Abstract
Ascorbate peroxidase (APX) is a key antioxidant enzyme that is involved in diverse developmental and physiological process and stress responses by scavenging H2O2 in plants. APX itself is also subjected to multiple posttranslational modifications (PTMs). However, redox-mediated PTM of APX in plants remains poorly understood. Here, we identified and confirmed that MaAPX1 interacts with methionine sulfoxide reductase B2 (MsrB2) in bananas. Ectopic overexpression of MaAPX1 delays the detached leaf senescence induced by darkness in Arabidopsis. Sulfoxidation of MaAPX1, i.e., methionine oxidation, leads to loss of the activity, which is repaired partially by MaMsrB2. Moreover, mimicking sulfoxidation by mutating Met36 to Gln also decreases its activity in vitro and in vivo, whereas substitution of Met36 with Val36 to mimic the blocking of sulfoxidation has little effect on APX activity. Spectral analysis showed that mimicking sulfoxidation of Met36 hinders the formation of compound I, the first intermediate between APX and H2O2. Our findings demonstrate that the redox state of methionine in MaAPX1 is critical to its activity, and MaMsrB2 can regulate the redox state and activity of MaAPX1. Our results revealed a novel post-translational redox modification of APX.
Keywords: ascorbate peroxidase, reactive oxygen species, methionine sulfoxide reductase, sulfoxidation, posttranslational modification
1. Introduction
Reactive oxygen species (ROS) are formed as natural byproducts during cellular metabolism and play important roles in cell signaling and homeostasis [1,2]. However, under stress conditions or during senescence, excess ROS accumulation can cause oxidative damage to macromolecules such as proteins, DNA, and lipids, which, in turn, results in loss of structure and function or even potential cell death [1,2,3,4]. To counteract oxidative stress, organisms have evolved complex protective systems, such as antioxidant systems and macromolecule repair systems [5].
The enzymatic antioxidant system constitutes the most critical protective mechanism in plants, including ascorbate peroxidase (APX), catalase, superoxide dismutase, glutathione peroxidase, and peroxidase [1,6,7]. In addition, some low molecular mass antioxidants (ascorbate, glutathione, tocopherols, and phenolic compounds) are involved in scavenging excess ROS in the plant [8]. Among the antioxidant enzymes, APX plays a crucial role in scavenging H2O2 by catalyzing the conversion of H2O2 to H2O and O2, using ascorbic acid as the electron donor [9]. APX has a higher affinity than does catalase for H2O2 and contributes maximally to H2O2 detoxification in chloroplasts, cytosol, mitochondria, and peroxisomes, as well as in the apoplastic space [10]. APXs are known to be involved in the physiological and developmental response, such as seed germination [11], leaf senescence [12], and programmed cell death [13]. APX also participates in environmental stresses in plants, including drought [14], salt [15], chilling [16,17], photo-oxidative stress [18], and high temperature [19].
Proteins are one of the main targets of oxidative damage. Oxidative stress can cause irreversible or reversible modification of proteins. Irreversible oxidation, such as carbonylation, generally leads to impairment of protein function [20]. Reversible oxidation, particularly at methionine and cysteine residues, can be repaired by the Msr and thioredoxin (Trx) systems, respectively. Met oxidation forms two diastereomers of methionine sulfoxide (Met-S-O and Met-R-O), which can be reduced by the methionine sulfoxide reductases, MsrA and MsrB, respectively. Previous studies on Msr have mainly focused on its role in resistance to oxidative stress in organisms [21,22,23,24], which is related to repair of oxidized proteins, such as GroEL [25], Fth [26], hERG [27], apolipoprotein A-I [28], CaMKII [29], TRPM6 channel [30], HypT [31], actin [32], CaM [33,34], GrpEL1/Mge1 [35], heme oxygenase [36], and high-density lipoprotein [37]. Growing evidence suggests that Msr may be implicated in the regulation of protein function by modifying the sulfoxidation of proteins in a similar manner to that of other protein modification, such as phosphorylation [38,39,40,41]. However, only a few potential targets of Msr in relation to sulfoxidation modification have been investigated in higher plants [23,34,42].
Seed-bearing fruits in flowering plants provide humans with an important food source. Senescence is a vital stage of fruit life and directly decreases fruit quality and reduces resistance to pathogens, which usually cause enormous economic losses. Most fruits after harvest undergo vigorous aerobic respiration, accompanied by the accumulation of ROS. Recent discoveries have revealed that fruit senescence is considered to be related to ROS accumulation and oxidative damage of protein [43]. Therefore, elimination of ROS and repair of oxidized proteins are crucial for the maintenance of the function of the proteins and regulation of senescence.
Banana (Musa acuminate L.) is one of the most economically important fruits, undergoing rapid ripening and senescence once harvested. Our preliminary study found that one APX (GSMUA_Achr5P07280_001), designated as MaAPX1, might be a target of MaMsrB2 by Co-IP and mass spectrum techniques in banana fruit [44]. Here, we evaluated the possible role of MaAPX1 in regulating senescence by heterologous expression of MaAPX1 in Arabidopsis thaliana. We then validated that MaAPX1 was a substrate of MaMsrB2 and reduction of oxidized Met in MaAPX1 by MaMsrB2 reversibly switched on the MaAPX1 activity. In addition, mimicking oxidation of Met36 in MaAPX1 further confirmed that the redox state of Met36 can influence MaAPX1 activity. Furthermore, the mechanism underlying MaMsrB2-mediated regulation of MaAPX1 activity was further investigated by spectroscopic analysis. Our results revealed a novel mechanism of post-translational redox modification of APX.
2. Materials and Methods
2.1. Plant Materials and Treatments
Green mature banana (Musa acuminata L.) fruit (approximately 110 days after anthesis) were harvested from an orchard in Guangzhou, Guangdong province, China. Fruit with uniform shape, color, and size were selected and grouped into two lots. One group was stored under 60% oxygen concentration at 25 °C and 85-90% relative humidity. The other group was stored under normal air conditions as the control. During storage, samples were periodically taken to measure the related physiological parameters.
2.2. Measurement of Physiological Parameters
Chlorophyll fluorescence was measured as previously described [45]. The protein carbonyl content was spectrophotometrically quantified using a carbonyl-specific reagent, 2, 4-dinitrophenylhydrazine. Malondialdehyde (MDA) content and total APX activity were determined using corresponding assay kits (Comin Biotechnology Co., Ltd., Suzhou, China) in accordance with the manufacturer’s instructions.
2.3. RNA Extraction, Gene Isolation, and Expression Analyses
Total RNA was extracted as previously described [46] and then subjected to reverse transcription-PCR. MaMsrB2 (GSMUA_Achr6T13640_001) and MaAPX1 (GSMUA_Achr5P07280_001) were isolated from a transcriptome database obtained using a SolexaHiSeqTM 2000 sequencing system. The gene-specific primers used for gene cloning are listed in Table S1. The PCR products were subcloned into a pMD20-T vector (TaKara) and then transformed into E. coli DH5α (TaKara) in accordance with the manufacturer’s protocol. The sequences were verified by further cloning and resequencing. Sequence alignments were carried out using ClustalX (version 1.83).
The qRT-PCR reactions for MaMsrB2 and MaAPX1 genes were carried out in the ABI 7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) with SYBR Green Real-Time PCR Master Mix (TOYOBO Co., Ltd.). The conditions were as followed: 30 s at 95 °C, 40 cycles of 5 s at 95 °C, and 34 s at 58 °C. MaActin was selected as the reference gene. qRT-PCR reactions were normalized using the Ct value corresponding to that of the reference gene. The relative expression levels of target genes were calculated using the formula 2−ΔΔCT. Three independent biological replicates were performed in the analysis. The primers used for gene expression analyses are listed in Table S1.
2.4. Generation of Transgenic Lines and Dark Treatment
The MaAPX1 cDNA was sub-cloned in to a pCambia-1302 vector and then transformed into E. coli DH5α (TaKara) following the manufacturer’s protocol. The validated destination vectors were transformed into the Agrobacterium tumefaciens strain GV3101, and then transferred into the apx1-2 Arabidopsis via the floral-dip method [47]. Homozygous transgenic progeny lines were obtained through hygromycin-resistance tests and RT-PCR analysis, and they were validated by analyzing the transcript levels of MaAPX1-GFP in the T3 generations. The gene-specific primers are listed in Table S1. Dark treatment of the detached leaves was performed as previously described [44]. Chlorophyll fluorescence was determined as per the above-mentioned method.
2.5. Subcellular Localization Analysis of MaMsrB2 and MaAPX1
The coding sequence fragments of MaMsrB2 and MaAPX1 without the stop codon were inserted into the pCambia1302 vector. Then, pCambia1302-MaMsrB2, pCambia1302-MaAPX1, or pCambia1302 (as the control) were transferred to Arabidopsis mesophyll protoplasts as previously described [48]. The green fluorescent protein (GFP) fluorescence was observed by a florescence microscope (Zeiss 510 Meta) after 18 h of incubation at 22 °C. All the transient expression assays were repeated at least three times.
2.6. Site-Directed Mutagenesis of Met36 Residues
The following site-directed mutagenesis in MaAPX1 were performed: Met36 to glutamine and valine using a PCR method [33]. The mutations were verified by DNA sequencing.
