Significance
T cells recognize their targets via their T-cell receptors (TCRs), which in the case of CD8+ T cells bind to MHC-I:antigen complexes on the surface of target cells. Many cancer cells evade immune recognition and killing by down-regulating MHC-I AgPPM. Here, we show how the histone acetyl transferases p300/CBP together with NF-κB epigenetically regulate expression of MHC-I molecules, immunoproteasome subunits, and peptide transporter to enable proper MHC-I antigen presentation. Notably, this pathway is frequently disrupted in human cancers. We now show that certain chemotherapeutics can augment MHC-I antigen presentation via NF-κB and p300/CBP activation, thereby enhancing cancer cell recognition and killing by effector CD8+ CTLs.
Keywords: histone acetylation, NF-κB, MHC-I, antigen presentation, immune checkpoint inhibitors
Abstract
Many cancers evade immune rejection by suppressing major histocompatibility class I (MHC-I) antigen processing and presentation (AgPP). Such cancers do not respond to immune checkpoint inhibitor therapies (ICIT) such as PD-1/PD-L1 [PD-(L)1] blockade. Certain chemotherapeutic drugs augment tumor control by PD-(L)1 inhibitors through potentiation of T-cell priming but whether and how chemotherapy enhances MHC-I–dependent cancer cell recognition by cytotoxic T cells (CTLs) is not entirely clear. We now show that the lysine acetyl transferases p300/CREB binding protein (CBP) control MHC-I AgPPM expression and neoantigen amounts in human cancers. Moreover, we found that two distinct DNA damaging drugs, the platinoid oxaliplatin and the topoisomerase inhibitor mitoxantrone, strongly up-regulate MHC-I AgPP in a manner dependent on activation of nuclear factor kappa B (NF-κB), p300/CBP, and other transcription factors, but independently of autocrine IFNγ signaling. Accordingly, NF-κB and p300 ablations prevent chemotherapy-induced MHC-I AgPP and abrogate rejection of low MHC-I–expressing tumors by reinvigorated CD8+ CTLs. Drugs like oxaliplatin and mitoxantrone may be used to overcome resistance to PD-(L)1 inhibitors in tumors that had “epigenetically down-regulated,” but had not permanently lost MHC-I AgPP activity.
Immune checkpoint inhibitor therapy (ICIT) had transformed cancer treatment (1–4), but even in ICIT-responsive metastatic melanoma and nonsmall cell lung cancer (NSCLC), response rates rarely exceed 40% (5). Other malignances, including prostate cancer (PCa) and pancreatic ductal adenocarcinoma (PDAC), are ICIT refractory (6–9). For a given neoplasm to respond to immune checkpoint inhibition, in particular PD-(L)1 blockade, it needs to be populated by cytotoxic T cells (CTLs) that recognize tumor antigens (4). However, even CTL-populated tumors can evade immune elimination either through activation of immunosuppressive mechanisms that induce CD8+ T-cell suppression or restrain their entry into tumors (10), down-regulation of major histocompatibility class I (MHC-I) AgPP (11, 12), or antigen editing and loss (13). Various strategies have been used to enhance ICIT responsiveness, including induction of immunogenic cells death (ICD) by radiotherapy and chemotherapy (14). By enhancing the release of damage associated molecular patterns (DAMPs) and other molecules, ICD stimulates tumor antigen uptake by antigen-presenting cells (APCs) that prime T cells against tumor antigens, as demonstrated by vaccination experiments (15). Primed T cells may accumulate in the tumor and lead to immune rejection as long as they can recognize and kill their targets (16). Such strategies are ineffective in cancers with low MHC-I or HLA-A/B/C expression (11, 16–19).
PCa is a typical ICIT-refractory cancer, presumably due to low expression of HLA-A/B/C molecules that together with β2 microglobulin form MHC-I heterodimers, which present tumor antigens to CD8+ CTL (20, 21). Using mouse models of PCa we found that the platin-based DNA-crosslinker oxaliplatin (Oxali) potentiates immune rejection of autochthonous or engrafted tumors after genetic or pharmacological depletion of PD-L1–expressing immunosuppressive IgA+ plasmocytes, which cause CTL exhaustion (22). Low-dose Oxali also enhances mouse PCa regression in response to anti–PD-L1 treatment (22). Similar results were obtained with low-dose Oxali or photodynamic therapy in other cancer models (23, 24) but the underlying mechanisms have not been explored. As Oxali is known to induce ICD and T-cell priming, we investigated whether its ability to potentiate the immune rejection of IgA+ plasmocyte-depleted or anti–PD-L1–treated low MHC-I prostate tumors also entails effects on the recognition and killing step of the cancer-immunity cycle, which depends on CTL–MHC-I interactions (16, 25). Here we show that Oxali and the structurally unrelated topoisomerase II inhibitor mitoxantrone (Mito) transcriptionally up-regulate expression of MHC-I molecules and their cognate antigen presentation and processing machinery (AgPPM). This response, which takes place in human and mouse cancers, depends on activation of nuclear factor kappa B (NF-κB) and nuclear translocation of the closely related histone (and lysine) acetyltransferases p300 and CREB binding protein (CBP). Whereas p300 ablation abrogated MHC-I AgPP induction and the synergy between low-dose Oxali and PD-(L)1 blockade, it had no effect on induction of antitumor immunity by Oxali-killed PCa cells used as an immunogen.
Results
Oxaliplatin and Mitoxantrone Induce MHC-I AgPPM Genes.
To determine the effect of Oxali and related drugs on gene expression in PCa cell lines used in our previous study (22), Myc-CaP cells were treated with different drugs at doses that induce no more than 10% cell death, and vital cells (SI Appendix, Fig. S1 A and B) were analyzed by whole genome RNA sequencing (RNA-seq) and assay for transposase-accessible chromatin (ATAC-seq) (26). Since CTL reinvigoration by anti–PD-L1 induces IFNγ production (27, 28), we also examined the effect of IFNγ alone or together with chemotherapy. Low-dose chemotherapy, in particular Oxali, induced marked changes in gene expression and chromatin accessibility depicted as differentially expressed genes (DEGs) and differentially accessible DNA regions (DARs) (Fig. 1A and SI Appendix, Fig. S1 C–F). The platinoid-induced changes were usually augmented by IFNγ, although the effects of Oxali were broader than that of IFNγ, which mainly enhanced gene expression magnitude rather than breadth. Some of the Oxali or IFNγ-inducible gene sets were common to both agents (Fig. 1A). Pathway enrichment analysis (Fig. 1B) identified the most significantly enriched pathways, activated by Oxali (red, e.g., epithelial-mesenchymal transition, TP53), IFNγ (blue, e.g., Myc), or Oxali + IFNγ together (purple, e.g., IFN type I and II, AgPPM). Notably, while either Oxali or IFNγ significantly enriched genes involved in MHC-I AgPP and IFNγ signaling, these effects were strongly enhanced when Oxali and IFNγ were combined (Fig. 1B and SI Appendix, Fig. S2 A–C). However, Oxali did not induce IFNγ expression, indicating that its ability to induce MHC-I AgPPM components was not due to autocrine IFNγ signaling.
Fig. 1.
Chemotherapy induces MHC-I AgPPM genes. (A) Heatmap showing all DEGs identified in bulk RNA-seq of Myc-CaP cells treated with IFNγ (0.2 or 2 ng/mL), Oxali (2 μM), or both (combo) (Left). Venn diagram shows overlapping DEGs between IFNγ (2 ng/mL), Oxali, and combo treatment groups relative to Ctrl (Middle) and heatmap shows all DARs identified in bulk ATAC-seq of Myc-CaP cells treated as indicated relative to control (Right). (B) Gene set enrichment analysis (GSEA) was applied to expression profiles specific to each treatment group relative to Ctrl. Top 30 significantly enriched pathways for each respective comparison are shown. Some pathways were considered both IFNγ and Oxali driven (purple), while others were specific to either IFNγ (blue) or Oxali (red). (C) Functional enrichment was applied to genes classified with additive response to combination therapy. The top 20 enriched REACTOME pathways are shown.
