Significance
A panel of 80 human IgA monoclonal antibodies isolated after seasonal influenza virus vaccination was interrogated. We have discovered an IgA monoclonal antibody binding to an epitope for H1N1 viruses at the head–stalk interface that displays antibody-dependent cell-mediated cytotoxicity activity. Also, a mechanism of antibody Fc-mediated effector function activation was refined for influenza virus, which describes that low-affinity Fc receptors require an additional point of contact between influenza HA and its cognate sialic acid-bearing receptor for activation, while high-affinity Fc receptors can be activated without this contact.
Keywords: influenza, IgA, Fc receptor
Abstract
In this study, we utilized a panel of human immunoglobulin (Ig) IgA monoclonal antibodies isolated from the plasmablasts of eight donors after 2014/2015 influenza virus vaccination (Fluarix) to study the binding and functional specificities of this isotype. In this cohort, isolated IgA monoclonal antibodies were primarily elicited against the hemagglutinin protein of the H1N1 component of the vaccine. To compare effector functionalities, an H1-specific subset of antibodies targeting distinct epitopes were expressed as monomeric, dimeric, or secretory IgA, as well as in an IgG1 backbone. When expressed with an IgG Fc domain, all antibodies elicited Fc-effector activity in a primary polymorphonuclear cell-based assay which differs from previous observations that found only stalk-specific antibodies activate the low-affinity FcγRIIIa. However, when expressed with IgA Fc domains, only antibodies targeting the stalk domain showed Fc-effector activity in line with these previous findings. To identify the cause of this discrepancy, we then confirmed that IgG signaling through the high-affinity FcγI receptor was not restricted to stalk epitopes. Since no corresponding high-affinity Fcα receptor exists, the IgA repertoire may therefore be limited to stalk-specific epitopes in the context of Fc receptor signaling.
Influenza viruses cause substantial morbidity and mortality in the human population around the world. Unfortunately, current vaccines are not ideal in the immunity conferred, as strain mismatches, poor seroconversion rates, and vaccine hesitancy prevent optimal protection from being achieved (1). Vaccination with current inactivated seasonal influenza virus vaccines predominantly elicit antibodies against the hemagglutinin (HA) glycoprotein on the surface of the influenza virion (2). Antibodies which sterilize influenza virus infection through interruption of the receptor binding functionality of the HA are classically considered the predominant protective class of antibodies (2, 3). Additionally, antibodies which bind to HA and neutralize the virus through alternate mechanisms have been shown to contribute to protection (4–6). In recent years, antibodies that are protective through Fc-mediated effector functions have also gained interest in the field (7, 8). Importantly, influenza virus-specific antibody functionalities have been studied extensively in the context of the human immunoglobulin (Ig) G isotype, but less is known of the role of human IgA isotype antibodies.
IgA isotype antibodies can be found in multiple forms: as monomeric IgA, multimeric IgA with a joining-chain between monomers, and secretory IgA with a polymeric Ig receptor cleavage product attached after transcytosis of multimeric IgA (9). Human IgA antibodies are abundant at mucosal sites in the body, including the respiratory tract, where influenza virus establishes infection (10). IgA1-isotype antibodies are found predominantly in serum and the upper respiratory tract, while IgA2 isotype antibodies are most often found in the mucosal tissues of the gut and reproductive tract (11). Influenza virus-specific IgA antibodies have been shown to neutralize similarly to their IgG counterparts, and the ability of these antibodies to interact with Fc receptors to elicit Fc-mediated effector functions has been examined (12–15).
In this study, we have screened a panel of human monoclonal antibodies (mAbs) derived from IgA-secreting plasmablasts to characterize the functionality of the antibody response after inactivated influenza virus vaccination. An investigation into the Fc receptor usage of IgA antibodies compared to their IgG counterparts revealed distinct epitope-dependent Fc receptor-mediated activity. Surprisingly, utilization of a primary human polymorphonuclear cell (PMN)-based assay revealed a lack of epitope dependence on stimulation of Fc-mediated effector functions by all IgG antibodies tested. The presence of a high-affinity Fcγ receptor not typically involved in natural killer-based or reporter-based Fc-receptor stimulation assays was speculated to explain this phenomenon. The observed epitope restriction of IgA antibodies may be owed to the requirement of stabilizing interactions through an additional point of contact between the HA receptor binding site (RBS) and sialic acid on the effector cell due to the lack of a high-affinity Fcα receptor.
Results
Isolated Influenza Virus Vaccine-Specific Human IgA mAbs Predominantly Bind H1.
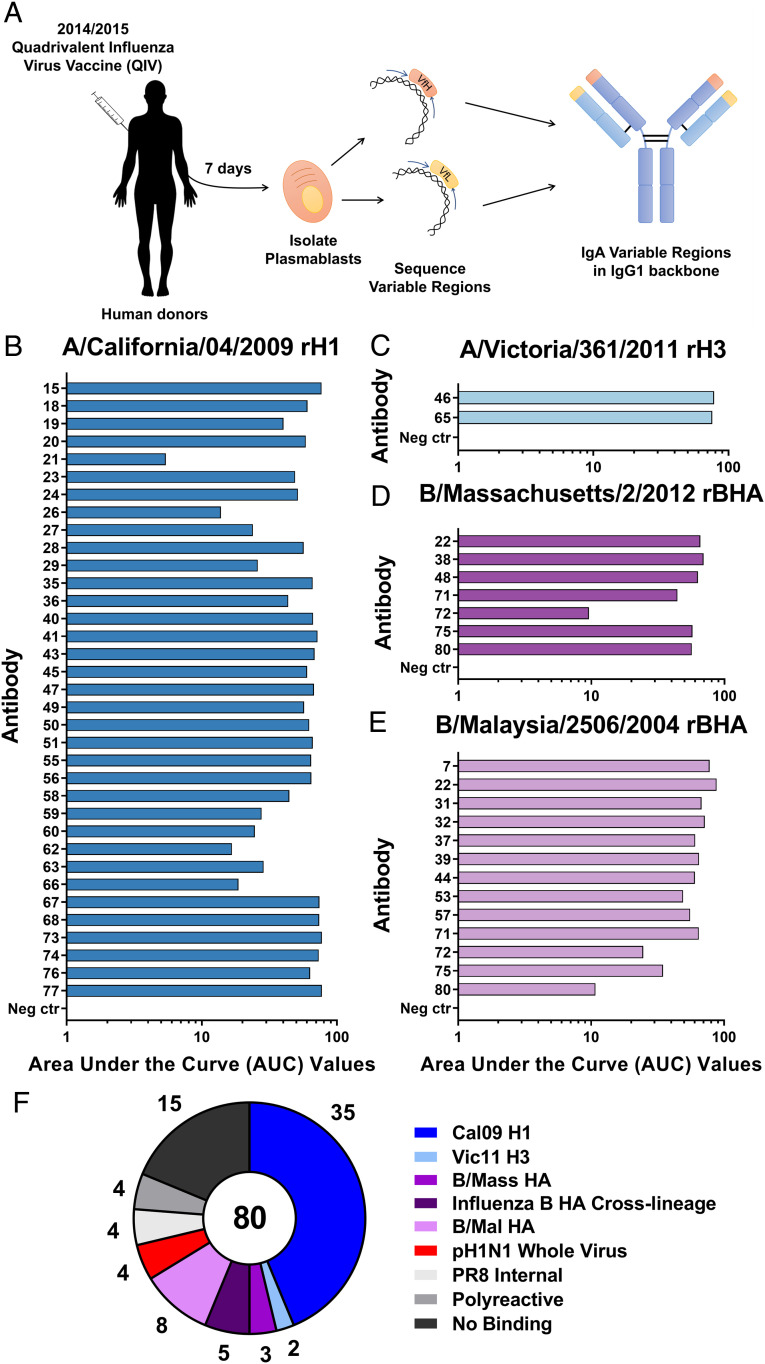
To assess the repertoire of IgA antibodies elicited after seasonal influenza virus vaccination, plasmablasts were isolated from healthy human donors (24 to 34 y old, 50% male/female) 7 d after immunization with the 2014/2015 Fluarix formulation of the quadrivalent influenza virus vaccine (QIV; Fig. 1A and SI Appendix, Fig. S1) (16). Variable region sequences were obtained, and mAbs were expressed and purified as IgG1 isotype antibodies for initial production. mAbs were then screened using an enzyme-linked immunosorbent assay (ELISA) to determine binding to the 2014/2015 Fluarix QIV.
Fig. 1.
Human IgA antibodies after 2014/2015 inactivated influenza virus vaccination primarily bind to H1 HA. (A) Eight human donors were vaccinated with the recommended dose of the 2014/2015 QIV. Seven days after vaccination, plasmablasts were isolated from peripheral blood, and sequencing was performed to specifically isolate IgA variable regions from single-cell suspensions. Cloning of IgA variable regions into an IgG1 backbone was performed prior to expression and purification of mAbs. ELISAs were performed using recombinant HA proteins: (B) A/California/04/2009 H1, (C) A/Victoria/361/2011 H3, (D) B/Massachusetts/2/2012 BHA, and (E) B/Malaysia/2506/2004 BHA. AUC values are reported as a representation of each binding curve. (F) The number of mAbs which bound to each antigen tested by ELISA is reported for the entire antibody panel. Negative control is assay run without antibody present.
