Significance
Dermatitis is a major inflammatory skin condition that is associated with pruritus and affects millions of people worldwide. However, there is still little knowledge about the contribution of the cutaneous sensory neurons to the chronic disease. Here we show that chronic, but not acute, exposure to allergens/pruritogens induces robust itch in the dermatitis-prone Grhl3PAR2/+ transgenic mouse line, in part due to profound transcriptional changes that are induced in trigeminal ganglion neurons that innervate the allergen-treated skin. Most upregulated genes were concentrated in unmyelinated primary afferents that express TRPV1, a known pruritoceptor. As comparable transcriptional changes were recorded in a second dermatitis model, we conclude that a common genetic priming program in sensory neurons contributes to dermatitis.
Keywords: itch, dermatitis, trigeminal neurons, PAR2, RNA sequencing
Abstract
Pruritus is a common symptom of inflammatory skin conditions, including atopic dermatitis (AD). Although primary sensory neurons that transmit pruritic signals are well-cataloged, little is known about the neuronal alterations that occur as a result of skin disruption in AD. To address this question, we examined the molecular and behavioral consequences of challenging Grhl3PAR2/+ mice, which overexpress PAR2 in suprabasal keratinocytes, with serial topical application of the environmental allergen house dust mite (HDM). We monitored behavior and used RNA sequencing, qPCR, and in situ hybridization to evaluate gene expression in trigeminal ganglia (TG), before and after HDM. We found that neither Grhl3PAR2/+ nor wild-type (WT) mice exhibited spontaneous scratching, and pruritogen-induced acute scratching did not differ. In contrast, HDM exacerbated scratching in Grhl3PAR2/+ mice. Despite the absence of scratching in untreated Grhl3PAR2/+ mice, several TG genes in these mice were up-regulated compared to WT. HDM treatment of the Grhl3PAR2/+ mice enhanced up-regulation of this set of genes and induced additional genes, many within the subset of TG neurons that express TRPV1. The same set of genes was up-regulated in HDM-treated Grhl3PAR2/+ mice that did not scratch, but at lesser magnitude. Finally, we recorded comparable transcriptional changes in IL31Tg mice, demonstrating that a common genetic program is induced in two AD models. Taken together, we conclude that transcriptional changes that occur in primary sensory neurons in dermatitis-susceptible animals underlie a genetic priming that not only sensitizes the animal to chronic allergens but also contributes to pruritus in atopic skin disease.
Itch is a major criterion for clinical diagnosis of atopic dermatitis (AD), a chronic inflammatory skin condition with complex genetic and environmental underpinnings (1–3). In adults with AD, skin barrier disruption, exposure to environmental allergens, and other stressors elicit pruritus flares (4). Among the major environmental allergens implicated in atopic disease exacerbations (5) are the common house dust mites (HDM) Dermatophagoides pteronyssinus and Dermatophagoides farinae. HDM protein detection in mattress dust correlates with asthma and AD activity (6), and reduction in household HDM allergen Der p 1 reduces AD severity (7). HDM exposure also triggers a multipronged allergic immune response. For example, Der p 1 and Der p 3 activate T cells directly, leading to local Th2 cytokine production, including interleukin (IL)-4 and IL-31 (8). HDM proteases activate protease-activated receptors (PARs), G protein-linked receptors expressed by neurons, endothelial cells, keratinocytes, and mast cells (9, 10). PAR2 is activated by HDM protein Der p 1 and endogenous proteases including tryptase and cathepsin S, which are released in inflamed skin (11, 12).
PAR2-expressing primary sensory neurons are a subset of TRPV1+ cutaneous C-fiber sensory afferents that transmit pain (nociceptors) and itch (pruritoceptors) (13). PAR2 signaling in pain is well-established (14, 15), and PAR2 is overexpressed in the skin of AD patients (16) who experience both itch and pain (17). Grhl3PAR2/+ transgenic mice, which overexpress PAR2 in suprabasal keratinocytes, spontaneously develop scratching, epidermal hyperplasia, and brisk dermal infiltrates suggestive of dermatitis (18).
Despite the abundance of PAR2-activating proteases in inflamed atopic skin, there is limited information as to the PAR-mediated mechanisms that activate primary sensory neurons (pruritoceptors) and generate allergic itch (19–21). Using RNA sequencing (RNA-Seq) and in situ hybridization (ISH) in trigeminal ganglion (TG) sensory neurons of the Grhl3PAR2/+ and wild-type (WT) mice, we characterized the transcriptional changes induced following HDM exposure (22). Unexpectedly, development of the dermatitis phenotype was stochastic; only 40% of Grhl3PAR2/+ mice developed visible inflammation, spontaneous scratching, and skin damage. We found significant transcriptional alterations in the TG of these “responder” mice, which were concentrated in the TRPV1+ subset of unmyelinated primary afferents. In contrast, we found minimal scratching in untreated Grhl3PAR2/+ mice or HDM-treated WT mice that correlated with limited transcriptional changes in the TG. Taken together, we conclude that Ghrl3PAR2/+ mice are in a primed state that can be triggered by chronic allergen challenge.
Results
HDM Treatment Produces a Dermatitis-Like Phenotype in Grhl3PAR2/+ Mice.
Many mouse models of chronic itch exhibit significant spontaneous scratching (e.g., transgenic overexpression of cytokine IL-31 or genetic deficiency in transcription factor Bhlhb5; refs. 23 and 24). Although Frateschi et al. (18) reported that PAR2 overexpression provokes spontaneous scratching, skin lesions, and dermatitis, in our colony we did not observe a difference in spontaneous scratching between the Grhl3PAR2/+ mice and WT littermates. Furthermore, with the exception of ringtails (Fig. 1A), Grhl3PAR2/+ mice in our colony rarely developed appreciable skin findings. Although the mechanisms underlying these phenotypic differences remain to be determined, it is most likely that they are due to different genetic background and housing conditions. Therefore, to induce dermatitis, we treated cheek skin of WT and Grhl3PAR2/+ mice for 9 wk with topical sodium dodecyl sulfate (SDS), which disrupts the skin barrier, followed by HDM ointment, a significant source of proteases. We restricted the SDS and HDM treatment to one side of the face; contralateral tissue served as an internal, untreated control. Other control groups included both WT and Grhl3PAR2/+ mice treated with SDS followed by Vaseline rather than HDM ointment.
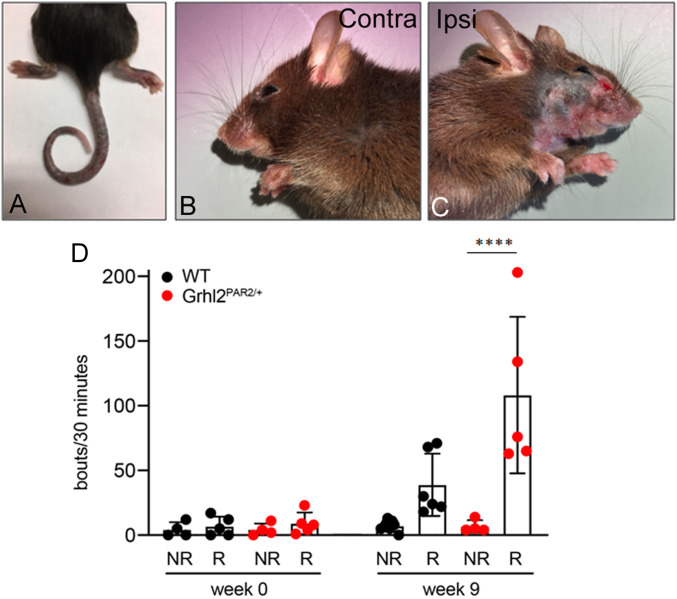
Fig. 1.
