Abstract
Listeria monocytogenes is a Gram-positive bacterial pathogen and the causative agent of listeriosis, a severe foodborne infection. L. monocytogenes is notorious for its ability to persist in food processing environments (FPEs) via a variety of adaptive traits. Even though traits such as cold tolerance, biofilm formation and sanitizer resistance have been extensively investigated for their roles in persistence of L. monocytogenes in FPEs, much less is known about resistance to bacteriophages. Previous studies explored phage resistance mechanisms in laboratory-created mutants but it is imperative to investigate phage resistance that is naturally exhibited in FPE-derived strains. Here, we integrated the analysis of whole genome sequence data from a panel of serotype 1/2a strains of sequence types 321 and 391 from turkey processing plants, with the determination of cell surface substituents required for phage adsorption and phage infection assays with the four wide-host-range phages A511, P100, 20422-1 and 805405-1. Using a specific set of recombinant phage protein probes, we discovered that phage-resistant strains lacked one or both of the serogroup 1/2-specific wall teichoic acid carbohydrate decorations, N-acetylglucosamine and rhamnose. Furthermore, these phage-resistant strains harbored substitutions in lmo1080, lmo1081, and lmo2550, which mediate carbohydrate decoration of the wall teichoic acids.
Keywords: Listeria, bacteriophage, phage resistance, whole genome sequencing, food processing plant, InlA, serotype 1/2a, wall teichoic acid
1. Introduction
Listeria monocytogenes is a Gram-positive facultative intracellular pathogen found ubiquitously in nature and is notorious for its capacity to persist in food processing environments (FPEs). Contamination of FPEs is critical for contamination of ready-to-eat foods by L. monocytogenes, with the potential to lead to outbreaks of the severe and potentially life-threatening foodborne disease listeriosis [1,2,3]. L. monocytogenes has the ability to persist in FPEs via multiple adaptations, including its ability to grow at low temperatures, to form biofilms and tolerate sanitizers [4,5,6,7,8]. Even though such adaptive traits have been extensively investigated, the potential roles of bacteriophage (phage) resistance in the persistence of this pathogen in FPEs remains poorly understood.
Listeria-specific phage have been approved as a biocontrol tool against Listeria in FPEs and foods, e.g., phage P100 in Listex P100 [9]. Repeated interactions between L. monocytogenes and phage exert selective pressures that may select for, and eventually result in, phage resistance. Phage resistance in L. monocytogenes can be mediated by failure of the phage to adsorb to its specific receptors via receptor loss or modification, and various post-infection intracellular resistance mechanisms [10,11,12]. In L. monocytogenes and other bacteria, the latter can include prophages, bacteriophage exclusion (BREX) systems, defense island system associated with restriction-modification (DISARM) systems and clustered, regularly interspaced, short palindromic repeats (CRISPR) systems [13,14,15,16,17,18,19].
Investigations of phage resistance established under laboratory conditions have shown that certain genes of L. monocytogenes are critical both for normal wall teichoic acid (WTA) decoration and for phage susceptibility [10,11,20,21,22]. In strains of serogroup 1/2, the WTA decorations required for susceptibility to broad-host-range phages such as A511 and P100 are N-acetylglucosamine and rhamnose [23,24,25,26]. Many of the genes mediating phage susceptibility in serotype 1/2a are found in two chromosomal operons, lmo1079-lmo1084 and lmo2549-lmo2550 [10,25,27]. A previous study showed that spontaneous phage-resistant mutants of L. monocytogenes 10403S harbored mutations concentrated in two loci containing seven genes in total (lmo1079-lmo1084 and lmo2549-lmo2550). Furthermore, serotype 1/2a isolates from seafood industries frequently harbored non-synonymous mutations in lmo2549 or lmo2550 [14]. However, reports that integrate phage resistance, WTA decoration analysis and underlying genetic alterations in L. monocytogenes under field conditions, such as prevailing in FPEs, are largely lacking.
In a previous study, we characterized L. monocytogenes from different turkey processing plants in the United States for their resistance to a panel of phages [12]. The majority of the phage-resistant strains were of serotype 1/2a, followed by 1/2b and 1/2c, i.e., serotypes which are commonly encountered in food and food processing ecosystems [12,28,29,30,31]. In the current study, our objective was to integrate whole genome sequence analysis, phage adsorption assays and phenotypic characterization of WTA decorations in order to elucidate mechanisms mediating phage resistance in FPE-derived serotype 1/2a strains. To address this objective, we utilized whole genome sequence data to assess genomic differences between phage-resistant and phage-susceptible strains, as well as to identify sequence alterations in a targeted panel of genes previously implicated in WTA biosynthesis. These data were further correlated with the WTA decoration patterns revealed by a novel set of glycotyping protein probes [32] and with phage adsorption and infection assays.
2. Results and Discussion
Earlier investigations characterized isolates of L. monocytogenes from turkey processing plants in the US for resistance to phage as well as to benzalkonium chloride and the heavy metals cadmium and arsenic [12,33]. A subset of 10 strains of serotype 1/2a that were previously screened for resistance to three wide-host-range phages, i.e., 20422-1, 805405-1 and A511 and were also resistant to benzalkonium chloride and cadmium were chosen for whole genome sequencing (Table 1). In-silico multilocus sequence typing (MLST) revealed two sequence types (STs), ST321 and ST391 (Table 1). Strains of the same ST were repeatedly isolated from the same processing plant over a 30-month period (Table 1), reflecting potential persistence in the FPE. In a previous study of South African food products, 9.7% of L. monocytogenes isolates belonged to ST321, while in another study 33 of 42 isolates of L. monocytogenes from a cold-smoked salmon processing facility belonged to ST or CC321 [34]. Analysis of the ST321 strains used in the current study (Table 1) using the NCBI pathogen detection pipeline [35] identified numerous similar strains from food and environmental isolates (data not shown). In contrast to ST321, little is known about ST391 in the food processing environment. Analysis of the ST391 strains analyzed in this study (Table 1) using the NCBI pathogen detection pipeline identified only a small number of closely-related strains, from ready-to-eat foods [35].
Table 1.