2.7. Preparation of Recombinant Proteins
The encoding sequence fragments of MaMsrB2, MaAPX1, or MaAPX1 mutants were subcloned into the pGEX-4T-3 (Amersham Biosciences) or pET-28a vector (Novagen), respectively. MaMsrB2-GST, MaAPX1-His, MaAPX1-M36Q-His, and MaAPX1-M36V-His were induced and expressed in the E. coli BL21 (DE3) strain, and then were purified with glutathione sepharose 4B (GE Healthcare) and nickel-nitrilotriacetic acid agarose (Qiagen), respectively, following the manufacturer’s instructions (Figure S1).
2.8. Yeast Two-Hybrid (Y2H) Assay
The coding sequences of MaAPX1 and MaMsrB2 were subcloned into pGBKT7-BD or pGADT7-AD vectors to create bait and prey constructs. Then, the paired DNA-binding domain (BD) and transcription-activating domain (AD) constructs were co-transformed into the yeast strain AH109 (BioMed) using the lithium acetate method. Yeast cells were grown on minimal synthetic defined-double-dropouts (SD-Leu/-Trp) medium (Clontech) for 3 days. The co-transformants were then transferred onto quadruple dropout (QDO) (SD-Leu/-Trp/-Ade/-His) medium to test the possible interactions based on their growth status. The ability of yeast cells to grow on QDO medium was scored as a positive interaction.
2.9. Bimolecular Fluorescence Complementation (BiFC) Assay
The coding sequence fragments of MaAPX1 and MaMsrB2 without stop codons were inserted into pUC-pSPYCE or pUC-pSPYNE vectors. The fusion constructs were used for transient assays through a polyethylene glycol transfection of Arabidopsis mesophyll protoplasts, as described earlier [48]. The yellow fluorescent protein (YFP) fluorescence was observed by a fluorescence microscope (Zeiss 510 Meta) after 18 h of incubation at 22 °C.
2.10. GST Pull-down Assay
Equal volumes of GST-MaMsrB2 fusion protein or GST protein alone were incubated with the His-MaAPX1 fusion protein in pull-down buffer at 4 °C for 4 h, followed by incubation with GST beads for another 4 h. After washing with pull-down buffer three times, the pull-down proteins were boiled and subjected to SDS-PAGE and Western blot analysis using the anti-His antibody (TransGen Biotech). The signal was detected using a SuperSignal® West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
2.11. Oxidation and Reduction of MaAPX1
MaAPX1 protein (1 mg) was oxidized using H2O2 (10 mM) at 22 °C for 3 h in 50 mM PBS (pH 7.0). H2O2 was removed by centrifugal filter units (Amicon Ultra-10, Millipore). The repair of oxidized MaAPX1 was conducted by incubating oxidized proteins (2 μM of OX-MaAPX1) with MaMsrB2 (2 μM) and 10 mM DTT at 37 °C for 3 h. DTT was cleaned by centrifugal filter units. The proteins were collected and used for SDS-PAGE. Different redox statuses of MaAPX1 proteins were digested by trypsin, and then analyzed by LC-MS/MS on a C18 reverse-phase column. The redox status of each Met-containing peptide was analyzed using methods previously described by Jiang et al. [49].
2.12. Enzymatic Activity Measurement
Recombinant MaAPX1 activity was measured by spectrophotometry as previously described [50]. The activity was also evaluated by the nitroblue tetrazolium (NBT) method as previously described [51]. For activity analysis in vivo, the coding sequences of MaAPX1, MaAPX1-M36Q, and MaAPX1-M36V were cloned into the pCambia1302 vector. pCambia1302-MaAPX1, pCambia1302-MaAPX1-M36Q, pCambia1302-MaAPX1-M36V, or pCambia1302 (as the control) were transferred to Arabidopsis mesophyll protoplasts. After incubation for 18 h at 22 °C, the protoplasts were collected to analyze APX activity.
2.13. Spectroscopic Analysis
According to the method of Hugo et al. [52], UV-visible spectra were recorded at 25 °C from Asc-free MaAPX1, MaAPX1-M36Q, and MaAPX1-M36V (1 μg/μL) proteins in the presence or absence of an equimolar H2O2 concentration in potassium phosphate buffer (50 mM, pH 7.0).
2.14. Statistical Analysis
The data are presented as the mean ± SE of three biological replicates. Differences among different treatments were determined by ANOVA, followed by Dunnett’s test. Statistical analysis was performed using SPSS version 7.5 (SPSS, Inc., Chicago, IL, USA).
3. Results
3.1. Ripening Characteristics and Redox Status of Harvested Banana Fruit under High Oxygen Stress
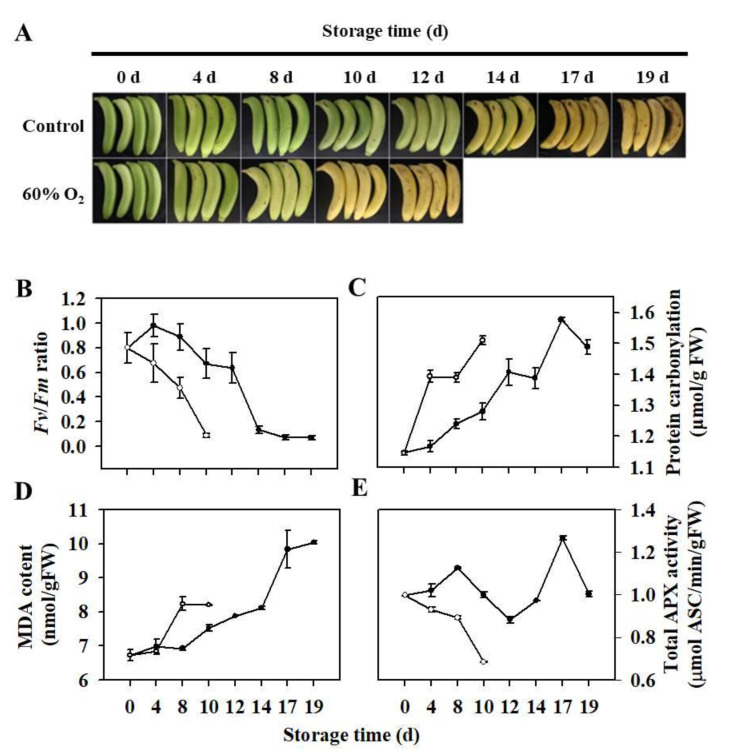
Banana is a typical climacteric fruit characterized by a pre-climacteric phase followed by a peak in ethylene production that initiates ripening-associated processes. Of these processes, the peel turning yellow is one of the most important characteristics. As shown in Figure 1A, control fruit turned yellow at 14 d after harvest. The Fv/Fm indicates the efficiency in the energy transfer process and chloroplast activity, and it decreases in plants when subjected to abiotic stresses or during senescence. In this study, Fv/Fm in control fruit after 12 d of storage rapidly decreased (Figure 1B), implying thylakoid membrane damage, possibly due to senescence or oxidative stress. Protein carbonylation and MDA levels are important indexes of lipid and protein oxidation, respectively, which are usually used to evaluate oxidative stress in organisms. It was found that protein carbonylation level (Figure 1C) and MDA content (Figure 1D) in control fruit significantly increased with fruit ripening. Overall, harvested banana fruit ripening was accompanied by oxidative damage of proteins and lipids.
Figure 1.
Ripening and senescence characteristics, and redox status of banana fruit stored under 60% O2 concentration. Fruit were stored under normal air conditions as the control. Changes in ripening phenotype (A), Fv/Fm (B), protein carbonylation (C), malondialdehyde (MDA) content (D), and ascorbate peroxidase (APX) activity (E) of banana fruit during storage.
We further mimicked oxidative stress by storing banana fruit at 60% oxygen concentration. High oxygen concentration accelerated ripening of harvested banana fruit, accompanied by more rapid decreases in Fv/Fm (Figure 1B). Compared with control fruit, fruit stored at 60% oxygen concentration exhibited more severe oxidative damage of proteins (Figure 1C) and lipids (Figure 1D). In addition, high oxygen concentration resulted in low APX activity in banana fruit (Figure 1E), which possibly results in a high accumulation of H2O2. It seemed that banana fruit ripening exhibited some oxidative stress-like attributes, implying that ROS played a role in banana fruit ripening.
MaMsrB2 and MaAPX1 were isolated from a banana transcriptome database. Their transcript levels in the peel during fruit ripening and senescence were investigated by quantitative real-time PCR (qRT-PCR). The transcript level of MaAPX1 initially increased and then decreased as ripening and senescence proceeded, whereas that of MaMsrB2 tended to increase (Figure S2). High oxygen concentration accelerated the change in the transcript level of MaAPX1 and MaMsrB2.