Grhl3PAR2/+ mice treated with HDM develop severe skin lesions and exacerbated spontaneous scratching. In our colonies, with the exception of ring tails (A), the Grhl3PAR2/+ mice that overexpress PAR2 in suprabasal keratinocytes are phenotypically indistinguishable from their WT littermates. Application of HDM to the cheek of the Grhl3PAR2/+ mice twice per week, for nine consecutive weeks, produced severe skin lesions on the treated cheek (C) compared to the contralateral cheek (B) and increased spontaneous scratching of the treated cheek (D; responders, R). Note that 40% of the HDM-treated Grhl3PAR2/+ did not develop skin lesions and scratching did not increase (D; nonresponders, NR). Scratching bouts before and after HDM treatment were compared within each group using a Student’s t test (****P < 0.001).
After 9 wk, 40% of the HDM-treated Grhl3PAR2/+ mice (hereafter designated “responders”) exhibited severe spontaneous scratching and skin changes consistent with dermatitis, including erythema and scaling. Skin changes only occurred in the HDM-treated cheek (Fig. 1 B–D). Another 40% of HDM-treated mice developed neither skin pathology nor elevated scratching (thus designated “nonresponders”). The remaining 20% of HDM-treated mice had an intermediate phenotype, with scratching but no gross skin pathology. Finally, although some HDM-treated WT mice treated with HDM (∼46%; 5 of 13) displayed spontaneous, albeit limited, scratching, none developed skin lesions and none of the Vaseline-only treated mice developed scratching or skin pathology over the 9-wk treatment course.
HDM Produces More Severe Skin Histopathology in Grhl3PAR2/+ than in WT Mice.
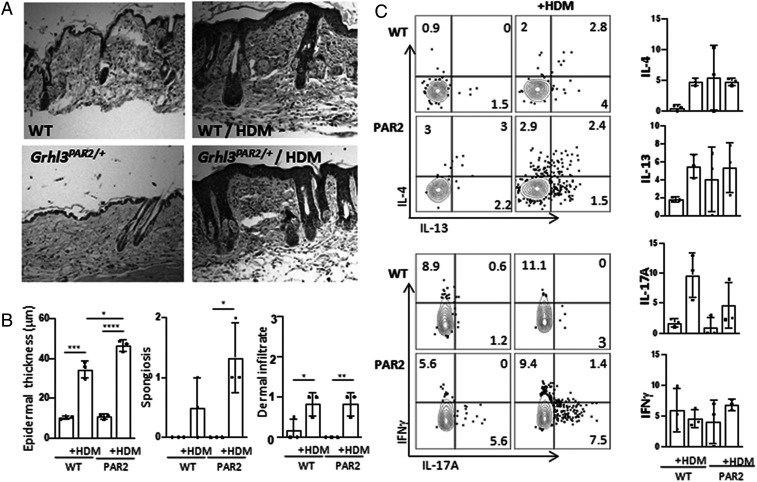
In our colony, the skin of Grhl3PAR2/+ animals was histologically indistinguishable from that of their WT littermates, with no acanthosis (epidermal thickening), spongiosis (epidermal edema), or inflammatory infiltrates (Fig. 2A). Because of chronic scratching-induced skin changes observed in 9-wk HDM-treated Grhl3PAR2/+ mice, we performed histopathologic analyses on Grhl3PAR2/+ and WT skin after 3 wk of HDM treatment, a time point at which the HDM-treated animals displayed scratching and hallmarks of skin inflammation but no skin lesions. Following HDM treatment, both the Grhl3PAR2/+ mice and WT littermate controls developed marked acanthosis and spongiosis as well as a mild inflammatory infiltrate (Fig. 2B). The magnitude of HDM-induced acanthosis was greater in the Grhl3PAR2/+ mice. Fluorescence-activated cell sorting (FACS) analysis of skin T lymphocyte subsets demonstrated that HDM treatment induced increased cytokine expression by CD4+ T-cells but similar levels of IL-4, IL-13, and IL-17 in both Grhl3PAR2/+ and WT mice (Fig. 2C).
Fig. 2.
HDM treatment produces dermatitis in both WT and Grhl3PAR2/+ mice. (A) H&E staining of skin biopsies illustrates that topical application of HDM extract to the cheek twice a week, for 3 wk, induced hallmarks of inflammation ipsilateral to the treated side, in both WT and Grhl3PAR2/+ mice. (B) The dermatitis manifest with acanthosis (epidermal thickening), spongiosis (epidermal edema), and increased dermal infiltrates. The magnitude of HDM-induced acanthosis was greater in the Grhl3PAR2/+ than in the WT mice. (C) FACS analysis of cheek skin CD4 T lymphocytes demonstrated that HDM treatment provoked expression of IL-4, IL-13, and IL-17, but not interferon gamma (IFNγ), in both Grhl3PAR2/+ and WT mice. Data are presented as mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001 (Student’s unpaired t test).
RNA-Seq in Responder and Nonresponder Grhl3PAR2/+ Mice.
To identify patterns of gene expression that differentiate responder and nonresponder mice, we performed RNA-Seq on TG that innervate ipsilateral (treated) and contralateral (untreated) cheek skin. TG samples collected from Grhl3PAR2/+ mice included HDM-treated responder (ipsi and contra), HDM-treated nonresponder (ipsi and contra), and Vaseline-treated (ipsi and contra). We also collected samples from untreated WT mice.