A panel of 80 positive IgA-derived mAbs (79 IgA1 and 1 IgA2) were selected for further screening against matched individual protein components of the 2014/2015 Fluarix QIV. The majority of responses observed for this panel were toward the matched H1 recombinant protein, with almost half (44.3%) of antibodies binding this protein (Fig. 1B and SI Appendix, Fig. S2 A–D). In contrast, only two antibodies bound to the H3 protein (Fig. 1C and SI Appendix, Fig. S2E). Specificity to influenza B virus HAs was also less common compared to influenza A virus H1, with 6 mAbs binding to the B/Yamagata16/1988-like lineage-matched HA and 13 antibodies binding to the B/Victoria/2/1987-like lineage-matched HA (Fig. 1 D and E and SI Appendix, Fig. S2 F–H). A fraction of the influenza B virus HA-binding mAbs were able to bind across lineages (5 of a total of 16 influenza B virus HA binding mAbs in this panel), while no influenza A virus cross-group antibodies were detected (Fig. 1F). Though cross-group influenza A HA-binding mAbs have been previously found after seasonal vaccination (17, 18), this lack of broadly reactive influenza A virus antibodies is not uncommon after QIV administration due to the limited amino acid similarity between the H1 and H3 subtypes (19, 20). Cross-lineage influenza B virus-specific mAbs are more commonly elicited and confer protection from natural infection through multiple mechanisms (21, 22). Polyclonal serum samples isolated from the blood of donors were also tested before vaccination and weekly until 1 mo postvaccination against QIV-matched recombinant HA proteins (SI Appendix, Fig. S3). Responses were found to slightly skew toward H1 and BHA components of the vaccine and to be lower for H3.
Initial screening of mAbs through QIV-based ELISA was performed to detect antibodies binding to any influenza virus protein present in the vaccine. Monoclonal antibodies specific to recombinant neuraminidase proteins were not discovered in this panel, which supports the notion that natural infection is superior to seasonal influenza virus vaccination at eliciting this class of antibodies (23). Antibodies specific to whole-virion preparations of both the matched H1N1 virus and backbone virus were also discovered in this panel, confirming that antibodies specific to internal influenza virus proteins or egg-derived proteins are elicited by seasonal vaccination (Fig. 1F) (24).
H1-Specific Antibodies Show Distinct Binding Sites and Functional Profiles.
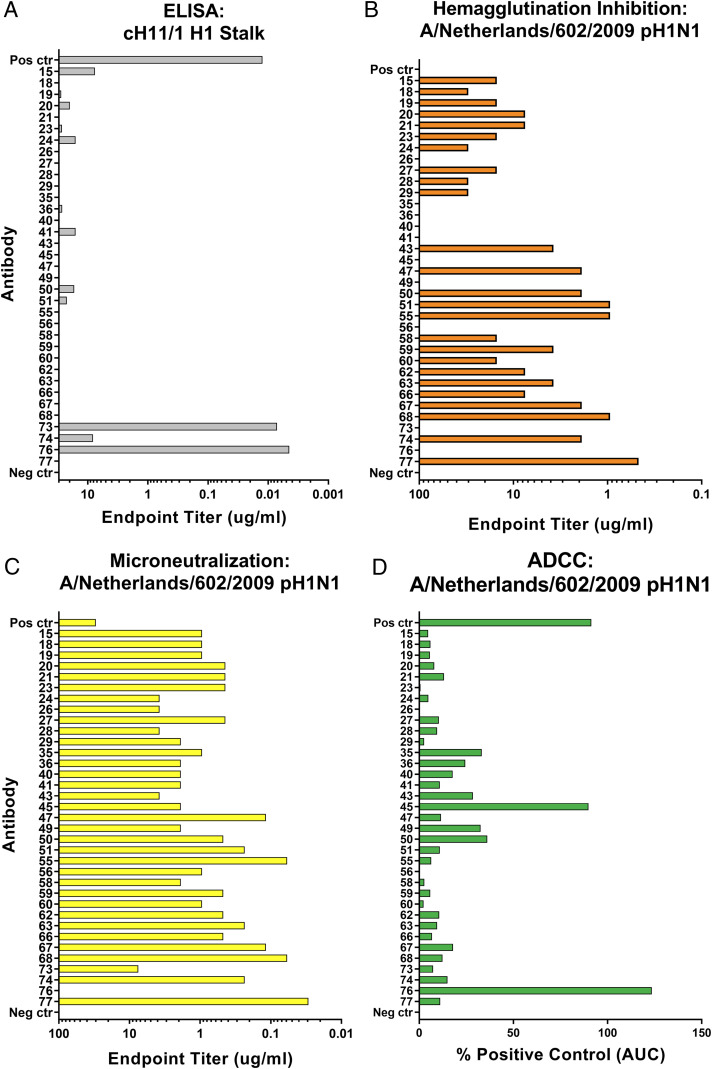
Further examination of the H1-specific IgA-derived mAbs in an IgG1 backbone was performed to determine the diversity present in binding characteristics and functionalities. A chimeric HA molecule expressing an avian H11 head (to which humans are naïve) and an H1 stalk was utilized in ELISAs to discriminate between head- and stalk-binding mAbs. Only 2 antibodies of 35 tested were found to bind the stalk (mAbs 73 and 76), which is consistent with previous reports that seasonal influenza virus vaccines are poor inducers of antibodies targeting the immunosubdominant stalk domain (Fig. 2A and SI Appendix, Fig. S4) (19, 20).
Fig. 2.
Human IgA-derived influenza H1-specific mAbs show distinct binding and functionalities. (A) Recombinant chimeric HA protein bearing the avian A/shoveler/Netherlands/18/99 H11N9 head domain and the A/California/04/2009 H1 stalk domain was utilized to determine stalk-binding mAbs. Reactivity is reported as endpoint titer. (B) Hemagglutination inhibition assay endpoint titers display the ability of mAbs to interfere with receptor binding activity. (C) A multicycle MNT assay revealed neutralizing activity of mAbs in addition to blocking of receptor binding activity, reported as endpoint titer. (D) Fc-mediated functionality was examined through the use of a Promega reporter assay. Engagement by mAbs to FcγRIIIa-bearing effector cells is assayed through luminescent output, a function of luciferase expression. AUC is determined, and values are reported as percent of positive control AUC. Positive control antibody is 3B05 (A–C) or CR9114 (D). Negative control is assay run without antibody present.
To examine functionality, classical hemagglutination inhibition (HI) and microneutralization (MNT) assays were performed. HI assays are restricted to only measuring antibodies which disrupt the interaction between the HA RBS and the sialic acid receptor (25). In our antibody panel, 25 of 35 mAbs were HI-active, albeit to various levels of potency (Fig. 2B). Both stalk-specific antibodies were not HI-active, as the distance of stalk epitopes from the RBS precludes these antibodies from disrupting receptor binding. A multicycle MNT assay is less restricted in detecting the neutralizing activity of mAbs, as antibodies which neutralize through mechanisms outside of interfering with receptor binding can also be discovered (26). These antibodies may neutralize through inhibition of HA fusion, blocking of viral egress, or disruption of neuraminidase activity through steric interference (5, 6, 27–31). When our panel was tested in an MNT assay, only a single antibody (mAb 76) lacked neutralization potential at the tested concentrations (Fig. 2C).
Additionally, stalk-specific antibodies have been reported to protect from influenza virus infection through Fc-mediated effector functions such as antibody-dependent cell-mediated cytotoxicity (ADCC) (7, 8). Classical HI active head-specific antibodies have not been shown to elicit potent ADCC responses due to a requirement for two points of contact, one each between the antibody Fc and Fc receptor as well as the viral RBS and sialic acid bearing receptor, being necessary to provide optimal stabilization of this interaction (32). To assess the ability of our H1-specific antibody panel to elicit Fc-mediated effector functions, ADCC reporter assays were performed utilizing a transgenic Jurkat cell line engineered to express the human low-affinity Fcγ receptor IIIa (FcγRIIIa; Promega). This cell line expresses luciferase under control of a nuclear factor of activated T cells (NFAT) promoter, which becomes active upon cross-linking of the cell-surface FcγRIIIa molecules. H1-specific mAbs were incubated with infected Madin–Darby canine kidney (MDCK) cells and effector cells, after which luciferase expression was measured (Fig. 2D and SI Appendix, Fig. S5). One of the stalk-specific antibodies (mAb 76) was able to induce strong ADCC reporter activity, and an antibody (mAb 45) which did not bind the stalk was also able to induce a strong ADCC signal. This antibody did not show HI activity, though, implying that two points of contact may still be present during the interaction.
