Abstract
The prebiotic activity of a commercially available oat product and a novel oat ingredient, at similar β-glucan loads, was tested using a validated in vitro gut model (M-SHIME®). The novel oat ingredient was tested further at lower β-glucan loads in vitro, while the commercially available oat product was assessed in a randomised, single-blind, placebo-controlled, and cross-over human study. Both approaches focused on healthy individuals with mild hypercholesterolemia. In vitro analysis revealed that both oat products strongly stimulated Lactobacillaceae and Bifidobacteriaceae in the intestinal lumen and the simulated mucus layer, and corresponded with enhanced levels of acetate and lactate with cross-feeding interactions leading to an associated increase in propionate and butyrate production. The in vitro prebiotic activity of the novel oat ingredient remained at lower β-glucan levels, indicating the prebiotic potential of the novel oat product. Finally, the stimulation of Lactobacillus spp. was confirmed during the in vivo trial, where lactobacilli abundance significantly increased in the overall population at the end of the intervention period with the commercially available oat product relative to the control product, indicating the power of in vitro gut models in predicting in vivo response of the microbial community to dietary modulation.
Keywords: Bifidobacterium, Lactobacillus, beta-glucan, intestine, microbiota, in vitro–in vivo correlation
1. Introduction
Prebiotics are defined as non-digestible substrates that are selectively utilised by the gut microbiome, thereby conferring beneficial effects for the host [1]. Oat products contain several soluble components capable of exerting prebiotic properties, such as β-glucans, arabinoxylans, arabinogalactans, resistant starch, and polyphenolic compounds. However, many of the health-promoting effects related to the consumption of oat products have been attributed to the intake of β-glucans. These non-starch polysaccharides consist of β-linked chains of D-glucose monomers, differing in their branching structure, viscosity, solubility, and molecular weight [2], and they have been associated with a reduction of systemic cholesterol levels [3], regulation of blood glucose concentrations, improved weight management [4], and modulation of immune function [2].
In order to exert their health-related properties, β-glucans need to be released from the cell wall of the oat groats, a process which is highly affected by the applied processing techniques. Indeed, hydrothermal processing has been linked with reduced release of oat β-glucans [5]. Mechanical processing, on the other hand, results in increased extractability of β-glucans from the oat groats, probably by increasing the exposed surface area [6]. Recently, Van den Abbeele et al. [7] reported an association between the β-glucan content and the prebiotic potential of different oat ingredients that were produced by different mechanical processing techniques, suggesting that different processing techniques might impact the potential functional properties of the final oat ingredients by affecting β-glucan extractability.
Several human studies have explored the prebiotic activity of oat products [8,9,10,11]. Connolly et al. [11], for instance, investigated the prebiotic activity of whole-grain oat granola, containing 1.3g β -glucan per dose in a human population with mild to moderate hypercholesterolemia. It was shown that intake of whole-grain oat granola selectively enhanced the abundance of Bifidobacterium and Lactobacillus species and total bacterial population compared to baseline levels, accompanied by reduced systemic levels of total and LDL cholesterol. Indeed, numerous studies have linked the consumption of oat products with hypocholesterolaemic effects [12,13,14]. While their cholesterol-lowering activity has been associated with increased excretion of bile acids and cholesterol in faeces due to the enhancement of viscosity of the intestinal content [15], recent research suggests a potential role of the gut microbial community [16]. Indeed, modulation of the intestinal microbiota might impact bile acid metabolism by stimulating bacterial bile salt hydrolase activity of certain bacterial groups, thereby affecting cholesterol excretion [17,18]. Furthermore, microbial production of propionate has been associated with a reduction of cholesterol levels [19,20]. As several in vitro and in vivo studies have reported modulation of the gut microbiota upon oat supplementation [11,21,22], a potential mechanism of action of oat products in controlling cholesterol homeostasis might be related to its prebiotic properties.
Assessments of the prebiotic properties of dietary fibres are often performed during in vitro studies. In vitro approaches offer an appropriate alternative to clinical trials because interactions between the gut microbial community and dietary fibres can be investigated at the site of action by strict control of environmental parameters [23,24].
As the prebiotic properties of oat ingredients might be involved in the lowering of cholesterol levels, the initial aim of this study was to compare the prebiotic activity of a commercially available oat product and a novel oat ingredient, standardised to provide similar levels of β-glucan, in a model of the human gastrointestinal tract of different healthy individuals with elevated cholesterol levels, making use of the validated Mucosal Simulator of the Human Intestinal Microbial Ecosystem (M-SHIME®, ProDigest and Ghent University, Ghent, Belgium) [25], in order to analyse potential similarity in prebiotic response of the microbial community following oat consumption. Moreover, it was assessed if the potential prebiotic properties of the novel oat product remained at lower β-glucan levels in terms of effects on microbial metabolic activity and community composition of the luminal and mucosal gut microbiome in vitro. Finally, a clinical trial, with a corresponding human population, was carried out with the commercially available oat product in order to determine if in vitro observations translated in vivo to provide evidence that the use of in vitro gut models can be used to predict dietary outcomes following supplementation of novel ingredients in vivo.
2. Results
2.1. Altered Microbial Metabolic Activity in Response to Oat Treatment in vitro
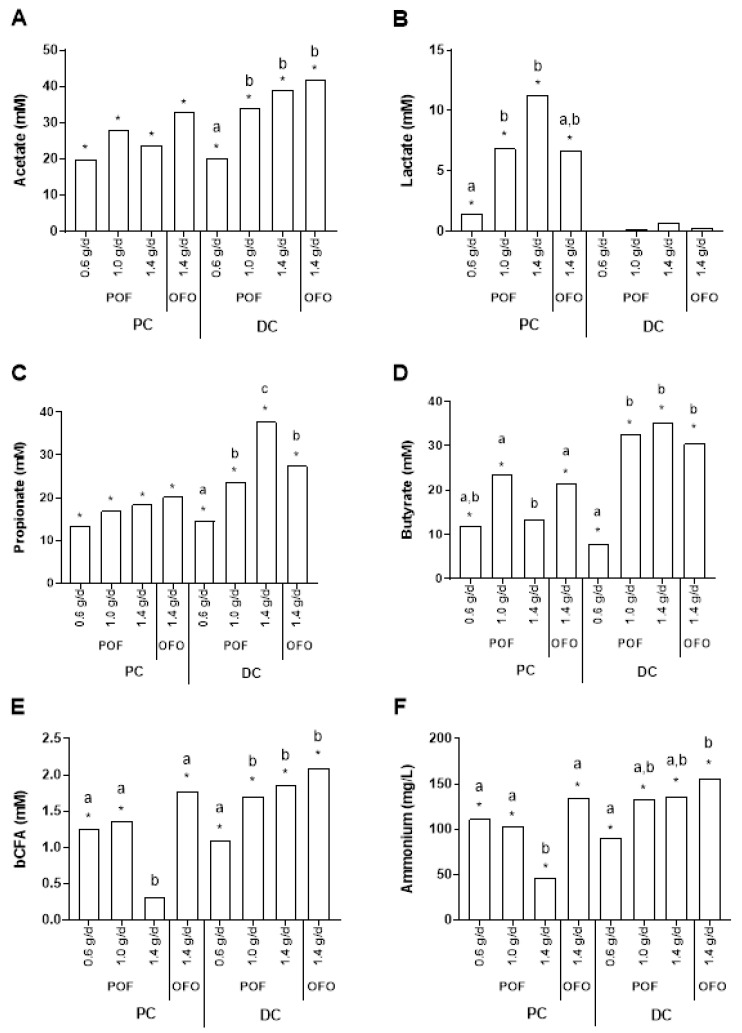
The short-chain fatty acids (SCFAs) detected consisted mainly of acetate, propionate, and butyrate (Figure 1) and trace amounts of branched-chain fatty acid (bCFA) (Figure 1). First, all test conditions significantly increased acetate (p < 0.0001), propionate (p < 0.0001), and butyrate (p < 0.0001) levels in both colon regions as compared to the control period. The only exception was noted in the proximal (PC) upon supplementation of pre-cooked oat flour (POF) at a concentration of 1.4 g β-glucan/day, where a marked, but not statistically significant (p = 0.211), increase in butyrate levels was still observed (+13.2 mM). The latter could be due to the fact that lactate most strongly accumulated (Figure 1) for this condition (i.e., an average increase in lactate levels of 11.3 mM). While acetate and propionate levels were not significantly different as a consequence of the supplementation of old-fashioned oats (OFO) relative to POF at a concentration of 1.4 g β-glucan/day in the PC, butyrate levels were significantly higher upon treatment with OFO in the PC (p < 0.0001), with an additional average increase of 8.3 mM upon OFO fermentation as compared to POF. In the DC, no significant differences in acetate and butyrate production were observed between supplementation of OFO and POF at a concentration of 1.4g β-glucan/day, whereas POF more strongly enhanced propionate production as compared to OFO (i.e., on average an additional increase of 10.2 mM) in this colonic region (p = 0.007). With respect to the different doses of POF, acetate, propionate, and butyrate levels increased in a dose-dependent manner in the distal colon (DC), with the highest final SCFA levels observed upon supplementation of the highest concentration of POF.