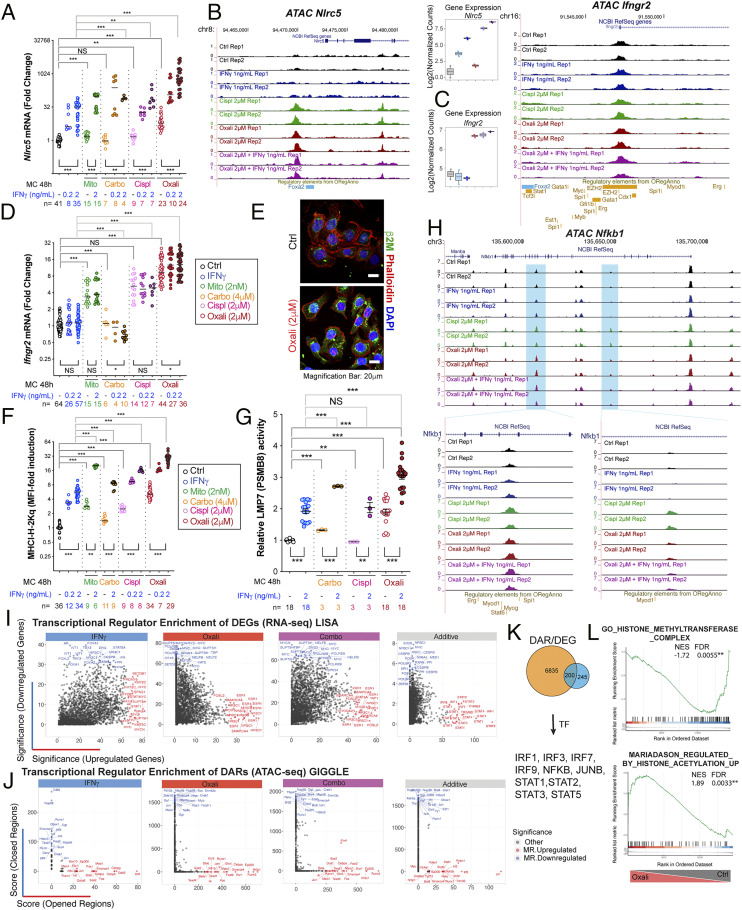
Pathway enrichment analysis of DEGs that were responsive to Oxali plus IFNγ revealed strong induction of genes related to type I and II IFN signaling and MHC-I AgPPM components, involved in protein folding, MHC-I complex assembly, and peptide loading, as well as genes involved in the endoplasmic reticulum (ER)-phagosome pathway and antiviral responses (Fig. 1C and SI Appendix, Fig. S2D). Most of these genes were also induced by Oxali alone. To understand how these genes were induced we examined the ATAC-seq patterns of a gene cluster on mouse chromosome 17 harboring the Psmb9, Tap1, Psmb8, and Tap2 genes, coding for immunoproteasome components and peptide transporters (SI Appendix, Figs. S1 E and F and S3A). Low-dose Oxali, and to a lesser extent cisplatin (Cispl), increased transcription factor (TF) accessibility at several sites within this locus (SI Appendix, Fig. S3A). Surprisingly, IFNγ alone had little effect, if any, on chromatin structure (SI Appendix, Fig. S1F). qRT-PCR analysis further confirmed induction of AgPPM genes by Oxali and Cispl and to a lesser extent by Mito, alone or together with IFNγ (SI Appendix, Fig. S3 B–D). A similar response pattern was displayed by the Nlrc5 gene coding for NLRC5/CITA, the master activator of the MHC-I AgPPM (Fig. 2A). ATAC-seq revealed increased Nlrc5 chromatin accessibility after low-dose Oxali or Cispl, but hardly any change after IFNγ treatment (Fig. 2B). Mito, Cispl, and Oxali, but not IFNγ, induced Ifngr2 mRNA, but had little effect on chromatin accessibility of its gene (Fig. 2 C and D). Of note, the chromosome 17 region opened up by Oxali contains binding motifs recognized by BORIS and CTCF (SI Appendix, Fig. S3A), general TF responsible for chromatin opening (29). Low-dose Oxali also increased β2 microglobulin (β2M), and all tested chemotherapeutics induced surface and mRNA expression of H-2Kq, the predominant MHC-I molecule in Myc-CaP cells (Fig. 2 E and F and SI Appendix, Fig. S3D). Low-dose Oxali increased immunoproteasome activity measured with an LMP7/PSMB8-specific substrate, Ac-ANW-AMC, an effect that was potentiated by IFNγ (Fig. 2G).
Fig. 2.
Transcriptional regulators of Oxali-induced MHC-I AgPPM genes. (A) RNAs from Myc-CaP cells incubated as indicated with IFNγ, Mito, Oxali, carboplatin (Carbo), or Cispl for 48 h were analyzed by qRT-PCR using Nlrc5 primers. (B and C) Candidate genomic loci for Nlrc5 (B) and Ifngr2 (C), showing library-size normalized read pair pileup profiles determined by ATAC-seq across samples. Expression of respective genes determined by RNA-seq is also shown. (D) RNAs from Myc-CaP cells incubated as indicated were analyzed by qRT-PCR using Ifngr2 primers. (E) Myc-CaP cells incubated with Oxali for 12 h were stained with β2M antibody (green) and phalloidin (red) and counterstained with DAPI. (Scale bar: 20 μm.) (F) Myc-CaP cells treated as indicated were analyzed for surface MHC-I (H-2Kq) expression by flow cytometry. (G) Myc-CaP cells were incubated as indicated and lysed. LMP7 (PSMB8) immunoproteasome activity was measured using a fluorogenic LMP7-specific substrate peptide Ac-ANW-AMC. (H) Candidate genomic locus for Nfkb1 showing read density profiles determined by ATAC-seq across samples. (I) LISA was applied to DEGs identified in comparisons of IFNγ, Oxali, and both (combo)-treated cells relative to control, as well as to genes classified with additive response (top 500 up-regulated, down-regulated DEGs). The top 20 enriched regulators of up-regulated (red) and down-regulated (blue) DEGs are noted. (J) GIGGLE applied to DARs identified in comparisons of IFNγ-, Oxali-, and combo-treated cells relative to control, as well as to regions classified with additive response. The top 20 enriched regulators of opened (red) and closed (blue) DARs are noted. (K) Two hundred common genes were identified by comparing TFs found by DEG and DAR analysis. (L) Gene set enrichment analysis (GSEA) was applied to expression profiles determined in Oxali-treated cells relative to control. The literature-curated known regulatory elements from ORegAnno database are shown. Candidate enrichment plots for representative pathways related to histone methylation (Top) and acetylation (Bottom) are shown. Two-sided t test (means ± SEM), and Mann–Whitney test (median) were used to determine significance between two groups. One-way ANOVA analysis (A, D, F, G) and multiple comparison confirmed the results. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. Specific n values are shown in A, D, F, and G, each experiment includes at least three biological replicates.
Putative Transcriptional Regulators of MHC-I AgPPM Induction.
We searched for signaling pathways and TF-mediating MHC-I AgPPM and IFNγR2 induction by low-dose Oxali. RNA-seq and pathway enrichment analyses suggested involvement of IRF, STAT, NF-κB, MYC family members, and androgen receptor (AR) (Fig. 2 H–K and SI Appendix, Fig. S3 E and F). Whereas the IRF, STAT, and NF-κB pathways were up-regulated by Oxali and potentiated by IFNγ, the MYC and to a lesser extent the AR pathway, both of which participate in PCa tumorigenesis (30–32), were down-regulated after Oxali + IFNγ treatment. Among IRF family members, IRF1, 7, and 9 were stimulated by Oxali and IFNγ, IRF2 was induced by Oxali, and IRF8 mainly responded to IFNγ (SI Appendix, Fig. S3F). Similarly, STAT1 and 2 were stimulated by Oxali, whereas IFNγ induced STAT1 and 3. JUN, ATF3, UBA7, CREB3, NFE2L1, and SOCS1 were induced by low-dose Oxali, along with NF-κB1 (p105) and NF-κB2 (p100) (SI Appendix, Fig. S3F). ATAC-seq confirmed that Oxali, but not IFNγ, enhanced chromatin accessibility of the Nfkb1 locus (Fig. 2H).
We employed two additional analytic approaches to identify master TF-mediating treatment-induced expression changes [LISA (33)] and TF binding enrichment within regions of differential chromatin accessibility [GIGGLE (34)]. These analyses predicted the master regulators (MRs) most likely to influence the DEGs (Fig. 2I) and DARs (Fig. 2J) by leveraging the complete set of TF binding datasets available from the CistromeDB collection. The results further highlighted treatment-related directional TF associations (up/down-regulated DEGs, open/closed DARs); Oxali: ESR1, NR3C1, and JUN; IFNγ: MYC, STAT1, ATF4, and FOS; Oxali + IFNγ: STAT1, YAP1, ESR1, IRF1, RELA, and IRF8 (Fig. 2 I and J). DEG and DAR integration revealed 200 common TFs, including IRFs and STATs (Fig. 2K). Notably, Oxali treatment elicited marked changes in histone methylation- and acetylation-related gene signatures (Fig. 2L) in agreement with the ATAC-seq data (SI Appendix, Fig. S1 E and F). Subset analyses focusing on histone modifying factors implicated the involvement of the histone acetyltransferases (HATs) p300 and CBP and several histone deacetylases (HDACs) (SI Appendix, Fig. S4A).
