Cai H., et al. Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dacheux L., Moreau A., Ataman-Onal Y., Biron F., Verrier B., Barin F. Evolutionary dynamics of the glycan shield of the human immunodeficiency virus envelope during natural infection and implications for exposure of the 2G12 epitope. J. Virol. 2004;78:12625–12637. doi: 10.1128/JVI.78.22.12625-12637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian J., Lopez C.A., Derdeyn C.A., Jones M.S., Pinter A., Korber B., Gnanakaran S. Effect of Glycosylation on an Immunodominant Region in the V1V2 Variable Domain of the HIV-1 Envelope gp120 Protein. PLoS Comput. Biol. 2016;12:e1005094. doi: 10.1371/journal.pcbi.1005094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stewart-Jones G.B., Soto C., Lemmin T., Chuang G.Y., Druz A., Kong R., Thomas P.V., Wagh K., Zhou T., Behrens A.J., et al. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei X., Decker J.M., Wang S., Hui H., Kappes J.C., Wu X., Salazar-Gonzalez J.F., Salazar M.G., Kilby J.M., Saag M.S., et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 69.Lemmin T., Soto C., Stuckey J., Kwong P.D. Microsecond Dynamics and Network Analysis of the HIV-1 SOSIP Env Trimer Reveal Collective Behavior and Conserved Microdomains of the Glycan Shield. Structure. 2017;25:1631–1639. doi: 10.1016/j.str.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 70.Yang M., Huang J., Simon R., Wang L.X., MacKerell A.D., Jr. Conformational Heterogeneity of the HIV Envelope Glycan Shield. Sci. Rep. 2017;7:4435. doi: 10.1038/s41598-017-04532-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rudd P.M., Dwek R.A. Glycosylation: Heterogeneity and the 3D structure of proteins. Crit. Rev. Biochem. Mol. Biol. 1997;32:1–100. doi: 10.3109/10409239709085144. [DOI] [PubMed] [Google Scholar]
- 72.Keele B.F., Giorgi E.E., Salazar-Gonzalez J.F., Decker J.M., Pham K.T., Salazar M.G., Sun C., Grayson T., Wang S., Li H., et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shaw G.M., Hunter E. HIV transmission. Cold Spring Harb Perspect. Med. 2012;2 doi: 10.1101/cshperspect.a006965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wawer M.J., Gray R.H., Sewankambo N.K., Serwadda D., Li X., Laeyendecker O., Kiwanuka N., Kigozi G., Kiddugavu M., Lutalo T., et al. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J. Infect. Dis. 2005;191:1403–1409. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- 75.Joseph S.B., Swanstrom R., Kashuba A.D., Cohen M.S. Bottlenecks in HIV-1 transmission: Insights from the study of founder viruses. Nat. Rev. Microbiol. 2015;13:414–425. doi: 10.1038/nrmicro3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kariuki S.M., Selhorst P., Arien K.K., Dorfman J.R. The HIV-1 transmission bottleneck. Retrovirology. 2017;14:22. doi: 10.1186/s12977-017-0343-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frost S.D., Liu Y., Pond S.L., Chappey C., Wrin T., Petropoulos C.J., Little S.J., Richman D.D. Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J. Virol. 2005;79:6523–6527. doi: 10.1128/JVI.79.10.6523-6527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bar K.J., Li H., Chamberland A., Tremblay C., Routy J.P., Grayson T., Sun C., Wang S., Learn G.H., Morgan C.J., et al. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J. Virol. 2010;84:6241–6247. doi: 10.1128/JVI.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chohan B., Lang D., Sagar M., Korber B., Lavreys L., Richardson B., Overbaugh J. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J. Virol. 2005;79:6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Y., Curlin M.E., Diem K., Zhao H., Ghosh A.K., Zhu H., Woodward A.S., Maenza J., Stevens C.E., Stekler J., et al. Env length and N-linked glycosylation following transmission of human immunodeficiency virus Type 1 subtype B viruses. Virology. 