3.2. Overexpression of MaAPX1 in apx1-2 Delays Senescence under Dark Condition
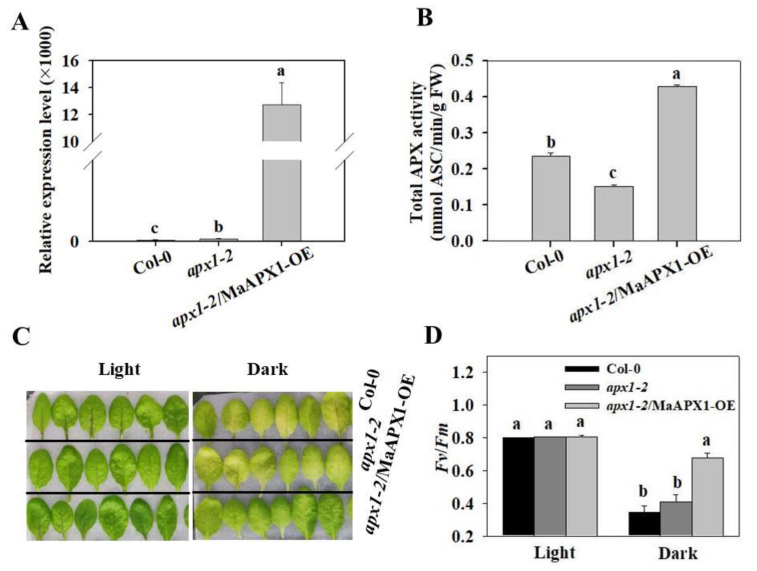
The APX (GSMUA_Achr5P07280_001) cDNA, designated as MaAPX1, was isolated from banana fruit. MaAPX1 was predicted to encode a 249-amino-acid protein with the characteristic sequence APLMLRLAWHSA in the active site, as well as binding sites of heme, substrate (AsA), and K+ (Figure S3). A previous study showed that APX1 in Arabidopsis is a central component of the reactive oxygen gene network, and the absence of APX1 led to an increase of H2O2 and an accumulation of oxidized proteins [53]. The apx1-2 mutant showed various developmental defects, which are fully rescued by APX1 transgene complementary [50]. In the present study, we found that MaAPX1 had a high homology (83.9%) with Arabidopsis APX1. Homologs may have similar biological functions. We speculated that MaAPX1 may play a role in regulating senescence and oxidative stress. Therefore, we generated transgenic Arabidopsis plants overexpressing MaAPX1 in the apx1-2 mutant (Figure 2A). Enzyme activity analysis showed that the leaves of apx1-2/MaAPX1-OE had higher APX activity than those of apx1-2 and Col-0 (Figure 2B). We further examined the senescence phenotypes of the detached leaf of Col-0, apx1-2, and apx1-2/MaAPX1-OE under dark conditions. As shown in Figure 2C, the leaves of the apx1-2 and Col-0 were seriously yellow under dark conditions for 5 d, whereas only a slight yellow was observed in the apx1-2/MaAPX1-OE. Moreover, the Fv/Fm value of apx1-2/MaAPX1-OE was significantly higher than those of apx1-2 and Col-0 (Figure 2D). These results indicated that ectopic overexpression of MaAPX1 delays the detached leaf senescence induced by dark in Arabidopsis.
Figure 2.
Ectopic overexpression of MaAPX1 delays the detached leave senescence induced by darkness in Arabidopsis. (A) Expression of MaAPX1 in Col-0, apx1-2, and apx1-2/MaAPX1-OE seedlings. (B) Total APX activity in Col-0, apx1-2, and apx1-2/MaAPX1-OE seedlings. (C,D) Phenotype and Fv/Fm of detached leaves from three different genotypes incubated under dark or light (control) conditions for 5 d. Different letters above the bars indicate statistically significant differences between the samples (Student’s t test; p < 0.05).
3.3. MaAPX1 Interacts Physically with MaMsrB2
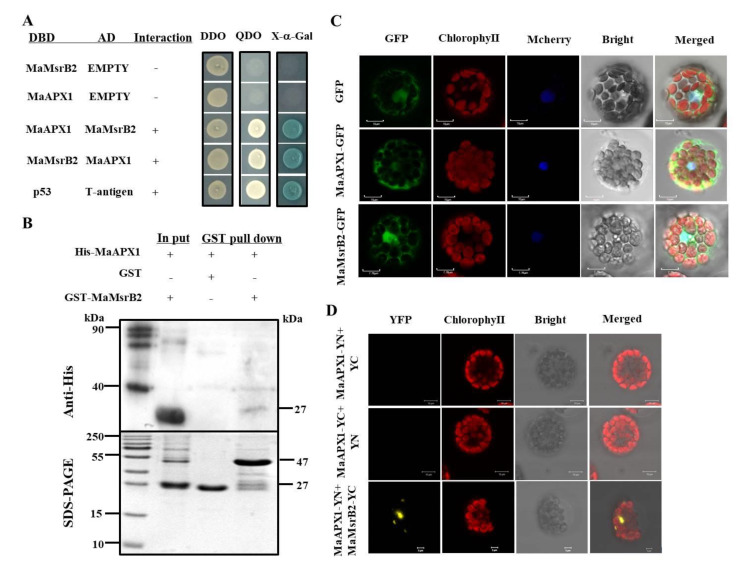
Three methods were applied to verify the interaction between MaAPX1 and MaMsrB2, including yeast two-hybrid (Y2H), bimolecular fluorescence complementation (BiFC), and pull-down assay. Y2H analysis showed that the yeast cells that were co-transformed with DNA-binding domain (DBD)-MaAPX1/activation domain (AD)-MaMsrB2 and DBD-MaMsrB2/AD-MaAPX1 grew well on minimal synthetic defined quadruple dropout (QDO) medium, while the DBD-MaAPX1/MaMsrB2 with AD did not grow (Figure 3A), indicating that MaAPX1 physically interact with MaMsrB2. Moreover, a pull-down assay showed that GST-MaMsrB2, but not GST alone, pulled down recombinant His-MaAPX1, indicating that MaAPX1 interacts with MaMsrB2 in vitro (Figure 3B).
Figure 3.
MaAPX1 interacts physically with MaMsrB2. (A) Interaction between MaAPX1 and MaMsrB2 in the yeast two-hybrid (Y2H) assay. (B) Interaction between MaAPX1 and MaMsrB2 in GST pull-down assay. The molecular weight of MaAPX1-His and MaMsrB2 is about 27-kDa and 47-kDa, respectively. (C) Subcellular localization of MaAPX1 and MaMsrB2. A green signal indicates green fluorescent protein (GFP) fluorescence; A red signal indicates chlorophyll auto fluorescence; The merged images represent a digital combination of chlorophyll auto fluorescence and GFP fluorescent images. (D) Interaction between MaAPX1 and MaMsrB2 in the bimolecular fluorescence complementation (BiFC) assay. A yellow signal indicates yellow fluorescent protein (YFP) fluorescence; a red signal indicates chlorophyll autofluorescence; the merged images represent a digital combination of the chlorophyll autofluorescence and YFP fluorescent images.
The subcellular localization analysis showed that MaAPX1 was mainly located in cytosol. In addition, MaAPX1 might also be distributed in the nucleus (Figure 3C). Furthermore, BiFC analysis further confirmed the interaction between MaAPX1 and MaMsrB2 in Arabidopsis mesophyll protoplasts. The interaction occurred in the nucleus and cytosol (Figure 3D), which was consistent with the subcellular localization of the two enzymes (Figure 3C).
3.4. MaMsrB2 Regulates Redox State and Activity of MaAPX1
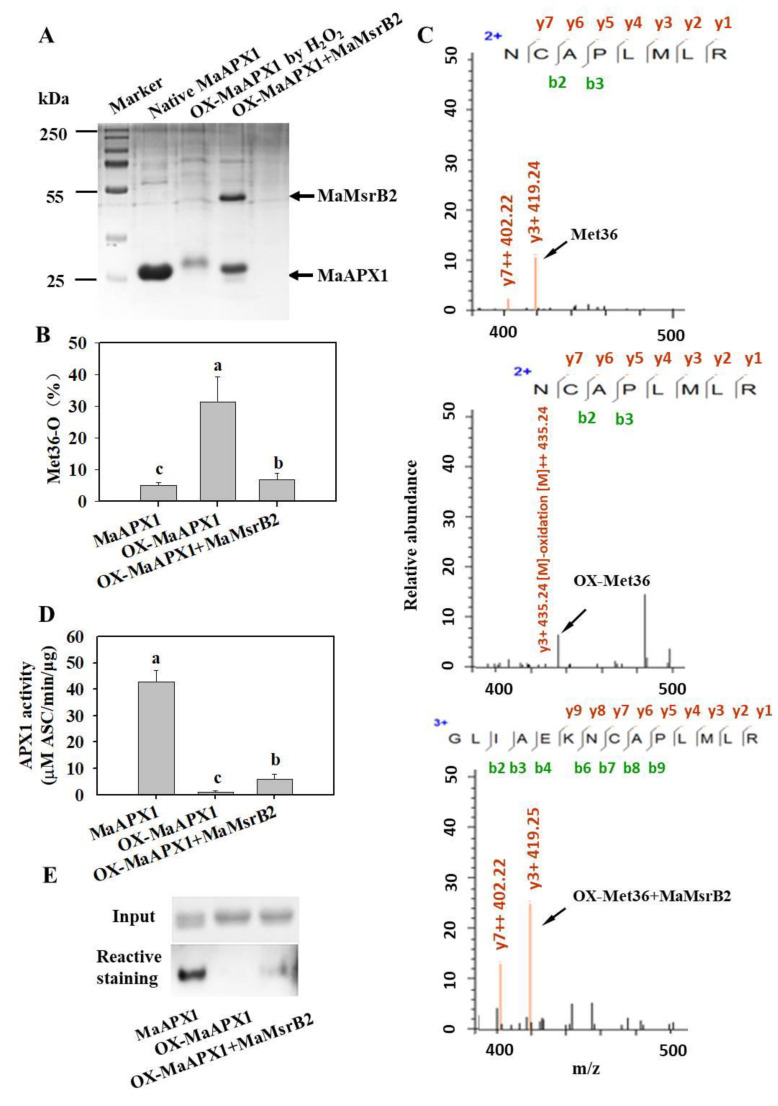
Recombinant MaAPX1 and MaMsrB2 proteins were prepared to determine whether MaMsrB2 can repair the oxidized MaAPX1. After oxidization with H2O2, the band of MaAPX1 in SDS-PAGE shifted to a higher molecular weight. When MaMsrB2 was added, the band of oxidized MaAPX1 almost moved back to the position of the native MaAPX1, suggesting that MaMsrB2 can reduce the oxidized MaAPX1 (Figure 4A).