To confirm induction and/or activation of some of the above TFs, PCa (Myc-CaP, TRAMP-C2/TRC2), and colon cancer (MC38) cell lines were treated as above with or without IFNγ. Protein immunoblotting (IB) and flow cytometric analyses confirmed induction of ER stress (P-eIF2α, CHOP) and DNA damage (p-p53, γH2AX, and p-ATM) markers, IRFs (IRF1, p-IRF3, and IRF7), STATs (P-STAT1 and STAT1), CREB1, JUNB, type I IFN-inducing proteins (cGAS and STING), PSMB9, and NF-κB signaling components (IκBα, RELA/p65, and P-p65) (SI Appendix, Fig. S4 B–G).
Chemotherapy Stimulates HAT Nuclear Localization and Activity.
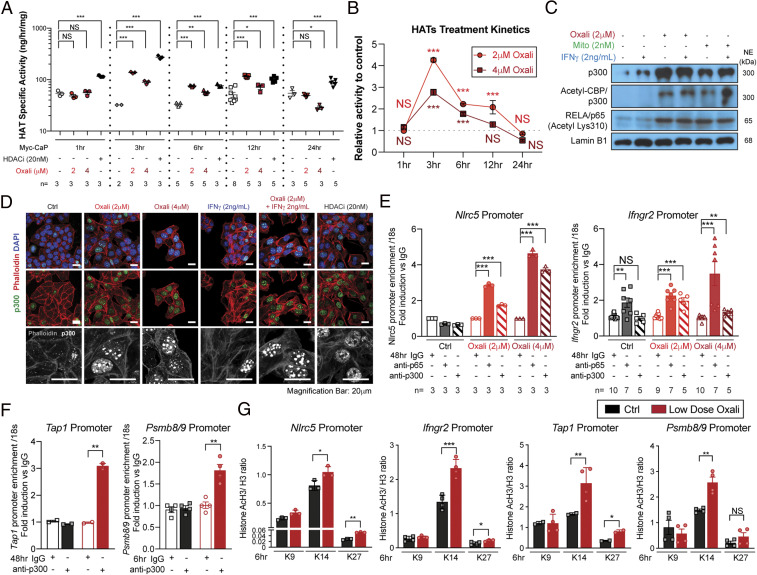
Chromatin structure opening, as revealed by ATAC-seq analysis, depends on histone acetylation (35). Importantly, Oxali treatment of Myc-CaP cells, increased HAT enzymatic activity within 3 h and its effect was comparable to that of a HDAC inhibitor (HDACi) (Fig. 3 A and B). Oxali and Mito also increased total p300, acetylated CBP/p300, and K310-acetylated RELA/p65 nuclear amounts (Fig. 3C). IFNγ also increased nuclear p300, but its effect was considerably weaker than that of Oxali (Fig. 3 C and D). Both Oxali and HDACi induced p300 nuclear translocation in murine PCa cells (Fig. 3D) and human PCa organoids (SI Appendix, Fig. S5A). Chromatin immunoprecipitation (ChIP) experiments showed that Oxali induced p300 and RELA/p65 recruitment to the Nlrc5 and Ifngr2 promoters and p300 recruitment to the Tap1 and Psmb8/9 promoters (Fig. 3 E and F). These promoter regions also exhibited increased H3K14 and K27 acetylation after Oxali treatment (Fig. 3G). Oxali-induced H3K14 acetylation at nuclear foci, similar to those revealed by p300 antibody staining, was also observed by immunofluorescence (IF) analysis (SI Appendix, Fig. S5B). Increased RELA/p65 K310 acetylation, which was attenuated after treatment by p300/CBP inhibitors, was confirmed by IB and IF analyses (SI Appendix, Fig. S5 C and D). Using HA- or Myc-tagged p300 and Flag-tagged Stat1 expression vectors followed by immunoprecipitation (IP), we confirmed binding of p300 to endogenous RELA/p65 and transfected STAT1 (SI Appendix, Fig. S5E), an interaction that stimulates p300 acetyltransferase activity (36). To investigate the basis for p300 nuclear translocation, we examined induction of HLA-B–associated transcript 3 gene product, BAT3, which controls intracellular p300 distribution (37). IF analysis confirmed Oxali-induced nuclear translocation of both p300 and BAT3 (SI Appendix, Fig. S5F).
Fig. 3.
Oxaliplatin and mitoxantrone stimulate HAT activity and nuclear localization. (A and B) Myc-CaP cells incubated with Oxali (2 or 4 μM) or the HDACi panobinostat (LBH589; 20 nM) for the indicated times were lysed and analyzed for HAT activity using H3 as a substrate. (C) Nuclear extracts of Myc-CaP cells treated with Oxali, Mito, and/or IFNγ were IB analyzed for p300, acetylated-CBP/p300, acetylated-RELA/p65 (lysine K310), and lamin B1 (loading control). (D) Myc-CaP cells treated as indicated for 12 h were stained with anti-p300 (green) and phalloidin (red; actin cables). Nuclei were counterstained with DAPI (blue). (Scale bar: 20 μm.) (E–G) Untreated and Oxali-treated Myc-CaP cells were subjected to ChIP analysis with control IgG and antibodies to p65/RelA, p300 as indicated (E and F) or acetylated H3 (lysine K9, K14, and K27) (G). Precipitation of the indicated promoter regions was determined by PCR. Two-sided t test (means ± SEM), and Mann–Whitney test (median) were used to determine significance. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. Specific n values are shown in A and E; each experiment includes at least three biological replicates.
p300 and CBP Control MHC-I AgPPM Expression and Neoantigen Amounts.
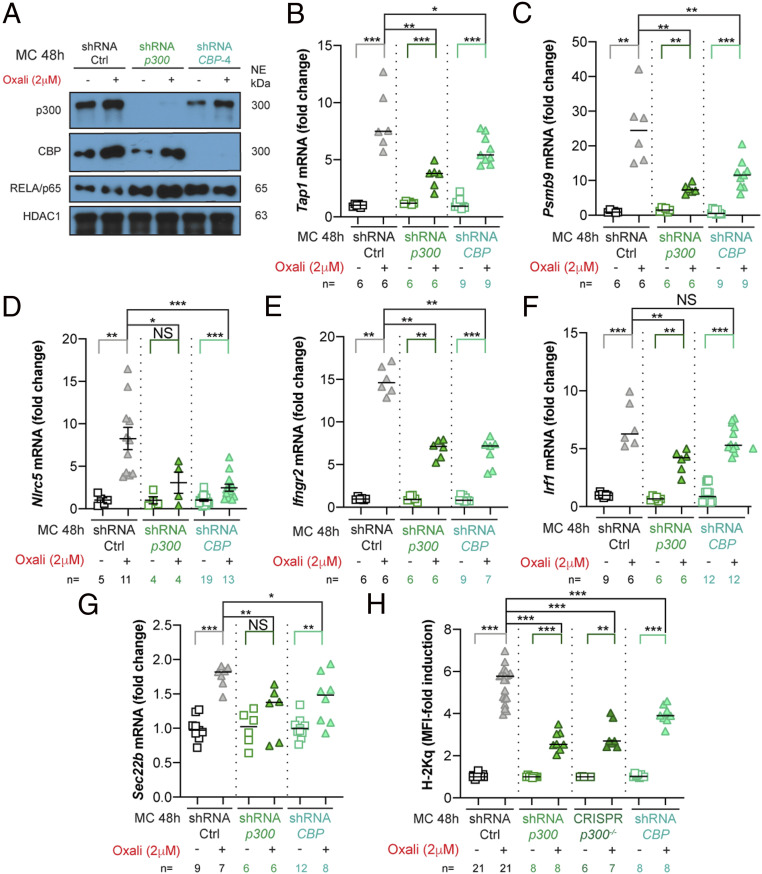
We generated cell lines deficient in p300 or CBP (Fig. 4A and SI Appendix, Fig. S6B). p300Δ-Myc-CaP cells expressed CBP and up-regulated its expression upon Oxali treatment, and CBPΔ-Myc-CaP cells behaved similarly. Notably, p300Δ-Myc-CaP cell viability did not differ from that of parental cells and neither p300 nor CBP ablation reduced total or Oxali-induced total RELA/p65 protein or Nfkbia mRNA (Fig. 4A and SI Appendix, Fig. S6 A–C). However, Tap1, Psmb9, Nlrc5, Infgr2, and Ifna mRNA inductions were attenuated by both p300 or CBP ablations, whereas Irf1 and Erap1 mRNA inductions were only reduced in p300Δ-Myc-CaP cells (Fig. 4 B–F and SI Appendix, Fig. S6 D and E). Both the p300 and CBP deficiencies attenuated induction of Sec22b mRNA (Fig. 4G), coding for a vesicle-trafficking protein that regulates phagosomal maturation and antigen cross-presentation (38). Consequently, both deficiencies hampered Oxali- and Mito-induced H-2Kq mRNA and surface expression (Fig. 4H and SI Appendix, Fig. S6 F and G). A p300/CBP inhibitor also attenuated H-2Kq protein, and Psmb9 and Tap1 mRNA inductions (SI Appendix, Figs. S6 H and I). Conversely, treatment of Myc-CaP cells with nonlethal doses of the HDACi panobinostat (LBH589) induced Nlrc5, Psmb9, and Tap1 mRNAs and surface H-2Kq (SI Appendix, Fig. S6 J–L).
