Abstract
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related death worldwide. Analogous to the border customs, liver mainly functions as a filter to detoxify chemicals and metabolite administered orally or intravenously. Besides, liver cancer cells overexpress the drug exporters which cause high drug effluxion from liver cancer cells, leading to chemoresistance and a diminished chemotherapeutic effect on liver cancer. Recently, we found that RNA nanoparticles display rubber-like property that can rapidly deliver therapeutics to tumor site efficiently and the rest of the RNA nanoparticle were cleared by renal excretion within half hour after systemic injection. Therefore, we designed a new multivalent RNA nanoparticle harboring three copies of hepatocyte targeting-ligands, one copy of miR122, and 24 copies of Paclitaxel to overcome the drug effluxion and chemoresistance thus, synergistically treat HCC. The hepatocyte targeting ligands introduce tumor specificity to the RNA nanoparticles as they selectively bind and internalize into liver cancer cells. The rubber-like RNA nanoparticles allow for enhanced targeting ability to the HCC tumors. The RNA nanoparticles carrying miR122 and PTX were delivered to the liver cancer cells efficiently due to their rubber-like property to enhance their EPR and transcytosis effect as well as the receptor-mediated endocytosis by hepatocyte targeting-ligands. The miR122 efficiently silenced the drug exporters and the oncogenic protein. The synergistic effect between miR122 and PTX was confirmed by HSA (Highest Single Agent) synergy model. IC50 was determined to be 460 nM. In vivo studies on mice xenografts revealed that the RNA nanoparticle predominantly accumulated in HCC tumor sites and efficiently inhibited the tumor growth after multiple IV injection. This demonstrates the potential of the rubber-like multivalent RNA nanoparticles to conquest the liver cancer, a currently incurable lethal disease.
Keywords: RNA nanotechnology, 6-way junction, Hepatocyte targeting ligands, Paclitaxel, MiRNA 122, Liver cancer therapy
INTRODUCTION
Liver cancer is one of the fastest-growing cancers, causing millions of deaths worldwide, which needs extreme attention.[1–3] Liver plays a vital role in metabolism, toxin clearance, and homeostasis[4]; however, failure in its function can lead to various liver diseases such as hepatitis, cirrhosis, fatty liver, and cancer. Hepatocellular carcinoma (HCC) is a primary liver cancer that arises due to hepatitis chronic viral infections, alcohol, exposure to carcinogens, and genetic diseases.[5, 6] Small-drugs such as Cabozantinib[7], Docetaxel[8], Lenvatinib[9], Nivolumab[10], Regorafenib[11], Sorafenib[12] have been used for the treatment of liver cancer, but these drugs lead to noxious side effects compromising patients quality of life.[13] Moreover, HCC often develops resistance to small-molecule drugs, resulting in the escalation of treatment dosage as well as adverse effects.[14–16] Therefore, various nanoparticle formulations, including peptides [17], lipids[18], gold[19, 20], iron[21] and silicon[22], have been investigated to reduce the side effects by inducing the Enhanced Permeability and Retention (EPR) effect.[23] Efficient therapeutic effects have been achieved in xenograft model[24–34], however, challenges has been encountered when translate these nanoparticles to clinical use. Most of these efforts did not proceed to clinical use due to their unfavorable pharmacokinetic/pharmacodynamic (PK/PD) profiles and resulted toxicity. Therefore, there is an urgent need to develop an effective therapeutic strategy applicable in clinical use.
We previously developed various RNA nanoparticles based on phi29 3-way junction (3WJ) RNA motif which have been used as drug carriers for various cancer types.[35–37] The RNA nanoparticles are thermodynamically and biologically stable with the inclusion of 2’-fluoro modifications.[38–40] The size of RNA nanoparticles ranges from ~8.0 to 20.0 nm leading to favorable PK/PD profiles.[41] Moreover, this size range enables penetration into tumor tissues via the EPR effect and allow for receptor-mediated endocytosis when targeting ligands are included.[42, 43] The RNA nanoparticles are proven to be non-immunogenic with low toxicity.[44] Furthermore, the hydrophilic RNA nanoparticles could significantly improve the solubility of hydrophobic small molecules conjugated to RNA nanoparticles.[45]
Recently the Guo Lab has utilized optical tweezers and in vivo fluorescence imaging technologies to discern the rubber-like property of RNA nanoparticles.[46] This rubber-like property have been used to build RNA nanoparticles with various shapes for drug delivery. RNA nanoarchitecture was stretchable and shrinkable by optical tweezer with multiple repeats while remaining intact. Compared to rigid nanoparticles, RNA nanoparticles display higher tumor-targeting abilities and less accumulation in non-target organs. As for exfiltration, RNA nanoparticles with the size up to 20 nm could pass through renal filtration, of which the upper limitation is 5.5-nm, and while retaining their original structure post excretion. These findings expound upon the two advantages of RNA nanotechnology: 1) The rubber property of RNA nanoparticles enable them to penetrate leaky tumor vasculature and enhance EPR effect, resulting in increased tumor targeting efficiency and therapeutic effect; 2) RNA nanoparticles can be swiftly cleared through the renal filtration process, leading to low accumulation within the body and therefore a limited toxicity. Considering the controllable shape, size, and rubbery property, RNA holds great potential for cancer treatment options with targeted delivery.
In this study, we developed and utilized a 6-way junction (6WJ) scaffold (Fig. 1) that possesses a higher thermodynamic stability and enables the conjugation of multiple copies of drug molecules along with various functional modules without affecting structural stability. The 6WJ nanoparticle was used as a vehicle to deliver one copy of miR122, 24-copies of PTX, and three copies of hepatocyte targeting ligands for HCC treatment. The multispecific strategy is crucial to treat HCC as it often develops drug resistance. Targeted delivery of the miR122 and PTX will enhance therapeutic effect by avoiding chemoresistance induced by a single drug strategy. PTX is a widely used chemotherapeutic drug for various cancers,[47] and its mechanism of action is well studied.[48, 49] Previous studies indicated the poor solubility of hydrophobic PTX contributing to severe side effects. To overcome the pitfall as well as increase the efficacy, PTX drug molecules are conjugated to the hydrophilic 6WJ RNA nanoparticles as prodrugs to improve their solubility and reduce their toxicity. However, using a single drug strategy to treat HCC often leads to chemoresistance and diminished therapeutic efficiency. It is widely reported that many miRNAs play important roles in inhibiting drug resistance and improving therapeutic effect.[50, 51] MiRNAs are small non-coding RNAs that regulate a broad spectrum of genes either by suppressing or inhibiting their target genes, thus regulating cell proliferation, migration and apoptosis.[52, 53] Hence, we anticipated that a combination therapy of a miRNA along with PTX would overcome the problem of drug resistance.[54]
Fig. 1. Design and construction of 6WJ/HTL/PTX/miR122 RNA nanoparticles.
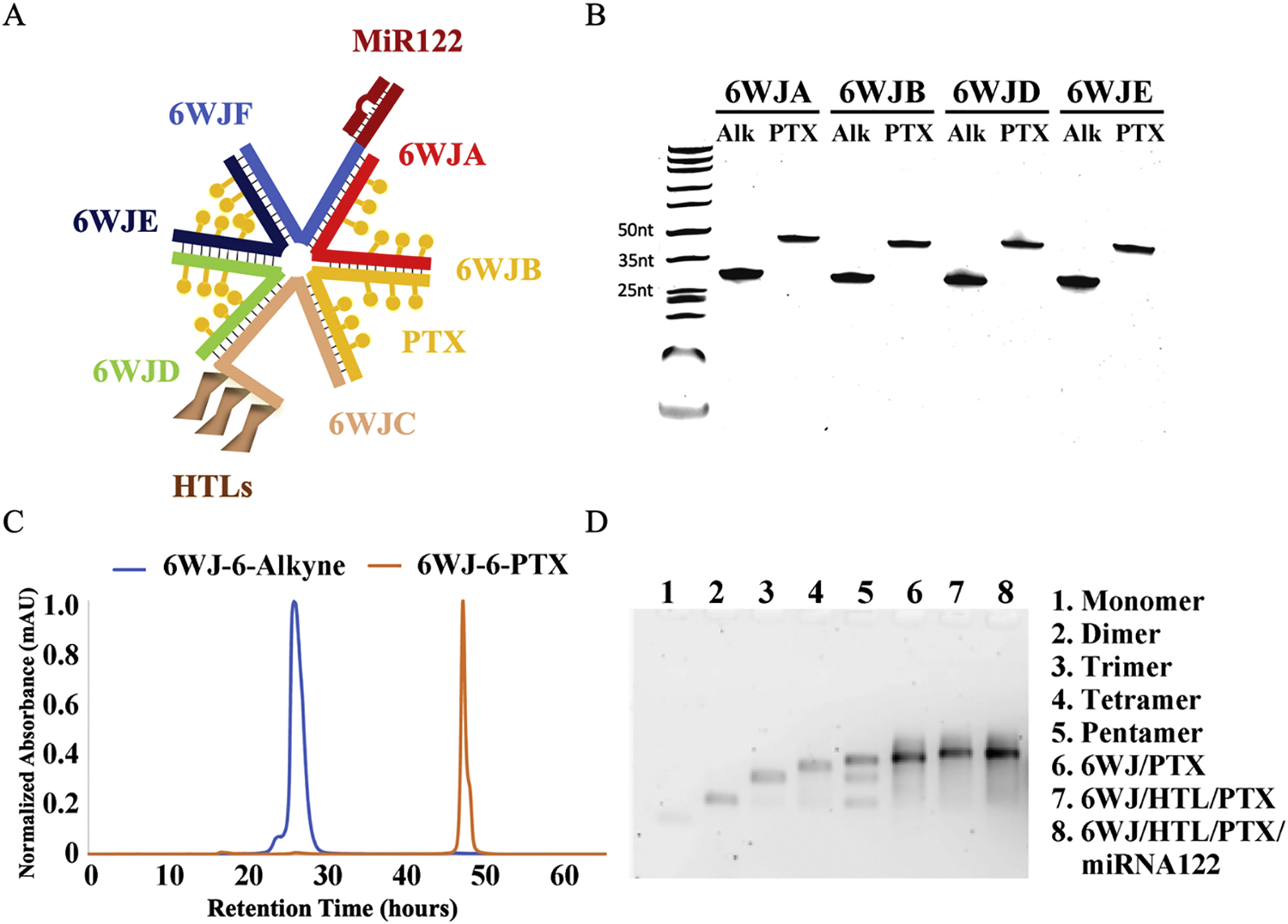
A. Schematic representation of 6WJ/HTL/PTX/miR122 RNA nanoparticle. B. Conjugation of Paclitaxel-azide to RNA-alkyne using copper catalyzed click chemistry and assayed for purity by 16% urea PAGE gel. C. HPLC profiles of 6WJ-6-alkyne and 6WJ-6-PTX RNA strands. D. Self-assembly of 6WJ/HTL, 6WJ/HTL/PTX and 6WJ/HTL/PTX/miR122 RNA nanoparticles, evaluated by 2% agarose gel.
MiR122 is a well-studied noncoding RNA that maintains liver integrity by regulating a set of genes, however, loss of miR122 in HCC compromises liver functions and promotes hepatocarcinogenesis.[55–58] The miR122 not only acts as antitumor drug for HCC by suppressing tumorigenic proteins but also as a drug effluxion inhibitor by down regulating drug efflux proteins such as P-glycoproteins.[50] Nonetheless, the higher therapeutic efficacy of the RNA nanoparticle formulation comes from higher specificity and binding affinity. Therefore, to increase the therapeutic effect and minimize the non-specific toxicity, we conjugated hepatocyte targeting ligand(HTL), composed of three galactosamine derivatives, to the RNA nanoparticle in a series to guide the nanoparticle formulation to the liver tumor site specifically. Hepatocytes uniquely express asialoglycoprotein receptors (ASGP-R) on their cell surface[59] and the galactosamine derivatives with triantennary structure bind to the ASGP-R expressing cells with high affinity.[60, 61] Thus, we hypothesized that the RNA-nanoparticles coupled with the galactosamine can carry the two anticancer drugs (miR122 and PTX) and deliver them to the liver tumor site with high affinity and specificity, leading to the inhibition of liver tumor growth without affecting healthy organs. In vitro studies on HepG2 cells revealed that the RNA nanoparticles selectively bind, internalize, and induce the highest toxicity to the hepatocyte cells as a result of the synergetic effect of the miR122 and PTX. Importantly, the miR122 functions as a suppressor of drug efflux and tumorigenic proteins, revealed from studying the expression levels of model proteins ADAM10 and MDR1 using western blot analysis in HepG2 cells. Particularly, the downregulation of drug efflux proteins helps keeping the PTX drug molecules in the cell cytosol longer and induced the highest cytotoxicity to the HepG2 cells. Biodistribution studies on tumor induced xenograft model revealed the RNA nanoparticles’ specificity to HCC tumor as they accumulated at the tumor site predominantly and minimized their retention in healthy organs. The therapeutic results on mice xenograft models further demonstrated that the RNA nanoparticle formulation exhibited the highest therapeutic efficacy due to their targeted delivery, longer retention times, and the synergistic effect between miR122 and PTX. Thus, in this study, we demonstrate that the RNA nanoparticles harboring combination drugs and targeting ligands selectively homing to the liver tumors and reduce their growth drastically due to their longer retention times and synergistic effects between miR122 and PTX. We anticipate that the developed RNA nanotechnology platform with a targeting ability will find wide use in delivering various drugs an/or imaging markers to various cancer types, enabling new advances in the fields of cancer and biomedical applications.
RESULTS AND DISCUSSION
Design, construction, and characterization of RNA nanoparticles harboring miR-122, PTX, and hepatocyte targeting ligands
The 6WJ RNA nanoparticle was constructed by a modular design and is composed of six component RNA strands. These RNA strands self-assembled into a globular structure with six RNA strands when mixed at an equimolar concentration using an annealing process (Fig. 1A). To improve the thermal stability and confer the RNase resistance, 2’-Fluoro modified pyrimidines were incorporated into the growing RNA strands during the solid phase synthesis. Each 6WJ-A, -B, -D, and -E strand were synthesized with 6-copies of alkyne functionalities for conjugating the prodrug PTX-azide. The PTX-azide was synthesized using an esterification reaction between the hydroxyl of the paclitaxel and the carboxyl of 6-azidohexanoic acid. The PTX-azide was successfully conjugated to the RNA-alkyne strands using Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) click reaction, evident from gel electrophoresis and HPLC (Fig. 1B & 1C). The 6WJ-C RNA strand was conjugated with hepatocyte targeting ligands (HTLs) and 6WJ-F RNA strand was extended with miR122 sequence (Fig. 1A). The multivalent RNA nanoparticle 6WJ/HTL/PTX/miR122 was constructed by mixing the RNA strands harboring miR122, PTX, and HTLs at a stoichiometric ratio using a bottom up self-assembly method. The RNA nanoparticles were assembled with high efficiency as shown in gel shift assay with a stepwise assembly of the complex (Fig. 1D).
The 6WJ/HTL/PTX/miR122 nanoparticles along with control nanoparticles 6WJ/HTL and 6WJ/HTL/PTX were measured for their average hydrodynamic diameter using Dynamic Light Scattering (DLS) and were shown to be 12.83 ± 4.3 nm, 13.33 ± 4.7 nm, and 17.27 ± 5.6 nm in size, respectively (Fig. 2A). The values indicated that the average size of the RNA nanoparticle increased from 6WJ/HTL to 6WJ/HTL/PTX/miR122, which is as expected due to additionally functional groups. The thermal stability of the nanoparticles was determined by melting temperature (Tm) by Temperature Gradient Gel Electrophoresis (TGGE) assay. The Tm values were found to be above 80 °C for the 6WJ/HTL, 6WJ/HTL/PTX nanoparticles and ~70 °C for 6WJ/HTL/PTX/miR122 nanoparticle (Fig. 2B). The thermal stability was further evaluated by annealing temperature (Ta) by thermal cycler at different concentrations. The Ta values were above 80 °C at 2.5 μM for all three nanoparticles while slight reduction in Ta was observed for 6WJ/HTL/PTX/miR122 at 1.0 μM (Ta = ~ 70 °C, Fig. 2C). Both the Tm and Ta values indicated that the RNA nanoparticles were stable enough to remain intact even at ultra-low concentrations. The results demonstrated that the multifunctional RNA nanoparticle with a defined size (17.7 nm), and higher thermodynamic stability could serve as a stable scaffold for targeted delivery of the combination therapeutics for the HCC treatment.
Fig. 2. Characterization of 6WJ/HTL/PTX/miR122 RNA nanoparticles.
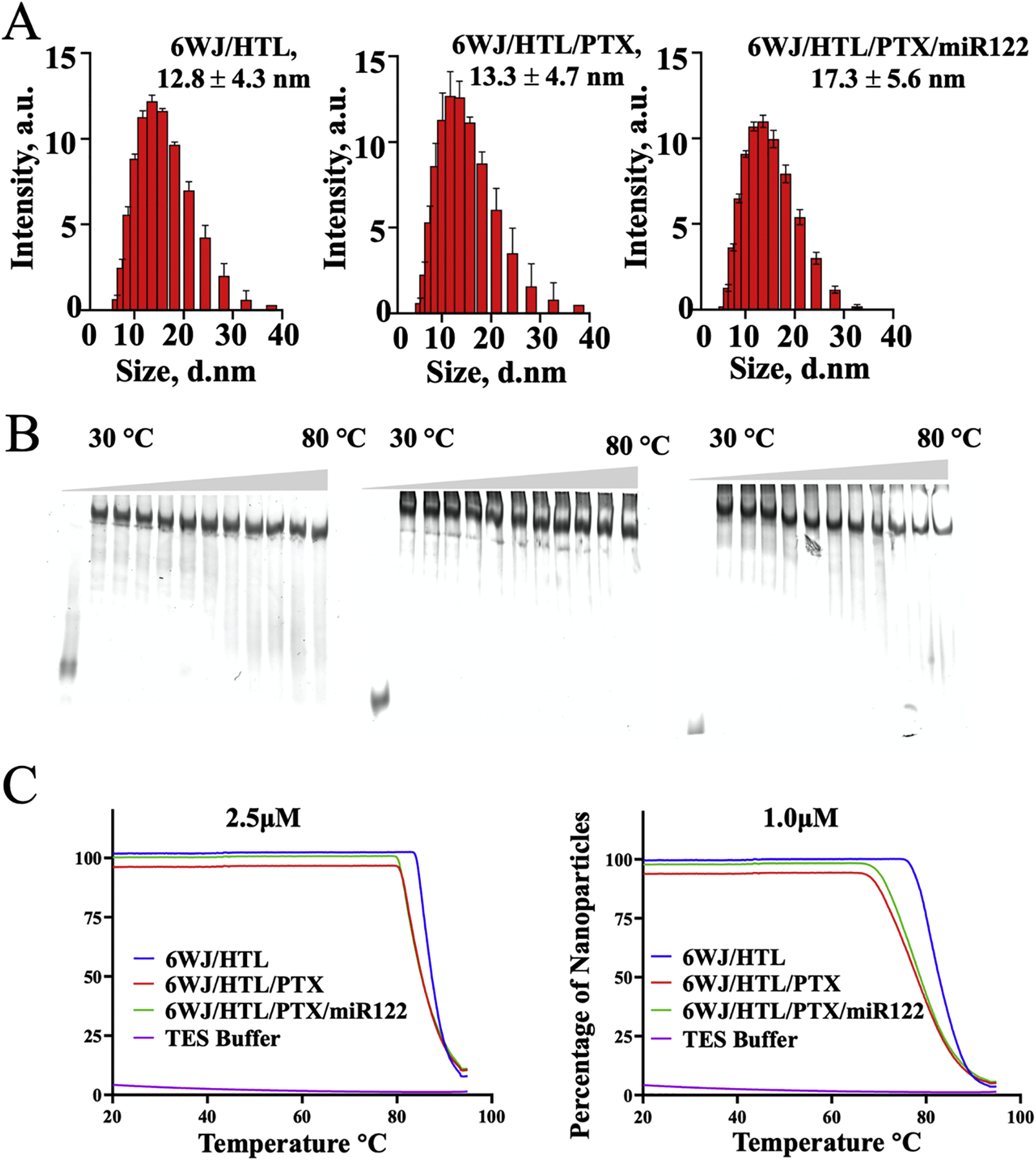
A. Size comparison of 6WJ/HTL, 6WJ/HTL/PTX, and 6WJ/HTL/PTX/miR122 in aqueous solution by DLS (n=3, mean ± SD). B. Thermal stability (Tm) of designed RNA nanoparticles (6WJ/HTL; 6WJ/HTL/PTX; and 6WJ/HTL/PTX/miR122) evaluated by 10% native TGGE (Temperature Gradient Gel Electrophoresis). C. Tm comparison of 6WJ/HTL, 6WJ/HTL/PTX, and 6WJ/HTL/PTX/miR122 by thermal cycler (n=3).
Binding affinity, specificity, and internalization of RNA nanoparticles to ASGP-R expressing HepG2 cells
Targeted delivery of anticancer drugs using nanoparticles not only depends on their ability to induce an EPR effect but also specifically target and internalize into targeted cancer cells. N-acetyl-galactosamine has been widely reported as an ASGP receptor targeting ligand overexpressed on the hepatocyte surface and facilitates cellular internalization.[62, 63] Therefore, we conjugated three galactosamine ligands to the RNA nanoparticles in a series and studied their binding affinity, specificity, and cellular uptake abilities by labeling a near infrared fluorophore (Alexa 647) as a marker onto the 6WJ. The flow cytometry results indicated that the RNA nanoparticles harboring targeting ligands exhibit higher binding affinity compared to nanoparticles with no ligands present (Fig. 3A). The dissociation constant (Kd) for the 6WJ/HTL nanoparticles was found to be around 0.4 μM for ASGP-R expressing HepG2 cells (Fig. 3B). Additionally, the RNA nanoparticles showed very little binding to ASGPR negative KB cells, indicating their specificity for hepatocytes (Fig. 3A). To further demonstrate that the binding affinity is attributed to the ASGP-R and ligand interaction, a binding inhibition assay was performed using a competitive inhibitor, free N-Acetylgalactosamine. The 6WJ/HTL nanoparticles binding for HepG2 cells was drastically inhibited as the free galactosamine concentration increased. In contrast, control experiments did not show any difference as concentration of free galactosamine increases (Fig. 3C), demonstrating the RNA nanoparticles specificity for ASGPR expressing HepG2 cells. These results demonstrated that RNA nanoparticles bind to ASGP-R expressing HepG2 cells with high specificity.
Fig. 3. In vitro binding affinity, specificity, and internalization studies of the 6WJ RNA nanoparticles harboring hepatocyte targeting ligands.
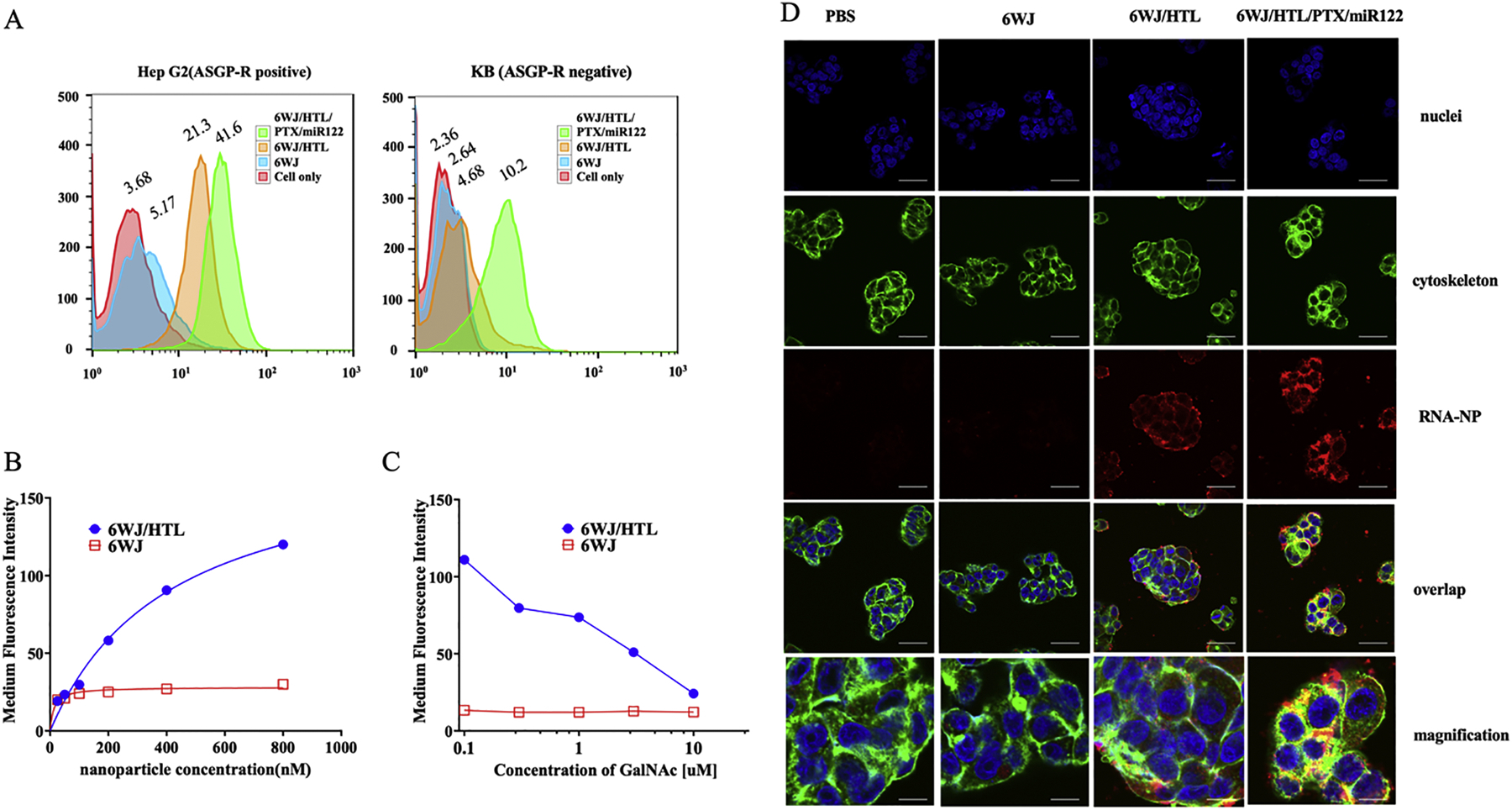
A. Binding assay of 6WJ/HTL & 6WJ/HTL/PTX/miR122 nanoparticles to ASGP-R positive HepG2 cells and ASGP-R negative KB cells as negative control. B. Effect of concentration on binding affinity of 6WJ (red) and 6WJ/HTL nanoparticles (blue) to ASGP-R positive HepG2 cells. C. Competitive assay of free galactosamine with 6WJ/HTL (blue) and 6WJ (red) nanoparticle against ASGP-R positive HepG2, respectively. D. Confocal imaging showing the internalization of 6WJ and 6WJ/HTL/PTX/miR122 nanoparticles into HepG2 cells, nuclei (blue), cytoskeleton (green) and RNA nanoparticles (red), 100 μm for original images, and 20 μm for magnified images.
Cellular internalization is the key to achieve the therapeutic effect of the RNA nanoparticles. Therefore, we further assessed the nanoparticles’ cellular internalization ability using confocal microscopy. The cellular internalization of the RNA nanoparticles is clearly evident by their strong co-localization with cytoplasm that resulted in a yellow color (overlaid image of red color form RNA nanoparticle & green color from cytoplasm, Fig. 3D). Whereas, cells incubated with control RNA nanoparticles (without targeting ligand) showe less nonspecific uptake. Overall, the results demonstrated that the RNA nanoparticles harboring hepatocyte targeting ligands bound strongly to ASGP-R expressing HepG2 cells with high specificity that favored their strong cell uptake efficiency through ASGP-Rs.
Downregulation of the oncogenic protein expression by RNA nanoparticles harboring miRNA122
The liver-specific miR122 regulates a set of genes and maintains liver integrity, however, miR122 is frequently suppressed in HCC leading to its proliferation and drug resistance. Therefore, the miR122 was delivered to HepG2 cells to suppress the tumorigenic proteins using the RNA nanoparticles. Downregulation of the oncogenic a disintegrin and metalloproteinase domain-containing protein 10 (ADAM10) expression was taken as a parameter to study the function of miR122 in HepG2 cells. The HepG2 cells were incubated with RNA nanoparticles harboring miR122 and control nanoparticles, then the cells were processed and measured for the expression of ADAM10 mRNA and protein expression using the real-time PCR and Western blot, respectively. The treatment groups with nanoparticles harboring miR122 showed significant suppression of the ADAM10 gene (Fig. 4A) as well as protein expression (~2–3 fold) whereas the cells treated with control nanoparticles did not show inhibited protein expression (Fig. 4B&C). The downregulation of the ADAM10 gene and protein expressions is a clear indication of the RNA nanoparticles ability to deliver the miR122 to hepatocyte cells and exercise its function in gene regulation.
Fig. 4. In vitro assay for the silencing of the oncogenic protein ADAM10 and the drug transporter MDR1 (P-gp) by 6WJ/HTL/miR122 nanoparticles.
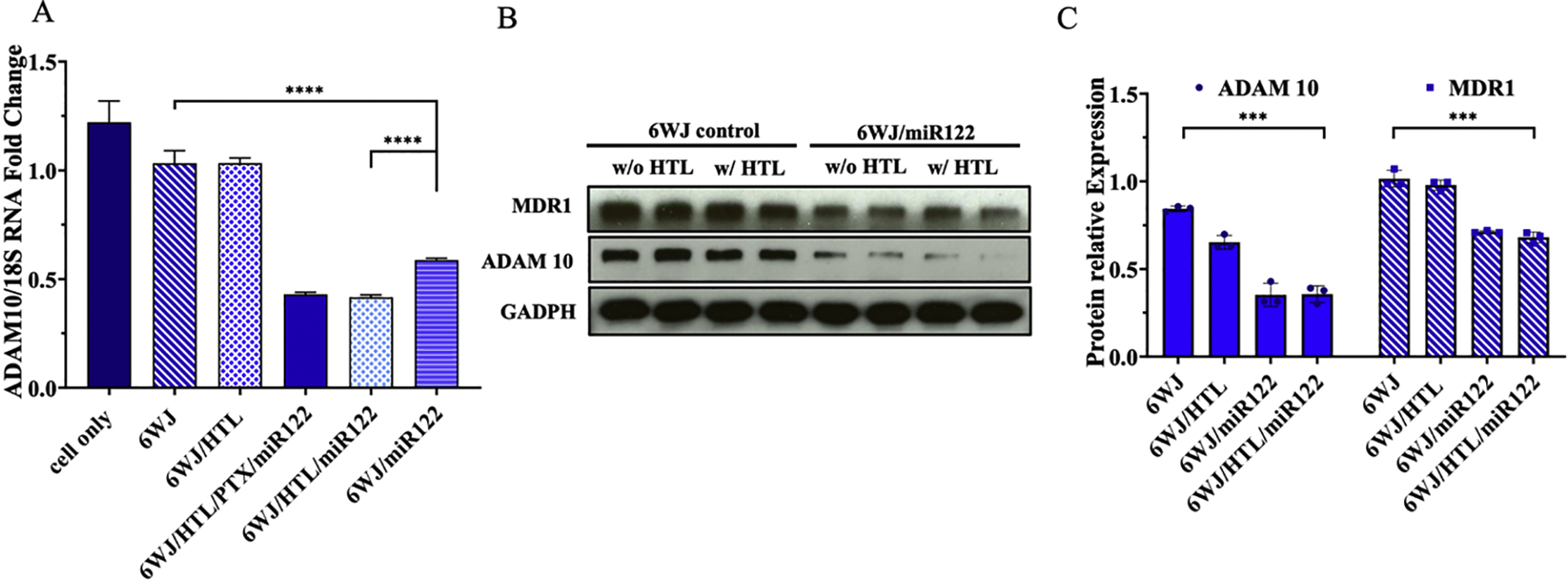
A. qRT-PCR showing effect of miR122 knock-down on ADAM 10 gene expression. B. Western blot showing the knock-down of ADAM10 and MDR1 proteins expression. C. Quantification of ADAM 10 & MDR1 proteins expression from western blot. (n = 3 biologically independent animals, statistics was calculated by two-tailed unpaired t-test presented as mean ± SD, *p < 0.05, **p < 0.01, ****p < 0.0001).
Downregulation of drug efflux transporters expression by RNA nanoparticles harboring miRNA122
The miR122 is also known to sensitize HCC tumor cells to chemical drugs by modulating multi-drug resistance (MDR) related genes that are associated with drug transporters such as multi-drug resistance protein 1 (MDR1). MDR1 is a drug efflux transporter also referred as P-glycoprotein (P-gp) that expels drugs from the cell cytosol. Western blot results indicated a significant reduction in MDR1 protein expression (~50%) in treatment groups compared to control groups (Fig. 4B&C). These results further demonstrated that the highest liver cancer cell inhibition of the RNA nanoparticles was not only a result of miR122 suppression of tumorigenic proteins such as ADAM10, but also a low effluxion of PTX drug from cell cytosol due to the down regulation of drug transporter MDR1.
Paclitaxel release from RNA nanoparticles
PTX conjugation to RNA as a prodrug was designed to improve its solubility and reduce its toxicity. Successful release of PTX from the RNA nanoparticle is vital to realize its’ antitumor effects. Therefore, we first studied the PTX release from PTX-RNA strands using 50% Fetal bovine serum (FBS), which is a well-known source of esterase enzyme, which cleaves ester bonds. The RNA-PTX strand was incubated with 50% FBS and the PTX was completely released from the RNA after 24 hrs of post incubation (Fig. 5A). The results indicated that the PTX was successfully released from the RNA strand due to ester bond cleavage by esterase. In addition, the release of PTX from 6WJ RNA nanoparticles was investigated through incubation with 50% FBS. The PTX released from the RNA nanoparticles was further assessed using LC/MS analysis. The results show that majority of the PTX remains intact with RNA nanoparticles and only ~20% of PTX releases even after 24 hrs of incubation (Fig. 5B). The difference of PTX release from single stranded RNA and RNA nanoparticles may be attributed to the steric hindrance of the compact RNA nanoparticles to esterase enzymes. The results indicated that the PTX remain conjugated to RNA nanoparticles during systemic circulation, however, the PTX molecules might release from the RNA nanoparticles once they are metabolized into single stranded RNAs, which was also evident from both in vitro and in vivo cancer inhibition results.
Fig. 5. Quantification of PTX release from RNA nanoparticles.
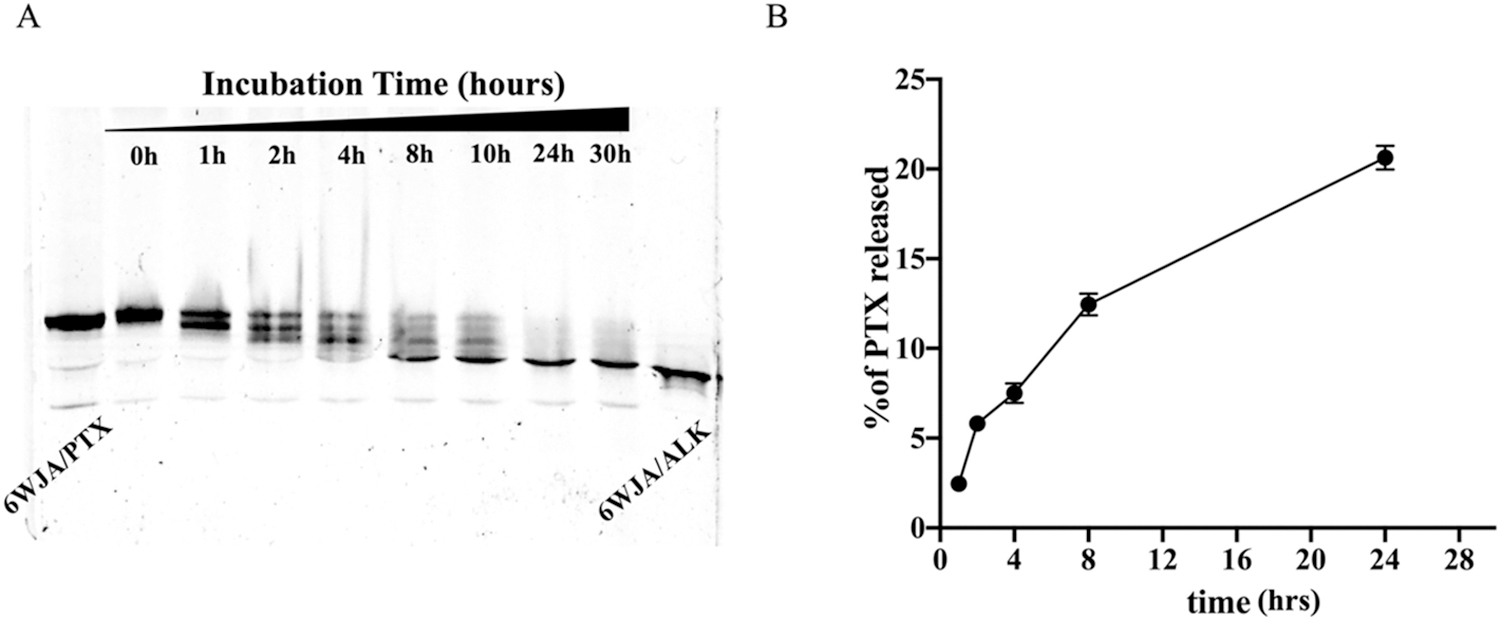
A. PTX release from single stranded RNA at 50% fetal bovine serum characterized by 16% urea PAGE gel. B. LC/MS quantification of PTX release from assembled 6WJ/HTL/PTX/miR122 nanoparticles at 50% fetal bovine serum.
Evaluating synergistic cytotoxic effect of the RNA nanoparticles harboring miR122 and PTX in vitro
The central theme of this novel study is that the targeted delivery of PTX and miR122 combination therapy by RNA nanoparticles results in enhanced toxicity to HCC cells due to their synergistic cytotoxic effect. To study the synergy between miR122 and PTX in cytotoxicity induction, an MTT cell viability assay was first performed. The RNA nanoparticles carrying miR122 and PTX showed the highest cell toxicity compared to the control groups harboring single treatment molecules (either PTX or miR122). Whereas 6WJ nanoparticles without treatment groups did not show any cancer cell inhibition effects, indicating the safety of RNA nanoparticles (Fig. 6A). A Caspase-3 activity assay was also used to further demonstrate the synergy between miR122 and PTX. The results showed that the treatment group with nanoparticles harboring both miR122 and PTX exhibited the highest apoptotic cell population compared to control groups using single treatments (Fig. 6B). Cell viability was used as an indicator to quantify the correlation of synergy between miR122 and PTX using Highest Single Agent (HSA) synergy modeling. The data was processed and quantified using Combenefit software to obtain the HSA synergy score between miR122 and PTX (Fig. 6C). In the matrix, the blue color at the middle of each drug concentration indicated the existing synergistic effect. The number represented the co-efficiency of synergy. The highest synergy between miR122 and PTX in cytotoxic induction was observed at 0.8 μM for miR122 and 3.2 μM for PTX. IC50 was determined to be around 460 nM.
Fig. 6. In vitro assay for cancer cell inhibition and synergistic effect of 6WJ/HTL/miR122 and 6WJ/HTL/PTX nanoparticles.
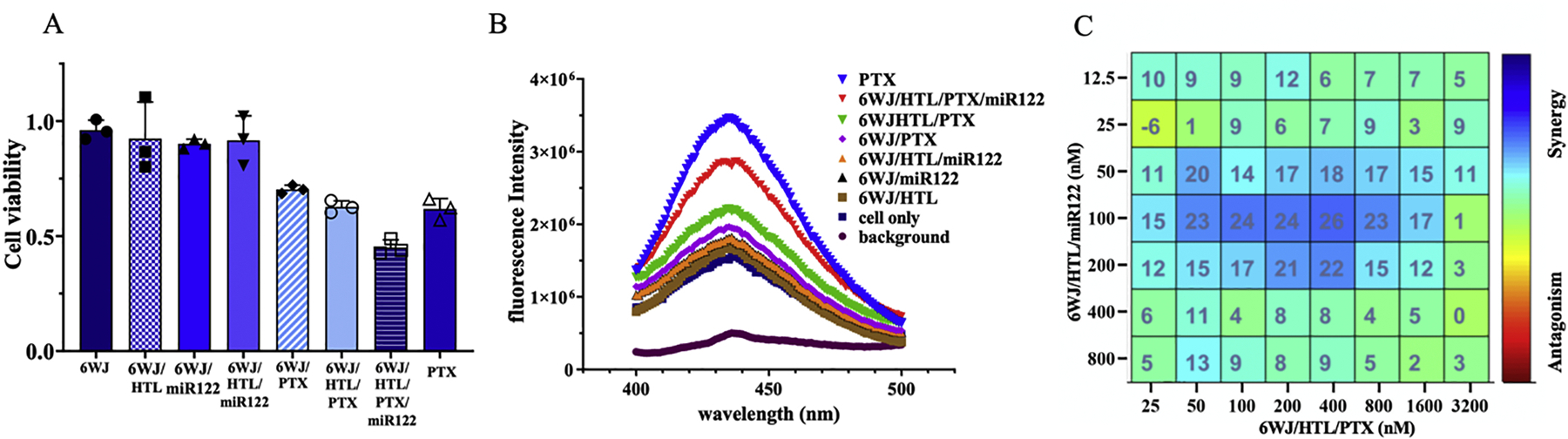
A. MTT assay (n=3; mean ± SD). B. Caspase-3 assay. C. Synergetic cytotoxic effect between 6WJ/HTL/miR122 and 6WJ/HTL/PTX nanoparticles were assayed using HSA synergy model.
In vivo biodistribution assay to assess tumor targeting effect and organ biodistribution of RNA nanoparticles
The Asialoglycoprotein receptor has been widely used as a liver-specific target for drug delivery in various liver diseases, which is also proven effective in clinical trial. To evaluate the RNA nanoparticles’ capabilities for tumor targeting in vivo, the HTL conjugated RNA nanoparticles were administered to HepG2 tumor bearing xenografts through IV injection. The mice were euthanized 8 hrs post-injection and organs were collected and imaged. The results showed that most of the HTL conjugated nanoparticles strongly accumulated in tumor, however, a slight accumulation in liver and kidney was also observed as the organs mainly function as drug filters. Mass normalized quantitative analysis of the organ images further indicated the higher tumor uptake ratio of 6WJ/HTL/PTX/miR122 compared to the negative control 6WJ (Fig. 7A). The biodistribution results proved that the hepatocyte targeting ligands could guide the RNA nanoparticles to tumor site and benefit the targeted drug delivery.
Fig. 7. Animal trials for liver cancer inhibition by 6WJ/HTL/PTX/miR122 nanoparticles.
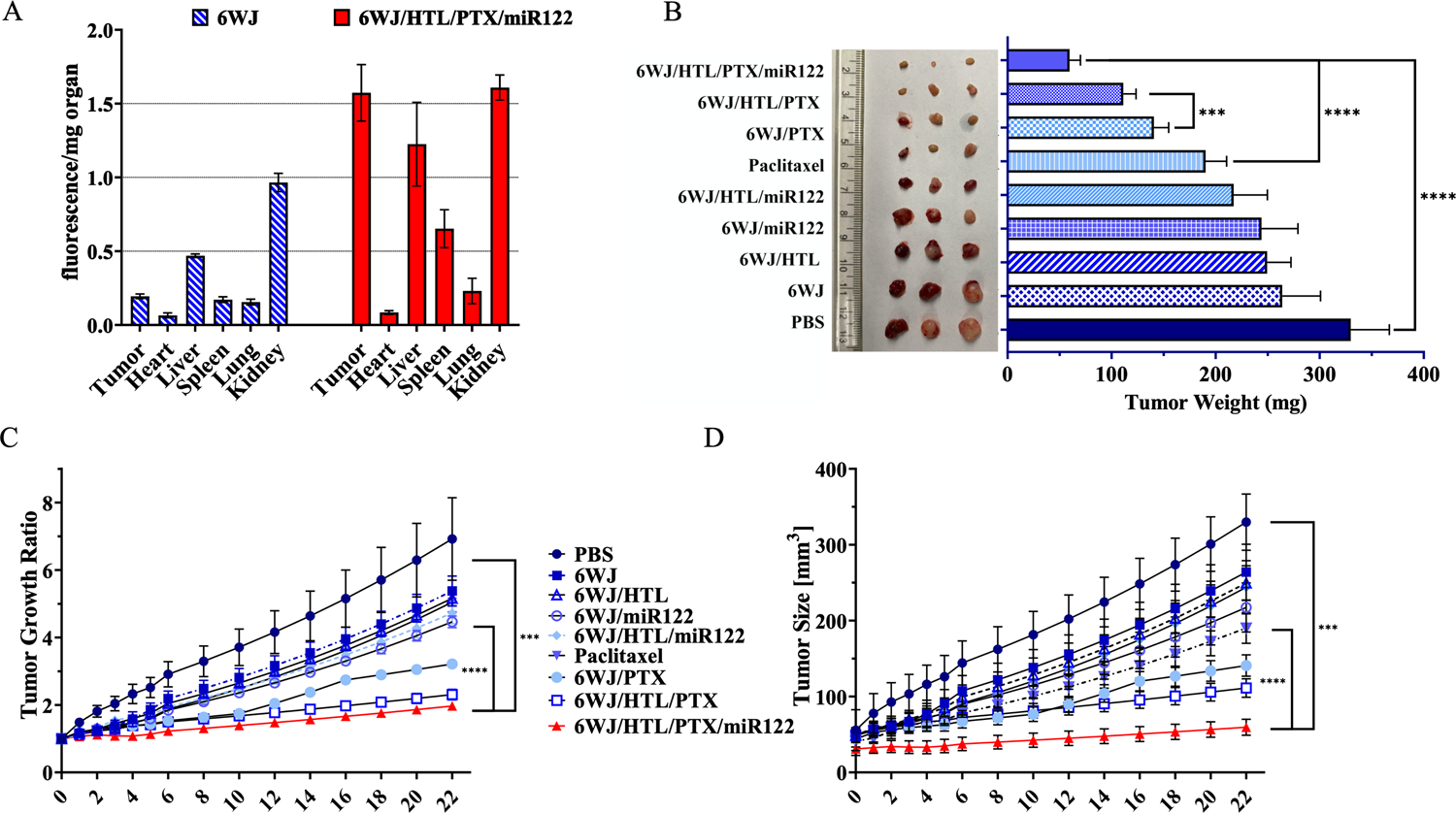
A. Quantitative analysis of RNA nanoparticle biodistribution in tumors and normal organs. B. Images of liver cancer tumors harvested from mice after treatments. C. D. Intravenous treatment of nude mice bearing Hep G2 xenografts with 6WJ/HTL/PTX/miR122 and control groups treated every three days for a total of 7 injections. Tumor size was monitored during the time course of treatments (n = 5 biologically independent animals, statistics was calculated by two-tailed unpaired t-test presented as mean ± SD, *p < 0.05, **p < 0.01, ****p < 0.0001).
Therapeutic efficacy of RNA nanoparticles carrying miR122 and PTX in animal trials.
After confirming the role of miR122 on the synergistic effect in combination with PTX in vitro, the therapeutic effect of the RNA nanoparticles was evaluated using HCC tumor bearing mice xenograft model. The RNA nanoparticles alongside controls were intravenously injected into mice at the clinically used dosage equivalent to 5 mg/kg (PTX dose per mouse weight) every 3 days for a total of 6 doses. Among all treatment groups, the mice administered with RNA nanoparticles harboring the combination therapy along with targeting ligands exhibited the highest tumor growth inhibition compared to other treatment groups carrying either PTX or miR122 with and without targeting ligands, which was evident from both tumor growth inhibition curves and reduced tumor weights (Fig. 7B–D). Additionally, the RNA nanoparticles harboring targeting ligands showed better therapeutic results than without ligand groups, demonstrating the tumor targeting specificity. Importantly, clinically used PTX formulation (PTX mixed with Cremophor/EtOH) exhibited only a marginal tumor inhibitory effect. Together, the tumor inhibition results suggested that the highest therapeutic effect of the RNA nanoparticles attributed to i) EPR effect; ii) targeted drug delivery; iii) low drug effluxion from cytosol due to the down regulation of drug transporters by miR122; iv) synergistic effect between miR122 and PTX.
Induction of pro-inflammatory factors by RNA nanoparticles
Safety is another important concern for translational medicine. The immunogenicity of RNA nanoparticles has been proved to be tunable to produce either a minimal immune response as safe therapeutic vectors, or a strong immune activation for cancer immunotherapy with the incorporation of specific immune stimulation sequences.[44, 64, 65] Here, we further studied the in vivo pro-inflammatory response after systemic injection of the nanoparticle formulation into the mice. We evaluated the production of four pro-inflammatory cytokines, including Tumor necrosis factor-α (TNF-α), Interleukin 6 (IL6), Interferon-gamma (IFNγ), and Cytokine IL-12, upon RNA nanoparticle treatment in an immune competent mice model. ELISA assay indicated that intravenous (IV) injection of RNA nanoparticles at the dose of 5 mg/kg induced undetectable or negligible cytokines production (Fig. 8). In contrast, PTX formulated in Cremophor EL/EtOH induced elevated production of these immune response indicators. The in vivo pro-inflammatory results demonstrated the safety of the developed RNA nanoparticles.
Fig. 8. In vivo immune-toxicity assay of 6WJ/HTL/PTX/miR122 nanoparticles.
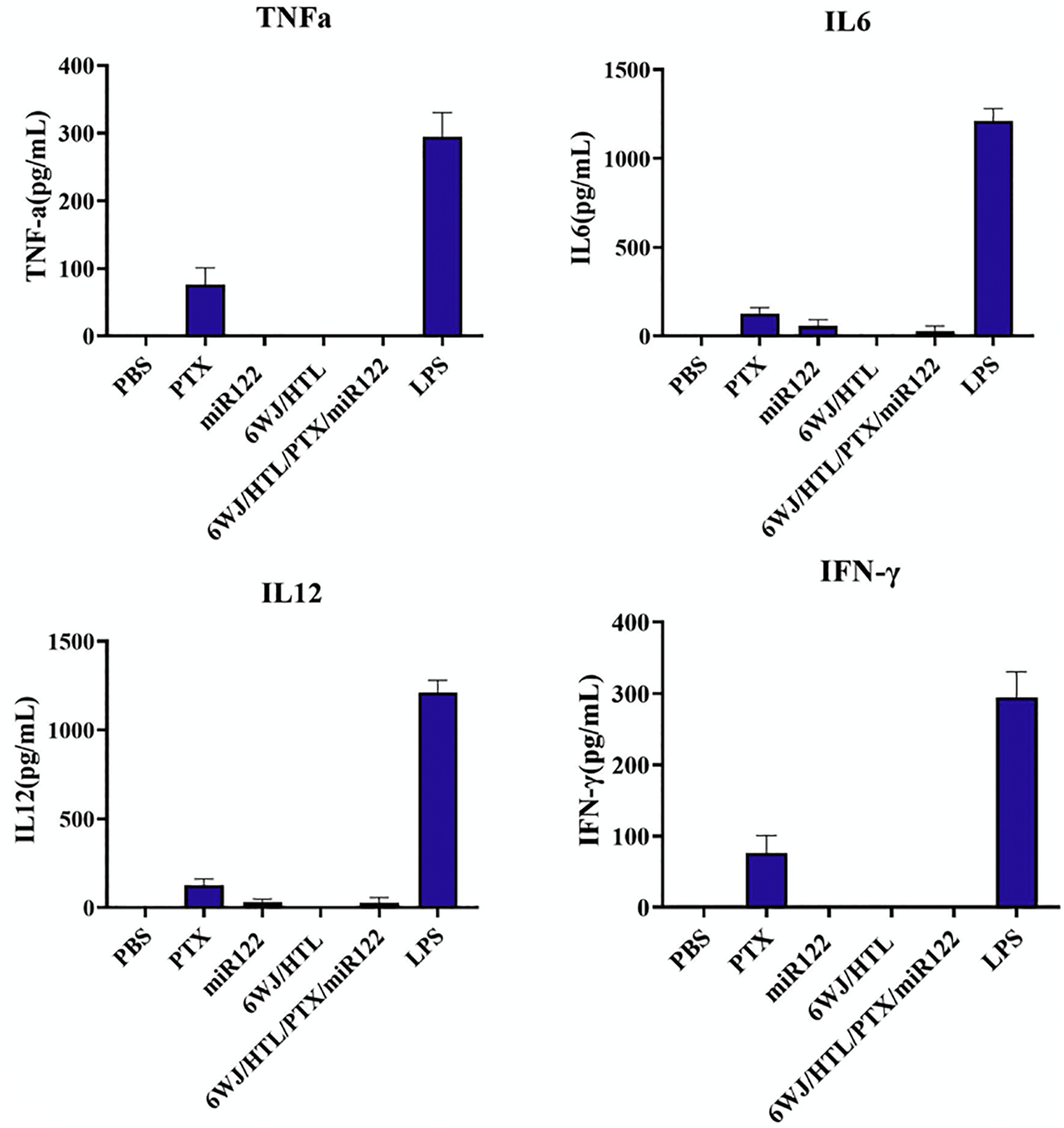
Evaluation of TNF-α, IL-6, IL-12, and IFN-γ secretion in mice after systemic injection of 6WJ/HTL/PTX/miR122 nanoparticles, evaluated by ELISA (n=3; mean ± SD).
DISSCUSION
Liver tumor targeting RNA nanoparticles were constructed for the co-delivery of miR122 and PTX which increased the therapeutic efficacy against liver cancer through both passive EPR effect and active tumor targeting by tumor specific ligands. Hydrophobic PTX was conjugated to RNA as pro-drugs with improved solubility, lower toxicity, and higher therapeutic efficiency. Exogeneous delivery of miR122 to HCC successfully downregulates tumorigenic proteins as well as inhibits tumor migration, proliferation and metastasis. More importantly, miR122 could suppress the drug efflux transporters expression, such as P-glycoproteins, leading to the synergetic effect with PTX by overcoming the HCC drug resistance to PTX as well as sensitizing the HepG2 cells to PTX treatment. The in vivo results reveal the high tumor targeting specificity of the RNA nanoparticle formulation as well as the strong tumor growth inhibition efficiency, arising from the synergistic effect between miR122 and PTX, which is superior than commercially used PTX formulation (Cremphol/EtOH). Overall, these findings demonstrate that the multivalent targeted drug delivery using 6WJ motif has a high potential to treat liver cancer effectively.
Although tumor targeted delivery can increase the therapeutic efficacy, there are some concerns that nanoparticles may also deliver the anti-cancer drugs to non-target organs and tissues, resulting in undesirable side effects and toxicity. The multivalent RNA nanoparticle conjugated with miR122, PTX, and HTLs show undetectable immunogenicity and toxicity in mice xenograft. This safety profile is attributed to the unique properties of RNA nanotechnology: 1) Optimal size and elasticity of RNA nanoparticles; Rubber-like deformation property of RNA nanoparticles allows for squeezing through tumor vasculature to improve the EPR effect. RNA structures with the size of 10–20 nm avoid reticuloendothelial system (RES) clearance in the liver, spleen, lung, and bone marrow, which induce non-specific uptake by innate immune cells such as macrophages. The unique size and elasticity of RNA carriers facilitate passing through direct glomerular filtration where larger molecules cannot be eliminated. The rapid excretion of RNA nanoparticles by direct kidney filtration thus avoiding retention in healthy organs and tissues. 2) Increased solubility of hydrophobic drugs conjugated to RNA prevents the nonspecific binding and accumulation into normal cells and eventual toxicity. Solubilizing the drugs also enhances the drug bioavailability and reduces the injection dose, further lowering the potential side effects. 3) High thermodynamic stability and chemical stability of RNA nanoparticles keep RNA structures intact in vivo, avoiding nonspecific release of toxic drugs. MiRNAs conjugated to RNA nanoparticles facilitate high stability, thus avoiding enzyme degradation and immunotoxicity.
Combining these advantages, we anticipate that the developed RNA nanotechnology platform with targeting ability will find extensive use in delivering various drugs/imaging markers to various cancer types as a safe delivery platform that enable new advances in the fields of cancer and biomedical applications.
CONCLUSIONS
The newly developed 6WJ RNA nanoparticles with defined size allows us to conjugate three liver targeting ligands, one copy of miR122 and 24-copies of PTX drug. The miR122 downregulated the tumor oncogenic factor and the drug efflux transporter, which in turn, inhibited the expulsion of the delivered drugs and sensitized tumor cells for PTX. The increased therapeutic efficacy in mice xenograft model is a combined effect of the multivalent RNA nanoparticles. The rubber-like property of the multivalent RNA nanoparticles enhances their tumor vascularization. The specific tumor cell targeting and entry effect is attributed to the three copies of the liver cell targeting ligands. The gene silencing and tumor suppression efficiency are attributed to the miR122 that silence the P-glycoproteins, which is a drug efflux transporter that leads to HCC resistance for drugs such as PTX. In vivo studies on mice xenografts revealed that the RNA nanoparticles predominantly accumulate in HCC tumor and efficiently inhibited the tumor growth after 22 days of intravenous (IV) administration. Most importantly, the nanoparticles are safe to use because of their tumor specificity, non-immunogenicity, fast renal clearance, undetectable liver accumulation, and fast tumor homing. Thus, the multivalent 6WJ nanoparticles are rapidly becoming a promising therapeutic for the treatment of the currently incurable liver cancer.
METHODS AND EXPERIMENTAL
Design and construction of 6WJ RNA nanoparticles
Multifunctional RNA nanoparticles were constructed using a bottom-up self-assembly approach. The 6WJ/HTL/PTX/miR122 contains seven RNA fragments (Fig. 2A) harboring ASGP-R targeting ligands, 24 copies of PTX and miR122. The controls include RNA nanoparticles without a targeting ligand (denoted as 6WJ/PTX/miR122); without a therapeutic module (denoted as 6WJ/HTL), or without therapeutic and targeting modules (denoted as 6WJ).
RNA strands were synthesized chemically using solid-phase synthesis. 2’-Fluoro (2’-F) modified cytosine (2’F-C) and uracil (2’F-U) nucleotides are incorporated in the RNA sequences to improve the RNA nanoparticle’s thermal stability and nuclease resistance. RNA sequences can be found in Supplementary information.
6WJ nanoparticle assembly
The 6WJ nanoparticles are synthesized by a bottom-up self-assembly of the single-stranded RNA fragments. The RNA strands were mixed at equal molar concentrations in an annealing Buffer (1X PBS), and heated to 95 °C for 5 minutes, then slowly cooled to 4 °C over 45 minutes. Self-assembly of the RNA nanoparticles was verified on a 2% agarose gel running in 1X TAE (89 mM Tris-Acetate, 2 mM EDTA) buffer and imaged using Typhoon FLA 7000.
Melting profile determination using TGGE method
The self-assembled nanoparticles were loaded on to 10% native PAGE gel and ran for 15 mins at 140 V prior to TGGE in 1X TBE buffer (100mM Tris-Borate, 1 mM EDTA). After running the RNA nanoparticle into the gel matrix, the gel was subjected to TGGE by increasing the temperature in a gradient manner perpendicular to the electrophoretic force from 30–80 °C. The gel run in 1X TBS at 100 V for 45 min then, imaged using Typhoon FLA 7000.
Annealing profile determined using thermal cycler
Pre-assembled RNA nanoparticles were added to a 96-well plate with a final concentration of 2.5 μM or 1 μM, respectively. One of the strands of the 6WJ nanoparticle was labeled with Alexa 647 prior to the assembly. Then, the RNA nanoparticles were mixed with SYBR Green II (as a reporter dye at a final concentration of 20X) at 2.5 μM and 1.0 μM concentrations. All samples were independently completed in triplicate. The RNA nanoparticle samples were heated to 95 °C for 5 min then slowly cooled to 20 °C at a rate of 0.11 °C/s using a Roche Lightcycler 480 machine. The RNA nanoparticle formation was monitored by measuring fluorescence levels at 480.0 nm excitation and plotted against temperature[66] and the Ta values were determined as the temperatures at which 50% of maximum SYBR green II fluorescence was detected.
Size measurement using DLS
The apparent hydrodynamic sizes for the assembled RNA nanoparticles (6WJ, 6WJ/HTL/PTX, and 6WJ/HTL/PTX/miR122) were measured at 25 °C by Zetasizer nano-ZS machine. The data was obtained from three independent measurements.
PTX release assay
The PTX release profile from the 6WJ/PTX/HTL/miR122 nanoparticle was studied by incubating them in 50% Fetal Bovine Serum (FBS) at 37 °C at a final concentration of 2 µM. The single strand PTX-RNA was incubated in FBS at different time intervals (0, 0.5, 1, 2, 4, 8, 12, 24, and 30 hrs), then, 10 µL of each sample was collected and subjected for a gel shift assay. The samples were ran on a 16% urea PAGE gel in 1X TBE at 160 V for 100 minutes and then imaged by Typhoon FLA 7000. In the study of PTX release from assembled RNA nanoparticles, the RNA nanoparticles were incubated in FBS at different time intervals (0, 0.5, 1, 2, 4, 8, 12 hrs). Then, 50 µL of each sample was collected and the released free PTX was extracted with 200 µl MTBE. After vortex, 200 µl MTBE was collected and dried by a speed vacuum. The PTX was resuspend in MeOH and subject to LC/MS.
Cell culture
Human hepatocellular carcinoma cells (HepG2) were obtained from the ATCC. Cells were grown and cultured in DMEM medium (ThermoFisher Scientific) containing 15% (v/v) Fetal Bovine Serum (FBS) in humidified air environment containing 5% CO2.
Confocal microscopy imaging
HepG2 cells were seeded on glass coverslips and cultured at 37 °C incubator overnight. Cells were treated with RNA nanoparticles conjugated with an Alexa 647 marker at 100 nM final concentration for 4 hrs at 37 °C. After incubation, cells were washed twice with cold PBS buffer, then fixed with 4% formaldehyde. Cells were then treated with 0.1% Triton X-100 (Sigma-Aldrich) in PBS buffer for 5 min then treated with cytoskeleton staining dye Alexa Fluor 488 phalloidin (ThermoFisher Scientific) for 30 min at room temperature. After rinsing with PBS buffer, the cells were stained by DAPI for cell nucleus staining and mounted with ProLong@ Gold Antifade Reagent (Life Technologies Corp., Carlsbad, CA. The slides were assayed on Olympus FV3000 confocal microscope (Olympus Corporation, Tokyo, Japan).
In vitro cytotoxicity assay
A CellTiter 96 Non-Radioactive Cell Proliferation Assay (Promega) was used for cell cytotoxicity study, following manufacturer instructions. Briefly, 5 × 103 HepG2 cells were seeded on a 96-well plate overnight. Triplicate wells were treated with RNA nanoparticles and free PTX were added at 400 nM. After incubation at 37 °C for 48 hrs in a 5% humidified CO2 environment, 15 μl of MTT Dye was aliquot to each well. The cells were incubated at 37 °C for 4 hrs following 50 μL of Solubilization Solution/Stop Mix was added to each well. The plate was incubated in the dark overnight on a shaker at room temperature to a uniformly colored solution and subjected to absorbance measurement at 570 nm by Synergy 4 microplate reader (Bio-Tek).
In vitro apoptosis assay
Caspase 3 Apoptosis Detection Kit (BD Pharmingen) was used for cell apoptosis following manufacturer instructions. HepG2 cells were seeded in a 24-well plate overnight and treated with 400 nM of the 6WJ/HTL/PTX/miR122 nanoparticles and its controls for another 24 hrs at 37 °C. Then, the cells were lysed using the cold cell lysis buffer, and 25 μL cell lysate of each treatment group was transferred to 0.6ml tube, 2 μL of reconstituted Ac-DEVD-AMC diluted in 80 μL of HEPES buffer was added to each tube and incubated at 37 °C for 1 hr. The fluorescent intensity of caspase-3-AMC substrate was measured by fluorometer at 400–500 nm window with an excitation of 380 nm.