Figure 4.
MaMsrB2 modifies the redox state and activity of MaAPX1. (A) Electrophoresis of native, oxidized, and reduced His-MaAPX1 in SDS-PAGE; (B) The percentage change of Met-O in the peptide containing Met36 with native, oxidized and reduced samples. Each bar represents the mean ± SE of three replicates. (C) Mass spectrometric analysis of the peptide containing Met36, Met36 oxidized by H2O2, and OX-Met36 reduced by MaMsrB2. (D,E) Activities of MaAPX1, oxidized MaAPX1, and MaMsrB2-repaired oxidized MaAPX1 by spectrophotometry (D) and reactive staining (E) methods. Different letters above the bars indicate statistically significant differences between the samples (Student’s t test; P < 0.05).
Then, native MaAPX1, oxidized MaAPX1 and MaMsrB2-repaired oxidized MaAPX1 were submitted to trypsin digestion. LC-MS/MS analysis showed that the oxidized form of methionine significantly increased in the peptide containing Met36 in oxidized MaAPX1. When subjected to repair by MaMsrB2, the oxidized form of the peptide was reduced (Figure 4B). The detailed mass data of the peptide with different redox statuses are shown in Figure 4C. Collectively, these results indicated that oxidized MaAPX1 is a direct substrate of MaMsrB2 in vitro.
We further investigated whether Met oxidation in MaAPX1 affects the activity in vitro using the spectrophotometric method. As shown in Figure 4D, native MaAPX1 had an activity of 43.5 μM Asc mol−1 min−1 μg−1, whereas oxidation resulted in almost total loss of the MaAPX1 activity. When repaired by MaMsrB2, the activity restored 20% of the native MaAPX1. Moreover, similar results were also observed using the reactive staining method (Figure 4E). These results demonstrated that the repair of oxidized MaAPX1 by MaMsrB2 permits the recovery of its activity.
3.5. Mimicked Oxidation of Met in MaAPX1 Decreases the Activity of MaAPX1 In Vitro and In Vivo
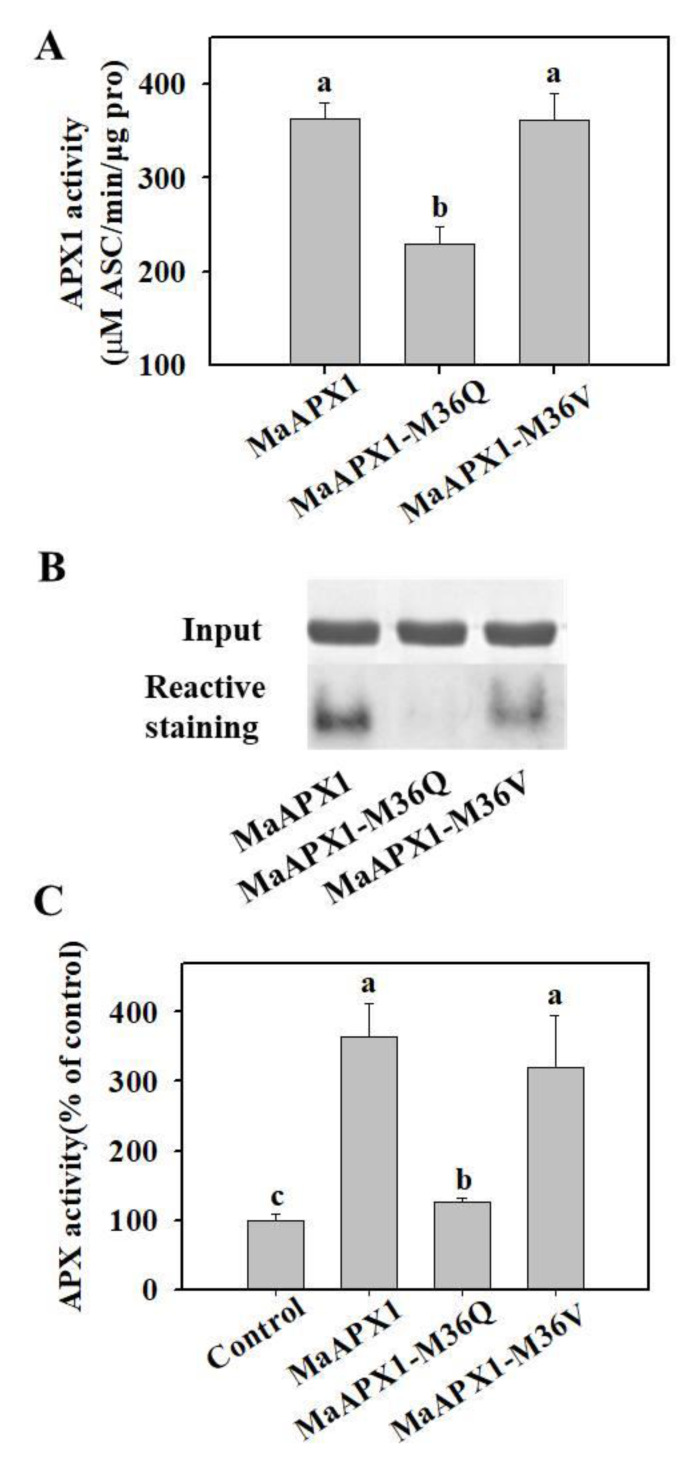
Met36 in MaAPX1 was highly conserved and located in the active site (Figure S3). We hypothesized that Met36 residue was an important site at which the redox status could influence the activity of MaAPX1. To confirm this hypothesis, we mutated Met36 residue to Gln36 to mimic methionine sulfoxidation, i.e., MaAPX1-M36Q, and mutated Met36 to Val36 to mimic the blocking of methionine sulfoxidation, i.e., MaAPX1-M36V. Recombinant MaAPX1, MaAPX1-M36Q, and MaAPX1-M36V proteins were prepared and their activities were analyzed. As shown in Figure 5A, mimicked oxidation of Met36 resulted in decreased APX activity, whereas there was no significant difference in APX activity between MaAPX1 and MaAPX1-M36V. The analysis by the reactive staining method also showed that mimicked oxidation of Met36 results in decreased APX activity (Figure 5B).
Figure 5.
Mimicking sulfoxidation decreases MaAPX1 activity. MaAPX1-M36Q is a mutated form of MaAPX1 in which Met36 is mutated to glutamine to mimic sulfoxidation, whereas Met36 is mutated to valine in MaAPX1-M36V to mimic the blocking of sulfoxidation. (A,B) Activities of purified recombinant MaAPX1, MaAPX1-M36Q, and MaAPX1-M36V proteins by spectrophotometry (A) and reactive staining (B) methods. (C) The MaAPX1, MaAPX1-M36Q, and MaAPX1-M36V constructs was transformed into Arabidopsis mesophyll protoplasts. After incubation overnight at 22 °C, the protoplasts were collected to analyze APX activity by spectrophotometry. Different letters above the bars indicate statistically significant differences between the samples (Student’s t test; p < 0.05).
Furthermore, MaAPX1, MaAPX1-M36Q, and MaAPX1-M36V proteins were expressed in Arabidopsis mesophyll protoplasts and their activities were analyzed. Compared with MaAPX1, mimicked oxidation of Met36 by mutant MaAPX1-M36Q significantly decreased APX activity, while mimicked blocking of Met36 sulfoxidation resulted in no significant influence on APX activity (Figure 5C).
