Abstract
Aim:
Acquired molecular changes in Lynch syndrome (LS) colorectal tumors have been largely unstudied. We identified methylated DNA markers (MDMs) for discrimination of colorectal neoplasia in LS and determined if these MDMs were comparably discriminant in sporadic patients.
Patients & methods:
For LS discovery, we evaluated DNA from 53 colorectal case and control tissues using next generation sequencing. For validation, blinded methylation-specific PCR assays to the selected MDMs were performed on 197 cases and controls.
Results:
OPLAH was the most discriminant MDM with areas under the receiver operating characteristic curve ≥0.97 for colorectal neoplasia in LS and sporadic tissues. ALKBH5, was uniquely hypermethylated in LS neoplasms.
Conclusion:
Highly discriminant MDMs for colorectal neoplasia in LS were identified with potential use in screening and surveillance.
Keywords: : biomarkers, colorectal neoplasms, colorectal neoplasms, colorectal prevention and control, hereditary nonpolyposis, DNA methylation, colorectal physiology
Lynch syndrome (LS) is the most common form of familial colorectal cancer (CRC), with population prevalence of 1 in 280 [1]. LS accounts for at least 3% of all newly diagnosed CRC cases and nearly 10–20% of early-onset CRC [2,3]. LS results from germline inactivation of mismatch repair (MMR) genes MLH1, MSH2, MSH6, PMS2 or loss of expression of MSH2 due to germline deletion in the EPCAM gene, increasing the risk of developing CRC and extra-colonic cancers including endometrial, small bowel, ureteral, renal pelvis, gastric, hepatobiliary and ovarian [4]. Without effective screening and surveillance, the lifetime risk of CRC in individuals with LS ranges between 10 and 80% and differs based on specific MMR alteration [5]. An international multicenter collaboration which aimed to prospectively determine the incidence of cancer and survival stratified by MMR pathogenic variant, age and gender showed that cumulative incidences at 75 years for CRC were 46, 43 and 15% in pathogenic MLH1, MSH2 and MSH6 carriers; for endometrial cancer 43, 57 and 46%; for ovarian cancer 10, 17 and 13%; for upper gastrointestinal (gastric, duodenal, bile duct or pancreatic) cancers 21, 10 and 7%; for urinary tract cancers 8, 25 and 11%; for prostate cancer 17, 32 and 18%; and for brain tumors 1, 5 and 1%, respectively [6]. Except for endometrial and prostate cancer, typical LS cancers were not diagnosed in pathogenic PMS2 carriers. Upper gastrointestinal, urinary tract and prostate cancers occurred predominantly at older ages and ovarian cancer occurred predominantly premenopausally [6], which is in contrast to ovarian cancers in carriers with a pathogenic variant in BRCA 1 and BRACA 2 genes.
The LS phenotype is characterized by a predominance of cancers on the right side of the colon and a predisposition for synchronous and metachronous CRCs [3]. Accelerated progression through the adenoma–carcinoma sequence or carcinoma arising from non-neoplastic crypt foci occurs in LS due to MMR deficiency that compromises the ability to repair base-pair (bp) mismatches in DNA and confers predisposition to CRC [3,7,8]. Consequently, screening and surveillance guidelines recommend colonoscopy every 1–2 years [9], and such intensive surveillance has been found to be effective in reducing CRC mortality in LS [9,10]. However, CRCs develop within surveillance programs [11], related to accelerated carcinogenesis, operator-dependent colonoscopy with variable adenoma detection rates [12–14], suboptimal effectiveness of current approaches to detect proximal lesions [10,15–19], and reported suboptimal compliance rates of surveillance colonoscopy in LS patients (as low as 50%) [20].
For patients with hereditary risk for CRC, such as LS, there are no current options for effective non-invasive screening. Accurate noninvasive detection tools may have value as complements to colonoscopic surveillance in LS kindreds and mitigate the impact of noncompliance to colonoscopy. Such tests would target markers which highly discriminate the presence of CRC and precursor lesions. Unlike the well-established germline pathogenic variants in MMR which help identify the syndrome, much less is known about acquired molecular alterations in precursor colorectal neoplasms which might serve as early detection markers for screening and surveillance. However, several studies have demonstrated differences in the profiles of acquired genetic and epigenetic changes in colorectal neoplasms between LS and sporadic patients [21–24].
A multi-target stool DNA test (MT-sDNA), clinically available as Cologuard® (Exact Sciences, WI, USA), has emerged as an accurate noninvasive molecular approach for detection of sporadic CRC and high risk precancers, and is approved by the US FDA for average risk population-based screening. MT-sDNA targets two aberrantly methylated genes (BMP3 and NDRG4) and seven KRAS mutations, normalized to β-actin, plus hemoglobin [25]. In two blinded cross-sectional screening studies [26,27], the test achieved point sensitivities of 94–100% for early stage CRC and 40–42%, 62–66% and 68–80% for adenomas >1 cm, >2 cm and >3 cm, respectively. Simple model estimates suggest that cumulative polyp detection rates by MT-sDNA with repeated testing at 3 year intervals may compare favorably to those of colonoscopy done at 10 year intervals [28]. However, it is not known whether MT-sDNA would perform similarly in detecting colorectal neoplasms from LS patients. Based on a preliminary study [29], the methylated DNA markers BMP3 and NDRG4 targeted by the MT-sDNA panel did not appear to discriminate CRC or adenomas in LS tissues as well as they did in sporadic colorectal tissues.
Since the development of the MT-sDNA test, our group has identified highly discriminant methylated DNA markers (MDMs) for detection of sporadic colorectal neoplasia based on extensive unbiased next generation sequencing and tissue validation [30,31]. Using selected MDM candidates from this effort, we found that a panel of MDMs alone applied to stool can detect CRC and high-grade dysplasia in inflammatory bowel disease patients with sensitivities and specificities above 90% [30]. These novel MDMs, however, have not been tested on colorectal neoplasms from LS patients and, to our knowledge, sequencing-based discovery to identify MDM candidates in LS has not been done.
Thus, the aims in the present study were to identify MDM candidates for detection of colorectal neoplasia in LS based on discovery by methylome sequencing with subsequent validation in independent tissues, evaluate discrimination accuracy for colorectal neoplasms by top candidate MDMs selected from the LS discovery and from our prior discovery in sporadic tissues using well-characterized colorectal tissues from LS and sporadic patients, and compare discrimination accuracy for the detection of colorectal neoplasia in LS and sporadic colorectal tissues by these novel MDM candidates with that of methylated BMP3 and NDRG4, the MDMs targeted by MT-sDNA.
Materials & methods
Study overview
This investigation comprised two sequential case-control studies, both using well-characterized archival paraffin embedded colorectal tissues. In the first, candidate MDMs to discriminate colorectal neoplasia in LS were identified by reduced representation bisulfite sequencing (RRBS) on DNA extracted from well-characterized case (adenomas and CRCs) and control (normal appearing mucosa) tissues. In the second, a blinded biological validation study was conducted. Top MDM candidates from the above discovery effort in LS samples, selected novel MDM candidates from a prior whole methylome discovery effort in sporadic samples [30,31], and the two MDMs (BMP3 & NDRG4) in the MT-sDNA test panel were assayed in an independent set comprising case and control tissues from both LS and sporadic patients. This study was approved by the Mayo Clinic Institutional Review Board.
Study population & sample sources
All patient tissues from this study were obtained from Mayo Clinic archives. By convention [2,3], LS patients were classified as “definite” (genetically-confirmed) if testing showed a germline pathogenic variant in MMR genes (MLH1, MSH2, MSH6 and PMS2) as well as the gene EPCAM. LS patients were classified as “suspected” if tissue immunohistochemistry (IHC) showed absence of MSH2, MSH6 and/or PMS2 proteins and germline testing was not available to confirm a pathogenic variant in the MMR genes. Cases with only tissue IHC showing absence of MLH1 but lacking confirmation of a germline pathogenic variant were excluded since these may represent biallelic somatic methylation induced silencing of the MLH1 promoter. Given the retrospective nature of the study, we had to rely on data available in the medical record.
