Abstract
Introduction
The absence of a reliable, universal biomarker is a significant limitation in neuroendocrine neoplasia (NEN) management. We prospectively evaluated two CgA assays, (NEOLISA, EuroDiagnostica) and (CgA ELISA, Demeditec Diagnostics (DD)) and compared the results to the NETest.
Methods
NEN cohort (n = 258): pancreatic, n = 67; small intestine, n = 40; appendiceal, n = 10; rectal, n = 45; duodenal, n = 9; gastric, n = 44; lung, n = 43. Image-positive disease (IPD) (n = 123), image & histology- negative (IND) (n = 106), and image-negative and histology positive (n = 29). CgA metrics: NEOLISA, ULN: 108 ng/mL, DD: ULN: 99 ng/mL. Data mean ± s.e.m. NETest: qRT-PCR – multianalyte analyses, ULN: 20. All samples de-identified and assessed blinded. Statistics: Mann–Whitney U-test, Pearson correlation and McNemar-test.
Results
CgA positive in 53/258 (NEOLISA), 32 (DD) and NETest-positive in 157/258. In image- positive disease (IPD, n = 123), NEOLISA-positive: 33% and DD: 19%. NETest-positive: 122/123 (99%; McNemar’s Chi2= 79–97, P < 0.0001). NEOLISA was more accurate than DD (P = 0.0003). In image- negative disease (IND), CgA was NEOLISA-positive (11%), DD (8%), P = NS, and NETest (33%). CgA assays could not distinguish progressive (PD) from stable disease (SD) or localized from metastatic disease (MD). NETest was significantly higher in PD (47 ± 5) than SD (29 ± 1, P = 0.0009). NETest levels in MD (35 ± 2) were elevated vs localized disease (24 ± 1.3, P = 0.008).
Conclusions
NETest, a multigenomic mRNA biomarker, was ~99% accurate in the identification of NEN disease. The CgA assays detected NEN disease in 19–33%. Multigenomic blood analysis using NETest is more accurate than CgA and should be considered the biomarker standard of care.
Keywords: NETs, CgA, diagnostic, ELISA, NETest, biomarker, genomic analysis
Introduction
Neuroendocrine neoplasms (NENs) constitute a heterogeneous group of neoplasia whose diagnosis is established by symptomology, biomarkers, multimodal imaging and histopathology (1). Diagnosis is difficult since symptomatology is protean and often evanescent while functional imaging requires sophisticated technology and is expensive and not widely available. Tissue pathology involves invasive biopsy and provides a random, one-time assessment of a heterogeneous neoplasm (2). Given these limitations, there remains a critical unmet need to identify an accurate circulating general NEN tumour biomarker. This would facilitate diagnosis, establish recurrence or residual disease after surgery, enable accurate monitoring of disease progression, evaluate response to therapy and refine prognosis (2). Although CgA in blood has been considered ‘effective’ for more than four decades, it is now known to have significant limitations in clinical utility due to assay variability, non-specific elevations and low specificity (1). At a scientific level, this reflects the fact that CgA is a secretory protein (monoanalyte) which does not capture the panoply of diverse regulatory genomic mechanisms that define tumour biology ‘the hallmarks of cancer’ (3). In terms of biometrics, CgA does not meet the minimum NIH criteria for a clinical biomarker and is regarded by the National Comprehensive Cancer Network (NCCN) as a Type III biomarker (negligible utility) (4).
The introduction of radioimmunoassays for various peptide hormones in the mid-1960s enabled the identification of secretory NEN for example, gastrinoma, insulinoma, etc. The first RIA for CgA in plasma was established in 1984 (5) and was proposed as a potential general biomarker for NENs. Initial studies were enthusiastic in supporting CgA since it exhibited the widest distribution amongst the chromogranin family and was ubiquitous in neuroendocrine tissues (6). Based upon its biological role in exocytosis and its co-secretion with bioactive peptides and amines in the neurosecretory granules, it was considered an effective tissue and circulating marker of either functional, or non-functional NENs (7). CgA is a valuable immunohistochemical marker in NEN (8). The initial surge of support for its clinical utility in blood has, in the last decade, waned significantly as assay variabilities, non-specific elevations and correlation with disease status proved difficult to validate (9, 10, 11).
A critical feature of a clinical biomarker is the requirement to provide reproducible and robust measurement (1). Currently, no international standard for CgA assay and its measurement has been recognized. A variety of different assays for intact CgA and cleavage products are available (12). These include enzyme linked immunosorbent assay (ELISA), IRMA, or RIA, and more recently an immunofluorescent assay based on Time-Resolved Amplified Cryptate Emission (TRACE) (9). The variety of assays reflect the presence of several CgA-related peptides and the recognition that CgA is differentially processed in diverse neuroendocrine cells/tissue (12). Similarly, the disparate methodology of individual CgA assays, or antibodies (monoclonal vs polyclonal) that detect different epitopes of the protein surface, affect the individual assay metrics (9). Clinical interpretation has been confounded by both differences in assay detection of NENs, varied levels from individual tumour sites (1, 7, 9, 10, 11, 12, 13) and the variations related to extent of tumour burden (13, 14, 15) or tumour grade (16).
In the oncology field, the recognition of the limitations of tissue biopsy, monoanalyte biomarkers and the complexities of imaging have resulted in a shift of focus to the development of ‘liquid biopsies’. This strategy provides real-time, multidimensional genomic information about the tumour and avoids repetitive, invasive random biopsy of a heterogeneous neoplasm. Liquid biopsy can assess multianalyte genomic biomarkers in the blood and can be used to provide a detailed assessment of the molecular biological status of an individual tumour in real-time (17). The implementation of such tools have proven to be of substantial clinical utility in breast, lung, colon and prostate cancer (18, 19, 20). In the neuroendocrine field, this concept has been investigated using a novel multigene mRNA test (NETest) blood test (21). Synchronous multigenomic quantification involves the mathematical algorithmic analysis of a series of 14 specific ‘omic clusters’ that are defined by 51 separate genes. This measurement captures the biology of an individual neuroendocrine tumour (NET) and defines the evolution of the tumour and the effect of therapy on it (21). Numerous clinical studies (n = 25) and an independent meta-analysis have demonstrated the NETest has an overall accuracy of >90% (22, 23, 24, 25, 26, 27) and provides real-time information that identifies residual tumour, progression or response to therapy (26, 27, 28, 29, 30, 31, 32). Furthermore, direct head-to-head comparison studies indicate that not only is it significantly more accurate than CgA but that the NETest detects lesions up to a year prior to their identification by imaging (31, 33).
To objectively assess the continuing concerns about the CgA accuracy as a NEN biomarker, we sought to perform a comparative analysis of CgA measurements and investigate their performance vs a multigenomic liquid biopsy strategy, the NETest. CgA measurements were undertaken using two separate CgA assays (NEOLISA, EuroDiagnostica, IBL-America, CLIA-certified laboratory, and CgA ELISA, Demeditec Diagnostics, Germany) performed independently in two clinically certified laboratories: one in North America, and one European – in an ENETS Center of Excellence.
Materials and methods
Institutional (Medical University of Silesia) Ethics Committee approved the study protocol. All study participants provided written consent.
Cohorts
The study cohort comprised 258 NENs, including gastroenteropancreatic (GEP) NENs (n = 215): pancreatic, PNENs, n = 67; small intestine (midgut), SINENs, n = 40; rectal, RNENs, n = 45; gastric, GNENs, n = 44; appendiceal, ANENs, n = 10; duodenal, DNENs, n = 9, and bronchopulmonary carcinoids (BPC, n = 43). The cohort was grouped into image-positive disease (IPD, n = 123), and image-negative disease (IND, n = 135) based on evidence vs no evidence of disease on anatomical and/or functional imaging. Amongst IND, there were 29 GEPNENs positive on histology (biopsy or a resection margin), these comprised GNENs Type 1 (n = 20), and RNENs (n = 9). The majority of the cohort was well-differentiated (NETs) (92.5% PNENs, 100% SINENs, 89% DNENs, 95% GNENs, 100% RNENs, and all lung NENs, Table 1).
Table 1.