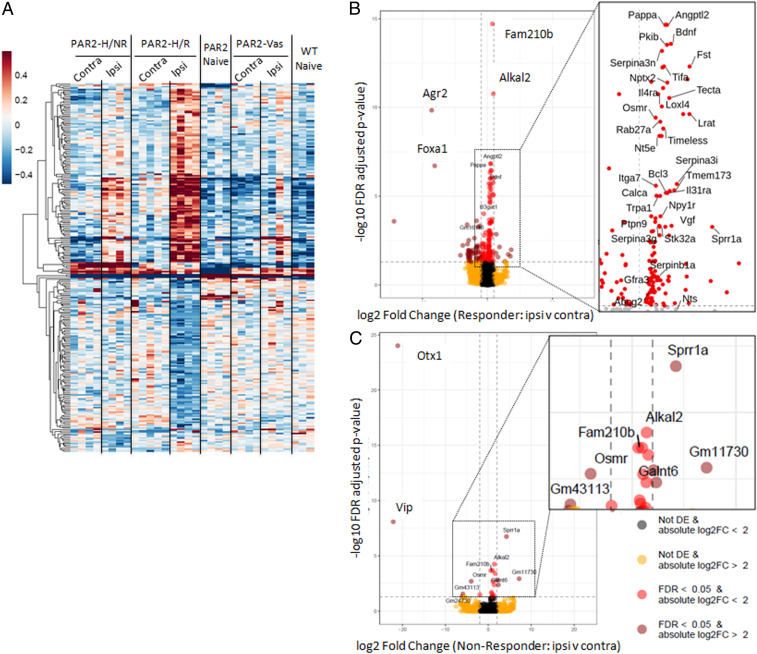
Consistent with our finding of minimal histopathological differences between the skin of untreated Grhl3PAR2/+ and WT mice, supervised hierarchical clustering of sequenced TG revealed few significant gene expression differences. In sharp contrast, the magnitude of changes in the ipsilateral TG of the HDM-treated responder mice differed strikingly from all other samples and included profoundly up- and down-regulated genes (Fig. 3 and SI Appendix, Fig. S1 A–C). In fact, the pattern of TG gene expression in the other groups, including the contralateral TG of the HDM-treated Grhl3PAR2/+ mice, was minimally altered in the absence of HDM treatment. SDS/Vaseline treatment up-regulated transcription of only two genes (Sprr1a and Nts) (SI Appendix, Fig. S2). Thirteen additional genes (Bdnf, Il31ra, Pkib, Osmr, Galnt6, Alkal2, Tmem173, Fam210b, Accsl, Serpina3i, Nkain1, Itk, and Gm11730) were selectively induced in the ipsilateral TG of HDM-treated nonresponder mice (SI Appendix, Fig. S1C). These genes were also induced in the ipsilateral TG of the HDM-treated responder group, but the fold change was considerably greater.
Fig. 3.
HDM treatment modulates gene expression in TG of Grhl3PAR2/+ mice. Nine weeks after HDM treatment, we performed RNA-Seq of ipsilateral (ipsi) and contralateral (contra) TG from responder (PAR2_HDM.R) and nonresponder (PAR2_HDM.NR) Grhl3PAR2/+ mice. We also sequenced samples from Vaseline-treated (ipsi and contra) as well as from untreated Grhl3PAR2/+ (PAR2_Naive) and untreated WT (WT_Naive) mice. (A) Heat map of overall transcriptional changes in the TG illustrates the magnitude of gene expression changes in all samples. Note that changes in the ipsilateral TG of the HDM-treated responder Grhl3PAR2/+ mice (PAR2_HDM.R) differed strikingly from all other samples and included profound up- (dark red) and down- (dark blue) regulated genes. (B) Volcano plot illustrates that the top up-regulated hits (red) included genes that encode secreted peptides (e.g., calca, which encodes calcitonin gene-related peptide [CGRP]), neurotrophic factors (e.g., brain-derived neurotrophic factor; Bdnf), G proteins (e.g., neuropeptide Y receptor 1; Npy1r), and other signaling molecules. (C) This volcano plot illustrates that a subset of these genes were also up-regulated in the ipsilateral TG of the nonresponder mice, but less so than in the responder mice. (See also SI Appendix, Figs. S1 and S2 for a complete list of up- and down-regulated genes from both responder and nonresponder TG samples).
Next, we focused on genes that were differentially expressed (up- or down-regulated) in the ipsilateral vs. contralateral TG of HDM-treated responder mice. Out of 210 genes that were differentially expressed in the ipsilateral TG of HDM-treated skin, 118 (56.2%) were up-regulated and 92 (43.8%) were down-regulated (Fig. 3 and SI Appendix, Fig. S1 A–C). Top up-regulated hits included genes that encode secreted peptides, neurotrophic factors, G proteins, and other signaling molecules. These included brain-derived neurotrophic factor (Bdnf), which is an established regulator of PAR2-mediated scratching (25) and has been associated with AD (26). In the TG of responder animals we also recorded enrichment of the genes that encode IL-31 receptor subunits (Il31ra and Osmr) and the transient receptor potential ankyrin 1 (Trpa1), two receptors implicated in pruritoception (4, 27–29).
To determine in which subpopulations of sensory neurons the top 40 up-regulated genes in HDM responders are expressed, we used the single-cell sequencing-based dorsal root ganglion (DRG) neuron classification of Usoskin et al. (30). The majority of up-regulated genes we identified map to peptidergic (PEP) and nonpeptidergic unmyelinated primary afferent subpopulations (Fig. 4A, red rectangle), which include many pruritoceptors (29).
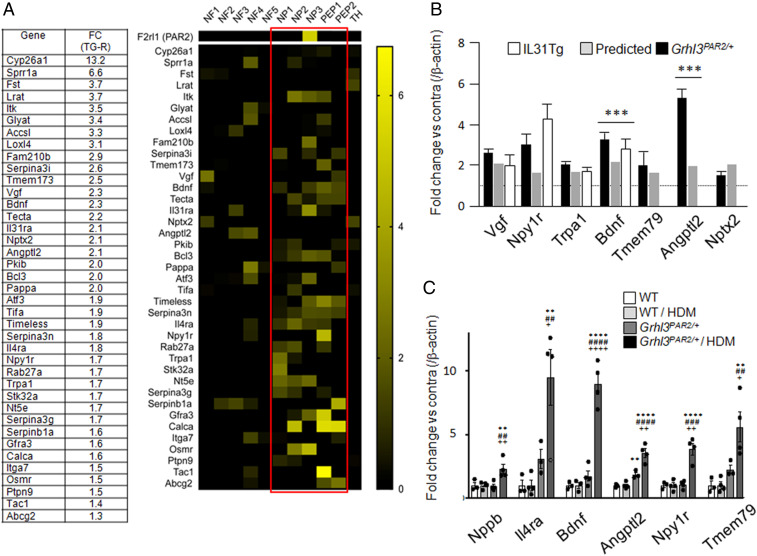
Fig. 4.
Distribution of up-regulated genes in subsets of TG neurons and qPCR validation of RNA-Seq hits. (A) Table: Fold change (FC) of the highest up-regulated TG genes in the responder Grhl3PAR2/+ mice (TG-R). Heat map: most up-regulated genes are enriched in the peptidergic (PEP1-2) and nonpeptidergic (NP1-3) sensory neurons (red rectangle), compared to the myelinated (NF1-5) and tyrosine hydroxylase (TH)-expressing primary afferents. (B) qPCR confirmed the transcriptional changes detected by RNA-Seq in the HDM-treated Grhl3PAR2/+ mice (black bars) compared to WT (controls; dotted line at 1) for Vgf nerve growth factor inducible (Vgf), neuropeptide Y receptor 1 (Npy1r), transient receptor potential ankyrin 1 (TrpA1), brain-derived neurotrophic factor (Bdnf), transmembrane protein 79 (Tmem79), angiopoietin-related protein 2 (Angptl2), and pentraxin2 (Nptx2). Comparable changes were detected in the TG of IL31Tg mice, another model of dermatitis (white bars). The fold changes calculated by qPCR were comparable to those obtained by RNA-Seq (gray bars). (C) qPCR validation of six representative neuronal genes in naïve WT, HDM-treated WT (WT/HDM), untreated Grhl3PAR2/+, and HDM-treated Grhl3PAR2/+ mice. Note that baseline expression of Il4ra and Angptl2 increased in the TG of untreated Grhl3PAR2/+ mice (dark gray bars), with no change in the other four genes. HMD treatment of the Grhl3PAR2/+ mice significantly up-regulated all six genes (black bars). Data are presented as mean ± SEM; one-way ANOVA; *compared to WT; #compared to WT/HDM and +compared to Grhl3PAR2/+. */#/+P < 0.05; **/##/++P < 0.01; ***/###/+++P < 0.005; ****/####/++++P < 0.001.