Food processing environment (FPE)-derived strains of L. monocytogenes used in this study.
| Strain 1 | CFSAN # | Serotype | ST | Source 2 | Date 3 | InlA 4 | Phage 5 | GlcNac 6 | Rham 6 | Gene(s) | SNP 7 | Mutation 8 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 176b-1 * | CFSAN095955 | 1/2a | 321 | A | 4/04 | T730 | R | - | - |
|
|
|
| 206a-5 * | CFSAN095958 | 1/2a | 321 | A | 6/04 | T730 | R | - | + |
|
|
|
| 210b-1 * | CFSAN095959 | 1/2a | 321 | A | 6/04 | T730 | R | - | + |
|
|
|
| 339b-5 | CFSAN095964 | 1/2a | 321 | A | 12/04 | T730 | R | + | - |
|
|
|
| 494b-1 | CFSAN095969 | 1/2a | 321 | A | 3/06 | T730 | R | + | - |
|
|
|
| L1624a | CFSAN095967 | 1/2a | 321 | B | 7/05 | T730 | S | + | + | |||
| 171b-1 | CFSAN095954 | 1/2a | 391 | A | 4/04 | FL | R | + | - | lmo1080 | Nt267 (G to A), Nt 1,095,828 | PMSC |
| 231b-1 | CFSAN095960 | 1/2a | 391 | A | 8/04 | FL | R | - | + | lmo2550 | Nt479 (G to A), Nt 2,580,368 | NS |
| 506a-1 * | CFSAN095970 | 1/2a | 391 | A | 3/06 | FL | R | + | - | lmo1081 | Nt116 (C to T), Nt 1,097,565 | NS |
| #24 | CFSAN095953 | 1/2a | 391 | A | 9/03 | FL | S | + | + |
1 Strains tested for phage adsorption are marked with *. Strain 176b-1 was previously tested for phage adsorption using phage 20422-1 [12], while strains 206a-5, 210b-1 and 506a-1 were tested in this study with phage P100. 2 Strains were from different sites in two different turkey processing plants (A and B) in the United States, as previously described [12]. 3 Date is in month/year, as previously described [12]. 4 InlA length is indicated as either full length (FL; 800 AA) or by the position of the premature stop codon in the deduced polypeptide. 5 R and S indicate phage resistance and susceptibility, respectively, to the four wide-host-range phages A511, P100, 20422-1 and 805405-1. 6 N-acetylglucosamine and rhamnose are indicated by GlcNac and Rham, respectively. Their presence or absence is indicated by + and -, respectively. 7 Genes harboring mutations. SNPs in the corresponding gene are indicated by the SNP location in the ORF followed by location in the entire chromosome. 8 NS and PMSC indicate non-synonymous mutation and premature stop codon, respectively.
The previously-reported phage resistance profiles [12] were confirmed for all strains, which were additionally tested for their susceptibility to the broad-host-range phage P100. While phages 20422-1 and 805405-1 were isolated from turkey processing plants in the United States in the same study as the L. monocytogenes strains investigated here [12], phages A511 and P100 were isolated in Germany in the 1990s [36,37,38,39]. All tested strains were either resistant (R) or susceptible (S) to all four phages, with only one strain of each ST being susceptible (strains L1624a and #24 in ST321 and ST391, respectively) (Table 1). Hereafter, the terms phage resistance and susceptibility pertain to the observed resistance or susceptibility, respectively, towards the four wide-host-range phages that were employed, i.e., A511, P100, 20422-1 and 805405-1.
The WGS data of all strains were also analyzed for the presence of premature stop codons (PMSCs) in inlA, previously reported to be common among serotype 1/2a strains from FPEs and associated with hypovirulence [27,29,40,41]. All six strains of ST321 harbored a shared PMSC (T730) in inlA, regardless of whether they were phage-resistant or susceptible (Table 1). PMSCs in inlA were also found in ST321 strains from the South African food study as well as a cold-smoked salmon processing facility [34,42], suggesting that this is a clonal trait potentially reflecting adaptation of these strains to the processing plant environment. None of the ST391 strains harbored PMSCs in inlA (Table 1).