Demographic and clinicopathological characteristics of the study cohort.
| Tumour location (n) | Gastroenteropancreatic neuroendocrine neoplasia (n = 215) | Lung NETs (n = 43) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variable | Category | PNEN (n = 67) | SINEN (n = 40) | DNEN (n = 9) | GNEN G1/2 (n = 41) | GNEN G3 (n = 3) | RNEN (n = 45) | ANEN (n = 10) | BPC (n = 43) | |
| Age | Mean | 56 | 58 | 48 | 55 | 60 | 57 | 41 | 61 | |
| Range | 19–87 | 27–77 | 29–62 | 28–87 | 39–84 | 37–78 | 19–65 | 31–78 | ||
| Gender | M:F | 24:43:00 | 22:18 | 5:04 | 10:31 | 2:01 | 24:21:00 | 5:05 | 14:29 | |
| Function status | NF:F | 58:09:00 | 25:15:00 | 9:00 | 41:00:00 | 3:00 | 45:00:00 | 10:00 | 41:02:00 | |
| Grade | G1a | 33 | 31 | 7 | 31 | 0 | 42 | 9 | 27 | |
| G2b | 27 | 8 | 1 | 10 | 0 | 3 | 1 | 16 | ||
| G3 NET | 2 | 1 | 0 | 0 | 1 | 0 | 0 | N/A | ||
| G3 NEC# | 3 | 0 | 1 | 0 | 2 | 0 | 0 | N/A | ||
| No data | 2 | 0 | 0 | 0 | 0 | 0 | 0 | N/A | ||
| TNM stage | Localized | IND | 23 | 2 | 2 | 24 | 1 | 37 | 9 | 19 |
| IPD | 11 | 0 | 5 | 15 | 1 | 1 | 0 | 6 | ||
| Regional metastatic | IND | 2 | 8 | 0 | 2 | 1 | 1 | 1 | 2 | |
| IPD | 8 | 6 | 2 | 0 | 0 | 1 | 0 | 8 | ||
| Distant metastatic | IND | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| IPD | 23 | 23 | 0 | 0 | 0 | 5 | 0 | 8 | ||
| Disease status (RECIST 1.1) | Progressive | 14 | 2 | 2 | 0 | 0 | 3 | N/A | 4 | |
| Stable | 28 | 27 | 5 | 24 | 3 | 4 | N/A | 18 | ||
| Therapy at blood draw | SSA | 27 | 22 | 0 | 0 | 0 | 5 | 0 | 7 | |
| MTT | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Cx | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | ||
| Previous therapy | SSA | – | 1 | 0 | 0 | 0 | 0 | 0 | – | |
| Primary resection/LRT | 44/6 | 27/4 | 3 | 33 | 2 | 45 | 9 | 39/1 | ||
| PRRT | 11 | 11 | 0 | 0 | 0 | 1 | 0 | 2 | ||
| Cx | 6 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | ||
| Rx | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | ||
| MTT | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
aTC for BPC; bAC for BPC.
ANEN, appendiceal NEN; BPC, bronchopulmonary carcinoids; Cx, chemotherapy; DNEN, duodenal NEN;Function status: NF, non-functioning; F, functioning; Gender: m, Male; F, female; GNEN, gastric NEN; IND, image-negative disease; IPD, image-positive disease; LRT, locoregional therapy; MTT, molecular targeted therapy; N/A, not applicable; PRRT, peptide receptor radionuclide therapy; PNEN, pancreatic NEN;Rx, radiotherapy; RNEN, rectal NEN; SINEN, small intestine (midgut) NEN; SSA, somatostatin analogues.
Methods
Strategy
We examined circulating CgA levels using two different ELISA assays (NEOLISA, EuroDiagnostica, IBL-America, CLIA-certified laboratory, and CgA ELISA, Demeditec Diagnostics GmbH (DD), Germany). CgA results for the entire study cohort (n = 258) were compared with the NETest measurement evaluated at the same time-point. Assay sensitivity was assessed to detect image-positive or microscopic disease and specificity in the disease-free cohort. Histological and radiological parameters were established by the independent analysis of an expert radiologist and dedicated neuroendocrine pathologist. The determination provided: no evidence of disease on imaging (image-negative disease) and no evidence of tumour by histological assessment. Diagnostic metrics (accuracy, sensitivity, specificity) were calculated. Intergroup analyses were undertaken using two-tailed non-parametric tests (Mann-Whitney U-test). Pearson correlation was used for assessment of concordance between the CgA assays.
Sample collection
Plasma/serum for CgA measurement
Peripheral blood samples were collected in S-Monovette K3-EDTA tubes and centrifuged per standard protocol to obtain plasma for NEOLISA CgA evaluation and in S-Monovette serum tubes with clot activator to obtain serum for DD CgA evaluation. Plasma samples for the NEOLISA measurements were frozen (at −80°C) within 30 min of collection and sent frozen in batches, on dry ice, to Wren Laboratories, Branford, Connecticut, USA. All samples were de-identified and anonymized prior to transport. Test analysis data were provided in numeric coded form to the Medical University of Silesia and the blinded data independently evaluated by the study authors. Serum samples for the DD measurements were frozen and stored at −30°C within 30 min of collection until the in-house CgA assay measurement in the ENETS CoE clinical (hospital) laboratory as a part of the standard clinical work-up. Samples were collected at the same time as for NEOLISA measurements.
Blood for NETest measurement
Peripheral blood samples (3 mL) were collected in EDTA tubes, mixed, and stored on ice. Tubes were de-identified and anonymously coded and stored at −80°C within 2 h of collection (34). De-identified blood samples were sent to a central laboratory (Wren Laboratories, Branford, Connecticut, USA). Test analysis data were provided in numeric coded form to the Medical University of Silesia and the blinded data independently evaluated by the study authors.
Radiological evaluation of NEN disease
Disease extent was determined by anatomical imaging: CT or MRI, and/or functional: 68Ga-DOTA-TATE PET/CT in well-differentiated NETs or 18F-FDG PET/CT in G2/G3 NENs. Gastric and rectal NENs were also assessed endoscopically (by gastroscopy or colonoscopy), and by endoscopic ultrasound (EUS). IPD was defined as either CT or MRI or 68Ga-DOTA-TATE/18F-FDG PET/CT-positive. IND was anatomical (CT/MRI), endoscopic/EUS and/or functional 68Ga-DOTA-TATE PET/CT negative.
Disease status
Progressive disease was defined based on anatomical imaging and RECIST 1.1 criteria. Parameters were at least a 20% increase in the sum of diameters of the target lesions (min. 5 mm) measured on anatomical imaging (CT) or detection of new lesions by imaging of the same modality when subsequently performed (35). The time interval of follow-up was a median of 7.5 months (range: 1.5–17 months).
Histological diagnosis
All NEN patients had histologically confirmed NEN disease, reported by an independent expert NEN pathologist in accordance with the WHO 2017 and TNM 8th edition classifications for NENs (36, 37, 38). All biopsy specimens were evaluated (H and E, immunohistochemistry) and reviewed by the same CoE pathologist.
Biomarker measurement
CgA measurement
NEOLISA TM kit (EuroDiagnostica) is an ELISA for detection of CgA in human plasma or serum. The measurement was performed in accordance with the assay manual. Principle of the assay: In a separate dilution plate samples, calibrators and controls are diluted five times in diluent. The diluted material is transferred to the microtiter wells and incubated at room temperature for 60 min. During this first incubation, a MAB captures the CgA to the well surface. After washing to remove unbound material a second, horseradish peroxidase (HRP) labelled MAB is added to detect CgA bound to the well. After incubation for 30 min, the wells are washed and a colour substrate (chromogenic tetramethylbenzidine (TMB) solution) added and re-incubated. The colour development was stopped using 0.5M H2SO4 after 15 min and the colour measured in a spectrophotometer (450 nm). The colour is directly proportional to the amount of CgA bound to the well and the amount of CgA determined by comparison with the calibrator sample colour development. ULN: 108 ng/mL. Measuring range: 10–1450) ng/mL.
Demeditec diagnostics (Germany, DD-Assay) is an ELISA for the quantitative determination of human CgA in serum. The measurement was performed in accordance with the assay manual. Principle of the assay: In the initial step, CgA in the sample binds to CgA-specific antibodies fixed to a 96-well microtiter plate. After incubation and following washing steps, a sandwich is formed by adding CgA antibodies conjugated to horseradish peroxidase. After incubation the wells are thoroughly washed, and the complex bound to the solid phase is detected by using TMB as a substrate. The reaction is monitored at 450 nm. the CgA concentrations in the samples are determined by comparison with a pre-set standard curve. ULN: 99 ng/mL. Measuring range: 7.4–700 ng/mL.
NETest measurement
Details of the PCR methodology, mathematical analysis and validation have been published in detail (30, 34). Assays were undertaken using de-identified samples in a central USA clinically and Federal Government certified laboratory (Wren Laboratories CL-0704, CLIA 07D2081388). In brief, mRNA was isolated from EDTA-collected whole blood samples (mini blood kit, Qiagen) and real-time PCR performed on pre-spotted plates (39). Target transcript levels are normalized and quantified vs a population control (30, 34, 39). Final results are expressed as an activity index (NETest score) from 0 to 100 (30, 34, 39). Normal score cut-off: 20.
Statistical analysis
The sample size needed to detect significant differences in CgA levels per assay (from previously published mean ± s.e.m., using a power of 0.8 and α = 0.05) was calculated to be a minimum of 210 individuals. Intergroup analyses were undertaken using two-tailed non-parametric tests (Mann-Whitney U- test), Pearson correlation to evaluate concordance between the CgA assays output and McNemar-test to compare the different assays with each other. MedCalc statistical calculator was used to calculate the diagnostic accuracy, sensitivity and specificity. Prism 8.4.2 for Windows (GraphPad Software, www.graphpad.com) was utilized. Statistical significance was defined at a P value < 0.05. Data are mean ± s.e.m.