2008;374:229–233. doi: 10.1016/j.virol.2008.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Finzi A., Pacheco B., Xiang S.H., Pancera M., Herschhorn A., Wang L., Zeng X., Desormeaux A., Kwong P.D., Sodroski J. Lineage-specific differences between human and simian immunodeficiency virus regulation of gp120 trimer association and CD4 binding. J. Virol. 2012;86:8974–8986. doi: 10.1128/JVI.01076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gnanakaran S., Bhattacharya T., Daniels M., Keele B.F., Hraber P.T., Lapedes A.S., Shen T., Gaschen B., Krishnamoorthy M., Li H., et al. Recurrent signature patterns in HIV-1 B clade envelope glycoproteins associated with either early or chronic infections. PLoS Pathog. 2011;7:e1002209. doi: 10.1371/journal.ppat.1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Helseth E., Olshevsky U., Furman C., Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J. Virol. 1991;65:2119–2123. doi: 10.1128/JVI.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Derdeyn C.A., Decker J.M., Bibollet-Ruche F., Mokili J.L., Muldoon M., Denham S.A., Heil M.L., Kasolo F., Musonda R., Hahn B.H., et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 85.Kafando A., Martineau C., El-Far M., Fournier E., Doualla-Bell F., Serhir B., Kazienga A., Sangare M.N., Sylla M., Chamberland A., et al. HIV-1 Envelope Glycoprotein Amino Acids Signatures Associated with Clade B Transmitted/Founder and Recent Viruses. Viruses. 2019;11:1012. doi: 10.3390/v11111012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baalwa J., Wang S., Parrish N.F., Decker J.M., Keele B.F., Learn G.H., Yue L., Ruzagira E., Ssemwanga D., Kamali A., et al. Molecular identification, cloning and characterization of transmitted/founder HIV-1 subtype A, D and A/D infectious molecular clones. Virology. 2013;436:33–48. doi: 10.1016/j.virol.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.De Wolf F., Hogervorst E., Goudsmit J., Fenyo E.M., Rubsamen-Waigmann H., Holmes H., Galvao-Castro B., Karita E., Wasi C., Sempala S.D., et al. Syncytium-inducing and non-syncytium-inducing capacity of human immunodeficiency virus type 1 subtypes other than B: Phenotypic and genotypic characteristics. WHO Network for HIV Isolation and Characterization. AIDS Res. Hum. Retrovir. 1994;10:1387–1400. doi: 10.1089/aid.1994.10.1387. [DOI] [PubMed] [Google Scholar]
- 88.Wilen C.B., Parrish N.F., Pfaff J.M., Decker J.M., Henning E.A., Haim H., Petersen J.E., Wojcechowskyj J.A., Sodroski J., Haynes B.F., et al. Phenotypic and immunologic comparison of clade B transmitted/founder and chronic HIV-1 envelope glycoproteins. J. Virol. 2011;85:8514–8527. doi: 10.1128/JVI.00736-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chandramouli B., Chillemi G., Abbate I., Capobianchi M.R., Rozera G., Desideri A. Importance of V3 loop flexibility and net charge in the context of co-receptor recognition. A molecular dynamics study on HIV gp120. J. Biomol. Struct. Dyn. 2012;29:879–891. doi: 10.1080/07391102.2012.10507416. [DOI] [PubMed] [Google Scholar]
- 90.Kaleebu P., Nankya I.L., Yirrell D.L., Shafer L.A., Kyosiimire-Lugemwa J., Lule D.B., Morgan D., Beddows S., Weber J., Whitworth J.A. Relation between chemokine receptor use, disease stage, and HIV-1 subtypes A and D: Results from a rural Ugandan cohort. J. Acquir. Immune Defic. Syndr. 2007;45:28–33. doi: 10.1097/QAI.0b013e3180385aa0. [DOI] [PubMed] [Google Scholar]