HSA synergy modeling
Cell viability was studied following MTT protocol in cytotoxicity assay. The result was presented by Prism 8.0 (Graph Pad). The viability result was entered into HSA synergy modeling by Combenefit and plotted by Prism 8.0.
qRT-PCR assay to study downstream gene expression
qRT-PCR Assay was performed following the procedure (Life Technologies) to study the ADAM10 mRNA expression. HepG2 cells were cultured with treatment groups at 400 nM final concentration for 48 hrs at 37 °C before the total RNA was extracted by TRIzol reagent (Life Technologies). Next, the cDNA strand was reverse transcript from total RNA (1 μg) using SuperScript III First-Strand Synthesis System (Life Technologies). qRT-PCR was performed in a final 20 μL volume using Taqman Universal PCR Master mix, primers and probe, and cDNA. The primer and probe for human ADAM10 and GADPH (housekeeping gene) were purchased from Life Technologies. PCR was performed on StepOnePlus systems (Applied Biosystem). The data were analyzed by the ΔΔCT method.
Western blot assay to study downstream gene expression
Western blot assay was used to investigate the ADAM10 and MDR1 protein expression. The HepG2 cells were cultured on a 24-well plate overnight at 37 °C. Then, cells were treated with 400 nM of the RNA nanoparticles following incubation for 72 hrs, and were lysed in RIPA buffer with protease inhibitor. Total protein concentration was quantified by BCA Protein Assay Kit. A total of 10 μg of protein was loaded into a 10% SDS PAGE. The gel was transferred to polyvinylidene fluoride membrane and followed by blocking in 5% fat-free milk on shaker at room temperature for 2 hrs. The membrane was then stained with primary antibody (rabbit-ADAM 10: 1:1000; mouse-GADPH: 1:10 000) at 4 °C overnight and washed with TBST buffer three times for 5 minutes each time. The membrane was then stained with secondary antibody (goat pAb to rabbit IgG and goat pAb to mouse IgG: 1:10 000) at room temperature for 1 hour and washed with TBST buffer three times for a total of 15 min. Membranes were then incubated with ECL substrate, exposed to Amersham HyperfilmTM together, and processed with a Series 2000A Processor film developer.
Subcutaneous tumor xenograft animal model
All animal procedures were housed and operation was performed in accordance with the subcommittee on Research Animal Care of The Ohio State University guidelines approved by the Institutional Review Board. To generate the Hepatocellular carcinoma xenograft model, female athymic nu/nu mice, 3–4 weeks old purchased from Charles River Laboratories were subcutaneously injected 2 × 106 HepG2 cells/site resuspended in sterile PBS into the mammary fat pads of nude mice.
In vivo biodistribution study
Alexa 647 labeled 6WJ nanoparticles (100 µl, 20 µM) were administered by IV injection into HepG2 tumor bearing mouse xenograft. PBS treatment was used as background control. The mice were euthanized 8 hrs post-injection by the inhalation of CO2 followed by cervical dislocation, and major organs (heart, liver, spleen, lungs, kidneys, tumor) were harvested and subjected to imaging by IVIS (XMRS) with excitation at 640 nm and emission at 680 nm. Fluorescence imaging was analyzed and quantified by Living Imaging (Perkin Elmer).
In vivo tumor inhibition by RNA nanoparticles
HepG2 tumor bearing mouse xenograft were randomly divided into nine groups (n=5 each group). Samples were administrated by IV injection in a total of 6 doses (5 mg/kg, PTX/body weight) every 3 days for 21 days. Tumor volume was monitored every day by caliper, calculated as (length × width2)/2, and mice weight was monitored every day. On day 21, the mice were sacrificed, and tumors were extracted. Data were statistically analyzed by unpaired t-test and presented as mean ± SD; ***p < 0.005, ****p < 0.0005.
In vivo cytokines induction evaluation
CD-1 mice (4–5 weeks old) were purchased from Charles River Laboratories. RNA nanoparticles and controls were administered into mice with three biological replicates via IV injection at 5 mg/kg (PTX/body weight). Three hours post-injection, blood samples were harvested from mice by cardiac puncture and serum was separated by centrifugation at 12,800 × g for 10 min. Concentrations of cytokine TNF-α, IL-6, IL-12 and IFN-γ in serum supernatant were examined in triplicates using Mouse ELISA MAX Deluxe sets (BioLegend), following manufacturer provided protocols.
Statistics
Each experiment for each tested sample was repeated at least three times independently and the results were presented as mean ± standard deviation (S.D.). Statistical differences were evaluated using unpaired t-test with GraphPad software, and p<0.05 was considered statistically significant.
Supplementary Material
Highlights.
Rubber-like 6WJ RNA nanoparticle carries 24 PTX, one miR122 and one chemical ligand
RNA nanoparticle displaying hepatocyte targeting ligand specifically target to tumor
Co-delivery of PTX and miR-122 exhibit synergetic therapeutic effect to liver cancer
The targeted RNA nanoparticles inhibited liver cancer growth in liver cancer xenograft
ACKNOWLEDGEMENT
The work was supported by NIH grant CA207946 to P.G. and CA016058 to OSU campus Microscopy and Imaging Facility. We thank the Analytical Cytometry Shared Resource, the Campus Microscopy and Imaging Facility, and the University Laboratory Animal Resources Facilitates at The Ohio State University Comprehensive Cancer Center for supporting flow cytometry analysis, confocal microscopy, and animal facility, respectively. We appreciate Dr. Daniel Binzel and Nicolas Burns in manuscript preparation. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. The funding to Peixuan Guo’s Endowed Chair in Nanobiotechnology position is from the William Fairish Endowment Fund.
Footnotes
CONFLICT OF INTEREST
P.G. is the consultant of Oxford Nanopore Technologies; the cofounder of Shenzhen P&Z Bio-medical Co. Ltd, as well as cofounder of ExonanoRNA, LLC and its subsidiary Weina Biomedical LLC in Foshan. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Balogh J, Victor D 3rd, Asham EH, Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM, Monsour HP Jr., Hepatocellular carcinoma: a review, J Hepatocell Carcinoma, 3 (2016) 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bruix J, Han K-H, Gores G, Llovet JM, Mazzaferro V, Liver cancer: approaching a personalized care, Journal of hepatology, 62 (2015) S144–S156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Raza A, Sood GK, Hepatocellular carcinoma review: current treatment, and evidence-based medicine, World journal of gastroenterology: WJG, 20 (2014) 4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Trefts E, Gannon M, Wasserman DH, The liver, Curr Biol, 27 (2017) R1147–R1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Anstee QM, Reeves HL, Kotsiliti E, Govaere O, Heikenwalder M, From NASH to HCC: current concepts and future challenges, Nat. Rev. Gastroenterol. Hepatol, 16 (2019) 411–428. [DOI] [PubMed] [Google Scholar]
- [6].Villanueva A, Hepatocellular Carcinoma, N. Engl. J. Med, 380 (2019) 1450–1462. [DOI] [PubMed] [Google Scholar]
- [7].Abou-Alfa GK, Meyer T, Cheng A-L, El-Khoueiry AB, Rimassa L, Ryoo B-Y, Cicin I, Merle P, Chen Y, J.-W.J.N.E.J.o.M. Park, Cabozantinib in patients with advanced and progressing hepatocellular carcinoma, 379 (2018) 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].CAO H, Phan H, Yang L-X, Improved chemotherapy for hepatocellular carcinoma, Anticancer research, 32 (2012) 1379–1386. [PubMed] [Google Scholar]
- [9].Kudo M, Finn RS, Qin S, Han K-H, Ikeda K, Piscaglia F, Baron A, Park J-W, Han G, Jassem JJTL, Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial, 391 (2018) 1163–1173. [DOI] [PubMed] [Google Scholar]
- [10].Finkelmeier F, Waidmann O, J.J.E.r.o.a.t. Trojan, Nivolumab for the treatment of hepatocellular carcinoma, 18 (2018) 1169–1175. [DOI] [PubMed] [Google Scholar]
- [11].Kudo M.J.L.c., A new era of systemic therapy for hepatocellular carcinoma with regorafenib and lenvatinib, 6 (2017) 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Peck-Radosavljevic M, Drug therapy for advanced-stage liver cancer, Liver Cancer, 3 (2014) 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Deng G-L, Zeng S, Shen H.J.W.j.o.h., Chemotherapy and target therapy for hepatocellular carcinoma: New advances and challenges, 7 (2015) 787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, Sarkar S, Drug resistance in cancer: an overview, Cancers, 6 (2014) 1769–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rudalska R, Dauch D, Longerich T, McJunkin K, Wuestefeld T, Kang T-W, Hohmeyer A, Pesic M, Leibold J, Von Thun A, In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer, Nature medicine, 20 (2014) 1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gottesman MM, Mechanisms of cancer drug resistance, Annu. Rev. Med, 53 (2002) 615–627. [DOI] [PubMed] [Google Scholar]
- [17].Nobuoka D, Yoshikawa T, Sawada Y, Fujiwara T, Nakatsura T.J.H.v., immunotherapeutics, Peptide vaccines for hepatocellular carcinoma, 9 (2013) 210–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gao J, Xia Y, Chen H, Yu Y, Song J, Li W, Qian W, Wang H, Dai J, Guo YJN, Polymer–lipid hybrid nanoparticles conjugated with anti-EGF receptor antibody for targeted drug delivery to hepatocellular carcinoma, 9 (2014) 279–293. [DOI] [PubMed] [Google Scholar]
- [19].Ashokkumar T, Prabhu D, Geetha R, Govindaraju K, Manikandan R, Arulvasu C, Singaravelu G, Apoptosis in liver cancer (HepG2) cells induced by functionalized gold nanoparticles, Colloids and Surfaces B: Biointerfaces, 123 (2014) 549–556. [DOI] [PubMed] [Google Scholar]
- [20].Shaat H, Mostafa A, Moustafa M, Gamal-Eldeen A, Emam A, El-Hussieny E, Elhefnawi M, Modified gold nanoparticles for intracellular delivery of anti-liver cancer siRNA, International journal of pharmaceutics, 504 (2016) 125–133. [DOI] [PubMed] [Google Scholar]