Results
Study cohort
Image-positive disease (n = 123)
101 GEPNENs (42 PNENs, 29 SINENs, 7 DNENs, 15 GNENs Type 1, 1 GNEN Type 3, 7 RNENs), and 22 BPC (Table 1).
Image-negative disease with no evidence of positive histology (biopsy or resection margin) (n = 106)
85 GEPNENs (25 PNENs, 11 PNENs, 2 DNENs, 17 GNENs Type 1, 2 GNECs, 18 RNENs, 10 ANENs), and 21 BPC.
Image-negative disease with positive histology (biopsy or resection margin) (n = 29)
9 GNENs Type 1, and 20 RNENs.
CgA assay concordance
In all patients (n = 258), NEOLISA assay CgA levels were significantly (P < 0.0001) higher (98.6 ± 11 ng/mL) than the DD-assay (76 ± 8 ng/mL) (Figs 1A and 2). The assays exhibited an acceptable concordance in output (Pearson r = 0.84, P < 0.0001) (Fig. 1B), however there were 12% (n = 32) discordant results (Fig. 1C): 22 in IPD, 5 in IND, and 5 in microscopic disease. The discordant results comprised: in IPD, NEOLISA-positive and DD-negative (n = 20), NEOLISA-negative and DD-positive (n = 2); in IND, there were NEOLISA-positive/DD-negative (n = 4), and DD-positive/ NEOLISA-negative (n = 1); and in microscopic disease, all five discordant cases were NEOLISA-positive and DD-negative. Overall, the NEOLISA assay detected more CgA-positive samples.
Figure 1.
Concordance between the CgA Assays (NEOLISA and Demeditec diagnostics). (A) In the entire cohort (n = 258), NEOLISA assay CgA levels were significantly (P < 0.0001) higher (98.6 ± 11 ng/mL) than the DD-assay (76 ± 8 ng/mL). (B) The assays exhibited a concordance of r = 0.84 (Pearson, P < 0.0001). (C) There were 12% (n = 32) discordant results with the NEOLISA assay detecting more CgA-positive samples. The majority of the discordant results comprised IPD.
Figure 2.
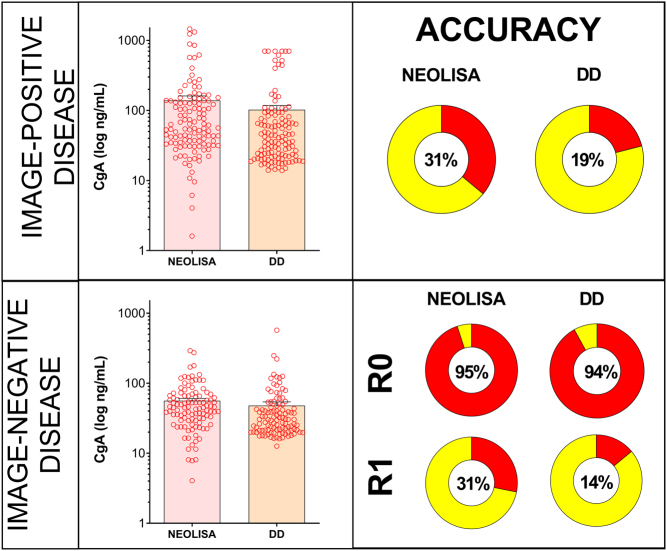
Distribution of CgA measurements of NEOLISA and DD assays compared to imaging. In image-positive disease, CgA levels were significantly elevated in the NEOLISA (139 ± 22) compared to the DD-assay (102 ± 15, P < 0.0016). The accuracy for disease detection ranged between 19and 31%. In image-negative disease, CgA levels were significantly elevated in the NEOLISA (56 ± 4.5) vs the DD-assay (P < 0.0011). The overall accuracy for IND ranged 89–92%. Individual accuracies for R0 were 94–95% while they were 14–31% for R1 disease.
Relationship to disease detection
CgA and image-positive disease (n = 123)
CgA-positives were detected in 41/123 (33%, NEOLISA) vs 23 (19%, DD-assay). McNemar’s Chi2=13.1, P=0.0003 OR: 0.1 (95% CI 0. 0.1–0.41). The NEOLISA was significantly more accurate for detecting NEN disease (Table 2).
Table 2.
Assay results in image-positive and image-negative disease.
| Site | Image positive disease (n = 123) | Image-negative disease and R0 (n = 106) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | True positive | False negative | Acc. | Total | True negative | False positive | Acc. | ||
| NEOLISA | Gastroenteropancreatic | 101 | 36 | 65 | 36% (36/101) | 85 | 74 | 11 | 87% (74/85) |
| PNEN | 42 | 15 | 27 | 25 | 23 | 2 | |||
| SINEN | 29 | 9 | 20 | 11 | 11 | 0 | |||
| DNEN | 7 | 0 | 7 | 2 | 2 | 0 | |||
| GNET Type 1 | 15 | 11 | 4 | 17 | 10 | 7 | |||
| GNEN G3 | 1 | 1 | 0 | 2 | 2 | 0 | |||
| RNET | 7 | 0 | 7 | 18 | 17 | 1 | |||
| ANEN | 0 | - | - | 10 | 9 | 1 | |||
| Bronchopulmonary | 22 | 5 | 17 | 23% (5/22) | 21 | 20 | 1 | 95% (20/21) | |
| All NENs (GEP+BP) | 123 | 41 | 82 | 33%(41/123) | 106 | 94 | 12 | 89%(94/106) | |
| DD | Gastroenteropancreatic | 101 | 21 | 80 | 21% (21/101) | 85 | 77 | 8 | 91% (77/85) |
| PNEN | 42 | 8 | 34 | 25 | 24 | 1 | |||
| SINEN | 29 | 4 | 25 | 11 | 10 | 1 | |||
| DNEN | 7 | 0 | 7 | 2 | 2 | 0 | |||
| GNET Type 1 | 15 | 8 | 7 | 17 | 11 | 6 | |||
| GNEN G3 | 1 | 1 | 0 | 2 | 2 | 0 | |||
| RNET | 7 | 0 | 7 | 18 | 18 | 0 | |||
| ANEN | 0 | - | - | 10 | 10 | 0 | |||
| Bronchopulmonary | 22 | 2 | 20 | 9% (2/22) | 21 | 20 | 1 | 95% (20/21) | |
| All NENs (GEP+BP) | 123 | 23 | 100 | 19%(23/123) | 106 | 97 | 9 | 92%(97/106) | |
| NETest | Gastroenteropancreatic | 101 | 100 | 1 | 99% (100/101) | 85 | 69 | 16 | 81% (69/85) |
| PNEN | 42 | 42 | 0 | 25 | 22 | 3 | |||
| SINEN | 29 | 28 | 1 | 11 | 9 | 2 | |||
| DNEN | 7 | 7 | 0 | 2 | 1 | 1 | |||
| GNET Type 1 | 15 | 15 | 0 | 17 | 10 | 7 | |||
| GNEN G3 | 1 | 1 | 0 | 2 | 2 | 0 | |||
| RNET | 7 | 7 | 0 | 18 | 18 | 0 | |||
| ANEN | 0 | - | - | 10 | 7 | 3 | |||
| Bronchopulmonary | 22 | 22 | 0 | 100% (22/22) | 21 | 2 | 19 | 9.5% (2/21) | |
| All NENs (GEP+BPC) | 123 | 122 | 1 | 99%(122/123) | 106 | 71 | 35 | 67%(71/106) | |
Acc., accuracy; ANEN, appendiceal NEN; BP, bronchopulmonary; BPC, bronchopulmonary carcinoid; DNEN, duodenal NEN; GEP, gastroenteropancreatic; GNEN, gastric NEN; PNEN, pancreatic NEN, RNEN, rectal NEN; SINEN, small intestine (midgut) NEN.
CgA and image-negative disease with no evidence of disease on histology (n = 106)
GEPNENs (n = 85)
NEOLISA was true negative (negative in subjects without the disease) in 74 (87%) and DD in 77 (91%). The majority (63.6–75%) of false positives (positive test in non-detectable disease) were associated with gastric NETs, 7/11 (63.6%) for NEOLISA, and 6/8 (75%) for DD assay. The remainder of false positives for NEOLISA were: PNENs, n = 2, RNEN, n = 1, ANEN, n = 1; for DD: PNEN, n = 1, and SINEN, n = 1 (Table 2).
BPC (n = 21)
NEOLISA and DD were both true negative in 20/21 (95%).
CgA and image-negative but histology-positive group (microscopic disease or R1)
This group comprised 20 RNENs and 9 GNENs Type 1 which were image-negative but positive on biopsy (GNENs, n = 4), or had a positive resection margin (GNENs, n = 5; RNENs, n = 20).
CgA-positives were detected in 7/9 (78%) GNENs Type 1, and 2/20 (10%) RNENs by NEOLISA; 4/9 (44%) GNENs Type 1, and 0/20 (0%) RNENs by DD assay and 9/9 (100%) GNENs Type 1 (Table 3).
Table 3.