Figure 1.
Effects on primary microbial metabolites. The average increase in (A) acetate (mM), (B) lactate (mM), (C) propionate (mM), (D) butyrate (mM), and (E) branched-chain fatty acid (bCFA; mM) and (F) ammonium (mg/L) levels during the treatment period as compared to the control period upon treatment with old-fashioned oats (OFO) at a dose of 1.4 g/d and pre-cooked oat flour (POF) at a dose of 0.6 g/d, 1.0 g/d, and 1.4 g/d in the proximal (PC) and distal colon (DC) of the human gastrointestinal tract for three human donors tested. For optimal observation of consistent effects over the different donors tested, the average of the three donors is presented (n = 3). Statistically significant differences relative to the control period are indicated with *, whereas statistically significant differences between the different test conditions, in the proximal and distal colon, respectively, are indicated with different letters (p < 0.05).
In terms of lactate production (Figure 1), a significant increase in lactate levels was observed in the PC in response to the different treatments, whereas lactate levels remained around the detection limit in the DC. In both colon regions, lactate levels were not significantly different between OFO and POF dosed at a concentration of 1.4 g β-glucan/day. Furthermore, lactate levels increased in a dose-dependent manner in the PC for the different doses of POF, with the highest lactate levels being observed upon supplementing the highest dose of POF.
With respect to markers of proteolytic fermentation (Figure 1), all treatments significantly increased ammonium and bCFA levels in response to the treatment in both colon regions, except for bCFA levels in the PC upon supplementation of POF at a concentration of 1.4 g β-glucan/day (p = 0.279). Overall, significantly lower ammonium and bCFA levels were observed in the PC for POF versus OFO at a concentration of 1.4 g β-glucan/day (p < 0.0001). Furthermore, in the PC, supplementation of POF at the highest concentration tested resulted in the lowest production of bCFA and ammonium, indicating the presence of a dose-response effect. In the DC, an opposite trend was observed.
2.2. Altered Microbial Composition in Response to Oat Treatment in vitro
To compare the prebiotic properties of the different test products and conditions, qPCR analysis of specific health-related groups (Bifidobacterium spp. and Lactobacillus spp.) was performed (Table 1). First, luminal Lactobacillus levels increased significantly in both colon regions upon treatment with the different test products. Overall, the strongest increase was observed upon supplementation of POF at a concentration of 1.4 g β-glucan/day, resulting in significantly higher Lactobacillus levels as compared to the other test conditions (only POF at a concentration of 1.0 g β-glucan/day resulted in similar increases in the luminal DC). Similar effects were observed for Bifidobacterium levels, which significantly increased in the luminal environment of both colon regions upon treatment with all the different test products. It was noted that OFO more strongly stimulated Bifidobacterium levels than POF at a concentration of 1.4 g β-glucan/day in the PC. Overall, effects on mucosal microbes (Table 1) were similar to the changes reported for the luminal microbiota, even though less pronounced, showing that the test products were able to stimulate the luminal and the mucosal levels of bifidobacteria and lactobacilli.
Table 1.
Effects on microbial community composition as assessed through qPCR. The average increase in Lactobacillus and Bifidobacterium levels (16S rRNA gene copies/mL for lumen and 16S rRNA gene copies/g for mucus) during the treatment period as compared to the control period upon treatment with OFO at a dose of 1.4 g/d and POF at a dose of 0.6 g/d, 1.0 g/d, and 1.4 g/d in the luminal and mucosal environment of the proximal (PC) and distal colon (DC) of the human gastrointestinal tract for three human donors tested. For optimal observation of consistent effects over the different donors tested, the average of the three donors is presented (n = 3). The intensity of the shading correlates with the absolute abundance, which is normalised for each of the different bacterial groups in the different colonic environments, respectively. Statistically significant differences relative to the control period are indicated with underlining (p < 0.05). Statistically significant differences between the different test conditions within a colonic region are indicated with different letters (a, b, c), with conditions not sharing any similar letter being significantly different from each other (p < 0.05).
| PC | DC | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| POF 0.6 g/d |
POF 1.0 g/d |
POF 1.4 g/d |
OFO 1.4 g/d |
POF 0.6 g/d |
POF 1.0 g/d |
POF 1.4 g/d |
OFO 1.4 g/d |
||||||||||
| Lactobacillus | Lumen | 3.5 × 108 | a | 1.4 × 109 | a | 6.2 × 109 | b | 4.3 × 108 | a | 1.5 × 108 | a | 9.1 × 108 | b,c | 5.6 × 109 | b | 2.8 × 108 | a,c |
| Mucus | 2.4 × 109 | a | 4.0 × 109 | a,b | 3.3 × 109 | b | 1.2 × 109 | a,b | 6.0 × 107 | a | 7.8 × 107 | a,b | 1.3 × 109 | b | 8.7 × 107 | a | |
| Bifidobacterium | Lumen | 4.5 × 109 | a,b | 9.0 × 109 | a | 4.5 × 109 | b | 7.2 × 109 | a | 1.4 × 109 | a | 4.2 × 109 | b | 4.3 × 109 | b | 3.6 × 109 | b |
| Mucus | 5.3 × 109 | a,b | 1.4 × 1010 | a | 9.2 × 109 | b | 1.0 × 1010 | a | 2.3 × 108 | 5.8 × 108 | 1.1 × 109 | 5.6 × 108 | |||||
In order to investigate more extensively the effects of treatment with OFO and POF at a dose of 1.4 g β-glucan per day on microbial community composition, microbiome profiling of the luminal PC and DC compartment was performed using 16S-targeted Illumina sequencing. At phylum level (Figure 2), it was noted that at the main site of fermentation, i.e., the lumen of the PC, both test products strongly increased Actinobacteria levels. Similar observations were made in the DC. Additionally, a consistent increase in Firmicutes levels was observed upon treatment with POF in the PC and DC, while supplementation of OFO resulted in enhanced Proteobacteria levels.
Figure 2.
Microbial community composition as assessed via 16S-targeted Illumina sequencing. Abundance (%) at microbial phylum level in the luminal environment of the proximal (PC) and distal colon (DC) of the human gastrointestinal tract at the end of the control (C; n = 6/donor) and the treatment (TR; n = 3/donor) period upon treatment with OFO at a dose of 1.4 g/d and POF at a dose of 1.4 g/d for three human donors tested. For optimal observation of consistent effects over the different donors tested, the average of the three donors is presented (n = 3).
At the family level, the primary focus was on the treatment effects of POF and OFO, at a concentration of 1.4 g β-glucan/day, at the main site of fermentation, i.e., the lumen of the PC (Table 2). For the luminal DC (Table 3), similar observations were made and therefore only specific and distinct changes versus the PC referred to here. First, treatment with POF and OFO strongly increased Bifidobacteriaceae levels (p < 0.0001). At the operational taxonomic unit (OTU) level, this was mainly attributed to a significant increase in Bifidobacteriaceae OTU 1 (related to Bifidobacterium adolescentis). To a lesser extent, Bifidobacteriaceae OTU 41 (related to Bifidobacterium bifidum) and Bifidobacteriaceae OTU 47 (related to Bifidobacterium longum) were stimulated. A significant increase in the abundance of Lactobacillaceae was observed upon treatment with both test products, with the strongest effects for POF supplementation. The increased abundance of Lactobacillaceae was mainly attributed to the increased abundance of Lactobacillaceae OTU 5 (related to Pediococcus acidilactici). Furthermore, upon treatment with both test products, Enterococcaceae, Enterobacteriaceae (p < 0.0001), and Prevotellaceae were significantly enriched in the luminal PC, while Akkermansiaceae (p = 0.0031 for POF and p = 0.0118 for OFO) and Enterobacteriaceae (p < 0.0001) were enhanced in the luminal DC. For the latter bacterial family, treatment with OFO resulted in a stronger enrichment as compared to POF supplementation in both the PC (p < 0.0001) and DC (p < 0.0001). For POF, an increased abundance of Veillonellaceae was observed upon treatment (p < 0.0001 in PC and p = 0.0007 in DC), which was mainly attributed to an increase in Veillonellaceae OTU 22 (related to Veillonella parvula) and Veillonellaceae OTU 11 (related to Veillonella ratti). Furthermore, treatment with POF consistently decreased Bacteroidaceae (p < 0.0001) and Lachnospiraceae (p < 0.0001) levels in the luminal PC, resulting in significantly (p < 0.0001) lower levels as compared to treatment with OFO. Treatment with OFO, on the other hand, resulted in increased Acidaminococcaceae levels in the PC, while in the DC, Bacteroidaceae levels decreased (p = 0.0063) at the expense of Prevotellaceae (p = 0.0019). Finally, the butyrate-producing Ruminococcaceae family was stimulated significantly in the PC environment upon treatment with the different test products, while decreased abundance was observed in the DC (p = 0.0009 for POF and p < 0.0001 for OFO).