Discovery
Formalin-fixed paraffin-embedded (FFPE) colorectal samples were selected from the institutional tissue registries at Mayo Clinic (Rochester, MN, USA) and were reviewed by an expert gastrointestinal pathologist (T.C.S.) to confirm correct histological classification. LS case tissues comprised CRCs (all adenocarcinomas) and adenomas (classical or sessile serrated ≥1 cm); LS control tissues included histologically normal colorectal epithelium from LS patients. An effort was made to match cases and controls on age and sex. Tissue specimens had been collected after informed consent from patients undergoing colonoscopy or colectomy at Mayo Clinic and were procured at the time of these procedures. The Mayo Component Laboratory contributed de-identified buffy coat samples to serve as leukocyte controls.
Biological validation
An independent set of case and control FFPE colorectal tissues from LS patients plus a set of case and control FFPE tissues from sporadic patients were obtained from the Mayo Clinic Tissue Registry and histologically confirmed as above. Case and control definitions were per Discovery set.
Technical procedures
Sample processing
Following micro-dissection of FFPE tissues, DNA was extracted using the QIAamp FFPE DNA Tissue Kit (Qiagen, Hilden Germany). Buffy coat sample DNA was purified with the QIAamp DNA Mini kit. Quantification of total DNA was performed using the Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific, MA, USA).
Reduced representation bisulfite sequencing
Library preparation
DNA samples were repurified and size selected (>500 bp) using 0.6X AMPure XP (Beckman Coulter, CA, USA). Sequencing libraries were prepared as we have previously described [30,32,33].
Sequencing & bioinformatics
Sequencing was performed by the Mayo Medical Genomics Facility using the HiSeq 2000 (Illumina, CA, USA). Reads were processed by Illumina pipeline modules for image analysis and base calling. Secondary analysis was performed using SAAP-RRBS [29]. Briefly, reads were cleaned-up using Trim-Galore and aligned to the GRCh37/hg19 reference genome build with BSMAP. Methylation ratios were determined by calculating C/(C+T) or conversely, G/(G+A) for reads mapping to reverse strand, for CpGs with coverage ≥10× and base quality score ≥20.
PCR assays for tissue validation
Quantitative methylation-specific PCR
Primers for each marker were designed to target bisulfite-modified methylated CpGs within each target sequence. A CpG agnostic region in the β-actin gene (ACTB) was used as a reference for total DNA. Designs were created using either Methprimer software [34] or by manual methods. Assays were tested and optimized by quantitative methylation-specific PCR on universally methylated and unmethylated genomic DNA controls. Annealing temperatures were determined empirically.
FFPE sample DNA (purification described above) was bisulfite-converted using the Zymo EZ-96 DNA Methylation kit (Zymo Research, CA, USA) and amplified (10 ng/marker) with SYBR Green dye (Thermo Fisher Scientific) detection using the LightCycler 480 instrument and reagents (Roche Diagnostics, IN, USA).
Quantitative allele specific real-time target & signal amplification
Several of our sporadic MDM assays had been previously configured in the quantitative allele specific real-time target & signal amplification format, which is a similar amplification-based platform to quantitative methylation-specific PCR and run on the same instrument [35]. These assays were also optimized using the same controls and ACTB as a reference gene. Sample DNA was prepared and used as above. Details of the assay methods have been reported [33].
Statistical methods
Discovery
From the RRBS data, individual CpGs which had a minimum of 10× vertical read coverage were ranked by hypermethylation ratio, namely the number of methylated cytosines at a given locus over the total cytosine count at that site. For cases, the ratios were required to be ≥20% but ≤5% for controls. CpGs that did not meet these criteria were discarded. Subsequently, candidate CpGs were binned by genomic location into differentially methylated regions (DMRs) ranging from approximately 60–200 bp in length with a minimum cut-off of five CpGs per region. DMRs with excessively high CpG density (>30%) were excluded to avoid GC-related amplification problems in the validation phase. For each candidate region, a 2D was created which compared individual CpGs in a sample-to-sample fashion for both cases and controls. These were then compared back to the reference sequence to assess whether neighboring CpGs had been discarded during the initial filtering. From this subset of regions, final selections required coordinated and contiguous hypermethylation of individual CpGs across the DMR sequence on a per sample level in cases. Conversely, control samples had to have at least tenfold less methylation than cases and the CpG pattern had to be more random and less coordinated. The most discriminant DMRs meeting these criteria were selected as candidate MDMs for testing in subsequent biological validation. Assuming an average read depth of 40 per CpG, 18 patients per group provided 80% power to detect a minimum difference of 4% in methylation rates (e.g., 2 vs 6%) with a two-sided significance level of 5%.
MDMs from previous sporadic discovery/validation studies & MT-sDNA
For the biological validation, we also selected 12 additional markers to evaluate in the validation set. These included ten high performing methylated markers (ARHGEF4, SFMBT2_897, OPLAH, LRRC4, ELMO1, VAV3, DAB2IP, PDGFD, CHST2_7889, AK055957) from earlier RRBS and independent validation studies on sporadic colorectal tissue samples that had not been assessed in the context of LS. Each of these MDMs had demonstrated excellent discrimination (AUC 0.90–0.98) when comparing both sporadic CRC and adenoma to normal colonic mucosa [30,31]. In addition, we tested two MDMs (BMP3 and NDRG4) incorporated into the FDA approved and clinically available MT-sDNA (Cologuard®, Exact Sciences) [25].
Biological tissue validation
Individual marker distributions were displayed using boxplots. Marker combinations were then studied using recursive partitioning trees (rPart). Briefly, rPart was used to first select a single MDM that provided the greatest separation between cases and controls (branch split). Once split, rPart searched for additional MDMs that provided the greatest separation between cases and controls under each branch. The rPart technique was applied to the entire MDM set, upon which an rPart predicted probability of colorectal neoplasia was calculated. Discrimination accuracy was evaluated using the area under the receiver operating characteristic curve (AUC) with corresponding 95% confidence interval. The effect of age and sex on the diagnostic accuracy of MDMs and panels was investigated by comparing stratified AUC values.
Results
Marker discovery in LS
Patient & lesion characteristics
For whole methylome sequencing by RRBS, we selected 53 well-characterized colorectal tissues from LS patients; samples comprised 18 normal mucosa, 18 adenomas ≥1 cm and 17 adenocarcinomas. For discovery, 15/18 (83%) of control, 10/18 (56%) of adenoma and 12/17 (71%) of CRC tissues came from LS patients classified as ‘definite’ and the remainder came from those classified as ‘suspected’. The clinico-pathologic characteristics are summarized in Table 1.
Table 1. . Patient and lesion characteristics for discovery and validation phases.
| Patient/lesion characteristic | Discovery |
Biological validation | |
|---|---|---|---|
| Lynch | Sporadic | Lynch | |
| Normal mucosa: | |||
| n | 18 | 35 | 23 |
| Definite LS | 15 (83%) | n/a | 13 (57%) |
| Age, median (IQR) | 53 (44.8–62.5) | 64 (53.5–71.5) | 50 (37.5–57) |
| Sex, % female | 61% | 49% | 57% |
| Advanced adenoma: | |||
| n | 18 | 38 | 39 |
| Definite LS | 10 (56%) | n/a | 26 (67%) |
| Age, median (IQR) | 57 (53.3–66.3) | 64.5 (56–76.3) | 59 (50.5–69) |
| Sex, % female | 33% | 40% | 59% |
| Site, % proximal | 55% | 76% | 51% |
| Tubular adenoma | 18 | 13 | 37 |
| Sessile serrated polyp | 0 | 25 | 2 |
| Adenocarcinoma: | |||
| n | 17 | 36 | 26 |
| Definite LS | 12 (71%) | n/a | 14 (54%) |
| Age, median (IQR) | 59 (54–71) | 68.5 (57.8–80.5) | 46.5 (36.5–58.8) |
| Sex, % female | 41% | 50% | 46% |
| Site, % proximal | 6159% | 47% | 46% |
IQR: Interquartile range; LS: Lynch syndrome; n: Number of patients.