Assay positivity in microscopic disease (image-negative but histology positive).
| Assay | Site | Image negative disease but histology positive (R1) (n = 29) | |||
|---|---|---|---|---|---|
| Total | True positive | False negative | Acc. | ||
| NEOLISA | GNEN Type 1 | 9 | 7 | 2 | 78% (7/9) |
| RNEN | 20 | 2 | 18 | 10% (2/20) | |
| DD | GNEN Type 1 | 9 | 4 | 5 | 44% (4/9) |
| RNEN | 20 | 0 | 20 | 0% (0/20) | |
| NETest | GNEN Type 1 | 9 | 9 | 0 | 100% (9/9) |
| RNEN | 20 | 4 | 16 | 20% (4/20) | |
Acc., accuracy; GNEN, gastric NEN; RNEN, rectal NEN.
In image-negative disease, CgA levels were significantly elevated in the NEOLISA (56±4.5) vs the DD-assay (P<0.0011). The overall accuracy for IND ranged from 89 to 92%. Individual accuracies for R0 were 94–95% while they were 14–31% for R1 disease.
Comparison with the NETest Data
Overall, the NETest was positive in 122/123 (99%; McNemar’s Chi2=79–97, P<0.0001 vs NEOLISA and DD-assay).
GEPNENs
NEOLISA was true positive (positive in subjects with confirmed disease) in 36/101 (36%), DD in 21/101 (21%), and the NETest in 100/101 (99%). The false negative results (negative tests in subjects with the disease), were most common (79%) in DD (80/101), 64% (65/101) in NEOLISA, and only 1% (1/101) in the NETest (Fig. 3).
Figure 3.
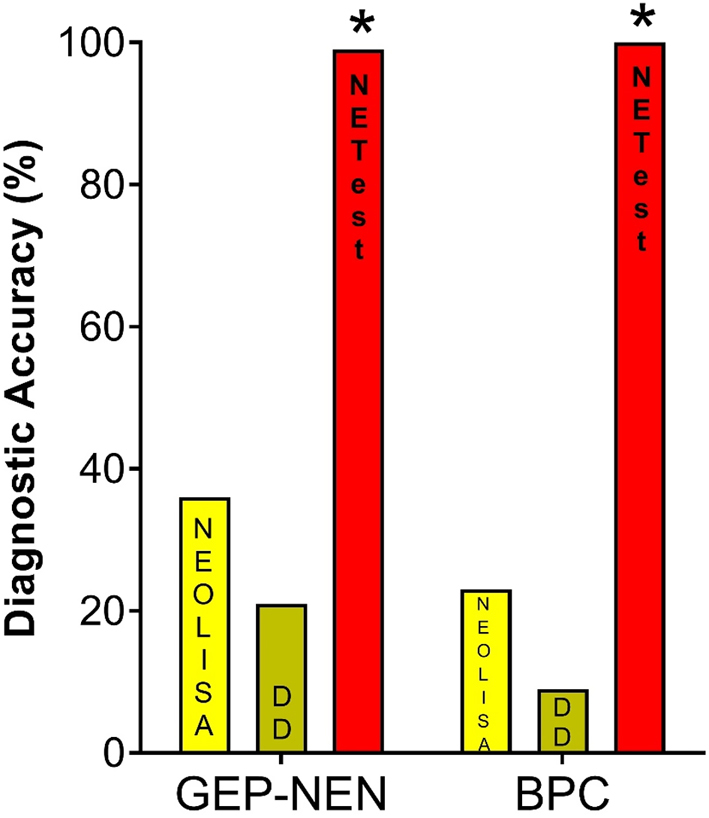
Comparison between the NETest and CgA assays for disease detection. In GEP-NENs (n = 101) test accuracy: NEOLISA 36%, DD 21% and NETest 99%. In BPC (n = 22) test accuracy, NEOLISA 23%, DD 9% and NETest 100% *P<0.0001 vs either CgA assay. GEP-NEN, gastroenteropancreatic NEN; BPC, bronchopulmonary carcinoids.
BPC
NEOLISA was true positive in 5/22 (23%), DD in 2/22 (9%), and the NETest in all (100%). The false negative results were most common (91%) in DD (20/22), 77% (17/22) in NEOLISA. There were no false negatives using the NETest (Fig. 3).
Diagnostic accuracy NETest vs CgA assays
Diagnostic accuracy in the NEN (GEPNEN+BPC) cohort was: 58% for NEOLISA, and 51% for DD compared to 84% for NETest. The metrics are included in Table 4. The NETest was significantly more accurate than NEOLISA (Chi2=32.2, P<0.0001) and the DD-assay (Chi2=49.0, P<0.0001). The NETest has a higher sensitivity (99%) to detect disease in image-detectable NENs than CgA (18 or 32%). In the absence of image-detectable disease, NETest specificity is spuriously lower than CgA since the NETest is known to be positive in image-negative microscopic disease (40). Thus, in low burden disease CgA can be falsely negative since it is less sensitive than the NETest for detection of microscopic disease. The overall accuracy analysis related to imaging, demonstrates the NETest is significantly more accurate (84%) as a diagnostic than CgA (51 or 57%).
Table 4.
The NETest and CgA assays metrics in image-detectable disease demonstrate the limitations of imaging and CgA in the detection of biochemical recurrence.
| Sensitivity (95% CI) | Specificity (95% CI) | Accuracy (95% CI) | |
|---|---|---|---|
| NETest | 99% 96–100%) | 65% (55–74%) | 84% (88–88%) |
| NEOLISA | 32% (24–41%) | 89% (81–94%) | 57% (51–64%) |
| DD-assay | 18% (12–26%) | 92% (85–96%) | 51% (44–58%) |
Disease burden NETest vs CgA
Assay levels in disease positive (macroscopic or microscopic disease) between clinical stages of the disease were analysed.
(A) Localized disease (n = 68) including image-positive, n = 39 (GEPNENs, n = 33, BPC, n = 6); and microscopic GEPNETs (RNENs, n = 20, GNENs T1, n = 9).
(B) Regional metastatic (n = 24, GEPNENs, n = 16, BPC, n = 8).
(C) Distant metastatic (n = 60, GEPNENs, n = 52, BPC, n = 8).
NEOLISA levels were not different between localized, regional or distant metastatic stage (142±30 vs 82±24 vs 133±30, P = 0.67). For DD, levels in localized stage were significantly higher than in regional metastatic stage (100.5±18 vs 69±28, P = 0.02) but not different to distant metastatic levels (101.7±22, P = 0.19). NETest levels for localized disease (24±1.3) were significantly lower than regional metastatic (30±1.5) and distant metastatic (37±3), or combined metastatic disease (35±2) P = 0.015, P = 0.026, P = 0.008 respectively). Number of image-positive BPC (n = 22, localized, n = 6, regional metastatic, n = 8, distant metastatic, n = 8), was insufficient for a separate analysis in lung NENs.
Tumour grade NETest vs CgA
In the disease positive group (n = 152), image-positive NENs comprised the following: NETs G1, n = 61, NETs G2, n = 32, NENs G3, n = 5 (including three NECs, two NETs), microscopic disease, NETs G1, n = 28, NETs G2, n = 1.
Levels between grades G1–G3 were compared. Neither of the CgA assays (NEOLISA, P = 0.48, DD; ANOVA, P = 0.45) identified a difference. Using the NETest, G2 levels (35±3.5) were significantly higher than in G1 (27±1.8, P = 0.012). The number of NENs G3 was insufficient for adequately powered statistical analysis, but levels were higher (48±12) than for G1 or G2.
Stable vs progressive disease status NETest vs CgA
In the image-positive cohort (n = 123 subjects), 25 were progressive (GEPNENs, n = 21 including PNENs, n = 14, SINENs, n = 2, RNENs, n = 3, DNENs, n = 2, and 4 BPC). Radiological stable disease was identified in 98/123.
Neither CgA assay identified a difference between PD and SD: NEOLISA- PD: 87±16.5 vs SD 151±26.8 (P = 0.61); DD- PD: 93±33 vs SD 104.5±17.5 (P = 0.76). Overall, similar proportions of subjects irrespective of disease status (PD or SD) exhibited an elevated CgA >2× ULN (10–14%, P = NS). NETest levels in PD (47±5) were significantly higher than in SD (29±1), P = 0.0009. In contrast to CgA, a higher proportion of PD (52%) exhibited an elevated NETest (>40 or 2× ULN) than SD (20%, Chi2=9.4, P = 0.002, Fig. 4).
Figure 4.
