Significance
Auxin is a key phytohormone influencing multiple aspects of plant development, including meristem maintenance, primordia initiation, floral organogenesis, and vascular differentiation. Local auxin biosynthesis and polar auxin transport are essential to establish and maintain auxin gradients that ensure proper plant development. Here, we demonstrate that CsDFB1, a member of the plant cystatin superfamily, which was previously implicated in defense responses, plays a critical role in regulating local auxin distribution and thus influences floral organogenesis in cucumber. Genetic and biochemical assays suggest that CsDFB1 affects local auxin distribution by acting as an attenuator that interacts with CsPHB and modulates CsPHB-mediated transcriptional control of CsYUC2 and CsPIN1. Our results shed light on the fine tuning of local auxin distribution in plants.
Keywords: auxin distribution, transcriptional regulation, CsDFB1, CsPHB, floral organogenesis
Abstract
Plant cystatins are cysteine proteinase inhibitors that play key roles in defense responses. In this work, we describe an unexpected role for the cystatin-like protein DEFORMED FLORAL BUD1 (CsDFB1) as a transcriptional regulator of local auxin distribution in cucumber (Cucumis sativus L.). CsDFB1 was strongly expressed in the floral meristems, floral primordia, and vasculature. RNA interference (RNAi)-mediated silencing of CsDFB1 led to a significantly increased number of floral organs and vascular bundles, together with a pronounced accumulation of auxin. Conversely, accompanied by a decrease of auxin, overexpression of CsDFB1 resulted in a dramatic reduction in floral organ number and an obvious defect in vascular patterning, as well as organ fusion. CsDFB1 physically interacted with the cucumber ortholog of PHABULOSA (CsPHB), an HD-ZIP III transcription factor whose transcripts exhibit the same pattern as CsDFB1. Overexpression of CsPHB increased auxin accumulation in shoot tips and induced a floral phenotype similar to that of CsDFB1-RNAi lines. Furthermore, genetic and biochemical analyses revealed that CsDFB1 impairs CsPHB-mediated transcriptional regulation of the auxin biosynthetic gene YUCCA2 and the auxin efflux carrier PIN-FORMED1, and thus plays a pivotal role in auxin distribution. In summary, we propose that the CsDFB1-CsPHB module represents a regulatory pathway for local auxin distribution that governs floral organogenesis and vascular differentiation in cucumber.
The phytohormone auxin plays fundamental roles in various processes involved in plant growth and development, including early embryo patterning (1, 2), meristem development (3), vascular differentiation (4, 5), floral organogenesis and flower fertility (5–7), fruit development (8, 9), and root patterning (2, 10). Auxin is synthesized locally in shoots and roots and then transported in a polar manner to other sites (7). Therefore, local auxin biosynthesis and polar auxin transport are critical for the establishment and maintenance of auxin gradients and are involved in regulating multiple developmental processes in plants (5, 7, 11–15).
Indole-3-acetic acid (IAA), the most common natural auxin in plants, is mainly synthesized from a tryptophan (Trp)-dependent pathway in a two-step reaction: Trp is first converted into indole-3-pyruvate (IPyA) by the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1) family of transaminases, and then IPyA is converted into IAA by the YUCCA (YUC) family of flavin-containing monooxygenases (7, 16–19). In its protonated form, IAA can enter the cell through passive diffusion or through active uptake by influx carriers of the AUXIN1/LIKE-AUX1 (AUX1/LAX) family (20). Auxin efflux is mainly mediated by PIN-FORMED (PIN) efflux carriers that regulate auxin distribution in plant tissues (21). In the past two decades, the regulatory mechanisms behind auxin have been dissected in great detail (17, 22). Several critical transcription factors contribute to the regulation of local auxin distribution by modulating the expression levels and patterns of YUC and PIN genes. For example, the HD-ZIP III transcription factor REVOLUTA (REV) is the direct upstream activator of YUC5 and TAA1 transcription (23). In agreement with this result, proper REV function is required for polar auxin transport in the shoot (24). Likewise, the MADS-box transcription factor AGAMOUS (AG) works together with the YABBY-type transcription factor CRABS CLAW (CRC) to regulate YUC4 expression (25). Similarly, SUPERMAN (SUP) plays a role in boundary formation of floral organs by regulating the expression of YUC1, YUC4, PIN3, and PIN4 (26). The cucumber (Cucumis sativus L.) MADS-box transcription factor FRUITFULL1 (CsFUL1) regulates fruit length by repressing CsPIN1 and CsPIN7 transcription (8). Therefore, transcriptional regulation of YUCs and PINs plays an important role in plant organ development.