MDM candidates
Among DMRs observed, the nine that met pre-established filtering criteria were selected as candidate MDMs. These candidates were located on USP44, STK32B, CBLN2, ADCY4, CNTFR, PITX1, ANTRX1, ALKBH5 and ADM genes. For both CRC and adenomas, fold-change increases ranged from 13 to >200. Discrimination metrics for each MDM selected are summarized in Supplementary Table 1.
Biological validation & comparison in colorectal tissues from LS & sporadic patients
In this study phase, the nine MDM candidates listed in Table 2 from the above LS discovery, ten MDM candidates from prior sequencing on sporadic colorectal neoplasia (ARHGEF4, SFMBT2_897, OPLAH, LRRC4, ELMO1, VAV3, DAB2IP, PDGFD, CHST2_7889, AK055957) and the two MDMs (BMP3 and NDRG4) in the MT-sDNA test were assayed in blinded fashion on independent tissues not previously tested from LS and sporadic patients.
Table 2. . Biological validation of methylated DNA marker candidates in independent colorectal tissues from Lynch syndrome and sporadic patients.
| Discovery method | MDM | Adenoma | Cancer | ||||
|---|---|---|---|---|---|---|---|
| Lynch | Sporadic | p-value | Lynch | Sporadic | p-value | ||
| Lynch discovery | PITX1 | 0.86 | 0.97 | 0.034 | 0.98 | 1 | 0.201 |
| CBLN2 | 0.86 | 0.95 | 0.11 | 0.97 | 0.82 | 0.025 | |
| USP44 | 0.85 | 0.96 | x0.066 | 0.96 | 0.97 | 0.671 | |
| STK32B | 0.79 | 0.9 | 0.15 | 0.95 | 0.95 | 0.932 | |
| CNTFR | 0.79 | 0.93 | 0.031 | 0.92 | 0.87 | 0.378 | |
| ALKBH5 | 0.68 | 0.44 | 0.015 | 0.89 | 0.45 | <0.001 | |
| ADCY4 | 0.81 | 0.75 | 0.508 | 0.88 | 0.93 | 0.415 | |
| ADM | 0.63 | 0.62 | 0.872 | 0.81 | 0.42 | <0.001 | |
| ANTXR1 | 0.53 | 0.96 | <0.001 | 0.71 | 0.79 | 0.378 | |
| Sporadic discovery | ARHGEF4 | 0.89 | 0.97 | 0.127 | 0.99 | 0.97 | 0.554 |
| SFMBT2_897 | 0.80 | 0.96 | 0.007 | 0.99 | 0.9 | 0.082 | |
| OPLAH | 0.99 | 0.99 | 0.83 | 0.97 | 1 | 0.416 | |
| ELMO1 | 0.78 | 0.85 | 0.368 | 0.92 | 0.84 | 0.25 | |
| LRRC4 | 0.86 | 0.88 | 0.795 | 0.92 | 0.98 | 0.188 | |
| VAV3 | 0.73 | 0.95 | 0.001 | 0.87 | 0.89 | 0.829 | |
| DAB2IP | 0.78 | 0.89 | 0.134 | 0.81 | 0.87 | 0.478 | |
| PDGFD | 0.72 | 0.9 | 0.02 | 0.79 | 0.88 | 0.293 | |
| CHST2_7889 | 0.68 | 0.88 | 0.013 | 0.77 | 0.91 | 0.08 | |
| AK055957 | 0.62 | 0.87 | 0.003 | 0.62 | 0.76 | 0.192 | |
| MT-sDNA test | BMP3 | 0.57 | 0.86 | <0.001 | 0.77 | 0.8 | 0.746 |
| NDRG4 | 0.60 | 0.93 | <0.001 | 0.69 | 0.9 | 0.02 | |
MDM: Methylated DNA marker; MT-sDNA test: Multi-target stool DNA test.
Patient & lesion characteristics
We evaluated 197 independent paraffin-embedded colorectal tissues, which included 88 from LS patients (23 normal mucosa, 39 advanced adenomas [37 classical, two sessile serrated) and 26 adenocarcinomas] and 109 from sporadic patients (35 normal mucosa, 38 advanced adenomas [13 classical, 25 sessile serrated] and 36 adenocarcinomas). In the LS validation set, 13/23 (57%) of normal, 26/39 (67%) of adenoma and 14/26 (54%) of cancer tissues came from LS patients classified as ‘definite’ and the remainder from those classified as ‘suspected’. The clinico-pathologic characteristics are shown in Table 1.
Marker distributions
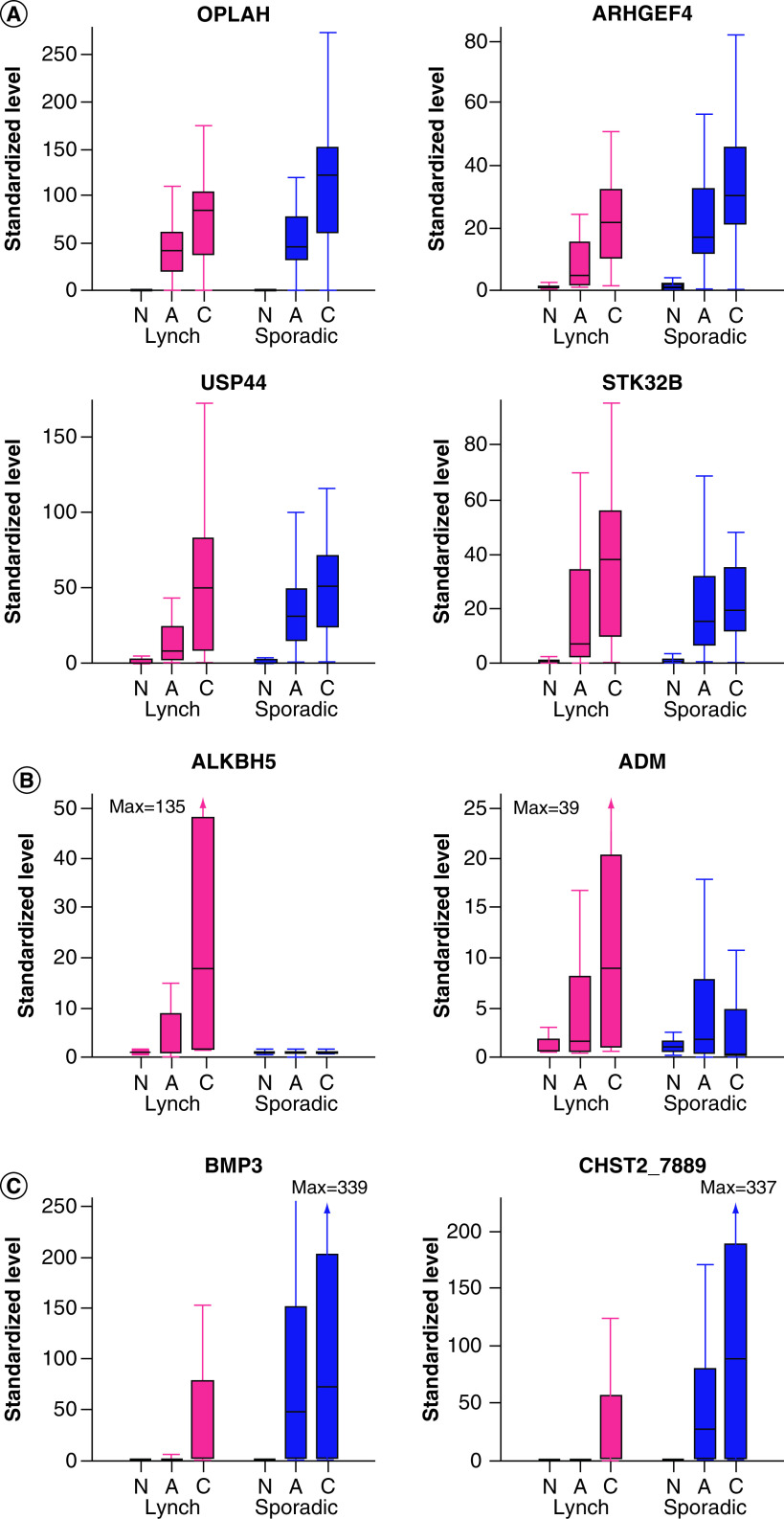
Numerous MDM candidates from each discovery pathway showed excellent separation between normal mucosa and colorectal neoplasms in tissues from both LS and sporadic patients, as illustrated with boxplots of OPLAH, ARHGEF4, USP44 and STK32B (Figure 1A). With these MDMs, tissue levels were negligible or minimal in normal mucosa and consistently elevated with substantial fold changes in both adenomas and cancers from LS and sporadic patients.