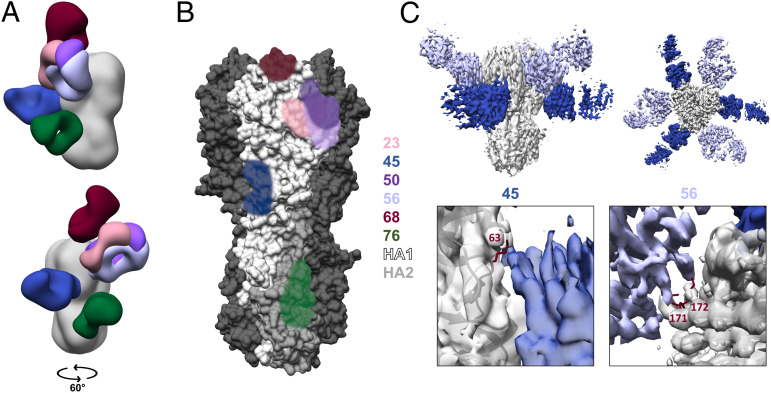
A deeper exploration of epitope-level binding and functionality was limited to a select panel of mAbs displaying distinct and diverse characteristics (Table 1). Epitope-level binding properties were interrogated through the use of single-particle microscopy. Antigen-binding fragments (Fabs) of six antibodies were produced and complexed to a stabilized form of recombinant H1 protein (33). Resulting negative-stain three-dimensional reconstructions were compiled to represent each individual Fab binding to a single protomer of the HA trimer (Fig. 3A and SI Appendix, Fig. S6 A and B). These data provided binding footprints of each antibody in the selected panel and confirmed the stalk-binding nature of mAb 76 and a range of head-binding specificities of mAbs 23, 50, 56, and 68 (Fig. 3B). Interestingly, mAb 45 was found to bind an epitope at the head–stalk interface, which has been previously described for other influenza subtypes but is also an epitope for the H1 subtype (34–38). Additionally, competition ELISAs were performed to determine the extent to which these antibodies overlap in the epitopes bound (SI Appendix, Fig. S7). mAbs 45 and 76 were found to lack competition with other mAbs in the panel, while the head-specific antibodies overlapped to varying extents with one another. Denaturing Western blots were also performed to determine if antibodies bound linear or conformational epitopes (SI Appendix, Fig. S8). mAbs 45 and 76 were found to lack binding to denatured recombinant H1 protein, and binding was concluded to be restricted to conformational epitopes.
Table 1.
Selection of a panel of functionally diverse IgA antibodies for detailed characterization
| mAb | Stalk | HI | MNT | ADCC | IgG kDa | IgA kDa |
| 23 | — | + | ++ | — | 3.07 × 10−9 | 5.24 × 10−8 |
| 45 | — | — | + | + | 1.59 × 10−9 | 4.93 × 10−8 |
| 50 | — | ++ | ++ | — | 3.71 × 10−11 | 4.18 × 10−8 |
| 56 | — | — | ++ | — | 8.14 × 10−10 | 4.66 × 10−8 |
| 68 | — | +++ | +++ | — | 1.21 × 10−10 | 4.48 × 10−8 |
| 76 | + | — | — | + | 2.32 × 10−10 | 2.26 × 10−8 |
A subset of H1-specific IgA mAbs was selected for detail analysis. Binding and functional activities were ranked based on the following schemes. Stalk cH11/1 binding endpoint titer, <1 μg/mL +; HI endpoint titer, <100 μg/mL +, <10 μg/mL ++, <1 μg/mL +++; MNT endpoint titer, <10 μg/mL +, <1 μg/mL ++, <0.1 μg/mL +++; ADCC, >50% positive control +. Antibody affinity is reported as dissociation constant (kDa) for IgG and IgA isotype mAbs binding to recombinant A/California/04/2009 H1.
Fig. 3.
Electron microscopy reveals binding sites of selected IgA antibodies. (A) Composite model of Fabs produced from selected IgA mAbs, modeled onto the HA trimer (Protein Data Bank ID code 4M4Y) resulting from individual 3D negative stain reconstructions. (B) Footprints of mAb binding sites are represented on a single HA monomer and colored for each corresponding mAb. (C) High-resolution cryo-EM reconstruction of Fabs 45 and 56 binding to the H1 trimer (Upper). Zoomed view of specific contacts with escape mutants modeled in maroon (Lower). Amino acid position is numbered following H3 numbering convention.
The breadth of reactivity of this antibody panel was investigated through use of a selection of recombinant HA proteins representative of the diversity within the H1 subtype and including an H5 protein to test heterologous binding within Group 1 (SI Appendix, Fig. S9). All selected antibodies were found to bind postpandemic H1 recombinant proteins as both IgG and IgA isotypes. Only mAb 76 was able to bind to all recombinant proteins tested, which confirms the broadly reactive nature of stalk-specific antibodies. Differences in the breadth of the antibody response were not altered by the isotype of the tested antibody.
To provide additional evidence for the binding sites of these antibodies and examine the neutralizing pressure applied by this select panel of mAbs, escape mutants were generated. Influenza viruses were cultured in the presence of increasing amounts of neutralizing antibody until 100 times the median inhibitory concentration for each antibody was no longer effective at preventing replication. High resolution cryo-electron microscopy (cryo-EM) reconstructions of mAbs 45 and 56 bound to HA reveal that the escape mutations are located at critical contact sites for each mAb (Fig. 3C and SI Appendix, Figs. S6C and S10). Due to the lack of neutralizing activity, mAb 76 could not be utilized for escape mutant production. To remedy this issue, modeling was performed, which compared mAb 76 to other stalk-binding antibodies, mAb CR9114 and mAb 39.29 (30, 39). The epitope bound by mAb 76 was found to be similar to these antibodies, and fitting previously modeled Fabs into the electron density revealed a proper alignment for mAb 39.29 and alignment with some rotational discrepancy for mAb CR9114 (SI Appendix, Fig. S11).
Optimal Activation of Low-Affinity, but Not High-Affinity, Fc Receptors Requires a Second Point of Contact.
Further investigation of the functional activity of the selected mAbs required cloning the antibody variable regions back into a human IgA1 backbone. Antibodies were then expressed in multiple forms as monomeric, dimeric, and secretory IgA through cotransfection of human J-chain alone or in addition to human secretory component during expression. mAbs were then purified and confirmed by ELISA (SI Appendix, Fig. S12).
IgG and IgA antibodies were then utilized in a primary human polymorphonuclear cell (PMN) assay to determine the ability of mAbs to activate primary human cells through Fc-mediated effector functions (40). PMNs were isolated from whole blood and incubated with purified virion and respective mAbs to induce reactive oxidative species (ROS) burst, which is detected through luminescence emitted by the small molecule luminol. IgA antibodies showed a pattern of Fc-mediated activation similar to that seen in the low-affinity FcγRIIIa ADCC reporter assay for IgG antibodies, whereby only stalk-specific IgA mAbs were able to stimulate PMNs (Fig. 4A and SI Appendix, Fig. S13). All IgA forms expressed were able to stimulate Fc effector mechanisms to a similar degree and were not compared further. These findings were confirmed by testing PMNs from two additional human donors (SI Appendix, Fig. S14). Interestingly, head-specific IgG antibodies were able to elicit responses in this assay, albeit to a lesser degree. This action of IgG antibodies was inconsistent with the current model of an additional contact being necessary to optimally stimulate Fc-mediated effector functions and demanded further investigation.
Fig. 4.
Fc-mediated effector functions elicited through low-affinity Fc receptors require stabilization provided by an additional point of contact. (A) Primary human PMNs were isolated to assay for Fc-receptor functionality of IgG and IgA isotype mAbs. PMNs were incubated with 2.5 μg of respective antibody in either IgG, monomeric IgA, dimeric IgA, or secretory IgA form complexed to 10 μg of purified influenza virion. Maximum assay signal after kinetic examination of ROS burst is reported as the average of experimental duplicates plus SD. (B) Activation of a high-affinity FcγRI was assessed using a Promega reporter assay. Respective mAbs were incubated with influenza virus-infected cells and FcγRI-bearing cells engineered to express luciferase upon activation. AUC is calculated for each luminescent dose titration curve, normalized to the positive control AUC, and represented as the average of experimental duplicates plus SD. Positive control antibody is CR9114, isotype control is AA12 (Zika virus-specific), and negative control is assay run without antibody present.