The plant cystatin protein superfamily contains a distinctive cystatin-like domain ([LVI]-[AGT]-[RKE]-[FY]-[AS]-[VI]-x-[EDQV]-[HYFQ]-N) in its N terminus (27). Plant cystatins were originally identified in rice seeds (28) and have since been characterized in monocots and dicots, including maize (Zea mays) (29), barley (Hordeum vulgare) (30), Arabidopsis (Arabidopsis thaliana) (31, 32), tobacco (Nicotiana tabacum) (33), and soybean (Glycine max) (34). They may play roles in defense responses to biotic and abiotic stresses (29–31, 35, 36), programmed cell death (32, 33), and regulation of protein turnover during seed development and germination (37). For instance, Arabidopsis CYSTATIN1 (CYS1) was preferentially expressed in the vasculature of all organs and in response to abiotic stresses such as high temperature and wounding (31). The silencing of tobacco CYS by RNA interference (RNAi) induced precocious cell death in the basal cell of the embryo, resulting in embryonic arrest and seed abortion (33). In pumpkin (Cucurbita maxima Duch.), PHLOEM PROTEIN1 (CmPP1) contains four copies of the cystatin-like domain, and the CmPP1 mRNA was localized in phloem companion cells at early stages of vascular differentiation (35). To date, plant cystatins have been generally considered as defense proteins and regulators of protein turnover, with little information suggesting that they might have additional roles during plant development.
Here, we report on the unexpected finding that the cucumber cystatin-like protein DEFORMED FLORAL BUD1 (CsDFB1) functions as a transcriptional regulator to attenuate auxin accumulation, and thus is indispensable for proper floral organogenesis and vascular differentiation. CsDFB1 was highly expressed not only in the floral meristems but also in the vascular tissues, and both overexpression and silencing of CsDFB1 caused defects in floral organogenesis and vascular differentiation. Further exploration showed that CsDFB1 interacts with the HD-ZIP III transcription factor PHABULOSA (CsPHB) and impairs CsPHB-mediated regulation of CsYUC2 and CsPIN1 expression. Therefore, the CsDFB1-CsPHB module controls local auxin distribution and auxin-dependent organ development. These results reveal a pathway that controls floral organ development by maintaining the delicate auxin homeostasis in cucumber.
Results
Csa7G067350 Is Highly Expressed in the Floral Meristems, Floral Primordia, and Vasculature in Cucumber.