Figure 1. . Box plot distributions of selected methylated DNA marker candidates in colorectal tissues from Lynch syndrome and sporadic patients from biological validation phase.
(A) MDMs showing similarly high neoplasm discrimination across Lynch syndrome and sporadic patients. (B) MDMs showing relatively higher neoplasm discrimination in Lynch syndrome patients. (C) MDMs showing relatively higher discrimination in sporadic patients.
A: Adenoma; C: Colorectal cancer; MDM: Methylated DNA marker; N: Normal.
In contrast, the degree of separation in tissue levels between normal and neoplastic tissues differed in LS and sporadic groups with a few MDMs. Two candidate MDMs identified from the LS discovery effort, ALKBH5 and ADM, showed greater separation between tissue types in LS patients than in sporadic patients (Figure 1B); this was particularly striking with ALKBH5, where levels were clearly higher in neoplasms than normal mucosa from LS patients yet there was essentially no separation seen between tissue types from sporadic patients. Likewise, some of the MDMs studied showed strikingly better separation between normal and neoplastic tissues in sporadic patients than in LS patients, as exemplified in boxplots of BMP3 and CHST2_7889 (Figure 1C).
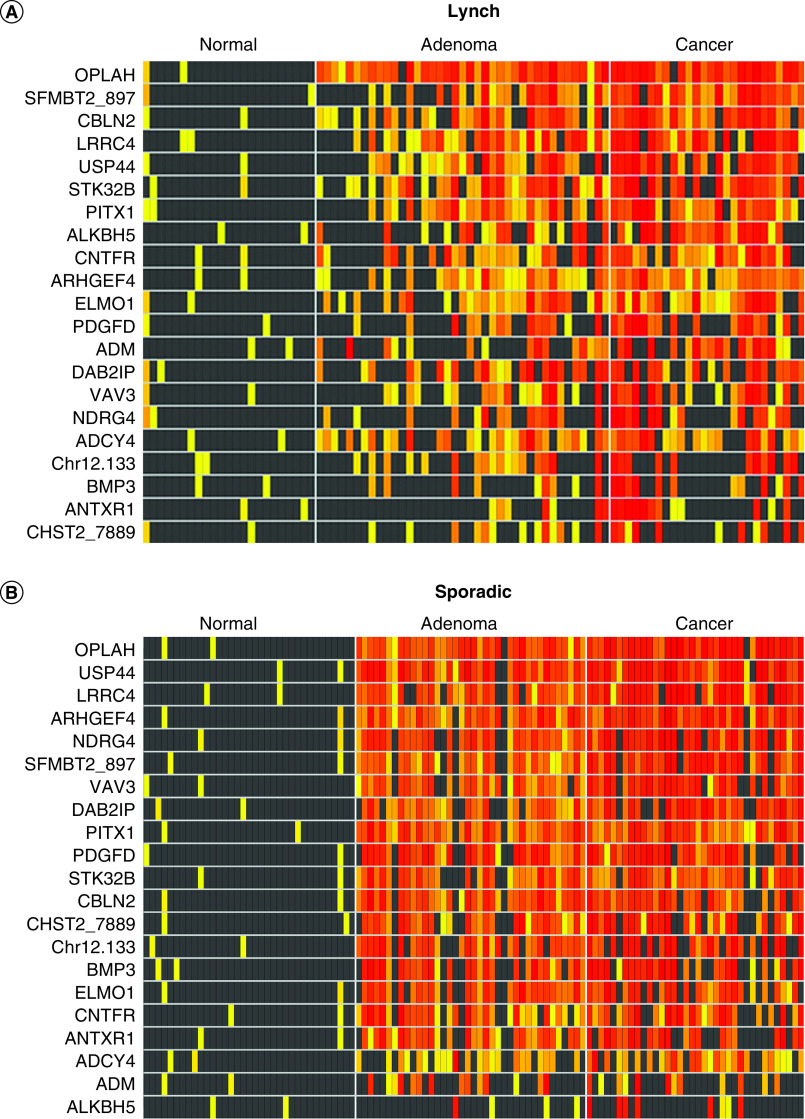
The degree of separation across tissue types for each MDM can also be illustrated by heat matrices, which show individual MDM presence and signal intensity for each patient in case and control tissues (Figure 2). Overall separation and differences in signal density between normal and neoplastic tissues are generally high for most candidate MDMs across both LS patients (Figure 2A) and sporadic patients (Figure 2B). Distributions of these MDMs were not significantly different on the basis of ‘suspected’ versus ‘definite’ LS; representative plots are shown in Supplementary Figure 1.
Figure 2. . Heat matrices: methylation intensity of methylated DNA marker candidates in independent colorectal tissues from biological validation phase.
(A) Lynch syndrome and (B) sporadic tissues. Increasing intensity of yellow-red color spectrum in boxes indicates methylation strand counts in deciles above the 90th percentile values for the control groups (histologically normal mucosa) of each candidate methylated DNA marker (rows) in each tissue sample (columns). Black boxes indicate values falling below the 90th percentile in controls.
Neoplasm discrimination
Discrimination for colorectal adenomas and CRC as assessed by AUCs is shown for each candidate MDM in Table 2. Of the nine MDMs carried forward from the discovery phase in LS, AUCs >0.79 were observed on biological validation in independent LS tissues with six MDMs for adenomas and eight MDMs for cancers; eight MDMs were above this AUC threshold in sporadic adenomas and seven MDMs in sporadic cancers. Of the ten MDMs selected from the prior discovery effort in sporadic tissues, four were above the AUC threshold of >0.79 in LS adenomas and in cancers; all (100%) were above this threshold in sporadic adenomas and all except AK055957 were above this AUC threshold for sporadic cancers. Individually, neither of the MT-sDNA MDMs had AUCs >0.79 in LS neoplasms but both were above this threshold in sporadic neoplasms.
Overall, the individual MDM found to be most discriminant for adenomas and cancer in both LS and sporadic patients was OPLAH. With OPLAH, the AUCs for adenomas were 0.99 (95% CI: 0.97–1.00) in LS and 0.99 (0.96–1.00) in sporadic patients and for cancers were 0.97 (0.92–1.00) and 1 (0.99–1.00), respectively. Other candidate MDMs found to be highly discriminant for adenomas and cancers in both LS and sporadic patients included ARHGEF4, PITX1, USP44, LRRC4 and STK32B. A dramatic exception was ALKBH5, which as suggested by the distributions in Figure 1B, was substantially and significantly more discriminant for both adenomas (p = 0.015) and cancers (p < 0.001) in LS patients compared with sporadic patients (Table 2).