By comparison, only 15 genes were significantly up-regulated in the ipsilateral TG of HDM-treated nonresponder mice (SI Appendix, Fig. S1C), but at lesser magnitude than in responder mice. We conclude that up-regulation of these genes is not sufficient to drive scratching after HDM treatment and that scratching is not required for their up-regulation. Rather, a threshold of up-regulated gene expression establishes a primed state that can be engaged by HDM treatment to drive scratching.
Validation of RNA-Seq.
To validate several of the significant changes from the responder group we used qPCR, focusing on seven of the transcripts with the highest TG enrichment (Vgf, Npy1r, Trpa1, Bdnf, Tmem79, Angptl2, and Nptx2). For five of these transcripts, the fold changes comparing ipsilateral and contralateral TG from HDM-treated Grhl3PAR2/+ mice calculated by qPCR did not differ statistically from those obtained by RNA-Seq (Fig. 4B). For two of the seven transcripts (Bdnf and Angptl2), values obtained by qPCR were significantly greater than those predicted by RNA-Seq.
We next used qPCR to determine whether genes up-regulated in the TG of the responder mice were driven by PAR2 overexpression, HDM treatment, or the combination. Here, we quantified the expression of several up-regulated TG transcripts (Npy1r, Bdnf, Tmem79, Angptl2, and Il4ra) across four groups of mice: naïve WT, HDM-treated WT, naïve Grhl3PAR2/+, and HDM-treated Grhl3PAR2/+. Because of its reported contribution to itch (31), we also examined expression of neuropeptide natriuretic polypeptide b (Nppb). In fact, none of these six transcripts was significantly up-regulated in WT mice after HDM treatment. Thus, HDM treatment alone does not alter expression of these genes (Fig. 4C). In contrast, HDM treatment significantly up-regulated all tested genes in the ipsilateral TG of the Grhl3PAR2/+ mice. We also observed elevated baseline expression of Il4ra and Angptl2 in TG of untreated Grhl3PAR2/+ mice, albeit lower than in HDM-treated Grhl3PAR2/+ mice. We conclude that transgenic overexpression of PAR2 in Grhl3PAR2/+ mice is sufficient to significantly alter TG gene expression. The transcriptional changes in the TG of the Grhl3PAR2/+ mice likely reflect basal PAR2 activation, presumably by endogenous proteases; HDM enhances these effects.
Trigeminal Distribution of Pruritus-Associated Genes.
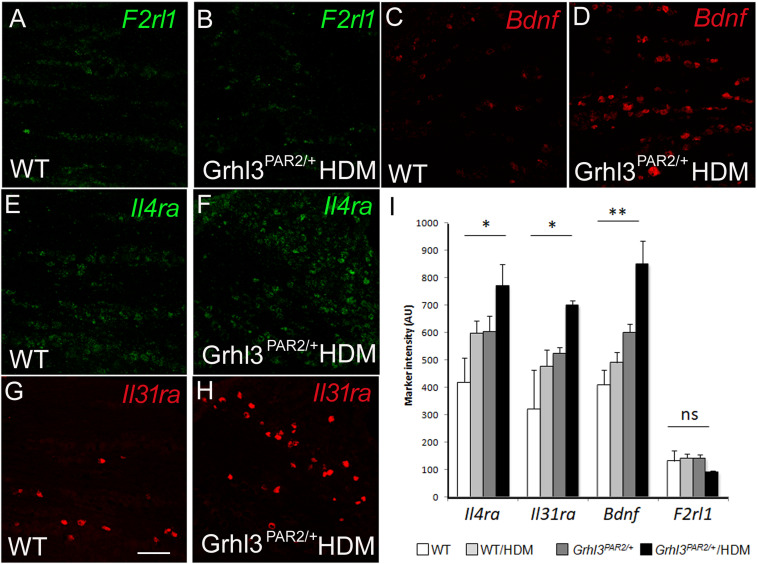
Neither RNA-Seq nor qPCR can distinguish the TG cell types in which these transcriptional changes occur (e.g., neurons or nonneuronal cells). To address directly this question and to assess how HDM treatment and PAR2 overexpression influence the neuronal distribution of genes implicated in pruritus, including PAR2, we used ISH on TG tissue sections from WT, HDM-treated WT, Grhl3PAR2/+, and HDM-treated Grhl3PAR2/+ mice. Although PAR2 messenger RNA (mRNA) was readily detected by in situ in TG neurons in WT mice, the number of PAR2-positive neurons was low and did not change in the Grhl3PAR2/+ mice, before or after HDM treatment (Fig. 5). On the other hand, and in agreement with RNA-Seq data, we recorded significant transcriptional up-regulation of several pruritus-associated genes (Bdnf, Il4ra, Trpa1, and Il31ra) in neurons of the HDM-treated Grhl3PAR2/+ mice but no evidence of their up-regulation in nonneuronal cells.
Fig. 5.
Neuronal expression of up-regulated genes in HDM-treated Grhl3PAR2/+ mice. (A–I) ISH illustrates F2rl1 (PAR2), Bdnf, Il4ra, and Il31ra mRNA in the TG of naive (WT: A, C, E, and G) and HDM-treated Grhl3PAR2/+ mice (B, D, F, and H). HDM treatment significantly up-regulated expression of Bdnf, Il4ra, and Il31ra but not F2rl1. Data are presented as mean ± SEM; n = 3; one-way ANOVA; *P < 0.05; **P < 0.001; ns: not significant. (Scale bar: 100 μm.)
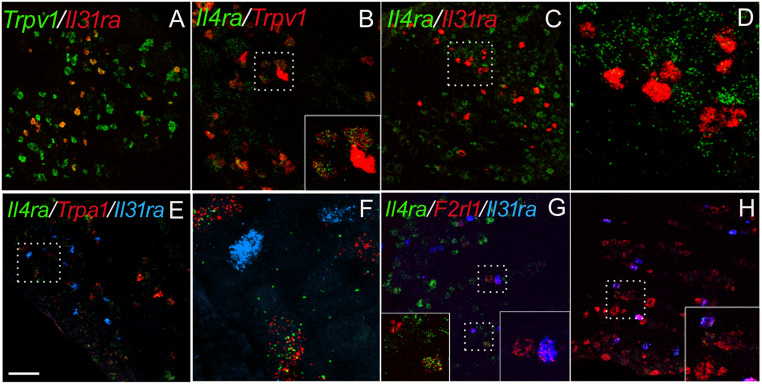
Next, we focused on gene expression within the TRPV1 subset of TG neurons, which is pruritoceptive, transmitting both histaminergic and nonhistaminergic signals (32). Using immunocytochemistry, we previously demonstrated BDNF, IL31RA, and OSMRβ expression in subsets of TRPV1+ TG neurons (27, 33). Here, we confirmed these findings by ISH and observed overlap in the expression of Bdnf, Il31ra, and F2rl1 (the transcript that encodes for PAR2) within the TRPV1+ population (Fig. 6) (100% Il31ra+ neurons were Trpv1+; Il31ra+/Trpv1+ neurons: 12.3 ± 1.5%; Trpv1+/F2rl1+ neurons: 68.6 ± 3.4%; F2rl1+/Trpv1+ neurons: 7.7 ± 1.0%; n = 3). Interestingly, although HDM treatment increased the number of Il31ra-expressing TG neurons all remained within the Trpv1+ subset, regardless of genotype.