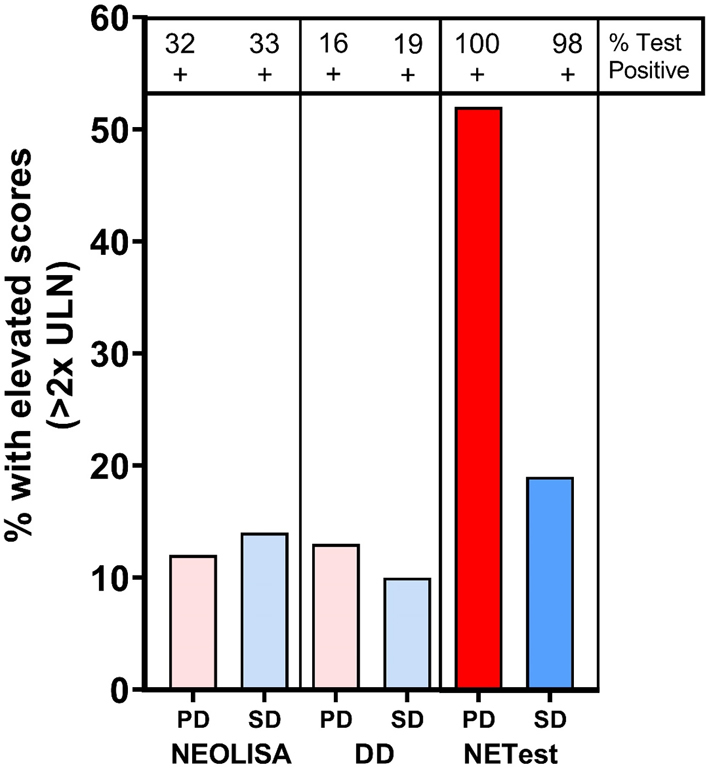
Stable vs progressive disease status identification. Analysis of percentage subjects with assay levels <2× ULN with comparison between PD (n = 25) and SD (n = 98). Similar proportions of patients exhibited elevated CgA levels irrespective of disease status. For NEOLISA this was 12–14% and for DD it was 10–13% (P = NS). For the NETest, significantly more PD exhibited elevated levels than SD (Chi2 = 9.4, P = 0.002). PD, progressive disease; SD, stable disease; ULN, upper limit of normal.
Discussion
Biomarkers of oncological disease are well-recognized as important adjunctive parameters in the management of the therapy of different cancer types. In NET disease, secretory markers such as gastrin, insulin, glucagon and VIP have proved valuable clinical tools for specific tumours (41). Ideally, however, a universal NEN biomarker should capture both secretory and non-secretory tumours. The introduction of CgA seemed promising. However, despite widespread initial enthusiasm, there is now growing recognition that CgA has significant clinical limitations (9, 10, 42, 43). Concerns range from lack of sensitivity to difficulties with different assay systems and the lack of standardized values (11, 42, 44). This presumably reflects the fact that a secretory protein has little relevance to the cellular mechanisms that constitute the hallmarks of cancer and define growth, invasion and metastasis (45). CgA and NETest therefore represent divergent approaches to quantification of NEN disease activity. CgA reflects the secretory characteristic of some tumours, while the NETest assesses molecular genomic regulators that determine the specific biological behaviour of all NETs.
Given the panoply of problems described with CgA, we undertook a prospective evaluation of CgA effectiveness in real-world practice. We compared two different CgA assays performed in two independent, clinically approved laboratories, a CLIA-certified laboratory (USA, NEOLISA assay) and a general hospital laboratory at the ENETS Center of Excellence (DD assay) to assess diagnostic accuracy and clinical utility. We then compared CgA to the NETest.
Our study was performed prospectively in a large NEN cohort (n = 258), comprising 83% GEPNENs (n = 214), and 17% of BPC (n = 43) to assure adequately powered analysis. To ensure rigorous analysis, all samples were de-identified and head-to-head comparison of the three assays undertaken. All the assays were evaluated using biological material obtained at the same blood draw. The NEOLISA CgA analysis and NETest assays were undertaken blinded at an independent CLIA-certified USA facility on processed samples frozen at -80°C.
The strengths of our study include the large number of patients enrolled; the head-to-head comparison of two commonly used CgA assays undertaken in independent, certified institutions and the blinded assessment of their efficacy compared to a novel molecular genomic assay strategy. Our study has some limitations. In particular, the small numbers in some subgroups, such as duodenal, appendiceal, NENs G3, limit the ability to derive definitive conclusions. A further limitation is that 3% patients (8/258) were not available for follow-up. Overall, these numbers represent a real-world assessment of the clinical material encountered in an ENETS CoE.
CgA levels were measured by two assays. Each assay had a different upper limit of normal (108 vs 99 ng/mL) and disparate measuring ranges (≤1450 vs 700 ng/mL). Although there was an adequate correlation (0.84) between the output of the two CgA assays, NEOLISA exhibited significantly higher levels than DD consistent with known differences in plasma vs serum measurements (9). Discordant results were related to IPD. The NEOLISA detected 15% more true positive samples than DD but also had a higher number of false positive results in disease-free individuals (3%).
In IPD, NEOLISA exhibited higher sensitivity (33%) than DD (19%). Both CgA assays were positive in 21 individuals (of 123 IPDs), half of which were metastatic. Both CgA assays were negative in 80 IPD individuals. The majority of this group was metastatic (70%, regional: 21%, distant: 49%). Overall, both CgA assays failed to accurately detect metastatic disease irrespective of primary site. Seventy percent of the NEOLISA-positive but DD-negative were metastatic. CgA measurements therefore failed to provide an accurate reflection of NET disease. The optimal metrics proposed for NEN biomarkers have been recommended to exceed 80% for sensitivity (1). In comparison to the 19–33% sensitivity for the CgA assays, the NETest sensitivity was 99%.
We next evaluated the assay specificity in the IND with no evidence of disease on histology (n = 106). DD had the highest specificity (91%), NEOLISA (87%), and the NETest (81%) in GEPNENs. The majority (44–75%) of false positives were associated with gastric NETs. Of note, all PPI-treated patients had medication discontinued for at least 10 days at blood draw; PPIs are therefore not a confounder (46). CgA identifies the secretory propensity of the fundic ECL cells while the NETest identifies molecular evidence of ECL cell neoplastic transformation not identifiable by endoscopy or imaging. Random histological biopsy is unable to identify molecular alterations that occur during the transformation of proliferating ECL cells to a neoplastic phenotype (47). The NETest and CgA assay therefore accurately recapitulate what is known about the natural history of ECL cell neoplastic transformation (48).
Overall, there were 43 image/histology-negative subjects that tested positive for either CgA (n = 12) assay and/or the NETest (n = 35). Overall, eight subjects were lost to follow-up. Three subjects positive on all three assays were GNENs T1, and one of them at 11-month FU exhibited a local recurrence. The remaining two (at 1 year, and 4-months FU) remained radiologically and histologically disease- free.
Two GEPNENs (1 PNET, 1 GNET T1) who tested NEOLISA/DD-positive but NETest negative, remained disease- free at 1.2 ± 1 years. Amongst five NEOLISA-positive and DD/NETest-negative, three were lost to follow-up, the remaining two were image-negative at 7 ± 5 months. One SINET was DD-positive and NEOLISA/NETest-negative and remained image-negative at 1.3 years. A GNET T1 which was NEOLISA/NETest-positive and DD -negative, had a recurrence at 2.3 years.
Thirty individuals were NETest-positive only (11 GEPNENs, and 19 BPC), with 8 borderline positive scores of 20, 17 of 27, 2 of 33, 2 of 40, and 1 of 46.7. Five were lost to follow-up. FU in the GEPNENs was 1.3 ± 0.9 years; one PNET (NETest: 33) had a recurrence in the liver at 2.5 years FU, and one GNET T1 (NETest: 40) was diagnosed with a meningioma. Amongst 19 BPC (1.3 ± 0.6 years FU), two patients (NETest: 20 and 27) developed recurrence (at 0.3 and 1.2 years), one (NETest: 40) developed a new lesion in the liver at 5-month follow- up. There was one patient with DIPNECH confirmed on histology (NETest: 20). These data are consistent with previous reports documenting that image identification of disease may lag 1–2 years behind NETest identification of residual/recurrent tumour (30, 34). We consider that the most likely explanation for the positive scores in image- negative individuals to represent residual disease not yet identifiable by imaging.
NETest-positive cases occurred in 19/21 BPC considered disease-free after surgery. CgA was negative in 95% these cases. Positive cases likely reflect the higher sensitivity of the molecular detection of microscopic disease not yet identifiable by imaging or by secretory markers. Pathology studies report that 50% of resected specimens demonstrate tumour deposits despite being undetectable by imaging (49, 50). Limits of detection for imaging range from 2–4 mm (MRI/ CT) to 4–6 mm with 68Ga-SSA PET/CT or 18F-Fluorodeoxyglucose PET/CT) (51). There is therefore a delay in tumour detection until sufficient tumour volume or adequate level of detectable receptor expression is achieved. Since a 5 mm lesion comprises 108 tumour cells and a 10 mm lesion at least 109 cells, a comparison can be made with NETest sensitivity. Spike-in studies (blood with NET tumour cells) demonstrate that measurement of circulating NET genes identify one tumour cell/millilitre of blood (52). By extrapolation, blood gene measurement can be considered ~ 120,000 times more sensitive than imaging. Thus, micrometastatic disease is more likely to be identified by molecular amplification (gene expression–PCR) techniques in blood than by imaging alone. Consistent with this observation is a report that 50% of histology-negative nodes from small bowel resections are mRNA-positive using PCR approaches (53). A recent report documents that 2 mm liver lesions, not visible on imaging, were detected by the NETest and their presence validated by biopsy (40). Molecular techniques (PCR-based) therefore are more sensitive than standard approaches to detect microscopic disease and will likely detect early metastatic disease and facilitate accurate staging. In our study, the NETest detected the highest number of microscopic disease cases (45%) compared to either CgA assay (14–31%) confirming its utility as a marker of microscopic disease. Proof-of-principle for the assay is provided by the longitudinal follow-up. Three of fourteen NETest-positive but R0-IND GEPNENs (two were lost to follow-up) demonstrated a recurrence while 3/16 R0-IND BPC (three were lost to follow-up) developed recurrences within a time frame between 3 and 30 months. Only two of the six recurrences were CgA-positive (one NEOLISA/DD-positive, and one NEOLISA-positive and DD-negative).