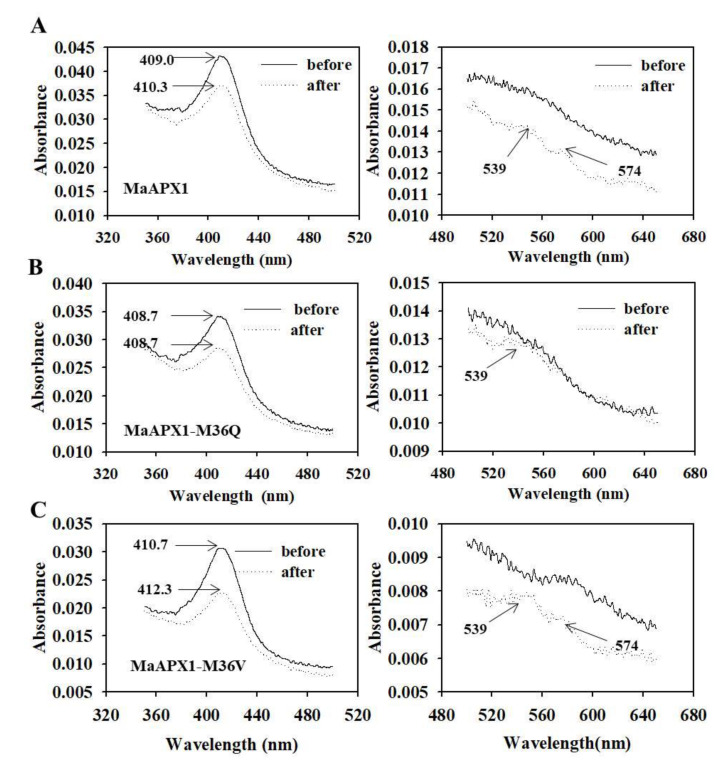
3.6. Visible Spectral Analysis of MaAPX1, MaAPX1-M36Q, and MaAPX1-M36V Reacting with H2O2
The optical spectra of MaAPX1, MaAPX1-M36Q, and MaAPX1-M36V (from 350 to 650 nm) were recorded before and after the addition of equimolar concentrations of H2O2. MaAPX1, MaAPX1-M36Q, and MaAPX1-M36V exhibited different Soret peaks at 409.0, 408.7, and 410.7 nm, respectively, indicating some structural perturbation at the heme microenvironment due to the amino acid replacement [52]. As shown in Figure 6, addition of H2O2 cause red shifts at the Soret region, which are associated with the formation of a compound I-like product [52,54,55]. The red shifts were observed in MaAPX1 and MaAPX1-M36V, accompanied by two humps at 539 and 574 nm. However, there was no shift at the Soret region for the mutant MaAPX1-M36Q, with only the 539 nm hump present (Figure 6).
Figure 6.
Absorption spectra of MaAPX1, MaAPX1-M36Q, and MaAPX1-M36V before and after H2O2 addition. (A) MaAPX1; (B) MaAPX1-M36Q; (C) MaAPX1-M36V. Left, spectra from 500 to 650 nm. Arrows indicate the Soret peak of the resting enzyme after the H2O2 reaction. Spectral shifts of the Soret peak are indicative of compound I-like formation. Right, spectra from 500 to 650 nm.
4. Discussion
APX is a crucial antioxidant enzyme in regulating redox homeostasis. APX itself may also be subjected to multiple post-translational redox modifications, including S-nitrosylation, nitration, Cys oxidation, metal nitrosylation, glutathionylation, and carbonylation. Here, we characterized the role of MaAPX1 in regulating senescence by heterogeneous expression and elucidated a novel mechanism that is involved in post-translational redox modification of APX.
4.1. MaAPX 1 Might Be Involved in Ripening and Senescence in Relation to Oxidative Stress
Low oxygen concentration is the basis of modified-atmosphere or controlled-atmosphere storage for fruits and vegetables. High oxygen-modified atmospheres have been considered as an alternative preservation technique to classical low oxygen-modified atmosphere packaging. High O2 concentrations may retard, have no effect, or prompt fruit and vegetable ripening/senescence or deterioration, depending on the commodity, O2 concentration, and storage time [56,57,58,59,60,61,62,63]. In the present study, 60% O2 concentration resulted in lower APX activity, lipid peroxidation, and more severe protein oxidation, and accelerated fruit ripening in bananas. Qin et al. [43] reported that protein carbonylation increase in mitochondria during the senescence of peach fruit, and reducing ROS accumulation by low temperature inhibited protein carbonylation and retarded fruit senescence, whereas H2O2 treatment had the opposite effect. Li et al. [64] found that application of L-cysteine hydrochloride, a reactive oxygen species scavenger, suppressed aril breakdown and maintained fruit quality by reducing H2O2 accumulation and alleviating protein oxidation. Therefore, redox homeostasis is of importance for maintenance of fruit quality. Our results suggested that accelerated fruit ripening and senescence under high oxygen is associated with decreased APX activity, lipid peroxidation, and protein oxidation.
APXs are heme-containing peroxidases that catalyze reduction of H2O2 using ascorbate as the source of reducing power. APXs play key roles in regulating the precise localized concentration of H2O2 and are involved in physiological and developmental processes. Overexpression of APX in plants increases resistance to abiotic stresses, such as drought [65], salt [66], high temperature [67], and chilling injury [68]. Mutations of APX lead to increased sensitivity to oxidative stresses [69]. In the present study, genetic transformation of apx1-2 mutant Arabidopsis with MaAPX1 obviously delayed the detached leave senescence induced by darkness, compared with the wild-type line. It is suggested that MaAPX1 plays a role in regulating senescence in plants.
4.2. MaAPX1 Is a Target of MaMsrB2
APXs in combination with other antioxidative enzymes and antioxidants regulate the homeostasis of ROS, especially H2O2. In plants, APXs are encoded by a multigene family. Arabidopsis, rice, and tomato contain nine, eight, and seven APX genes, respectively [70]. According to subcellular location, APX isozymes are classified into four categories, namely cytosolic, mitochondrial, chloroplastic, and peroxisomal APX [71]. In the present study, MaAPX1 was mainly localized in the cytoplasm and therefore belongs to cytosolic APX. Surprisingly, MaAPX1 was also detected in the nucleus. Under stress conditions or during aging, excessive accumulation of ROS leads to oxidative damage of proteins. Reversible oxidation of methionine in protein can be repaired by Msrs. In plants, MsrBs belong to one of two types, the 1-Cys type and the 2-Cys type based on the number of redox-active cysteines. Arabidopsis thaliana possesses nine MsrB genes and only AtMsrB1 belongs to the 1-Cys type [72]. MaMsrB2 had higher homology with AtMsrB1 than with other A. thaliana MsrBs (Figure S4). Consistently, MaMsrB2 has only one redox-active cysteine and is classified as the 1-Cys type (Figure S5). In A. thaliana, MsrBs are predicted to be located in cytosol and plastidial. Surprisingly, MaMsrB2 was distributed in cytosol and nucleus. The similar subcellular location enables the interaction between MaMsrB2 and MaAPX1.
Msr-mediated repair of oxidized methionine in proteins is of importance for organisms in protecting against oxidative stress [34,38,39]. A great many proteins have been found through proteomics to be subjected to methionine oxidation [73,74]. However, only a few potential proteins have been validated as Msr targets in organisms [25,26,27,28,29,30,31,32,33,34,35,36,37]. Recently, Jiang et al. [49] found that E4/SlMsrB2 interacts with NON-RIPENING (NOR), an important ripening regulator, to regulate fruit ripening in tomato. In the present study, we verified using Y2H, BiFC, and pull-down assays that MaAPX1 interacted with MaMsrB2. Moreover, MaMsrB2 could reduce the oxidized methionine in MaAXP1. Interestingly, most of the MetO36 was reduced back to methionine. MaMsrB2 is expected to be stereospecific for the R-diastereomer. It seems that Met36 oxidation mainly resulted in the formation of R-diastereomer. These results indicated that MaAPX1 is a direct substrate of MaMsrB2 in bananas. Similarly, Tarrago et al. [75] used affinity chromatography to identify catalase, another enzyme that removes H2O2, as a potential target of AtMsrB1 in Arabidopsis.
4.3. MaMsrB2 Modifies the Redox Status of MaAPX1 and Affects Its Activity
Msr is implicated in sulfoxidation modification of proteins by reducing methionine sulfoxide to methionine. Previous studies on sulfoxidation modification have been mainly related to aging [76] and resistance to oxidative stresses [38,39]. Recently, it was considered that Msr-mediated redox modifications of proteins played a role as a post-translation modification [77]. The oxidation of methionine in proteins suppresses or improves their function, which can be reversed by Msr-mediated reduction [35,36,78]. More recently, Jiang et al. [49] reported that Met sulfoxidation in NOR, an important ripening-related transcriptional factor, or simulated sulfoxidation impairs its function in vitro, whereas E4 and SlMsrB2 partially reduce oxidized NOR and restore its function. Interestingly, methionine sulfoxidation in CaMKII activates its activity, whereas MsrA-mediated reduction of methionine sulfoxidation suppresses its activity in mice [29]. Similarly, simulated sulfoxidation of the hypochlorite-specific transcription factor HypT leads to the activated transcriptional activity in E. coli cells when subjected to HOCl stress, but the activity is suppressed by Msr-mediated reduction [31]. Therefore, Msr-mediated sulfoxidation modification in proteins is implicated in regulating their biological function.
APXs function as important regulators of redox balance in plant cells. However, APXs are susceptible to multiple redox-related post-translational modifications. Yang et al. [50] reported that S-nitrosylation of the Arabidopsis cytosolic ascorbate peroxidase1 (APX1) at cysteine (Cys)-32 enhances its activity of scavenging hydrogen peroxide, leading to the increased resistance to oxidative stress. Yamazaki et al. [79] identified cAPX as a potential target of Trx, and Gelhaye et al. [80] found that incubation of pea cAPX with reduced poplar Trxh, or reduced glutathione and dithiothreitol, significantly inactivate its activity, suggesting that activation of cAPX is related to Cys oxidation. Carbonylation of cAPX leads to an irreversible inhibition of APX activity [81]. Therefore, APX activity is regulated by multiple post-translation redox modification in plants.