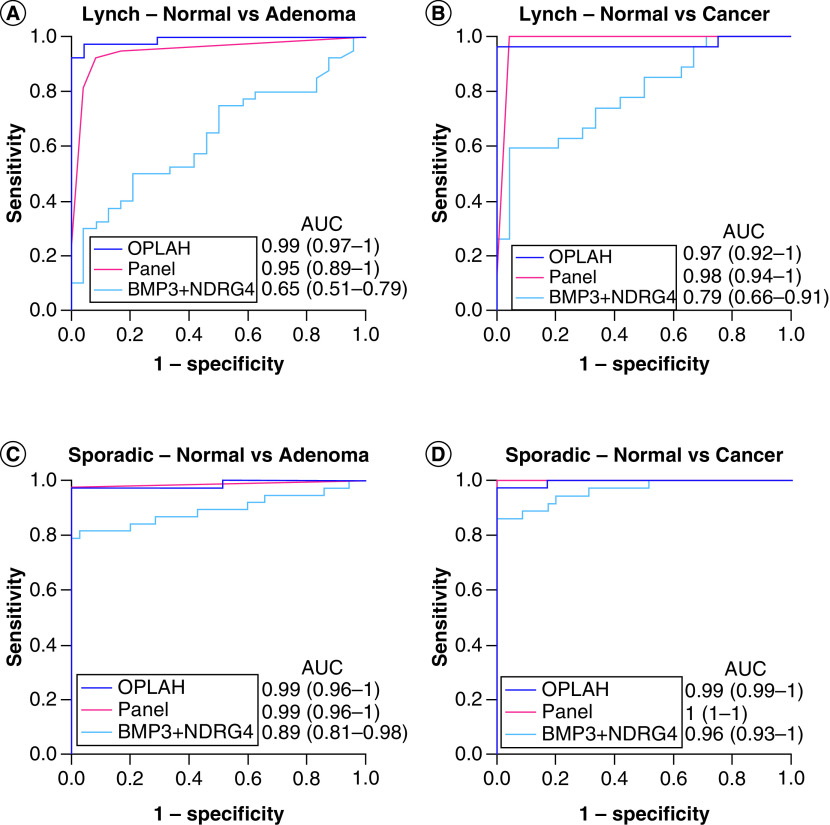
Within and across clinical groupings, the best candidate MDMs from the present LS and prior sporadic discovery pathways were compared with the combination of BMP3 and NDRG4, the MT-sDNA MDMs (Figure 3). For the combination of BMP3 and NDRG4, discrimination was better for sporadic than LS adenomas with respective AUCs of 0.89 and 0.65 (p = 0.003) and for sporadic than LS cancers with respective AUCs of 0.96 and 0.79 (p = 0.008), which is consistent with our preliminary observations [29]. OPLAH alone showed superior discrimination for neoplasia in all clinical groups compared with the combination of BMP3 and NDRG4. For LS adenomas, the AUC was 0.99 for OPLAH versus 0.65 for the combination of BMP3 and NDRG4 (p < 0.001); for LS cancers, AUCs were respectively 0.97 versus 0.79 (p = 0.008); for sporadic adenomas, AUCs were respectively 0.99 versus 0.89 (p = 0.04); and for sporadic cancers, AUCs were respectively 0.99 versus 0.96 (p = 0.12). At 100% specificity, sensitivity for LS adenomas was 92% for OPLAH versus 10% for the combination of BMP3 and NDRG4 (p < 0.001). Sensitivities for LS cancers were respectively 96 versus 27% (p < 0.001); sensitivities for sporadic adenomas were respectively 97 versus 79% (p = 0.03), and sensitivities for sporadic cancers were respectively 97 versus 86% (p = 0.2). Several combinations of novel MDMs achieved high discrimination comparable to that of OPLAH; for example, a panel of four markers from the rPart analysis (ARHGEF4, LRRC4, ANTXR1 and PITX1) showed high AUCs essentially identical to those of OPLAH (Figure 3).
Figure 3. . Discrimination of selected methylated DNA marker candidates for colorectal neoplasia as assessed by receiver operator curves.
AUC in each graph are shown for OPLAH alone, a panel of novel methylated DNA markers without OPLAH (ARHGEF4, LRRC4, ANTXR1, PITX1) and the combination of BMP3 + NDRG4 with (A) Lynch adenomas, (B) Lynch cancers, (C) sporadic adenomas and (D) sporadic cancers.
AUC: Area under the curve.
Covariates were explored. For the overall most discriminant MDMs, patient variables of age and sex did not affect detection accuracy for either LS or sporadic neoplasms, nor did cancer variables of stage or site or did adenoma variables of site or histologic type. Within the sporadic pre-cancer subset, OPLAH detected 100% of tubular adenomas and 92% of sessile serrated adenomas at 100% specificity (p = 0.5). While there were not enough sessile serrated adenomas in the LS group to make such a comparison, it is noteworthy that each of the two sessile serrated adenomas in LS was detected by OPLAH. Moreover, neoplasm detection rates by OPLAH did not differ significantly between LS-Definite and LS-Suspected patients (p > 0.8 for both adenoma and CRC comparisons).
Discussion
In this tissue-based discovery and validation study, we identified novel MDMs highly discriminant of adenomas and CRC in LS patients. Several individual MDMs (e.g. OPLAH) and MDM combinations (e.g., ARHGEF4, LRRC4, ANTXR1, PITX1) achieved almost perfect discrimination of adenomas and CRC across both LS and sporadic colorectal tissues. OPLAH alone exhibited not only AUCs approaching 1.0 but also very high fold change in marker levels between neoplasms and normal mucosa which translates into strong signal strength to facilitate its detection in media such as stool or blood. These novel MDMs that we found to be highly discriminant in tissue have potential for application as a novel screening modality that can complement colonoscopy to optimize early detection of colorectal neoplasia in LS.
While some have used chip-based and other discovery methods to evaluate methylation differences in LS compared with sporadic colorectal neoplasms [21–24], to our knowledge, this is the first study to identify candidate MDMs for detection of LS colorectal neoplasms through discovery by unbiased next-generation methylation sequencing followed by independent tissue validation. All MDM candidates in our study were intentionally selected to represent aberrantly hyper-methylated sequences, as neoplasia-associated hypomethylation is a more subtle effect relative to the normal state and thus more difficult to measure in distant media. The ratio of case to control methylation was more than 20% for cases and <5% for controls. Regions of hypomethylation did not reach this degree of aberrancy. Most of the MDM candidates we describe from this LS discovery effort and those carried over from our previous discovery in sporadic colorectal neoplasia have not been previously described in LS [30,31].
Of importance and supporting potential functional effects of identified aberrant methylation, nearly all of the candidate MDM sequences discovered in LS tissues are harbored by genes known to be associated with cancer [36–53]. Several of our RRBS discovered markers were found on genes known to be pivotal in tumorigenesis, cell signaling and differentiation and transcriptional regulation. While none has been implicated to our knowledge in colorectal neoplasms with LS, genetic or epigenetic aberrations of all nine genes have been described in sporadic cancers from multiple organ sites (Table 3). Of note in this study, aberrant hypermethylation of ALKBH5 was uniquely seen with LS colorectal adenomas and cancers but absent in sporadic neoplasms. This gene encodes for an RNA demethylase which appears to influence pathways related to DNA damage reversal and DNA double-strand break repair [39,54–56]. Diseases associated with ALKBH5 include retinitis pigmentosa, and aberrant expression of ALKBH5 gene has been reported with several cancer types, but has not been reported with LS neoplasms to our knowledge [39,54–56]. The single most discriminant MDM for all neoplasm subgroups in both LS and sporadic tissues was OPLAH, which was identified in our prior whole methylome discovery effort in sporadic tissues, [30,31] the OPLAH gene appears to be involved in glutathione salvage and abnormal expression has been reported in several cancer types [43].