2.1. Absence of WTA Substituents in Phage-Resistant FPE-Derived Strains of ST321 and ST391 Is Accompanied by Phage Resistance
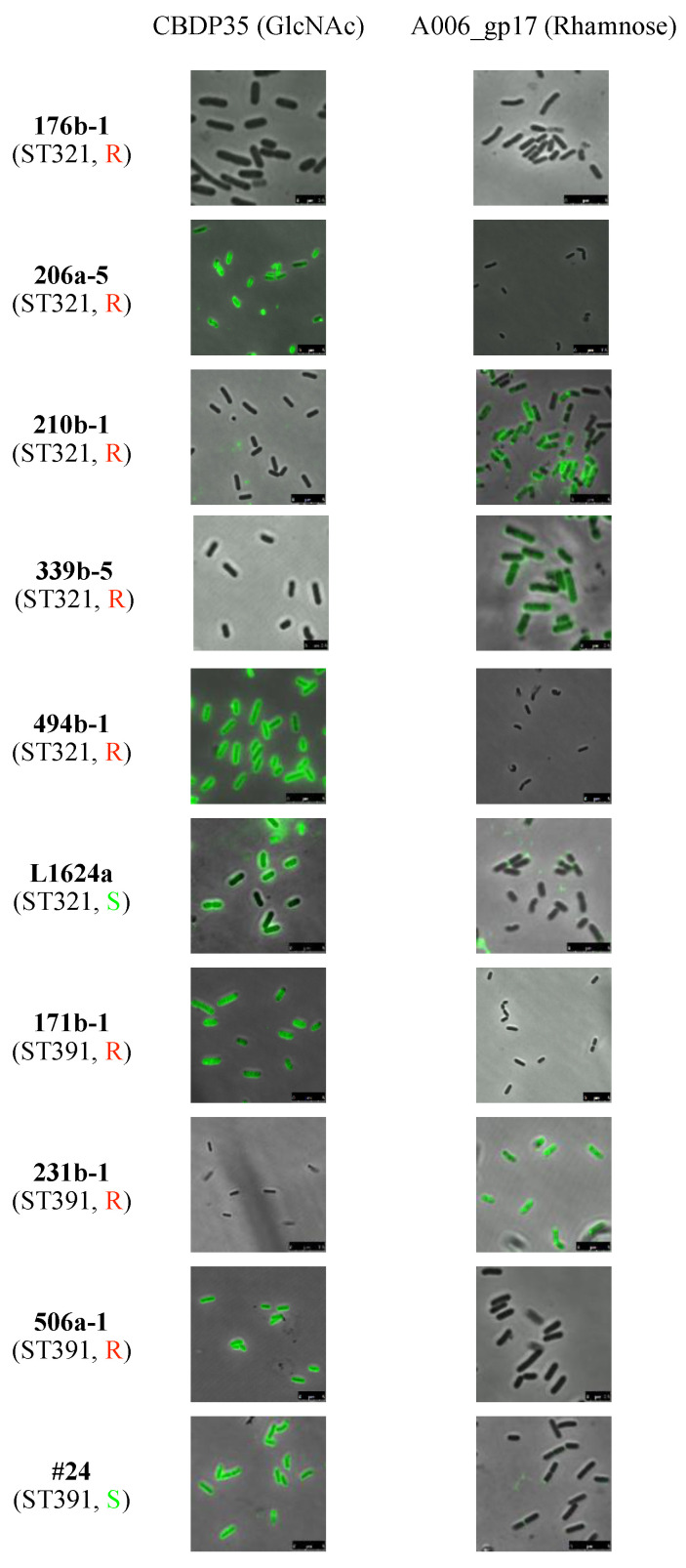
Previous studies have shown that N-acetylglucosamine (GlcNAc) and rhamnose substituents on WTA are critical for adsorption of phages A118, A511 and P35 to L. monocytogenes of serotype 1/2a, with loss of either of these conferring phage resistance [39,43,44,45]. Employment of a glycotyping assay specific to WTA-associated GlcNAc and rhamnose [32] revealed that the phage-susceptible strains L1624a and #24 (ST321 and, ST391, respectively) were positive for both GlcNAc and rhamnose, with the latter exhibiting relatively weak binding concentrated at the polar ends of the cell (Table 1 and Figure 1). In contrast, all phage-resistant strains of ST321 and ST391 lacked at least one of the WTA substituents. Specifically, the ST321 strains 339b-5 and 210b-1 and the ST391 strain 231b-1 were missing GlcNAc, while the ST321 strains 206a-5 and 494b-1 and the ST391 strains 171b-1 and 506a-1 were lacking rhamnose. Interestingly, one strain, 176b-1 (ST321) was missing both of these WTA substituents (Table 1 and Figure 1). The absence of both GlcNAc and rhamnose in the WTA of this strain was surprising. There is no obvious selective pressure for the loss of both WTA substituents, as the absence of just one is sufficient for resistance to broad-host-range phage [43,44]. Further studies are needed to elucidate the potential selective pressures that may render the loss of both WTA substituents advantageous to this strain.
Figure 1.
Glycotyping of serotype 1/2a L. monocytogenes strains of ST321 and ST391 strains investigated in this study. N-acetylglucosamine (GlcNAc) and rhamnose (Rhamn) were detected using GFP-labeled CBDP35 and A006_gp17 [32]. The green signal indicates the presence of the respective WTA substituent. Sequence type (ST) designations are in parentheses underneath the strain designation. R (red font) and S (green font) indicates phage resistance and susceptibility, respectively, to the four wide-host-range phages employed in the study, i.e., A511, P100, 20422-1 and 805405-1. The glycotyping assay was done as described in Materials and Methods.
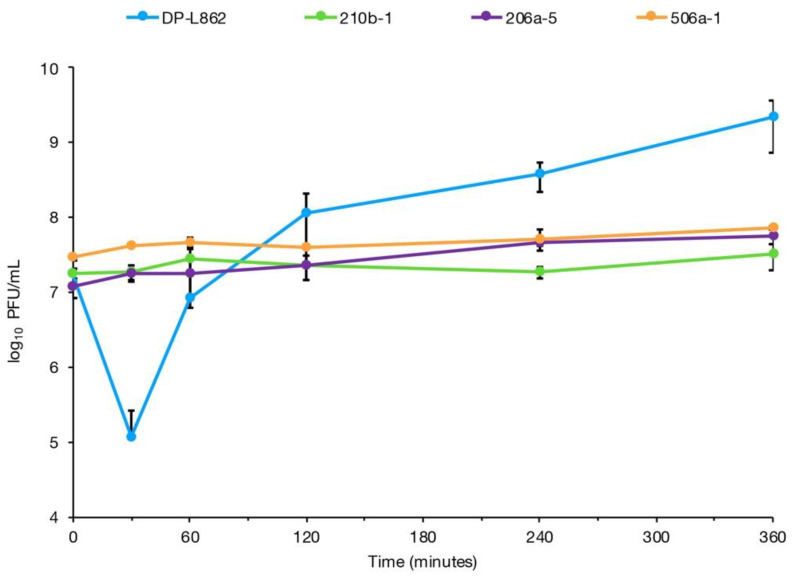
Previous work reported that phage 20422-1 failed to adsorb on strain 176b-1 (ST321) [12]. This was supported by the glycotyping data discussed above, which showed absence of both GlcNAc and rhamnose in this strain (Figure 1, Table 1). Testing phage P100 adsorption on additional strains of each ST representing the remaining glycotyping profiles (Table 1) confirmed that the phage failed to adsorb on phage-resistant strains that lacked either GlcNAc or rhamnose (Figure 2).
Figure 2.
Failure of phage to adhere to representative phage-resistant serotype 1/2a L. monocytogenes strains of ST321 and ST391. The phage-resistant strains were exposed to phage P100 for 360 min and PFU/mL in the supernatant was enumerated at specific time points as described in Materials and Methods. The phage-susceptible serotype 1/2a strain L. monocytogenes DP-L862 (blue) was used as positive control and the phage-resistant strains are 210b-1 (ST321, green), 206-5 (ST321, purple) and 506a-5 (ST391, orange). Error bars represent standard deviation.