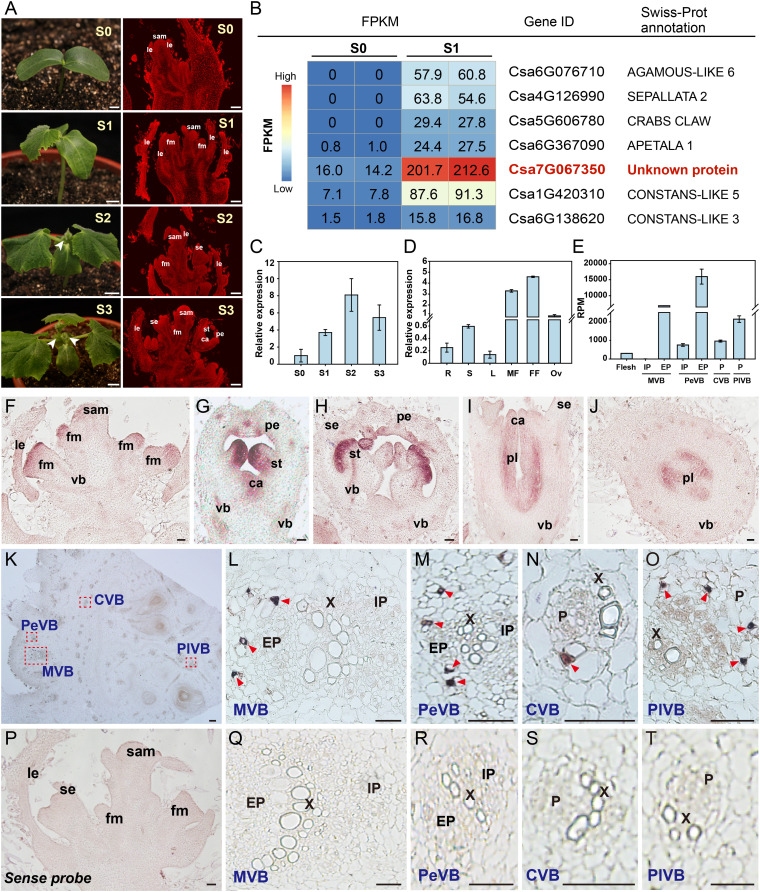
Floral organogenesis is an important developmental process that directly affects fruit yield and quality in cucumber. After a very short juvenile stage, the indeterminate growth habit of cucumber plants is accompanied by the parallel production of both vegetative and reproductive structures (38). We observed floral buds in the axil of leaves in seedlings at the two-leaf (S2) and three-leaf (S3) stages (Fig. 1 A, Left). An anatomical analysis of shoot tips showed that floral meristems initiated at the one-leaf stage (S1) and subsequently formed floral primordia at and after S2 (Fig. 1 A, Right). To generate a floral meristem-specific gene-expression atlas, we collected the shoot tips of seedlings at the S0 (cotyledons only) and S1 stages for transcriptome deep-sequencing (RNA-seq) analysis. As expected, transcripts for several known floral-specific genes such as AGAMOUS-LIKE6 (AGL6), SEPALLATA2 (SEP2), CRABS CLAW (CRC), APETALA1 (AP1), CONSTANS-LIKE3 (COL3), and COL5 were abundant in S1 shoot tips but barely detectable in S0 shoot tips (Fig. 1B and Dataset S1) (39, 40). Notably, we noticed that the Csa7G067350 gene, encoding an unknown protein, exhibited high expression in S1 shoot tips as well (Fig. 1B). RT-qPCR indicated that Csa7G067350 transcript levels gradually increased from the S0 to the S2 in shoot tips, and then decreased at the S3 (Fig. 1C). We then determined the expression levels of Csa7G067350 in various organs and tissues: Csa7G067350 expression was relatively high in reproductive organs, including male flowers, female flowers, and ovaries (Fig. 1D). Notably, the vascular bundles (VBs) in the ovary/fruit of cucumber are arranged in four systems, two collateral and two bicollateral (with internal and external phloem located on both sides of the xylem) (41). Accordingly, based on our previous transcriptome of fruit phloem during early development (41), Csa7G067350 was found to show a significantly higher expression in the phloem of those four VB systems when compared to the fruit flesh (Fig. 1E).
Fig. 1.
