Ma and Wang (1) tested our recently reported severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) main protease (Mpro) noncovalent inhibitors (2) using their in vitro assays, and they obtained negligible or much lower inhibitory activities compared to ours. Knowing the discrepancy, we first carefully rechecked our original experimental records, and we did not find any potential concern with our data that had been repeated by multiple coauthors. Notably, the fluorescence resonance energy transfer (FRET)-based enzymatic assay used by Ma and Wang (1) is different from our FRET-based enzymatic assay in the Mpro protein (discussed below) and the FRET substrate [longer than the more popularly used substrate (3, 4) utilized in our assay].
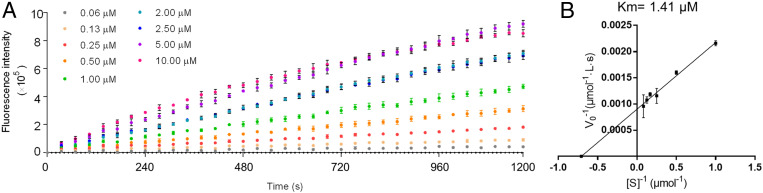
Particularly for Mpro, Ma and Wang (1) incorrectly state that a GST-tagged Mpro was used in our assay. Actually, as described in our paper (2), the GST tag was cleaved with thrombin; the GST tag was used only for conveniently isolating Mpro from the culture medium. So, the Mpro protein used in our assay is the true wild-type Mpro with native N and C termini. In comparison, the FRET-based enzymatic assay described by Ma et al. (5) used a C-terminal His-tagged Mpro protein. As noted correctly by Ma and Wang (1), Mpro requires a native N terminus to form the enzymatically active dimer. In fact, both the N and C termini of Mpro are very close to the active-site cavity in the dimer according to available X-ray crystal structures (Protein Data Bank [PDB] ID code 7BUY) (6), including one (PDB ID code 6WTT) shown by Ma et al. (5). Thus, an additional tag on the N or C terminus could interfere with Mpro binding with a ligand (substrate or inhibitor). So, a given ligand could have a lower binding affinity with the His-tagged Mpro. In fact, we obtained Michaelis constant (Km) = 1.41 μM (Fig. 1) for the Mpro protein without any tag, and our reported catalytic efficiency (2) is close to the previously reported value (catalytic constant kcat/Km = 28,500 M−1⋅s−1) (4). However, Km = 28.2 μM for the His-tagged Mpro (5). So, the His-tagged Mpro has a ∼20-fold lower binding affinity with the substrate compared to the tag-free Mpro. In other words, the activity determined by using the assay with a His-tagged Mpro might not reflect the actual activity with native Mpro.
Fig. 1.
Michaelis–Menten kinetics of SARS-CoV-2 Mpro (100 nM) against substrate MCA-AVLQSGFR-Lys(Dnp)-Lys-NH2 at various concentrations. (A) Original data obtained. (B) Lineweaver–Burk plot used to determine the catalytic parameters.
Ma and Wang (1) also used native mass spectrometry (MS) and thermal shift assays (TSA) to detect the protein–ligand binding. For binding driven by hydrophobic interaction the protein–ligand complex will most likely dissociate in MS (7). For TSA, false negatives are also known to occur (8, 9). Both assays might not be suitable for analyzing noncovalent inhibitors of Mpro.
Finally, GC-376, a covalent inhibitor identified in their earlier reports, was used as a positive control to validate their assays by Ma and Wang (1). However, it is difficult to understand why their results show half-maximum inhibitory concentration (IC50) = 28 or 33 nM when the enzyme concentration was 100 nM. Their data, if validated, would imply that each GC-376 molecule inactivated multiple Mpro protein molecules through an unusual mechanism.
Acknowledgments
We acknowledge the National Key R&D Program of China (2017YFB0202600), National Natural Science Foundation of China (81903542, 81522041, and 21877134), Science Foundation of Guangdong Province (2020A111128007), NSF grant CHE-1111761, Taishan Scholars Program (tsqn201909170), and Innovative Leader of Qingdao Program (19-3-2-26-zhc).
Footnotes
The authors declare no competing interest.
References
- 1.Ma C., Wang J., Dipyridamole, chloroquine, montelukast sodium, candesartan, oxytetracycline, and atazanavir are not SARS-CoV-2 main protease inhibitors. Proc. Natl. Acad. Sci. U.S.A., 10.1073/pnas.2024420118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z., et al., Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. Proc. Natl. Acad. Sci. U.S.A. 117, 27381–27387 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai W., et al., Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 368, 1331–1335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin Z., et al., Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Ma C., et al., Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 30, 678–692 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin Z., et al., Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 27, 529–532 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Pedro L., Quinn R. J., Native mass spectrometry in fragment-based drug discovery. Molecules 21, 984 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larsson A., Jansson A., Åberg A., Nordlund P., Efficiency of hit generation and structural characterization in fragment-based ligand discovery. Curr. Opin. Chem. Biol. 15, 482–488 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Savitski M. M., et al., Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 346, 1255784 (2014). [DOI] [PubMed] [Google Scholar]