2.2. FPE-Derived Strains Are Closely Related Despite Differences in Phage Resistance and Glycotyping Profiles
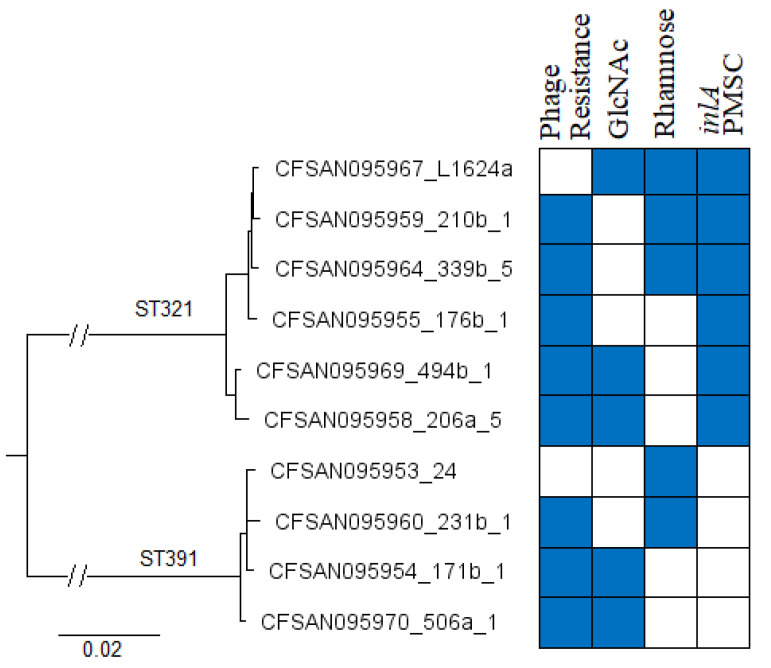
Analysis of the WGS data revealed that the FPE strains of the same ST were closely related (Figure 3). Analysis of the core genome (1748 genes) revealed only four to 18 cgMLST differences among the ST321 strains, and even fewer (four to eight cgMLST differences) among those of ST391. Interestingly, these strains did not segregate phylogenetically by their phage resistance phenotypes or glycotyping profiles (Figure 3). For instance, the ST321 strains L1624a and 210b-1 that were phage susceptible and resistant, respectively, only had four cgMLST allele differences (lmo0135, lmo0947, lmo1711 and lmo2518).
Figure 3.
Phylogenetic tree of FPE-derived phage-resistant and phage susceptible L. monocytogenes strains investigated in this study. Phage resistance (blue) and susceptibility (white), presence (blue) or absence (white) of N-acetylglucosamine (GlcNAc) and rhamnose in the WTA, and presence (blue) or absence (white) of an inlA premature stop codon (PMSC) are indicated in the heat map to the right of the tree. The strain designations are included after the CFSAN designation (with hyphens replaced by _), e.g. the top first and second strains in the tree correspond to strains L1624a and 210b-1, respectively. The tree was based on 1748 L. monocytogenes core genes from the Institute Pasteur multilocus sequence typing (MLST) database and was constructed as described in Materials and Methods.
WTA substituents are serotype-specific [23] and the genes involved in WTA glycosylation, e.g., lmo1079-lmo1084 and lmo2549-lmo2550, are not part of the L. monocytogenes core genome. Therefore, whole genome MLST (wgMLST) was also employed to determine allele differences among the strains. A relatively small number of differences were also found using wgMLST analysis, with 14–106 wgMLST differences among ST321 strains and only 16–46 wgMLST differences among those of ST391. Based on wgMLST there are 66 variable genes among the ST321 and 35 among the ST391 strains, excluding missing and incomplete alleles (Supplementary Table S1, Supplementary Table S2). Comparison of the alleles found to be variable by wgMLST revealed only three genes that were variable in both STs: lmo1080, lmo1081 and lmo2550. As indicated earlier, these three genes have all been implicated in WTA glycosylation, either with rhamnose (lmo1080 and lmo1081) or N-acetylglucosamine (lmo2550) [10,14,25,43].
2.3. SNPs in Wall Teichoic Acid Genes Contribute to Phage Resistance in FPE-Derived Strains
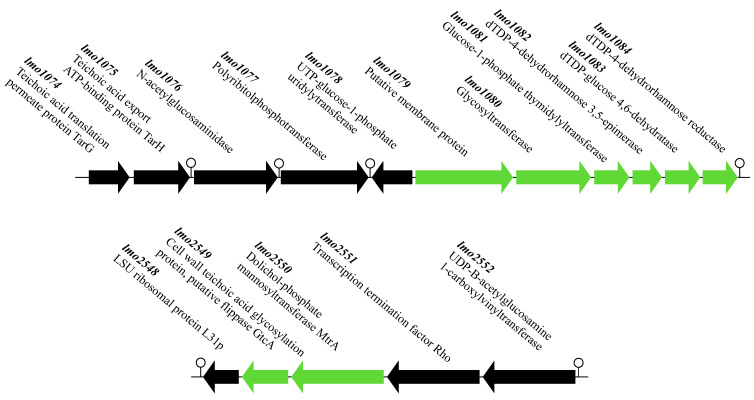
In a previous study, phage resistance of L. monocytogenes 10403S selected under laboratory conditions was found to be accompanied by SNPs in a panel of genes mediating glycosylation of the WTA with rhamnose and N-acetylglucosamine [10]. These genes are organized in two operons, lmo1079-1084 and lmo2549-lmo2550, in the chromosome of L. monocytogenes serotype 1/2a (Figure 4). Interestingly, the only three genes found to be variable in both STs via the wgMLST analysis discussed above, i.e., lmo1080, lmo1081 and lmo2550, belonged to these two operons.
Figure 4.
Genomic organization of lmo1079-lmo1084 (top) and lmo2549-lmo2550 (bottom) in the serotype 1/2a reference strain L. monocytogenes EGD-e [46]. Arrows indicate the direction of transcription. Green indicates ORFs previously described to be involved in glycosylation of WTA in L. monocytogenes serotype 1/2a with N-acetylglucosamine or rhamnose [10,14]. Lollipop symbols indicate putative terminators.