Expression profile of Csa7G067350 in wild-type cucumber. (A) Morphology of cucumber plants from S0 to S3 (Left) and corresponding longitudinal paraffin sections of shoot tips (Right). White arrowheads indicate floral buds. (Scale bars, 1 cm at Left and 100 μm at Right.) (B) Heatmap representation of relative expression values of several differentially expressed genes between S0 and S1 shoot tips. (C and D) Expression analysis of Csa7G067350 by RT-qPCR in shoot tips from S0 to S3 (C) and different organs (D). Error bars indicate SD of three biological replicates from different plants. (E) Expression of Csa7G067350 by laser capture microdissection (LCM)-derived RNA-seq analysis in phloem systems of cucumber ovary/fruit during early development. For detailed experimental methods, please refer to Sui et al. (41). (F–T) In situ hybridization detection of Csa7G067350 transcripts in the shoot tip (F–J and P) and ovary/fruit (K–O and Q–T). (F) Floral meristem. (G) Floral bud at early stage. (H) Male floral bud. (I and J) Female floral bud. (K) Cross-section of the ovary/fruit. (L–O) Close-up views of different types of vascular bundles, indicated by the red dashed boxes in K. (P–T) Negative control for in situ hybridization, using Csa7G067350 sense probe in the shoot tip (P) and ovary/fruit (Q–T). Purple staining in F–J and red triangles in L–O indicate positive signal. (Scale bar, 50 μm.) Abbreviations: S0, cotyledon stage; S1, one-leaf stage; S2, two-leaf stage; S3, three-leaf stage; FPKM, fragments per kilobase of transcript per million mapped reads; R, root; S, stem; L, leaf; MF, male flower; FF, female flower; Ov, ovary; sam, shoot apical meristem; le, leaf or leaf primordia; fm, floral meristem; vb, vascular bundle; se, sepal primordia; pe, petal primordia; st, stamen primordia; ca, carpel primordia; pl, placenta; MVB, PeVB, CVB, and PlVB, main, peripheral, carpel, and placental vascular bundle, respectively; EP, external phloem; X, xylem; IP, internal phloem; and P, phloem.
Next, we conducted in situ hybridization to validate the spatial expression pattern of Csa7G067350 in shoot tips and ovaries/fruits (Fig. 1 F–T). In agreement with the RNA-seq and RT-qPCR results above, we observed strong signals in floral meristems, floral primordia, and the vasculature (Fig. 1 F–J). Specifically, Csa7G067350 was expressed throughout the floral meristems at the early stage (Fig. 1F), later accumulating in petal, stamen, and carpel primordia within floral buds (Fig. 1 G–J). In addition, Csa7G067350 transcripts specifically accumulated in the phloem region of the main (MVB), peripheral (PeVB), carpel (CVB), and placenta (PlVB) vascular bundles in the ovary/fruit (Fig. 1 K–O). These results suggested that Csa7G067350 might function in floral organogenesis and vascular development.
Phylogenetic analysis indicated that the Csa7G067350 protein belongs to group D of the plant cystatins, which is a new group consisting of proteins from Cucurbitaceae (SI Appendix, Fig. S1 A and B) (35, 42). Csa7G067350 shared high similarity with PHLOEM PROTEIN1 (PP1), previously identified in pumpkin and cucumber (SI Appendix, Fig. S1) (35, 42). Interestingly, as for several other group D members such as cucumber PP1 (CsPP1) and pumpkin PP1 (CmPP1), the predicted protein sequence of Csa7G067350 lacked the highly conserved QxVxG motif normally associated with cysteine proteinase inhibitor activity (SI Appendix, Fig. S1C) (30, 35, 36). Consistent with the absence of this conserved motif, in vitro inhibitory assays indicated that the protein encoded by Csa7G067350 did not inhibit the activity of the cysteine proteinase papain (SI Appendix, Fig. S2). Further analysis of conserved motifs established that Csa7G067350 contained only one copy of the [LVI]-[AGT]-[RKE]-[FY]-[AS]-[VI]-x-[EDQV]-[HYFQ]-N motif, while CsPP1 presented two copies of this motif 1 and CmPP1 had eight copies (35). As compared to CsPP1 and CmPP1, Csa7G067350 contained conserved motifs 6, 7, and 9, but lacked motif 2 (SI Appendix, Fig. S1 B–D).