Isolation of PMNs from peripheral blood results predominantly in the purification of human neutrophils, as these cells constitute around 40 to 60% of circulating white blood cells (41). Basophils and eosinophils may also be isolated through this method, but constitute roughly 0.5 to 1% or 1 to 4% of peripheral white blood cells, respectively (41). Human neutrophils express multiple Fc receptors, including the low-affinity Fcα receptor (FcαR), FcγRIIa, and FcγRIIIa, as well as the high-affinity receptor FcγRI (42, 43). The high-affinity FcγRI is generally sparsely expressed on the cell surface, but is quickly induced in response to inflammatory signals (43). Considering the lack of a high-affinity Fcα receptor and the absence of activation of PMNs by head-specific IgA isotype antibodies, we explored whether IgG isotype antibodies were able to stimulate Fc-mediated effector functions through the high-affinity FcγRI. We utilized a Promega antibody-dependent cell-mediated phagocytosis reporter assay similar to the ADCC reporter assay but with the high-affinity FcγRI stably expressed on the Jurkat cell surface. Incubation of IgG isotype antibodies with infected cells and FcγRI-expressing effector cells revealed a similar pattern observed with human PMNs, whereby stalk-specific antibodies elicited strong responses and head-specific antibodies elicited moderate responses (Fig. 4B and SI Appendix, Fig. S15). Correlations were subsequently performed to determine the role of antibody affinity in activation of Fc-mediated effector functions through these assays (SI Appendix, Fig. S16). A strong correlation was found between antibody affinity and the activation of PMNs, suggesting that the stability of antibody binding impacts the resulting ROS burst. Interestingly, this correlation did not stand when Fc receptor-based reporter assays were utilized with either low- or high-affinity FcγRs. We therefore conclude that the previously described “two-contact hypothesis” applies to low-affinity Fcγ and Fcα receptors, but not to the high-affinity FcγRI.
Discussion
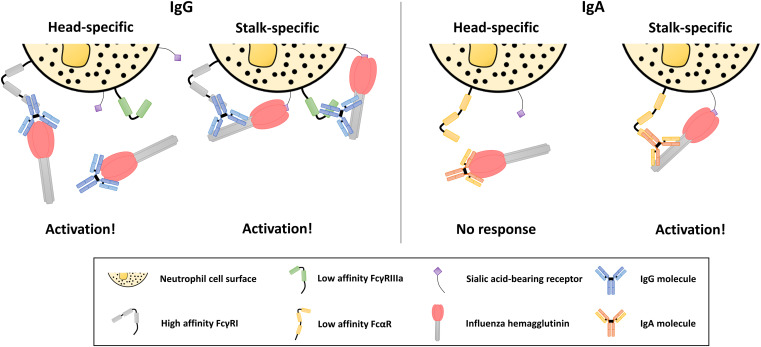
Based on the results of this study, we posit the following mechanism (Fig. 5). Influenza virus HA stalk-specific antibodies are potent activators of Fc-mediated effector functions due to their ability to bind strongly to the HA without occluding the RBS. This epitope specificity allows for the RBS to bind sialic acid-bearing receptors on immune cell surfaces, which acts to stabilize the immunological synapse and increase signaling of low-affinity Fc receptor-dependent pathways, leading to functional activity (32). In the context of influenza virus HA head-specific antibodies, this interaction is blocked, and therefore the overall strength of the immunological synapse is diminished, preventing activation. This phenomenon is altered in the presence of a high-affinity Fc receptor. The affinity of the interaction between the receptor and the antibody Fc region is strong enough to allow activation in the absence of a second point of contact. The additional interaction between the RBS and sialic acid-bearing receptor does still act to strengthen the synapse and therefore leads to a more potent response.
Fig. 5.
Activation of Fc receptors by IgA antibodies is restricted to stalk epitopes due to lack of a high-affinity FcαR and requires a second point of contact. Activation of Fc receptors by antibodies is governed by the epitope present on the antigen (HA), the affinity of the Fc receptor engaged, and the availability of the sialic acid-binding site on the HA, which acts to stabilize the interaction between immune cells and the virus (HA). This second point of contact conferred by the sialic acid-binding site of influenza viruses is necessary for the engagement of low-affinity Fc receptors by both IgG and IgA isotype antibodies. Antibodies which occlude the RBS disrupt this interaction and are therefore only able to activate immune cells through a high-affinity receptor. IgA isotype antibodies lack a corresponding high-affinity receptor and thus are restricted to antibodies which do not interfere with sialic acid-binding activity to stimulate Fc-mediated effector functions. Not drawn to scale.
Activation of the high-affinity FcγRI by head-specific IgG antibodies is surprising, as Fc-mediated functionality was previously thought to be restricted to stalk-specific antibodies. Assays utilized to test antibody Fc effector functionality rarely use systems which contain FcγRI expression, and therefore the Fc-mediated functionality of head-specific IgG antibodies may have been missed. Further investigation should be performed to determine the role this specific aspect of anti-influenza virus immunity has on disease progression or protection.
Interestingly, the isolated mAb which binds to an H1 epitope at the head–stalk interface (mAb 45) appears to follow these rules in reporter assays where target cells are infected and the Fc receptor of interest is overexpressed on the cell surface, but does not act to activate the low-affinity FcαR when primary cells are interacting with purified virion. This implies that the mechanism described above requires further refinement and that edge cases (such as this unique antibody) point to further effects such as receptor abundance, steric hinderance, or potentially an additional point of contact between effector cells and infected cells playing a role in Fc receptor activation.
IgA isotype antibodies have been described to inhibit influenza virus infection through many of the same classical mechanisms described for IgG-isotype antibodies (13, 15, 40). Additionally, it has been shown that IgA antibodies are able to neutralize influenza viruses through sialic acid termini on glycans located near the C terminus of the Fc region of the molecule (44). IgA antibodies have also been shown to prevent influenza virus transmission more effectively than their IgG counterparts (45). Though both of these isotypes are able to engage and function through Fc-mediated effector functions, the underlying Fc receptor biology governing these interactions is starkly different for IgG versus IgA. IgG antibodies can engage with a variety of Fc receptors, including both high-affinity (FcγRI) and low-affinity receptors (FcγRIIa, FcγRIIb, FcγRIIIa, and FcγRIIIb) (46). This diversity in receptor usage allows for activating and inhibitory receptors expressed on a variety of immune cells, which results in a complex arrangement of the underlying immunology. IgA antibodies engage only two Fc receptors, the low-affinity FcαR and the moderate-affinity Fcα/μR, the latter of which is mostly involved in immune complex capture by antigen-presenting cells and is not thought to be involved in directly combating infection through eliciting effector functions (47–49). Furthering this point, the common laboratory mouse (Mus musculus) lacks an FcαR entirely, though it still generates a substantial amount of IgA antibody in response to a variety of infections (50). The reason for this absence is unclear; however, this deletion appears to be the exception rather than the rule, but may indicate that FcR-mediated functionality is more limited for IgA antibodies on mucosal surfaces (51).
The distribution of IgA mAbs in our panel binding to each of the four components present in the seasonal QIV was heavily skewed toward the H1 component of the vaccine. This phenomenon could be due to an inherent immunodominance of the H1 protein relative to the other strains or may also be due to “original antigenic sin,” given the younger age of individual donors in this cohort and assuming that individuals were primed by infection with H1N1 viruses (52, 53). IgA antibodies have also been found to be primarily strain-specific in younger individuals after seasonal influenza virus vaccination, which could explain this skewing of the plasmablast response toward the H1 component of the vaccine (54). The presence of cross-lineage reactive anti-influenza B virus HA antibodies may be the result of formulating seasonal influenza virus vaccines to contain representatives of both B/Victoria/2/1987-like and B/Yamagata/16/1988-like lineage influenza B virus strains. The diversity seen between influenza B virus strains is far less than that seen for influenza A viruses, explaining the difficulty in eliciting cross-group influenza A antibodies by seasonal vaccination (19, 20).
It has been found that IgG and IgA isotype vaccine-stimulated plasmablasts are largely similar in terms of transcriptional profile, suggesting that functionality may largely rely on cell-specific interactions governed by Fc–Fc receptor pairing (55). The IgA repertoire assessed reflects the serum antibody response to vaccination, which may be different from the mucosal or tissue immunity present in the body after infection due to differences in B cell repertoires present at those sites (56). Live attenuated influenza virus vaccines have been described to potently stimulate local IgA responses in the upper respiratory tract due to their administration directly to the mucosa (57). Though the stimulated response is potent in children, adults respond poorly to these vaccines due to preexisting immunity, dampening replication of the vaccine virus (58). Strategies to overcome these obstacles to immunogenicity in adults may lead to improved vaccines, which stimulate local IgA responses that are functional at preventing infection with less overall pathology (14).
The immunological response to influenza virus is complex and incorporates facets of protection from simple neutralization to Fc-mediated effector functions acting through various immune cells. Comprehension of this network of interactions is necessary to inform the rational design of vaccines, which act to elicit responses that correlate with protection from infection and transmission. Vaccines which elicit broadly reactive antibodies that act primarily through Fc-mediated effector functions are currently being pursued in the search for universal influenza virus vaccines. Though drawbacks have been proposed to utilizing vaccine constructs which stimulate only stalk-specific responses, such as the potential for escape mutation (59), vaccines which elicit this class of antibodies have shown promise in human clinical trials (60, 61). Understanding the mechanisms of protection and interactions governing the response to influenza virus is critical for furthering the pursuit of more efficacious influenza virus vaccines.
