Significance
A hallmark of pancreatic ductal adenocarcinoma (PDAC) is its extensively reprogrammed metabolic network, in which NAD and its reduced form NADH are critical cofactors. Here, we show that IFN signaling, present in a subset of PDAC tumors, increases consumption of NAD(H) through upregulation of PARP9, PARP10, and PARP14. This NAD(H) consumption results in increased dependence upon NAMPT for the recycling of NAM to salvage NAD pools, thus sensitizing these tumor cells to treatment with pharmacologic NAMPT inhibition by decreasing PDAC cell proliferation and invasion in vitro and suppressing orthotopic tumor growth and liver metastases in vivo through this mechanism.
Keywords: pancreatic cancer, interferon, NAD, NAMPT, PARP
Abstract
Emerging evidence suggests that intratumoral interferon (IFN) signaling can trigger targetable vulnerabilities. A hallmark of pancreatic ductal adenocarcinoma (PDAC) is its extensively reprogrammed metabolic network, in which nicotinamide adenine dinucleotide (NAD) and its reduced form, NADH, are critical cofactors. Here, we show that IFN signaling, present in a subset of PDAC tumors, substantially lowers NAD(H) levels through up-regulating the expression of NAD-consuming enzymes PARP9, PARP10, and PARP14. Their individual contributions to this mechanism in PDAC have not been previously delineated. Nicotinamide phosphoribosyltransferase (NAMPT) is the rate-limiting enzyme in the NAD salvage pathway, a dominant source of NAD in cancer cells. We found that IFN-induced NAD consumption increased dependence upon NAMPT for its role in recycling NAM to salvage NAD pools, thus sensitizing PDAC cells to pharmacologic NAMPT inhibition. Their combination decreased PDAC cell proliferation and invasion in vitro and suppressed orthotopic tumor growth and liver metastases in vivo.
Pancreatic ductal adenocarcinoma (PDAC) is the major type of pancreatic cancer, with a median overall survival of less than 1 y (1). In addition to an aggressive tumor biology and late stage at diagnosis, the poor prognosis of PDAC is due to its resistance to current therapies. In recent years, studies have profiled its extensively reprogrammed metabolic network and characterized its extreme tumor microenvironment (2–5). These studies have identified PDAC cells’ dependence on glycolysis (2, 6), lipogenesis (2, 7), glutamine metabolism (8, 9), alanine metabolism, and tricarboxylic acid cycle/oxidative phosphorylation (10, 11). Strategies targeting each of these specific metabolic pathways have been attempted but have not been successfully translated into clinical therapeutics. However, all of these rewired metabolic pathways rely on nicotinamide adenine dinucleotide (NAD) or its reduced form, NADH, as cofactors.
NAD is central to cellular bioenergetic and metabolic functions. NAD is synthesized via three major pathways: the de novo biosynthesis kynurenine pathway, the Preiss–Handler pathway, and the salvage pathway (12). The kynurenine pathway starts with the catabolism of the amino acid tryptophan that is then converted via two steps to the intermediate kynurenine, which can generate NAD, kynurenic acid, or xanthurenic acid. The Preiss–Handler pathway and the salvage pathway synthesize NAD from pyridine bases. The Preiss–Handler pathway synthesizes NAD from nicotinic acid (NA) in three steps via the intermediate NA adenine dinucleotide. The NAD salvage pathway starts from the recycling of nicotinamide (NAM) to nicotinamide mononucleotide (NMN) by intracellular nicotinamide phosphoribosyltransferase (NAMPT), followed by the conversion of NMN into NAD via the NMN adenylyltransferases (NMNATs) (12). A recent quantitative analysis revealed that liver cells actively synthesize NAD de novo from tryptophan, releasing NAM into the blood, thereby supporting NAD biosynthesis in the rest of the body, whereas both tumor cells and most other tissues use NAM as the main NAD source (13). NAMPT is the rate-limiting step by which tumor cells utilize NAM in the synthesis of NAD. It is also important to note that, while there are many metabolic enzymes that use NAD or NADH as cofactors and affect the NAD/NADH ratio, NAD(H) is consumed and broken down into NAM by the poly(ADP ribose) polymerase (PARP) family proteins, the sirtuin (SIRT) family proteins, and CD38 (14).
NAMPT inhibitors (NAMPTis) showed promising potency in a variety of preclinical tumor models, including pancreatic cancer (15–18). Two NAMPTis, FK866 (19) and CHS-828 (20, 21), have been tested in clinical trials, where lack of objective responses and dose-limiting toxicity suggest it is necessary to identify subsets of tumors with high sensitivity to NAMPT inhibition. Multiple studies have examined intracellular factors that affect cancer cell sensitivity to NAMPT inhibition, such as NAMPT levels (22), NA phosphoribosyltransferase levels (23), PPM1D mutations (15), and CD38 levels (24). We hypothesized that, in addition to intracellular factors, the tumor microenvironment also impacts PDAC cell NAD(H) levels and thus the sensitivity to NAMPTis.
In this study, we found that type I interferons (IFNs) lower PDAC cell NAD(H) levels through the up-regulation of NAD(H)-consuming enzymes PARP9, PARP10, and PARP14 (PARP9/10/14). Unlike their better studied relatives, PARP1 and PARP2, which catalyze poly-ADP ribosylation, PARP9/10/14 catalyze the transfer of a single unit of ADP ribose to their targets, a process referred to as monoADP ribosylation (25). The roles of PARP9/10/14 in mediating NAD depletion in PDAC cells have not been previously explored to the authors’ knowledge. We further hypothesized that IFN-mediated NAD depletion caused by the up-regulation of PARP9/10/14 sensitizes PDAC cells to NAMPTis, which we confirmed in both in vitro and in vivo PDAC models.
Results
Type I IFN in the Tumor Microenvironment Reduces NAD(H) Levels in PDAC Cells.
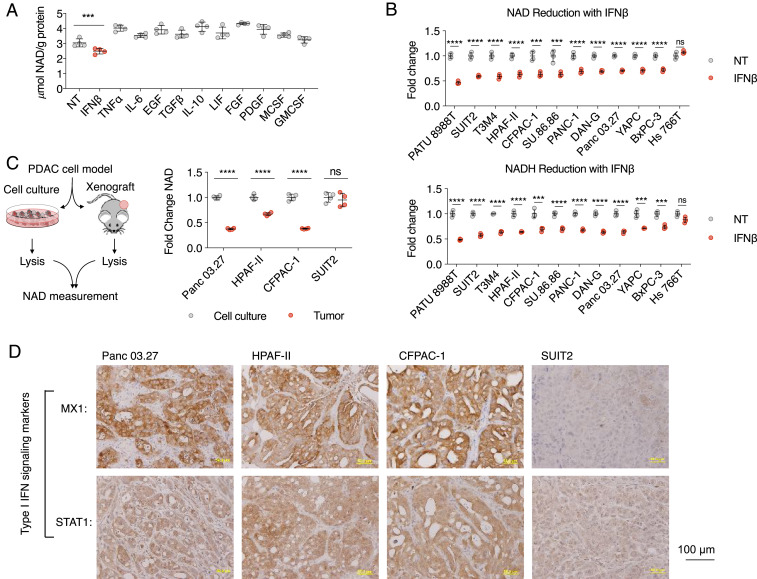
We hypothesized that, in a subset of PDAC tumors, certain cytokine(s) in the PDAC tumor microenvironment impact tumor NAD(H) levels. To test this hypothesis, we cultured Panc 03.27 cells with the supplementation of individual cytokines that have been previously reported as present in the PDAC tumor microenvironment (26–30) (Fig. 1A). Among them, only IFNβ supplementation significantly reduced NAD levels in PDAC cells. To further examine the effect of IFNβ on reducing NAD levels, we supplemented IFNβ in the culture medium of a panel of PDAC cell lines (Fig. 1B). With the exception of Hs 766T cells, all PDAC cell lines tested showed significantly reduced NAD levels. In addition, IFNβ also significantly reduced NADH, the reduced form of NAD, in our panel of PDAC cell lines (again with the exception of Hs 766T). It is important to note that, while IFNβ reduced the total abundance of NAD(H), it did not significantly affect the NAD/NADH ratio (SI Appendix, Fig. S1A).
Fig. 1.