Fig. 4.
p300 and CBP control Oxali-induced MHC-I AgPPM genes. (A) Parental (shRNA-Ctrl) and p300 or CBP silenced Myc-CaP cells were incubated with Oxali for 48 h. Nuclear extracts were IB analyzed for p300, CBP, p65/RELA, and HDAC1 (loading control). (B–G) RNA expression in above cells was analyzed by qRT-PCR with the indicated primers. (H) Parental and gene-edited Myc-CaP cells were incubated with Oxali and analyzed for surface MHC-I (H-2Kq) expression by flow cytometry. Two-sided t test (means ± SEM), and Mann–Whitney test (median) were used to determine significance unless indicated otherwise. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. Specific n values are shown in B–H. Each experiment includes at least three biological replicates.
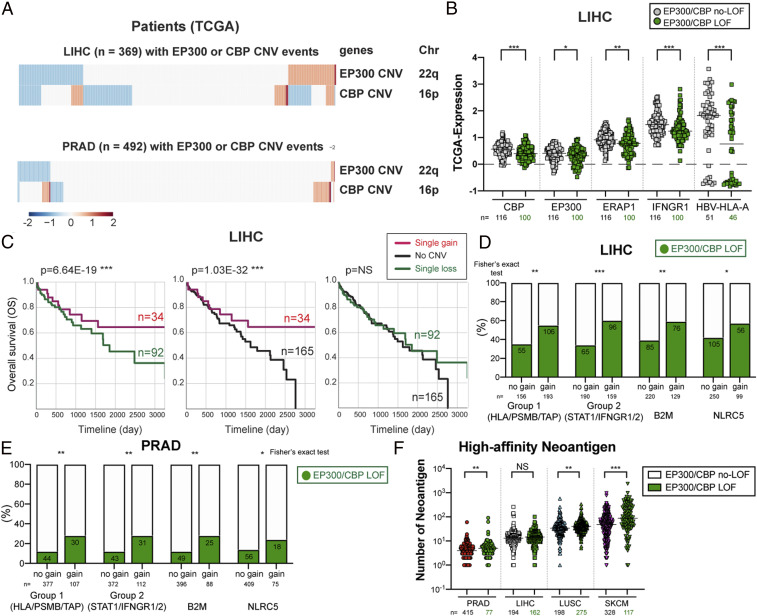
To gather information about p300 and CBP in human cancer, we examined The Cancer Genome Atlas (TCGA) dataset and found significant correlations between EP300 or CBP mRNAs and genes identified by our integrative RNA-seq and ATAC-seq analyses, including RELA, STAT1, NFKB1, IFNGR2, and NLRC5 (Fig. 5 A and B and SI Appendix, Fig. S7A). Similar correlations were found between histone modifiers and genes involved in MHC-I AgPP and T-cell inflammation, particularly in human liver cancer (SI Appendix, Fig. S7 B and C). Cancers with EP300/CBP loss of function (LOF; deletion and/or copy number variants [CNV] loss) showed lower ERAP1 and IFNGR1 or HLA-A expression (Fig. 5B). Curiously, EP300 and CBP were described both as oncogenes and oncosuppressors (39, 40). Phenotypes associated with heterozygous alterations were described in B cell lymphoma and Rubinstein-Taybi syndrome 1 (41, 42), suggesting dosage-dependent EP300/CBP function. We found that multiple CNVs, both gains and losses, affected EP300 and CBP in liver hepatocellular carcinoma (LIHC) and prostate adenocarcinoma (PRAD) (Fig. 5A and SI Appendix, Table S1) and correlated with their expression (SI Appendix, Fig. S7D). Interestingly, increased EP300 and CBP copy numbers correlated with improved LIHC patient survival (Fig. 5C). We also analyzed CNVs of genes involved in MHC-I AgPPM and IFNγ signaling (Fig. 5 D and E and SI Appendix, Fig. S8A and Table S1). We found that LIHC and PRAD patients with gains in MHC-I AgPPM genes showed more frequent EP300/CBP LOF (Fig. 5 D and E and SI Appendix, Table S1), suggesting a compensatory mechanism that allows cancers with elevated MHC-I AgPPM to evade immune recognition. We also analyzed the number and the fraction of neoantigens in different cancers (SI Appendix, Fig. S8 B and C). Remarkably, cancers with EP300/CBP LOF showed higher neoantigen amount (Fig. 5F and SI Appendix, Fig. S8D), supporting the notion that tumor immunoediting shapes the neoantigen landscape (43, 44) and that EP300/CBP may be part of this process.
Fig. 5.
p300/CBP control MHC-I AgPPM expression in human cancers. (A) EP300 and CBP CNV losses and gains in LIHC (n = 369) and PRAD (n = 492). Key: dark blue (homozygous deletion), light blue (one copy loss), white (no CNV), pink (one copy gain), and red (high amplification). (B) Comparison of immune gene expression between LIHC EP300/CBP LOF and non-LOF groups. Nonviral patients are included for comparison of CBP, EP300, ERAP1, and IFNGR1 expression. HLA-A expression was compared between EP300/CBP LOF and non-LOF HBV-infected groups. (C) Kaplan–Meier survival curves of LIHC patients with single gain (n = 34), no CNV (n = 165), and single loss (n = 92) events in EP300/CBP. P values are based on log rank test. (D and E) Enrichment comparison of LIHC (D) and PRAD (E) with or without EP300/CBP LOF or no CNV versus CNV gain in the indicated gene groups: MHC-I AgPPM (HLA/PSMBs/TAPs), STAT1/IFNGR1/IFNGR2, β2M, and NLRC5. Fisher’s exact test was used to determine significance. (F) Landscape of high-affinity neoantigens in four tumor types: PRAD (n = 492), LIHC (n = 356), lung squamous cell carcinoma (LUSC; n = 473), skin cutaneous melanoma (SKCM; n = 445). Groups were separated into EP300/CBP LOF (green dots) and non-LOF for neoantigen analysis. Mutations receiving a rank score of <2 and <0.5 were considered binding and strong binding, respectively. P values are based on Wilcoxon rank-sum test. Expression of neoantigens was determined from TCGA mRNA-seq reads. Two-sided t test (means ± SEM), and Mann–Whitney test (median) were used to determine significance unless indicated otherwise. One-way ANOVA analysis and multiple comparison confirmed the results. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. Specific n values are shown in A–F. Each experiment includes at least three biological replicates.
NF-κB Signaling and MHC-I AgPPM Induction.