Neither of the CgA assays identified any significant differences between disease stages or grades. This differs from previous reports where it has been suggested that CgA could provide a stratification (13, 14, 15). It seems biologically unlikely that a secretory protein level could be an effective biomarker of disease extent and proliferative activity of a tumour. We have accordingly interpreted this as evidence that disease burden may have been a confounding variable in such studies. In our study, NETest levels correlated effectively with disease stage and higher grade (G2 vs G1).
In terms of disease status, an effective clinical biomarker should identify disease progression and correlate with it. We therefore compared assays levels between radiologically progressive (n = 25) vs stable (n = 98). For both CgA assays, levels between PD and SD were not different. NETest levels, however, were significantly higher in PD than in SD consistent with a multigene biomarker that assessed molecular genomic regulators of disease activity. These observations of the NETest confirm a recent prospective study in an ENETS CoE (54), and are consistent with a number of other studies that demonstrate that the NETest is an accurate marker of disease status (28, 31, 34).
Our study highlights that the different methodology of individual CgA assays affect metrics and results obtained are often not comparable. Others have identified a 36% discordance rate between IRMA and ELISA assays (55). Separately, an external quality control study reported a five-fold inter-laboratory variation rate between IRMA and ELISA results (11) and other reports have noted differences in CgA levels between plasma and serum (higher in plasma) (9). There are also differences between measuring CgA fragments vs the intact molecule and protein folding are likely responsible for different antibody recognition proteins (6). In this study, we confirm the lack of utility of CgA measurement in NEN disease using two standard assays (12).
CgA is a monoanalyte assay that captures a one-dimensional data point as opposed to multianalyte assays that measure numerous data points for example, transcripts. Hanahan and Weinberg in their two seminal publications (2000 and 2011) (3, 56) identified that tumours are not homogeneous but represent multifactorial biological entities integrated as a neoplastic unit. It is noteworthy that secretion is not considered a neoplastic hallmark. Thus many neuroendocrine disease assays which measure CgA, insulin, gastrin, serotonin do not define the cancer hallmarks. Multianalyte markers, in contrast to secretory markers, have the capacity to measure multiple biologically relevant cancer hallmarks simultaneously for example, Mammaprint. The latter is a metastatic breast cancer assay which has prognostic utility and is used for treatment stratification (57, 58). In multianalyte assays such as the NETest output is a score that is based on multi-algorithmic analyses, usually machine learning or neural network models (59, 60). The NETest is a blood-based equivalent that is based on transcriptomic evaluation of NETs per gene discovery. The basis of the assay is mRNA isolation from whole blood with subsequent cDNA production with target genes measured by PCR. Gene expression is evaluated using machine learning algorithms and the output scaled 0–100 based on omic gene expression for example, levels of expression of the proliferome, RAS-RAF-signaling axis and epigenome (multi-analyte measurements of tumour pathobiology). Final results are expressed as an activity index (NETest score) from 0 to 100 (30, 34, 39). Cut-off points were derived from prospective analyses of receiver operator curves (ROC) and correlate with RECIST-defined disease stability or progression (28). The normal score cut-off is ≤20 while values 21–40% are considered ‘stable’ disease and 41–100 reflect ‘progressive disease. Clinical superiority was assessed by AUC comparison of the NETest (0.95–0.98) vs the monoanalytes CgA (0.67), neurokinin A (0.66) and pancreastatin (0.56) (61). We envisage that such an approach may in the future evolve to provide functions other than as a diagnostic. Thus, identification of the functional components of a tumour may enable a modified assay to be used as a prognostic (62) or to accurately stratify patients for different treatments as ha recently been shown for peptide receptor radionuclide therapy (PRRT) by Bodei and colleagues (63).
Overall, our study demonstrates that a molecular genomic assay that measures NET-specific genes in blood is significantly more effective than two clinically approved assays of CgA in blood. In contradistinction to the NETest, CgA levels were not clinically useful in the diagnosis of GEPNEN or BPC and identification of disease grade or progress. We are of the opinion that monoanalyte biomarkers such as CgA fail to provide multidimensional insight into the complex processes inherent in neoplasia. This head-to-head comparison of the two monoanalyte (CgA) assays vs multianalyte demonstrates the superiority of the NETest. Based on our study, we would propose that the NETest be included for clinical evaluation of NET patients.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported. Wren Laboratories CT, USA measured samples using NEOLISA for CgA and NETEST®. Wren Laboratories were provided with deidentified samples. Wren laboratories were not involved in data analysis or manuscript writing.
Funding
This work did not receive any specific grant from any funding agency in the public, commercial or not-for-profit sector.
Author contribution statement
A Malczewska: responsible for study design, data analysis and manuscript writing. B Kos-Kudla: study design, clinical data analysis and manuscript writing. K Oberg: study design, CgA assay assessment, clinical data analysis, and manuscript writing and discussion, critique analysis.
Acknowledgement
The authors gratefully acknowledge the generosity of Wren Laboratories for providing sample measurement pro bono.
References
- 1.Oberg K, Modlin IM, De Herder W, Pavel M, Klimstra D, Frilling A, Metz DC, Heaney A, Kwekkeboom D, Strosberg Jet al. Consensus on biomarkers for neuroendocrine tumour disease. Lancet Oncology 2015. 16 e435–e446. ( 10.1016/S1470-2045(15)00186-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capdevila J, Bodei L, Davies P, Gorbounova V, Jensen RT, Knigge UP, Krejs GJ, Krenning E, O'Connor JM, Peeters Met al. Unmet medical needs in metastatic lung and digestive neuroendocrine neoplasms. Neuroendocrinology 2019. 108 18–25. ( 10.1159/000493319) [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA.Hallmarks of cancer: the next generation. Cell 2011. 144 646–674. ( 10.1016/j.cell.2011.02.013) [DOI] [PubMed] [Google Scholar]
- 4.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM.Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics. Reporting recommendations for tumor marker prognostic studies (REMARK). Journal of the National Cancer Institute 2005. 97 1180–1184. ( 10.1093/jnci/dji237) [DOI] [PubMed] [Google Scholar]
- 5.O'Connor DT, Bernstein KN.Radioimmunoassay of chromogranin A in plasma as a measure of exocytotic sympathoadrenal activity in normal subjects and patients with pheochromocytoma. New England Journal of Medicine 1984. 311 764–770. ( 10.1056/NEJM198409203111204) [DOI] [PubMed] [Google Scholar]
- 6.Bartolomucci A, Possenti R, Mahata SK, Fischer-Colbrie R, Loh YP, Salton SR.The extended granin family: structure, function, and biomedical implications. Endocrine Reviews 2011. 32 755–797. ( 10.1210/er.2010-0027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofland J, Zandee WT, de Herder WW.Role of biomarker tests for diagnosis of neuroendocrine tumours. Nature Reviews: Endocrinology 2018. 14 656–669. ( 10.1038/s41574-018-0082-5) [DOI] [PubMed] [Google Scholar]
- 8.Kyriakopoulos G, Mavroeidi V, Chatzellis E, Kaltsas GA, Alexandraki KI.Histopathological, immunohistochemical, genetic and molecular markers of neuroendocrine neoplasms. Annals of Translational Medicine 2018. 6 252. ( 10.21037/atm.2018.06.27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marotta V, Zatelli MC, Sciammarella C, Ambrosio MR, Bondanelli M, Colao A, Faggiano A.Chromogranin A as circulating marker for diagnosis and management of neuroendocrine neoplasms: more flaws than fame. Endocrine-Related Cancer 2018. 25 R11–R29. ( 10.1530/ERC-17-0269) [DOI] [PubMed] [Google Scholar]