Table 2.
Effects on microbial community composition at the family level in the proximal colon. Abundance (log counts/mL) of microbial families in the luminal environment of the proximal colon (PC) of the human gastrointestinal tract at the end of the control (C; n = 3/donor) and the treatment (TR; n = 3/donor) period upon treatment with OFO at a dose of 1.4 g/d and POF at a dose of 1.4 g/d for three human donors tested. For optimal observation of consistent effects over the different donors tested, the average of the three donors is presented (n = 3). The intensity of the shading correlates with the absolute abundance, which was normalised for each of the different families. Statistically significant differences relative to the control period are indicated in bold (p < 0.05). Statistically significant differences between the different test products (i.e., TR–POF versus TR–OFO) are indicated with different letters (a, b), with conditions not sharing a similar letter being significantly different from each other (p < 0.05).
| Proximal Colon | |||||
|---|---|---|---|---|---|
| POF | OFO | ||||
| C | TR | C | TR | ||
| Actinobacteria | Bifidobacteriaceae | 6.83 | 8.03 | 6.94 | 8.12 |
| Coriobacteriaceae | <LOD | 5.76 | <LOD | 5.10 | |
| Bacteroidetes | Bacteroidaceae | 7.53 | 6.01 | 7.55 | 7.85 |
| Muribaculaceae | <LOD | <LOD | <LOD | 4.37 | |
| Prevotellaceae | 5.04 | 7.65 | 4.73 | 7.12 | |
| Rikenellaceae | 4.32 | 4.20 | 4.34 | 5.41 | |
| Tannerellaceae | 5.64 | 4.67 | 5.92 | 6.58 | |
| Firmicutes | Acidaminococcaceae | 6.53 | 5.36 | 5.20 | 7.25 |
| Enterococcaceae | 3.36 | 5.34 | 3.53 | 7.53 | |
| Erysipelotrichaceae | <LOD | 4.11 | 4.64 | 4.72 | |
| Lachnospiraceae | 7.33 | 5.86 | 7.37 | 7.76 | |
| Lactobacillaceae | 3.45 | 7.78 | 3.51 | 6.32 | |
| Ruminococcaceae | 3.60 | 4.09 | 3.71 | 4.80 | |
| Veillonellaceae | 6.96 | 7.93 | 7.14 | 7.20 | |
| Fusobacteria | Fusobacteriaceae | <LOD | <LOD | 4.21 | 4.89 |
| Proteobacteria | Burkholderiaceae | 5.15 | 5.62 | 5.62 | 6.65 |
| Desulfovibrionaceae | 5.40 | 4.41 | 5.66 | 4.93 | |
| Enterobacteriaceae | 4.91 | 5.99 | 5.27 | 7.50 | |
| Pseudomonadaceae | 4.75 | 4.14 | 5.20 | 4.59 | |
| uncultured | <LOD | <LOD | 3.24 | 4.20 | |
| Synergistetes | Synergistaceae | 4.59 | 3.88 | 3.81 | 4.70 |
| Verrucomicrobia | Akkermansiaceae | <LOD | <LOD | 3.61 | 3.94 |
Table 3.
Effects on microbial community composition at the family level in the distal colon. Abundance (log counts/mL) of microbial families in the luminal environment of the distal colon (DC) of the human gastrointestinal tract at the end of the control (C; n = 3/donor) and the treatment (TR; n = 3/donor) period upon treatment with OFO at a dose of 1.4 g/d and POF at a dose of 1.4 g/d for three human donors tested. For optimal observation of consistent effects over the different donors tested, the average of the three donors is presented (n = 3). The intensity of the shading correlates with the absolute abundance, which was normalised for each of the different families. Statistically significant differences relative to the control period are indicated in bold (p < 0.05). Statistically significant differences between the different test products (i.e., TR–POF versus TR–OFO) are indicated with different letters (a, b), with conditions not sharing a similar letter being significantly different from each other (p < 0.05).
| Distal Colon | |||||
|---|---|---|---|---|---|
| POF | OFO | ||||
| C | TR | C | TR | ||
| Actinobacteria | Bifidobacteriaceae | 7.39 | 8.26 | 7.45 | 8.13 |
| Coriobacteriaceae | 5.61 | 5.31 | 5.66 | 4.93 | |
| Bacteroidetes | Bacteroidaceae | 8.35 | 8.20 | 8.30 | 8.04 |
| Muribaculaceae | 5.26 | 6.23 | 4.87 | 6.02 | |
| Prevotellaceae | 6.55 | 6.95 | 6.27 | 7.22 | |
| Rikenellaceae | 6.29 | 5.74 | 6.12 | 6.10 | |
| Tannerellaceae | 7.26 | 7.84 | 7.28 | 7.64 | |
| Firmicutes | Acidaminococcaceae | 6.61 | 7.22 | 6.79 | 6.94 |
| Enterococcaceae | 4.39 | 6.11 | 4.34 | 7.88 | |
| Erysipelotrichaceae | 5.64 | 4.86 | 5.65 | <LOD | |
| Lachnospiraceae | 8.23 | 8.28 | 8.16 | 8.27 | |
| Lactobacillaceae | 4.53 | 8.25 | 4.45 | 6.76 | |
| Ruminococcaceae | 6.64 | 6.12 | 6.74 | 5.90 | |
| Veillonellaceae | 7.37 | 8.04 | 7.34 | 7.36 | |
| Fusobacteria | Fusobacteriaceae | 5.62 | 6.34 | 5.70 | 7.66 |
| Proteobacteria | Burkholderiaceae | 6.48 | 6.73 | 6.52 | 6.31 |
| Desulfovibrionaceae | 6.58 | 6.79 | 6.76 | 6.94 | |
| Enterobacteriaceae | 5.42 | 6.36 | 5.45 | 7.63 | |
| Pseudomonadaceae | 6.57 | 5.52 | 6.44 | 6.48 | |
| uncultured | 5.89 | 6.51 | 6.05 | 6.33 | |
| Synergistetes | Synergistaceae | 6.93 | 7.52 | 7.16 | 7.27 |
| Verrucomicrobia | Akkermansiaceae | 7.24 | 7.67 | 7.41 | 7.77 |
2.3. In vitro–in vivo Comparison of Microbial Response to OFO Treatment
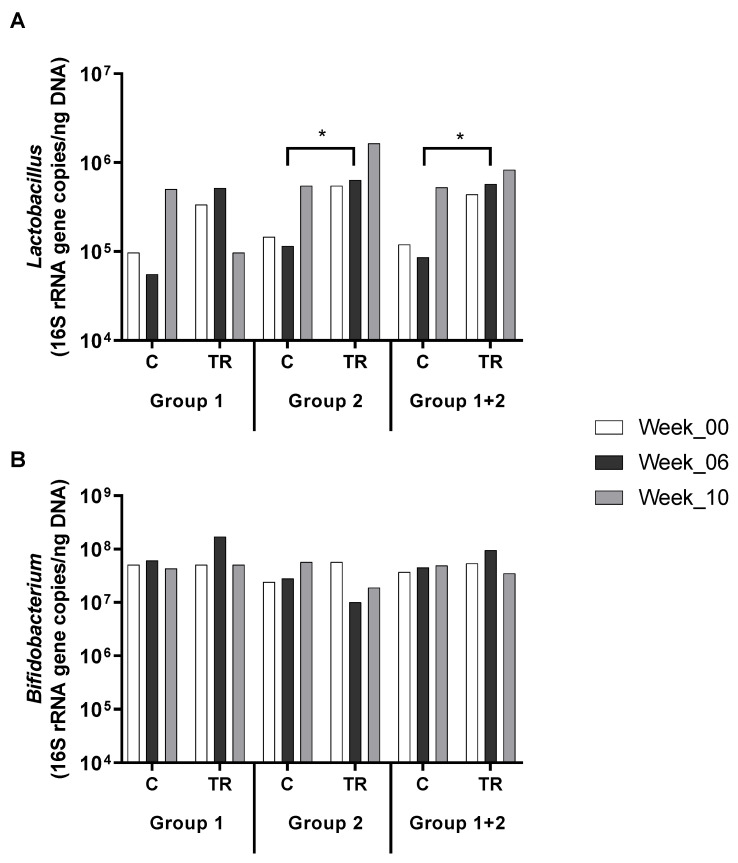
In vitro, the prebiotic activity of OFO was characterised by a significant stimulation of lactobacilli and bifidobacteria in both the luminal and mucosal environment (Table 1). Therefore, selective primers were selected to analyse the levels of Lactobacillus spp. and Bifidobacterium spp. in the faecal samples collected during the in vivo trial by means of quantitative polymerase chain reaction (qPCR). During the in vivo trial, stimulation of Lactobacillus spp. was observed (Figure 3A). Indeed, significant stimulation of Lactobacillus species was observed at the end of the intervention period with OFO as compared to the control test product for the overall population (p = 0.037) and in the population designated as ‘Group 2’, i.e., the group that received the investigational product during the second intervention period (p = 0.017; Figure 3A). For Bifidobacterium levels (Figure 3B), no overall significant differences were observed between intervention with OFO and the control test product, although a trend towards increased Bifidobacterium levels was observed upon intervention with OFO in the ‘Group 1’ population (i.e., the group that received the investigational product during the first intervention period) (p = 0.299). With respect to plasma SCFA levels, no statistically significant differences were observed between intervention with OFO and the control test product.