To determine whether mutations in the genes of the lmo1079-1084 and lmo2549-lmo2550 operons were associated with phage resistance in the FPE-derived-resistant strains in the current study, we compared the sequence of each gene between phage resistant and phage-susceptible strains of the same ST. Two phage-resistant strains, 494b-1 (ST321) and 506a-1 (ST391), were found to have non-synonymous SNPs in lmo1080 and to lack rhamnose in the WTA (Table 1, Figure 1). Disruption of lmo1080, as well as the dTDP-L-rhamnose biosynthesis genes (lmo1081-lmo1084), have been shown to result in loss of rhamnose in the WTA, as well as phage resistance [10,25]. In addition, phage P100 failed to adsorb to strain 506a-1 (Figure 2). Four strains, i.e., the ST321 strains 176b-1, 339b-5 and 210b-1 and the ST391 strain 231b-1, were found to have non-synonymous SNPs in lmo2550 and also lacked GlcNAc in the WTA (Table 1, Figure 1). Previous and current adsorption assays with two of these strains, 176b-1 and 210b-1, indicated failure of phage to adsorb to 176b-1 [12] and 210b-1 (Figure 2). Mutations in lmo2550 in serogroup 1/2 strains have been known to be accompanied with lack of GlcNAc in the WTA, both in laboratory mutants and seafood industry strains, even though resistance of the latter to phage was not reported [14,43].
Two strains, 206a-5 (ST321) and 171b-1 (ST391), were found to harbor PMSCs in lmo1080 and were missing rhamnose from the WTA. While no other SNPs were found in 171b-1 there was a second SNP in 206a-5 (lmo1084). The latter SNP is not expected to cause the loss of rhamnose as it was also found in four other strains, two of which had rhamnose in the WTA (Table 1, Figure 1). While not causing the absence of rhamnose or GlcNAc from the WTA, this non-synonymous substitution at nt 599 of lmo1084 was found in all five phage-resistant ST321 strains.
The SNP and glycotyping data, together with findings from the previous literature [10,14], allow us to postulate that PMSCs and non-synonymous SNPs in lmo1080 and lmo1081 may cause absence of rhamnose from the WTA, while non-synonymous SNPs in lmo2550 may cause absence of GlcNAc from the WTA in L. monocytogenes colonizing food processing plants, leading to resistance to wide-host-range phages. Phage adsorption data for strains 206a-5, 506a-1, 210b-1 and 176b-1 suggest linkage between mutations in lmo1080, lmo1081, and lmo2550 and failure of P100 to adsorb to the cell (Figure 2). Interestingly, the SNPs in these genes were at different locations than those reported previously [10], suggesting that mutations in multiple locations of these genes can alter WTA decoration and lead to phage resistance.
2.4. Further Studies
Exposure to phage in the FPEs and other environments may select for the loss of teichoic acid decorations in L. monocytogenes. The resulting resistance to phage may contribute to the apparent FPE persistence of the ST321 and ST391 strains investigated here, which were closely related and recovered from the same FPE over more than two years. However, in addition to serving as phage receptors, WTA glycosylation is increasingly recognized for its importance in other functions including surface adhesion, biofilm formation, anchoring of virulence determinants to the cell surface and resistance to antimicrobial peptides [14,19,47,48,49]. It will be of interest to investigate such potential trade-offs with the strains investigated here and other FPE-derived phage-resistant strains of serotype 1/2a. WTA glycosylation profiles differ noticeably among different serotypes of L. monocytogenes [23,24], and the fitness impacts of the loss of WTA decorations may exhibit serotype-specific traits.
3. Materials and Methods
3.1. Bacterial Strains and Growth Conditions
The L. monocytogenes strains investigated in this study are listed in Table 1. Unless otherwise noted, L. monocytogenes strains were grown in brain heart infusion broth (BHI; Becton, Dickinson & Co., Sparks, MD, USA) at 37 °C or on Luria–Bertani (LB) supplemented with 1.2% agar (LBA; Becton, Dickinson & Co.) and 10 mM calcium chloride (CaCl2) at 25 °C.
3.2. Listeria Phage Collection and Propagation, Phage Susceptibility and Adsorption Assays
Listeria phages used in this study are listed in Table 2. Phage propagation was as described [12] using L. monocytogenes DP-L862 as the propagating strain, resulting in phage titers of approximately 1.0 × 109 plaque forming units (PFU)/mL. Strains were screened for phage susceptibility in 96-well plates, as described [12], with minor modifications. Specifically, each strain was tested using six dilutions of phage, ranging from undiluted (~ 1.0 × 109 PFU/mL) to 10−5 (~1.0 × 104 PFU/mL). The 96-well plates were then incubated at 37 °C for 30 min, well contents transferred on to LBA-10mM CaCl2 using a stainless-steel replicator and incubated overnight at 25 °C.
Table 2.
Wide-host-range bacteriophages used in this study.
| Phage | Characteristics 1 | Source [Reference] | Date |
|---|---|---|---|
| 20422-1 | ND | Processing Plant, North Carolina, USA [12] | 2004 |
| 805405-1 | ND | Processing Plant, Virginia, USA [12] | 2005 |
| A511 | Virulent, Myoviridae | Sewage, Germany [40] | 1990 |
| P100 | Virulent, Myoviridae | Sewage, dairy plant, southern Germany [40] | 1997 |
1 ND, not determined, as these phages have not been sequenced or fully characterized.
Phage adsorption assays were done as described [12] with minor modifications. Specifically, L. monocytogenes DP-L862 was used to enumerate filtrate dilutions for each time point after phage infection (0, 0.5, 1, 2, 4 and 6 h). L. monocytogenes DP-L862 was used as positive control for phage adsorption and replication. Strains were tested in at least two independent trials.
3.3. Whole Genome Sequencing and Analysis
Genomic DNA was extracted using a DNeasy blood and tissue kit (Qiagen, Valencia, CA) from strains grown overnight at 37 °C in BHI broth (Becton, Dickinson & Co.). Libraries were prepared using 1 ng of genomic DNA with a Nextera XT DNA library preparation kit (Illumina, San Diego, CA, USA), and the genomes were sequenced using either a NextSeq 500 sequencer with the NextSeq 500/550 high-output kit v2.5 (300 cycles, 2 × 150 bp) (Illumina) or a MiSeq desktop sequencer with the Miseq kit v2 (500 cycles, 2 × 250 bp) (Illumina) according to the manufacturer’s instructions. The raw sequencing reads were then quality-trimmed and assembled de novo using Spades v.3.14.1 [50]; the assemblies were then quality-assessed using QUAST v.4.6.4 [51]. Default parameters were used for all software.