- 10.Pulvirenti A, Rao D, McIntyre CA, Gonen M, Tang LH, Klimstra DS, Fleisher M, Ramanathan LV, Reidy-Lagunes D, Allen PJ.Limited role of chromogranin A as clinical biomarker for pancreatic neuroendocrine tumors. HPB 2018. 21 612–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verderio P, Dittadi R, Marubini E, Pizzamiglio S, Gion M, De Apollonia L, Paradiso A.Italian Network for Quality Assessment of Tumor Biomarkers (INQAT) Group. An Italian program of external quality control for chromogranin A (CgA) assay: performance evaluation of CgA determination. Clinical Chemistry and Laboratory Medicine 2007. 45 1244–1250. ( 10.1515/CCLM.2007.251) [DOI] [PubMed] [Google Scholar]
- 12.Oberg K, Couvelard A, Delle Fave G, Gross D, Grossman A, Jensen RT, Pape UF, Perren A, Rindi G, Ruszniewski Pet al. Enets consensus guidelines for standard of care in neuroendocrine tumours: biochemical markers. Neuroendocrinology 2017. 105 201–211. ( 10.1159/000472254) [DOI] [PubMed] [Google Scholar]
- 13.Gkolfinopoulos S, Tsapakidis K, Papadimitriou K, Papamichael D, Kountourakis P.Chromogranin A as a valid marker in oncology: clinical application or false hopes? World Journal of Methodology 2017. 7 9–15. ( 10.5662/wjm.v7.i1.9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eriksson B, Oberg K, Stridsberg M.Tumor markers in neuroendocrine tumors. Digestion 2000. 62 (Supplement 1) 33–38. ( 10.1159/000051853) [DOI] [PubMed] [Google Scholar]
- 15.Campana D, Nori F, Piscitelli L, Morselli-Labate AM, Pezzilli R, Corinaldesi R, Tomassetti P.Chromogranin A: is it a useful marker of neuroendocrine tumors? Journal of Clinical Oncology 2007. 25 1967–1973. ( 10.1200/JCO.2006.10.1535) [DOI] [PubMed] [Google Scholar]
- 16.Korse CM, Taal BG, Vincent A, van Velthuysen ML, Baas P, Buning-Kager JC, Linders TC, Bonfrer JM.Choice of tumour markers in patients with neuroendocrine tumours is dependent on the histological grade. A marker study of chromogranin A, Neuron specific enolase, progastrin-releasing peptide and cytokeratin fragments. European Journal of Cancer 2012. 48 662–671. ( 10.1016/j.ejca.2011.08.012) [DOI] [PubMed] [Google Scholar]
- 17.Siravegna G, Marsoni S, Siena S, Bardelli A.Integrating liquid biopsies into the management of cancer. Nature Reviews: Clinical Oncology 2017. 14 531–548. ( 10.1038/nrclinonc.2017.14) [DOI] [PubMed] [Google Scholar]
- 18.Hodara E, Morrison G, Cunha A, Zainfeld D, Xu T, Xu Y, Dempsey PW, Pagano PC, Bischoff F, Khurana Aet al. Multiparametric liquid biopsy analysis in metastatic prostate cancer. JCI Insight 2019. 4 e125529. ( 10.1172/jci.insight.125529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald BR, Contente-Cuomo T, Sammut SJ, Odenheimer-Bergman A, Ernst B, Perdigones N, Chin SF, Farooq M, Mejia R, Cronin PAet al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Science Translational Medicine 2019. 11 eaax7392. ( 10.1126/scitranslmed.aax7392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wills B, Gorse E, Lee V.Role of liquid biopsies in colorectal cancer. Current Problems in Cancer 2018. 42 593–600. ( 10.1016/j.currproblcancer.2018.08.004) [DOI] [PubMed] [Google Scholar]
- 21.Modlin IM, Drozdov I, Kidd M.The identification of gut neuroendocrine tumor disease by multiple synchronous transcript analysis in blood. PLoS ONE 2013. 8 e63364. ( 10.1371/journal.pone.0063364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Toubah TE, Cives M, Valone T, Blue K, Strosberg JR.Sensitivity and specificity of the NETest: a validation study. Neuroendocrinology 2020. 37 222–222. ( 10.1159/000509866) [DOI] [PubMed] [Google Scholar]
- 23.Pavel M.Translation of molecular pathways into clinical trials of neuroendocrine tumors. Neuroendocrinology 2013. 97 99–112. ( 10.1159/000336089) [DOI] [PubMed] [Google Scholar]
- 24.Peczkowska M, Cwikla J, Kidd M, Lewczuk A, Kolasinska-Cwikla A, Niec D, Michalowska I, Prejbisz A, Januszewicz A, Chiarelli Jet al. The clinical utility of circulating neuroendocrine gene transcript analysis in well-differentiated paragangliomas and pheochromocytomas. European Journal of Endocrinology 2017. 176 143–157. ( 10.1530/EJE-16-0727) [DOI] [PubMed] [Google Scholar]
- 25.van Treijen MJC, Korse CM, van Leeuwaarde RS, Saveur LJ, Vriens MR, Verbeek WHM, Tesselaar MET, Valk GD.Blood transcript profiling for the detection of neuroendocrine tumors: results of a large independent validation study. Frontiers in Endocrinology 2018. 9 740. ( 10.3389/fendo.2018.00740) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malczewska A, Oberg K, Bodei L, Aslanian H, Lewczuk A, Filosso PL, Wojcik-Giertuga M, Rydel M, Zielinska-Les I, Walter Aet al. NETest liquid biopsy is diagnostic of lung neuroendocrine tumors and identifies progressive disease. Neuroendocrinology 2019. 108 219–231. ( 10.1159/000497037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malczewska A, Witkowska M, Makulik K, Bocian A, Walter A, Pilch-Kowalczyk J, Zajecki W, Bodei L, Oberg KE, Kos-Kudla B.NETest liquid biopsy is diagnostic of small intestine and pancreatic neuroendocrine tumors and correlates with imaging. Endocrine Connections 2019. 1 19, -0030. ( 10.1530/EC-19-0030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Öberg K, Califano A, Strosberg JR, Ma S, Pape U, Bodei L, Kaltsas G, Toumpanakis C, Goldenring JR, Frilling Aet al. A meta-analysis of the accuracy of a neuroendocrine tumor mRNA genomic biomarker (NETest) in blood. Annals of Oncology 2020. 31 202–212. ( 10.1016/j.annonc.2019.11.003) [DOI] [PubMed] [Google Scholar]
- 29.Bodei L, Kidd M, Modlin IM, Severi S, Drozdov I, Nicolini S, Kwekkeboom DJ, Krenning EP, Baum RP, Paganelli G.Measurement of circulating transcripts and gene cluster analysis predicts and defines therapeutic efficacy of peptide receptor radionuclide therapy (PRRT) in neuroendocrine tumors. European Journal of Nuclear Medicine and Molecular Imaging 2016. 43 839–851. ( 10.1007/s00259-015-3250-z) [DOI] [PubMed] [Google Scholar]
- 30.Cwikla JB, Bodei L, Kolasinska-Cwikla A, Sankowski A, Modlin IM, Kidd M.Circulating transcript analysis (NETest) in GEP-NETs treated with somatostatin analogs defines therapy. Journal of Clinical Endocrinology and Metabolism 2015. 100 E1437–E. ( 10.1210/jc.2015-2792) [DOI] [PubMed] [Google Scholar]
- 31.Liu E, Paulson S, Gulati A, Freudman J, Grosh W, Kafer S, Wickremesinghe PC, Salem RR, Bodei L.Assessment of NETest clinical utility in a US Registry-based study. Oncologist 2019. 24 783–790. ( 10.1634/theoncologist.2017-0623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Modlin IM, Frilling A, Salem RR, Alaimo D, Drymousis P, Wasan HS, Callahan S, Faiz O, Weng L, Teixeira Net al. Blood measurement of neuroendocrine gene transcripts defines the effectiveness of operative resection and ablation strategies. Surgery 2016. 159 336–347. ( 10.1016/j.surg.2015.06.056) [DOI] [PubMed] [Google Scholar]
- 33.van Treijen MJC, van der Zee D, Heeres BC, Staal FCR, Vriens MR, Saveur LJ, Verbeek WHM, Korse CM, Maas M, Valk GDet al. Blood molecular genomic analysis predicts the disease course of GEP NET patients: a validation study of the predictive value of the NETest ®. Neuroendocrinology 2020. ( 10.1159/000509091) [DOI] [PubMed] [Google Scholar]
- 34.Pavel M, Jann H, Prasad V, Drozdov I, Modlin IM, Kidd M.NET blood transcript analysis defines the crossing of the clinical Rubicon: when stable disease becomes progressive. Neuroendocrinology 2017. 104 170–182. ( 10.1159/000446025) [DOI] [PubMed] [Google Scholar]
- 35.de Mestier L, Dromain C, d'Assignies G, Scoazec JY, Lassau N, Lebtahi R, Brixi H, Mitry E, Guimbaud R, Courbon Fet al. Evaluating neuroendocrine tumors progression and therapeutic response: state of the art. Endocrine-Related Cancer 2013. 18 18. [DOI] [PubMed] [Google Scholar]
- 36.Delle Fave G, O'Toole D, Sundin A, Taal B, Ferolla P, Ramage JK, Ferone D, Ito T, Weber W, Zheng-Pei Zet al. ENETS consensus guidelines update for gastroduodenal neuroendocrine neoplasms. Neuroendocrinology 2016. 103 119–124. ( 10.1159/000443168) [DOI] [PubMed] [Google Scholar]