Materials and Methods
Cells, Viruses, and Recombinant Proteins.
Human embryonic kidney (HEK) 293T cells (American Type Culture Collection [ATCC]), MDCK cells (ATCC), and human alveolar basal epithelial A549 cells (ATCC) were grown at 37 °C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes; Gibco), 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco). HEK Expi293F cells were grown in Expi293 expression medium (Gibco) at 37 °C and 8% CO2, shaking at 125 rpm.
Influenza A virus A/Netherlands/602/2009 pH1N1 (Neth09) was grown in 10-d-old embryonated chicken eggs (Charles River) at 37 °C for 48 h before harvesting allantoic fluid. Hemagglutination positive samples were pooled, aliquoted, and frozen at −80 °C to produce a viral stock. To make a purified virus stock, allantoic fluid was layered onto 5 mL of a 30% sucrose solution. After spinning at 100,000 × g for 2 h at 4 °C, supernatant was removed, the remaining pellet was dissolved in phosphate-buffered saline (PBS), and protein concentration was determined by a Bradford assay. The stock was aliquoted and frozen at −80 °C.
Recombinant influenza virus glycoproteins A/California/04/2009 H1, A/Michigan/45/2015 H1, A/New Caledonia/20/1999 H1, A/Puerto Rico/8/1934 H1, A/Victoria/361/2011 H3, A/Texas/50/2012 H3, A/Indonesia/05/2005 H5, B/Massachusetts/2/2012 BHA, B/Malaysia/2506/2004 BHA, A/California/04/2009 N1, A/Texas/12/2007 N2, B/Wisconsin/1/2010 BNA, B/Massachusetts/2/2012 BNA, and cH11/1 (A/shoveler/Netherlands/18/99 H11N9 head domain and the A/California/04/2009 pH1N1 stalk domain) were produced in a baculovirus system as previously described (62). Purified proteins were stored at −80 °C until use.
Isolation and Sequencing of Human mAbs.
Human mAbs were generated as previously described (63). In brief, peripheral blood was obtained from eight healthy human donors (24 to 34 y old, 50% male/female) in acid citrate dextrose tubes approximately 7 d after vaccination with the 2014/2015 Fluarix QIV (Glaxo SmithKline), and RosetteSep Human B Cell Enrichment Mixture (STEMCELL Technologies) was added at a ratio of 125 μL per 50 mL whole blood. After incubation for 20 min at room temperature (RT), blood was diluted 1:1 in PBS with 2% bovine serum albumin (BSA; Sigma). Diluted blood (25 mL) was layered onto 25 mL of Lymphoprep separation media (STEMCELL Technologies) and spun at 800 × g for 20 min without break. The resulting B cell layer, which resides just above the Lymphoprep media, was moved to a new tube and washed twice with PBS with 2% BSA before spinning at 500 × g for 5 min at 4 °C. Cells were filtered through a 70-μm cell strainer and brought to ∼1 × 107 cells per milliliter in PBS with 2% BSA. Cells were stained with α-human CD3 FITC (clone 7D6, 1:50; Invitrogen), α-human CD19 Pacific Blue (clone H1B19, 1:100; Biolegend), α-human CD27 PE (clone O323, 1:100; Biolegend), and α-human CD38 AF647 (clone HIT2, 1:200; Biolegend) for 30 min on ice in the dark. Cells were washed with PBS with 2% BSA and filtered through a 35-μm filter-cap FACS tube (Thermo Fisher). IgG+ and IgA+ plasmablasts (CD3−CD19+CD27hiCD38hi) were FACS-sorted into 500 μL Roswell Park Memorial Institute (RPMI) medium (Invitrogen), supplemented with 1% penicillin-streptomycin (Gibco), 1% Hepes (Invitrogen), 1% L-glutamine (Gibco), and 10% heat-inactivated FBS (Gibco). Further single-cell sorting (BD FACSAria Fusion) into 96-well plates was performed into 10 μL of catch buffer (nuclease-free water [Invitrogen], 1% 1 M Tris⋅HCl, pH 8 [Thermo Fisher], 2.5% RNase inhibitor [Clontech]). Reverse transcription master mix was prepared by adding 3 μL 5× buffer mix (Thermo Fisher), 1.5 μL 5% IGEPAL (Sigma-Aldrich), and 1.5 μL Maxima Enzyme Mix (Thermo Fisher) together per well. Master mix (6 µL) was added to each well, and plates were spun at 700 × g for 10 s at RT. Reverse transcription was performed by incubating at 25 °C for 10 min, then 50 °C for 30 min, 85 °C for 5 min, and a final hold at 4 °C. Immunoglobulin heavy- and light-chain genes were then amplified by PCR (primer details have been previously described in detail [63]); sequenced; cloned into AbVec human IgG1 heavy-, human kappa light-, or human lambda light-chain expression vectors (14); and cotransfected into HEK 293T cells. Antibody isotypes were confirmed using Vgenes, an in-house interface built on IgBlast (National Center for Biotechnology Information), and by single-cell RNA sequencing.
Expression and Purification of Human mAbs.
To express antibodies for screening, an HEK 293T-based system was utilized. Cells were plated overnight at 2.5 × 106 cells in 10 mL in a 10-cm tissue culture-treated dish. Twenty-four hours later, 15 μg of antibody heavy-chain and 15 μg of antibody light-chain plasmid were mixed in 1.5 mL of Opti-MEM (Gibco). Trans-IT LT1 (75 µL; Mirus) was added to 1.5 mL of Opti-Mem, and the DNA and transfection reagent were mixed and allowed to sit at RT for 30 min undisturbed. Media was removed from the cells, and a rinse with PBS was performed. Ten milliliters of 9:1 Opti-MEM:cDMEM was added to the cells. The transfection mixture was added drop-wise, and the cells were then placed at 37 °C for 5 d. Supernatant was then harvested and spun at 4,000 × g for 30 min at 37 °C and filtered through a 0.22-μm filter before proceeding to purification.
To express IgG and IgA antibodies for selected analyses, an HEK 293F system was utilized. Cells were seeded at a concentration of 2 × 106 cells per milliliter 1 d prior to transfection. The following day, cells were counted, and 30 mL of cells at 3 × 106 cells per milliliter were added to a 125-mL tissue culture-treated Erlenmeyer flask (Corning). Transfection reagents were prepared as described above. After adding transfection mixture drop-wise, cells were incubated overnight at 37 °C and 8% CO2 with shaking at 125 rpm. ExpiFectamine 293 Enhancers 1 and 2 (Thermo Fisher) were added 24 h post transfection per the manufacturer’s instructions. The cells were allowed to continue incubating for an additional 4 d. The supernatant was then harvested and filtered as described above before proceeding to purification.
IgG isotype antibodies were purified using gravity flow over a bed of protein G Sepharose (Invitrogen). In short, the column was washed with three column volumes (CVs) of PBS before application of the supernatant. Flowthrough was run over the column an additional two times before washing with three CVs of PBS. Elution was performed into 2 M Tris⋅HCl, pH 10, by addition of 0.1 M glycine at pH 2.5 to the column at a ratio of 1:9. Eluate was stored at 4 °C until buffer exchange. The column was then washed with three CVs of PBS and stored in 20% ethanol in PBS.
IgA isotype antibodies were purified using an Äkta high-performance liquid chromatography (HPLC) system (GE Healthcare) with a 1-mL IgM HiTrap column (GE Healthcare). The IgM HiTrap utilizes an ammonium sulfate precipitation followed by purification with a 2-mercaptopyridine thiophilic resin useful for isolation of polymeric immunoglobulins. Before application, supernatant was brought to 1 M of ammonium sulfate through slow addition and shaking. The column was washed with 10 CVs of IgM HiTrap buffer (20 mM sodium phosphate, 5:24 monobasic:dibasic, with 1 M ammonium sulfate; Sigma), sample was applied, and the column was washed with an additional 10 CVs of IgM HiTrap buffer. Elution was performed using IgM HiTrap buffer lacking ammonium sulfate. Peak fractions were pooled and buffer-exchanged to PBS using an Amicon Ultra-15 filter with 30-kDa cutoff (Mirus). Protein concentration was determined by nanodrop with absorbance at 280 nm or Bradford assay read at 595 nm.
Antibody expression yields were as follows: 293T transfection followed by Protein G Spin Trap purification gave roughly 0.2 to 0.6 mg antibody per milliliter of supernatant, 293F transfection followed by gravity-flow Protein G purification gave around 0.3 to 0.8 mg antibody per milliliter of supernatant, and IgM HiTrap column gave a yield of roughly 1 mg antibody per milliliter of supernatant.
ELISAs.