Figure 3.
Effects on microbial community composition in vivo. Effect of intervention with OFO (TR) on faecal (A) Lactobacillus and (B) Bifidobacterium levels as compared to an intervention with the control test product (C) during a randomised, single-blind, cross-over study in healthy individuals with elevated cholesterol levels for the per protocol (PP) population, with samples taken at baseline (week_00), at the end of the intervention period (week_06) and at the end of the washout period (week_10). Results are presented for Group 1 (received OFO during the first intervention period), Group 2 (received OFO during the second intervention period), and the overall (Group 1 + 2) population. Results are presented as mean (16S rRNA gene copies/ng DNA). * indicates statistically significant differences between control and treatment within a specific experimental week for Group 1, Group 2, and Group 1 + 2 (p < 0.05).
3. Discussion
In the present study, the potential prebiotic effects of prolonged administration of the commercially available oat product OFO were assessed in the human gastrointestinal tract of different healthy individuals with mild hypercholesterolemia. For this purpose, the validated in vitro M-SHIME® model [25] was utilised, allowing analysis of an intestinal microbial community that is fully stable prior to treatment [26]. It followed that the prebiotic activity of OFO was characterised by strong stimulation of Lactobacillus and Bifidobacterium species in the intestinal lumen and the simulated mucus layer of the PC and DC. Lactobacilli [27] and bifidobacteria [28] are saccharolytic gut microbes and are both capable of producing high levels of lactic acid, thereby exerting antimicrobial properties [29] and stimulating trophic interactions with other bacteria resulting in the production of secondary metabolites such as butyric acid [30]. Due to their saccharolytic metabolism, Lactobacillus and Bifidobacterium species mainly thrive in the proximal regions of the colon [31]. Therefore, the effects of prebiotic compounds on these bacterial groups are often more difficult to observe in in vivo trials. However, during the current study, the stimulation of Lactobacillus spp. by OFO was also observed in the faecal samples of human subjects with mild hypercholesterolemia during the randomised, single-blind, placebo-controlled, cross-over study. This is a striking observation as in vivo studies are inherently confounded by large variability due to interindividual differences among individuals and a strong influence of external factors such as diet [32]. Moreover, the investigated in vivo samples considered faecal samples that are only to a limited extent representative for the microbial changes in the PC. For Bifidobacterium levels, no significant differences were observed between intervention with OFO and the control test product, although in the population that received OFO during the first intervention period a trend towards increased Bifidobacterium levels was observed upon intervention with OFO. Overall, existing scientific evidence and the preliminary results in this study show the significant stimulatory effects of OFO on lactobacilli in human subjects with mild hypercholesterolemia, which points to the prebiotic potential of OFO. Because it has been reported extensively that lactobacilli and bifidobacteria exert hypocholesterolaemic effects both in both animals and humans [33,34,35,36], further in vivo studies are warranted to explore this effect for OFO. Furthermore, as stimulation of Lactobacillus spp. by OFO in vivo correlated with the obtained in vitro data, the current study shows that in vitro gut models might be an interesting tool in predicting in vivo response of the microbial community to dietary modulation.
The in vitro component of the current study also aimed to assess the prebiotic potential of a novel oat product, i.e., POF, relative to OFO. It was revealed that both exerted a highly similar prebiotic activity. First, both test products resulted in significantly higher levels of acetate and lactate, which correlated with the strongly enhanced levels of Lactobacillaceae and Bifidobacteriacea species [31], indicating the involvement of these bacterial groups in primary substrate degradation upon supplementation of the different oat products. At the OTU level, the main changes were attributed to a significant increase in an OTU related to Bifidobacterium adolescentis. It has been previously reported that fermentation of oat bran stimulates the growth of Bifidobacterium adolescentis in vitro [37], while also fermentation of β-glucans has been associated with this bacterial species [38]. The significant stimulation of Lactobacillaceae upon treatment with the different test products was linked with increased abundance of an OTU related to Pediococcus acidilactici, which has been associated with immune-enhancing effects [39]. Furthermore, both test products enhanced Akkermansiaceae levels in the DC. The only representative of Akkermansiaceae in the gut is the mucin-degrading Akkermansia muciniphila, which has been correlated with several health benefits, such as the inverse relationship between colonisation of Akkermansia muciniphila and inflammatory conditions [40]. Additionally, significantly enhanced levels of propionate and butyrate were observed upon administration of both test ingredients. Administration of oat products has previously been linked with increased levels of butyrate [41,42]. Indeed, Knudsen et al. [42] reported that addition of oat bran to the diet of pigs increased butyrate concentrations in the luminal environment of the porcine colon. However, administration of β-glucan enriched oat fractions to the porcine diet did not result in butyrate enrichment, indicating that other dietary components (e.g., arabinoxylans) are responsible for the observed increase in butyrate concentration upon oat supplementation. β-glucans on the other hand have been shown to selectively stimulate propionate levels in the colonic environment [7,43,44]. It was proposed that the hypocholesterolaemic effect of oat fibres [12,13,14] might be associated with this propionogenic response. Indeed, upon its production, propionate is transported to the liver, where it impacts cholesterol and fatty acid synthesis [19,20]. Moreover, next to the reduction of cholesterol levels, health-beneficial effects of propionate include weight management by stimulation of satiety [45], regulation of immune function in adipose tissue [46,47], and protection against cancer development [48]. The stimulation of SCFA production by the different oat ingredients therefore suggests their prebiotic potential.
In addition, some differences were observed between the fermentation of OFO and POF. For instance, supplementation of POF resulted in the strongest stimulation of Lactobacillus levels in the PC and DC, while OFO administration more strongly enhanced butyrate production and Bifidobacterium concentrations in the PC. Previous in vitro research by Van den Abbeele et al. [7] revealed that product-specific microbial pathways were boosted upon administration of structurally different oat ingredients. For instance, while all products resulted in a significant increase in Bifidobacterium levels, oat bran showed the strongest bifidogenic effect of the six oat ingredients investigated, indicating that slight differences in prebiotic response exist among oat-derived products, as observed in the current study.
Moreover, in vitro investigations were carried out to determine if the potential prebiotic properties of the novel oat product POF persisted at lower test doses. It was observed that acetate, propionate, and butyrate levels increased in a dose-dependent manner in the DC with the highest final SCFA levels observed upon supplementation of the highest concentration of POF. However, even at the lowest concentration tested, POF stimulated SCFA production and Lactobacillus and Bifidobacterium levels, indicating that the prebiotic activity of POF remained at lower β-glucan levels. Overall, the obtained results demonstrate the prebiotic potential of the novel oat product POF, even when administered at lower concentrations.
In order to further investigate the impact on the luminal microbial community composition upon supplementation of POF and OFO at similar β-glucan load, 16S-targeted Illumina sequencing was performed. Treatment with both test products increased Prevotellaceae levels, mainly in the PC. Saccharolytic fermentation by members of this bacterial family results in the production of acetate and succinate [49], with the latter being a substrate for succinate-converting, propionate producing micro-organisms such as Bacteroides and Veillonella species [50]. In the current study, an increased abundance of Veillonellaceae was observed upon treatment with POF, but not upon OFO supplementation. However, it might be that specific members of the Veillonellaceae family were stimulated upon OFO administration, while others reduced, resulting in the absence of effects at the family level. Finally, both test products increased the abundance of Enterobacteriaceae. This bacterial family contains several opportunistic pathogenic species, however, also many commensals able to ferment proteins are members of this bacterial group [51]. The strongest effects were observed upon OFO supplementation, which correlated with the more strongly increased levels of branched SCFA and ammonium, which are both markers of proteolysis that were observed upon fermentation of OFO as compared to POF. Overall, these results indicate that in vitro gut models combined with accurate molecular techniques have the potential to highlight specific microbial taxonomic changes and functional activities pathways upon dietary intervention.
From this study, it can be concluded that the novel oat product POF and the commercially available OFO, when administered at similar β-glucan load, exerted equivalent prebiotic activity in the human gastrointestinal tract in vitro, with profound effects being observed on Lactobacillus and Bifidobacterium levels. The stimulation of Lactobacillus spp. observed in vitro was confirmed during an in vivo trial investigating the effect of OFO in human subjects with mild hypercholesterolemia. Moreover, the in vitro prebiotic activity of POF remained at lower β-glucan levels, demonstrating the potent prebiotic potential of the novel oat product even when administered at lower concentrations.