Whole genome analysis including an in-house BLAST of target genes was conducted using the Pathosystems Resource Integration Center (PatricBRC) and Artemis [52,53]. Briefly, chromosomal nucleotide locations of previously-described single-nucleotide polymorphisms (SNPs) were taken from Denes et al. [10] and localized in the L. monocytogenes 10403S genome using the nucleotide search function in Artemis. This was converted into whole-genome nucleotide locations for each of the genomes as well as ORF nucleotide locations for the target genes, yielding both whole-genome nucleotide location and nucleotide location within the relevant ORF (Table 1). We extracted the wild-type alleles of the genes of interest (lmo1079-lmo1084, lmo2549-lmo2550) from the phage-susceptible strains of each ST and used BLAST to identify their counterparts in phage-resistant strains of the same ST. Whole genome (wgMLST) and core genome (cgMLST) multilocus allele differences were identified using BIGSdb PasteurMLST Genome Comparator [54]. A phylogenetic tree of the strains was constructed by importing the 1748 core genes from the PasteurMLST into Ridom SeqSphere+, as previously described [55].
3.4. Glycotyping Protein Toolkit Analysis
Glycotyping with a pair of GFP-tagged phage proteins including A006_gp17 (to identify rhamnose) and CBDP35 (to identify N-acetylglucosamine) was performed as previously described [32]. Briefly, Listeria cells from log phase cultures (OD600nm = ~0.5) were harvested by centrifugation (10,000 g, 1 min), and resuspended in 1/5 volume of PBS (pH 7.4). The cell suspension (100 µL) was incubated with 5 µL of 1 mg/mL of the GFP-fused proteins and incubated for 5 min at room temperature. The cells were centrifuged, washed twice in PBS and finally resuspended in PBS. The samples were transferred onto a glass slide with a cover slip and examined by a confocal laser scanning microscope (Leica Microsystems GmbH, Germany) equipped with a HCX PL FLUOTAR 100 × 1.30 oil objective. Image analysis was performed in Leica Suite Software (Bitplane AG, Zurich, Switzerland). For ease of visualization, contrast in red and green channels was enhanced. Each strain was tested in at least two independent trials.
3.5. Genome Sequence Accession Numbers
The whole genome sequence for strains used in this study can be found under the following accession numbers under the BioProject PRNJA215355: 176b-1 (SRR13521630), 206a-5 (SRR13521680), 210b-1 (SRR13521681), 339b-5 (SRR13521793), 494b-1 (SRR13521790), L1624a (SRR13521632), 171b-1 (SRR13521825), 231b-1 (SRR13521718), 506a-1 (SRR13521795) and #24 (SRR13521631).
Acknowledgments
We would like to acknowledge the sequencing effort by the CFSAN GenomeTrackr sequencing team. We thank James Jackson for technical assistance and all members of our laboratory for support.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/10/2/199/s1, Table S1: ST321 wgMLST variable alleles, Table S2: ST391 wgMLST variable alleles.
Author Contributions
Conceptualization, S.K., P.B.; formal analysis, P.B. funding acquisition, S.K., C.P., M.J.L.; investigation, P.B., Y.C., Y.S., E.B.; project administration, S.K.; visualization, P.B., Y.C., Y.S.; writing—original draft, P.B.; writing—review and editing, P.B., S.K., Y.C., C.P., M.J.L., and Y.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by award 2018-07464 from the USDA National Institute of Food and Agriculture. Any opinions, findings, conclusions or recommendations expressed are those of the author and do not necessarily reflect the view of the USDA.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.De Noordhout C.M., Devleesschauwer B., Angulo F.J., Verbeke G., Haagsma J., Kirk M., Havelaar A., Speybroeck N. The global burden of listeriosis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014;14:1073–1082. doi: 10.1016/S1473-3099(14)70870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Painter J., Slutsker L. Listeriosis in humans. In: Ryser E.T., Marth E.H., editors. Listeriosis in Humans. Listeria, Listeriosis, and Food Safety. 3rd ed. CRC Press; Boca Raton, FL, USA: 2007. pp. 85–110. [Google Scholar]
- 3.Scallan E., Hoekstra R.M., Angulo F.J., Tauxe R.V., Widdowson M.A., Roy S.L., Jones J.L., Griffin P.M. Foodborne illness acquired in the United States--major pathogens. Emerg. Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Augustin J.C., Zuliani V., Cornu M., Guillier L. Growth rate and growth probability of Listeria monocytogenes in dairy, meat and seafood products in suboptimal conditions. J. Appl. Microbiol. 2005;99:1019–1042. doi: 10.1111/j.1365-2672.2005.02710.x. [DOI] [PubMed] [Google Scholar]
- 5.Hua Z., Korany A.M., Shinawy E.S.H., Zhu M.J. Comparative evaluation of different sanitizers against Listeria monocytogenes biofilms on major food-contact surfaces. Front. Microbiol. 2019;10:2462. doi: 10.3389/fmicb.2019.02462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Junttila J.R., Niemelä S.I., Hirn J. Minimum growth temperatures of Listeria monocytogenes and non-haemolytic Listeria. J. Appl. Bacteriol. 1988;65:321–327. doi: 10.1111/j.1365-2672.1988.tb01898.x. [DOI] [PubMed] [Google Scholar]
- 7.Kovacevic J., Ziegler J., Zacharska W.E., Reimer A., Kitts D.D., Gilmour M.W. Tolerance of Listeria monocytogenes to quaternary ammonium sanitizers is mediated by a novel efflux pump encoded by emrE. Appl. Environ. Microbiol. 2015;82:939–953. doi: 10.1128/AEM.03741-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee B.H., Cole S., Berchoux B.S., Guillier L., Felix B., Krezdorn N., Hébraud M., Bernardi T., Sultan I., Piveteau P. Biofilm formation of Listeria monocytogenes strains under food processing environments and pan-genome-wide association study. Front. Microbiol. 2019;10:2698. doi: 10.3389/fmicb.2019.02698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soni K.A., Nannapaneni R. Removal of Listeria monocytogenes biofilms with bacteriophage P100. J. Food. Prot. 2010;73:1519–1524. doi: 10.4315/0362-028X-73.8.1519. [DOI] [PubMed] [Google Scholar]