- 91.Zolla-Pazner S., Cardozo T. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat. Rev. Immunol. 2010;10:527–535. doi: 10.1038/nri2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pejchal R., Doores K.J., Walker L.M., Khayat R., Huang P.S., Wang S.K., Stanfield R.L., Julien J.P., Ramos A., Crispin M., et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou T., Georgiev I., Wu X., Yang Z.Y., Dai K., Finzi A., Kwon Y.D., Scheid J.F., Shi W., Xu L., et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McNearney T., Hornickova Z., Markham R., Birdwell A., Arens M., Saah A., Ratner L. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc. Natl. Acad. Sci. USA. 1992;89:10247–10251. doi: 10.1073/pnas.89.21.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bar K.J., Tsao C.Y., Iyer S.S., Decker J.M., Yang Y., Bonsignori M., Chen X., Hwang K.K., Montefiori D.C., Liao H.X., et al. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog. 2012;8:e1002721. doi: 10.1371/journal.ppat.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moore P.L., Gray E.S., Morris L. Specificity of the autologous neutralizing antibody response. Curr. Opin. HIV AIDS. 2009;4:358–363. doi: 10.1097/COH.0b013e32832ea7e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bonsignori M., Kreider E.F., Fera D., Meyerhoff R.R., Bradley T., Wiehe K., Alam S.M., Aussedat B., Walkowicz W.E., Hwang K.K., et al. Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aai7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Montfort T., Eggink D., Boot M., Tuen M., Hioe C.E., Berkhout B., Sanders R.W. HIV-1 N-glycan composition governs a balance between dendritic cell-mediated viral transmission and antigen presentation. J. Immunol. 2011;187:4676–4685. doi: 10.4049/jimmunol.1101876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gringhuis S.I., den Dunnen J., Litjens M., van der Vlist M., Geijtenbeek T.B. Carbohydrate-specific signaling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori. Nat. Immunol. 2009;10:1081–1088. doi: 10.1038/ni.1778. [DOI] [PubMed] [Google Scholar]
- 100.Jan M., Arora S.K. Innate Sensing of HIV-1 by Dendritic Cell-Specific ICAM-3 Grabbing Nonintegrin on Dendritic Cells: Degradation and Presentation Versus Transmission of Virus to T Cells Is Determined by Glycan Composition of Viral Envelope. AIDS Res. Hum. Retrovir. 2017;33:765–767. doi: 10.1089/aid.2016.0290. [DOI] [PubMed] [Google Scholar]
- 101.van Liempt E., Bank C.M., Mehta P., Garcia-Vallejo J.J., Kawar Z.S., Geyer R., Alvarez R.A., Cummings R.D., Kooyk Y., van Die I. Specificity of DC-SIGN for mannose- and fucose-containing glycans. FEBS Lett. 2006;580:6123–6131. doi: 10.1016/j.febslet.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 102.Geijtenbeek T.B., Kwon D.S., Torensma R., van Vliet S.J., van Duijnhoven G.C., Middel J., Cornelissen I.L., Nottet H.S., KewalRamani V.N., Littman D.R., et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/S0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 103.Yolitz J., Schwing C., Chang J., Van Ryk D., Nawaz F., Wei D., Cicala C., Arthos J., Fauci A.S. Signal peptide of HIV envelope protein impacts glycosylation and antigenicity of gp120. Proc. Natl. Acad. Sci. USA. 2018;115:2443–2448. doi: 10.1073/pnas.1722627115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.von Heijne G. Signal sequences. The limits of variation. J. Mol. Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 105.Li Y., Luo L., Thomas D.Y., Kang C.Y. Control of expression, glycosylation, and secretion of HIV-1 gp120 by homologous and heterologous signal sequences. Virology. 1994;204:266–278. doi: 10.1006/viro.1994.1531. [DOI] [PubMed] [Google Scholar]
- 106.Li Y., Bergeron J.J., Luo L., Ou W.J., Thomas D.Y., Kang C.Y. Effects of inefficient cleavage of the signal sequence of HIV-1 gp 120 on its association with calnexin, folding, and intracellular transport. Proc. Natl. Acad. Sci. USA. 1996;93:9606–9611. doi: 10.1073/pnas.93.18.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li Y., Luo L., Rasool N., Kang C.Y. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J. Virol. 1993;67:584–588. doi: 10.1128/JVI.67.1.584-588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Land A., Braakman I. Folding of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum. Biochimie. 2001;83:783–790. doi: 10.1016/S0300-9084(01)01314-1. [DOI] [PubMed] [Google Scholar]