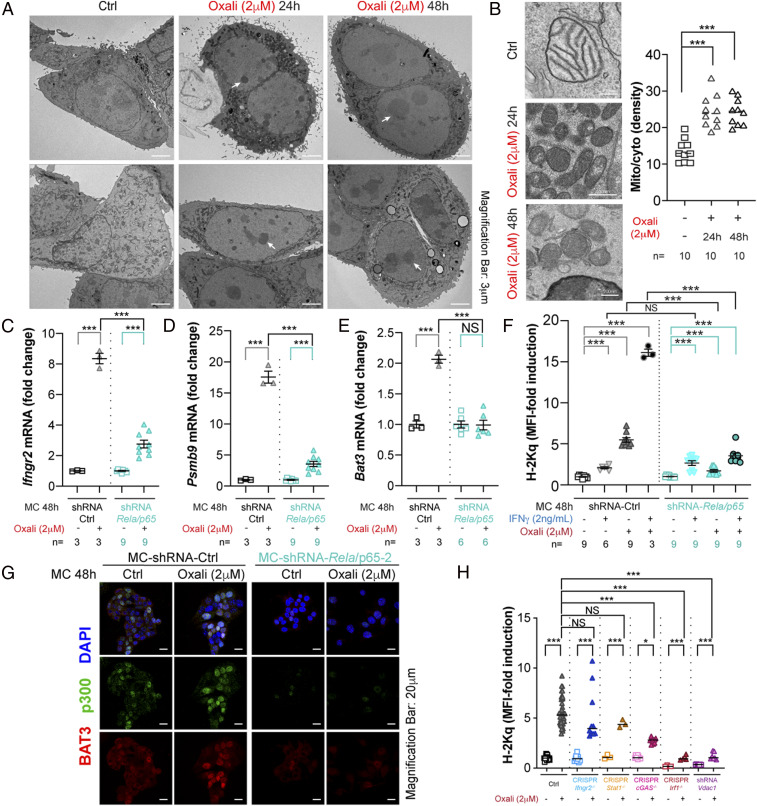
Electron microscopy (EM) suggested that Oxali-treated cells underwent nucleolar/ribosomal and mitochondrial stress indicated by the condensed appearance of both organelles (Fig. 6 A and B and SI Appendix, Fig. S9A). However, the absence of nuclear or mitochondrial fragmentation confirmed that most of the stressed cells remained viable. Oxali-induced nucleolar/ribosomal stress (45, 46), which was confirmed by mass spectrometry (MS) and RNA-seq analyses (SI Appendix, Fig. S9 B–E), can account for NF-κB activation (47). To determine NF-κB’s role in the response to Oxali, we generated RELA/p65-deficient cell lines (SI Appendix, Fig. S10 A and B). Consistent with the ChIP experiments shown above (Fig. 3E), RELA/p65 was needed for full induction of Ifngr2, Tap1, Psmab9, Nlrc5, and Bat3 mRNAs by low-dose Oxali (Fig. 6 C–E and SI Appendix, Fig. S10 C and D). RELA/p65 ablation strongly inhibited surface H-2Kq induction by Oxali but barely affected the response to IFNγ (Fig. 6F). BAT3 and p300 nuclear translocation was also attenuated in RELA-deficient cells (Fig. 6G and SI Appendix, Fig. S10E). Treatment of Myc-CaP cells with IKKβ inhibitors also reduced H-2Kq surface expression (SI Appendix, Fig. S10F). We also generated IRF1-, STING/cGAS-, STAT1-, IFNγR2-, and VDAC1-deficient Myc-CaP cells (Fig. 6H and SI Appendix, Fig. S11 A–E). VDAC1 (voltage-dependent anion channel 1) was recently shown to be required for the cytoplasmic release of mitochondrial (mt) DNA (48), which is considerably elevated in Oxali-stressed cells (SI Appendix, Fig. S11 E and F). Notably, VDAC-1 ablation strongly reduced Rela, p300, and Irf-1 mRNA induction by Oxali (SI Appendix, Fig. S11G). Ablation of VDAC1 and IRF1, but not STAT1 or IFNγR2, abrogated Oxali-induced expression of surface H-2Kq and MHC-I AgPPM genes (Fig. 6H and SI Appendix, Fig. S11 H–K). Ablation of cGAS led to a small decrease in H-2Kq expression and no effect on induction of most AgPPM genes (Fig. 6H and SI Appendix, Fig. S11 H–K). Not surprisingly, STAT1, IRF-1, and IFNγR2 as well as VDAC1 and cGAS were required for H-2Kq surface expression in Myc-CaP cells treated with IFNγ alone or IFNγ + Oxali (SI Appendix, Fig. S11L). Oxali treatment also led to modest induction of PD-L1, a response that was enhanced by exogenous IFNγ and was IRF1 dependent (SI Appendix, Fig. S11M), which has previously been shown to contribute to efficacy of ICIT (49). PD-L1 induction was not affected by TAP1 ablation, which completely prevented H-2Kq surface expression.
Fig. 6.
Role of NF-κB signaling in Oxali-induced MHC-I AgPPM expression. (A and B) Myc-CaP cells treated as indicated were fixed and examined by electron microscopy. Magnification bars are indicated in each image. White arrows indicate nucleolar stress. Ten representative images from each treatment group were analyzed and mitochondria/cytoplasm ratios were determined (B, Right). (C–E) Parental (shRNA-Ctrl) or Rela/p65-silenced Myc-CaP cells were incubated with Oxali as indicated. RNAs were analyzed by qRT-PCR with the indicated primers. The results were confirmed using three different Rela/p65 shRNAs. (F) Myc-CaP cells described in C were treated as indicated and analyzed for surface MHC-I (H-2Kq) expression. (G) Parental or Rela/p65-silenced Myc-CaP cells were treated as indicated and stained with mouse anti-p300 (green) and rabbit anti-BAT3 (red). Nuclei were counterstained with DAPI (blue) (n = 3). (Scale bar: 20 μm.) (H) Parental and gene edited (Ctrl, CRISPR-Cas9) or shRNA silenced Myc-CaP cells were treated as indicated and analyzed for surface MHC-I (H-2Kq) expression. (B, F, and H) Two-sided t test (means ± SEM), and Mann–Whitney test (median) were used to determine significance. One-way ANOVA and multiple comparisons were used to confirm significance. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. Specific n values are shown in B–F. Each experiment includes at least three biological replicates.
Chemotherapy-Induced Functional Antigen Presentation.
To confirm that Oxali stimulates neoantigen presentation, we used MS to determine the peptidomes of H-2Kb and H-2Db molecules isolated from MC-38 cells after treatments, as described previously (50). Treatment with IFNγ + Oxali induced higher amounts (based on area under the curve) of H-2Kb–bound peptides relative to Oxali or IFNγ alone (SI Appendix, Fig. S12A). Although IFNγ led to higher amounts of H-2Db–bound peptides than Oxali in this particular cell line, chosen for its high MHC-I expression, some peptides were more efficiently presented after IFNγ + Oxali treatment.
The T-cell activating ability of the Oxali-induced MHC-I bound peptides was confirmed using TRC2 PCa cells expressing high-, medium-, and low-affinity ovalbumin (Ova) variants. Oxali treatment stimulated H-2Kb presentation of the SIINFEKL epitope, especially in TRC2-N4 cells made to express the high-affinity (wild-type [WT]) variant (SI Appendix, Fig. S12B). When incubated with OT-I CD8+ T cells, whose T-cell receptor (TCR) is SIINFEKL specific, Oxali-treated TRC2-N4 cells were more readily killed by activated OT-I T cells (SI Appendix, Fig. S12 C and D). OT-I T cells enhanced presentation of the WT SIINFEKL epitope by TRC2-N4 cells in the absence of Oxali but had no effect on cells expressing the medium (TRC2-G4)- or low (TRC2-E1)–affinity variants. These results are consistent with a previous publication showing that only the high-affinity SIINFEKL epitope induces IFNγ secretion by OT-I cells (51), and further establish that the effect of Oxali is mechanistically distinct from that of IFNγ and dependent on neoantigen affinity and TCR activation.
We examined mouse and human cancer cell lines that differ in basal MHC-I expression. As described above, cells with high basal MHC-I such as MC-38 and B16 melanoma showed a weak response to platinoids alone but that response, including Nlrc5 mRNA and surface MHC-I, was augmented by IFNγ (SI Appendix, Fig. S13 A and B). In other cancer cells, e.g., the mouse melanoma YUMM cell lines, we observed a considerable variation in the response (SI Appendix, Fig. S13C). Strong Oxali-induced MHC-I surface expression was detected in the human PC3 PCa cell line, PCSD1 cells, a three-dimensional (3D) organoid culture from a patient-derived xenograft (PDX) of bone metastatic PCa, certain primary melanoma cells, and MIA PaCa-2 cells, representing ICIT-refractory PDAC (SI Appendix, Fig. S13 D–G).
Activation of p300/CBP and NF-κB Is Needed for Oxali + Anti–PD-L1 Synergy.
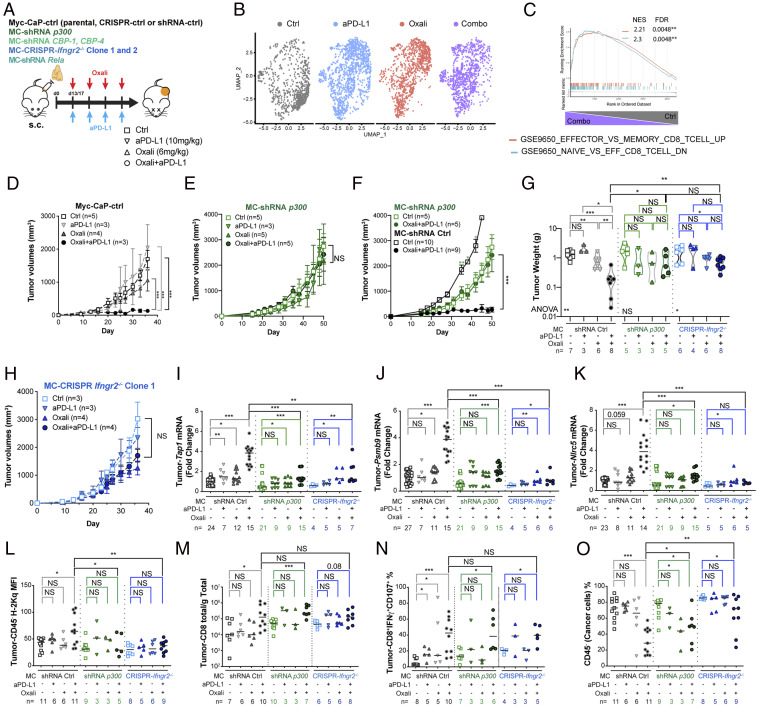
We sorted tumor-infiltrating CD8+ T cells (TI-CD8+) from subcutaneous (s.c.) Myc-CaP tumors, treated with either Oxali, anti–PD-L1, Oxali + anti–PD-L1 (combo), or left untreated (control [Ctrl]) and performed single-cell (sc)RNA-seq (Fig. 7A and SI Appendix, Fig. S14A). Several clusters of TI-CD8+ cells with distinguishable gene expression and cluster-specific pathway enrichment patterns were detected (Fig. 7B, and SI Appendix, Fig. S14 B–H). Notably, elevated Gzmb, Gzam, Prf1, and Tbx21(Tbet) mRNAs were detected in TI-CD8+ from combo-treated mice (SI Appendix, Fig. S14E). Only combo therapy was associated with a significantly higher Teff signature (Fig. 7C and SI Appendix, Fig. S14 F–H).