Immulon 4 HBX flat-bottomed 96-well dishes (Thermo Fisher) were coated with 100 ng of recombinant protein in PBS and kept overnight at 4 °C. The next day, the plates were washed three times with PBS plus 0.1% Tween 20 (PBST; Fisher) and blocked with 0.5% milk and 3% goat serum (Gibco) in PBST for 1 h at RT. mAbs or polyclonal sera were diluted in blocking buffer using threefold serial dilutions and added to the blocked plates for 2 h at RT. Plates were washed three times with PBST, then horseradish peroxidase (HRP)-linked secondary antibody was diluted to the appropriate concentration (anti-human IgG, Fab-specific, 1:3,000 [Sigma, A0293]; anti-human IgA, 1:3,000 [Sigma, A0295]; anti-human secretory component, 1:750 [Nordic-MUbio, GAHu/SC]) and added to each well for 1 h at RT. Plates were washed four times with PBST, and 100 μL of ο-phenylenediamine dihydrochloride substrate (Sigma) was added for 10 min before quenching with 3 M HCl (Fisher). Plates were read on a Synergy H1 hybrid multimode microplate reader (BioTek) at an absorbance of 490 nm. Data were analyzed using GraphPad 8 (Prism), and area under the curve (AUC) was determined using a background cutoff of the average of negative wells plus three times the SD or 0.07, whichever value was higher. AUC values under 1 were adjusted to 1.
Competition ELISAs used a similar setup, with some modification. After blocking, 10 times the EC80 of each blocking mAb was diluted in blocking buffer, and 100 μL was added to each well and incubated at RT for 2 h. In the meantime, detection antibodies were biotinylated using EZ-Link NHS-PEG4-Biotin, No-Weigh Format (Thermo Fisher) by incubating 2 μL of biotin working stock with 100 μL of mAb at 1 mg/mL at RT for 30 min. mAbs were buffer-exchanged into PBS as described above and measured for concentration using a nanodrop. Plates were washed, and detection mAb was added at 30 μg/mL and serially diluted threefold in blocking buffer. Plates were incubated at RT in the dark for 2 h before washing. Pierce high-sensitivity streptavidin-HRP (Thermo Fisher) was diluted 1:3,000, and 50 μL was added to each well and incubated for 1 h in the dark. Plates were developed as described above. Data were analyzed using GraphPad 8 (Prism), and half-maximal effective concentration was determined after fitting a nonlinear regression curve.
Hemagglutination Inhibition Assay.
Influenza virus A/Netherlands/602/2009 pH1N1 hemagglutination activity was determined by serial-diluting virus twofold in PBS across a V-bottom 96-well dish, leaving 50 μL in each well. Chicken red blood cells (RBCs; Lampire) were diluted to 0.5% in PBS, and 50 μL was added to each well. The virus/RBC mixture was allowed to develop at 4 °C for 1 h. Wells in which RBC pellets were not observed were considered positive, and the last dilution at which the virus agglutinated was considered the viral hemagglutination titer.
Influenza virus A/Netherlands/602/2009 pH1N1 was diluted to a titer of four HA units in PBS and kept on ice. mAbs were serial-diluted twofold in PBS and mixed at a ratio of 1:1 with diluted virus. The mixture was shaken at RT for 1 h. Virus/mAb mixture (50 μL) was added to a V-bottom 96-well dish, and 50 μL of 0.5% chicken RBCs (Lampire) in PBS was added and agitated gently to mix. The plates were allowed to develop at 4 °C for 1 h. Wells which lacked agglutination were deemed HI-positive, and the concentration of mAb in the final well showing a lack of agglutination was determined as the endpoint titer.
Microneutralization Assay.
MDCK cells were plated in tissue culture-treated flat-bottom 96-well dishes to a final count of 2.5 × 104 cells per well and allowed to culture overnight at 37 °C and 5% CO2. The next morning, mAbs were diluted twofold in infection media (Ultra MDCK media [Lonza] supplemented with 6-[1-tosylamido-2-phenyl] ethyl chloromethyl ketone [TPCK]-treated trypsin at a concentration of 1 μg/mL) and added at a 1:1 ratio to 100 times the median tissue culture infectious dose (TCID50) of influenza virus A/Netherlands/602/2009 pH1N1 diluted in infection media. The mixture was shaken at RT for 1 h. MDCK cells were rinsed with PBS, 100 μL of virus/mAb mixture was added, and the plate was placed at 33 °C for 1 h. Cells were then rinsed with PBS, and wells were filled with 100 μL mAbs diluted a further twofold with infection media. Infection was allowed to proceed for 72 h at 33 °C before an HA readout was performed by mixing 50 μL of supernatant from each well with 50 μL of 0.5% chicken RBCs (Lampire) and allowing development at 4 °C for 1 h. Endpoint was determined as the concentration of mAb in the final well showing a lack of agglutination activity.
Antibody-Dependent Cell-Mediated Cytotoxicity Reporter Assay.
MDCK cells or A549 cells were plated in white-walled, tissue culture-treated, flat-bottomed, 96-well dishes (Costar no. 3917) at a concentration of 2.5 × 105 cells per milliliter in 100 μL of complete DMEM and cultured overnight at 37 °C and 5% CO2. The following morning, cells were washed with PBS and infected with influenza virus Neth09 at a multiplicity of infection of 5 in UltraMDCK media (Lonza). Infection was allowed to proceed for 24 h in the absence of TPCK-treated trypsin. Assay buffer was prepared as RPMI 1640 media supplemented with 4% Ultra-Low IgG FBS (Gibco). mAbs were serial-diluted threefold in assay buffer. Media was removed from infected cells, and 25 μL of assay buffer was added, along with 25 μL of diluted mAbs. Promega human effector cells (Jurkat cells expressing human FcγRIIIa or FcγRI with NFAT-driven luciferase reporter gene) were diluted in warm assay buffer to a final concentration of 3 × 106 cells per milliliter, and 25 μL was added to the infected cells. The reaction was allowed to incubate for 6 h at 37 °C and 5% CO2 before removing the plate to RT for 15 min. BioGlo luciferase substrate (75 μL; Promega) was added to each well, and luminescence was read immediately using a Synergy H1 hybrid multimode microplate reader (BioTek). AUC was determined using Prism 8 (GraphPad) with a baseline cutoff as the average plus three times the SD of negative control wells. Percent positive AUC was determined through normalizing AUC values to the average of positive control wells.
Generation of Escape Mutants.
MDCK cells were seeded in a tissue culture-treated six-well dish to a final count of 5 × 105 cells per well and incubated overnight at 37 °C and 5% CO2. Influenza virus Neth09 was diluted to 100× TCID50 in PBS and maintained on ice. mAbs were diluted to 1/10 the neutralization endpoint titer in PBS as determined by MNT assay as described above. A 1:1 mixture of virus:mAb was prepared and shaken at RT at 300 rpm for 1 h. MDCK cells were washed with PBS, and 250 μL of the virus/mAb mixture was added and incubated at 37 °C and 5% CO2 for 1 h with periodic shaking to prevent the cells from drying out. Cells were then washed with PBS to remove unbound virus, and 2 mL of Ultra MDCK media (Lonza) supplemented with TPCK-treated trypsin at 1 μg/mL and mAb at the same neutralizing concentration as incubated with the virus was added to each well. Infection was allowed to proceed for 48 h at 37 °C and 5% CO2, after which a hemagglutination readout was performed to determine if virus escaped antibody pressure. Wells with HA-positive virus were passaged with increasing concentrations of antibody until ∼100 times the neutralizing endpoint titer was reached. Control virus was passaged in the presence of an isotype control antibody (2B9 anti-influenza H3, 18003 anti-HRV, AA12 anti-Zika virus) or in the absence of antibody in parallel for each mAb. RNA was extracted from escape mutant viruses and controls using the QIAamp Viral RNA Mini Kit (Qiagen), and PCR products were produced using the SuperScript III One-Step RT-PCR with Platinum Taq HiFi kit (Invitrogen). Sanger sequencing was performed using A/Netherlands/602/2009 pH1N1 HA-specific primers (forward, CAACCGCAAATGCAGACACA; and reverse, ACAAGGGTGTTTTTCTCATGCT) to determine the escape mutation after removal of any additional mutations found in the control virus.
mAb Digestion and Immune Complexing.
Antibody Fabs were produced through incubation of full-length mAbs with activated papain (Sigma P3125) in digestion buffer (10 mM L-cysteine, 100 mM Tris⋅HCl, 2 mM ethylenediaminetetraacetic acid [EDTA] in H2O) and allowed to incubate at 37 °C for 4 h, followed by quenching with 10 mM iodoacetamide. Digested mAbs were run by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) to confirm cleavage, then purified by size-exclusion chromatography using a Superdex 200 Increase 10/300 column operated on an Äkta HPLC system (GE Healthcare). Isolated Fabs were buffer-exchanged into PBS using an Amicon Ultra filter (Millipore). Fabs were then complexed with recombinant HA protein (A/California/04/2009 with stabilizing mutation E47K in HA2) at a 3:1 molar ratio for at least 30 min at RT (33).