We then tested the subcellular localization of Csa7G067350 by transiently expressing Csa7G067350 fused to the Green Fluorescent Protein (GFP) gene and placed under the control of the Super promoter, in both cucumber protoplasts and Nicotiana benthamiana leaves. We detected green fluorescence in the cytoplasm and the nucleus (SI Appendix, Fig. S3), which is similar to the localization of the homologous barley (H. vulgare) proteins CYSTEINE PROTEASE INHIBITOR1 (HvCPI1) and HvCPI4 (30). We repeated this experiment with the endogenous Csa7G067350 promoter driving the GFP fusion construct and observed a similar pattern, validating the localization of the protein (SI Appendix, Fig. S3).
CsDFB1 Governs Floral Organ Development and Vascular Patterning.
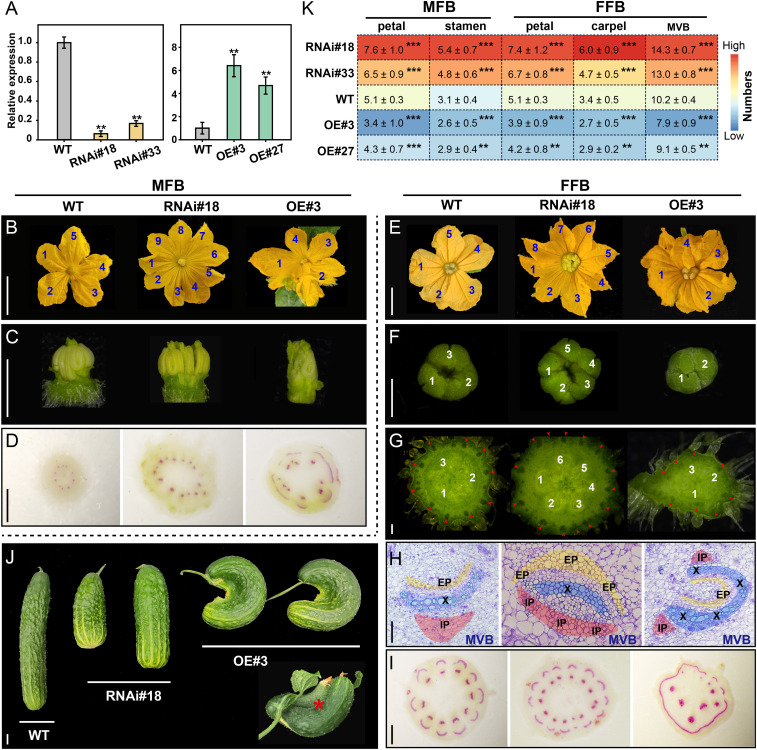
To investigate the molecular function of Csa7G067350 in cucumber, we generated transgenic cucumber lines with higher (overexpression) and lower (RNAi) levels of Csa7G067350. We obtained more than 40 primary (T0) transformants in total and selected two independent RNAi T2 lines (RNAi#18 and RNAi#33) and two overexpression T2 lines (OE#3 and OE#27) based on their Csa7G067350 expression levels for further study. RT-qPCR confirmed that Csa7G067350 transcript levels were dramatically reduced in the floral buds of RNAi lines, but increased in OE lines, as compared to wild-type (WT) plants (Fig. 2A).
Fig. 2.