- 109.Owji H., Nezafat N., Negahdaripour M., Hajiebrahimi A., Ghasemi Y. A comprehensive review of signal peptides: Structure, roles, and applications. Eur. J. Cell Biol. 2018;97:422–441. doi: 10.1016/j.ejcb.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 110.Broome-Smith J.K., Gnaneshan S., Hunt L.A., Mehraein-Ghomi F., Hashemzadeh-Bonehi L., Tadayyon M., Hennessey E.S. Cleavable signal peptides are rarely found in bacterial cytoplasmic membrane proteins (review) Mol. Membr. Biol. 1994;11:3–8. doi: 10.3109/09687689409161023. [DOI] [PubMed] [Google Scholar]
- 111.Snapp E.L., McCaul N., Quandte M., Cabartova Z., Bontjer I., Kallgren C., Nilsson I., Land A., von Heijne G., Sanders R.W., et al. Structure and topology around the cleavage site regulate post-translational cleavage of the HIV-1 gp160 signal peptide. Elife. 2017;6 doi: 10.7554/eLife.26067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Land A., Zonneveld D., Braakman I. Folding of HIV-1 envelope glycoprotein involves extensive isomerization of disulfide bonds and conformation-dependent leader peptide cleavage. FASEB J. 2003;17:1058–1067. doi: 10.1096/fj.02-0811com. [DOI] [PubMed] [Google Scholar]
- 113.Li Y., Luo L., Thomas D.Y., Kang C.Y. The HIV-1 Env protein signal sequence retards its cleavage and down-regulates the glycoprotein folding. Virology. 2000;272:417–428. doi: 10.1006/viro.2000.0357. [DOI] [PubMed] [Google Scholar]
- 114.Hiss J.A., Schneider G. Architecture, function and prediction of long signal peptides. Brief. Bioinform. 2009;10:569–578. doi: 10.1093/bib/bbp030. [DOI] [PubMed] [Google Scholar]
- 115.Asmal M., Hellmann I., Liu W., Keele B.F., Perelson A.S., Bhattacharya T., Gnanakaran S., Daniels M., Haynes B.F., Korber B.T., et al. A signature in HIV-1 envelope leader peptide associated with transition from acute to chronic infection impacts envelope processing and infectivity. PLoS ONE. 2011;6:e23673. doi: 10.1371/journal.pone.0023673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rapoport T.A., Li L., Park E. Structural and Mechanistic Insights into Protein Translocation. Annu. Rev. Cell Dev. Biol. 2017;33:369–390. doi: 10.1146/annurev-cellbio-100616-060439. [DOI] [PubMed] [Google Scholar]
- 117.von Heijne G. The signal peptide. J. Membr. Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 118.da Silva J.X., Franco O.L., Lemos M.A., Gondim M.V., Prosdocimi F., Arganaraz E.R. Sequence variations of Env signal peptide alleles in different clinical stages of HIV infection. Peptides. 2011;32:1800–1806. doi: 10.1016/j.peptides.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 119.Yue L., Pfafferott K.J., Baalwa J., Conrod K., Dong C.C., Chui C., Rong R., Claiborne D.T., Prince J.L., Tang J., et al. Transmitted virus fitness and host T cell responses collectively define divergent infection outcomes in two HIV-1 recipients. PLoS Pathog. 2015;11:e1004565. doi: 10.1371/journal.ppat.1004565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sagar M., Laeyendecker O., Lee S., Gamiel J., Wawer M.J., Gray R.H., Serwadda D., Sewankambo N.K., Shepherd J.C., Toma J., et al. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J. Infect. Dis. 2009;199:580–589. doi: 10.1086/596557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Upadhyay C., Feyznezhad R., Yang W., Zhang H., Zolla-Pazner S., Hioe C.E. Alterations of HIV-1 envelope phenotype and antibody-mediated neutralization by signal peptide mutations. PLoS Pathog. 2018;14:e1006812. doi: 10.1371/journal.ppat.1006812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pfeiffer T., Pisch T., Devitt G., Holtkotte D., Bosch V. Effects of signal peptide exchange on HIV-1 glycoprotein expression and viral infectivity in mammalian cells. FEBS Lett. 2006;580:3775–3778. doi: 10.1016/j.febslet.2006.05.070. [DOI] [PubMed] [Google Scholar]