Negative-Stain Electron Microscopy.
Samples of immune complexes at ∼20 μg/mL were deposited onto glow-discharged carbon-coated 400 mesh copper grids (EMS) for 8 s. After wicking away liquid with Whatman paper, grids were negatively stained with 2% wt/vol uranyl formate in two applications of ∼30 s each. Grids were imaged on a Tecnai T20 microscope (FEI) with a CMOS 4k camera (TVIPS) operating at 200 kV with a nominal magnification of 62,000×. Micrographs were collected using Leginon, particles were picked and stacked using a difference-of-gaussians picker and Appion, and particles were subjected to reference-free two-dimensional (2D) classification and three-dimensional (3D) reconstruction using Relion3.0 (64–68). Maps were evaluated and figures were made in University of California, San Francisco (UCSF), Chimera and Adobe Photoshop (69).
Cryo-EM.
Samples at 800 μg/mL were incubated with 5 μM lauryl maltose neopentyl glycol (Anatrace) and deposited onto glow-discharged 1.2/1.3 Quantifoil 400 grids (EMS). Using a Vitrobot (FEI), grids were plunge-frozen in liquid ethane and stored in liquid nitrogen. Grids were imaged on a Talos Arctica electron microscope (FEI) operating at 200 kV and 36,000× nominal magnification with a CETA 4K CMOS camera (FEI) and Gatan K2 Summit detector in counting mode. A total of 2,270 micrographs were collected using Leginon, frames were aligned using MotionCor2, and contrast transfer function estimation was performed using GCTF-beta in CryoSPARC2 (64, 70, 71). Subsets of particles were manually picked and used as references for template picking of total micrographs using CryoSPARC2. Particles were subjected to multiple rounds of reference-free 2D classification, 3D classification, and 3D refinement using CryoSPARC2 and Relion3.0 (68, 71). Maps were evaluated and figures were made in Prism 8 (GraphPad), UCSF Chimera, and Adobe Photoshop (69).
Western Blot.
Recombinant A/California/04/2009 H1 protein was diluted in Laemmli buffer containing β-mercaptoethanol and boiled for 5 min. One microgram of denatured protein was run per well of a 4 to 20% gradient Mini-Protean TGX gel (BioRad) for SDS/PAGE. Ten microliters of Novex Sharp Pre-Stained Protein Standard ladder (Invitrogen) was run to determine size of the resulting bands. The protein was transferred to a 0.2-μm polyvinylidene difluoride membrane using a Trans-Blot Turbo Transfer Pack (BioRad). Blots were rinsed with water and blocked with 3% milk powder in PBST for 1 h with shaking at RT. Primary antibody was added at a concentration of 1 μg/μL for mAbs or 1:5,000 for anti-HIS (6xHis Monoclonal Antibody; Takara) in 3% milk in PBST overnight at 4 °C with shaking. Blots were washed with PBST and secondary antibody (goat anti-human IgG γ chain, 1:3,000; Millipore; or goat anti-mouse IgG Fc, 1:1,000; Abcam) in 3% milk for 1 h with shaking at RT. Blots were washed with PBST, then developed with a 1:1 mixture of Pierce ECL Western Blotting Substrate (Thermo Scientific). Blots were imaged on a ChemiDoc XRS+ machine (BioRad).
Biolayer Interferometry for Antibody Affinity.
Antibody affinity was determined using an Octet Red96 machine. Recombinant A/California/04/2009 H1 was diluted to 10 μg/mL in kinetics buffer (PBS with 0.1% BSA [Sigma] and 0.02% Tween 20 [Fisher]). mAbs were diluted to 60 μg/mL in kinetics buffer and added to a black-walled 96-well dish (Greiner Bio). Nickel-nitrilotriacetic acid–coated sensors (Forté Bio) were moved to kinetics buffer-containing wells, and a baseline was measured for 3 min. Sensors were then moved to HA-containing wells, and binding was allowed to proceed for 5 min before moving back to buffer for 2 min. Then, sensors were dipped in antibody-containing wells for 5 min to determine the on-rate (Kon) of the specific mAb. Sensors were then moved to unused buffer for 15 min to determine the off-rate (Koff). Curves were fit to the resulting data after subtraction of reference wells lacking antibody. KD was determined as Koff divided by Kon.
Primary Human PMN-Based ROS Burst Assay.
Peripheral blood was taken from participants enrolled in the Mount Sinai Personalized Virology Initiative. The study protocol for the collection of peripheral blood mononuclear cells from healthy volunteers was reviewed and approved by the Mount Sinai Hospital Institutional Review Board (no. 16–01199). Participants provided written informed consent prior to donating blood, and specimens were coded by the clinical team prior to processing. Blood was collected in EDTA-coated Vacutainer tubes (BD) and immediately processed for isolation of PMNs by drop-wise addition onto a two-layer Histopaque gradient (Sigma) consisting of 3 mL of Histopaque 1077 on top of 3 mL of Histopaque 1119. This mixture was spun at 800 × g for 30 min with no brake to separate PMNs. After aspirating the serum and buffy-coat layers, the cell-bearing layer immediately above the RBC layer was pooled from each tube and washed with 25 mL of PMN buffer (Hanks buffered salt solution [Gibco] containing 300 μM EDTA [Gibco] and 0.5% bovine serum albumin [Sigma-Aldrich]). Cells were spun at 400 × g for 5 min, and supernatant was removed before resuspending in 5 mL of ACK Lysing Buffer (Gibco) for 5 min before quenching with 20 mL of PMN buffer. Cells were spun and washed with PMN buffer two additional times before resuspending in a low volume of PMN buffer and counting. Cells were diluted to 1 to 1.5 × 107 cells per milliliter in PMN buffer for assay use.
The assay was prepared in a white-walled, tissue culture-treated, flat-bottomed, 96-well dish (Costar no. 3917). mAb was added to purified influenza virus to final concentrations of 2.5 μg per well and 10 μg per well, respectively, and the mixture was allowed to incubate at 4 °C for 30 min. Luminol in PMN buffer was then added, and finally primary human neutrophils were added to a final concentration of 5 × 105 cells per well. Luminescence was immediately read in 1-min intervals for 1 to 2 h on a Filter Max F3 multimode plate reader (Molecular Devices). Maximum assay signal was determined as the peak value of the kinetic curve.
Supplementary Material
Acknowledgments
We thank Fatima Amanat for recombinant HA protein production, Yunping Huang for assistance in cloning mAbs for this study, and María Carolina Bermúdez González for coordination with the Personalized Virology Initiative. This study was supported by CEIRS (Centers for Excellence for Influenza Research and Surveillance) Contract HHSN272201400008C (to F.K., R.N., and P.P.), Collaborative Influenza Vaccine Innovation Centers Contract 75N93019C00051 (to F.K., R.N., A.B.W., P.C.W., and P.P.), Public Health Service Institutional Research Training Award T32 AI07647 (to A.W.F.), CEIRS Training Program award (Option 21E) TR430967F (to A.W.F.), NIH Grant P01AI097092—Toward a Universal Influenza Virus Vaccine (to P.P.), NIH Grant R01 AI145870 (to P.P.), National Institute of Allergy and Infectious Diseases (NIAID) 2 T32 AI007244-36 (to J.H.), and National Heart, Lung, and Blood Institute 2 T32HL007605-35 (to J.J.G.). UCSF Chimera was used to analyze electron microscopy maps and generate figures; UCSF Chimera developers received support from NIH R01-GM129325 and P41-GM103311 grants and the Office of Cyber Infrastructure and Computational Biology (NIAID).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018102118/-/DCSupplemental.
Data Availability
The authors declare that all data supporting the findings of this study are available within the paper and its SI Appendix information files. Dataset S1 describes the unique antibody identifiers for numbered antibodies in this manuscript, as well as information about antibody variable regions. The range of Electron Microscopy Data Bank accession ID codes for microscopy images is as follows: EMD-23313–23319.
References
- 1.CDC , Seasonal Influenza Vaccine Effectiveness, 2005-2020 (2020). https://www.cdc.gov/flu/vaccines-work/past-seasons-estimates.html. Accessed 8 February 2021.