Fig. 7.
Oxali + anti–PD-L1 synergy depends on p300/CBP and IFNγR2 expression. (A) Schematic description of in vivo experiments. Mice bearing s.c. Myc-CaP tumors were allocated into four treatment groups: 1) control, 2) Oxali (6 mg/kg; weekly), 3) anti-PD-L1 (10 mg/kg; weekly), and 4) Oxali + anti–PD-L1 (weekly). After four treatment cycles, during which tumor size was measured, the mice were killed and analyzed. (B) Uniform manifold approximation and projection (UMAP) representation of total T-cell populations profiled by scRNA-seq. Eleven distinct clusters were identified (SI Appendix, Fig. S14 B and C). Proportional contributions of each cluster to sample-specific T-cell populations is shown. Total cell numbers in each cluster are noted in SI Appendix, Fig. S14D. (C) Enrichment plots for candidate pathways defining CD8+ Teff over Tmem and Tnaïve cells. Enrichment plots for the comparison between control and combo treatment is shown (other comparisons are shown in S14H). (D, F, and H) Mice bearing s.c. Myc-CaP tumors generated by control (D), p300-silenced (E and F), and Ifngr2 ablated (clone 1) (H) cells were treated as described in A. Specific n values are shown in each panel. Transient Cas9 expression and stable shRNA transfectants were used to avoid immune responses. Each dot is a treatment group, mean ± SEM. (G) Tumor weights for the indicated experimental groups (n = 3 to 8). (D and H) Two-sided t test (means ± SEM) and two-way ANOVA were used to determine significance. (I–K) Total tumor RNA was analyzed by qRT-PCR for expression of the indicated genes. (L–N) Single-cell tumor suspensions were analyzed for H-2Kq (L) expression on CD45− cells, total number of TI-CD8+ (M) and effector CD8+IFNγ+CD107+ T-cell subsets (N). (O) Solid tumor composition (% cancer cells) was determined by gating on CD45− cells. (G and I–O) Each dot represents a mouse. Mann–Whitney test (median) was used to determine significance. One-way ANOVA and multiple comparisons were used to confirm significance. Specific n values are shown in D–O.
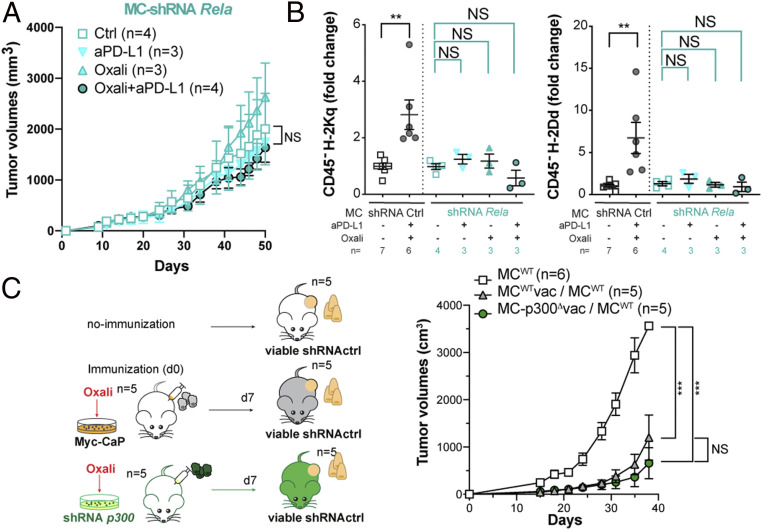
Next, we examined the involvement p300, CBP, IFNγR2, and NF-κB/RelA in Oxali-enhanced and CTL-mediated rejection of Myc-CaP tumors (Fig. 7A). The synergistic inhibition of tumor growth by Oxali + anti–PD-L1 was completely abrogated by p300 and CBP ablation in Myc-CaP cells (Fig. 7 D–G and SI Appendix, Fig. S15A). IFNγR2 ablation also abolished the response to Oxali + anti–PD-L1 (Fig. 7 G and H and SI Appendix, Fig. S15B). As found in vitro, low-dose Oxali induced expression of Ifngr2, Tap1, Psmb9, and Nlrc5 mRNA in Myc-CaP tumors (Fig. 7 I–K and SI Appendix, Fig. S15C). PD-L1 blockade did not affect Ifngr2 mRNA expression, although it potentiated Tap1, Psmb9, and Nlrc5 mRNA induction by Oxali, probably through IFNγ secretion by reinvigorated CTLs (Fig. 7 I–K and SI Appendix, Fig. S15C). Indeed, IFNγR2 ablation had little effect on the response to Oxali alone while abrogating the response to Oxali + anti–PD-L1. Tap1, Psmb9, and Nlrc5 induction by Oxali or Oxali + anti–PD-L1 was abrogated after p300 ablation (Fig. 7 I–K). IFNγR2 and p300 ablation also attenuated therapy-induced MHC-I (H-2Kq and H-2Dd) surface expression on CD45− cancer cells (Fig. 7L and SI Appendix, Fig. S15 D and E), but had no effect on PD-L1 expression (SI Appendix, Fig. S15F). IFNγR2 and p300 ablations also had no effect on H-2Kq expression by tumor-infiltrating CD11c+ dendritic cells (SI Appendix, Fig. S15G). In accordance with scRNA-seq data, the Oxali + anti–PD-L1 combo increased the percentage and/or total numbers of tumor-infiltrating CD8+ and CD4+ T cells, CD107+IFNγ+ CTLs, IFNγ+ CD8+, and TNF+IFNγ+ CD8+ T cells, CD8+CD44+ Teff cells, and CD8+CD44+PD1+TIM-3+ T cells analyzed by flow cytometry (Fig. 7 M and N and SI Appendix, Fig. S16 A and C–H). Similar results were obtained for splenic CD8+ T cells (SI Appendix, Fig. S16 B and I–K). Of note, p300 or IFNγR2 ablation had little effect on tumor-infiltrating effector CD8+ T cells, whose numbers were similarly increased after Oxali + anti–PD-L1 treatment in p300- or IFNγR2-expressing and nonexpressing tumors (Fig. 7 M and N and SI Appendix, Fig. S16 C–H). By contrast, the Oxali + anti–PD-L1 combo decreased the fractions of each tumor occupied by CD45− “cancer” cells, an effect that was most pronounced in WT tumors relative to p300 or IFNγR2 ablated tumors (Fig. 7O). NF-κB/RelA ablation also abolished the response to Oxali + anti–PD-L1 (Fig. 8A), consistent with its requirement for MHC-I and IFNγR2 induction (Fig. 8B and SI Appendix, Fig. S17A), Thus, Oxali-induced up-regulation of MHC-I AgPPM genes in malignant cells is important for the final recognition and killing stage of the cancer-immunity cycle (25) but has no role in ICIT-induced CTL reinvigoration.
Fig. 8.
Oxali-enhanced immune rejection requires NF-κB signaling. (A) Mice bearing s.c. Myc-CaP tumors generated by control and RelA-silenced cells were treated and analyzed as described in Fig. 7A. Each dot represents a treatment group mean ± SEM. (B) Single tumor cell suspensions were analyzed for H-2Kq (Left) and H-2Dd (Right) expression on CD45− cells. (C) Scheme of vaccination experiments (Left). Two groups of mice were immunized with lysates of Oxali-killed shRNA-ctrl (MCwt) or p300-silenced (MC-p300Δ) Myc-CaP cells. After 7 d, mice were s.c. inoculated with live shRNA-ctrl (MCwt) cells. Live shRNA-ctrl (MCwt) cells were also implanted into nonimmunized mice as a control. Tumor growth curves are shown (Right). (A–C) Two-sided t test (means ± SEM) and two-way ANOVA were used to determine significance. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. Specific n values are shown in A–C.