Type I IFN in the tumor microenvironment reduces NAD(H) levels in PDAC cells. (A) Identification of IFNβ as the key cytokine implicated in the reduction of NAD levels. NAD levels in Panc0327 cells cultured with indicated cytokines were measured (n = 4): IFNβ 100 U/mL, TNFɑ 10 ng/mL, IL-6 10 ng/mL, EGF 50 ng/mL, TGFβ 10 ng/mL, IL-10 250 pg/mL, LIF 500 pg/mL, FGF 100 ng/mL, PDGF 50 ng/mL, MCSF 5 ng/mL, and GMCSF 100 ng/mL. (B) Effects of IFNβ on NAD(H) levels across a panel of PDAC cell lines. NAD and NADH levels in indicated PDAC cells were measured after 24 h culture with and without 100 U/mL IFNβ. (C) Supporting evidence that the in vivo tumor microenvironment in a subset of PDAC tumors decreases cancer cell NAD levels. NAD levels from indicated PDAC cells collected from cell culture and from xenograft tumors were compared (n = 4). (D) Type I IFN signaling is present in a subset of PDAC tumors. IHC staining of type I IFN signaling markers MX1 and STAT1 was performed in xenograft tumors collected in the experiment described in C (***P < 0.001, ****P < 0.0001). ns, not significant.
To further support these findings, we extracted cellular metabolites of four PDAC cell lines (Panc 03.27, HPAF-II, CFPAC-1, and SUIT2) from cultured cells and xenograft tumors (Fig. 1C). NAD levels were significantly lower in xenograft tumors compared to cultured cells (Panc 03.27, HPAF-II, and CFPAC-1; Fig. 1C) regardless of two-dimensional (2D) or three-dimensional (3D) culture methods (SI Appendix, Fig. S1B). No significant difference in NAD levels was detected between cultured SUIT2 cells and SUIT2 xenograft tumors. We performed immunohistochemistry (IHC) staining of IFNβ signaling markers MX1 and STAT1 in our xenograft tumors derived from PDAC cell lines Panc 03.27, HPAF-II, CFPAC-1, and SUIT2 (Fig. 1D). While Panc 03.27, HPAF-II, and CFPAC-1 tumors were positive for MX1 and STAT1, SUIT2 tumors were negative for the IFNβ markers. The PDAC tumors in which IFNβ signaling is present correlated with lower measured NAD levels. Given the lack of interspecies IFN cross-reactivity, we are able to conclude that the IFNβ-positive xenograft PDAC tumors engaged in autocrine type I IFN signaling. We recognize that SUIT2 cells represent a subset of PDAC tumors that do not produce IFNβ in vivo (Fig. 1D) but do have an intact IFNβ signaling pathway and respond to supplemented IFNβ in vitro (Fig. 1B).
IFNβ Increases the Expression of NAD(H)-Consuming Enzymes PARP9, PARP10, and PARP14, Leading to a Reduction in Cellular NAD(H) Levels.
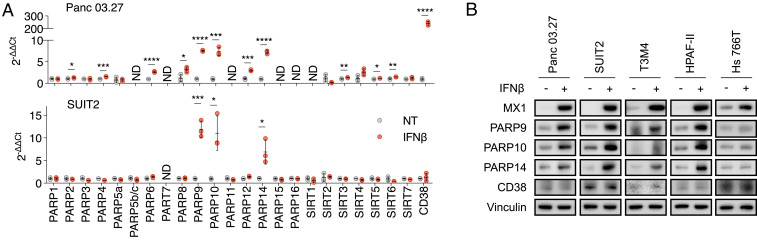
IFNs signal by stimulating the expression of certain genes, known as IFN-stimulated genes (ISGs) (31). We hypothesized that IFNβ reduced NAD(H) levels through up-regulating the expression of genes for NAD(H)-consuming enzymes. To test this hypothesis, we profiled the mRNA levels of all reported NAD(H)-consuming enzymes, including 16 PARP family members, 7 SIRT family members, and CD38, in two PDAC cell lines (Panc 03.27 and SUIT2) with and without IFNβ supplementation (Fig. 2A). In both cell lines, the mRNA levels of PARP9, PARP10, and PARP14 were substantially up-regulated by IFNβ, and the mRNA level of CD38 was increased only in Panc 03.27 cells (Fig. 2A). We examined the effect of IFNβ on the protein levels of PARP9, PARP10, PARP14, and CD38 in Panc 03.27 and SUIT2 cells, as well as three additional PDAC cell lines (T3M4, HPAF-II, and Hs766T; Fig. 2B). PARP9, PARP10, and PARP14 protein levels were increased by IFNβ in all PDAC cell lines tested except for Hs766T, and CD38 protein levels were not affected by IFNβ (Fig. 2B). These data suggest that the up-regulation of PARP9/10/14 expression mediates the reduction of NAD(H) levels induced by IFNβ. The cell line Hs 766T, which did not show reduced NAD(H) levels with IFNβ supplementation (Fig. 1D), also did not demonstrate up-regulation of PARP9/10/14 protein levels (though these cells expressed the marker ISG MX1 in response to IFNβ, demonstrating an intact IFNβ signaling pathway; Fig. 2B).
Fig. 2.
IFNβ elevates the expression of NAD(H)-consuming enzymes PARP9, PARP10, and PARP14. (A) PARP9, PARP10, and PARP14 transcription levels were significantly up-regulated after exposure to IFNβ. Panc0327 and Suit2 cells were cultured with 100 U/mL IFNβ prior to quantification of PARP9, PARP10, and PARP14 mRNA levels (n = 3). ND, not detected. (B) PARP9, PARP10, and PARP14 protein levels were measured after exposure to IFNβ. Indicated PDAC cells were cultured with 100 U/mL IFNβ prior to immunoblot analysis of PARP9, PARP10, and PARP14 protein levels in whole-cell lysates (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
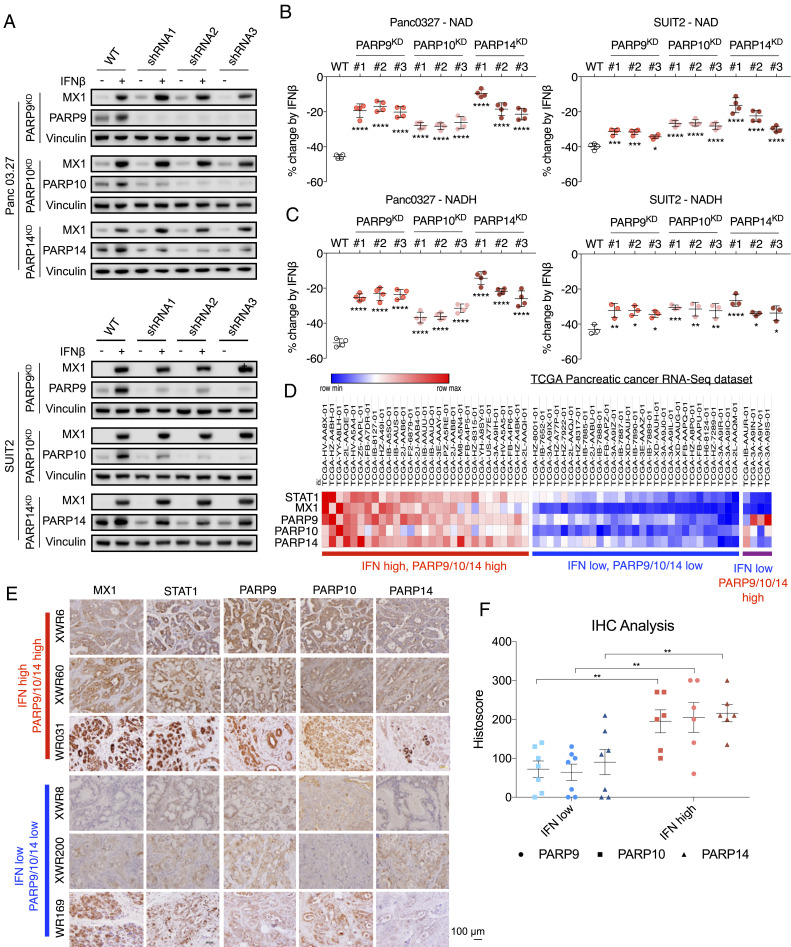
To evaluate the discrete contributions of PARP9, PARP10, and PARP14 to IFN-induced NAD depletion, we knocked down their expression individually in both Panc 03.27 and SUIT2 cells using short hairpin RNA (shRNA). Analysis of protein levels after knockdown demonstrated an intact IFNβ signaling pathway but appropriate lack of up-regulation of each of the corresponding enzymes (Fig. 3A). We found that knockdown of each gene partially mitigated the effect of IFNβ treatment on NAD/NADH depletion, but did not completely rescue either NAD or NADH levels, indicating that these ISGs may function in concert to deplete NAD(H) pools in PDAC cells (Fig. 3B).