- 10.Denes T., Bakker D.H.C., Tokman J.I., Guldimann C., Wiedmann M. Selection and characterization of phage-resistant mutant strains of Listeria monocytogenes reveal host genes linked to phage adsorption. Appl. Environ. Microbiol. 2015;81:4295–4305. doi: 10.1128/AEM.00087-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran H.L., Fiedler F., Hodgson D.A., Kathariou S. Transposon-induced mutations in two loci of Listeria monocytogenes serotype 1/2a result in phage resistance and lack of N-acetylglucosamine in the teichoic acid of the cell wall. Appl. Environ. Microbiol. 1999;65:4793–4798. doi: 10.1128/AEM.65.11.4793-4798.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J.W., Siletzky R.M., Kathariou S. Host ranges of Listeria-specific bacteriophages from the turkey processing plant environment in the United States. Appl. Environ. Microbiol. 2008;74:6623–6630. doi: 10.1128/AEM.01282-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denomy B.J., Qian J., Westra E.R., Buckling A., Guttman D.S., Davidson A.R., Maxwell K.L. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016;10:2854–2866. doi: 10.1038/ismej.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brauge T., Faille C., Sadovskaya I., Charbit A., Benezech T., Shen Y., Loessner M.J., Bautista J.R., Bourdin M.G. The absence of N-acetylglucosamine in wall teichoic acids of Listeria monocytogenes modifies biofilm architecture and tolerance to rinsing and cleaning procedures. PLoS ONE. 2018;13:e0190879. doi: 10.1371/journal.pone.0190879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di H., Ye L., Yan H., Meng H., Yamasak S., Shi L. Comparative analysis of CRISPR loci in different Listeria monocytogenes lineages. Biochem. Biophys. Res. Commun. 2014;454:399–403. doi: 10.1016/j.bbrc.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 16.Goldfarb T., Sberro H., Weinstock E., Cohen O., Doron S., Amikam C.Y., Afik S., Ofir G., Sorek R. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 2015;34:169–183. doi: 10.15252/embj.201489455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hupfeld M., Trasanidou D., Ramazzini L., Klumpp J., Loessner M.J., Kilcher S. A functional type II-A CRISPR-Cas system from Listeria enables efficient genome editing of large non-integrating bacteriophage. Nucleic Acids Res. 2018;46:6920–6933. doi: 10.1093/nar/gky544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J.W., Dutta V., Elhanafi D., Lee S., Osborne J.A., Kathariou S. A novel restriction-modification system is responsible for temperature-dependent phage resistance in Listeria monocytogenes ECII. Appl. Environ. Microbiol. 2012;78:1995–2004. doi: 10.1128/AEM.07086-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ofir G., Melamed S., Sberro H., Mukamel Z., Silverman S., Yaakov G., Doron S., Sorek R. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat. Microbiol. 2018;3:90–98. doi: 10.1038/s41564-017-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sumrall E.T., Shen Y., Keller A.P., Rismondo J., Pavlou M., Eugster M.R., Boulos S., Disson O., Thouvenot P., Kilcher S., et al. Phage resistance at the cost of virulence: Listeria monocytogenes serovar 4b requires galactosylated teichoic acids for InlB-mediated invasion. PLoS Pathog. 2019;15:8032. doi: 10.1371/journal.ppat.1008032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei X.H., Fiedler F., Lan Z., Kathariou S. A novel serotype-specific gene cassette (gltA-gltB) is required for expression of teichoic acid-associated surface antigens in Listeria monocytogenes of serotype 4b. J. Bacteriol. 2001;183:1133–1139. doi: 10.1128/JB.183.4.1133-1139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Promadej N., Fiedler F., Cossart P., Dramsi S., Kathariou S. Cell wall teichoic acid glycosylation in Listeria monocytogenes serotype 4b requires gtcA, a novel, serogroup-specific gene. J. Bacteriol. 1999;181:418–425. doi: 10.1128/JB.181.2.418-425.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uchikawa K., Sekikawa I., Azuma I. Structural studies on teichoic acids in cell walls of several serotypes of Listeria monocytogenes. J. Biochem. 1986;99:315–327. doi: 10.1093/oxfordjournals.jbchem.a135486. [DOI] [PubMed] [Google Scholar]
- 24.Fiedler F. Biochemistry of the cell surface of Listeria strains: A locating general view. Infection. 1988;2:S92–S97. doi: 10.1007/BF01639729. [DOI] [PubMed] [Google Scholar]
- 25.Eugster M.R., Morax L.S., Hüls V.J., Huwiler S.G., Leclercq A., Lecuit M., Loessner M.J. Bacteriophage predation promotes serovar diversification in Listeria monocytogenes. Mol. Microbiol. 2015;97:33–46. doi: 10.1111/mmi.13009. [DOI] [PubMed] [Google Scholar]
- 26.Shen Y., Boulos S., Sumrall E., Gerber B., Rodero J.A., Eugster M.R., Fieseler L., Nyström L., Ebert M.O., Loessner M.J. Structural and functional diversity in Listeria cell wall teichoic acids. J. Biol. Chem. 2017;292:17832–17844. doi: 10.1074/jbc.M117.813964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rismondo J., Percy M.G., Gründling A. Discovery of genes required for lipoteichoic acid glycosylation predicts two distinct mechanisms for wall teichoic acid glycosylation. J. Biol. Chem. 2018;293:3293–3306. doi: 10.1074/jbc.RA117.001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurpas M., Osek J., Moura A., Leclercq A., Lecuit M., Wieczorek K. Genomic Characterization of Listeria monocytogenes isolated from ready-to-eat meat and meat processing environments in Poland. Front. Microbiol. 2020;11:1412. doi: 10.3389/fmicb.2020.01412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maury M.M., Tsai Y.H., Charlier C., Touchon M., Francisque C.V., Leclercq A., Criscuolo A., Gaultier C., Roussel S., Brisabois A., et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016;48:308–313. doi: 10.1038/ng.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moura A., Criscuolo A., Pouseele H., Maury M.M., Leclercq A., Tarr C., Björkman J.T., Dallman T., Reimer A., Enouf V., et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2017;2:16185. doi: 10.1038/nmicrobiol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H., Chen W., Wang J., Xu B., Liu H., Dong Q., Zhang X. 10-Year Molecular Surveillance of Listeria monocytogenes Using whole-genome sequencing in Shanghai, China, 2009–2019. Front. Microbiol. 2020;11:3037. doi: 10.3389/fmicb.2020.551020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sumrall E.T., Röhrig C., Hupfeld M., Selvakumar L., Du J., Dunne M., Schmelcher M., Shen Y., Loessner M.J. Glycotyping and specific separation of Listeria monocytogenes with a novel bacteriophage protein tool kit. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.00612-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mullapudi S., Siletzky R.M., Kathariou S. Diverse cadmium resistance determinants in Listeria monocytogenes isolates from the turkey processing plant environment. Appl. Environ. Microbiol. 2010;76:627–630. doi: 10.1128/AEM.01751-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrand A.S., Jagadeesan B., Baert L., Wiedmann M., Orsi R.H. Evolution of Listeria monocytogenes in a food processing plant involves limited single-nucleotide substitutions but considerable diversification by gain and loss of prophages. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.02493-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The NCBI Pathogen Detection Project [Internet] [(accessed on 26 January 2021)]; Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/pathogens.