4. Materials and Methods
4.1. Chemicals and Test Product
Unless otherwise stated, all chemicals were obtained from Sigma-Aldrich (Overijse, Belgium). PepsiCo, Inc. (Barrington, IL, USA) provided the different oat ingredients, including Quaker whole grain pre-cooked oat flour (POF) and old-fashioned oats (OFOs). OFOs were produced by steaming the whole groats to make them soft and pliable and then pressed to flatten them obtaining a particle size of 0.51−0.76 mm. POF was produced using PepsiCo’s proprietary process providing a ready-to-use format with improved dispersibility. The granulation range for POF ranged from 50–250 μm with the targeting of 178–250 μm. The daily dose of both oat ingredients during the in vitro experiment was standardised to provide 1.4g β-glucan once per day. For POF, two additional test doses were investigated—standardised to provide 0.3 g β-glucan twice per day (to total 0.6 g β-glucan per day) and 1.0 g β-glucan once per day. In order to mimic the in vivo conditions as close as possible, the test ingredient OFO was cooked according to the manufacturer’s instructions prior to use in the in vitro experiment. During the in vivo study, dry OFO was supplied to the subjects and cooked according to the manufacturer’s instructions prior to consumption. The consumption of cooked OFO was investigated during an intervention period of six weeks, providing a daily one serving dose of 40 g oats containing 1.4 g β-glucan. The control test product during the in vivo trial was Cream of Rice (Nabisco, East Hanover, NJ, USA), a cooked cereal containing no fibre or β-glucan and was supplemented at a dose of 40 g per day.
4.2. Simulator of the Human Intestinal Microbial Ecosystem (SHIME®)
The reactor setup simulating the human gastrointestinal tract was derived from the SHIME® (ProDigest and Ghent University, Ghent, Belgium) as described by Molly et al. [23]. To optimally address the research questions, the SHIME® setup was adapted from a single SHIME configuration (including one SHIME® arm) to a TripleSHIME® configuration (including three SHIME® arms). Each arm of the TripleSHIME® consisted of a succession of three reactors simulating the different regions of the gastrointestinal tract. The first reactor mimicked the upper gastrointestinal tract with the subsequent simulation of a gastric and small intestinal phase. The two subsequent colonic reactors simulated the proximal colon (PC), operated at pH 5.6–5.9 with a retention time of 20 h, and the distal colon (DC), operated at pH 6.6–6.9 with a retention time of 32 h. In order to simulate both the luminal and mucus-associated microbial community in the colonic reactors, mucin-covered beads were included as described by Van den Abbeele et al. [25]. Inoculum preparation, temperature settings, feeding regime, and reactor feed composition were adopted from Possemiers et al. [52]. To evaluate the properties of four different test ingredients and/or concentrations, four parallel TripleSHIME® experiments were executed using the microbiota of three healthy adult human donors with elevated total and low-density lipoproteins (LDL) cholesterol levels (total ≥ 5.5 mmol/L and <7 mmol/L and LDL ≥ 3.4 mmol/L and ≤ 4.9 mmol/L) and a BMI between 20 kg/m2 and 25 kg/m2 (donor A: F, 25 y; donor B: F, 30 y; Donor C: M, 35 y), meaning that each test ingredient/concentration was tested in a separate TripleSHIME® experiment including the microbial community of three different donors. Upon inoculation with the faecal inoculum from the different donors, a two-week stabilisation period was initiated allowing the faecal microbiota to differentiate in the colonic reactors depending on the local environmental conditions. Subsequently, the baseline microbial community composition and activity were determined in the PC and DC during a two-week control period, followed by an evaluation of the effects of repeated daily administration of the test products during a three-week treatment period. The test products were pre-digested prior to administration to the colonic reactors in order to produce relevant product fractions that would reach the colonic environment. Pre-digestion was performed as previously described by Van den Abbeele et al. [7], with some minor modifications. During the oral phase, the test ingredients were diluted to obtain a concentration mimicking the daily test doses of interest, corrected for moisture content. Furthermore, the intestinal dialysis approach included a 4.5-hour incubation, during which the dialysis fluid was replaced once every 45 minutes with fresh fluid.
4.3. In vivo Study
This was a randomised, single-blind, placebo-controlled, and cross-over study to assess the effect of hot cooked OFO on a range of variables including faecal bacterial composition and plasma SCFAs, in a sample of healthy adults with elevated cholesterol levels within the same range as the in vitro study. There were six visits in total with two phases to data collection (Figure S1). Each phase consisted of a 10-week period. The trial phase comprised of a six-week intervention period (Week 0–Week 6) after which subjects completed a four-week washout (Week 10).
In total, 34 randomised subjects (aged 18–65 years old, BMI of 18.5–30 kg/m2, fasting blood glucose (FBG) of 3.0–6.0 mmol/L) were included in the analysis. To enrol in the trial, subjects provided written informed consent, and then completed the required screening procedures to evaluate their eligibility for the trial. At screening (Visit 1), fasting blood samples were collected to examine potential subject’s safety and blood lipid profiles, the subjects were also administered a food frequency questionnaire (FFQ); this information determined eligibility. The study participants were required to have a low to moderate consumption of dietary fibre based on data from the 2008–2010 Irish National Adult Nutrition Survey [53] (Low- fibre diet: ≥9.9 g/d and ≤17 g/d for males; 8.2 g/d and ≤14.3 g/d for females; moderate extends to 25.1 g/d for males and to 22.3 g/d for females), and elevated total and LDL cholesterol levels (total cholesterol ≥ 5.5 and <7 mmol/L and LDL ≥ 3.4 mmol/L and ≤ 4.9 mmol/L). The trial included a two-week run-in period to washout possible pre-trial prebiotics prior to baseline assessment. Eligible subjects were then randomised in a 1:1 ratio at Visit 2 to receive either treatment or control product in Phase 1 (Baseline: Visit 2; Week 0—End of Phase 1: Visit 3 at Week 6; Washout period: Visit 4 at Week 10), and they would then cross-over to consume the alternative product for Phase 2 (Baseline: Visit 4 at Week 10; Week 10—End of Phase 2: Visit 5 at Week 16; Washout period: Visit 6 at Week 20). Group 1 received OFO during the first intervention period and Cream of Rice during the second intervention period, while Group 2 received Cream of Rice during the first intervention period and OFO during the second intervention period. Subjects were provided with a stool collection kit at visits 1, 2, 3, 4, and 5 and instructed to collect a sample at home (24 hours prior to their visit) and bring it to the clinic at their next visit. Samples were kept chilled preceding analysis.
The subjects consumed the study product once daily, at breakfast, for six weeks starting on Day 1, the day after Visit 2. The subjects were asked not to consume any other prebiotic/probiotics and fibre supplements or whole-grain oat products throughout the duration of the trial (22 weeks). They were also asked to maintain their habitual lifestyle in relation to physical activity level and diet.
Ethical approval was granted by the Clinical Research Ethics Committee, Cork Ireland, prior to the study starting with code ECM 5 (6) 17/01/18. The study was conducted according to the principles of ICH-GCP and the Declaration of Helsinki.
4.4. Microbial Metabolic Activity
During the in vitro SHIME® experiment, samples for analysis of microbial metabolic activity were collected three times per week during the control and treatment period from each colonic reactor. Short-chain fatty acid (SCFA) measurements were performed as described by De Weirdt et al. [54] and included acetate, propionate, butyrate, and branched-chain fatty acids (isobutyrate, isovalerate, and isocaproate; bCFA). Lactate levels were determined using a commercially available enzymatic assay kit (R-Biopharm, Darmstadt, Germany) according to the manufacturer’s instructions. Ammonium determination was conducted as previously reported by Duysburgh et al. [55].
During the in vivo experiment, samples for analysis of plasma SCFA were collected at each scheduled visit, starting from Visit 2. Four plasma samples were collected in total per participant, i.e., one sample per baseline and endpoint of Phase 1 and Phase 2 of the in vivo experiment. Samples were prepared, extracted, and subjected to Ultra Performance Liquid Chromatography-Tandem Mass Spectrometry (UPLC-MS; Waters Ltd. Herts, United Kingdom) and analysed according to Brown et al. [56].
4.5. DNA Extraction
During the control and treatment period of the in vitro SHIME® experiment, samples for microbial community analysis were collected once per week from each colonic reactor. Total DNA was isolated as described by Boon et al. [57], with some minor modifications as previously reported by Duysburgh et al. [55]. For the in vivo study, metagenomic DNA was extracted from all faecal samples by Teagasc (Cork, Ireland) using a modified version of the QIAGEN FAST Stool extraction kit (QIAGEN, Manchester, UK). The modification included an additional bead-beating step at the start of the procedure. Briefly, faeces were added to the buffer as described by the manufacturer’s instructions, after which homogenisation with a bead beater device was performed, followed by the protocol for Gram-positive bacteria as described by manufacturer’s instructions.