Fig. 6.
Trigeminal neuron subtype expression of up-regulated genes in Grhl3PAR2/+ mice. (A–H) ISH illustrates that HDM-induced genes predominated in the TrpV1-expressing subset of TG neurons, including Il31ra (A; red) and Il4ra (B; green). (C and D) Although all Il31ra+ TG neurons colabeled for TrpV1, coexpression of Il31ra (red) and Il4ra (green) was rare. High magnification of the box in C is shown in D. (E and F) Although TrpA1+ (red) neurons coexpress Il4ra (green), none colabeled with Il31ra (blue). High magnification of the box in E is shown in F. (G) By contrast, both Il31ra (blue) and Il4ra (green) overlapped with F2rl1 (red). (H) We also recorded F2rl1 expression (green) in Bdnf-positive neurons (red), many of which expressed Il31ra (blue). Insets in B, G, and H show high magnification of the boxed areas in these images. (Scale bar: 100 μm.)
In contrast, both small- and large-diameter sensory neurons expressed Il4ra (Fig. 6), and Il4ra+ and F2rl1+ populations partially overlapped. Furthermore, although many Il4ra+ neurons coexpressed Trpv1, the subset of Il31ra+/Trpv1+ neurons coexpressing Il4ra was small (∼13 ± 0.8%; n = 4), and those expressing Il4ra did so at very low levels. We also found that the great majority of Trpa1+ neurons, another subset of the Trpv1+ population, express Il4ra (95.9 ± 0.4%; n = 3), but none coexpressed Il31ra (0.9 ± 0.1%; n = 3) (Fig. 6 E–G). We conclude, therefore, that IL31RA+ neurons and IL4RA+/TRPA1+ neurons represent two separate subsets of TRPV1+ TG neurons.
Activation of Stress-Related TG Genes.
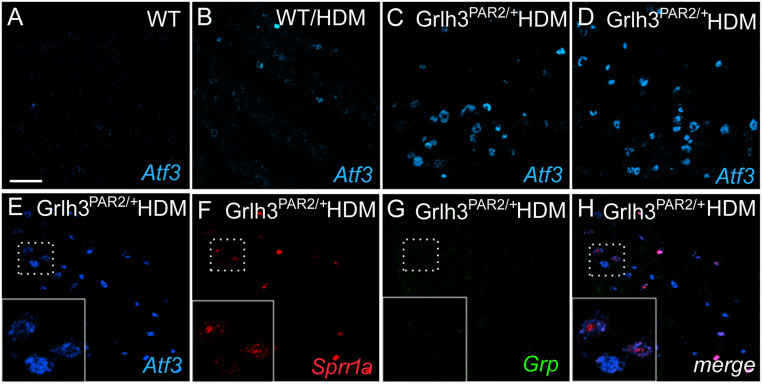
As some genes up-regulated by HDM are also observed in sensory neurons after peripheral nerve injury (e.g., Bdnf and Sprr1a; refs. 34 and 35), we also used in situ to evaluate expression of Atf3, a marker of stress (36) or peripheral nerve damage (37). Consistent with our RNA-Seq analysis, we recorded de novo expression of Atf3 in TG neurons of HDM-treated Grhl3PAR2/+ mice and HDM-treated WT mice (Fig. 7 A–D). As expected, most of the Sprr1a+ neurons were also Atf3-positive (Fig. 7 E–H). Unexpectedly, we also recorded large numbers of Atf3-positive neurons in untreated Grhl3PAR2/+ animals. These results suggest that chronic allergen (HDM) exposure and/or genetic overexpression of PAR2 (Grhl3PAR2/+ mice) activates a stress response pathway within the TG. Importantly, however, neither Sprr1a nor Atf3 induction is sufficient to drive scratching in untreated Grhl3PAR2/+ mice—HDM treatment is critical.
Fig. 7.
HDM treatment and PAR2 overexpression independently induce stress-related genes in TG neurons. ISH illustrates de novo induction of activating transcription factor 3 (Atf3; blue; A–E), a sensory neuron marker of stress and/or peripheral nerve damage, in TG neurons after HDM treatment (B), PAR2 overexpression (C), or the combination (D). Another stress-related gene, Sprr1a (red, F) is also induced under these conditions; gastrin-releasing peptide (GRP; green, G), a gene that we reported to be induced in DRG neurons after peripheral nerve injury, was not induced (45). H shows the merged picture of E–G. (Scale bar: 100 μm.)
Transcriptional Changes across Different Models of Dermatitis.
To evaluate our findings in another model of dermatitis we examined gene expression in cervical DRG from IL31Tg mice, which constitutively overexpress the pruritogenic cytokine IL-31 in T lymphocytes and exhibit spontaneous scratching/itch phenotypes, including skin lesions (23, 38). Many transcripts that were altered in the HDM-treated Grhl3PAR2/+ responder mice also changed significantly in the DRGs that innervate lesional skin of the IL31Tg animals, compared to DRGs from WT skin (Fig. 4B). As the transcriptional changes were comparable in magnitude to those exhibited by HDM-treated PAR2 responders, we suggest that a common sensory neuron gene expression program is induced across multiple dermatitis models.
Grhl3PAR2/+ Mice Are Not Sensitized to Acute Pruritogens.
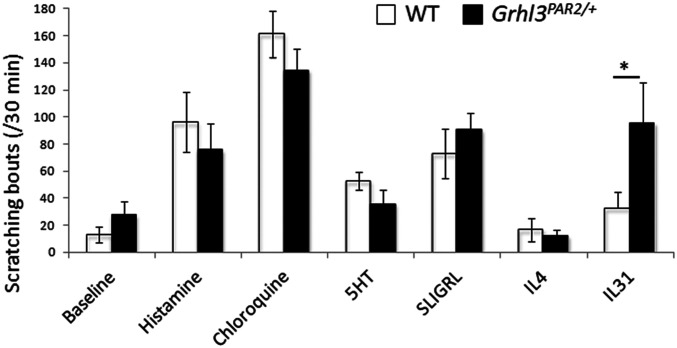
Our RNA-Seq analyses revealed that TG mRNA expression of several pruritogen receptors (notably Il31ra) increased significantly in the Grhl3PAR2/+ mice (Fig. 3). Therefore, we next tested the hypothesis that overexpression of PAR2 in keratinocytes and consequent up-regulation of IL31RA in the TG—while not driving scratching in the absence of HDM—could nevertheless sensitize the Grhl3PAR2/+ mice to multiple dermatitis-associated pruritogens. We injected the nape of the neck of WT and Grhl3PAR2/+ mice with histaminergic and nonhistaminergic pruritogens: recombinant IL-31, histamine, chloroquine, SLIGRL, α-methyl-5-hydroxytryptamine, and IL-4. The Grhl3PAR2/+ mice did exhibit a threefold increase in scratching bouts in response to IL-31, which correlates with increased TG expression of IL31RA in these mice (Fig. 8). However, we observed no increase in responsiveness (scratching) to SLIGRL, α-methyl-5-hydroxytryptamine, or IL-4 in Grhl3PAR2/+ compared to WT mice, despite overexpression of PAR2 in keratinocytes and up-regulation of IL4RA in the TG. These results indicate that, unlike other genetic models of chronic itch that display increased acute responses to pruritogen injections (24, 27), there is not a generalized sensitization to pruritogens in the Grhl3PAR2/+ mice.