Fig. 3.
PARP9, PARP10, and PARP14 induction by IFNβ reduces NAD(H) levels. (A) PARP9, PARP10, and PARP14 were knocked down by three shRNAs per gene in Panc03.27 and SUIT2 cells. MX1, PARP9, PARP10, and PARP14 protein levels were measured with and without exposure to 100 U/mL IFNβ. (B and C) Knockdown of PARP9, PARP10, or PARP14 significantly rescued NAD (B) and NADH (C) levels decreased by IFNβ (n = 4). NAD and NADH levels were measured in Panc03.27 and SUIT2 cells following 24 h exposure to 100 U/mL IFNβ. (D) Subsets of PDAC tumors within the TCGA pancreatic cancer dataset with high and low expression levels of type I IFN signaling (STAT1 and MX1) and PARP9, PARP10, and PARP14. (E) PDAC PDX and primary tumor models with high or low levels of PARP9, PARP10, and PARP14 correlate with high or low levels of ISGs MX1 and STAT1, respectively. IHC staining of MX1, STAT1, PARP9, PARP10, and PARP14 in tumor sections from PDAC PDX models XWR6, XWR60, XWR8, and XWR200 and primary tumors WR031 and WR169. (F) The histoscores for PARP9, PARP10, and PARP14 between the IFN-marker high and IFN-marker low groups, as defined by their MX1 and STAT1 histoscores, were all significantly different. The images corresponding to the remaining histoscores not shown here are shown in SI Appendix, Fig. S2B (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Among the PDAC tumor samples in The Cancer Genome Atlas (TCGA) dataset, we observed a correlation between IFNβ signaling marker mRNA levels (STAT1 and MX1) and PARP9/10/14 mRNA levels (Fig. 3D). There is a minor subset of PDAC tumors with high PARP9, PARP10, or PARP14 levels but without high STAT1/MX1 levels, suggesting that PARP9/10/14 can occasionally be induced by factors other than type I IFN signaling. A total of 16% of all TCGA PDAC tumors exhibit high PARP9/10/14 levels in association with high STAT1/MX1 levels (Fig. 3D). There was no significant difference in disease progression between the PARP9/10/14 high and low subsets of PDAC patients (SI Appendix, Fig. S2A). We also confirmed the association between high or low levels of ISGs MX1 and STAT1 and high or low levels of PARP9, PARP10, and PARP14 within cancer cells in PDAC patient-derived xenograft (PDX), cell line, and primary tumor models using IHC staining (Fig. 3E and SI Appendix, Fig. S2B). Histoscores were calculated for all of these samples, and the histoscores for PARP9, PARP10, and PARP14 were compared between the IFN-marker high and IFN-marker low groups, as defined by their MX1 and STAT1 histoscores, and were found to be significantly different (Fig. 3F). We therefore hypothesized that IFN signaling-induced up-regulation of PARP9/10/14 causing increased NAD consumption would sensitize these tumor cells to further NAD-depleting therapies.
NAD(H) Consumption via Up-Regulation of PARP9/10/14 Results in Increased Dependency of These Tumors on NAMPT, Therefore Sensitizing Them to NAMPT Inhibition.
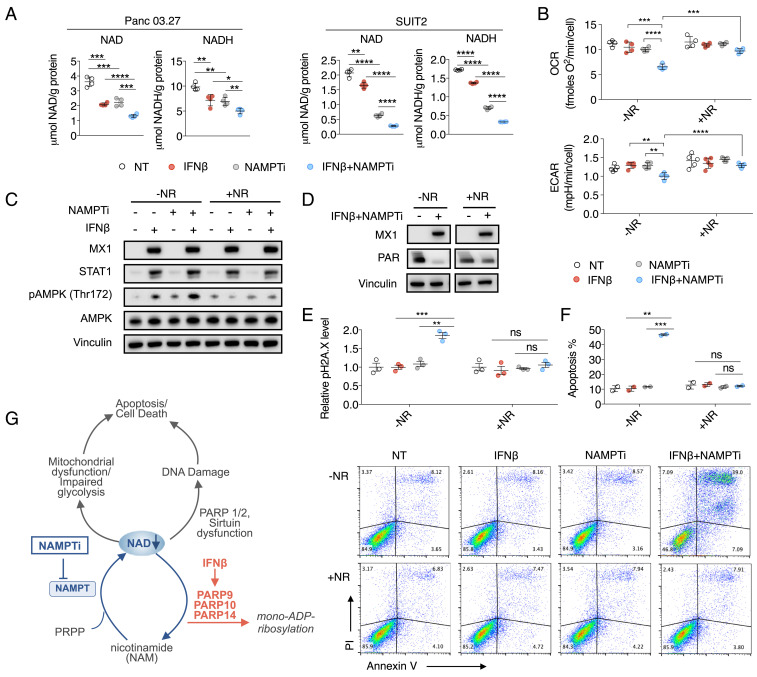
NAMPT mediates the rate-limiting step in the NAD(H) salvage pathway, which is the dominant source of NAD(H) supply in cancer cells and most normal tissues (13). We hypothesized that type I IFN signaling, which increases NAD(H) consumption through up-regulating PARP9/10/14 expression, increases the dependency of these tumors on NAMPT to recycle NAM and regenerate NAD(H), thus sensitizing them to treatment with NAMPTis. To test this hypothesis, we first examined the effect of combining IFNβ and an NAMPTi, FK866, on NAD and NADH levels in PDAC cells. In both Panc 03.27 and SUIT2, the combination of IFNβ and NAMPTi reduced NAD and NADH levels to significantly lower levels compared to either IFNβ or NAMPTi alone, supporting our hypothesis that these cells are reliant upon NAMPT to maintain NAD pools via NAM recycling in the setting of PARP9/10/14 up-regulation by IFNβ (Fig. 4A).
Fig. 4.
Type I IFN signaling results in NAD(H) consumption through up-regulation of PARP9/10/14, increasing PDAC cells’ dependency on NAMPT and sensitizing them to NAMPTi in vitro. (A) IFNβ enhanced the NAD(H)-depleting effect of NAMPTi. Panc 03.27 and SUIT2 cells were cultured with 100 U/mL IFNβ and 8 nM NAMPTi FK866 for 24 h prior to NAD and NADH measurements. (B) IFNβ and NAMPTi inhibited mitochondrial respiration and decreased glycolytic reserve in Panc 03.27 cells. Cells were incubated with 100 U/mL IFNβ and 8 nM NAMPTi FK866 for 24 h prior to measuring OCR and ECAR. (C) Western blot demonstrated increased pAMPK expression in combination-treated PDAC cells compared to either treatment alone, which is rescued by NR supplementation. SUIT2 cells were incubated with or without 100 U/mL IFNβ ± 8 nM FK866 ± 500 μM NR for 48 h. (D) NAD(H) reduction by IFNβ/NAMPTi suppressed PARP activity in DNA repair. Panc 03.27 cells were exposed to indicated treatments for 48 h prior to immunoblot analysis of MX1 and PAR (IFNβ, 100 U/mL; NAMPTi FK866, 4 nM; NR, 500 μM). (E) IFNβ enhanced the effect of NAMPTi on inducing DNA damage. Panc 03.27 cells were incubated with or without 100 U/mL IFNβ ± 8 nM FK866 ± 500 μM NR for 48 h prior to pH2A.X quantification by flow cytometry. (F) IFNβ enhanced the effect of NAMPTi on inducing apoptosis. Panc 03.27 cells were incubated with or without 100 U/mL IFNβ ± 8 nM FK866 ± 500 μM NR for 48 h prior to Annexin V/PI quantification by flow cytometry. Apoptosis was quantified by the amount of Annexin V-positive cells. (G) Proposed model of NAD depletion induced by inhibition of NAMPT in combination with IFNβ signaling, thus blocking salvage of the NAM product of PARP9/10/14 (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). ns, not significant.