4.6. Microbial Community Analysis through qPCR
qPCR assays to quantify Lactobacillus spp. and Bifidobacterium spp. were completed using a QuantStudio 5 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). Each sample was analysed in technical triplicate and outliers (more than 1 CT difference) were removed. The qPCR assay, including primer sequences and amplification program, for Lactobacillus spp. was conducted as reported by Furet et al. [58], while the qPCR for Bifidobacterium spp. was previously described by Rinttilä et al. [59].
4.7. Microbial Community Analysis through 16S-Targeted Illumina Sequencing
In order to compare effects on microbial community composition of treatment with OFO and POF at a dose of 1.4 g β-glucan per day, microbial community profiling of the luminal PC and DC compartment of the in vitro SHIME® experiment was performed using 16S-targeted Illumina sequencing. Library preparation and sequencing on an Illumina MiSeq platform with v3 chemistry (2 × 300bp) were conducted by LGC Genomics GmbH (Berlin, Germany) using the primers as reported by Klindworth et al. [60], with modification of the reverse primer (785Rmod; 5’-GAC TAC HVG GGT ATC TAA KCC-3’) to increase coverage.
4.8. Flow Cytometric Determination
Samples were also collected once per week from each colonic reactor for enumeration of bacterial cells via flow cytometry. A 10-fold dilution series was prepared initially in phosphate-buffered saline. Assessment of the viable, non-viable, and total population of the microbial community was done by staining the appropriate dilutions with SYTO 24 and propidium iodide. Samples were analysed on a BD FACSVerse (BD Biosciences, Erembodegem, Belgium). The samples were run using the high flow rate. Bacterial cells were separated from medium debris and signal noise by applying a threshold level of 200 on the SYTO channel. Proper parent and daughter gates were set to determine all populations. Results were obtained as log counts/mL.
4.9. Data and Statistical Analysis
For the analysis of 16S-targeted Illumina sequence data, read assembly and clean-up was adopted from the MiSeq procedure [61,62]. Briefly, mothur (v. 1.40.5, University of Michigan, Ann Arbor, MI, USA) was applied to assemble reads into contigs, perform alignment-based quality filtering (alignment to the mothur-reconstructed SILVA SEED alignment, v. 123), remove chimeras, assign taxonomy using a naïve Bayesian classifier [63] and SILVA NR v132 and cluster contigs into operational taxonomic units (OTUs) at 97% sequence similarity. All sequences classified as Archaea, Chloroplasts, Eukaryota, and Mitochondria were omitted, in addition to sequences that could not be classified at all. For each OTU, representative sequences were picked as the most abundant sequence within that OTU. To identify related bacterial species, the obtained sequences were matched using the SeqMatch tool in the Ribosomal Database Project software (Michigan State University, East Lansing, MI, USA) [64]. Results were presented as proportional values versus the total amount of sequences within each sample. Combining the proportional values of the 16S-targeted Illumina together with the enumeration of the total cell count obtained through flow cytometry, as was previously described by Vandeputte et al. [65], provided quantitative abundances of the different taxonomic entities inside the colonic reactors. These quantitative abundances were obtained by multiplying the proportional values of the 16S-targeted Illumina with the total log cell count obtained by flow cytometry.
With respect to microbial parameters during the in vitro SHIME® study, consistent findings were made for the three different donors in response to the treatment with the different oat products so that the averages of all parameters over the three donors were calculated per colon compartment for optimal visualisation of donor-independent treatment effects. Statistical analyses were carried out using Statistical Analysis Software version 9.4 (SAS Institute, Cary, NC, USA). Data were analysed by comparing the averages over the three donors for the control period as compared to the treatment period, and for the differences between the test product during the treatment period and this for both PC and DC separately, using Bonferroni correction. For 16S-targeted Illumina sequence data, statistical analysis was only performed for those bacterial families with complete data from at least two donors to capture both the noise within each donor and variability between donors. The significance level for all statistical tests was set at α = 0.05. Shapiro–Wilk tests were conducted to assess the normality of the data. Log-transformation was applied where appropriate.
For the in vivo study, outcomes were analysed on the per-protocol population (PP), i.e., all subjects with complete data and at least 80% compliance in terms of the amount of product consumed. Statistical analysis was performed using the SPSS Statistics software (IBM, Armonk, NY, USA), version 25. Normality of data and equality of the variances were determined with a Shapiro–Wilk test and a Levene’s test, respectively. For normally distributed data with equal variances, a one-way ANOVA with a Bonferroni post hoc test was used. For normally distributed data with unequal variances, a Welch test with a Games–Howell post hoc test was conducted. For non-normally distributed data, a Kruskal–Wallis one-way ANOVA test with multiple post hoc pairwise comparisons were performed. In terms of statistics, the differences indicated by ‘p < 0.05’ were significant. All data were normalised to the DNA concentration of the extracts obtained from the faecal samples.
Acknowledgments
The work was financially supported by PepsiCo, Inc. The views expressed in this manuscript are those of the authors and do not necessarily reflect the position or policy of PepsiCo, Inc. The authors would like to thank Fiona Crispie, Calum Walsh, Karina Quilter, and Alvaro Lopez Gallardo for their assistance in the in vivo study analysis. The authors would also like to thank Kevin O’Regan for his assistance with the in vivo study management.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/10/2/235/s1, Figure S1: Cross-over study design.
Author Contributions
Conceptualisation, P.V.d.A., M.M. (Massimo Marzorati), P.D.C., S.A.J., A.K., Y.C., and L.F.; methodology, P.V.d.A., P.D.C., and S.A.J.; formal analysis, C.D., W.B., O.O., P.D.C., K.Q., S.A.J., M.M. (Mike Murphy), G.D., and T.G.D.; investigation, C.D., W.B., O.O., P.D.C., S.A.J., M.M. (Mike Murphy), G.D., and T.G.D.; data curation, C.D., P.J.D.C., Y.C., W.B., O.O., P.D.C., S.A.J., M.M. (Mike Murphy), G.D., and T.G.D.; writing—original draft preparation, C.D. and P.V.d.A.; writing—review and editing, C.D., P.V.d.A., A.K., L.F., P.D.C., S.A.J., P.J.D.C., and G.D.; supervision, P.V.d.A., M.M. (Massimo Marzorati), and Y.C.; project administration, M.M. (Massimo Marzorati), A.K., and T.G.D.; funding acquisition, A.K., Y.C., and L.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of University College Cork, Ireland (study code AFCRO-083, approved on and 17/01/2018).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data available on request due to restrictions.