Fig. 8.
Grhl3PAR2/+ mice are not sensitized to acute pruritogens. We injected the nape of the neck of WT (white bars) and Grhl3PAR2/+ (black bars) mice with an array of histaminergic and nonhistaminergic pruritogens (histamine, chloroquine, α-methyl-5-hydroxytryptamine [5HT], SLIGRL, IL-4, and IL-31) and video-recorded scratching over 30 min after the pruritogen administration. With the exception of IL-31, for which we recorded a threefold increase of scratching, these pruritogens did not provoke increased scratching in the Grhl3PAR2/+ mice compared to the WT mice. Data are presented as mean ± SEM; n = 5; Student’s t test; *P < 0.05.
Discussion
Using molecular, anatomical, and behavioral assays, we found that concurrent genetic priming/sensitization and environmental allergen exposure are required for Grhl3PAR2/+ mice to develop dermatitis and pruritus. Unexpectedly, appearance of the dermatitis phenotype after HDM treatment was stochastic, with only some mice developing spontaneous scratching and skin damage (responders); others never scratched (nonresponders). In nonresponder mice, we recorded only 15 up-regulated and 6 down-regulated genes in the TG ipsilateral to the HDM treatment. In responder mice, we recorded more than 100 up-regulated genes, and the 15 genes up-regulated in the nonresponder mice were detected at much greater fold induction. Many of these genes are associated with pruritus and dermatitis (27, 39), including Bdnf, Il31ra, and Osmr. Others such as Sprr1a have not previously been implicated in pruritus. We conclude that HDM induces a dermatitis-relevant transcriptional program in the TG of Grhl3PAR2/+ mice, but a threshold of transcriptional induction is required for the scratching phenotype to manifest.
Transcriptional Profiles of TG in Responder Grhl3PAR2/+ Mice.
Although the TG contains satellite cells, resident macrophages, and neurons, our ISH studies demonstrated that the majority of genes modulated in HDM-responder mice are expressed in the unmyelinated primary afferents. Whether genes are modulated across different subsets of unmyelinated neurons remains to be determined. However, our results indicate that most of the up-regulated genes are found at high levels in the TRPV1+ population of sensory neurons, namely the NP3 subclassification of Usoskin et al. (30). Responder Ghrl3PAR2/+ mice exhibited higher Il31ra and Osmr transcription, which accords with an increased behavioral response to nape-of-the-neck injection of recombinant IL-31 in the Ghrl3PAR2/+ mice (scratching) compared to WT mice. In responder Ghrl3PAR2/+ mice, we also recorded increased neuronal Il4ra, which Oetjen et al. (39) implicated in chronic itch and sensitization of sensory neurons to acute pruritogens. Although we confirmed that capsaicin-responsive (TRPV1+) sensory neurons coexpressed Il4ra and Il31ra, these two receptors marked separate subsets of TRPV1+ neurons, one that expresses IL31RA and another that coexpresses IL4RA and TRPA1. This expression pattern differs from a calcium imaging study (39) which found that 89% of IL-4–responsive neurons responded to both IL-31 and capsaicin, suggesting extensive coexpression of IL4RA, IL31RA, and TRPV1. Conceivably, very low IL31RA expression in IL4RA-expressing neurons (below in situ detection thresholds) is sufficient to confer responsiveness to IL-31. Differential sensitivity of calcium imaging and ISH may also explain the different conclusions as to coexpression of IL31RA and TRPA1 (27).
The TG of responder mice also exhibited high transcriptional induction of Sprr1a, a gene most commonly associated with skin barrier (40). We found that Sprr1a, which increases in axotomized DRG neurons and has been implicated in axonal regeneration (34), colocalized in TG neurons with Atf3, a transcription factor also induced in axotomized neurons (37, 41). However, as we observed Atf3 induction in TG neurons of untreated Ghrl3PAR2/+ mice and in HDM-treated WT mice, we suggest that the Atf3 up-regulation we observed represents a stress response that can be induced by the Ghrl3PAR2/+ transgene and/or HDM. The absence of other well-documented nerve injury-induced genes (e.g., NPY; ref. 42) supports this conclusion. Furthermore, the coinduction of Sprr1a and Atf3 illustrates that despite the absence of a scratching phenotype in the Ghrl3PAR2/+ mice, keratinocyte overexpression of PAR2 nevertheless exerts a profound influence on sensory neurons that innervate skin.
Ghrl3PAR2/+ Mice Are Primed to Develop Robust Itch Responses to Allergens.
HDM treatment produced more scratching in responder Ghrl3PAR2/+ than in untreated Ghrl3PAR2/+ or HDM-treated WT mice. This result correlates well with the gene expression analyses that revealed larger transcriptional changes in the TG of HDM-treated Ghrl3PAR2/+ mice compared to untreated Ghrl3PAR2/+ or HDM-treated WT mice. Together, our findings indicate that skin overexpression of PAR2 induces transcriptional changes in the TG which, by themselves, do not influence behavior. Specifically, we found no increase in baseline scratching and, with the exception of IL-31, no increase in acute pruritogen-induced scratching. On the other hand, chronic challenge with exogenous pruritogens (HDM) potentiated the PAR2-induced TG transcriptional changes and induced strong behavioral indices of itch in the Ghrl3PAR2/+ mice. We conclude, therefore, that pruritoceptors in Ghrl3PAR2/+ mice are in a sensitized state that mirrors that of genetically susceptible AD patients. Specifically, PAR2 overexpression in the skin primes the neurosensory circuits through which environmental allergen exposure modulates pruritoception, thereby fueling the itch–scratch cycle that characterizes AD.
Materials and Methods
Animals.
Animal experiments were approved by the University of California, San Francisco (UCSF) Institutional Animal Care and Use Committee and were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals (43). Grhl3PAR2/+ mice (18) were generously provided by Shaun Coughlin, UCSF. IL31Tg mice were generated as previously described (23).
HDM Treatment.
Twice per week for 9 wk, the right cheek of each mouse was shaved, then detergent (SDS, 4%) was applied to the exposed skin under isoflurane (2%) anesthesia. Two hours later, mice were reanesthetized and HDM extract (Biostir AD; Biostir) was massaged into cheek skin as previously described (44). Control mice were treated with SDS followed by Vaseline.
Skin Histopathology.
Six-millimeter punch biopsies were harvested from HDM-treated and vehicle-treated cheek skin, formalin-fixed, and paraffin-embedded (FFPE). Five-micrometer FFPE skin sections were stained with hematoxylin and eosin (H&E) and photographed with a 10× objective (Zeiss Axioplan). We quantified epidermal thickness for each condition by averaging six different measurements along the epidermis using the ImageJ line tool. Acanthosis and dermal infiltrate severity were determined by blind scoring of three H&E sections per mouse.