We hypothesized that the NAD(H) depletion induced by this combination therapy would cause dysfunction of known NAD(H)-requiring metabolic pathways. NAD(H) is a cofactor of complex I for mitochondrial respiration and glyceraldehyde-3-P dehydrogenase and lactate dehydrogenase in glycolysis. We examined the mitochondrial oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR), a measure of glycolytic reserve, in PDAC cells exposed to IFNβ and NAMPTi, both alone and in combination. The combination of IFNβ and NAMPTi significantly reduced OCR and ECAR more than either IFNβ or NAMPTi alone (Fig. 4B). Additionally, this reduction in OCR and ECAR was rescued by supplementation with nicotinamide riboside (NR; Fig. 4B), which bypasses the NAMPT step in the salvage pathway for NAD(H) synthesis (14), demonstrating the on-target effect of this therapy. A consequence of NAD(H) depletion, reduced OCR, and decreased glycolytic reserve is ATP depletion. To determine ATP depletion, we examined the Thr172 phosphorylation of AMPK, a cellular energy sensor. IFNβ and NAMPTi together triggered AMPK phosphorylation, and this was again rescued by NR supplementation (Fig. 4C).
Yet another consequence of NAD(H) depletion is DNA damage, because the activities of the critical DNA repair enzymes in the PARP and SIRT families are NAD(H)-dependent. The NAD(H)-dependent PARP and SIRT families are critical players in DNA repair (32, 33). PARP family activity can be monitored by measuring PAR abundance (34). PAR abundance was substantially reduced by IFNβ and NAMPTi, and this reduction was rescued by NR supplementation (Fig. 4D). We observed that IFNβ and NAMPTi together increased H2A.X phosphorylation, a marker of DNA damage, which was also rescued by NR supplementation (Fig. 4E). Given these observations, we hypothesized that the NAD(H) reduction by IFNβ and NAMPTi together would result in increased PDAC cell apoptosis, which was confirmed by Annexin V/propidium iodide (PI) quantification by flow cytometry (Fig. 4F). This was also rescued by NR supplementation (Fig. 4F). Finally, it has been demonstrated that NAD(H) depletion results in decreased malignant cell invasion (35). We found that IFNβ and NAMPTi together inhibited the invasion of PDAC cells, whereas neither IFNβ nor NAMPTi significantly affected PDAC cell invasion as single agents (SI Appendix, Fig. S3A), and, at the molecular level, IFNβ and NAMPTi down-regulated the protein levels of epithelial cell markers E-cadherin and N-cadherin (SI Appendix, Fig. S3B).
Taken together, our results confirmed that IFNβ sensitized PDAC cells to NAMPTi, with NAD(H) depletion resulting in cytotoxicity and metabolic dysfunction due to a multifactorial model including DNA damage, impaired DNA repair activity of the PARP and SIRT family members, and decreased mitochondrial respiration and glycolytic reserve (Fig. 4G).
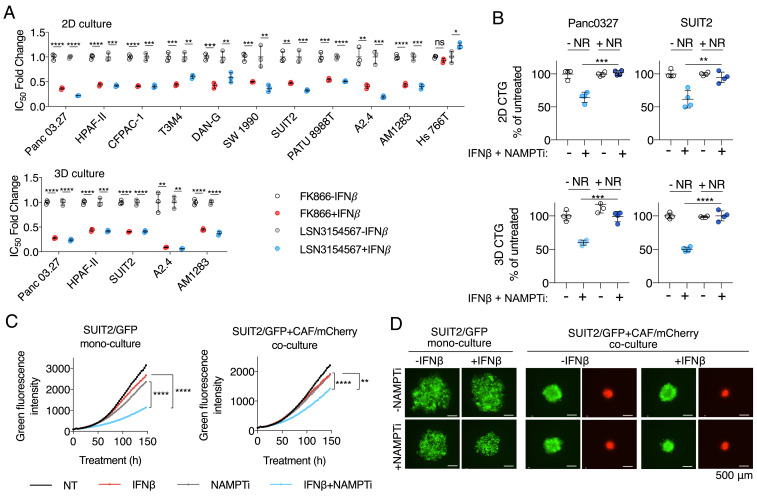
To further support our finding that increased NAD(H) consumption by IFNβ sensitizes PDAC cells to NAMPTi, we tested two chemically different NAMPTi inhibitors, FK866 and LSN3154567 (17), in a viability assay in a panel of PDAC cell lines and primary PDAC cultures (A2.4 and AM1283) with and without IFNβ supplementation. Except for Hs 766T, which we previously found does not up-regulate PARP9/10/14 and lower NAD(H) levels when supplemented with IFNβ (Figs. 2B and 1D), all PDAC cell lines and primary cultures showed lower half maximal inhibitory concentration (IC50) values of FK866 and LSN3154567 in the presence of IFNβ, both in standard 2D culture and anchorage-independent 3D culture (Fig. 5A and SI Appendix, Table S1). In both 2D and 3D cultures, the increased cytotoxicity of NAMPTi by IFNβ was rescued by NR supplementation (Fig. 4B), confirming that this phenotype was due to on-target NAD(H) depletion.
Fig. 5.
Type I IFN signaling sensitizes PDAC cells to NAMPTis in vitro. (A) IFNβ enhances the potency of NAMPTi in both 2D and 3D cultures. PDAC cell lines and primary PDAC cultures (A2.4 and AM1283) were treated with NAMPTi FK866 or LSN3154567 for 72 h with and without 100 U/mL IFNβ supplementation. Cell viability was measured by CellTiter-Glo assay, and IC50 values were determined using GraphPad Prism 7. (B) Cytotoxicity of the combination of IFNβ and NAMPTi in PDAC cells was rescued by NR supplementation. PDAC cells were incubated with or without 8 nM FK866 ± 100 U/mL IFNβ ± 500 μM NR in 2D or 3D cultures. Cell viability was determined by CellTiter-Glo assay. (C) IFNβ sensitizes cells to NAMPTi in both PDAC monoculture and coculture with PDAC CAFs. SUIT2/GFP cell spheroids with and without CAF/mCherry were treated with or without 8 nM FK866 ± 100 U/mL IFNβ. Green and red fluorescence were monitored by IncuCyte every 3 h for a 7-d period. (D) Representative fluorescence images of spheroids at the experiment endpoint in C (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). ns, not significant.
PDAC is characterized by the presence of abundant desmoplastic stroma primarily composed of cancer-associated fibroblasts (CAFs), which support PDAC cell survival and chemoresistance (36–39). Therefore, we tested the effect of the IFNβ/NAMPTi combination side by side on the growth of spheroids with SUIT2/GFP cancer cells alone and spheroids with both SUIT2/GFP cancer cells and CAF/mCherry stromal cells. In both spheroid models, the IFNβ/NAMPTi combination suppressed spheroid growth better than either IFNβ or NAMPTi alone (Fig. 5 C and D and Movies S1 and S2). Taken together, our results indicate that the presence of IFNβ sensitizes PDAC cells to the cytotoxicity of NAMPTi in an NAD(H)-dependent manner.
Inactivation of Type I IFN Signaling Promotes Resistance to NAMPT Inhibitors.