Conflicts of Interest
A.K., L.F., P.J.D.C., and Y.C. are employees of PepsiCo, Inc.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gibson G.R., Hutkins R., Sanders M.E., Prescott S.L., Reimer R.A., Salminen S.J., Scott K., Stanton C., Swanson K.S., Cani P.D., et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 2.Daou C., Zhang H. Oat Beta-Glucan: Its Role in Health Promotion and Prevention of Diseases. Compr. Rev. Food Sci. Food Saf. 2012;11:355–365. doi: 10.1111/j.1541-4337.2012.00189.x. [DOI] [Google Scholar]
- 3.Braaten J.T., Wood P.J., Scott F.W., Wolynetz M.S., Lowe M.K., Bradley-White P., Collins M.W. Oat beta-glucan reduces blood cholesterol concentration in hypercholesterolemic subjects. Eur. J. Clin. Nutr. 1994;48:465–474. [PubMed] [Google Scholar]
- 4.El Khoury D., Cuda C., Luhovyy B.L., Anderson G.H. Beta Glucan: Health Benefits in Obesity and Metabolic Syndrome. J. Nutr. Metab. 2012;2012:28. doi: 10.1155/2012/851362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang D., Doehlert D.C., Moore W.R. Rheological Properties of (1→3),(1→4)-β-d-Glucans from Raw, Roasted, and Steamed Oat Groats. Cereal Chem. 1998;75:433–438. doi: 10.1094/CCHEM.1998.75.4.433. [DOI] [Google Scholar]
- 6.Grundy M.M.L., Quint J., Rieder A., Ballance S., Dreiss C.A., Butterworth P.J., Ellis P.R. Impact of hydrothermal and mechanical processing on dissolution kinetics and rheology of oat β-glucan. Carbohydr. Polym. 2017;166:387–397. doi: 10.1016/j.carbpol.2017.02.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van den Abbeele P., Kamil A., Fleige L., Chung Y., De Chavez P., Marzorati M. Different Oat Ingredients Stimulate Specific Microbial Metabolites in the Gut Microbiome of Three Human Individuals in Vitro. ACS Omega. 2018;3:12446–12456. doi: 10.1021/acsomega.8b01360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson M.L., Nobaek S., Berggren A., Nyman M., Bjorck I., Ahrne S., Jeppsson B., Molin G. Survival of Lactobacillus plantarum DSM 9843 (299v), and effect on the short-chain fatty acid content of faeces after ingestion of a rose-hip drink with fermented oats. Int. J. Food Microbiol. 1998;42:29–38. doi: 10.1016/S0168-1605(98)00055-5. [DOI] [PubMed] [Google Scholar]
- 9.Mårtensson O., Biörklund M., Lambo A.M., Dueñas-Chasco M., Irastorza A., Holst O., Norin E., Welling G., Öste R., Önning G. Fermented, ropy, oat-based products reduce cholesterol levels and stimulate the bifidobacteria flora in humans. Nutr. Res. 2005;25:429–442. doi: 10.1016/j.nutres.2005.03.004. [DOI] [Google Scholar]
- 10.Coman M.M., Verdenelli M.C., Cecchini C., Silvi S., Vasile A., Bahrim G.E., Orpianesi C., Cresci A. Effect of buckwheat flour and oat bran on growth and cell viability of the probiotic strains Lactobacillus rhamnosus IMC 501(R), Lactobacillus paracasei IMC 502(R) and their combination SYNBIO(R), in synbiotic fermented milk. Int. J. Food Microbiol. 2013;167:261–268. doi: 10.1016/j.ijfoodmicro.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Connolly M.L., Tzounis X., Tuohy K.M., Lovegrove J.A. Hypocholesterolemic and Prebiotic Effects of a Whole-Grain Oat-Based Granola Breakfast Cereal in a Cardio-Metabolic “At Risk” Population. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiwari U., Cummins E. Meta-analysis of the effect of beta-glucan intake on blood cholesterol and glucose levels. Nutrition. 2011;27:1008–1016. doi: 10.1016/j.nut.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Whitehead A., Beck E.J., Tosh S., Wolever T.M. Cholesterol-lowering effects of oat beta-glucan: A meta-analysis of randomized controlled trials. Am. J. Clin. Nutr. 2014;100:1413–1421. doi: 10.3945/ajcn.114.086108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho H.V., Sievenpiper J.L., Zurbau A., Blanco Mejia S., Jovanovski E., Au-Yeung F., Jenkins A.L., Vuksan V. The effect of oat beta-glucan on LDL-cholesterol, non-HDL-cholesterol and apoB for CVD risk reduction: A systematic review and meta-analysis of randomised-controlled trials. Br. J. Nutr. 2016;116:1369–1382. doi: 10.1017/S000711451600341X. [DOI] [PubMed] [Google Scholar]
- 15.Wolever T.M., Tosh S.M., Gibbs A.L., Brand-Miller J., Duncan A.M., Hart V., Lamarche B., Thomson B.A., Duss R., Wood P.J. Physicochemical properties of oat beta-glucan influence its ability to reduce serum LDL cholesterol in humans: A randomized clinical trial. Am. J. Clin. Nutr. 2010;92:723–732. doi: 10.3945/ajcn.2010.29174. [DOI] [PubMed] [Google Scholar]
- 16.Jonsson A.L., Backhed F. Role of gut microbiota in atherosclerosis. Nat. Rev. Cardiol. 2017;14:79–87. doi: 10.1038/nrcardio.2016.183. [DOI] [PubMed] [Google Scholar]
- 17.Jones M.L., Tomaro-Duchesneau C., Martoni C.J., Prakash S. Cholesterol lowering with bile salt hydrolase-active probiotic bacteria, mechanism of action, clinical evidence, and future direction for heart health applications. Expert Opin. Biol. Ther. 2013;13:631–642. doi: 10.1517/14712598.2013.758706. [DOI] [PubMed] [Google Scholar]
- 18.Joyce S.A., Kamil A., Fleige L., Gahan C.G.M. The Cholesterol-Lowering Effect of Oats and Oat Beta Glucan: Modes of Action and Potential Role of Bile Acids and the Microbiome. Front. Nutr. 2019;6 doi: 10.3389/fnut.2019.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berggren A.M., Nyman E.M., Lundquist I., Bjorck I.M. Influence of orally and rectally administered propionate on cholesterol and glucose metabolism in obese rats. Br. J. Nutr. 1996;76:287–294. doi: 10.1079/BJN19960032. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y., Vonk R.J., Slooff M.J., Kuipers F., Smit M.J. Differences in propionate-induced inhibition of cholesterol and triacylglycerol synthesis between human and rat hepatocytes in primary culture. Br. J. Nutr. 1995;74:197–207. doi: 10.1079/BJN19950123. [DOI] [PubMed] [Google Scholar]
- 21.Connolly M.L., Lovegrove J.A., Tuohy K.M. In Vitro evaluation of the microbiota modulation abilities of different sized whole oat grain flakes. Anaerobe. 2010;16:483–488. doi: 10.1016/j.anaerobe.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Ryan P.M., London L.E.E., Bjorndahl T.C., Mandal R., Murphy K., Fitzgerald G.F., Shanahan F., Ross R.P., Wishart D.S., Caplice N.M., et al. Microbiome and metabolome modifying effects of several cardiovascular disease interventions in apo-E−/− mice. Microbiome. 2017;5:30. doi: 10.1186/s40168-017-0246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molly K., Vande Woestyne M., Verstraete W. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 1993;39:254–258. doi: 10.1007/BF00228615. [DOI] [PubMed] [Google Scholar]
- 24.Minekus M., Smeets-Peeters M., Bernalier A., Marol-Bonnin S., Havenaar R., Marteau P., Alric M., Fonty G., Huis in′t Veld J.H. A computer-controlled system to simulate conditions of the large intestine with peristaltic mixing, water absorption and absorption of fermentation products. Appl. Microbiol. Biotechnol. 1999;53:108–114. doi: 10.1007/s002530051622. [DOI] [PubMed] [Google Scholar]
- 25.Van den Abbeele P., Roos S., Eeckhaut V., MacKenzie D.A., Derde M., Verstraete W., Marzorati M., Possemiers S., Vanhoecke B., Van Immerseel F., et al. Incorporating a mucosal environment in a dynamic gut model results in a more representative colonization by lactobacilli. Microb. Biotechnol. 2012;5:106–115. doi: 10.1111/j.1751-7915.2011.00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L., Firrman J., Tanes C., Bittinger K., Thomas-Gahring A., Wu G.D., Van den Abbeele P., Tomasula P.M. Establishing a mucosal gut microbial community in vitro using an artificial simulator. PLoS ONE. 2018;13:e0197692. doi: 10.1371/journal.pone.0197692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pridmore R.D., Berger B., Desiere F., Vilanova D., Barretto C., Pittet A.C., Zwahlen M.C., Rouvet M., Altermann E., Barrangou R., et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA. 2004;101:2512–2517. doi: 10.1073/pnas.0307327101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pokusaeva K., Fitzgerald G.F., van Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011;6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raybaudi-Massilia R.M., Mosqueda-Melgar J., Soliva-Fortuny R., Martín-Belloso O. Control of Pathogenic and Spoilage Microorganisms in Fresh-cut Fruits and Fruit Juices by Traditional and Alternative Natural Antimicrobials. Compr. Rev. Food Sci. Food Saf. 2009;8:157–180. doi: 10.1111/j.1541-4337.2009.00076.x. [DOI] [PubMed] [Google Scholar]
- 30.Duncan S.H., Louis P., Flint H.J. Lactate-Utilizing Bacteria, Isolated from Human Feces, That Produce Butyrate as a Major Fermentation Product. Appl. Environ. Microbiol. 2004;70:5810. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macfarlane G.T., Gibson G.R. Carbohydrate Fermentation, Energy Transduction and Gas Metabolism in the Human Large Intestine. In: Mackie R.I., White B.A., editors. Gastrointestinal Microbiology: Volume 1 Gastrointestinal Ecosystems and Fermentations. Springer US; Boston, MA, USA: 1997. pp. 269–318. [DOI] [Google Scholar]