Table 3. . Functions of genes methylated in Lynch syndrome colorectal cancers.
| Gene | Protein | Function | Cancer associations/mechanism† |
|---|---|---|---|
| ADCY4 | Adenylate cyclase 4 | Signal transduction | Prostate cancer/hypermethylation |
| ADM | Adrenomedullin | Signal transduction | CRC, PDAC, renal cell cancer/upregulation |
| ALKBH5 | RNA demethylase ALKBH5 | Cell differentiation | Glioblastoma, breast cancer/upregulation |
| ANTXR1 | Anthrax toxin receptor 1 | Cell attachment and migration | CRC, lung cancer, breast cancer/upregulation |
| CBLN2 | Cerebellin 2 precursor | Synapse assembly | CRC/hypermethylation |
| CNTFR | Ciliary neurotrophic factor receptor | Cellular adhesion/signal transduction | Glioma/upregulation |
| PITX1 | Pituitary homeobox 1 | Transcriptional regulation | CRC, PDAC, gastric, bronchial, prostate, oral cancers/downregulation |
| STK32B | Serine/threonine kinase 32B | Signal transduction | Oral squamous cell cancer, breast cancer/upregulation |
| USP44 | Ubiquitin carboxyl-terminal hydrolase 44 | Cell cycle regulation | CRC/transcriptional silencing, hypermethylation |
Information on table based on literature review [32–49].
CRC: Colorectal cancer; PDAC: Pancreatic ductal adenocarcinoma.
While most of the top MDM candidates brought to validation were comparably discriminant for colorectal neoplasia in LS and sporadic tissues, there were significant differences between groups with some MDMs. A key finding was that hyper-methylation of ALKBH5 (a gene from the ALKBH family of demethylases that repair both DNA and RNA by removing alkyl lesions) [39,54–58] was exclusively observed in LS neoplasms and absent in sporadic neoplasms. In a syndrome characterized by hallmark germline pathogenic variants in DNA MMR genes, this novel finding of an acquired epigenetic change also related to DNA repair may have significant functional significance. Given the uniform presence of aberrantly methylated ALKBH5 in LS-related colorectal neoplasms and its absence in sporadic neoplasms, this MDM could potentially serve as a useful acquired somatic marker to identify LS associated colorectal neoplasia.
Supporting our earlier preliminary observation [29], we confirmed that the two MDMs (BMP3 and NDRG4) targeted in the MT-sDNA panel are significantly less discriminant for LS-related colorectal neoplasms than for sporadic ones. This difference between LS and sporadic groups was particularly marked with detection of adenomas, highlighting fundamental differences in pathway to CRC in LS and sporadic. In contrast, the novel MDMs described in the present study achieved high discrimination for neoplasia (advanced adenomas and CRCs) in both LS and sporadic colorectal tissues, and represent attractive targets for an early detection tool in LS patients that may complement current screening modalities. However, it should be emphasized that the MT-sDNA test also targets mutant KRAS and hemoglobin [25], and it cannot be concluded that these MDM findings would translate into reduced neoplasm detection by MT-sDNA in LS patients without prospective clinical observations on stool.
While this study focused on identification of candidate MDMs for detection of colorectal neoplasms in LS, this approach to marker discovery can be applied to other gastrointestinal and extra-intestinal organs that are at increased risk for cancer development in LS [33]. Testing a common distant medium, like stool or blood, to simultaneously screen multiple organs could have added value by aggregating tumor prevalence and increasing the efficiency and potential yield of a single screening intervention [59]. We have found highly discriminant MDMs through whole methylome discoveries in sporadic patients with pancreatic [33], esophageal [60], gastric [32], hepatic [61], endometrial [62] and ovarian (unpublished observations) neoplasms. Candidate MDMs from these studies and those of others could be tested in LS-associated tumors from these commonly involved organs to evaluate their discrimination.
Methodologic strengths of the present discovery and blinded validation study have been mentioned, but several study limitations merit acknowledgement. First, only FFPE tissues were available for the RRBS discovery in LS tissues. As formalin fixation damages DNA [63], the amount of high quality DNA with extraction is less than that obtained from frozen tissues. However, the none candidate MDMs from the LS discovery phase led to several highly discriminant MDMs on biological validation. MDM candidates carried over from the discovery effort in sporadic tissues were based on DNA extracted from frozen tissues [31,33]. MSP assays used in the validation phase tend to perform well on FFPE tissue based on our previous observations [31–33,60,61]. Second, not all patients studied had confirmation of diagnosis of LS by germline testing. Although genetic testing can have several advantages including avoiding unnecessary surveillance programs for non-carriers and for carriers the opportunity for tailored screening and surveillance recommendations, improving survival through early detection, there are differing perceptions and attitudes toward genetic testing. A study by Keogh et al. which aimed to identify factors affecting the decision to decline genetic testing, included individuals who had been offered genetic testing for LS or bi-allelic MUTYH mutations. Factors for declining genetic testing included lack of knowledge of the availability of genetic testing, lack of trust in genetic test information; a desire to see a stronger benefit from genetic testing before proceeding; and a sense that there may be more negative than positive outcomes from genetic testing [64]. Other studies have shown that the most common reasons for refraining from genetic testing include anticipating problems with life insurance and mortgage, not experiencing any physical complaints and being content with life as it is [65]. Our study was a retrospective study. All patient tissues from this study were obtained from Mayo Clinic archives. Therefore, we had to rely on data available in the medical record. Suspected LS patients, defined as having absence of MSH2, MSH6, PMS2, or EPCAM proteins on tumor IHC with no germline testing available to confirm a pathogenic variant in the MMR genes, were included in the study. It is possible that the source of defective MMR in these patients may be secondary to having two somatic pathogenic variants in any DNA MMR gene (Lynch-like syndrome) or rarely constitutional methylation of MLH1. Cases with only tissue IHC showing absence of MLH1 without germline testing were excluded, as it is possible that defective MMR may be secondary biallelic somatic methylation induced silencing of the MLH1 promoter. Third, since not all individuals underwent confirmation of LS diagnosis with germline testing, MDM results were not stratified by individual germline pathogenic variant. However, as discrimination for colorectal neoplasms was so universally high with the top MDM candidates across LS tissues, it is neither likely that specific germline subsets, nor ‘definite versus suspected’ status would have influenced central results. Fourth, few sessile serrated adenomas/polyps could be found in our archival LS tissues for use in validation. These lesions serve as important precursor lesions in sporadic CRC and have been shown to exfoliate MDMs in sporadic patients, leading to their detection in stool [27,66]. Several studies suggest that sessile serrated polyps are uncommon in LS and that traditional adenomas are the predominant precursor to CRC in LS [67,68]. Of note, we found that the top MDMs detected classical and sessile serrated adenomas with equally high discrimination in sporadic tissues and each of the two sessile serrated adenomas included in the LS group were detected as well. Fifth, MMR or IHC was not performed on adenomas and it is unknown whether these lesions are going through an MMR pathway. However, detection rate was high for adenomas and sporadic adenomas are important lesions to detect in Lynch patients. Finally, despite that the discriminatory accuracy of these MDMs was only validated in tissue, our findings are novel and lay the groundwork for a future prospective study with a larger number of LS cases and individuals with deficient MMR expression but lacking mutation (Lynch-like syndrome) to validate the performance of these markers in other biological samples such as stool.
Conclusion
We have identified novel MDMs that highly discriminate colorectal neoplasia in LS. These MDMs have potential for testing in media such as stool or blood for application in CRC screening or surveillance in this high-risk group. Larger prospective studies to validate these MDMs in media such as stool will first require targeted capture and assays to be developed, optimized and evaluated in carefully designed feasibility studies. Given the high risk of developing interval cancer among individuals with LS, availability of accurate noninvasive early detection tools could complement current screening and surveillance approaches with the potential to improve early detection and reduce morbidity and mortality from CRC.
Future perspective
In this tissue-based discovery and validation study we identified novel highly discriminant methylated markers for the detection of LS associated neoplasia. Once we validate these markers in other media such as stool in larger prospective studies, we may develop a panel of top performing markers. A Lynch specific marker panel could potentially be used to complement colonoscopy to optimize early detection of colorectal neoplasia and improve compliance in these high-risk patients. Furthermore, this approach to marker discovery can also be applied to other gastrointestinal and extra-intestinal organs that are at increased risk for cancer development in LS.
Summary points.
Without effective screening and surveillance, the lifetime risk of colorectal cancer (CRC) in individuals with Lynch syndrome (LS) ranges between 10 and 80% and differs based on specific mismatch repair alteration.
Accelerated progression through the adenoma-carcinoma sequence or carcinoma arising from non-neoplastic crypt foci occurs in LS due to mismatch repair deficiency and confers predisposition to CRC.
Patient compliance with colonoscopic surveillance is suboptimal.
For patients with hereditary risk for CRC, such as LS, there are no current options for effective non-invasive screening.
Accurate noninvasive tools may complement current screening and surveillance approaches with the potential to improve early detection and reduce morbidity and mortality.
Novel methylated DNA markers, specifically OPLAH, and methylated DNA marker combinations identified in this discovery and validation study achieved almost perfect discrimination of colorectal neoplasia across LS and sporadic tissues.
Aberrantly methylated ALKBH5, which was exclusively observed in LS neoplasms, could potentially serve as an acquired somatic marker to identify LS associated colorectal neoplasia.
Our findings are novel and lay the groundwork for a future larger prospective study to validate the performance of these markers in other biological samples such as stool.
Supplementary Material
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/epi-2020-0132
Financial & competing interests disclosure
Funding was provided by Eugene and Eva Lane, the Mayo Foundation, the Hispanic Clinical and Translational Research Education and Career Development (HCTRECD) Award R25MD007607 and Exact Sciences. Additional support was provided by CA214679 (to JBK). The Clinical Core of the Mayo Clinic for Cell Signaling in Gastroenterology (P30DK084567) provided a subset of the biospecimens. During course of this study and data analyses, DA Ahlquist & JB Kisiel, Messrs. Taylor & Mahoney and TC Yab were employees of Mayo Clinic and are listed as inventors in an intellectual property agreement between Mayo Clinic and Exact Sciences, under which they could receive royalties, in accordance with Mayo Clinic policy. DA Ahlquist currently serves as senior scientific advisor to Exact Sciences. GP Lidgard is Chief Science Officer at Exact Sciences. Exact Sciences provided partial funding and valuable assay reagents but played no role in the design, conduct, or data analyses for this study. CR Boland has given lectures for Ambry Genetics. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
- 1.Win AK, Jenkins MA, Dowty JG et al. Prevalence and penetrance of major genes and polygenes for colorectal cancer. Cancer Epidemiol Biomarkers Prev. 26(3), 404–412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boland PM, Yurgelun MB, Boland CR. Recent progress in Lynch syndrome and other familial colorectal cancer syndromes. CA Cancer J. Clin. 68(3), 217–231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinicrope FA. Lynch syndrome-associated colorectal cancer. N. Engl. J. Med. 379(8), 764–773 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Watson P, Vasen HFA, Mecklin JP et al. The risk of extra-colonic, extra-endometrial cancer in the Lynch syndrome. Int. J. Cancer 123(2), 444–449 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kastrinos F, Stoffel EM. History, genetics, and strategies for cancer prevention in Lynch syndrome. Clin. Gastroenterol. Hepatol. 12(5), 715–727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moller P, Seppala TT, Bernstein I et al. Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: a report from the Prospective Lynch Syndrome Database. Gut 67(7), 1306–1316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahadova A, Gallon R, Gebert J et al. Three molecular pathways model colorectal carcinogenesis in Lynch syndrome. Int. J. Cancer 143(1), 139–150 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Edelstein DL, Axilbund J, Baxter M et al. Rapid development of colorectal neoplasia in patients with Lynch syndrome. Clin. Gastroenterol. Hepatol. 9(4), 340–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giardiello FM, Allen JI, Axilbund JE et al. Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US multi-society task force on colorectal cancer. Am. J. Gastroenterol. 109(8), 1159–1179 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Baxter NN, Goldwasser MA, Paszat LF, Saskin R, Urbach DR, Rabeneck L. Association of colonoscopy and death from colorectal cancer. Ann. Intern. Med. 150(1), 1–8 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Engel C, Vasen HF, Seppala T et al. No difference in colorectal cancer incidence or stage at detection by colonoscopy among 3 countries with different Lynch syndrome surveillance policies. Gastroenterology 155(5), 1400–1409 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Kahi CJ, Hewett DG, Norton DL, Eckert GJ, Rex DK. Prevalence and variable detection of proximal colon serrated polyps during screening colonoscopy. Clin. Gastroenterol. Hepatol. 9(1), 42–46 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Kaminski MF, Regula J, Kraszewska E et al. Quality indicators for colonoscopy and the risk of interval cancer. N. Engl. J. Med. 362(19), 1795–1803 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Corley DA, Jensen CD, Marks AR et al. Adenoma detection rate and risk of colorectal cancer and death. N. Engl. J. Med. 370(14), 1298–1306 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giardiello FM, Allen JI, Axilbund JE et al. Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US multi-society task force on colorectal cancer. Dis. Colon. Rectum. 57(8), 1025–1048 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Johnson DH, Kisiel JB, Burger KN et al. Multitarget stool DNA test: clinical performance and impact on yield and quality of colonoscopy for colorectal cancer screening. Gastrointest. Endosc. 85(3), 657–665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishihara R, Wu K, Lochhead P et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N. Engl. J. Med. 369(12), 1095–1105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samadder NJ, Curtin K, Tuohy TM et al. Characteristics of missed or interval colorectal cancer and patient survival: a population-based study. Gastroenterology 146(4), 950–960 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Brenner H, Hoffmeister M, Arndt V, Stegmaier C, Altenhofen L, Haug U. Protection from right- and left-sided colorectal neoplasms after colonoscopy: population-based study. J. Natl. Cancer Inst. 102(2), 89–95 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Newton K, Green K, Lalloo F, Evans DG, Hill J. Colonoscopy screening compliance and outcomes in patients with Lynch syndrome. Colorectal. Dis. 17(1), 38–46 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Sahnane N, Magnoli F, Bernasconi B et al. Aberrant DNA methylation profiles of inherited and sporadic colorectal cancer. Clin. Epigenetics 7, 131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goel A, Xicola RM, Nguyen TP et al. Aberrant DNA methylation in hereditary nonpolyposis colorectal cancer without mismatch repair deficiency. Gastroenterology 138(5), 1854–1862 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaz A, Kim YH, Dzieciatkowski S et al. Evidence for the role of aberrant DNA methylation in the pathogenesis of Lynch syndrome adenomas. Int. J. Cancer 120(9), 1922–1929 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Chen CH, Sheng Jiang S, Hsieh LL et al. DNA Methylation identifies loci distinguishing hereditary nonpolyposis colorectal cancer without germ-line MLH1/MSH2 mutation from sporadic colorectal cancer. Clin. Transl. Gastroenterol. 7(12), e208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lidgard GP, Domanico MJ, Bruinsma JJ et al. Clinical performance of an automated stool DNA assay for detection of colorectal neoplasia. Clin. Gastroenterol. Hepatol. 11(10), 1313–1318 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Imperiale TF, Ransohoff DF, Itzkowitz SH et al. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 370(14), 1287–1297 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Redwood DG, Asay ED, Blake ID et al. Stool DNA Testing for screening detection of colorectal neoplasia in Alaska native people. Mayo. Clin. Proc. 91(1), 61–70 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Sweetser S, Ahlquist DA. Multi-target stool DNA test: is the future here? Curr. Gastroenterol. Rep. 18(6), 30 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Ballester V, Giakoumopoulos M, Yab TC et al. Sa1921 molecular detection of colorectal neoplasia: do markers that target acquired DNA alterations in sporadic cases also discriminate Lynch syndrome cases? Gastroenterology 148(4), S355 (2015). [Google Scholar]
- 30.Kisiel JB, Klepp P, Allawi HT et al. Analysis of DNA methylation at specific loci in stool samples detects colorectal cancer and high-grade dysplasia in patients with inflammatory bowel disease. Clin. Gastroenterol. Hepatol. 17(5), 914–921 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor WR, Kisiel JB, Yab TC et al. 109 Discovery of novel DNA methylation markers for the detection of colorectal neoplasia: selection by methylome-wide analysis. Gastroenterology 146(5), S30 (2014). [Google Scholar]
- 32.Anderson BW, Suh YS, Choi B et al. Detection of gastric cancer with novel methylated DNA markers: discovery, tissue validation, and pilot testing in plasma. Clin. Cancer Res. 24(22), 5724–5734 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kisiel JB, Raimondo M, Taylor WR et al. New DNA methylation markers for pancreatic cancer: discovery, tissue validation, and pilot testing in pancreatic juice. Clin. Cancer Res. 21(19), 4473–4481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MethPrimer2.0. (2020). http://www.urogene.org/methprimer2/ (Accessed 15 October 2015).