- 37.Kos-Kudla B, Blicharz-Dorniak J, Strzelczyk J, Baldys-Waligorska A, Bednarczuk T, Bolanowski M, Boratyn-Nowicka A, Borowska M, Cichocki A, Cwikla JBet al. Diagnostic and therapeutic guidelines for gastro-entero-pancreatic neuroendocrine neoplasms (recommended by the Polish Network of Neuroendocrine Tumours). Endokrynologia Polska 2017. 68 79–110. ( 10.5603/EP.2017.0015) [DOI] [PubMed] [Google Scholar]
- 38.Lipinski M, Rydzewska G, Foltyn W, Andrysiak-Mamos E, Baldys-Waligorska A, Bednarczuk T, Blicharz-Dorniak J, Bolanowski M, Boratyn-Nowicka A, Borowska Met al. Gastroduodenal neuroendocrine neoplasms, including gastrinoma – management guidelines (recommended by the Polish Network of Neuroendocrine Tumours). Endokrynologia Polska 2017. 68 138–153. ( 10.5603/EP.2017.0016) [DOI] [PubMed] [Google Scholar]
- 39.Kidd M, Drozdov IA, Matar S, Gurunlian N, Ferranti NJ, Malczewska A, Bennett P, Bodei L, Modlin IM.Utility of a ready-to-use PCR system for neuroendocrine tumor diagnosis. PLoS ONE 2019. 14 e0218592. ( 10.1371/journal.pone.0218592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malczewska A, Bodei L, Kidd M, Modlin IM.Blood mRNA measurement (NETest) for neuroendocrine tumor diagnosis of image-negative liver metastatic disease. Journal of Clinical Endocrinology and Metabolism 2019. 104 867–872. ( 10.1210/jc.2018-01804) [DOI] [PubMed] [Google Scholar]
- 41.Falconi M, Eriksson B, Kaltsas G, Bartsch DK, Capdevila J, Caplin M, Kos-Kudla B, Kwekkeboom D, Rindi G, Kloppel Get al. ENETS consensus guidelines update for the management of patients with functional pancreatic neuroendocrine tumors and non-functional pancreatic neuroendocrine tumors. Neuroendocrinology 2016. 103 153–171. ( 10.1159/000443171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gut P, Czarnywojtek A, Fischbach J, Baczyk M, Ziemnicka K, Wrotkowska E, Gryczynska M, Ruchala M.Chromogranin A – unspecific neuroendocrine marker. Clinical utility and potential diagnostic pitfalls. Archives of Medical Science 2016. 12 1–9. ( 10.5114/aoms.2016.57577) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marotta V, Nuzzo V, Ferrara T, Zuccoli A, Masone M, Nocerino L, Del Prete M, Marciello F, Ramundo V, Lombardi Get al. Limitations of chromogranin A in clinical practice. Biomarkers 2012. 17 186–191. ( 10.3109/1354750X.2012.654511) [DOI] [PubMed] [Google Scholar]
- 44.Stridsberg M, Eriksson B, Oberg K, Janson ET.A comparison between three commercial kits for chromogranin A measurements. Journal of Endocrinology 2003. 177 337–341. ( 10.1677/joe.0.1770337) [DOI] [PubMed] [Google Scholar]
- 45.Modlin IM, Bodei L, Kidd M.Neuroendocrine tumor biomarkers: From monoanalytes to transcripts and algorithms. Best Practice and Research: Clinical Endocrinology and Metabolism 2016. 30 59–77. ( 10.1016/j.beem.2016.01.002) [DOI] [PubMed] [Google Scholar]
- 46.Grozinsky-Glasberg S, Alexandraki KI, Angelousi A, Chatzellis E, Sougioultzis S, Kaltsas G.Gastric carcinoids. Endocrinology and Metabolism Clinics of North America 2018. 47 645–660. ( 10.1016/j.ecl.2018.04.013) [DOI] [PubMed] [Google Scholar]
- 47.Lawrence B, Kidd M, Svejda B, Modlin I.A clinical perspective on gastric neuroendocrine neoplasia. Current Gastroenterology Reports 2011. 13 101–109. ( 10.1007/s11894-010-0158-4) [DOI] [PubMed] [Google Scholar]
- 48.Malczewska A, Procner A, Walter A, Kusnierz K, Zajecki W, Aslanian H, Kos-Kudla B.The NETest liquid biopsy is diagnostic for gastric neuroendocrine tumors: observations on the blood-based identification of microscopic and macroscopic residual diseaseOK. BMC Gastroenterology 2020. 20 235. ( 10.1186/s12876-020-01348-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elias D, Lefevre JH, Duvillard P, Goere D, Dromain C, Dumont F, Baudin E.Hepatic metastases from neuroendocrine tumors with a ‘thin slice’ pathological examination: they are many more than you think. Annals of Surgery 2010. 251 307–310. ( 10.1097/SLA.0b013e3181bdf8cf) [DOI] [PubMed] [Google Scholar]
- 50.Gibson WE, Gonzalez RS, Cates JMM, Liu E, Shi C.Hepatic micrometastases are associated with poor prognosis in patients with liver metastases from neuroendocrine tumors of the digestive tract. Human Pathology 2018. 79 109–115. ( 10.1016/j.humpath.2018.05.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bodei L, Sundin A, Kidd M, Prasad V, Modlin IM.The status of neuroendocrine tumor imaging: from darkness to light? Neuroendocrinology 2015. 101 1–17. ( 10.1159/000367850) [DOI] [PubMed] [Google Scholar]
- 52.Modlin IM, Kidd M, Malczewska A, Drozdov I, Bodei L, Matar S, Chung KM.The NETest: the clinical utility of multigene blood analysis in the diagnosis and management of neuroendocrine tumors. Endocrinology and Metabolism Clinics of North America 2018. 47 485–504. ( 10.1016/j.ecl.2018.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lawrence B, Kenney B, Svejda B, Schimmack S, Alaimo D, Barbieri A, Jedrych J, Kidd M, Modlin I.Comparison of PCR-based detection of chromogranin A mRNA with traditional histological lymph node staging of small intestinal neuroendocrine neoplasia. BMC Research Notes 2012. 5 318. ( 10.1186/1756-0500-5-318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malczewska A, Witkowska M, Wójcik-Giertuga M, Kuśnierz K, Bocian A, Walter A, Rydel M, Robek A, Pierzchała S, Malczewska Met al. Prospective evaluation of the NETest as a liquid biopsy for gastroenteropancreatic and bronchopulmonary neuroendocrine tumours: an ENETS Centre of Excellence Experience. Neuroendocrinology 2020. ( 10.1159/000508106) [DOI] [PubMed] [Google Scholar]
- 55.Leon A, Torta M, Dittadi R, degli Uberti E, Ambrosio MR, Delle Fave G, De Braud F, Tomassetti P, Gion M, Dogliotti L.Comparison between two methods in the determination of circulating chromogranin A in neuroendocrine tumors (NETs): results of a prospective multicenter observational study. International Journal of Biological Markers 2005. 20 156–168. ( 10.1177/172460080502000303) [DOI] [PubMed] [Google Scholar]
- 56.Hanahan D, Weinberg RA.The hallmarks of cancer. Cell 2000. 100 57–70. ( 10.1016/s0092-8674(00)81683-9) [DOI] [PubMed] [Google Scholar]
- 57.Krop I, Ismaila N, Andre F, Bast RC, Barlow W, Collyar DE, Hammond ME, Kuderer NM, Liu MC, Mennel RGet al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology clinical practice guideline focused update. Journal of Clinical Oncology 2017. 35 2838–2847. ( 10.1200/JCO.2017.2874.0472) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cardoso F, van't Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, Pierga JY, Brain E, Causeret S, DeLorenzi Met al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. New England Journal of Medicine 2016. 375 717–729. ( 10.1056/NEJMoa1602253) [DOI] [PubMed] [Google Scholar]
- 59.Lee W, Huang DS, Han K.Constructing cancer patient-specific and group-specific gene networks with multi-omics data. BMC Medical Genomics 2020. 13 81. ( 10.1186/s12920-020-00736-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Modlin IM, Kidd M, Drozdov IA, Bodei L.The use of deep learning and neural networks in imaging – welcome to the new mathematical milieu of medicine. Neuroendocrinology 2020. 110 322–327. ( 10.1159/000504605) [DOI] [PubMed] [Google Scholar]
- 61.Modlin IM, Drozdov I, Alaimo D, Callahan S, Teixiera N, Bodei L, Kidd M.A multianalyte PCR blood test outperforms single analyte ELISAs (chromogranin A, pancreastatin, neurokinin A) for neuroendocrine tumor detection. Endocrine-Related Cancer 2014. 21 615–628. ( 10.1530/ERC-14-0190) [DOI] [PubMed] [Google Scholar]
- 62.Kidd M, Kitz A, Drozdov IA, Modlin IM.Neuroendocrine tumor omic gene cluster analysis amplifies the prognostic accuracy of the NETest. Neuroendocrinology 2020. ( 10.1159/000508573) [DOI] [PubMed] [Google Scholar]
- 63.Bodei L, Kidd MS, Singh A, van der Zwan WA, Severi S, Drozdov IA, Cwikla JB, Baum RP, Kwekkeboom DJ, Paganelli Get al. PRRT genomic signature in blood for prediction of 177Lu-octreotate efficacy. European Journal of Nuclear Medicine and Molecular Imaging 2018. 45 1155–1169. ( 10.1007/s00259-018-3967-6) [DOI] [PMC free article] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a