- 32.Lampe J.W., Navarro S.L., Hullar M.A.J., Shojaie A. Inter-individual differences in response to dietary intervention: Integrating omics platforms towards personalised dietary recommendations. Proc. Nutr. Soc. 2013;72:207–218. doi: 10.1017/S0029665113000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar M., Nagpal R., Kumar R., Hemalatha R., Verma V., Kumar A., Chakraborty C., Singh B., Marotta F., Jain S., et al. Cholesterol-lowering probiotics as potential biotherapeutics for metabolic diseases. Exp. Diabetes Res. 2012;2012:902917. doi: 10.1155/2012/902917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park S., Kang J., Choi S., Park H., Hwang E., Kang Y., Kim A., Holzapfel W., Ji Y. Cholesterol-lowering effect of Lactobacillus rhamnosus BFE5264 and its influence on the gut microbiome and propionate level in a murine model. PLoS ONE. 2018;13:e0203150. doi: 10.1371/journal.pone.0208294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuentes M.C., Lajo T., Carrión J.M., Cuñé J. Cholesterol-lowering efficacy of Lactobacillus plantarum CECT 7527, 7528 and 7529 in hypercholesterolaemic adults. Br. J. Nutr. 2013;109:1866–1872. doi: 10.1017/S000711451200373X. [DOI] [PubMed] [Google Scholar]
- 36.Jones M.L., Martoni C.J., Parent M., Prakash S. Cholesterol-lowering efficacy of a microencapsulated bile salt hydrolase-active Lactobacillus reuteri NCIMB 30242 yoghurt formulation in hypercholesterolaemic adults. Br. J. Nutr. 2011;107:1505–1513. doi: 10.1017/S0007114511004703. [DOI] [PubMed] [Google Scholar]
- 37.Kristek A., Wiese M., Heuer P., Kosik O., Schar M.Y., Soycan G., Alsharif S., Kuhnle G.G.C., Walton G., Spencer J.P.E. Oat bran, but not its isolated bioactive beta-glucans or polyphenols, have a bifidogenic effect in an in vitro fermentation model of the gut microbiota. Br. J. Nutr. 2019;121:549–559. doi: 10.1017/S0007114518003501. [DOI] [PubMed] [Google Scholar]
- 38.Zhao J., Cheung P.C. Fermentation of beta-glucans derived from different sources by bifidobacteria: Evaluation of their bifidogenic effect. J. Agric. Food Chem. 2011;59:5986–5992. doi: 10.1021/jf200621y. [DOI] [PubMed] [Google Scholar]
- 39.Takata K., Kinoshita M., Okuno T., Moriya M., Kohda T., Honorat J.A., Sugimoto T., Kumanogoh A., Kayama H., Takeda K., et al. The Lactic Acid Bacterium Pediococcus acidilactici Suppresses Autoimmune Encephalomyelitis by Inducing IL-10-Producing Regulatory T Cells. PLoS ONE. 2011;6:e27644. doi: 10.1371/journal.pone.0027644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneeberger M., Everard A., Gómez-Valadés A.G., Matamoros S., Ramírez S., Delzenne N.M., Gomis R., Claret M., Cani P.D. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 2015;5:16643. doi: 10.1038/srep16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staka A., Bodnieks E., Puķītis A. Impact of Oat-Based Products on Human Gastrointestinal Tract. Proc. Latv. Acad. Sci. 2015;69:145–151. doi: 10.1515/prolas-2015-0021. [DOI] [Google Scholar]
- 42.Knudsen K.E.B., Jensen B.B., Hansen I. Oat Bran but Not a β-Glucan-Enriched Oat Fraction Enhances Butyrate Production in the Large Intestine of Pigs. J. Nutr. 1993;123:1235–1247. doi: 10.1093/jn/123.7.1235. [DOI] [PubMed] [Google Scholar]
- 43.Kedia G., Vazquez J.A., Charalampopoulos D., Pandiella S.S. In Vitro fermentation of oat bran obtained by debranning with a mixed culture of human fecal bacteria. Curr. Microbiol. 2009;58:338–342. doi: 10.1007/s00284-008-9335-1. [DOI] [PubMed] [Google Scholar]
- 44.Hughes S.A., Shewry P.R., Gibson G.R., McCleary B.V., Rastall R.A. In Vitro fermentation of oat and barley derived beta-glucans by human faecal microbiota. FEMS Microbiol. Ecol. 2008;64:482–493. doi: 10.1111/j.1574-6941.2008.00478.x. [DOI] [PubMed] [Google Scholar]
- 45.Zhou J., Martin R.J., Tulley R.T., Raggio A.M., McCutcheon K.L., Shen L., Danna S.C., Tripathy S., Hegsted M., Keenan M.J. Dietary resistant starch upregulates total GLP-1 and PYY in a sustained day-long manner through fermentation in rodents. Am. J. Physiol. Endocrinol. Metab. 2008;295:E1160–E1166. doi: 10.1152/ajpendo.90637.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Lahham S.H., Roelofsen H., Priebe M., Weening D., Dijkstra M., Hoek A., Rezaee F., Venema K., Vonk R.J. Regulation of adipokine production in human adipose tissue by propionic acid. Eur. J. Clin. Investig. 2010;40:401–407. doi: 10.1111/j.1365-2362.2010.02278.x. [DOI] [PubMed] [Google Scholar]
- 47.Al-Lahham S., Roelofsen H., Rezaee F., Weening D., Hoek A., Vonk R., Venema K. Propionic acid affects immune status and metabolism in adipose tissue from overweight subjects. Eur. J. Clin. Investig. 2012;42:357–364. doi: 10.1111/j.1365-2362.2011.02590.x. [DOI] [PubMed] [Google Scholar]
- 48.Jan G., Belzacq A.S., Haouzi D., Rouault A., Metivier D., Kroemer G., Brenner C. Propionibacteria induce apoptosis of colorectal carcinoma cells via short-chain fatty acids acting on mitochondria. Cell Death Differ. 2002;9:179–188. doi: 10.1038/sj.cdd.4400935. [DOI] [PubMed] [Google Scholar]
- 49.Hayashi H., Shibata K., Sakamoto M., Tomita S., Benno Y. Prevotella copri sp. nov. and Prevotella stercorea sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2007;57:941–946. doi: 10.1099/ijs.0.64778-0. [DOI] [PubMed] [Google Scholar]
- 50.Hosseini E., Grootaert C., Verstraete W., Van de Wiele T. Propionate as a health-promoting microbial metabolite in the human gut. Nutr. Rev. 2011;69:245–258. doi: 10.1111/j.1753-4887.2011.00388.x. [DOI] [PubMed] [Google Scholar]
- 51.Kang E., Crouse A., Chevallier L., Pontier S., Alzahrani A., Silue N., Campbell-Valois F.-X., Montagutelli X., Gruenheid S., Malo D. Enterobacteria and host resistance to infection. Mamm. Genome. 2018;29 doi: 10.1007/s00335-018-9749-4. [DOI] [PubMed] [Google Scholar]
- 52.Possemiers S., Verthe K., Uyttendaele S., Verstraete W. PCR-DGGE-based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol. Ecol. 2004;49:495–507. doi: 10.1016/j.femsec.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 53.Alliance I.U.N. National Adult Nutrition Survey: Summary Report on Food and Nutrient Intakes, Physical Measurements, Physical Activity Patterns and Food Choice Motives. Trinity College Dublin, Irish Universities Nutrition Alliance; Dublin, Ireland: 2011. [Google Scholar]
- 54.De Weirdt R., Possemiers S., Vermeulen G., Moerdijk-Poortvliet T.C., Boschker H.T., Verstraete W., Van de Wiele T. Human faecal microbiota display variable patterns of glycerol metabolism. FEMS Microbiol. Ecol. 2010;74:601–611. doi: 10.1111/j.1574-6941.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- 55.Duysburgh C., Van den Abbeele P., Krishnan K., Bayne T.F., Marzorati M. A synbiotic concept containing spore-forming Bacillus strains and a prebiotic fiber blend consistently enhanced metabolic activity by modulation of the gut microbiome in vitro. Int. J. Pharm. X. 2019;1:100021. doi: 10.1016/j.ijpx.2019.100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown J.R.M., Flemer B., Joyce S.A., Zulquernain A., Sheehan D., Shanahan F., O’Toole P.W. Changes in microbiota composition, bile and fatty acid metabolism, in successful faecal microbiota transplantation for Clostridioides difficile infection. BMC Gastroenterol. 2018;18:131. doi: 10.1186/s12876-018-0860-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boon N., Top E.M., Verstraete W., Siciliano S.D. Bioaugmentation as a tool to protect the structure and function of an activated-sludge microbial community against a 3-chloroaniline shock load. Appl. Environ. Microbiol. 2003;69:1511–1520. doi: 10.1128/AEM.69.3.1511-1520.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furet J.P., Firmesse O., Gourmelon M., Bridonneau C., Tap J., Mondot S., Dore J., Corthier G. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 2009;68:351–362. doi: 10.1111/j.1574-6941.2009.00671.x. [DOI] [PubMed] [Google Scholar]
- 59.Rinttila T., Kassinen A., Malinen E., Krogius L., Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 2004;97:1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
- 60.Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glöckner F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schloss P.D., Westcott S.L. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 2011;77:3219–3226. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cole J.R., Wang Q., Fish J.A., Chai B., McGarrell D.M., Sun Y., Brown C.T., Porras-Alfaro A., Kuske C.R., Tiedje J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vandeputte D., Kathagen G., D′Hoe K., Vieira-Silva S., Valles-Colomer M., Sabino J., Wang J., Tito R.Y., De Commer L., Darzi Y., et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature. 2017;551:507–511. doi: 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available on request due to restrictions.