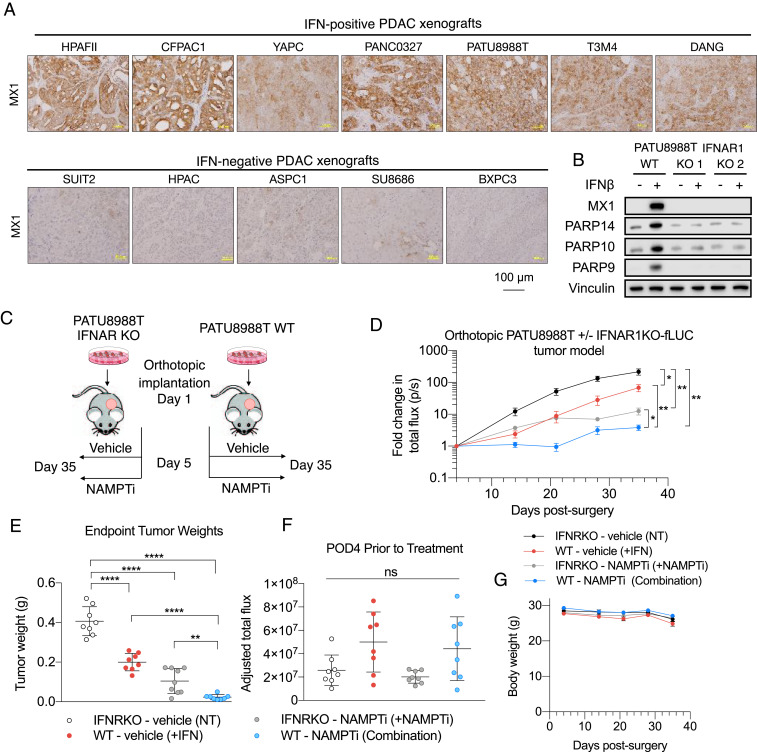
Having demonstrated the IFNβ-induced, PARP9/10/14-mediated sensitization to NAMPTi of PDAC cells in vitro, we hypothesized that type I IFN signaling is required to see this effect in vivo. We profiled a broad panel of PDAC xenograft models for in vivo type I IFN signaling based on IHC analyses of the type I IFN ISG MX1 (Fig. 6A). We found a subset of xenograft tumors that stained positive for MX1, which we termed IFN-positive PDAC xenografts, and another subset which did not stain for MX1, which we termed IFN-negative. We selected a representative IFN-positive PDAC xenograft PATU8988T and used CRISPR-Cas9 to knock out (KO) the type I IFN receptor IFNAR1 (SI Appendix, Fig. S4). To confirm that PARP9, PARP10, and PARP14 expression was mediated by type I IFN signaling, we also examined protein levels of those enzymes and found that IFNAR1 KO abolished the increased expression of MX1, PARP9, PARP10, and PARP14 after IFNβ treatment seen in WT cells (Fig. 6B).
Fig. 6.
Inactivation of type I IFN signaling promotes resistance to NAMPTis. (A) Profiling of a broad panel of PDAC xenograft models for in vivo type I IFN signaling based on IHC analyses of type I IFN signaling marker MX1. (B) A loss-of-function PDAC model of autocrine type I IFN signaling. PATU8988T cells underwent KO of the type I IFN receptor, and two of these KO models were chosen and were exposed to 100 U/mL IFNβ supplementation in cell culture for 48 h, prior to immunoblot analyses of type I IFN signaling marker MX1 and PARP 9/10/14. (C) Schematic of in vivo experimental design. (D) Curves of bioluminescence intensity of orthotopic PATU8988T WT or IFNAR1 KO-fLUC tumors in mice treated with vehicle control or 10 mg/kg daily intraperitoneal (i.p.) FK866. After confirmation of tumor establishment by BLI, treatments started on day 5 postsurgical implantation. Data are shown as mean ± SEM (n = 8 per group). (E) Endpoint tumor weights after harvest of orthotopic PATU8988T WT and IFNAR1 KO tumors in mice treated with either vehicle or 10 mg/kg FK866 (daily i.p.). Data are shown as mean ± SEM (n = 8 per group). (F) BLI imaging demonstrating no significant difference between tumor-adjusted total flux on postimplantation day 4 prior to the initiation of treatment. Data are shown as mean ± SEM (n = 8 per group). (G) Treatments with vehicle control or 10 mg/kg FK866 were well tolerated by mice bearing PATU8988T WT or IFNAR1 KO-fLUC orthotopic tumors (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). ns, not significant.
In order to compare the efficacy of NAMPTi in in vivo PDAC tumor models with and without type I IFN signaling, we utilized this PATU8988T model with its IFNAR1 KO isogenic line as a loss-of-function control. After orthotopic PATU8988T WT and IFNAR1 KO tumors were established, mice were randomized to either be treated with vehicle control or NAMPTi (Fig. 6C). Prior to initiation of treatment, there were no significant differences in tumor size between the groups (Fig. 6F). After 3 wk of treatment, PDAC tumors with intact type I IFN signaling and treated with NAMPTi (representing the combination group) were significantly smaller, both based on weekly bioluminescent imaging (BLI; Fig. 6D) and endpoint tumor weights (Fig. 6E), than both IFNAR1 KO tumors treated with NAMPTi (representing the NAMPTi-only treatment group) and WT untreated tumors (representing the type I IFN-only treatment group). Our results confirmed that increased PARP9/10/14 expression, NAD(H) reduction, growth inhibition, and enhanced sensitivity to NAMPTi in vivo are mediated by type I IFN signaling.
Increased IFN Signaling Downstream of STING Activation Sensitizes Tumors to NAMPTi.
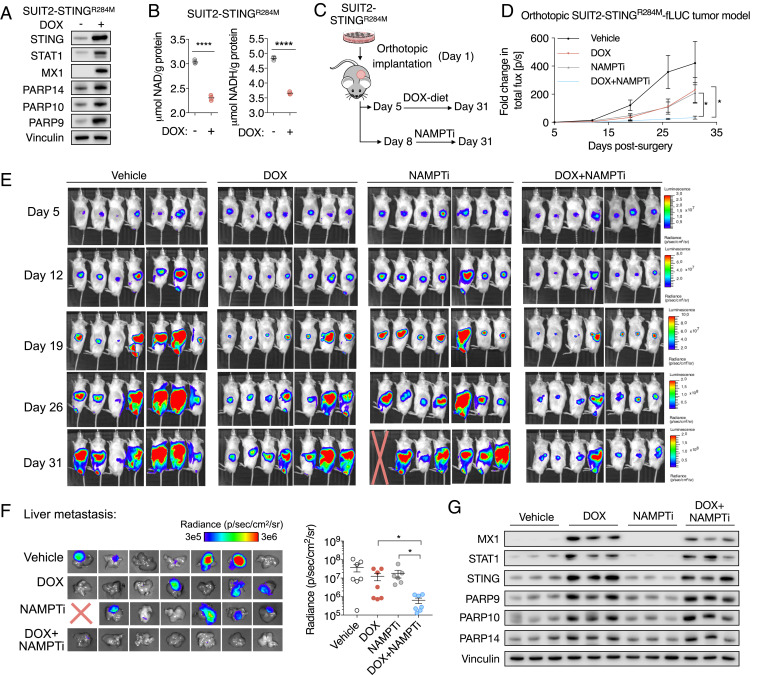
Type I IFN production by cancer cells is frequently driven by the activation of STING due to genomic instability (40). In order to compare the efficacy of NAMPTi in in vivo PDAC tumor models with controllable type I IFN signaling, we established a gain-of-function PDAC SUIT2 model of autocrine type I IFN signaling with doxycycline (DOX)-inducible expression of an active STINGR284M mutant (41). DOX exposure stimulated the expression of ISGs STAT1 and MX1 as well as PARP9, PARP10, and PARP14 (Fig. 7A), indicating the activation of type I IFN signaling in the presence of DOX. DOX exposure significantly lowered NAD(H) levels in SUIT2-STINGR284M cells (Fig. 7B). To confirm that PARP9, PARP10, and PARP14 expression was mediated by autocrine type I IFN signaling in these models, we used CRISPR-Cas9 to KO type I IFN receptor IFNAR1 (SI Appendix, Fig. S6A). In the presence of DOX, IFNAR1 KO abolished STINGR284M-mediated expression of STAT1, MX1, PARP9, PARP10, and PARP14 (SI Appendix, Fig. S6B). IFNAR1 KO did not significantly affect the proliferation of SUIT2-STINGR284M cells in the absence of DOX (SI Appendix, Fig. S6C). In the presence of DOX, IFNAR1 KO rescued the growth-inhibitory effect of STINGR284M and the effect of STINGR284M on sensitizing SUIT2 cells to NAMPTi (SI Appendix, Fig. S6 D and E).
Fig. 7.