- [21].Maeng JH, Lee D-H, Jung KH, Bae Y-H, Park I-S, Jeong S, Jeon Y-S, Shim C-K, Kim W, Kim J, Multifunctional doxorubicin loaded superparamagnetic iron oxide nanoparticles for chemotherapy and magnetic resonance imaging in liver cancer, Biomaterials, 31 (2010) 4995–5006. [DOI] [PubMed] [Google Scholar]
- [22].Tamarov KP, Osminkina LA, Zinovyev SV, Maximova KA, Kargina JV, Gongalsky MB, Ryabchikov Y, Al-Kattan A, Sviridov AP, Sentis M, Radio frequency radiation-induced hyperthermia using Si nanoparticle-based sensitizers for mild cancer therapy, Scientific reports, 4 (2014) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang X, Ng HLH, Lu A, Lin C, Zhou L, Lin G, Zhang Y, Yang Z, Zhang HJNN, Biology, Medicine, Drug delivery system targeting advanced hepatocellular carcinoma: Current and future, 12 (2016) 853–869. [DOI] [PubMed] [Google Scholar]
- [24].Lu J, Wang J, Ling D, Surface engineering of nanoparticles for targeted delivery to hepatocellular carcinoma, Small, 14 (2018) 1702037. [DOI] [PubMed] [Google Scholar]
- [25].Anirudhan TS, Dextran based nanosized carrier for the controlled and targeted delivery of curcumin to liver cancer cells, Int. J. Biol. Macromol, 88 (2016) 222–235. [DOI] [PubMed] [Google Scholar]
- [26].Maruyama-Tabata H, Harada Y, Matsumura T, Satoh E, Cui F, Iwai M, Kita M, Hibi S, Imanishi J, Sawada T, Effective suicide gene therapy in vivo by EBV-based plasmid vector coupled with polyamidoamine dendrimer, Gene Therapy, 7 (2000) 53–60. [DOI] [PubMed] [Google Scholar]
- [27].Poon RT, Borys N, Lyso-thermosensitive liposomal doxorubicin: a novel approach to enhance efficacy of thermal ablation of liver cancer, Expert Opin. Pharmacother, 10 (2009) 333–343. [DOI] [PubMed] [Google Scholar]
- [28].Zheng G, Zhao R, Xu A, Shen Z, Chen X, Shao J, Co-delivery of sorafenib and siVEGF based on mesoporous silica nanoparticles for ASGPR mediated targeted HCC therapy, Eur. J. Pharm. Sci, 111 (2018) 492–502. [DOI] [PubMed] [Google Scholar]
- [29].Qi L, Xu Z, Chen M, In vitro and in vivo suppression of hepatocellular carcinoma growth by chitosan nanoparticles, European journal of cancer, 43 (2007) 184–193. [DOI] [PubMed] [Google Scholar]
- [30].Sarika P, James NR, Kumar PA, Raj DK, Kumary T, Gum arabic-curcumin conjugate micelles with enhanced loading for curcumin delivery to hepatocarcinoma cells, Carbohydr. Polym, 134 (2015) 167–174. [DOI] [PubMed] [Google Scholar]
- [31].Lombardi G, Zustovich F, Farinati F, Cillo U, Vitale A, Zanus G, Donach M, Farina M, Zovato S, Pastorelli D, Pegylated liposomal doxorubicin and gemcitabine in patients with advanced hepatocellular carcinoma: results of a phase 2 study, Cancer, 117 (2011) 125–133. [DOI] [PubMed] [Google Scholar]
- [32].Baig B, Halim SA, Farrukh A, Greish Y, Amin A, Current status of nanomaterial-based treatment for hepatocellular carcinoma, Biomedicine & Pharmacotherapy, 116 (2019) 108852. [DOI] [PubMed] [Google Scholar]
- [33].Wang L, Su W, Liu Z, Zhou M, Chen S, Chen Y, Lu D, Liu Y, Fan Y, Zheng Y, CD44 antibody-targeted liposomal nanoparticles for molecular imaging and therapy of hepatocellular carcinoma, Biomaterials, 33 (2012) 5107–5114. [DOI] [PubMed] [Google Scholar]
- [34].Zhou Q, Ching AK-K, Leung WK-C, Szeto CY-Y, Ho S-M, Chan PK-S, Yuan Y-F, Lai PB-S, Yeo W, Wong N, Novel therapeutic potential in targeting microtubules by nanoparticle albumin-bound paclitaxel in hepatocellular carcinoma, International journal of oncology, 38 (2011) 721–731. [DOI] [PubMed] [Google Scholar]
- [35].Shu D, Li H, Shu Y, Xiong G, Carson WE, Haque F, Xu R, Guo P, Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology, ACS Nano, 9 (2015) 9731–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yin H, Xiong G, Guo S, Xu C, Xu R, Guo P, Shu D, Delivery of Anti-miRNA for Triple-Negative Breast Cancer Therapy Using RNA Nanoparticles Targeting Stem Cell Marker CD133, Mol Ther, (2019). [DOI] [PMC free article] [PubMed]
- [37].Guo P, The emerging field of RNA nanotechnology, Nature Nanotechnology, 5 (2010) 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shu D, Shu Y, Haque F, Abdelmawla S, Guo P, Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics, Nature Nanotechnology, 6 (2011) 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jasinski D, Haque F, Binzel DW, Guo P, Advancement of the Emerging Field of RNA Nanotechnology, ACS Nano, 11 (2017) 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Guo S, Huang Y, Jiang Q, Sun Y, Deng L, Liang Z, Du Q, Xing J, Zhao Y, Wang PC, Dong A, Liang XJ, Enhanced gene delivery and siRNA silencing by gold nanoparticles coated with charge-reversal polyelectrolyte, ACS Nano, 4 (2010) 5505–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Xu C, Haque F, Jasinski DL, Binzel DW, Shu D, Guo P, Favorable biodistribution, specific targeting and conditional endosomal escape of RNA nanoparticles in cancer therapy, Cancer letters, 414 (2018) 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li H, Lee T, Dziubla T, Pi F, Guo S, Xu J, Li C, Haque F, Liang X, Guo P, RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications, Nano Today, 10 (2015) 631–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jasinski DL, Li H, Guo P, The Effect of Size and Shape of RNA Nanoparticles on Biodistribution, Mol Ther, 26 (2018) 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Guo S, Li H, Ma M, Fu J, Dong Y, Guo P, Size, Shape, and Sequence-Dependent Immunogenicity of RNA Nanoparticles, Mol Ther. Nucleic Acids, 9 (2017) 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Guo S, Vieweger M, Zhang K, Yin H, Wang H, Li X, Li S, Hu S, Sparreboom A, Evers BM, Ultra-thermostable RNA nanoparticles for solubilizing and high-yield loading of paclitaxel for breast cancer therapy, Nature communications, 11 (2020) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ghimire C, Wang H, Li H, Vieweger M, Xu C, Guo P, RNA Nanoparticles as Rubber for Compelling Vessel Extravasation to Enhance Tumor Targeting and for Fast Renal Excretion to Reduce Toxicity, ACS Nano, (2020). [DOI] [PMC free article] [PubMed]
- [47].Singh S, Dash AK, Paclitaxel in cancer treatment: perspectives and prospects of its delivery challenges, Crit Rev. Ther. Drug Carrier Syst, 26 (2009) 333–372. [DOI] [PubMed] [Google Scholar]
- [48].Yardley D.A.J.J.o.C.R., nab-Paclitaxel mechanisms of action and delivery, 170 (2013) 365–372. [DOI] [PubMed] [Google Scholar]
- [49].Horwitz SB, Mechanism of action of taxol, Trends Pharmacol. Sci, 13 (1992) 134–136. [DOI] [PubMed] [Google Scholar]
- [50].Xu Y, Xia F, Ma L, Shan J, Shen J, Yang Z, Liu J, Cui Y, Bian X, Bie P, Qian C, MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest, Cancer Lett, 310 (2011) 160–169. [DOI] [PubMed] [Google Scholar]
- [51].Ma J, Dong C, Ji C, MicroRNA and drug resistance, Cancer Gene Ther, 17 (2010) 523–531. [DOI] [PubMed] [Google Scholar]
- [52].Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z, Cai F, Ma L, Yu Y, The Role of MicroRNAs in Hepatocellular Carcinoma, J Cancer, 9 (2018) 3557–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Rupaimoole R, Slack FJ, MicroRNA therapeutics: towards a new era for the management of cancer and other diseases, Nat Rev Drug Discov, 16 (2017) 203–222. [DOI] [PubMed] [Google Scholar]
- [54].Ding B, Lou W, Xu L, Fan W, Non-coding RNA in drug resistance of hepatocellular carcinoma, Biosci Rep, 38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Huang H, Zhu Y, Li S, MicroRNA-122 mimic transfection contributes to apoptosis in HepG2 cells, Mol Med Rep, 12 (2015) 6918–6924. [DOI] [PubMed] [Google Scholar]
- [56].Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K, MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib, J Biol Chem, 284 (2009) 32015–32027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bandiera S, Pfeffer S, Baumert TF, Zeisel MB, miR-122--a key factor and therapeutic target in liver disease, J Hepatol, 62 (2015) 448–457. [DOI] [PubMed] [Google Scholar]
- [58].Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, Hsu MT, Wu JC, Huang HD, Shiao MS, Hsiao M, Tsou AP, MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis, J. Clin. Invest, 122 (2012) 2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Shi B, Abrams M, Sepp-Lorenzino L.J.J.o.H., Cytochemistry, Expression of asialoglycoprotein receptor 1 in human hepatocellular carcinoma, 61 (2013) 901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Khorev O, Stokmaier D, Schwardt O, Cutting B, Ernst B, Trivalent, Gal/GalNAc-containing ligands designed for the asialoglycoprotein receptor, Bioorg Med Chem, 16 (2008) 5216–5231. [DOI] [PubMed] [Google Scholar]
- [61].Meier M, Bider MD, Malashkevich VN, Spiess M, Burkhard P, Crystal structure of the carbohydrate recognition domain of the H1 subunit of the asialoglycoprotein receptor, J Mol Biol, 300 (2000) 857–865. [DOI] [PubMed] [Google Scholar]
- [62].Hu J, Liu J, Yang D, Lu M, Yin J, Physiological roles of asialoglycoprotein receptors (ASGPRs) variants and recent advances in hepatic-targeted delivery of therapeutic molecules via ASGPRs, Protein and peptide letters, 21 (2014) 1025–1030. [DOI] [PubMed] [Google Scholar]
- [63].Monestier M, Charbonnier P, Gateau C, Cuillel M, Robert F, Lebrun C, Mintz E, Renaudet O, Delangle P, ASGPR-Mediated Uptake of Multivalent Glycoconjugates for Drug Delivery in Hepatocytes, ChemBioChem, 17 (2016) 590–594. [DOI] [PubMed] [Google Scholar]
- [64].Chandler M, Johnson MB, Panigaj M, Afonin KA, Innate immune responses triggered by nucleic acids inspire the design of immunomodulatory nucleic acid nanoparticles (NANPs), Curr. Opin. Biotechnol, 63 (2019) 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ke W, Hong E, Saito RF, Rangel MC, Wang J, Viard M, Richardson M, Khisamutdinov EF, Panigaj M, Dokholyan NV, Chammas R, Dobrovolskaia MA, Afonin KA, RNA-DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-kappaB in human cells, Nucleic Acids Res, In Press (2018). [DOI] [PMC free article] [PubMed]
- [66].Sukowati CH, Rosso N, Croce LS, Tiribelli C, Hepatic cancer stem cells and drug resistance: Relevance in targeted therapies for hepatocellular carcinoma, World J Hepatol, 2 (2010) 114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