- 123.Gonzalez M.W., DeVico A.L., Lewis G.K., Spouge J.L. Conserved molecular signatures in gp120 are associated with the genetic bottleneck during simian immunodeficiency virus (SIV), SIV-human immunodeficiency virus (SHIV), and HIV type 1 (HIV-1) transmission. J. Virol. 2015;89:3619–3629. doi: 10.1128/JVI.03235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Powell R.L.R., Totrov M., Itri V., Liu X., Fox A., Zolla-Pazner S. Plasticity and Epitope Exposure of the HIV-1 Envelope Trimer. J. Virol. 2017;91 doi: 10.1128/JVI.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zolla-Pazner S., Cohen S.S., Boyd D., Kong X.P., Seaman M., Nussenzweig M., Klein F., Overbaugh J., Totrov M. Structure/Function Studies Involving the V3 Region of the HIV-1 Envelope Delineate Multiple Factors That Affect Neutralization Sensitivity. J. Virol. 2016;90:636–649. doi: 10.1128/JVI.01645-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Berman P.W., Nunes W.M., Haffar O.K. Expression of membrane-associated and secreted variants of gp160 of human immunodeficiency virus type 1 in vitro and in continuous cell lines. J. Virol. 1988;62:3135–3142. doi: 10.1128/JVI.62.9.3135-3142.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Culp J.S., Johansen H., Hellmig B., Beck J., Matthews T.J., Delers A., Rosenberg M. Regulated expression allows high level production and secretion of HIV-1 gp120 envelope glycoprotein in Drosophila Schneider cells. Biotechnology (N Y) 1991;9:173–177. doi: 10.1038/nbt0291-173. [DOI] [PubMed] [Google Scholar]
- 128.Binley J.M., Sanders R.W., Clas B., Schuelke N., Master A., Guo Y., Kajumo F., Anselma D.J., Maddon P.J., Olson W.C., et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 2000;74:627–643. doi: 10.1128/JVI.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Herrera A.M., Musacchio A., Fernandez J.R., Duarte C.A. Efficiency of erythropoietin’s signal peptide for HIV(MN)-1 gp 120 expression. Biochem. Biophys. Res. Commun. 2000;273:557–559. doi: 10.1006/bbrc.2000.2974. [DOI] [PubMed] [Google Scholar]
- 130.Rantalainen K., Berndsen Z.T., Murrell S., Cao L., Omorodion O., Torres J.L., Wu M., Umotoy J., Copps J., Poignard P., et al. Co-evolution of HIV Envelope and Apex-Targeting Neutralizing Antibody Lineage Provides Benchmarks for Vaccine Design. Cell Rep. 2018;23:3249–3261. doi: 10.1016/j.celrep.2018.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Willey R.L., Bonifacino J.S., Potts B.J., Martin M.A., Klausner R.D. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. USA. 1988;85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alam S.M., Liao H.X., Tomaras G.D., Bonsignori M., Tsao C.Y., Hwang K.K., Chen H., Lloyd K.E., Bowman C., Sutherland L., et al. Antigenicity and immunogenicity of RV144 vaccine AIDSVAX clade E envelope immunogen is enhanced by a gp120 N-terminal deletion. J. Virol. 2013;87:1554–1568. doi: 10.1128/JVI.00718-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pitisuttithum P., Rerks-Ngarm S., Bussaratid V., Dhitavat J., Maekanantawat W., Pungpak S., Suntharasamai P., Vanijanonta S., Nitayapan S., Kaewkungwal J., et al. Safety and reactogenicity of canarypox ALVAC-HIV (vCP1521) and HIV-1 gp120 AIDSVAX B/E vaccination in an efficacy trial in Thailand. PLoS ONE. 2011;6:e27837. doi: 10.1371/journal.pone.0027837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rerks-Ngarm S., Pitisuttithum P., Nitayaphan S., Kaewkungwal J., Chiu J., Paris R., Premsri N., Namwat C., de Souza M., Adams E., et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 135.Nitayaphan S., Pitisuttithum P., Karnasuta C., Eamsila C., de Souza M., Morgan P., Polonis V., Benenson M., VanCott T., Ratto-Kim S., et al. Safety and immunogenicity of an HIV subtype B and E prime-boost vaccine combination in HIV-negative Thai adults. J. Infect. Dis. 2004;190:702–706. doi: 10.1086/422258. [DOI] [PubMed] [Google Scholar]