Increased type I IFN signaling downstream of STING activation sensitizes tumors to NAMPTis. (A) A gain-of-function PDAC model of DOX-inducible autocrine type I IFN signaling. SUIT2 cells with a DOX-inducible active STINGR284M mutant were exposed to 50 ng/mL DOX in cell culture for 4 d, prior to immunoblot analyses of type I IFN signaling markers. (B) DOX-induced type I IFN signaling significantly lowered NAD and NADH levels in SUIT2-STINGR284M cells. Cells were exposed to 50 ng/mL DOX for 5 d prior to NAD and NADH measurements (n = 3). (C) Schematic of in vivo experimental design. (D) Curves of bioluminescence intensity of orthotopic SUIT2-STINGR284M-fLUC tumors in mice with DOX diet and/or 10 mg/kg FK866 (daily i.p.). After confirmation of tumor establishment by BLI, treatments started on day 6 postsurgical implantation. Data are shown as mean ± SEM (n = 7 per group). (E) BLI images of disease progression in indicated experimental groups. BLI measurement in photons per second per square centimeter per steradian (p/s/cm2/sr) was translated to color to indicate disease activity. A mouse died on day 30 due to tumor invasion into the gastrointestinal tract. (F) BLI measurement of liver metastasis of SUIT2-STINGR284M-fLUC tumor cells. At 5 min after luciferin injection, mice were euthanized, and livers were harvested for BLI measurement. (Left) BLI images of liver metastasis. (Right) Quantification of BLI intensity in livers. (G) Immunoblot analysis of indicated proteins in tumor homogenates. Three representative tumors from each group were included for comparison.
We implanted orthotopic SUIT2-STINGR284M cells, and, when tumors were established, mice were randomized to either a vehicle control or DOX-containing diet (to activate type I IFN signaling) and also to be treated with either a vehicle control or NAMPTi (Fig. 7C). Prior to the initiation of treatment, there were no significant differences in tumor sizes between the groups (SI Appendix, Fig. S5G). After a 3-wk treatment, PDAC tumors with type I IFN signaling (DOX diet) and treated with NAMPTi were significantly smaller than IFN-negative (control diet) tumors treated with NAMPTi, as well as smaller than tumors with type I IFN signaling alone (Fig. 7 D and E). In addition to suppressing primary orthotopic tumor growth, NAMPTi and type I IFN signaling together resulted in a decreased number and size of liver metastases (Fig. 7F), a common feature of clinical PDAC. During the treatment period, the dosage of NAMPTi we used was well tolerated by the animals (SI Appendix, Fig. S6F). Immunoblot analyses of tumor homogenates revealed activation of autocrine type I IFN signaling and expression of PARP9/10/14 in samples collected from animals on the DOX diet (Fig. 7G). Our results indicate that NAMPTi is more effective in suppressing both tumor growth and liver metastases of PDAC tumors with active type I IFN signaling. Taken together, our results support our hypothesis that the sensitization to NAMPTi seen with type I IFN signaling is dependent upon PARP9/10/14-mediated NAD(H) depletion leading to an increased dependence upon NAMPT for salvage of NAD(H) pools.
Discussion
Chronic inflammation, a defining characteristic of PDAC tumors, has been linked to elevated levels of cytokines produced by tumor, stromal, and immune cells (42). Of the inflammatory cytokines in the PDAC tumor microenvironment (43, 44), IFNs are among the most important given their high prevalence and ability to regulate the expression of thousands of genes (45, 46) with both tumor-supportive (47–49) and suppressive phenotypes (45, 49). Emerging evidence suggests that IFN signaling also triggers targetable vulnerabilities in cancer cells (50). Unlike IFNγ that is mainly produced by activated T cells, type I IFNs can be produced by multiple cell types in the PDAC tumor microenvironment, including tumor cells themselves with autocrine circuitries (31, 51). While the signaling and immunomodulatory effects of type I IFNs have been described, their impact on tumor cell metabolism remains poorly understood. In this study, we identified an effect of type I IFN, which is present in a subset of PDAC tumors, on reducing tumor cell NAD(H) levels and sensitizing PDAC cells to NAMPTi through stimulating the expression of PARP9, PARP10, and PARP14.
PARP1 and PARP2 are the founding members in the PARP family and have been extensively studied for their DNA repair activity. Compared to PARP1 and PARP2, the roles of PARP9, PARP10, and PARP14 are less well characterized. Our data showed that silencing of PARP9, PARP10, or PARP14 partially rescued IFNβ-induced NAD(H) reduction. Both PARP10 and PARP14 have broad-spectra ADP ribosylation substrates identified in protein microarrays (52), which are consistent with their NAD(H) consumption in our observations. In contrast, PARP9 lacks catalytic activity (53), but it interacts with other PARP family members and regulates their expression and activity (54). In addition, PARP9 also promotes cellular response to IFNs (54). These indirect effects of PARP9 may explain our observation that PARP9 silencing reduced IFNβ-induced NAD(H) consumption.
Previous clinical trials of NAMPTi for cancer treatment suggest that its clinical success requires the identification of cancer subsets with high sensitivity to NAD(H) reduction. While the salvage pathway for NAD(H) supply has been extensively studied and explored for cancer treatment, the significance of NAD(H) consumption has not yet been comprehensively examined. We found that up-regulation of PARP9/10/14 by type I IFN lowered NAD(H) levels, increased dependency on NAMPT to prevent severe depletions of NAD pools, and sensitized PDAC cells to NAMPTi treatment, which then resulted in the suppression of both glycolysis and mitochondrial respiration, impaired DNA repair, and the induction of apoptosis.
Autocrine type I IFN signaling is regulated by STING. Our gain-of-function model of type I IFN signaling suggests that small-molecule STING agonists are potentially able to up-regulate PARP9/10/14 levels in IFN signaling-negative PDAC tumors and sensitize them to NAMPTi therapy. While many STING agonists require intratumoral injection (55), which cannot be practically applied for clinical PDAC treatment, a recent study reported a potent STING agonist with systemic anticancer activity (56). Regarding NAMPT, while previous inhibitors demonstrated a lack of objective responses and dose-limiting toxicities in clinical trials and there is a wide consensus that novel NAMPTis without these same toxicities are necessary, there is a recently developed NAMPTi (KPT-9274) that is distinguished from previous NAMPTs by impressive preliminary toxicology, an acceptable side-effect profile, and excellent oral bioavailability (57). It is being investigated currently in multiple clinical trials for the treatment of solid cancers and non-Hodgkin’s lymphoma (ClinicalTrials.gov ID codes NCT02702492, NCT04281420). Our research suggests that there may be utility in examining the use of systemic STING agonists as a strategy to sensitize tumors to next-generation NAMPTis.
In conclusion, our hypothesis-driven study implicates the up-regulation of PARP9/10/14 as the mechanism by which type I IFN signaling leads to increased NAD(H) consumption in PDAC tumors. This NAD(H) consumption results in increased dependence upon NAMPT for the recycling of NAM to salvage NAD pools, thus sensitizing these tumors to treatment with NAMPTi.
Materials and Methods
Cell Culture.
SI Appendix, Supplementary Methods, provides information regarding the acquisition of cell lines. Primary human CAFs were established from surgical PDAC specimens by a previously described protocol (58) and verified by wild-type KRAS status and α-smooth muscle actin expression as described previously (59).
Antibodies and Drugs.
Information regarding the acquisition of all antibodies and drugs is provided in SI Appendix, Supplementary Methods.
NAD/NADH Assay.
NAD/NADH levels were measured by the NAD/NADH-Glo Assay (Promega G9071). Cells were seeded in 96-well plates (2D culture) or poly(2-hydroxyethyl methacrylate)-coated (20 mg/mL in 95% ethanol; Sigma P3932) 96-well plates (3D culture) for 24 h. Then, cells were lysed with 50 μL of D-phosphate-buffered saline (PBS) solution and 50 μL of 0.2 N NaOH solution with 1% dodecyltrimethylammonium bromide. The lysates were centrifuged at 4 °C, 14,000 × g, and the supernatant was collected. A BCA Protein Assay Kit (Pierce no. 23227) was used to measure the protein concentration of the lysates. To measure NAD, 20 μL lysate was added to a 384-well plate (Greiner Bio-One no. 781098) treated with 10 μL 0.4 N HCL and heat-quenched at 60 °C for 15 min. This was then neutralized with 10 μL Trizma base solution. NADH samples alone were heat-quenched in the same manner as the NAD samples, followed by the addition of 20 μL HCL/Trizma solution. An equal volume of NAD/NADH-Glo Detection Reagent was added to each well and incubated at room temperature for 30 min. The luminescence was measured by Synergy H1 Hybrid Multi-Mode Reader (BioTek). The sample NAD/NADH levels based on luminescence intensity were calculated based on the NAD/NADH standard curves. The final data [μmol NAD(H) per gram protein] were adjusted based on BCA results.
pH2A.X Assay and 5-Ethynyl-2′-Deoxyuridine Cell-Cycle Profiling.