In this study, we found that methionine sulfoxidation in MaAPX1 inactivated its activity, which was reversed partially by MaMsrB2-mediated reduction of sulfoxidation (Figure 4D). Mimicking sulfoxidation of Met36, which is located in the active site, resulted in significantly decreased APX activity, whereas substitution of Met36 with Val36 to mimic the blocking of sulfoxidation had little effect on APX activity in vitro (Figure 5A,B). Similar results were also observed in Arabidopsis mesophyll protoplasts in vivo (Figure 5C). In APX-catalyzed reactions, APX first reacts with hydrogen peroxide to produce compound I, where the heme (iron V) is oxidized to the oxyferryl (Fe4+ = O) species, and then compound I returns to the resting ferric (Fe3+) state by two successive one-electron reactions with the substrate ascorbate [82]. Visible spectral analysis showed that mimicking sulfoxidation of Met36 hindered the formation of compound I, whereas substitution of Met36 with Val36 did not affect the formation of compound I. Our findings demonstrate that the redox state of methionine in MaAPX1 is of importance to its activity and MaMsrB2 can regulate the redox state and activity of MaAPX1 in bananas.
5. Conclusions
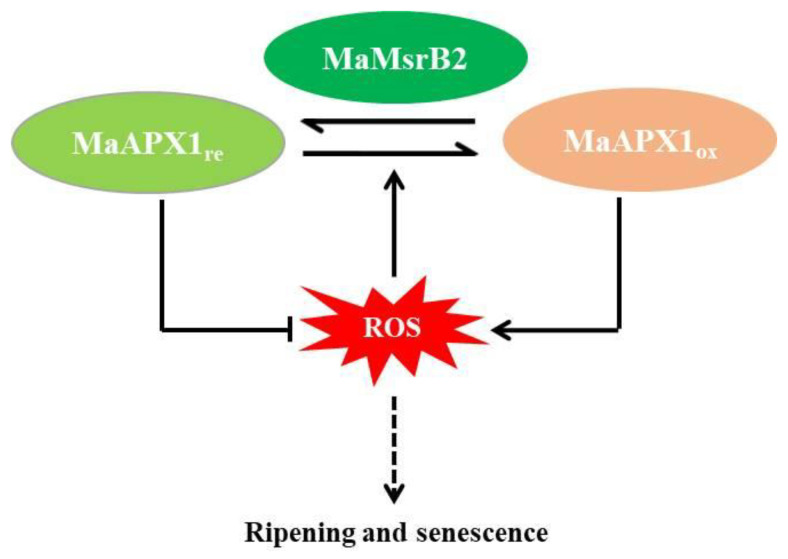
In conclusion, we identified and confirmed that MaAPX1 is a substrate of MaMsrB2 in banana fruit. Ectopic overexpression of MaAPX1 delays the detached leaf senescence induced by darkness in Arabidopsis. Methionine sulfoxidation in MaAPX1 suppresses its activity, which can be partially restored by MaMsrB2. It is suggested that the MaMsrB2-mediated sulfoxidation modification of MaAPX1 possibly regulates redox status of banana fruit during ripening and senescence (Figure 7). Our results revealed a novel post-translational redox modification of APX.
Figure 7.
A proposed model of the involvement of MaMsrB2-mediated redox modification of methionine in MaAPX1 in regulating ripening and senescence.
Acknowledgments
We thank Jianru Zuo (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing the mutant seeds (apx1-2).
Supplementary Materials
The supplementary are available online at https://www.mdpi.com/2076-3921/10/2/310/s1.
Author Contributions
Conceptualization, X.D., G.J., and L.X.; investigation, L.X., G.J., H.Y., and H.L.; writing-original draft preparation, L.X.; writing-review and editing, X.D.; supervision, X.D., G.J., X.S., and Y.J.; funding acquisition, X.D. and G.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Nos. 31830070, 31772041, and 31871856), China Postdoctoral Science Foundation (2020M682954), Joint Foundation of Basic, Applied Basic Research of Guangdong Province (2020A1515110100), and Science and Technology Planning Project of Guangzhou (No. 201804020041). The work was also supported by Guangdong Provincial Key Laboratory of Applied Botany and Key Laboratory of Post-Harvest Handling of Fruits, Ministry of Agriculture.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Choudhury F.K., Rivero R.M., Blumwald E., Mittler R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2016;90:856–867. doi: 10.1111/tpj.13299. [DOI] [PubMed] [Google Scholar]
- 2.Pitzschke A., Hirt H. Mitogen-activated protein kinases and reactive oxygen species signaling in plants. Plant Physiol. 2006;141:351–356. doi: 10.1104/pp.106.079160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Møller I.M., Jensen P.E., Hansson A. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 2007;58:459–481. doi: 10.1146/annurev.arplant.58.032806.103946. [DOI] [PubMed] [Google Scholar]
- 4.Rey P., Tarrago L. Physiological roles of plant methionine sulfoxide reductases in redox homeostasis and signaling. Antioxidants. 2018;7:114. doi: 10.3390/antiox7090114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine R.L., Moskovitz J., Stadtman E.R. Oxidation of methionine in proteins: Roles in antioxidant defense and cellular regulation. IUBMB Life. 2000;50:301–307. doi: 10.1080/15216540051081056. [DOI] [PubMed] [Google Scholar]
- 6.Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Singh A., Agrawal M. Reduction in metal toxicity by applying different soil amendments in agricultural field and its consequent effects on characteristics of radish plants (Raphanus sativus L.) J. Agric. Sci Technol. 2013;15:1553–1564. [Google Scholar]
- 8.Ferreira I.C.F.R., Barros L., Abreu R.M.V. Antioxidants in wild mushrooms. Curr. Med. Chem. 2009;16:1543–1560. doi: 10.2174/092986709787909587. [DOI] [PubMed] [Google Scholar]
- 9.Rosa S.B., Caverzan A., Teixeira F.K., Lazzarotto F., Silveira J.A., Ferreira-Silva S.L., Abreu-Neto J., Margis R., Margis-Pinheiro M. Cytosolic APx knockdown indicates an ambiguous redox responses in rice. Phytochemistry. 2010;71:548–558. doi: 10.1016/j.phytochem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Sofo A., Scopa A., Nuzzaci M., Vitti A. Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. Int. J. Mol. Sci. 2015;16:13561–13578. doi: 10.3390/ijms160613561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L., Jia J., Zhao X., Zhang M., Huang X., Ji E., Ni L., Jiang M. The ascorbate peroxidase APX1 is a direct target of a zinc finger transcription factor ZFP36 and a late embryogenesis abundant protein OsLEA5 interacts with ZFP36 to co-regulate OsAPX1 in seed germination in rice. Biochem. Biophys. Res. Commun. 2018;495:339–345. doi: 10.1016/j.bbrc.2017.10.128. [DOI] [PubMed] [Google Scholar]
- 12.Ribeiro C.W., Korbes A.P., Garighan J.A., Jardim-Messeder D., Carvalho F.E., Sousa R.H., Caverzan A., Teixeira F.K., Silveira J.A., Margis-Pinheiro M. Rice peroxisomal ascorbate peroxidase knockdown affects ROS signaling and triggers early leaf senescence. Plant Sci. 2017;263:55–65. doi: 10.1016/j.plantsci.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 13.De Pinto M.C., Locato V., Sgobba A., Romero-Puertas M.D.C., Gadaleta C., Delledonne M., De Gara L. S-Nitrosylation of ascorbate peroxidase is part of programmed cell death signaling in tobacco bright yellow-2 Cells. Plant Physiol. 2013;163:1766–1775. doi: 10.1104/pp.113.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J.-X., Feng K., Duan A.-Q., Li H., Yang Q.-Q., Xu Z.-S., Xiong A.-S. Isolation, purification and characterization of an ascorbate peroxidase from celery and overexpression of the AgAPX1 gene enhanced ascorbate content and drought tolerance in Arabidopsis. BMC Plant Biol. 2019;19:1–13. doi: 10.1186/s12870-019-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunajska-Ordak K., Skorupa-Kłaput M., Kurnik K., Tretyn A., Tyburski J. Cloning and expression analysis of a gene encoding for ascorbate peroxidase and responsive to salt stress in beet (Beta vulgaris) Plant Mol. Biol. Rep. 2014;32:162–175. doi: 10.1007/s11105-013-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J., Yang J., Duan X., Jiang Y., Zhang P. Increased expression of native cytosolic Cu/Zn superoxide dismutase and ascorbate peroxidase improves tolerance to oxidative and chilling stresses in cassava (Manihot esculenta Crantz) BMC Plant Biol. 2014;14:1–14. doi: 10.1186/s12870-014-0208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao G.-L., Liu Q., Li Y.-Q., Zhong M., Huang C.-H., Jia D.-F., Xu X.-B. Identification and expression profiling analysis of ascorbate peroxidase gene family in Actinidia chinensis (Hongyang) J. Plant Res. 2020;133:715–726. doi: 10.1007/s10265-020-01206-y. [DOI] [PubMed] [Google Scholar]
- 18.Kuo E.Y., Cai M.-S., Lee T.-M. Ascorbate peroxidase 4 plays a role in the tolerance of Chlamydomonas reinhardtii to photo-oxidative stress. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-70247-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Huang W., Yang Z., Liu J., Huang B. Transcriptional regulation of heat shock proteins and ascorbate peroxidase by CtHsfA2b from African bermudagrass conferring heat tolerance in Arabidopsis. Sci. Rep. 2016;6:28021. doi: 10.1038/srep28021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ugarte N., Petropoulos I., Friguet B. Oxidized mitochondrial protein degradation and repair in aging and oxidative stress. Antioxid. Redox Signal. 2010;13:539–549. doi: 10.1089/ars.2009.2998. [DOI] [PubMed] [Google Scholar]