- 136.Bekker L.G., Moodie Z., Grunenberg N., Laher F., Tomaras G.D., Cohen K.W., Allen M., Malahleha M., Mngadi K., Daniels B., et al. Subtype C ALVAC-HIV and bivalent subtype C gp120/MF59 HIV-1 vaccine in low-risk, HIV-uninfected, South African adults: A phase 1/2 trial. Lancet HIV. 2018;5:e366–e378. doi: 10.1016/S2352-3018(18)30071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gilbert P.B., Berger J.O., Stablein D., Becker S., Essex M., Hammer S.M., Kim J.H., Degruttola V.G. Statistical interpretation of the RV144 HIV vaccine efficacy trial in Thailand: A case study for statistical issues in efficacy trials. J. Infect. Dis. 2011;203:969–975. doi: 10.1093/infdis/jiq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cohen J. HIV/AIDS research. Beyond Thailand: Making sense of a qualified AIDS vaccine “success”. Science. 2009;326:652–653. doi: 10.1126/science.326_652. [DOI] [PubMed] [Google Scholar]
- 139.Desrosiers R.C. Protection against HIV Acquisition in the RV144 Trial. J. Virol. 2017;91 doi: 10.1128/JVI.00905-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zolla-Pazner S., Gilbert P.B. Revisiting the Correlate of Reduced HIV Infection Risk in the Rv144 Vaccine Trial. J. Virol. 2019;93 doi: 10.1128/JVI.00629-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rerks-Ngarm S., Pitisuttithum P., Excler J.L., Nitayaphan S., Kaewkungwal J., Premsri N., Kunasol P., Karasavvas N., Schuetz A., Ngauy V., et al. Randomized, Double-Blind Evaluation of Late Boost Strategies for HIV-Uninfected Vaccine Recipients in the RV144 HIV Vaccine Efficacy Trial. J. Infect. Dis. 2017;215:1255–1263. doi: 10.1093/infdis/jix099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Karnasuta C., Paris R.M., Cox J.H., Nitayaphan S., Pitisuttithum P., Thongcharoen P., Brown A.E., Gurunathan S., Tartaglia J., Heyward W.L., et al. Antibody-dependent cell-mediated cytotoxic responses in participants enrolled in a phase I/II ALVAC-HIV/AIDSVAX B/E prime-boost HIV-1 vaccine trial in Thailand. Vaccine. 2005;23:2522–2529. doi: 10.1016/j.vaccine.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 143.Karasavvas N., Billings E., Rao M., Williams C., Zolla-Pazner S., Bailer R.T., Koup R.A., Madnote S., Arworn D., Shen X., et al. The Thai Phase III HIV Type 1 Vaccine trial (RV144) regimen induces antibodies that target conserved regions within the V2 loop of gp120. AIDS Res. Hum. Retrovir. 2012;28:1444–1457. doi: 10.1089/aid.2012.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pollara J., Bonsignori M., Moody M.A., Liu P., Alam S.M., Hwang K.K., Gurley T.C., Kozink D.M., Armand L.C., Marshall D.J., et al. HIV-1 vaccine-induced C1 and V2 Env-specific antibodies synergize for increased antiviral activities. J. Virol. 2014;88:7715–7726. doi: 10.1128/JVI.00156-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Haynes B.F., Gilbert P.B., McElrath M.J., Zolla-Pazner S., Tomaras G.D., Alam S.M., Evans D.T., Montefiori D.C., Karnasuta C., Sutthent R., et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Laher F., Moodie Z., Cohen K.W., Grunenberg N., Bekker L.G., Allen M., Frahm N., Yates N.L., Morris L., Malahleha M., et al. Safety and immune responses after a 12-month booster in healthy HIV-uninfected adults in HVTN 100 in South Africa: A randomized double-blind placebo-controlled trial of ALVAC-HIV (vCP2438) and bivalent subtype C gp120/MF59 vaccines. PLoS Med. 2020;17:e1003038. doi: 10.1371/journal.pmed.1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mascola J.R., Stiegler G., VanCott T.C., Katinger H., Carpenter C.B., Hanson C.E., Beary H., Hayes D., Frankel S.S., Birx D.L., et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 148.Veselinovic M., Neff C.P., Mulder L.R., Akkina R. Topical gel formulation of broadly neutralizing anti-HIV-1 monoclonal antibody VRC01 confers protection against HIV-1 vaginal challenge in a humanized mouse model. Virology. 