Phenotypic analysis of CsDFB1 transgenic plants. (A) RT-qPCR analysis of CsDFB1 (Csa7G067350) expression in floral buds from WT plants, RNA interference lines (RNAi#18 and RNAi#33), and overexpression lines (OE#3 and OE#27). Error bars indicate SD of three biological replicates from different plants. (B–I) Detailed phenotypes of MFB (B–D) and FFB (E–I) from transgenic lines (RNAi#18 and OE#3) and WT plants as control. (B, C, E, and F) Morphological characteristics of petals from MFBs (B) and FFBs (E), stamens (C), and stigma (F). (G) Hand cross-section of ovary/fruit. (H) Paraffin cross-sections of MVBs in the ovary/fruit. Yellow, blue, and red regions indicate the EP, xylem, and IP, respectively. (D and I) Hand cross-sections of pedicels (D) and peduncles (I) stained with phloroglucinol. Red regions indicate the presence of lignified xylem cells. Blue and white numbers indicate the number of petals (B and E), stigma (F), and carpels (G), respectively; red arrows in G indicate the position of MVBs. (Scale bars, 1 cm in B and E; 1 mm in D, G, and I; 500 μm in C and F; and 100 μm in H.) (J) Morphology of cucumber fruits after 30 d of development. Red asterisk indicates organ (fruit) fusion in OE lines. (Scale bar, 2 cm.) (K) Summary of floral organ number and MVB number in the WT and transgenic plants. Error bars indicate SD of 50 flowers from around 10 individual plants. Significance analysis was conducted with the two-tailed Student’s t test (***P < 0.001; **P < 0.01). Abbreviations: DEFORMED FLORAL BUD1 (CsDFB1); MFB, male floral bud; FFB, female floral bud; EP, external phloem; X, xylem; IP, internal phloem; and MVB, main vascular bundles.
Cucumber plants bear both male and female flower buds (designated MFB and FFB, respectively). Importantly, both MFBs (Fig. 2B–D) and FFBs (Fig. 2E–I) displayed a deformed phenotype relative to wild-type flowers in RNAi and OE transgenic lines, which inspired the Csa7G067350 gene name DEFORMED FLORAL BUD1 (CsDFB1). Silencing of CsDFB1 by RNAi produced more floral organs per flower compared to WT plants (Fig. 2 B, C, and E–G), with an increase in organ number of about 20 to 80% for petals, stamens, and carpels (Fig. 2K). By contrast, OE lines had fewer floral organs than WT plants (Fig. 2 B, C, E, G, and K). Furthermore, overexpression of CsDFB1 could lead to the phenotype of organ fusion between whorls of MFB and FFB, including fused MFBs, fused FFBs, and even fused FFBs with MFB (SI Appendix, Fig. S4 A–I). This fusion defect was observed in the range of 25 to 70% in the MFBs of CsDFB1-OE lines, and 3 to 20% in the FFBs of the OE plants (SI Appendix, Fig. S4 J and K).
The determination and patterning of VBs in the floral buds of CsDFB1 transgenic lines was similarly disrupted compared to that in WT plants (Fig. 2 D and G–I). The number of pedicel/peduncle VBs (Fig. 2 D and I) and ovary/fruit MVBs (Fig. 2 G and K) was increased in RNAi lines but decreased in OE lines. Preparation of hand-cut sections followed by phloroglucinol staining confirmed the disruption of vascular distribution in the transgenic lines, especially in OE plants (Fig. 2 D and I). Moreover, in RNAi lines, we observed an abnormal pattern of increased differentiation of the phloem at the expense of the xylem in the MVB region of ovaries/fruits; conversely, OE lines exhibited xylem tissue partially surrounding the phloem (Fig. 2H). Accordingly, after 30 d of development, the fruits produced by RNAi lines were shorter and thicker than those of WT plants, whereas fruits from OE lines took on a hook-like shape, sometimes even bent and two-fused shape (Fig. 2J). These results demonstrated that CsDFB1 may participate in floral organogenesis and vascular differentiation in cucumber.
CsDFB1 Regulates Local Auxin Distribution.