- 21.Châtelain E., Satour P., Laugier E., Vu B.L., Payet N., Rey P., Montrichard F. Evidence for participation of the methionine sulfoxide reductase repair system in plant seed longevity. Proc. Natl. Acad. Sci. USA. 2013;110:3633–3638. doi: 10.1073/pnas.1220589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laugier E., Tarrago L., Dos Santos C.V., Eymery F., Havaux M., Rey P. Arabidopsis thaliana plastidial methionine sulfoxide reductases B, MSRBs, account for most leaf peptide MSR activity and are essential for growth under environmental constraints through a role in the preservation of photosystem antennae. Plant J. 2009;61:271–282. doi: 10.1111/j.1365-313X.2009.04053.x. [DOI] [PubMed] [Google Scholar]
- 23.Lee S.-H., Li C.-W., Koh K.W., Chuang H.-Y., Chen Y.-R., Lin C.-S., Chan M.-T. MSRB7 reverses oxidation of GSTF2/3 to confer tolerance of Arabidopsis thaliana to oxidative stress. J. Exp. Bot. 2014;65:5049–5062. doi: 10.1093/jxb/eru270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stolarska E., Bilska K., Wojciechowska N., Bagniewska-Zadworna A., Rey P., Kalemba E.M. Integration of MsrB1 and MsrB2 in the redox network during the development of orthodox and recalcitrant acer seeds. Antioxidants. 2020;9:1250. doi: 10.3390/antiox9121250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khor H.K., Fisher M.T., Schöneich C. Potential role of methionine sulfoxide in the inactivation of the chaperone GroEL by hypochlorous acid (HOCl) and peroxynitrite (ONOO–) J. Biol. Chem. 2004;279:19486–19493. doi: 10.1074/jbc.M310045200. [DOI] [PubMed] [Google Scholar]
- 26.Ezraty B., Aussel L., Barras F. Methionine sulfoxide reductases in prokaryotes. Biochim. Biophys. Acta Proteins Proteom. 2005;1703:221–229. doi: 10.1016/j.bbapap.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Su Z., Limberis J., Martin R.L., Xu R., Kolbe K., Heinemann S.H., Hoshi T., Cox B.F., Gintant G.A. Functional consequences of methionine oxidation of hERG potassium channels. Biochem. Pharmacol. 2007;74:702–711. doi: 10.1016/j.bcp.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao B., Cavigiolio G., Brot N., Oda M.N., Heinecke J.W. Methionine oxidation impairs reverse cholesterol transport by apolipoprotein A-I. Proc. Natl. Acad. Sci. USA. 2008;105:12224–12229. doi: 10.1073/pnas.0802025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erickson J.R., Joiner M.-L.A., Guan X., Kutschke W., Yang J., Oddis C.V., Bartlett R.K., Lowe J.S., O’Donnell S.E., Aykin-Burns N., et al. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133:462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao G., Lee K.P., van der Wijst J., De Graaf M., van der Kemp A., Bindels R.J.M., Hoenderop J.G.J. Methionine sulfoxide reductase B1 (MsrB1) recovers TRPM6 channel activity during oxidative stress. J. Biol. Chem. 2010;285:26081–26087. doi: 10.1074/jbc.M110.103655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drazic A., Miura H., Peschek J., Le Y., Bach N.C., Kriehuber T., Winter J. Methionine oxidation activates a transcription factor in response to oxidative stress. Proc. Natl. Acad. Sci. USA. 2013;110:9493–9498. doi: 10.1073/pnas.1300578110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee B.C., Péterfi Z., Hoffmann F.W., Moore R.E., Kaya A., Avanesov A., Tarrago L., Zhou Y., Weerapana E., Fomenko D.E., et al. MsrB1 and MICALs regulate actin assembly and macrophage function via reversible stereoselective methionine oxidation. Mol. Cell. 2013;51:397–404. doi: 10.1016/j.molcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang G., Wu F., Li Z., Li T., Gupta V.K., Duan X., Jiang Y. Sulfoxidation Regulation of Musa acuminata calmodulin (MaCaM) influences the functions of MaCaM-binding proteins. Plant Cell Physiol. 2018;59:1214–1224. doi: 10.1093/pcp/pcy057. [DOI] [PubMed] [Google Scholar]
- 34.Jiang G., Xiao L., Yan H., Zhang D., Wu F., Liu X., Su X., Dong X., Wang J., Duan X., et al. Redox regulation of methionine in calmodulin affects the activity levels of senescence-related transcription factors in litchi. Biochim. Biophys. Acta Gen. Subj. 2017;1861:1140–1151. doi: 10.1016/j.bbagen.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Allu P.K., Marada A., Boggula Y., Karri K.S., Krishnamoorthy T. Methionine sulfoxide reductase 2 reversibly regulates Mge1, a cochaperone of mitochondrial Hsp70, during oxidative stress. Mol. Biol. Cell. 2014;26:406–419. doi: 10.1091/mbc.E14-09-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding P., Fang L., Wang G., Li X., Huang S., Gao Y., Zhu J., Xiao L., Tong J., Chen F., et al. Wheat methionine sulfoxide reductase A4.1 interacts with heme oxygenase 1 to enhance seedling tolerance to salinity or drought stress. Plant Mol. Biol. 2019;101:203–220. doi: 10.1007/s11103-019-00901-2. [DOI] [PubMed] [Google Scholar]
- 37.Xu Y., Li F., Zhao X., Tan C., Wang B., Chen Y., Cao J., Wu D., Yu H. Methionine sulfoxide reductase A attenuates atherosclerosis via repairing dysfunctional HDL in scavenger receptor class B type I deficient mice. FASEB J. 2020;34:3805–3819. doi: 10.1096/fj.201902429R. [DOI] [PubMed] [Google Scholar]
- 38.Bigelow D.J., Squier T.C. Thioredoxin-dependent redox regulation of cellular signaling and stress response through reversible oxidation of methionines. Mol. BioSyst. 2011;7:2101–2109. doi: 10.1039/c1mb05081h. [DOI] [PubMed] [Google Scholar]
- 39.Gennaris A., Ezraty B., Henry C., Agrebi R., Vergnes A., Oheix E., Bos J., Leverrier P., Espinosa L., Szewczyk J., et al. Repairing oxidized proteins in the bacterial envelope using respiratory chain electrons. Nat. Cell Biol. 2015;528:409–412. doi: 10.1038/nature15764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang B., Adams Z., Moonah S., Shi H., Maupin-Furlow J., Moskovitz J. The antioxidant enzyme methionine sulfoxide reductase A (MsrA) interacts with Jab1/CSN5 and regulates its function. Antioxidants. 2020;9:452. doi: 10.3390/antiox9050452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee H.-J., Park J.S., Yoo H.J., Lee H.M., Lee B.C., Kim J.H. The selenoprotein MsrB1 Instructs dendritic cells to induce t-helper 1 immune responses. Antioxidants. 2020;9:1021. doi: 10.3390/antiox9101021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tossounian M.-A., Van Molle I., Wahni K., Jacques S., Gevaert K., Van Breusegem F., Vertommen D., Young D., Rosado L.A., Messens J. Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage. Biochim. Biophys. Acta Gen. Subj. 2018;1862:775–789. doi: 10.1016/j.bbagen.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 43.Qin G., Meng X., Wang Q., Tian S. Oxidative damage of mitochondrial proteins contributes to fruit senescence: A redox proteomics analysis. J. Proteome Res. 2009;8:2449–2462. doi: 10.1021/pr801046m. [DOI] [PubMed] [Google Scholar]
- 44.Yan H., Jiang G., Wu F., Li Z., Xiao L., Jiang Y., Duan X. Sulfoxidation regulation of MaNAC42 transcription factor influences its functions in relation to stress-induced fruit ripening in banana. J. Exp. Bot. 2020;10:682–699. doi: 10.1093/jxb/eraa474. [DOI] [PubMed] [Google Scholar]
- 45.Ueda H., Kusaba M. Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiol. 2015;169:138–147. doi: 10.1104/pp.15.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wan C.Y., Wilkins T.A. Isolation of multiple cDNAs encoding the vacuolar H+-ATPase subunit B from developing cotton (Cossypium hirsutum L.) ovules. Plant Physiol. 1994;106:393–394. doi: 10.1104/pp.106.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X., Henriques R., Lin S.-S., Niu Q.-W., Chua N.-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 48.Yoo S.-D., Cho Y.-H., Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 49.Jiang G., Zeng J., Li Z., Song Y., Yan H., He J., Jiang Y., Duan X. Redox regulation of the NOR Transcription factor is involved in the regulation of fruit ripening in tomato. Plant Physiol. 2020;183:671–685. doi: 10.1104/pp.20.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang H., Mu J., Chen L., Feng J., Huanjie Y., Lichao C., Zhou J.-M., Zuo J. S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 2015;167:1604–1615. doi: 10.1104/pp.114.255216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mittler R., Zilinskas B.A. Purification and characterization of pea cytosolic ascorbate peroxidase. Plant Physiol. 1991;97:926–968. doi: 10.1104/pp.97.