- 36.Carlton R.M., Noordman W.H., Biswas B., de Meester E.D., Loessner M.J. Bacteriophage P100 for control of Listeria monocytogenes in foods: Genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharmacol. 2005;43:301–312. doi: 10.1016/j.yrtph.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Guenther S., Huwyler D., Richard S., Loessner M.J. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 2009;75:93–100. doi: 10.1128/AEM.01711-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Habann M., Leiman P.G., Vandersteegen K., Van den Bossche A., Lavigne R., Shneider M.M., Bielmann R., Eugster M.R., Loessner M.J., Klumpp J. Listeria phage A511, a model for the contractile tail machineries of SPO1-related bacteriophages. Mol. Microbiol. 2014;92:84–99. doi: 10.1111/mmi.12539. [DOI] [PubMed] [Google Scholar]
- 39.Klumpp J., Loessner M.J. Listeria phages: Genomes, evolution, and application. Bacteriophage. 2013;3:6861. doi: 10.4161/bact.26861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manuel C.S., Van Stelten A., Wiedmann M., Nightingale K.K., Orsi R.H. Prevalence and distribution of Listeria monocytogenes inlA alleles prone to phase variation and inlA alleles with premature stop codon mutations among human, food, animal, and environmental isolates. Appl. Environ. Microbiol. 2015;81:8339–8345. doi: 10.1128/AEM.02752-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward T.J., Evans P., Wiedmann M., Usgaard T., Roof S.E., Stroika S.G., Hise K. Molecular and phenotypic characterization of Listeria monocytogenes from U.S. Department of Agriculture Food Safety and Inspection Service surveillance of ready-to-eat foods and processing facilities. J. Food Prot. 2010;73:861–869. doi: 10.4315/0362-028X-73.5.861. [DOI] [PubMed] [Google Scholar]
- 42.Matle I., Mafuna T., Madoroba E., Mbatha K.R., Magwedere K., Pierneef R. Population structure of non-ST6 Listeria monocytogenes isolated in the red meat and poultry value chain in South Africa. Microorganisms. 2020;8:1152. doi: 10.3390/microorganisms8081152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eugster M.R., Haug M.C., Huwiler S.G., Loessner M.J. The cell wall binding domain of Listeria bacteriophage endolysin PlyP35 recognizes terminal GlcNAc residues in cell wall teichoic acid. Mol. Microbiol. 2011;81:1419–1432. doi: 10.1111/j.1365-2958.2011.07774.x. [DOI] [PubMed] [Google Scholar]
- 44.Wendlinger G., Loessner M.J., Scherer S. Bacteriophage receptors on Listeria monocytogenes cells are the N-acetylglucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology. 1996;142:985–992. doi: 10.1099/00221287-142-4-985. [DOI] [PubMed] [Google Scholar]
- 45.Bielmann R., Habann M., Eugster M., Lurz R., Calendar R., Klumpp J., Loessner M.J. Receptor binding proteins of Listeria monocytogenes bacteriophages A118 and P35 recognize serovar-specific teichoic acids. Virology. 2015;477:110–118. doi: 10.1016/j.virol.2014.12.035. [DOI] [PubMed] [Google Scholar]
- 46.Glaser P., Frangeul L., Buchrieser C., Rusniok C., Amend A., Baquero F., Berche P., Bloecker H., Brandt P., Chakraborty T., et al. Comparative genomics of Listeria species. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 47.Meireles D., Pombinho R., Carvalho F., Sousa S., Cabanes D. Listeria monocytogenes wall teichoic acid glycosylation promotes surface anchoring of virulence factors, resistance to antimicrobial peptides, and decreased susceptibility to antibiotics. Pathogens. 2020;9:290. doi: 10.3390/pathogens9040290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carvalho F., Atilano M.L., Pombinho R., Covas G., Gallo R.L., Filipe S.R., Sousa S., Cabanes D. L-Rhamnosylation of Listeria monocytogenes wall teichoic acids promotes resistance to antimicrobial peptides by delaying interaction with the membrane. PLoS Pathog. 2015;11:4919. doi: 10.1371/journal.ppat.1004919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carvalho F., Sousa S., Cabanes D. L-Rhamnosylation of wall teichoic acids promotes efficient surface association of Listeria monocytogenes virulence factors InlB and Ami through interaction with GW domains. Environ. Microbiol. 2018;20:3941–3951. doi: 10.1111/1462-2920.14351. [DOI] [PubMed] [Google Scholar]
- 50.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carver T., Harris S.R., Berriman M., Parkhill J., McQuillan J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis J.J., Wattam A.R., Aziz R.K., Brettin T., Butler R., Butler R.M., Chlenski P., Conrad N., Dickerman A., Dietrich E.M., et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020;48:D606–D612. doi: 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jolley K.A., Bray J.E., Maiden M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y., Escalona G.N., Hammack T.S., Allard M.W., Strain E.A., Brown E.W. Core genome multilocus sequence typing for identification of globally distributed clonal groups and differentiation of outbreak strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2016;82:6258–6272. doi: 10.1128/AEM.01532-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.