2012;432:505–510. doi: 10.1016/j.virol.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Saunders K.O., Pegu A., Georgiev I.S., Zeng M., Joyce M.G., Yang Z.Y., Ko S.Y., Chen X., Schmidt S.D., Haase A.T., et al. Sustained Delivery of a Broadly Neutralizing Antibody in Nonhuman Primates Confers Long-Term Protection against Simian/Human Immunodeficiency Virus Infection. J. Virol. 2015;89:5895–5903. doi: 10.1128/JVI.00210-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Parren P.W., Ditzel H.J., Gulizia R.J., Binley J.M., Barbas C.F., 3rd, Burton D.R., Mosier D.E. Protection against HIV-1 infection in hu-PBL-SCID mice by passive immunization with a neutralizing human monoclonal antibody against the gp120 CD4-binding site. AIDS. 1995;9:F1–F6. doi: 10.1097/00002030-199506000-00001. [DOI] [PubMed] [Google Scholar]
- 151.Hessell A.J., Poignard P., Hunter M., Hangartner L., Tehrani D.M., Bleeker W.K., Parren P.W., Marx P.A., Burton D.R. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 2009;15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Barouch D.H., Liu J., Li H., Maxfield L.F., Abbink P., Lynch D.M., Iampietro M.J., SanMiguel A., Seaman M.S., Ferrari G., et al. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012;482:89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Deruaz M., Moldt B., Le K.M., Power K.A., Vrbanac V.D., Tanno S., Ghebremichael M.S., Allen T.M., Tager A.M., Burton D.R., et al. Protection of Humanized Mice From Repeated Intravaginal HIV Challenge by Passive Immunization: A Model for Studying the Efficacy of Neutralizing Antibodies In Vivo. J. Infect. Dis. 2016;214:612–616. doi: 10.1093/infdis/jiw203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Julg B., Tartaglia L.J., Keele B.F., Wagh K., Pegu A., Sok D., Abbink P., Schmidt S.D., Wang K., Chen X., et al. Broadly neutralizing antibodies targeting the HIV-1 envelope V2 apex confer protection against a clade C SHIV challenge. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Baba T.W., Liska V., Hofmann-Lehmann R., Vlasak J., Xu W., Ayehunie S., Cavacini L.A., Posner M.R., Katinger H., Stiegler G., et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 2000;6:200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 156.Mascola J.R., Lewis M.G., Stiegler G., Harris D., VanCott T.C., Hayes D., Louder M.K., Brown C.R., Sapan C.V., Frankel S.S., et al. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 1999;73:4009–4018. doi: 10.1128/JVI.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sun M., Li Y., Yuan Z., Lu W., Kang G., Fan W., Li Q. VRC01 antibody protects against vaginal and rectal transmission of human immunodeficiency virus 1 in hu-BLT mice. Arch. Virol. 2016;161:2449–2455. doi: 10.1007/s00705-016-2942-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gauduin M.C., Parren P.W., Weir R., Barbas C.F., Burton D.R., Koup R.A. Passive immunization with a human monoclonal antibody protects hu-PBL-SCID mice against challenge by primary isolates of HIV-1. Nat. Med. 1997;3:1389–1393. doi: 10.1038/nm1297-1389. [DOI] [PubMed] [Google Scholar]