Skin Digestion, Ex Vivo Activation of Skin Hematopoietic Cells, and Flow Cytometry.
We digested 6-mm-diameter cheek skin punches in RPMI supplemented with Type XI collagenase and bovine deoxyribonuclease I (Sigma), followed by mechanical dissociation (GentleMACS; Miltenyi) and filtration. The single-cell suspension was resuspended in Dulbecco’s modified Eagle’s medium (Sigma) containing fetal bovine serum (Omega Scientific), l-glutamine, nonessential amino acid solution, and sodium pyruvate (Sigma). Cells were stimulated by addition of phorbol 12-myristate 13-acetate, ionomycin, and Brefeldin A (Sigma) for 4 h at 37 °C, 10% CO2.
All skin cell suspensions were stained with fixable viability dye eFluor780 (Thermo Fisher) and fluorescence-conjugated antibodies to hematopoietic cell surface lineage markers including CD45-BV711 (clone 30-F11; BD Horizon), CD3-PE-Cf594 (145-2C11; Biolegend), γδTCR-BV605 (GL3; Biolegend), and CD4-PE-Cy7 (RM4-5; Biolegend). Cells were subsequently fixed in 4% paraformaldehyde (Electron Microscopy Sciences) and permeabilized with 0.5% saponin (Fisher Scientific). Antibodies used for intracellular cytokine detection were IL-4-APC (11B11; eBioscience), IL-13-PE-e710 (eBio13A; eBioscience), IFNγ-eFluor450 (XMG1.2; eBioscience), and IL-17A-Alexa Fluor 488 (eBio17B7; eBioscience).
RNA-Seq.
Mice were anesthetized and transcardially perfused with saline (10 mL). Trigeminal ganglia ipsilateral and contralateral to the HDM-treated cheek were removed. All samples were flash-frozen then homogenized in TRIzol (Ambion) and purified using the PureLink RNA Mini Kit (Ambion). Sample quality was measured using the RNA6000 Pico Kit (Agilent). For all sequencing libraries, input RNA was of high quality (RNA integrity number >8). Complementary DNA (cDNA) libraries were prepared using the Ovation Mouse RNA-Seq System (Nugen). Sequencing was performed on the Illumina HiSeq 2500 system in rapid run, paired-end, 2 × 100 base pairs mode. Final, filtered sequencing depth was between 50 and 90 million reads (SI Appendix, Table S1). Reads were aligned to the mouse genome GRCh38 and quantified using the STAR aligner software version 2.7.2b. Differential expression analysis was performed in the R computing environment version 3.6.1 using the software DESeq2 version 1.26. A P value <0.05 as well as a false discovery rate <0.05 was considered significant for differentially expressed gene analysis. A minimum differential expression of log2 fold change ± 1 was also used to determine significance.
RNA Preparation from IL31Tg Animals.
Cervical DRG samples from IL31Tg mice were prepared using the same RNA extraction protocol as for RNA-Seq. As lesions were bilateral, cervical DRGs from WT littermates were used as controls.
qPCR Confirmation of RNA-Seq Hits.
RNA was prepared as described above. cDNA was generated using the SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen). mRNA levels were quantified with the Bio-Rad CFX Connect System using PowerUp SYBR Green Master Mix (Applied Biosystems). Melting curves were generated to ensure the specificity of all primers. Transcripts were normalized to actin expression.
Primers.
See SI Appendix, Table S2 for the list of primers we used for qPCR and Grhl3PAR2/+ genotyping.
ISH.
We followed the Advanced Cell Diagnostics protocol for RNAscope ISH on fresh TG tissue processed as previously described (45).
Behavior.
For spontaneous scratching evaluation of HDM-treated and control animals, we habituated mice for 1 h in plastic cylinders then video-recorded behavior for the next 30 min. Hindpaw scratching was quantified as the total number of discrete scratching bouts throughout the recording. We defined a bout as a lift of the hindpaw, scratching the skin, and then replacing the hindpaw. Scratching bouts of the Grhl3PAR2/+ mice that were included in the RNA-Seq analysis are shown in SI Appendix, Table S3.
For acute itch studies, after habituation for 1 h in plastic cylinders the mice received a nape-of-the-neck injection of the pruritogen and then were video-recorded for the next 30 min. Injected pruritogens were as follows: histamine (500 μg; Sigma), chloroquine (200 μg; Sigma), SLIGRL (100 nmol; Peptides International Inc.), α-methyl-5-hydroxytryptamine (500 μg; Tocris), and IL-31 (100 ng; Peprotech). Hindpaw scratching was quantified as the total number of discrete scratching bouts for the 30-min recording period. In all behavioral analyses the investigator scoring the behavior was blind to treatment and codes were only revealed after scoring.
Statistical Analyses.
All statistical analyses were performed with Prism (Graph Pad). Anatomical and behavioral data are reported as mean ± SEM. An unpaired Student’s t test was used to compare WT vs. Grhl3PAR2/+ mice (effects of pruritogens; Fig. 8), WT vs. WT-HDM, or Grhl3PAR2/+ vs. Grhl3PAR2/+-HDM mice (spontaneous scratching; Fig. 1). For the in situ analyses we measured density of fluorescence with ImageJ in four groups of mice and compared results using a one-way ANOVA (Fig. 5). For RNA-Seq analysis we used Deseq2 software, which uses a negative binomial regression model for statistical testing.
Supplementary Material
Acknowledgments
This work was supported by NIH grants NS14627 (A.I.B.) and NSR35097306 (A.I.B.), a Dermatology Foundation Career Development Award (M.S.F.), National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant K08AR084556 (M.S.F.), and Merit Review Award BX000794 from the Veterans Health Administration Office of Research and Development (R.G.). We thank Dr. Shaun Coughlin for providing the Grhl3PAR2/+ mice and Andrew Schroeder for superb help with the RNA-Seq analysis.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021386118/-/DCSupplemental.
Data Availability
All study data are included in the article, SI Appendix, and Dataset S1.