Cells were harvested, fixed, and permeabilized, then fixed before the acquisition of data by flow cytometry as described in SI Appendix, Supplemental Methods. Cell-cycle durations were calculated using equations for multiple time-point measurements according to previously published methods (60).
Western Blot.
Cells were lysed in cold RIPA lysis buffer with protease and phosphatase inhibitors (Thermo Scientific). The lysates were subsequently normalized using a BCA assay, then diluted using RIPA buffer and 6× Laemmli loading dye. Protein extracts were resolved on sodium dodecyl sulfate polyacrylamide gel electrophoresis and then electrotransferred to an Immun-Blot Nitrocellulose membrane (Bio-Rad no. 1620115). After blocking using 5% nonfat milk in Tris-buffered saline (TBS) + 0.1% Tween-20 (TBS-T), membranes were probed with the indicated primary antibodies at 4 °C overnight. After incubation, the membranes were washed with TBS-T and then probed with horseradish peroxidase (HRP)-conjugated secondary antibodies at room temperature for 1 h. Blots were developed using Immobilon Forte Western HRP Substrate (Millipore WBLUF0500) and imaged on the LI-COR Odyssey imaging system.
Seahorse Respirometry Assay.
Respirometry assays were performed as described previously (61). Panc 03.27 and SUIT2 cells were plated and treated as described in SI Appendix, Supplemental Methods.
Kinetic Proliferation Assay.
The IncuCyte Zoom live-cell imaging system was used to track cell proliferation as described in SI Appendix, Supplemental Methods.
Knockdown.
For generation of stable knockdown cell lines, PDAC cells were transduced with a lentivirus harvested from HEK293FT cells in the presence of Polybrene. Following transduction, cells underwent antibiotic selection, and knockdown efficiency was confirmed using immunoblot analysis. For inducible PARP9, PARP10, and PARP14 knockdown, the oligonucleotide sequences and induction details are provided in SI Appendix, Supplemental Methods.
IFN Receptor KO.
Three sgRNA sites were designed for the IFNAR1 gene. The recombinant plasmids of lentiviral vector2‐IFNAR1‐sgRNA were constructed. The constructed vectors were transfected into PATU8988T and SUIT2-mSTING cells with Lipofectamine 3000. At 24 h later, cells were treated with puromycin for 3 d. The two guide RNAs (gRNAs; no. 1, GCACTAGGGTCGTCGCGCCC; no. 2, GCTCGTCGCCGTGGCGCCAT) with the best KO effect were selected. Following puromycin selection, cells were singly cloned. Single-cell cloning was performed with limited dilution of cells into 96-well plates. At least two single clones of each gRNA were confirmed as KO ones with genomic DNA PCR and Tracking of Indels by Decomposition (TIDE) in/del analysis after Sanger sequencing.
Real-Time qPCR.
Total RNA was isolated from cells using the Zymo Quick-RNA MiniPrep kit. Reverse transcription was performed using the High Capacity cDNA Reverse Transcription kit (Life Technologies). qPCR was performed using EvaGreen qPCR Master Mix (Lamda Biotech). RNA expression values were normalized and calculated as relative expression to control. Primer sequences used for real-time qPCR for each gene are provided in SI Appendix, Supplemental Methods.
Immunohistochemistry.
Formalin-fixed, paraffin-embedded tumor samples were incubated at 60 °C for 1 h, deparaffinized in xylene, and rehydrated with graded alcohol washes. Slides were then boiled in 0.01 M sodium citrate buffer for 15 min, followed by quenching of endogenous peroxidase with 3% hydrogen peroxide for antigen retrieval. After 1 h of blocking with 5% donkey serum at room temperature, primary antibodies were added and incubated overnight at 4 °C. Biotin-conjugated anti-rabbit secondary antibody (1:500; Jackson Labs) was added and developed using Elite Vectastain ABC kit.
Histoscore Calculation.
Histoscores for IHC samples were calculated according to a previously described method whereby the sample is first given a score of 0 to 3 based on staining intensity and then assigned a percent epithelial cell staining from 0 to 100% (62). These two components are then multiplied to give the final histoscore. The histoscores for PARP9, PARP10, and PARP14 between the IFN-marker high and IFN-marker low groups was defined by their MX1 and STAT1 histoscores.
Mice.
NCG mice (NOD-Prkdcem26Cd52Il2rgem26Cd22/NjuCrl) were purchased from Charles River. Mice used for orthotopic implantation were males 6 to 8 wk of age.
Orthotopic implantation and tumor imaging.
Cells were trypsinized and washed twice in PBS. Mice were anesthetized with Isothesia (isoflurane) solution (Henry Schein Animal Health no. 029405). After shaving and swabbing with a sterile alcohol pad followed by povidone-iodide scrub, a 0.5-cm incision was made on the left lateral abdomen. The tail of the pancreas was located. Cells (SUIT2, 3 × 104 cells; Panc 03.27, 2 × 105 cells) in 30 μL fetal bovine serum-free Dulbecco’s modified Eagle medium and Matrigel (1:1) were injected into the tail of the pancreas with an insulin syringe (BD no. 8290–3249-11). The abdominal and skin incisions were closed with 5–0 coated Vicryl sutures (Ethicon J385H). Carporfen (100 μg; Zoetis; Rimadyl no. 141–199) was injected immediately after surgery and on postoperative day 1. To monitor tumor growth, mice were injected intraperitoneally with 50 mg/kg D-Luciferin (BioVision no. 7903) and imaged with the IVIS Lumina III imaging system (PerkinElmer). Data were analyzed using Living Image v4.5 software.
Viability Assay.
CellTiter-Glo analysis was measured using a BioTek microplate luminescence reader as described in SI Appendix, Supplemental Methods.
Invasion Assay.
Cells were seeded in a Transwell plate (Corning BioCoat Matrigel Invasion Chamber; catalog no. 354480). Methods and data acquisition and analysis are described in SI Appendix, Supplemental Methods.
Study Approval.
All reported animal studies were approved by the University of California, Los Angeles, Animal Research Review Board.
Supplementary Material
Acknowledgments
We thank Dr. Christine A. Iacobuzio-Donahue (Memorial Sloan Kettering Cancer Center) for generously providing the PDX model A2.4 and the National Cancer Institute (NCI) for generously providing the PDX model AM1283. A.M.M., S.K, A.M.D, and I.E. were supported under NIH T32 Training Grant T32DK07180 and the HH Lee Grant. This work was supported by a seed grant provided by the Hirshberg Foundation for Pancreatic Cancer Research. A.Y. was supported by the David Geffen School of Medicine Short Term Training Program. T.R.D. is supported by funding from NIH/NCI R01 CA187678 and U01 CA198846, City of Hope/NIH/NCI R01CA174294, and the Jonsson Comprehensive Cancer Center.
Footnotes
Competing interest statement: C.G.R. is a cofounder of Sofie Biosciences and Trethera Corporation. He and the University of California (UC) hold equity in Sofie Biosciences and Trethera Corporation. The intellectual property developed by C.G.R. and licensed by UC to Sofie Biosciences and Trethera Corporation was not used in this study.