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hugo M., Martínez A., Trujillo M., Estrad D., Mastrogiovanni M., Linares E., Augusto O., Issoglio F., Zeida A., Estrín D.A., et al. Kinetics, subcellular localization, and contribution to parasite virulence of a Trypanosoma cruzi hybrid type A heme peroxidase (TcAPx-CcP) Proc. Natl. Acad. Sci. USA. 2017;114:E1326–E1335. doi: 10.1073/pnas.1618611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davletova S., Rizhsky L., Liang H., Shengqiang Z., Oliver D.J., Coutu J., Shulaev V., Schlauch K., Mittler R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell. 2004;17:268–281. doi: 10.1105/tpc.104.026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jasion V.S., Polanco J.A., Meharenna Y.T., Li H., Poulos T.L. Crystal structure of leishmania major peroxidase and characterization of the compound I tryptophan radical. J. Biol. Chem. 2011;286:24608–24615. doi: 10.1074/jbc.M111.230524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yadav R.K., Dolai S., Pal S., Adak S. Role of tryptophan-208 residue in cytochrome c oxidation by ascorbate peroxidase from Leishmania major-kinetic studies on Trp208Phe mutant and wild type enzyme. Biochim. Biophys. Acta Proteins Proteom. 2008;1784:863–871. doi: 10.1016/j.bbapap.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 56.Artés F., Allende A. Processing lines and alternative preservation techniques to prolong shelf-life of minimally fresh processed leafy vegetables. Eur. J. Hortic. Sci. 2005;70:231–245. [Google Scholar]
- 57.Jiang Y., Joyce D.C. Softening response of 1-methylcyclopropene-treated banana fruit to high oxygen atmospheres. Plant Growth Regul. 2003;41:225–229. doi: 10.1023/B:GROW.0000007506.38661.c6. [DOI] [Google Scholar]
- 58.Odriozola-Serrano I., Oliu G.O., Soliva-Fortuny R., Martín-Belloso O. Effect of high-oxygen atmospheres on the antioxidant potential of fresh-cut tomatoes. J. Agric. Food Chem. 2009;57:6603–6610. doi: 10.1021/jf900776j. [DOI] [PubMed] [Google Scholar]
- 59.Duan X., Liu T., Zhang D., Su X., Lin H., Jiang Y. Effect of pure oxygen atmosphere on antioxidant enzyme and antioxidant activity of harvested litchi fruit during storage. Food Res. Int. 2011;44:1905–1911. doi: 10.1016/j.foodres.2010.10.027. [DOI] [Google Scholar]
- 60.Liu X., Wang T., Lu Y., Yang Q., Li Y., Deng X., Liu Y., Du X., Qiao L., Zheng J. Effect of high oxygen pretreatment of whole tuber on anti-browning of fresh-cut potato slices during storage. Food Chem. 2019;301:125287. doi: 10.1016/j.foodchem.2019.125287. [DOI] [PubMed] [Google Scholar]
- 61.Dong Q., Li Y., Liu G., Zhang Z., Zhou H., Yang H. High oxygen treatments enhance the contents of phenolic compound and ganoderic acid, and the antioxidant and DNA damage protective activities of Ganoderma lingzhi fruiting body. Front. Microbiol. 2019;10:2363. doi: 10.3389/fmicb.2019.02363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Z., Wang X. Changes in color, antioxidant, and free radical scavenging enzyme activity of mushrooms under high oxygen modified atmospheres. Postharvest Biol. Technol. 2012;69:1–6. doi: 10.1016/j.postharvbio.2012.02.008. [DOI] [Google Scholar]
- 63.Van de Velde F., Méndez-Galarraga M.P., Pirovani M.E. Effect of enriched O2 and CO2 atmospheres on the overall quality and the bioactive potential of fresh blackberries. Postharvest Biol. Technol. 2020;164:111166. doi: 10.1016/j.postharvbio.2020.111166. [DOI] [Google Scholar]
- 64.Li T., Wu Q., Zhou Y., Yun Z., Duan X., Jiang Y. L-Cysteine hydrochloride delays senescence of harvested longan fruit in relation to modification of redox status. Postharvest Biol. Technol. 2018;143:35–42. doi: 10.1016/j.postharvbio.2018.04.011. [DOI] [Google Scholar]
- 65.Sun W.-H., Liu X.-Y., Wang Y., Hua Q., Song X.-M., Gu Z., Pu D.-Z. Effect of water stress on yield and nutrition quality of tomato plant overexpressing StAPX. Biol. Plant. 2013;58:99–104. doi: 10.1007/s10535-013-0360-y. [DOI] [Google Scholar]
- 66.Shafi A., Pal A.K., Sharma V., Kalia S., Kumar S., Ahuja P.S., Singh A.K. Transgenic potato plants overexpressing SOD and APX exhibit enhanced lignification and starch biosynthesis with improved salt stress tolerance. Plant Mol. Biol. Rep. 2017;35:504–518. doi: 10.1007/s11105-017-1041-3. [DOI] [Google Scholar]
- 67.Kim M.D., Kim Y.-H., Kwon S.-Y., Yun D.-J., Kwak S.-S., Lee H.-S. Enhanced tolerance to methyl viologen-induced oxidative stress and high temperature in transgenic potato plants overexpressing the CuZnSOD, APX and NDPK2 genes. Physiol. Plant. 2010;140:153–162. doi: 10.1111/j.1399-3054.2010.01392.x. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y., Wisniewski M., Meilan R., Cui M., Webb R., Fuchigami L. Overexpression of cytosolic ascorbate peroxidase in tomato confers tolerance to chilling and salt stress. J. Am. Soc. Hortic. Sci. 2005;130:167–173. doi: 10.21273/JASHS.130.2.167. [DOI] [Google Scholar]
- 69.Miller G., Suzuki N., Rizhsky L., Hegie A., Koussevitzky S., Mittler R. Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol. 2007;144:1777–1785. doi: 10.1104/pp.107.101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pandey S., Fartyal D., Agarwal A., Shukla T., James D., Kaul T., Negi Y.K., Arora S., Reddy M.K. Abiotic stress tolerance in plants: Myriad roles of ascorbate peroxidase. Front. Plant Sci. 2017;8:581. doi: 10.3389/fpls.2017.00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caverzan A., Passaia G., Rosa S.B., Ribeiro C.W., Lazzarotto F., Margis-Pinheiro M. Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 2012;35:1011–1019. doi: 10.1590/S1415-47572012000600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tarrago L., Laugier E., Rey P. Protein-repairing methionine sulfoxide reductases in photosynthetic organisms: Gene Organization, reduction mechanisms, and physiological roles. Mol. Plant. 2009;2:202–217. doi: 10.1093/mp/ssn067. [DOI] [PubMed] [Google Scholar]
- 73.Ghesquière B., Gevaert K. Proteomics methods to study methionine oxidation. Mass Spectrom. Rev. 2013;33:147–156. doi: 10.1002/mas.21386. [DOI] [PubMed] [Google Scholar]
- 74.Ghesquière B., Jonckheere V., Colaert N., Van Durme J., Timmerman E., Goethals M., Schymkowitz J., Rousseau F., Vandekerckhove J., Gevaert K. Redox proteomics of protein-bound methionine oxidation. Mol. Cell. Proteom. 2011;10:M110-006866. doi: 10.1074/mcp.m110.006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tarrago L., Kieffer-Jaquinod S., Lamant T., Marcellin M., Garin J., Rouhier N., Rey P. Affinity chromatography: A valuable strategy to isolate substrates of methionine sulfoxide reductases? Antioxid. Redox Signal. 2012;16:79–84. doi: 10.1089/ars.2011.4153. [DOI] [PubMed] [Google Scholar]
- 76.Oien D.B., Moskovitz J. Genetic regulation of longevity and age-associated diseases through the methionine sulfoxide reductase system. Biochim. Biophys. Acta Mol. Basis Dis. 2019;1865:1756–1762. doi: 10.1016/j.bbadis.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 77.Valverde H., Cantón F.R., Aledo J.C. MetOSite: An integrated resource for the study of methionine residues sulfoxidation. Bioinformatics. 2019;35:4849–4850. doi: 10.1093/bioinformatics/btz462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dos Santos S.L., Petropoulos I., Friguet B. The oxidized protein repair enzymes methionine sulfoxide reductases and their roles in protecting against oxidative stress, in ageing and in regulating protein function. Antioxidants. 2018;7:191. doi: 10.3390/antiox7120191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamazaki D., Motohashi K., Kasama T., Hara Y., Hisabori T. Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell. Physiol. 2004;45:18–27. doi: 10.1093/pcp/pch019. [DOI] [PubMed] [Google Scholar]
- 80.Gelhaye E., Navrot N., Macdonald I.K., Rouhier N., Raven E.L., Jacquot J.-P. Ascorbate peroxidase–thioredoxin interaction. Photosynth. Res. 2006;89:193–200. doi: 10.1007/s11120-006-9100-x. [DOI] [PubMed] [Google Scholar]
- 81.Bai X., Yang L., Tian M., Chen J., Shi J., Yang Y., Hu X. Nitric oxide enhances desiccation tolerance of recalcitrant Antiaris toxicaria seeds via protein S-nitrosylation and carbonylation. PLoS ONE. 2011;6:e20714. doi: 10.1371/journal.pone.0020714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anjum N.A., Sharma P., Gill S.S., Hasanuzzaman M., Khan E.A., Kachhap K., Mohamed A.A., Thangavel P., Devi G.D., Vasudhevan P., et al. Catalase and ascorbate peroxidase—representative H2O2-detoxifying heme enzymes in plants. Environ. Sci. Pollut. Res. 2016;23:19002–19029. doi: 10.1007/s11356-016-7309-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article or supplementary material.