Of note, ICD-mediated T-cell priming proceeded normally in the absence of p300. FVB mice were immunized with Oxali-killed p300-proficient or -deficient Myc-CaP cells and challenged 1 wk later with vital p300-proficient or -deficient Myc-CaP cells (Fig. 8C and SI Appendix, Fig. S17B). Myc-CaP cells grew significantly slower in FVB mice immunized with either p300-proficient or -deficient Myc-CaP cells compared to nonvaccinated mice, indicating that p300 has no effect on ICD-mediated T-cell priming.
Discussion
ICIT-induced tumor rejection depends on activation of the cancer-immunity cycle, initiated by priming of tumor-directed T cells and terminated by killing of the targeted cancer cells by effector CTLs (16, 25). T-cell priming can be enhanced by certain chemotherapeutic drugs capable of inducing ICD (14) and ICIT (4, 27). Nonetheless, even highly effective T-cell priming and ICIT do not ensure successful CTL-mediated tumor killing, which requires MHC-I–mediated presentation of tumor-specific antigens (2, 14, 17, 52). Many cancers, especially PCa (SI Appendix, Fig. S7E) (53), evade immune elimination by down-regulating MHC-I molecules or essential AgPPM components (11). Here we show that two different chemotherapeutic drugs, Oxali and Mito used at rather low concentrations, enhance CTL-mediated cancer cell recognition and killing through transcriptional induction of MHC-I AgPP (schematic summary, SI Appendix, Fig. S17C). Although induction of MHC-I antigen presentation by chemotherapy and radiotherapy was described (54–56), the underlying mechanisms were only partly explored and attributed to type I IFN signaling. However, recent reports showing that sustained type I IFN signaling contributes to anti–PD-(L)1 resistance (57, 58) cast doubt on the role played by type I IFN in chemotherapy- or radiotherapy-induced immune stimulation. Our results show that Oxali renders low MHC-I–expressing PCa cells responsive to anti–PD-(L)1 therapy through transcriptional activation of the MHC-I AgPPM by NF-κB and p300/CBP, but not via the IFN-responsive TF STAT1. Ablation of p300 (or CBP) or NF-κB/RelA abolished the ability of low-dose Oxali to synergize with anti–PD-L1 and induce rejection of Myc-CaP tumors. Consistent with their direct involvement in transcriptional activation of MHC-I AgPPM genes, ablation of p300 or RelA abrogated Tap1, Psmb9, and Nlrc5 induction in Myc-CaP tumors, but had no effect on tumor infiltration by effector CD8+ cells. Tumor-infiltrating CTLs, however, were strongly increased after anti–PD-L1 + Oxali treatment as indicated by scRNA-seq and flow cytometry. In contrast, ablation of p300 had no effect on the ability of Oxali-killed Myc-CaP cells to prime antitumor immunity, as indicated by vaccination experiments.
Oxali treatment triggers nucleolar/ribosomal stress (45, 46), possibly through its preferential interaction with rDNA or inhibition of rRNA synthesis, which represents almost half of the human genome (59). By virtue of its highly repetitive nature, rRNA integrity and expression are also sensitive to loss of topoisomerase II activity (60), a sequelae of Mito treatment. EM analysis of Oxali-treated Myc-CaP cells confirmed altered nucleolar morphology, consistent with nucleolar stress, which can trigger NF-κB activation (47). By inducing Bat3 transcription, RelA/NF-κB supports p300/CBP nuclear translocation, further increasing its own activity and stimulating histone acetylation. Oxali treatment can also enhance NF-κB activity via mitochondrial stress, whose presence in Myc-CaP cells is suggested by increased mitochondrial density and appearance of fragmented mtDNA in the cytosol. Ablation of VDAC1, through which mtDNA exits the mitochondrion (48), reduced p300 and Rela mRNA expression and abrogated induction of NLRC5 and different MHC-I AgPPM components. NF-κB is also needed for induction of IFNγR2. Although IFNγR2 ablation had no effect on Oxali-induced MHC-I surface expression in cultured cells, it abrogated the rejection of Myc-CaP tumors, and inhibited induction of MHC-I AgPPM genes in mice treated with anti–PD-L1 + Oxali. We postulate that IFNγR2 induction in Myc-CaP cells makes them more responsive to IFNγ secreted by tumor-infiltrating CTLs.
Neither Oxali nor Mito were developed as immunostimulatory drugs. It is therefore understandable that their immunogenic activity depends on multiple signaling pathways that are activated on induction of sublethal DNA damage and nucleolar and mitochondrial stress. Given the number of different signaling pathways activated by Oxali or Mito, it is rather surprising that ablation of either p300/CBP or RelA results in almost complete inhibition of the drug-induced immunogenic response. These findings parallel the cardinal importance of p300/CBP and NF-κB in activation of the MHC-I AgPP system. p300 plays a key role in assembly of the NLRC5 transcriptional activation complex and NF-κB recruitment to MHC-I genes (61). Notably, down-regulation of NLRC5 has been observed in multiple cancer types, resulting in evasion of immune elimination (12). Conversely, we found that cancers with NLRC5 gain were more likely to undergo CPB/EP300 loss. We also found that EP300 and CBP LOF mutations and CNVs are rather common in certain types of cancer, and that their presence correlates with reduced MHC-I AgPPM expression. These genetic alterations seem more common than HLA loss mutations. Moreover, hepatocellular carcinoma and PCa with gain of HLA, PSMB, and TAP genes, possibly due to chromosome 6p amplification (62), show higher frequency of CBP/EP300 loss, which may allow them to undergo immune evasion. Based on its loss in several types of cancer, EP300 was suggested to behave as a tumor suppressor gene (63, 64). We suggest that CBP/EP300 loss promotes tumor growth by enabling immune evasion. One way to restore recognition of tumors with monoallelic EP300/CBP loss is treatment with low-dose Oxali or Mito or more potent and specific EP300/CBP activators.
Materials and Methods
Detailed information about the animal models, in vivo and in vitro studies, flow cytometry, qRT-PCR, immunoblot analysis, bioinformatic analysis, statistics, and materials is provided in SI Appendix, Material and Methods and Table S2.
Supplementary Material
Acknowledgments
We thank N. T. Ryujin, A. Perkins, C. R. Lichtenstern, T. Deerinck, M. Mackey, D. T. Tam, S. Lee, E. Sanchez-Lopez, S. Pandit, K. Wong, and M. Muldong for technical support and help; and A. Birmingham, L. Delamarre, E. Fokas, and C. M. Rödel for discussions and advice. S.S. was supported by Prostate Cancer Foundation Young Investigator Award, Merck Investigator Studies Program 57917, and the National Institute on Alcohol Abuse and Alcoholism funded Southern California Research Center for Alcoholic Liver and Pancreatic Diseases & Cirrhosis (P50 AA011999). Work in the M.K. laboratory was supported by grants from the NIH (AI043477, CA128814, and CA211794), the Tower Cancer Research Foundation, San Diego National Cancer Institute’s Cancer Centers Council (C3), and Padres Pedal the Cause No. PTC2018. Additional support came from U01AA027681 (to S.S. and M.K.), P01 CA128814 (to M.K./Ze’ev Ronai [Cancer Center, Sanford Burnham Prebys Medical Discovery Institute] and M.B.), U24CA232979 (to S.L.), DP5‐OD017937 (to H.C.), 111 project (B16021) (to X.-J.L.), NIH National Library of Medicine Training Grant T15LM011271 (to M.D.), NIH Grant UL1TR001442 of Clinical and Translational Science Awards (to K.M.F.), NIH P41GM103412, R24GM137200, and S10OD021784 (to M.H.E.), Mary Kay Ash Breast Cancer Grant 047.16, and Moore Cancer Center award (to S.B. and J. E. Dixon [Department of Pharmacology, University of California San Diego]).
Footnotes
Competing interest statement: I.M., J.R.L., R.B., Q.T.P. and G.H. are employees of Genentech.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025840118/-/DCSupplemental.
Data Availability
This study did not generate new unique materials. The sequencing data are available in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) database: mouse ATAC-seq (GSE126287) and RNA-seq (GSE126274). Data from in vitro experiments are available under the GEO accession no. GSE126288. Single-cell RNA-seq data from live CD8+CD3+ tumor-infiltrating cells are available under the GEO accession no. GSE151611. The results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga.
Change History
March 2, 2021: The license for this article has been updated.