- 35.Zou H, Allawi H, Cao X et al. Quantification of methylated markers with a multiplex methylation-specific technology. Clin. Chem. 58(2), 375–383 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Brikun I, Nusskern D, Gillen D et al. A panel of DNA methylation markers reveals extensive methylation in histologically benign prostate biopsy cores from cancer patients. Biomark Res. 2(1), 25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaudhary A, Hilton MB, Seaman S et al. TEM8/ANTXR1 blockade inhibits pathological angiogenesis and potentiates tumoricidal responses against multiple cancer types. Cancer Cell 21(2), 212–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen YN, Chen H, Xu Y, Zhang X, Luo Y. Expression of pituitary homeobox 1 gene in human gastric carcinogenesis and its clinicopathological significance. World J. Gastroenterol. 14(2), 292–297 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 9(2), 124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamidov Z, Altendorf-Hofmann A, Chen Y, Settmacher U, Petersen I, Knosel T. Reduced expression of desmocollin 2 is an independent prognostic biomarker for shorter patients survival in pancreatic ductal adenocarcinoma. J. Clin. Pathol. 64(11), 990–994 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Knosel T, Chen Y, Hotovy S, Settmacher U, Altendorf-Hofmann A, Petersen I. Loss of desmocollin 1–3 and homeobox genes PITX1 and CDX2 are associated with tumor progression and survival in colorectal carcinoma. Int. J. Colorectal. Dis. 27(11), 1391–1399 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Kwok SC, Liu X, Mangel P, Daskal I. PTX1(ERGIC2)-VP22 fusion protein upregulates interferon-beta in prostate cancer cell line PC-3. DNA Cell Biol. 25(9), 523–529 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Hyde AS, Simpson MA, Barycki JJ. Emerging regulatory paradigms in glutathione metabolism. Adv. Cancer Res. 122, 69–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lord RV, Brabender J, Wickramasinghe K et al. Increased CDX2 and decreased PITX1 homeobox gene expression in Barrett's esophagus and Barrett's-associated adenocarcinoma. Surgery 138(5), 924–931 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Lu J, Ksendzovsky A, Yang C et al. CNTF receptor subunit alpha as a marker for glioma tumor-initiating cells and tumor grade: laboratory investigation. J. Neurosurg. 117(6), 1022–1031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakabayashi M, Osaki M, Kodani I et al. PITX1 is a reliable biomarker for predicting prognosis in patients with oral epithelial dysplasia. Oncol. Lett. 7(3), 750–754 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nikitenko LL, Leek R, Henderson S et al. The G-protein-coupled receptor CLR is upregulated in an autocrine loop with adrenomedullin in clear cell renal cell carcinoma and associated with poor prognosis. Clin. Cancer Res. 19(20), 5740–5748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nouguerede E, Berenguer C, Garcia S et al. Expression of adrenomedullin in human colorectal tumors and its role in cell growth and invasion in vitro and in xenograft growth in vivo. Cancer Med. 2(2), 196–207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Parris TZ, Aziz L, Kovacs A et al. Clinical relevance of breast cancer-related genes as potential biomarkers for oral squamous cell carcinoma. BMC Cancer 14, 324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qi DL, Ohhira T, Fujisaki C et al. Identification of PITX1 as a TERT suppressor gene located on human chromosome 5. Mol. Cell Biol. 31(8), 1624–1636 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramachandran V, Arumugam T, Hwang RF, Greenson JK, Simeone DM, Logsdon CD. Adrenomedullin is expressed in pancreatic cancer and stimulates cell proliferation and invasion in an autocrine manner via the adrenomedullin receptor, ADMR. Cancer Res 67(6), 2666–2675 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Sloane MA, Wong JW, Perera D et al. Epigenetic inactivation of the candidate tumor suppressor USP44 is a frequent and early event in colorectal neoplasia. Epigenetics 9(8), 1092–1100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei J, Li G, Zhang J et al. Integrated analysis of genome-wide DNA methylation and gene expression profiles identifies potential novel biomarkers of rectal cancer. Oncotarget 7(38), 62547–62558 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dixit D, Xie Q, Rich JN, Zhao JC. Messenger RNA methylation regulates glioblastoma tumorigenesis. Cancer Cell 31(4), 474–475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He L, Li J, Wang X et al. The dual role of N6-methyladenosine modification of RNAs is involved in human cancers. J. Cell Mol. Med. 22(10), 4630–4639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang S, Zhao BS, Zhou A et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31(4), 591–606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bjornstad LG, Meza TJ, Otterlei M, Olafsrud SM, Meza-Zepeda LA, Falnes PO. Human ALKBH4 interacts with proteins associated with transcription. PLoS ONE 7(11), e49045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fedeles BI, Singh V, Delaney JC, Li D, Essigmann JM. The AlkB family of Fe(II)/alpha-Ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 290(34), 20734–20742 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahlquist DA. Universal cancer screening: revolutionary, rational, and realizable. NPJ Precis. Oncol. 2, 23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin Y, Wu CW, Taylor WR et al. Discovery, validation, and application of novel methylated DNA markers for detection of esophageal cancer in plasma. Clin. Cancer Res. 25(24), 7396–7404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kisiel JB, Dukek BA, VSRK R et al. Hepatocellular carcinoma detection by plasma methylated DNA: discovery, Phase I pilot, and Phase II clinical validation. Hepatology 69(3), 1180–1192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bakkum-Gamez JN, Kisiel JB, Slettedahl SW et al. Discovery and validation of novel DNA methylation markers for the detection of endometrial cancer: selection by methylome-wide analysis. Gynecologic Oncol. 154, 71 (2019). [Google Scholar]
- 63.Peluso MEM, Munnia A, Tarocchi M et al. Oxidative DNA damage and formalin-fixation procedures. Toxicol. Res. 3(5), 341–349 (2014). [Google Scholar]
- 64.Keogh LA, Niven H, Rutstein A, Flander L, Gaff C, Jenkins M. Choosing not to undergo predictive genetic testing for hereditary colorectal cancer syndromes: expanding our understanding of decliners and declining. J. Behav. Med. 40(4), 583–594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leenen CH, Heijer M, Van Der Meer C, Kuipers EJ, Van Leerdam ME, Wagner A. Genetic testing for Lynch syndrome: family communication and motivation. Fam. Cancer 15(1), 63–73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heigh RI, Yab TC, Taylor WR et al. Detection of colorectal serrated polyps by stool DNA testing: comparison with fecal immunochemical testing for occult blood (FIT). PLoS One 9(1), e85659 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Andersen SH, Lykke E, Folker MB, Bernstein I, Holck S. Sessile serrated polyps of the colorectum are rare in patients with Lynch syndrome and in familial colorectal cancer families. Fam. Cancer 7(2), 157–162 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Snover DC. Update on the serrated pathway to colorectal carcinoma. Hum. Pathol. 42(1), 1–10 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.