- 159.Shingai M., Donau O.K., Plishka R.J., Buckler-White A., Mascola J.R., Nabel G.J., Nason M.C., Montefiori D., Moldt B., Poignard P., et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J. Exp. Med. 2014;211:2061–2074. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shibata R., Igarashi T., Haigwood N., Buckler-White A., Ogert R., Ross W., Willey R., Cho M.W., Martin M.A. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 1999;5:204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 161.Julg B., Liu P.T., Wagh K., Fischer W.M., Abbink P., Mercado N.B., Whitney J.B., Nkolola J.P., McMahan K., Tartaglia L.J., et al. Protection against a mixed SHIV challenge by a broadly neutralizing antibody cocktail. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aao4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Barnett S.W., Burke B., Sun Y., Kan E., Legg H., Lian Y., Bost K., Zhou F., Goodsell A., Zur Megede J., et al. Antibody-mediated protection against mucosal simian-human immunodeficiency virus challenge of macaques immunized with alphavirus replicon particles and boosted with trimeric envelope glycoprotein in MF59 adjuvant. J. Virol. 2010;84:5975–5985. doi: 10.1128/JVI.02533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stoddart C.A., Galkina S.A., Joshi P., Kosikova G., Long B.R., Maidji E., Moreno M.E., Rivera J.M., Sanford U.R., Sloan B., et al. Efficacy of broadly neutralizing monoclonal antibody PG16 in HIV-infected humanized mice. Virology. 2014;462–463:115–125. doi: 10.1016/j.virol.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Parren P.W., Marx P.A., Hessell A.J., Luckay A., Harouse J., Cheng-Mayer C., Moore J.P., Burton D.R. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 2001;75:8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gautam R., Nishimura Y., Pegu A., Nason M.C., Klein F., Gazumyan A., Golijanin J., Buckler-White A., Sadjadpour R., Wang K., et al. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature. 2016;533:105–109. doi: 10.1038/nature17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pegu A., Yang Z.Y., Boyington J.C., Wu L., Ko S.Y., Schmidt S.D., McKee K., Kong W.P., Shi W., Chen X., et al. Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Sci. Transl. Med. 2014;6:243ra288. doi: 10.1126/scitranslmed.3008992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pietzsch J., Gruell H., Bournazos S., Donovan B.M., Klein F., Diskin R., Seaman M.S., Bjorkman P.J., Ravetch J.V., Ploss A., et al. A mouse model for HIV-1 entry. Proc. Natl. Acad. Sci. USA. 2012;109:15859–15864. doi: 10.1073/pnas.1213409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hessell A.J., Rakasz E.G., Tehrani D.M., Huber M., Weisgrau K.L., Landucci G., Forthal D.N., Koff W.C., Poignard P., Watkins D.I., et al. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J. Virol. 2010;84:1302–1313. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Liu Y., Cao W., Sun M., Li T. Broadly neutralizing antibodies for HIV-1: Efficacies, challenges and opportunities. Emerg Microbes Infect. 2020;9:194–206. doi: 10.1080/22221751.2020.1713707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Joyce J.G., Krauss I.J., Song H.C., Opalka D.W., Grimm K.M., Nahas D.D., Esser M.T., Hrin R., Feng M., Dudkin V.Y., et al. An oligosaccharide-based HIV-1 2G12 mimotope vaccine induces carbohydrate-specific antibodies that fail to neutralize HIV-1 virions. Proc. Natl. Acad. Sci. USA. 2008;105:15684–15689. doi: 10.1073/pnas.0807837105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Barouch D.H., Tomaka F.L., Wegmann F., Stieh D.J., Alter G., Robb M.L., Michael N.L., Peter L., Nkolola J.P., Borducchi E.N., et al. Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13-19) Lancet. 2018;392:232–243. doi: 10.1016/S0140-6736(18)31364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Stamatatos L., Pancera M., McGuire A.T. Germline-targeting immunogens. Immunol. Rev. 2017;275:203–216. doi: 10.1111/imr.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Escolano A., Steichen J.M., Dosenovic P., Kulp D.W., Golijanin J., Sok D., Freund N.T., Gitlin A.D., Oliveira T., Araki T., et al. Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice. Cell. 2016;166:1445–1458.e1412. doi: 10.1016/j.cell.2016.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.