- 2.Wrammert J., et al., Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453, 667–671 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hobson D., Curry R. L., Beare A. S., Ward-Gardner A., The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J. Hyg. (Lond.) 70, 767–777 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imai M., Sugimoto K., Okazaki K., Kida H., Fusion of influenza virus with the endosomal membrane is inhibited by monoclonal antibodies to defined epitopes on the hemagglutinin. Virus Res. 53, 129–139 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Kosik I., Yewdell J. W., Influenza A virus hemagglutinin specific antibodies interfere with virion neuraminidase activity via two distinct mechanisms. Virology 500, 178–183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bangaru S., et al., A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nat. Commun. 9, 2669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiLillo D. J., Palese P., Wilson P. C., Ravetch J. V., Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Invest. 126, 605–610 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiLillo D. J., Tan G. S., Palese P., Ravetch J. V., Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 20, 143–151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandtzaeg P., Prydz H., Direct evidence for an integrated function of J chain and secretory component in epithelial transport of immunoglobulins. Nature 311, 71–73 (1984). [DOI] [PubMed] [Google Scholar]
- 10.Brokstad K. A., et al., High prevalence of influenza specific antibody secreting cells in nasal mucosa. Scand. J. Immunol. 54, 243–247 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Woof J. M., Kerr M. A., The function of immunoglobulin A in immunity. J. Pathol. 208, 270–282 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Taylor H. P., Dimmock N. J., Mechanism of neutralization of influenza virus by secretory IgA is different from that of monomeric IgA or IgG. J. Exp. Med. 161, 198–209 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muramatsu M., et al., Comparison of antiviral activity between IgA and IgG specific to influenza virus hemagglutinin: Increased potential of IgA for heterosubtypic immunity. PLoS One 9, e85582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renegar K. B., Small P. A. Jr, Boykins L. G., Wright P. F., Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract. J. Immunol. 173, 1978–1986 (2004). [DOI] [PubMed] [Google Scholar]
- 15.He W., et al., Broadly neutralizing anti-influenza virus antibodies: Enhancement of neutralizing potency in polyclonal mixtures and IgA backbones. J. Virol. 89, 3610–3618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeKosky B. J., et al., High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat. Biotechnol. 31, 166–169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J., et al., Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat. Med. 22, 1456–1464 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bangaru S., et al., A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell 177, 1136–1152.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nachbagauer R., Palese P., Is a universal influenza virus vaccine possible? Annu. Rev. Med. 71, 315–327 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Krammer F., The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 19, 383–397 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Liu Y., et al., Cross-lineage protection by human antibodies binding the influenza B hemagglutinin. Nat. Commun. 10, 324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Vries R. D., et al., Primary human influenza B virus infection induces cross-lineage hemagglutinin stalk-specific antibodies mediating antibody-dependent cellular cytoxicity. J. Infect. Dis. 217, 3–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y. Q., et al., Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 173, 417–429.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koroleva M., et al., Heterologous viral protein interactions within licensed seasonal influenza virus vaccines. NPJ Vaccines 5, 3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirst G. K., Adsorption of influenza hemagglutinins and virus by red blood cells. J. Exp. Med. 76, 195–209 (1942). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker D. L., Horsfall F. L. Jr, Lack of identity in neutralizing and hemagglutination-inhibiting antibodies against influenza viruses. J. Exp. Med. 91, 65–86 (1950). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashem A. M., et al., Universal antibodies against the highly conserved influenza fusion peptide cross-neutralize several subtypes of influenza A virus. Biochem. Biophys. Res. Commun. 403, 247–251 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Rajendran M., et al., Analysis of anti-influenza virus neuraminidase antibodies in children, adults, and the elderly by ELISA and enzyme inhibition: Evidence for original antigenic sin. MBio 8, e02281-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wohlbold T. J., et al., Hemagglutinin stalk- and neuraminidase-specific monoclonal antibodies protect against lethal H10N8 influenza virus infection in mice. J. Virol. 90, 851–861 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dreyfus C., et al., Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brandenburg B., et al., Mechanisms of hemagglutinin targeted influenza virus neutralization. PLoS One 8, e80034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leon P. E., et al., Optimal activation of Fc-mediated effector functions by influenza virus hemagglutinin antibodies requires two points of contact. Proc. Natl. Acad. Sci. U.S.A. 113, E5944–E5951 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong M., et al., Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J. Virol. 87, 12471–12480 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henry Dunand C. J., et al., Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe 19, 800–813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan G. S., et al., Broadly-reactive neutralizing and non-neutralizing antibodies directed against the H7 influenza virus hemagglutinin reveal divergent mechanisms of protection. PLoS Pathog. 12, e1005578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neu K. E., Henry Dunand C. J., Wilson P. C., Heads, stalks and everything else: How can antibodies eradicate influenza as a human disease? Curr. Opin. Immunol. 42, 48–55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fleury D., et al., A complex of influenza hemagglutinin with a neutralizing antibody that binds outside the virus receptor binding site. Nat. Struct. Biol. 6, 530–534 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Zhu X., et al., A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J. Virol. 87, 12619–12635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura G., et al., An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe 14, 93–103 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Mullarkey C. E., et al., Broadly neutralizing hemagglutinin stalk-specific antibodies induce potent phagocytosis of immune complexes by neutrophils in an Fc-dependent manner. MBio 7, e01624-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chernecky C. C., Differential Leukocyte Count (diff) - Peripheral Blood (Elsevier Saunders, St Louis, MO, ed. 6, 2013), pp. 440–446. [Google Scholar]
- 42.Worley M. J., et al., Neutrophils mediate HIV-specific antibody-dependent phagocytosis and ADCC. J. Immunol. Methods 457, 41–52 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Wang Y., Jönsson F., Expression, role, and regulation of neutrophil Fcγ receptors. Front. Immunol. 10, 1958 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maurer M. A., et al., Glycosylation of human IgA directly inhibits influenza A and other sialic-acid-binding viruses. Cell Rep. 23, 90–99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seibert C. W., et al., Recombinant IgA is sufficient to prevent influenza virus transmission in Guinea pigs. J. Virol. 87, 7793–7804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nimmerjahn F., Gordan S., Lux A., FcγR dependent mechanisms of cytotoxic, agonistic, and neutralizing antibody activities. Trends Immunol. 36, 325–336 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Morton H. C., van Egmond M., van de Winkel J. G., Structure and function of human IgA Fc receptors (Fc alpha R). Crit. Rev. Immunol. 16, 423–440 (1996). [PubMed] [Google Scholar]
- 48.Sakamoto N., et al., A novel Fc receptor for IgA and IgM is expressed on both hematopoietic and non-hematopoietic tissues. Eur. J. Immunol. 31, 1310–1316 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Shibuya A., et al., Fc alpha/mu receptor mediates endocytosis of IgM-coated microbes. Nat. Immunol. 1, 441–446 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Akula S., Mohammadamin S., Hellman L., Fc receptors for immunoglobulins and their appearance during vertebrate evolution. PLoS One 9, e96903 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parham P., The genetic and evolutionary balances in human NK cell receptor diversity. Semin. Immunol. 20, 311–316 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Francis T., On the doctrine of original antigenic sin. Proc. Am. Philos. Soc. 104, 572–578 (1960). [Google Scholar]
- 53.Abreu R. B., Kirchenbaum G. A., Clutter E. F., Sautto G. A., Ross T. M., Preexisting subtype immunodominance shapes memory B cell recall response to influenza vaccination. JCI Insight 5, 132155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abreu R. B., Clutter E. F., Attari S., Sautto G. A., Ross T. M., IgA responses following recurrent influenza virus vaccination. Front. Immunol. 11, 902 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neu K. E., et al., Spec-seq unveils transcriptional subpopulations of antibody-secreting cells following influenza vaccination. J. Clin. Invest. 129, 93–105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meng W., et al., An atlas of B-cell clonal distribution in the human body. Nat. Biotechnol. 35, 879–884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beyer W. E., Palache A. M., de Jong J. C., Osterhaus A. D., Cold-adapted live influenza vaccine versus inactivated vaccine: Systemic vaccine reactions, local and systemic antibody response, and vaccine efficacy. A meta-analysis. Vaccine 20, 1340–1353 (2002). [DOI] [PubMed] [Google Scholar]
- 58.He X. S., et al., Cellular immune responses in children and adults receiving inactivated or live attenuated influenza vaccines. J. Virol. 80, 11756–11766 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park J. K., et al., Pre-existing immunity to influenza virus hemagglutinin stalk might drive selection for antibody-escape mutant viruses in a human challenge model. Nat. Med. 26, 1240–1246 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernstein D. I., et al., Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine candidates: Interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect. Dis. 20, 80–91 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yassine H. M., et al., Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 21, 1065–1070 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Margine I., Palese P., Krammer F., Expression of functional recombinant hemagglutinin and neuraminidase proteins from the novel H7N9 influenza virus using the baculovirus expression system. J. Vis. Exp., e51112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guthmiller J. J., Dugan H. L., Neu K. E., Lan L. Y., Wilson P. C., An efficient method to generate monoclonal antibodies from human B cells. Methods Mol. Biol. 1904, 109–145 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Suloway C., et al., Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Lander G. C., et al., Appion: An integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Voss N. R., Yoshioka C. K., Radermacher M., Potter C. S., Carragher B., DoG picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zivanov J., et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pettersen E. F., et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its SI Appendix information files. Dataset S1 describes the unique antibody identifiers for numbered antibodies in this manuscript, as well as information about antibody variable regions. The range of Electron Microscopy Data Bank accession ID codes for microscopy images is as follows: EMD-23313–23319.