References
- 1.Hanifin J. M., Rajka G., Diagnostic feature of atopic dermatitis. Acta Derm. Venereol. 92, 44–47 (1980). [Google Scholar]
- 2.Dold S., Wjst M., von Mutius E., Reitmeir P., Stiepel E., Genetic risk for asthma, allergic rhinitis, and atopic dermatitis. Arch. Dis. Child. 67, 1018–1022 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strachan D. P., Wong H. J., Spector T. D., Concordance and interrelationship of atopic diseases and markers of allergic sensitization among adult female twins. J. Allergy Clin. Immunol. 108, 901–907 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Yosipovitch G., Berger T., Fassett M. S., Neuroimmune interactions in chronic itch of atopic dermatitis. J. Eur. Acad. Dermatol. Venereol. 34, 239–250 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paller A. S., Spergel J. M., Mina-Osorio P., Irvine A. D., The atopic march and atopic multimorbidity: Many trajectories, many pathways. J. Allergy Clin. Immunol. 143, 46–55 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Krämer U., et al., Seasonality in symptom severity influenced by temperature or grass pollen: Results of a panel study in children with eczema. J. Invest. Dermatol. 124, 514–523 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Tan B. B., Weald D., Strickland I., Friedmann P. S., Double-blind controlled trial of effect of housedust-mite allergen avoidance on atopic dermatitis. Lancet 347, 15–18 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Szegedi K., et al., House dust mite allergens Der f and Der p induce IL-31 production by blood-derived T cells from atopic dermatitis patients. Exp. Dermatol. 27, 393–395 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Asokananthan N., et al., House dust mite allergens induce proinflammatory cytokines from respiratory epithelial cells: The cysteine protease allergen, Der p 1, activates protease-activated receptor (PAR)-2 and inactivates PAR-1. J. Immunol. 169, 4572–4578 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Lin Y.-P., Nelson C., Kramer H., Parekh A. B., The allergen Der p 3 from house dust mite stimulates store-operated Ca2+ channels and mast cell migration through PAR4 receptors. Mol. Cell 70, 228–241.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Kempkes C., et al., “Role of PAR-2 in neuroimmune communication and itch” in Itch: Mechanisms and Treatment, Carstens E., Akiyama T., Eds. (CRC Press, Boca Raton, FL, 2014). [PubMed] [Google Scholar]
- 12.Reddy V. B., Lerner E. A., Plant cysteine proteases that evoke itch activate protease-activated receptors. Br. J. Dermatol. 163, 532–535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amadesi S., et al., Protease-activated receptor 2 sensitizes the capsaicin receptor transient receptor potential vanilloid receptor 1 to induce hyperalgesia. J. Neurosci. 24, 4300–4312 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinhoff M., et al., Agonists of proteinase-activated receptor 2 induce inflammation by a neurogenic mechanism. Nat. Med. 6, 151–158 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Hassler S. N., et al., The cellular basis of protease-activated receptor 2-evoked mechanical and affective pain. JCI Insight 5, e137393 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinhoff M., et al., Proteinase-activated receptor-2 mediates itch: A novel pathway for pruritus in human skin. J. Neurosci. 23, 6176–6180 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vakharia P. P., et al., Burden of skin pain in atopic dermatitis. Ann. Allergy Asthma Immunol. 119, 548–552.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frateschi S., et al., PAR2 absence completely rescues inflammation and ichthyosis caused by altered CAP1/Prss8 expression in mouse skin. Nat. Commun. 2, 161 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith L., et al., House dust mite-treated PAR2 over-expressor mouse: A novel model of atopic dermatitis. Exp. Dermatol. 28, 1298–1308 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Meng J., et al., New mechanism underlying IL-31-induced atopic dermatitis. J. Allergy Clin. Immunol. 141, 1677–1689.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Buddenkotte J., Steinhoff M., Pathophysiology and therapy of pruritus in allergic and atopic diseases. Allergy 65, 805–821 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Jeong S. K., et al., Mite and cockroach allergens activate protease-activated receptor 2 and delay epidermal permeability barrier recovery. J. Invest. Dermatol. 128, 1930–1939 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Dillon S. R., et al., Interleukin 31, a cytokine produced by activated T cells, induces dermatitis in mice. Nat. Immunol. 5, 752–760 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Ross S. E., et al., Loss of inhibitory interneurons in the dorsal spinal cord and elevated itch in Bhlhb5 mutant mice. Neuron 65, 886–898 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson S. R., et al., The epithelial cell-derived atopic dermatitis cytokine TSLP activates neurons to induce itch. Cell 155, 285–295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raap U., et al., Brain-derived neurotrophic factor is increased in atopic dermatitis and modulates eosinophil functions compared with that seen in nonatopic subjects. J. Allergy Clin. Immunol. 115, 1268–1275 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Cevikbas F., et al., A sensory neuron-expressed IL-31 receptor mediates T helper cell-dependent itch: Involvement of TRPV1 and TRPA1. J. Allergy Clin. Immunol. 133, 448–460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson S. R., et al., TRPA1 is required for histamine-independent, Mas-related G protein-coupled receptor-mediated itch. Nat. Neurosci. 14, 595–602 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakashima C., Otsuka A., Kabashima K., Interleukin-31 and interleukin-31 receptor: New therapeutic targets for atopic dermatitis. Exp. Dermatol. 27, 327–331 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Usoskin D., et al., Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18, 145–153 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Mishra S. K., Hoon M. A., The cells and circuitry for itch responses in mice. Science 340, 968–971 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imamachi N., et al., TRPV1-expressing primary afferents generate behavioral responses to pruritogens via multiple mechanisms. Proc. Natl. Acad. Sci. U.S.A. 106, 11330–11335 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dembo T., Braz J. M., Hamel K. A., Kuhn J. A., Basbaum A. I., Primary afferent-derived BDNF contributes minimally to the processing of pain and itch. eNeuro 5, ENEURO.0402-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonilla I. E., Tanabe K., Strittmatter S. M., Small proline-rich repeat protein 1A is expressed by axotomized neurons and promotes axonal outgrowth. J. Neurosci. 22, 1303–1315 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starkey M. L., et al., Expression of the regeneration-associated protein SPRR1A in primary sensory neurons and spinal cord of the adult mouse following peripheral and central injury. J. Comp. Neurol. 513, 51–68 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hai T., Wolfgang C. D., Marsee D. K., Allen A. E., Sivaprasad U., ATF3 and stress responses. Gene Expr. 7, 321–335 (1999). [PMC free article] [PubMed] [Google Scholar]
- 37.Bráz J. M., Basbaum A. I., Differential ATF3 expression in dorsal root ganglion neurons reveals the profile of primary afferents engaged by diverse noxious chemical stimuli. Pain 150, 290–301 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cevikbas F., et al., Synergistic antipruritic effects of gamma aminobutyric acid A and B agonists in a mouse model of atopic dermatitis. J. Allergy Clin. Immunol. 140, 454–464.e2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oetjen L. K., et al., Sensory neurons co-opt classical immune signaling pathways to mediate chronic itch. Cell 171, 217–228.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson N. A., Lapic S., Welter J. F., Eckert R. L., S100A11, S100A10, annexin I, desmosomal proteins, small proline-rich proteins, plasminogen activator inhibitor-2, and involucrin are components of the cornified envelope of cultured human epidermal keratinocytes. J. Biol. Chem. 272, 12035–12046 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Tsujino H., et al., Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: A novel neuronal marker of nerve injury. Mol. Cell. Neurosci. 15, 170–182 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Zhang X., Wiesenfeld-Hallin Z., Hökfelt T., Effect of peripheral axotomy on expression of neuropeptide Y receptor mRNA in rat lumbar dorsal root ganglia. Eur. J. Neurosci. 6, 43–57 (1994). [DOI] [PubMed] [Google Scholar]
- 43.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 44.Yamamoto M., et al., A novel atopic dermatitis model induced by topical application with dermatophagoides farinae extract in NC/Nga mice. Allergol. Int. 56, 139–148 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Solorzano C., et al., Primary afferent and spinal cord expression of gastrin-releasing peptide: Message, protein, and antibody concerns. J. Neurosci. 35, 648–657 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article, SI Appendix, and Dataset S1.