This article is a PNAS Direct Submission. D.A.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012469118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Daemen A., et al., Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc. Natl. Acad. Sci. U.S.A. 112, E4410–E4417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum R., Kloog Y., Metabolism addiction in pancreatic cancer. Cell Death Dis. 5, e1065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sousa C. M., Kimmelman A. C., The complex landscape of pancreatic cancer metabolism. Carcinogenesis 35, 1441–1450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halbrook C. J., Lyssiotis C. A., Employing metabolism to improve the diagnosis and treatment of pancreatic cancer. Cancer Cell 31, 5–19 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Ying H., et al., Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auciello F. R., et al., A stromal lysolipid-autotaxin signaling axis promotes pancreatic tumor progression. Cancer Discov. 9, 617–627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Son J., et al., Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biancur D. E., et al., Compensatory metabolic networks in pancreatic cancers upon perturbation of glutamine metabolism. Nat. Commun. 8, 15965 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sousa C. M., et al., Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viale A., et al., Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628–632 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiarugi A., Dölle C., Felici R., Ziegler M., The NAD metabolome–A key determinant of cancer cell biology. Nat. Rev. Cancer 12, 741–752 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Liu L., et al., Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 27, 1067–1080.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verdin E., NAD+ in aging, metabolism, and neurodegeneration. Science 350, 1208–1213 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Fons N. R., et al., PPM1D mutations silence NAPRT gene expression and confer NAMPT inhibitor sensitivity in glioma. Nat. Commun. 10, 3790 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Touat M., et al., DNA repair deficiency sensitizes lung cancer cells to NAD+ biosynthesis blockade. J. Clin. Invest. 128, 1671–1687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao G., et al., Discovery of a highly selective NAMPT inhibitor that demonstrates robust efficacy and improved retinal toxicity with nicotinic acid coadministration. Mol. Cancer Ther. 16, 2677–2688 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Chini C. C., et al., Targeting of NAD metabolism in pancreatic cancer cells: Potential novel therapy for pancreatic tumors. Clin. Cancer Res. 20, 120–130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holen K., Saltz L. B., Hollywood E., Burk K., Hanauske A. R., The pharmacokinetics, toxicities, and biologic effects of FK866, a nicotinamide adenine dinucleotide biosynthesis inhibitor. Invest. New Drugs 26, 45–51 (2008). [DOI] [PubMed] [Google Scholar]
- 20.von Heideman A., Berglund A., Larsson R., Nygren P., Safety and efficacy of NAD depleting cancer drugs: Results of a phase I clinical trial of CHS 828 and overview of published data. Cancer Chemother. Pharmacol. 65, 1165–1172 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Ravaud A., et al., Phase I study and pharmacokinetic of CHS-828, a guanidino-containing compound, administered orally as a single dose every 3 weeks in solid tumours: An ECSG/EORTC study. Eur. J. Cancer 41, 702–707 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Barraud M., et al., A pancreatic ductal adenocarcinoma subpopulation is sensitive to FK866, an inhibitor of NAMPT. Oncotarget 7, 53783–53796 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piacente F., et al., Nicotinic acid phosphoribosyltransferase regulates cancer cell metabolism, susceptibility to NAMPT inhibitors, and DNA repair. Cancer Res. 77, 3857–3869 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Grozio A., et al., CD73 protein as a source of extracellular precursors for sustained NAD+ biosynthesis in FK866-treated tumor cells. J. Biol. Chem. 288, 25938–25949 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fehr A. R., et al., The impact of PARPs and ADP-ribosylation on inflammation and host-pathogen interactions. Genes Dev. 34, 341–359 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Q., et al., Transforming growth factor-β in pancreatic diseases: Mechanisms and therapeutic potential. Pharmacol. Res. 142, 58–69 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Tsai S., et al., A serum-induced transcriptome and serum cytokine signature obtained at diagnosis correlates with the development of early pancreatic ductal adenocarcinoma metastasis. Cancer Epidemiol. Biomarkers Prev. 28, 680–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farajzadeh Valilou S., Keshavarz-Fathi M., Silvestris N., Argentiero A., Rezaei N., The role of inflammatory cytokines and tumor associated macrophages (TAMs) in microenvironment of pancreatic cancer. Cytokine Growth Factor Rev. 39, 46–61 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Batchu R. B., et al., Inhibition of interleukin-10 in the tumor microenvironment can restore mesothelin chimeric antigen receptor T cell activity in pancreatic cancer in vitro. Surgery 163, 627–632 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Li N., Grivennikov S. I., Karin M., The unholy trinity: Inflammation, cytokines, and STAT3 shape the cancer microenvironment. Cancer Cell 19, 429–431 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider W. M., Chevillotte M. D., Rice C. M., Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 32, 513–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carafa V., et al., Sirtuin functions and modulation: From chemistry to the clinic. Clin. Epigenetics 8, 61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chalkiadaki A., Guarente L., The multifaceted functions of sirtuins in cancer. Nat. Rev. Cancer 15, 608–624 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Bai P., Biology of poly(ADP-ribose) polymerases: The factotums of cell maintenance. Mol. Cell 58, 947–958 (2015). [DOI] [PubMed] [Google Scholar]
- 35.van Horssen R., et al., Intracellular NAD(H) levels control motility and invasion of glioma cells. Cell. Mol. Life Sci. 70, 2175–2190 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X., Song E., Turning foes to friends: Targeting cancer-associated fibroblasts. Nat. Rev. Drug Discov. 18, 99–115 (2019). [DOI] [PubMed] [Google Scholar]
- 37.von Ahrens D., Bhagat T. D., Nagrath D., Maitra A., Verma A., The role of stromal cancer-associated fibroblasts in pancreatic cancer. J. Hematol. Oncol. 10, 76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Öhlund D., et al., Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214, 579–596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feig C., et al., The pancreas cancer microenvironment. Clin. Cancer Res. 18, 4266–4276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li T., Chen Z. J., The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 215, 1287–1299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang E. D., Wang C. Y., Single amino acid change in STING leads to constitutive active signaling. PLoS One 10, e0120090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hausmann S., Kong B., Michalski C., Erkan M., Friess H., The role of inflammation in pancreatic cancer. Adv. Exp. Med. Biol. 816, 129–151 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Roshani R., McCarthy F., Hagemann T., Inflammatory cytokines in human pancreatic cancer. Cancer Lett. 345, 157–163 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Bellone G., et al., Cytokine expression profile in human pancreatic carcinoma cells and in surgical specimens: Implications for survival. Cancer Immunol. Immunother. 55, 684–698 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker B. S., Rautela J., Hertzog P. J., Antitumour actions of interferons: Implications for cancer therapy. Nat. Rev. Cancer 16, 131–144 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Samarajiwa S. A., Forster S., Auchettl K., Hertzog P. J., INTERFEROME: The database of interferon regulated genes. Nucleic Acids Res. 37, D852–D857 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benci J. L., et al., Opposing functions of interferon coordinate adaptive and innate immune responses to cancer immune checkpoint blockade. Cell 178, 933–948.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benci J. L., et al., Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell 167, 1540–1554.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mandai M., et al., Dual faces of IFNγ in cancer progression: A role of PD-L1 induction in the determination of pro- and antitumor immunity. Clin. Cancer Res. 22, 2329–2334 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Liu H., et al., Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat. Med. 25, 95–102 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Sistigu A., et al., Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat. Med. 20, 1301–1309 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Feijs K. L., et al., ARTD10 substrate identification on protein microarrays: Regulation of GSK3β by mono-ADP-ribosylation. Cell Commun. Signal. 11, 5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aguiar R. C., Takeyama K., He C., Kreinbrink K., Shipp M. A., B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 280, 33756–33765 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Iwata H., et al., PARP9 and PARP14 cross-regulate macrophage activation via STAT1 ADP-ribosylation. Nat. Commun. 7, 12849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheridan C., Drug developers switch gears to inhibit STING. Nat. Biotechnol. 37, 199–201 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Ramanjulu J. M., et al., Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature 564, 439–443 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Mitchell S. R., et al., Selective targeting of NAMPT by KPT-9274 in acute myeloid leukemia. Blood Adv. 3, 242–255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bachem M. G., et al., Pancreatic carcinoma cells induce fibrosis by stimulating proliferation and matrix synthesis of stellate cells. Gastroenterology 128, 907–921 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Kadera B. E., et al., MicroRNA-21 in pancreatic ductal adenocarcinoma tumor-associated fibroblasts promotes metastasis. PLoS One 8, e71978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Terry N. H., White R. A., Flow cytometry after bromodeoxyuridine labeling to measure S and G2+M phase durations plus doubling times in vitro and in vivo. Nat. Protoc. 1, 859–869 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Elliott I. A., et al., Lysosome inhibition sensitizes pancreatic cancer to replication stress by aspartate depletion. Proc. Natl. Acad. Sci. U.S.A. 116, 6842–6847 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moris M., et al., Plectin-1 as a biomarker of malignant progression in intraductal papillary mucinous neoplasms: A multicenter study. Pancreas 45, 1353–1358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.