References
- 1.Eggermont A. M. M., et al., Adjuvant pembrolizumab versus placebo in resected stage III melanoma. N. Engl. J. Med. 378, 1789–1801 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Gandhi L.et al.; KEYNOTE-189 Investigators , Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N. Engl. J. Med. 378, 2078–2092 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Hugo W., et al., Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165, 35–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Havel J. J., Chowell D., Chan T. A., The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 19, 133–150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conforti F., et al., Cancer immunotherapy efficacy and patients’ sex: A systematic review and meta-analysis. Lancet Oncol. 19, 737–746 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Guo S., Contratto M., Miller G., Leichman L., Wu J., Immunotherapy in pancreatic cancer: Unleash its potential through novel combinations. World J. Clin. Oncol. 8, 230–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonarakis E. S., et al., Pembrolizumab for treatment-refractory metastatic castration-resistant prostate cancer: Multicohort, open-label phase II KEYNOTE-199 study. J. Clin. Oncol. 38, 395–405 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hossain M. K., Nahar K., Donkor O., Apostolopoulos V., Immune-based therapies for metastatic prostate cancer: An update. Immunotherapy 10, 283–298 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Beer T. M., et al., Randomized, double-blind, phase III trial of ipilimumab versus placebo in asymptomatic or minimally symptomatic patients with metastatic chemotherapy-naive castration-resistant prostate cancer. J. Clin. Oncol. 35, 40–47 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Mariathasan S., et al., TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dersh D., Hollý J., Yewdell J. W., A few good peptides: MHC class I-based cancer immunosurveillance and immunoevasion. Nat. Rev. Immunol., 10.1038/s41577-020-0390-6 (2020). [Google Scholar]
- 12.Yoshihama S., et al., NLRC5/MHC class I transactivator is a target for immune evasion in cancer. Proc. Natl. Acad. Sci. U.S.A. 113, 5999–6004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumacher T. N., Scheper W., Kvistborg P., Cancer neoantigens. Annu. Rev. Immunol. 37, 173–200 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Galluzzi L., Buqué A., Kepp O., Zitvogel L., Kroemer G., Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 17, 97–111 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Galluzzi L., et al., Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer 8, e000337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen D. S., Mellman I., Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Chowell D., et al., Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy. Nat. Med. 25, 1715–1720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gettinger S., et al., Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov. 7, 1420–1435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Šmahel M., PD-1/PD-L1 blockade therapy for tumors with downregulated MHC class I expression. Int. J. Mol. Sci. 18, 1331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vitkin N., Nersesian S., Siemens D. R., Koti M., The tumor immune contexture of prostate cancer. Front. Immunol. 10, 603 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cresswell P., Ackerman A. L., Giodini A., Peaper D. R., Wearsch P. A., Mechanisms of MHC class I-restricted antigen processing and cross-presentation. Immunol. Rev. 207, 145–157 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Shalapour S., et al., Immunosuppressive plasma cells impede T-cell-dependent immunogenic chemotherapy. Nature 521, 94–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He C., et al., Core-shell nanoscale coordination polymers combine chemotherapy and photodynamic therapy to potentiate checkpoint blockade cancer immunotherapy. Nat. Commun. 7, 12499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfirschke C., et al., Immunogenic chemotherapy sensitizes tumors to checkpoint blockade therapy. Immunity 44, 343–354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen D. S., Mellman I., Oncology meets immunology: The cancer-immunity cycle. Immunity 39, 1–10 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Buenrostro J. D., Wu B., Chang H. Y., Greenleaf W. J., ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei S. C., Duffy C. R., Allison J. P., Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 8, 1069–1086 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Karachaliou N., et al., Interferon gamma, an important marker of response to immune checkpoint blockade in non-small cell lung cancer and melanoma patients. Ther. Adv. Med. Oncol. 10, 1758834017749748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hore T. A., Deakin J. E., Marshall Graves J. A., The evolution of epigenetic regulators CTCF and BORIS/CTCFL in amniotes. PLoS Genet. 4, e1000169 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernard D., Pourtier-Manzanedo A., Gil J., Beach D. H., Myc confers androgen-independent prostate cancer cell growth. J. Clin. Invest. 112, 1724–1731 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koh C. M., et al., MYC and prostate cancer. Genes Cancer 1, 617–628 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinlein C. A., Chang C., Androgen receptor in prostate cancer. Endocr. Rev. 25, 276–308 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Qin Q., et al., Lisa: Inferring transcriptional regulators through integrative modeling of public chromatin accessibility and ChIP-seq data. Genome Biol. 21, 32 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janky R., et al., iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol. 10, e1003731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shahbazian M. D., Grunstein M., Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Ortega E., et al., Transcription factor dimerization activates the p300 acetyltransferase. Nature 562, 538–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sebti S., et al., BAT3 modulates p300-dependent acetylation of p53 and autophagy-related protein 7 (ATG7) during autophagy. Proc. Natl. Acad. Sci. U.S.A. 111, 4115–4120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cebrian I., et al., Sec22b regulates phagosomal maturation and antigen crosspresentation by dendritic cells. Cell 147, 1355–1368 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Goodman R. H., Smolik S., CBP/p300 in cell growth, transformation, and development. Genes Dev. 14, 1553–1577 (2000). [PubMed] [Google Scholar]
- 40.Iyer N. G., Özdag H., Caldas C., p300/CBP and cancer. Oncogene 23, 4225–4231 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Roelfsema J. H., et al., Genetic heterogeneity in Rubinstein-Taybi syndrome: Mutations in both the CBP and EP300 genes cause disease. Am. J. Hum. Genet. 76, 572–580 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J., et al., The Crebbp acetyltransferase is a haploinsufficient tumor suppressor in B cell lymphoma. Cancer Discov. 7, 322–337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marty R., et al., MHC-I genotype restricts the oncogenic mutational landscape. Cell 171, 1272–1283.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castro A., et al., Elevated neoantigen levels in tumors with somatic mutations in the HLA-A, HLA-B, HLA-C and B2M genes. BMC Med. Genomics 12, 107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bruno P. M., et al., A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat. Med. 23, 461–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ozdian T., et al., Proteomic profiling reveals DNA damage, nucleolar and ribosomal stress are the main responses to oxaliplatin treatment in cancer cells. J. Proteomics 162, 73–85 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Chen J., Stark L. A., Insights into the relationship between nucleolar stress and the NF-κB pathway. Trends Genet. 35, 768–780 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Kim J., et al., VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science 366, 1531–1536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin H., et al., Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J. Clin. Invest. 128, 805–815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yadav M., et al., Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Denton A. E., et al., Affinity thresholds for naive CD8+ CTL activation by peptides and engineered influenza A viruses. J. Immunol. 187, 5733–5744 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Langer C. J.et al.; KEYNOTE-021 investigators , Carboplatin and pemetrexed with or without pembrolizumab for advanced, non-squamous non-small-cell lung cancer: A randomised, phase 2 cohort of the open-label KEYNOTE-021 study. Lancet Oncol. 17, 1497–1508 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ylitalo E. B., et al., Subgroups of castration-resistant prostate cancer bone metastases defined through an inverse relationship between androgen receptor activity and immune response. Eur. Urol. 71, 776–787 (2017). [DOI] [PubMed] [Google Scholar]
- 54.de Biasi A. R., Villena-Vargas J., Adusumilli P. S., Cisplatin-induced antitumor immunomodulation: A review of preclinical and clinical evidence. Clin. Cancer Res. 20, 5384–5391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khallouf H., et al., 5-Fluorouracil and interferon-α immunochemotherapy enhances immunogenicity of murine pancreatic cancer through upregulation of NKG2D ligands and MHC class I. J. Immunother. 35, 245–253 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Wan S., et al., Chemotherapeutics and radiation stimulate MHC class I expression through elevated interferon-beta signaling in breast cancer cells. PLoS One 7, e32542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J., et al., Type I IFN protects cancer cells from CD8+ T cell-mediated cytotoxicity after radiation. J. Clin. Invest. 129, 4224–4238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacquelot N., et al., Sustained Type I interferon signaling as a mechanism of resistance to PD-1 blockade. Cell Res. 29, 846–861 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warmerdam D. O., Wolthuis R. M. F., Keeping ribosomal DNA intact: A repeating challenge. Chromosome Res. 27, 57–72 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.French S. L., et al., Distinguishing the roles of Topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol. Cell. Biol. 31, 482–494 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meissner T. B., et al., NLRC5 cooperates with the RFX transcription factor complex to induce MHC class I gene expression. J. Immunol. 188, 4951–4958 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santos G. C., Zielenska M., Prasad M., Squire J. A., Chromosome 6p amplification and cancer progression. J. Clin. Pathol. 60, 1–7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Attar N., Kurdistani S. K., Exploitation of EP300 and CREBBP lysine acetyltransferases by cancer. Cold Spring Harb. Perspect. Med. 7, a026534 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gayther S. A., et al., Mutations truncating the EP300 acetylase in human cancers. Nat. Genet. 24, 300–303 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate new unique materials. The sequencing data are available in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) database: mouse ATAC-seq (GSE126287) and RNA-seq (GSE126274). Data from in vitro experiments are available under the GEO accession no. GSE126288. Single-cell RNA-seq data from live CD8+CD3+ tumor-infiltrating cells are available under the GEO accession no. GSE151611. The results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga.