Significance
Cell surface proteolysis is a key mechanism for cell-cell communication and cellular signaling that is commonly dysregulated in human disease. Proteolytic cleavage events at the cell surface often evade detection by conventional proteomics methods owing to their low relative abundance compared to cytoskeletal and cytoplasmic proteins. Here we address this limitation by developing a new enzymatic tool, subtiligase-TM, for targeted mapping of cell surface proteolysis. We combine subtiligase-TM with quantitative proteomics to map proteolytic cleavage sites on the cell surface with single amino acid resolution. Based on the importance of cell surface proteolysis in human health and disease, subtiligase-TM opens up new opportunities for identifying biomarkers and therapeutic targets.
Keywords: proteomics, proteolysis, N terminomics
Abstract
N terminomics is a powerful strategy for profiling proteolytic neo-N termini, but its application to cell surface proteolysis has been limited by the low relative abundance of plasma membrane proteins. Here we apply plasma membrane-targeted subtiligase variants (subtiligase-TM) to efficiently and specifically capture cell surface N termini in live cells. Using this approach, we sequenced 807 cell surface N termini and quantified changes in their abundance in response to stimuli that induce proteolytic remodeling of the cell surface proteome. To facilitate exploration of our datasets, we developed a web-accessible Atlas of Subtiligase-Captured Extracellular N Termini (ASCENT; http://wellslab.org/ascent). This technology will facilitate greater understanding of extracellular protease biology and reveal neo-N termini biomarkers and targets in disease.
Proteolysis is a key posttranslational modification that controls the function, localization, and degradation of nearly all proteins (1, 2). More than 500 proteases are encoded in the human genome, but many of their biological functions remain incompletely understood, because few of their substrates are known. Several methods for selective isolation of protein N-terminal peptides, including those arising from proteolytic cleavage events, have been developed (3–6), enabling global analysis of cellular proteolysis in response to biological perturbations. These strategies exploit the unique chemical structure and reactivity of the protein N terminus compared to other biological amines to isolate unblocked N termini either by positive enrichment or by depletion of internal peptides following protease digestion. The isolated N-terminal peptides can then be analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS), enabling sequencing of protease cleavage sites at single amino acid resolution. While these techniques have proven powerful to identify protease substrates, they provide limited coverage of cell surface proteins, which often escape detection by mass spectrometry due to their low abundance relative to cytoplasmic and cytoskeletal proteins (7, 8). Cell surface proteolysis is crucial for cell-cell communication and is often dysregulated in disease; thus, new techniques are needed for identifying protease cleavage sites in plasma membrane proteins to facilitate biomarker and target discovery.
Subtiligase is a rationally designed variant of the serine protease subtilisin that harbors two key mutations (S221C and P225A) that enable it to catalyze a ligation reaction between a peptide ester and the N-terminal α-amine of a peptide or protein (9). Based on this activity, subtiligase has been applied broadly as a tool for N-terminal modification to enable selective enrichment of N-terminal peptides and their identification and quantification by LC-MS/MS (3, 10). This strategy, known as subtiligase N terminomics, has uncovered thousands of protease cleavage events that are stimulated in the contexts of apoptosis (3), inflammation (11), bacterial (12) and viral infection (13), and protein trafficking (14). However, cell surface proteins are underrepresented in N terminomics datasets due to their low abundance (15). We sought to develop a method to target cell surface N termini while avoiding the low specificity and material losses that would result from traditional approaches to isolating the plasma membrane.
Here we present a strategy, termed subtiligase-TM, to target subtiligase activity to the surface of living cells where membranes, protein complexes, and spatial relationships remain intact. We demonstrate that the activity of subtiligase-TM is restricted to the plasma membrane, providing subcellular spatial resolution of proteolytic neo-N termini, as well as higher coverage of cell surface proteins in N terminomics datasets. We deployed this tool to quantify changes in the abundance of proteolytic neo-N termini in response to pervanadate treatment, which stimulates proteolytic remodeling of the cell surface proteome. Subtiligase-TM combines the advantages of methods that enable identification of the exact sites of proteolytic cleavage events with those that provide subcellular resolution, enabling detection of protease cleavage sites that evade capture by other strategies.
Results
Defining the Subcellular Distribution of Proteins Enriched in Subtiligase N Terminomics Experiments.
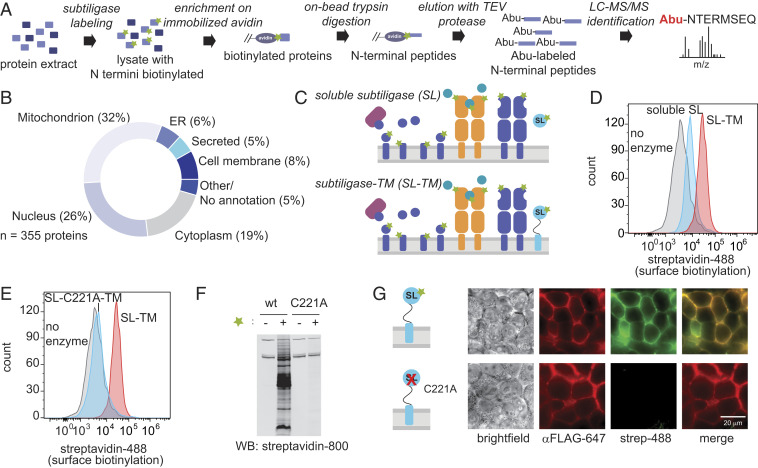
To determine the distribution of subcellular localizations of proteins typically captured in subtiligase N terminomics experiments, we enriched protein N termini from HEK293T cell lysate derived from 25 million cells using soluble subtiligase (Fig. 1A). Proteins biotinylated by subtiligase were enriched on neutravidin resin, on-bead trypsin digestion was performed to remove internal peptides, and N-terminal peptides were selectively eluted by cleavage of a TEV protease site incorporated into the subtiligase substrate. Following TEV cleavage, each subtiligase-modified N terminus retains an aminobutyric acid (Abu) mass tag for unequivocal identification. The enriched, eluted N-terminal peptides were then analyzed by LC-MS/MS. From three experiments, a total of 357 unique N termini from 355 unique proteins were identified (Fig. 1B and Dataset S1). Subcellular location annotations for these proteins in UniProt revealed that only 29 proteins (8%) were cell membrane proteins, while 18 (5%) were secreted proteins. The remaining proteins identified in the datasets were derived from the mitochondrion (113 proteins; 32%), nucleus (92 proteins; 26%), cytoplasm (66 proteins; 19%), endoplasmic reticulum (ER) (21 proteins; 6%), and other or unknown subcellular locations (16 proteins; 5%) (Fig. 1B and Dataset S2). Of the 29 proteins derived from the cell membrane, 4 (1.1% of the total) were enriched based on tagging of an N terminus predicted to reside in the portion of the protein on the extracellular surface.
Fig. 1.
Restricting subtiligase activity to the cell surface. (A) Workflow for subtiligase N terminomics in cell lysate. Biotinylated peptide ester is represented by a green star. (B) Subcellular locations of N termini identified in a subtiligase N terminomics experiment performed using soluble subtiligase in HEK293T cell lysate. (C) Approaches for labeling cell surface N termini. (Top) Addition of soluble subtiligase to live cells. (Bottom) Expression of subtiligase fused to the PDGFRβ chain (subtiligase-TM) in cells. (D) Streptavidin-488 flow cytometry demonstrates that subtiligase-TM (red) more efficiently biotinylates cell surface N termini compared to soluble subtiligase added to cells (cyan). No enzyme control is shown in gray. (E) Streptavidin-488 flow cytometry shows that robust cell surface biotinylation is observed with activate subtiligase-TM (red) but not with the catalytically inactive C221A mutant (cyan). (F) A Western blot of subtiligase-TM-expressing HEK293T cells shows that biotinylation activity is dependent on both active subtiligase and the presence of a biotinylated peptide ester substrate. No biotinylation is observed when the inactive subtiligase-C221A-TM mutant is used or in the absence of biotinylated peptide ester substrate. (G) Fluorescence microscopy shows that subtiligase-TM expression (red) and biotinylation activity (green) are colocalized at the cell surface. No biotinylation activity is observed when the inactivate subtiligase-C221A-TM mutant is expressed.
To address whether cell type or the amount of starting material used impacts coverage of cell surface N termini, we also performed subtiligase N terminomics using lysates from 250 million Jurkat cells (Dataset S3). Although a larger number of proteins were identified (885 proteins), the subcellular distribution of proteins in the datasets was similar to our observations in HEK293T cells, with 13% of enriched proteins annotated as cell membrane or secreted; 81% derived from the cytoplasm, nucleus, mitochondrion, or ER; and 5% derived from other intracellular compartments or having no annotation (SI Appendix, Fig. S1 and Dataset S4). Based on these results, we concluded that cell membrane proteins are underrepresented in N terminomics datasets.
Restricting Subtiligase Activity to the Extracellular Side of the Plasma Membrane.
We wondered whether restricting subtiligase activity to the cell surface could increase coverage of cell membrane N termini and specificity for extracellular N termini. We initially attempted to target subtiligase activity to the cell surface by adding the enzyme and a biotinylated subtiligase substrate to a suspension of living cells (Fig. 1 C, Top). Staining of the cells with streptavidin-Alexa Fluor 488 and analysis by flow cytometry and fluorescence microscopy revealed an approximately twofold increase in the biotinylation signal at the cell surface (Fig. 1D and SI Appendix, Fig. S2). We reasoned that the efficiency of cell surface N-terminal biotinylation could be increased by tethering subtiligase to the extracellular side of the plasma membrane (Fig. 1 C, Bottom). Therefore, we genetically targeted subtiligase to the desired location by fusing it to the transmembrane domain of the platelet-derived growth factor receptor beta chain (PDGFRβ-TM) to generate subtiligase-TM. We initiated labeling by adding the biotinylated substrate and measured cell surface biotinylation by flow cytometry. This experiment revealed a robust increase in biotinylation (by ∼10-fold) compared to the soluble enzyme (Fig. 1D). In contrast, a catalytically inactive mutant of subtiligase (C221A) did not show any cell surface biotinylation activity (Fig. 1E). Streptavidin blot analysis of cell lysate demonstrated that many proteins were biotinylated in a manner dependent on both subtiligase activity and the presence of biotinylated subtiligase substrate (Fig. 1F), and that maximal biotinylation was achieved within 10 min (SI Appendix, Fig. S3). Fluorescence microscopy analysis of subtiligase expression and biotinylation activity showed that biotinylation and subtiligase activity were colocalized at the cell surface (Fig. 1G).
Evaluating Subtiligase-TM for Cell Surface N Terminomics.
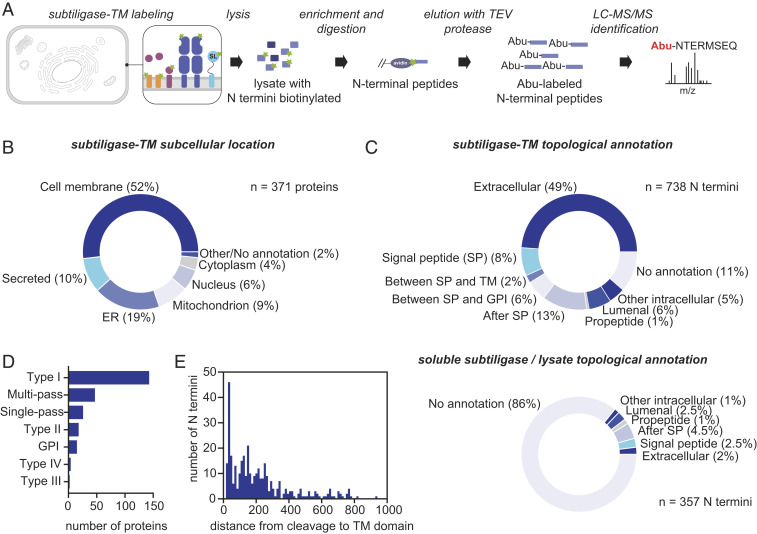
We next tested subtiligase-TM in an MS N terminomics experiment using 25 million HEK293T cells (Fig. 2A). Proteins biotinylated by subtiligase were enriched on neutravidin beads, an on-bead trypsin digestion was performed to remove internal peptides, and N-terminal peptides were selectively eluted by TEV protease cleavage. From four experiments, a total of 737 unique N termini from 371 unique proteins were identified (Dataset S5). Subcellular location annotations for these proteins in UniProt (16) revealed that our N-terminal dataset was highly enriched for cell membrane and secreted proteins, with 249 of the identified proteins (62%) having a subcellular location annotation (cell membrane or secreted) suggesting that they would be present at the cell surface (Fig. 2B and Dataset S6). To enable exploration of our datasets, we developed an Atlas of Subtiligase-Captured Extracellular N Termini (ASCENT; wellslab.org/ascent) for browsing and searching for subtiligase-captured cell surface N termini.
Fig. 2.
Cell surface N terminomics with subtiligase-TM. (A) Workflow for cell surface N terminomics with subtiligase-TM. (B) Subcellular locations of N termini identified in a subtiligase-TM N terminomics experiment performed in HEK293T cells. (C) Topological locations of N-terminal peptides identified in the subtiligase-TM N terminomics experiment (Top) or a subtiligase lysate experiment (Bottom). (D) Distribution of membrane protein types observed in the subtiligase-TM N terminomics experiment. (E) Distribution of distances between cleavage sites identified in the subtiligase N terminomics experiments and the corresponding transmembrane domains.
Based on the design of the subtiligase-TM construct, we expected subtiligase activity to be restricted to the extracellular side of the plasma membrane and thus to modify free N termini also located on the extracellular surface. To assess the specificity of subtiligase-TM for extracellular N termini, we mapped the N-terminal peptides captured with subtiligase-TM or with soluble subtiligase added to a lysate to topological domain annotations in UniProt (16) (Fig. 2 C, Top and Dataset S7). Of the 737 N termini that we identified in the subtiligase-TM dataset, 360 (49%) mapped to annotated extracellular domains, while 23 (3%) mapped to cytoplasmic domains. For the remaining N termini that did not map to annotated extracellular domains, we mined UniProt for other protein features to provide clues about their topological locations. The positions of many of the N termini relative to other protein features were consistent with an extracellular location. Among this group, 58 N termini corresponded to annotated signal peptide cleavages, 16 N termini were located between a signal peptide and a transmembrane domain, 42 N termini were located between a signal peptide and a GPI anchor, 6 N termini corresponded to annotated propeptide cleavage sites, and 92 N termini occurred between a signal peptide and the protein C terminus. The remaining N termini mapped to the ER lumen (45 sequences) or to other intracellular compartments (11 sequences) or had no annotation (84 sequences). In total, 79% of the identified N-terminal peptides were likely positioned on the extracellular surface during the live cell subtiligase-TM labeling experiment. In contrast, when we performed this analysis for N termini derived from the HEK293T lysate dataset, of the 357 N termini identified, 2% mapped to annotated extracellular domains, and 7% had relationships to other features suggesting that they could be derived from the extracellular surface (Fig. 2 C, Bottom and Dataset S8). The majority of N termini in the lysate dataset (86%) had no topological annotation, suggesting that they are derived from proteins located entirely within a single compartment and are not associated with cellular membranes that would provide a topological orientation. Similarly, in Jurkat lysate, of the 1,693 N termini identified, 1% mapped to annotated extracellular domains, 4% had relationships to other features suggesting that they are derived from the extracellular surface, and 92% had no topological annotation (SI Appendix, Fig. S4 and Dataset S9).
For subtiligase-TM to capture a neo-N terminus produced by cleavage of the extracellular domain of a protein, the neo-N terminus must remain associated with the cell surface. Therefore, we hypothesized that membrane protein types with their N-terminal domains on the extracellular surface (type I membrane proteins) would predominate over those with their C-terminal domains on the extracellular surface (type II membrane proteins). Consistent with this hypothesis, type I membrane proteins were overrepresented in our dataset compared to their abundance in the proteome (Fig. 2D and Dataset S10). However, type II, type III, type IV, multipass, GPI-anchored, and other single-pass proteins were also observed in our dataset, suggesting that some proteolytically cleaved fragments that are not tethered to the plasma membrane nonetheless may remain associated with the cell.
We also hypothesized that the N-terminal peptides captured by subtiligase-TM would depend on the reach of subtiligase-TM from the membrane. Taking into account the length of the linker between subtiligase and the PDGFRβ-TM domain (estimated at 23 to 27 Å on average; ref. 17), as well as the size of subtiligase itself (estimated radius 20 Å; ref. 18), we would expect that subtiligase should have the reach to modify even large extracellular domains up to ∼500 kDa (estimated radius 50 Å; ref. 18). Consistent with this estimate, we observed subtiligase modification of extracellular N termini 1 to 2,585 aa away from their TM domains (Fig. 2E). The distribution of distances between observed N-terminal modification sites and TM domains was similar to the distribution of extracellular domain lengths across the proteome (SI Appendix, Fig. S5 and Dataset S11).
Introduction of Subtiligase Specificity Mutants for Expanded Coverage of the Cell Surface N Terminome.
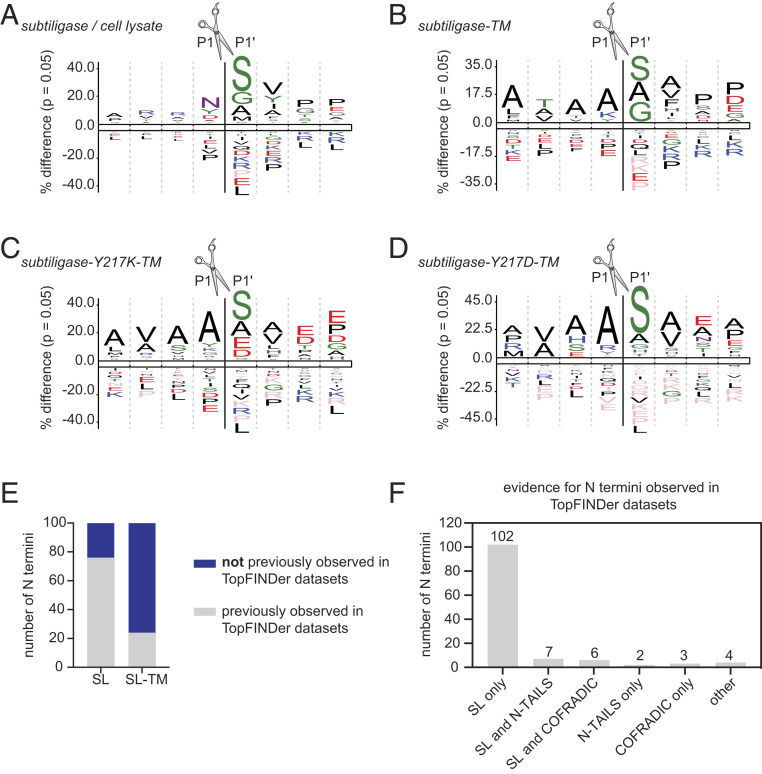
We next examined whether the use of subtiligase specificity mutants in the context of subtiligase-TM could increase coverage of N termini at the cell membrane in our experiment. We observed that the N-terminal specificity of subtiligase-TM was similar to that of subtiligase added to cell lysate (Fig. 3 A and B), with most of the N termini that were recovered having Ser, Gly, or Ala in the first position. However, when we introduced the Y217K mutation into subtiligase-TM, Glu and Asp N termini were also captured efficiently (Fig. 3C and SI Appendix, Fig. S6), while introduction of the Y217D mutation led to more efficient capture of His N termini (Fig. 3D and SI Appendix, Fig. S6). These results are consistent with our previous specificity data for these mutants (19) and increase the total number of cell surface N termini that we identified to 807 (Datasets S12 and S13).
Fig. 3.
Comparison of subtiligase-TM to subtiligase-TM mutants and to other N terminomics methods. (A–D) iceLogos for the N-terminal sequence specificity of subtiligase added to cell lysate (A), wild-type subtiligase-TM (B), subtiligase-Y217K-TM (C), and subtiligase-Y217D-TM (D). (E) Fraction of subtiligase-captured N termini observed in TopFINDer datasets (gray) or not observed in TopFINDer datasets (blue) for soluble subtiligase (Left) or subtiligase-TM (Right). (F) Evidence for subtiligase-TM–captured N termini that were previously observed in TopFINDer datasets.
Comparison of Subtiligase-TM with Other N Terminomics Technologies.
To assess the ability of subtiligase-TM to capture cell surface N termini in comparison with other N terminomics technologies, we searched for the N-terminal sequences identified in our datasets in the TopFIND 3.0 knowledge base (20), a database that includes 165,044 N termini that have been experimentally identified by various methods, including COFRADIC (5), N-TAILS (4), and subtiligase N terminomics (3) performed in lysate. For a representative subtiligase-TM dataset, we found that for wild-type subtiligase-TM, 398 of the 522 (76%) N termini that we identified had never been observed experimentally, while the other 124 (24%) had been previously identified by one or more experimental methods (Fig. 3E and Dataset S14). Among the previously observed N termini, 102 N termini were identified with subtiligase (20%), 2 were identified by N-TAILS (0.4%), 2 were identified by COFRADIC (0.6%), 4 were identified by other methods (0.8%), 7 were identified by both subtiligase and N-TAILS (1.3%), and 6 were identified by both subtiligase and COFRADIC (1.1%) (Fig. 3F). Similarly, for subtiligase-Y217K-TM, 120 of 174 (69%) of N termini had never been previously observed, while 54 N termini (31%) had been previously characterized by COFRADIC (n = 1; 0.6%), N-TAILS (n = 5; 2.9%), subtiligase in lysate (n = 53; 30%), or other methods (n = 24; 14%) (Dataset S15). For subtiligase-Y217D-TM, 42 of 72 N termini (58%) had never been previously observed, while 30 (42%) had been previously detected by N-TAILS (n = 2; 2.8%), subtiligase in lysate (n = 26; 36%), or other methods (n = 16; 22%) (Dataset S16). In contrast, for our lysate experiments in which data were collected on the same mass spectrometer as used for the subtiligase-TM experiments, 471 of 1,693 N termini (28%) did not appear in the TopFIND database, while 1,223 of the 1,693 (72%) had been previously observed by COFRADIC (n = 112; 6.6%), N-TAILS (n = 53; 3.1%), subtiligase in lysate (n = 1,185; 70%), or other methods (n = 400; 24%) (Dataset S17). The increase in N termini identified with subtiligase in the lysate experiment reported here compared to those appearing in the TopFIND database is likely attributable to improvements in MS technology since previous subtiligase datasets were collected. However, our subtiligase-TM datasets contain a much higher fraction (76%) of novel N termini compared to the new subtiligase lysate datasets (28%), suggesting that subtiligase-TM provides increased coverage of cell surface N termini compared to other N terminomics methods.
Quantitative Cell Surface N Terminomics Using Subtiligase-TM.
We assessed the utility of subtiligase-TM for quantifying changes in levels of cell surface N termini by treating subtiligase-TM–expressing HEK293T cells with pervanadate, a covalent tyrosine phosphatase inhibitor that stimulates numerous cell surface proteolysis events (21) (Fig. 4A), or a vehicle control. On addition of pervanadate, we observed an accumulation of phosphotyrosine by Western blot analysis (Fig. 4B). We isotopically encoded the pervanadate-treated and untreated samples using stable isotope labeling with amino acids in cell culture (SILAC), treated them with a cell-impermeable subtiligase substrate, and isolated biotinylated N-terminal peptides for identification and quantification by mass spectrometry. In total, we quantified 326 N termini (Dataset S18 and SI Appendix, Figs. S7–S9). We identified 38 N termini that were significantly up-regulated (greater than twofold increase, P < 0.05) on pervanadate treatment and 24 N termini that were significantly down-regulated (greater than twofold decrease, P < 0.05) on pervanadate treatment (Fig. 4C). Among the pervanadate up-regulated N termini were proteolytic products of proteins that are known sheddase targets, including cadherin-2 (CADH2), glypican-4 (GPC4), CD99, scavenger receptor class B member 1 (SCRB1/CD36), and CD166 (ALCAM) (22). Notably, although these proteins were previously known to be shed from the cell surface, in many cases, the exact sites of proteolytic cleavage that our method identified were not previously known.
Fig. 4.
Quantitative subtiligase-TM N terminomics in pervanadate-treated vs. untreated cells. (A) Pervanadate treatment leads to proteolytic shedding of cell surface proteins. (B) Western blot showing an accumulation of phosphotyrosine on pervanadate treatment of cells. (C) Volcano plot showing the log2-fold change of N termini measured by quantitative mass spectrometry in pervanadate-treated vs. untreated cells. The significance of the changes is indicated by the −log10 P value (y-axis). (D) Flow cytometry validation of changes in the N-terminal proteome observed in the mass spectrometry dataset. Monoclonal antibodies that bind epitopes N-terminal to the observed cleavage site were chosen such that a decrease in signal on pervanadate treatment was expected.
To validate our results, we used flow cytometry to examine loss of the domains that our MS data identified as cleaved. We selected three significantly up-regulated N termini derived from proteins for which monoclonal antibodies with defined epitopes were available: CADH2_386, CD99_92, and CD166_504 (Fig. 4D and SI Appendix, Figs. S10–S15). For the three proteins that we evaluated, flow cytometry confirmed the change that we observed in our MS dataset. Together, these results demonstrate that subtiligase-TM is a robust tool for capturing proteolytic neo-N termini on the extracellular surface.
Discussion
The cell surface is a hub for cell-cell communication and a frequent target for small-molecule, biologic, and cell-based therapeutics. While cell surface and extracellular proteases were once thought to function exclusively in remodeling the extracellular matrix, recent evidence demonstrates that they often play more complex roles in the cleavage of receptors, cell adhesion molecules, growth factors, cytokines, and kinases that regulate cell signaling (23), and that these proteolytic processes are often dysregulated in human diseases, such as cancer (24, 25). Subtiligase-TM is a genetically encoded enzymatic tool that enables unbiased mapping of proteolytic cleavage sites at the extracellular surface, opening up new opportunities to dissect the functions of these events in cellular signaling and to target them as biomarkers and therapeutics. Subtiligase-TM is broadly applicable to the study of a wide array of signaling processes and is generalizable to any system that can be genetically manipulated, including cell lines, primary cells, and whole animals. One limitation of subtiligase-TM is that it cannot be applied to systems that are not amenable to genetic manipulation. In the future, other targeting strategies may enable insertion of exogenously added subtiligase into the plasma membrane to nongenetically target its activity to the cell surface.
Subtiligase-TM combines the strengths of such N terminomics methods as terminal amine isotopic labeling of substrates (TAILS) (4), combined fractional diagonal chromatography (COFRADIC) (5), and traditional subtiligase N terminomics (3), which provide information about the exact sites of proteolytic cleavage events, with the strengths of such methods as secretome protein enrichment with click sugars (SPECS) (26), which increases coverage of cell surface proteins but does not provide positional information. These advantages make subtiligase-TM a transformative technique for identifying proteolytic cleavage sites in cell surface proteins at single amino acid resolution. Given the importance of extracellular proteolysis in health and disease, we expect that this tool will be widely adopted by protease biologists and translational scientists to discover new biomarkers or neoepitopes for immunotherapy.
Materials and Methods
Cell Lysate N Terminomics.
Cell lysate experiments were performed using either Jurkat E6-1 cells (250 million cells) or HEK293T cells (25 million cells). For each experiment, cells were collected by centrifugation for 5 min at 300 × g and then washed twice with 50 mL PBS. Cells were resuspended in lysis buffer (400 mM tricine pH 8, 4% [wt/vol] sodium dodecyl sulfate [SDS], 100 mM phenylmethylsulfonyl fluoride, 100 mM 4-benzenesulfonyl fluoride hydrochloride, and 2.5 mM EDTA) and lysed by probe ultrasonication (20% amplitude, 10 cycles of 5 s on/1 s off). Insoluble material was pelleted by centrifugation at 20,000 × g for 20 min at room temperature. The sample was boiled in the presence of 5 mM tris(2-carboxyethyl)phosphine (TCEP) for 15 min to reduce disulfide bonds. Free cysteines were then alkylated in the presence of 10 mM iodoacetamide at room temperature in the dark for 1 h. Iodoacetamide was quenched by the addition of dithiothreitol (DTT) to a final concentration of 25 mM. Triton X-100 was added to a final concentration of 2.5% (vol/vol), and the sample was diluted fourfold with water. Biotinylated subtiligase substrate Tev ester 6 (SI Appendix, Fig. S16) was added to a final concentration of 2.5 mM, and the reaction was initiated by the addition of stabiligase to a final concentration of 1 µM. The reaction mixture was incubated for 1 h at room temperature. After subtiligase labeling, biotinylated N-terminal peptides were enriched as described previously and then analyzed by LC-MS/MS.
Plasmid Construction.
Plasmids were constructed using Gibson cloning with Escherichia coli XL10 as the cloning host. KOD Hot Start Polymerase (EMD Millipore) was used for PCR amplifications with the oligonucleotides listed in SI Appendix, Table S2. Plasmids were verified by Sanger sequencing (Quintara Biosciences). Subtiligase-TM was initially cloned into pCDNA3.2-Igκ-FLAG-GFP-PDGF TM (J.A.W. laboratory) between the NdeI/BamHI sites. Subcloning was then performed as described below to construct vectors for lentiviral transduction.
pLX302-Igk-FLAG-subtiligase-PDGF TM.
A fragment encoding a fusion of the Igκ chain leader sequence, a Ca2+-independent variant of subtiligase, a 10× Gly-Ser linker, and the PDGF receptor β chain transmembrane domain was amplified from pCDNA3.2-Igκ-FLAG-SL-PDGF TM using Igκ-FLAG-SL-PDGF F1 and R1 (SI Appendix, Table S2). The fragment was inserted between the BsrGI and NheI sites of pLX302 (Addgene; 25896) using Gibson assembly.
Mutants of Igκ-FLAG-subtiligase-PDGF TM.
Subtiligase mutants were constructed using overlap PCR with oligonucleotides listed in SI Appendix, Table S2 and then inserted into pLX302 using Gibson assembly.
Construction of Cell Lines Expressing Subtiligase-TM.
To construct subtiligase-TM cell lines, HEK293T cells were lentivirally transduced. Lentivirus was produced by transfecting HEK293T cells at 80% confluence with a mixture of the transfer plasmid pLX302-Igκ-FLAG-subtiligase-PDGF TM and second-generation lentiviral packaging plasmids pMD2.g and pCMV-R8.91 using FuGENE HD transfection reagent (Promega). After 6 h, the supernatant was removed and replaced with complete Dulbecco's Modified Eagle Medium (DMEM). After 72 h, the virus-containing supernatant was passed through a 0.45-µm polyvinylidene difluoride filter and used directly for infection of HEK293T cells. HEK293T cells to be infected were grown to 80% confluence in a 6-well plate. The medium was then removed and replaced with a mixture of virus (1 mL), complete DMEM containing 8 µg/mL polybrene (1.5 mL), and complete DMEM (0.5 mL). After 24 h, the lentivirus-containing medium was removed and replaced with complete DMEM. Puromycin was added to 2 µg/mL at 72 h after transfection to select for transduced cells. Expression was validated by flow cytometry using Alexa Fluor 647-conjugated anti-DYKDDDDK tag antibody (BioLegend; 637315).
Flow Cytometry Analysis of Subtiligase-TM Activity.
Cells were dissociated by incubation with versene (0.04% EDTA in Ca2+/Mg2+-free PBS) and collected by centrifugation at 300 × g for 5 min. Cells were blocked with PBS containing 3% BSA and stained with Alexa Fluor 647–anti-DYKDDDDK (BioLegend) at 0.5 µg/mL or with Alexa Fluor 488-streptavidin (Life Technologies) at 1 µg/mL for 30 min at 4 °C. Cells were washed three times with PBS containing 3% BSA, resuspended in PBS, and analyzed on a Beckman Coulter CytoFlex flow cytometer. Data were analyzed using FlowJo software.
Immunofluorescence.
HEK293T cells expressing subtiligase-TM and control cells were plated on glass-bottom poly-d-lysine–coated imaging dishes. Dishes were incubated for 24 h at 37 °C in a 5% CO2 atmosphere. Cell were washed with PBS, fixed with 1% paraformaldehyde, and permeabilized with 0.1% Triton X-100. Cells were blocked with PBS containing 3% BSA, and Alexa Fluor 647-anti-DYKDDDDK (0.5 µg/mL) and Alexa Fluor 488-streptavidin (1 µg/mL) were added. After a 30-min incubation at room temperature, cells were washed three times with PBS containing 3% BSA, washed with PBS, and imaged on a Zeiss Axio Observer Z1 using a 63× oil objective. For the images presented in SI Appendix, Fig. S2, cells were imaged on a Revolve Echo epifluorescence microscope using a 20× objective. False-color images were produced using Fiji software.
Subtiligase-TM N Terminomics.
For each experiment, a 500-cm2 dish of HEK293T cells expressing subtiligase-TM or a specificity variant was grown to ∼80% confluency. The medium was removed, and cells were washed once with 50 mL of PBS. Versene (25 mL) was added, and plates were incubated at 37 °C for 10 min to allow cell dissociation to occur. Cells were harvested by centrifugation at 300 × g for 5 min, washed with 50 mL of PBS, and transferred into a 1.5-mL microcentrifuge tube. Cells were washed three times with 1 mL of labeling buffer (100 mM tricine pH 8 and 150 mM NaCl) and then resuspended in labeling buffer containing 2.5 mM Tev ester 6 (SI Appendix, Fig. S16). Subtiligase labeling was allowed to proceed for 1 h at 4 °C on a rotating mixer. Following labeling, cells were pelleted by centrifugation at 300 × g for 5 min and then washed three times with PBS. Cells were resuspended in RIPA lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with Halt Protease and Phosphatase Inhibitor Mixture (Thermo Fisher Scientific). Lysis was completed by probe ultrasonication (20% amplitude, 10 cycles of 5 s on/1 s off). Insoluble material was pelleted by centrifugation at 20,000 × g for 20 min at 4 °C. Biotinylated proteins were enriched from the supernatant on high-capacity NeutrAvidin agarose resin (Thermo Fisher Scientific; 0.5 mL of 50% resin slurry). The resin was washed with each of the following buffers: RIPA (10 × 800 mL), PBS with 1 M NaCl (10 × 800 mL), 100 mM ammonium bicarbonate (10 × 800 mL), and 100 mM ammonium bicarbonate with 2 M urea (10 × 800 mL). Resin was resuspended in 1 mL of 100 mM ammonium bicarbonate with 2 M urea and transferred to a 1.5-mL microcentrifuge tube. Then 1 M TCEP was added to a final concentration of 5 mM, and the sample was incubated at room temperature on a rotating mixer for 30 min, and 500 mM iodoacetamide was added to a final concentration of 10 mM, and the resin was incubated for 1 h at room temperature in the dark. Resin was pelleted at 500 × g, washed with 100 mM ammonium bicarbonate with 2 M urea (3 × 800 mL), and resuspended in 1 mL of 100 mM ammonium bicarbonate with 2 M urea. Sequencing grade modified trypsin (20 µg; Promega) was added, and digestion was allowed to proceed at room temperature on a rotating mixer overnight. The resin was then washed with each of the following buffers: 100 mM ammonium bicarbonate with 2 M urea (10 × 800 mL), 100 mM ammonium bicarbonate (10 × 800 mL), 4 M guanidinium hydrochloride (10 × 800 mL), 100 mM ammonium bicarbonate (10 × 800 mL), and TEV elution buffer (100 mM ammonium bicarbonate and 2 mM DTT; 10 × 800 mL). The resin was resuspended in 0.5 mL of TEV elution buffer and incubated with TEV protease (10 µg) overnight at room temperature to elute biotinylated peptides. The eluted peptides were collected by spinning through a spin filter to remove the resin. The eluate was adjusted to 5% trifluoroacetic acid, incubated on ice for 10 min, and then centrifuged at 20,000 × g at 4 °C to pellet precipitated TEV protease. Peptides were desalted on C18 spin tips (Thermo Fisher Scientific), dried by vacuum centrifugation, dissolved in 10 µL of 0.1% formic acid/2% acetonitrile, and analyzed by LC-MS/MS.
LC-MS/MS Analysis.
Peptides were injected onto an Acclaim PepMap rapid separation liquid chromatography column (75 µm × 15 cm, 2 µm particle size, 100 Å pore size; Thermo Fisher Scientific) and analyzed using a Dionex UltiMate 3000 RSLCnano liquid chromatography system and Q-Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Samples were loaded onto the column over 15 min at 0.5 µL/min in mobile phase A (0.1% formic acid). Peptides were eluted at 0.3 µL/min using a linear gradient from mobile phase A to 40% mobile phase B (0.1% formic acid, 80% acetonitrile) over 125 min. Data-dependent acquisition was performed scanning a mass range from 300 to 1,500 m/z using Xcalibur software (Thermo Fisher Scientific).
MS Data Analysis.
Thermo RAW files were converted to peaklists using MSConvert (ProteoWizard). Peaklists were searched against the human SwissProt database using ProteinProspector (University of California San Francisco), with a false discovery rate of <1%. The parent ion tolerance was set at 6 ppm, and the fragment ion tolerance was set at 20 ppm. Tryptic specificity was required only at the C terminus of peptides to enable identification of endogenous proteolytic cleavages, and two missed tryptic cleavages were allowed. Carbamidomethylation at Cys was set as a constant modification, and Abu at peptide N termini, acetylation at protein N termini, oxidation at Met, pyroglutamate formation at N-terminal Gln, and Met excision at protein N termini were set as variable modifications.
Subcellular Location, Topologic, and Membrane Protein Type Annotations.
Subcellular location annotations and Gene Ontology (GO) cellular component annotations were used to determine the subcellular locations of proteins identified by LC-MS/MS. Annotations were retrieved from the UniProt flat file using custom Python scripts. UniProt subcellular location annotations were used when available; if a UniProt subcellular location annotation was not available, GO cellular component annotations were used.
Analysis of Distance between TM Domains and Cleavage Sites.
Type I membrane proteins were analyzed to determine the distances between the cleavage sites observed in the subtiligase-TM dataset and the transmembrane domain annotated in UniProt using a custom Python script.
Sequence Logos for Subtiligase, Subtiligase-TM, and Specificity Variants.
N termini captured with subtiligase, subtiligase-TM, or subtiligase-TM specificity variants, as well as bioinformatically inferred sequences C terminal to the cleavage site, were analyzed for enrichment or de-enrichment of each amino acid using the standalone version of iceLogo software. The input datasets for the iceLogos shown in Fig. 3 A–D consisted of the combined set of unique N termini identified in nonquantitative subtiligase-TM experiments for each subtiligase-TM variant (Datasets S5, S12, and S13). The input datasets for the iceLogos shown in SI Appendix, Fig. S6 consisted of unique N termini identified in SILAC datasets for each of the mutants (Dataset S18). The human SwissProt database was used as a reference set with random sampling.
Pervanadate Treatment of Subtiligase-TM–Expressing HEK293T Cells.
Pervanadate treatment and SILAC analysis were performed for subtiligase-TM, subtiligase-Y217K-TM, and subtiligase-Y217D-TM. For each replicate of the SILAC experiment, one 500-cm2 dish of subtiligase-TM–expressing cells was grown in heavy SILAC DMEM, and one dish was grown in light SILAC DMEM. Two replicates were performed in which the light cells were treated with pervanadate, and two replicates were performed in which the heavy cells were treated with pervanadate. Before each experiment, a 50 mM stock solution of pervanadate was prepared fresh by mixing equal volumes of 100 mM sodium orthovanadate (New England Biolabs) and 100 mM hydrogen peroxide. Medium was removed from the cells and replaced with serum-free heavy or light DMEM. The pervanadate stock solution was diluted 1:1,000 into the serum-free medium for pervanadate treatment. For control cells, an equal volume of water was added. Treated and untreated cells were incubated for 2 h at 37 °C under a 5% CO2 atmosphere. After 2 h, the medium was removed, cells were washed with PBS, and versene was added to lift the cells. Cells were harvested by centrifugation at 300 × g for 5 min, resuspended in PBS, and counted. Equal numbers of light pervanadate-treated and heavy untreated cells or heavy pervanadate-treated and light untreated cells were mixed together and labeled using the subtiligase-TM method described above.
SILAC Data Analysis.
SILAC quantification was performed using Skyline software. The results of a Protein Prospector search were exported as a pepXML file. Because of inconsistencies in how Protein Prospector and Skyline assign the N-terminal Abu modification, the pepXML file was modified to be Skyline-compatible using a custom Python script. An mzXML format peaklist was generated from the Thermo RAW files corresponding to each experiment using MSConvert (ProteoWizard). The pepXML, mzXML, and RAW files were imported into Skyline, along with a FASTA format list of the identified peptides. The total MS1 area and the isotope dot product for each peptide were calculated using Skyline, and a report was exported for further analysis. The mean experiment-to-control ratio for each N-terminal peptide was calculated using a custom Python script. An isotope dot product of >0.95 for both the heavy and light MS1 peaks was required for quantification of an identified peptide. A t test was performed to evaluate the significance of the observed changes in the experiment-to-control ratio across the four replicate experiments, treating each experiment as an independent sample.
Flow Cytometry for SILAC Data Validation.
HEK293T cells were treated with pervanadate or vehicle as described above. Cells were dissociated by incubation with versene (0.04% EDTA in Ca2+/Mg2+-free PBS) and collected by centrifugation at 300 × g for 5 min. Cells were blocked with PBS containing 3% BSA and stained with either allophycocyanin conjugate anti-human CD99 antibody clone 3B2/TA8 (1:20 dilution; BioLegend, 341307), phycoerythrin anti-human CD166 clone 3A6 (1:20 dilution; BioLegend, 343903), or mouse monoclonal anti-N-cadherin clone GC-4, followed by Alexa Fluor 647-conjugated goat anti-mouse antibody (1:200 dilution; Thermo Fisher Scientific). Staining was performed for 30 min at 4 °C. Cells were washed three times with PBS containing 3% BSA, resuspended in PBS, and analyzed on a Beckman Coulter CytoFlex flow cytometer. Data were analyzed using FlowJo software.
Statistical Analysis.
Statistical tests for flow cytometry were performed using GraphPad Prism 8. To compare pervanadate-treated cells and untreated cells, the median fluorescence intensity (MFI) for the relevant channel was calculated using FlowJo. P values were calculated using the two-tailed unpaired t test. Error bars indicate the mean MFI ± SD for four cell culture replicates. Statistical tests for SILAC MS data were performed using a custom Python script. P values were calculated using scipy.stats.ttest_ind, a built-in function of the SciPy library. A two-sided test for the null hypothesis that the heavy and light samples have identical values was performed. Unless indicated otherwise, four cell culture replicates for each experiment were performed, with the light cells pervanadate-treated for two of the replicates and the heavy cells pervanadate-treated for two of the replicates. Fluorescence microscopy images are representative of experiments performed for three cell culture replicates.
Supplementary Material
Acknowledgments
We thank S. Coyle, M. Ravalin, D. Sashital, A. Dufour, and current and former members of the J.A.W. laboratory for helpful discussions. This work was supported by NIH Grant 5R01 GM081051-09, the Chan Zuckerberg Biohub Investigatorship, and the Harry and Dianna Hind Professorship in Pharmaceutical Sciences (to J.A.W.). J.R.B. was supported by an NIH Ruth L. Kirschstein National Research Service Award (5F32CA239417). A.M.W. was supported by a Helen Hay Whitney Postdoctoral Fellowship (F-1112) and a Career Award at the Scientific Interface from the Burroughs Wellcome Fund (1017065).
Footnotes
Competing interest statement: A.M.W. and J.A.W. and the Regents of the University of California have filed a patent application (US Provisional Patent Application 62/398,898) related to engineered subtiligase variants.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018809118/-/DCSupplemental.
Data Availability
All data generated or analyzed for this study are available in the main text and SI Appendix. In addition, raw MS data and search results have been deposited in the ProteomeXchange repository under the following accession numbers listed in SI Appendix, Table S1: PXD017664, PXD017667, PXD017668, PXD017669, PXD017685, PXD017686, and PXD017687. Python scripts used for data analysis are available in the GitHub repository (https://zenodo.org/record/3678926) under GNU General Public License v3.0.
References
- 1.Puente X. S., Sánchez L. M., Overall C. M., López-Otín C., Human and mouse proteases: A comparative genomic approach. Nat. Rev. Genet. 4, 544–558 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Rawlings N. D., et al., The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 46, D624–D632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahrus S., et al., Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134, 866–876 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleifeld O., et al., Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 28, 281–288 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Staes A., et al., Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat. Protoc. 6, 1130–1141 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Griswold A. R., et al., A chemical strategy for protease substrate profiling. Cell Chem. Biol. 26, 901–907.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wollscheid B., et al., Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 27, 378–386 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bausch-Fluck D., et al., A mass spectrometric-derived cell surface protein atlas. PLoS One 10, e0121314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abrahmsén L., et al., Engineering subtilisin and its substrates for efficient ligation of peptide bonds in aqueous solution. Biochemistry 30, 4151–4159 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Weeks A. M., Wells J. A., Subtiligase-catalyzed peptide ligation. Chem. Rev. 120, 3127–3160 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Agard N. J., Maltby D., Wells J. A., Inflammatory stimuli regulate caspase substrate profiles. Mol. Cell. Proteomics 9, 880–893 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roncase E. J., et al., Substrate profiling and high-resolution co-complex crystal structure of a secreted C11 protease conserved across commensal bacteria. ACS Chem. Biol. 12, 1556–1565 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill M. E., et al., The unique cofactor region of Zika virus NS2B-NS3 protease facilitates cleavage of key host proteins. ACS Chem. Biol. 13, 2398–2405 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Calvo S. E., et al., Comparative analysis of mitochondrial N-termini from mouse, human, and yeast. Mol. Cell. Proteomics 16, 512–523 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford E. D., et al., The DegraBase: A database of proteolysis in healthy and apoptotic human cells. Mol. Cell. Proteomics 12, 813–824 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The UniProt Consortium , UniProt: The universal protein knowledge base. Nucleic Acids Res. 45, D158–D169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Rosmalen M., Krom M., Merkx M., Tuning the flexibility of glycine-serine linkers to allow rational design of multidomain proteins. Biochemistry 56, 6565–6574 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milo R., Phillips R., Cell Biology by the Numbers (Garland Science, 2016). [Google Scholar]
- 19.Weeks A. M., Wells J. A., Engineering peptide ligase specificity by proteomic identification of ligation sites. Nat. Chem. Biol. 14, 50–57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fortelny N., Yang S., Pavlidis P., Lange P. F., Overall C. M., Proteome TopFIND 3.0 with TopFINDer and PathFINDer: Database and analysis tools for the association of protein termini to pre- and post-translational events. Nucleic Acids Res. 43, D290–D297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reiland J., et al., Pervanadate activation of intracellular kinases leads to tyrosine phosphorylation and shedding of syndecan-1. Biochem. J. 319, 39–47 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lichtenthaler S. F., Lemberg M. K., Fluhrer R., Proteolytic ectodomain shedding of membrane proteins in mammals—hardware, concepts, and recent developments. EMBO J. 37, 826 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Otín C., Bond J. S., Proteases: Multifunctional enzymes in life and disease. J. Biol. Chem. 283, 30433–30437 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sevenich L., Joyce J. A., Pericellular proteolysis in cancer. Genes Dev. 28, 2331–2347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mason S. D., Joyce J. A., Proteolytic networks in cancer. Trends Cell Biol. 21, 228–237 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn P.-H., et al., Systematic substrate identification indicates a central role for the metalloprotease ADAM10 in axon targeting and synapse function. eLife 5, 1174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed for this study are available in the main text and SI Appendix. In addition, raw MS data and search results have been deposited in the ProteomeXchange repository under the following accession numbers listed in SI Appendix, Table S1: PXD017664, PXD017667, PXD017668, PXD017669, PXD017685, PXD017686, and PXD017687. Python scripts used for data analysis are available in the GitHub repository (https://zenodo.org/record/3678926) under GNU General Public License v3.0.