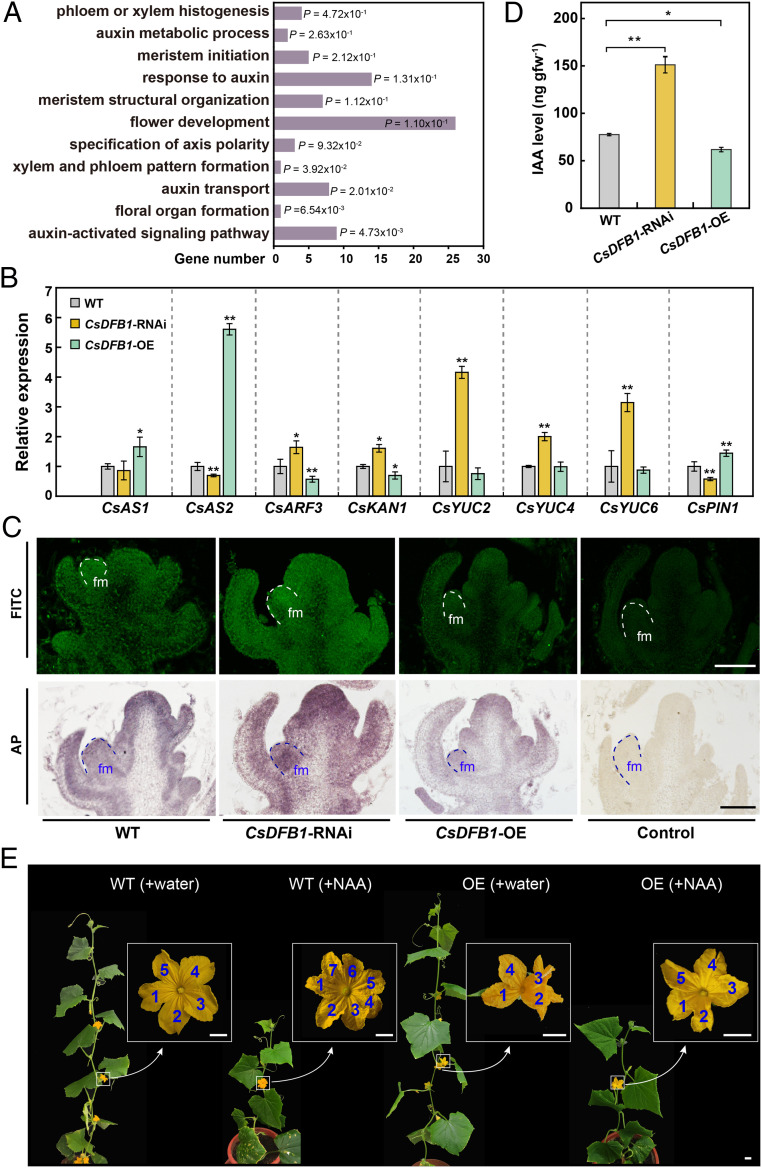
To explore the function of CsDFB1 during floral organ development and vascular formation, we conducted an RNA-seq analysis to access the complement of differentially expressed genes (DEGs) between WT and CsDFB1-RNAi transgenic plants in shoot tips. We identified 320 down-regulated and 366 up-regulated genes in the RNAi lines relative to WT plants (Dataset S2), using a false discovery rate (FDR) of 0.05 and fold change (FC) of at least 1.5 as selection criteria. Gene Ontology (GO) term enrichment analysis revealed that a number of biological processes related to floral organ and VB development were significantly enriched, such as phloem or xylem histogenesis/pattern formation, meristem structural organization, specification of axis polarity, and floral organ formation (Fig. 3A), providing further support to our earlier observations. In agreement with the GO results, we determined that the expression of several polarity- or boundary-related genes, such as ASYMMETRIC LEAVES1 (AS1), AS2, AUXIN RESPONSE TRANSCRIPTION FACTOR3 (ARF3), and KANADI1 (KAN1), was markedly altered in RNAi and OE lines, in opposite directions (Fig. 3B) (43, 44).
Fig. 3.
CsDFB1 transgenic plants have disrupted auxin distribution. (A) Significantly enriched GO pathways in CsDFB1-RNAi plants based on RNA-seq analysis. (B) RT-qPCR analysis of genes related to polarity or meristem development (e.g., CsAS1, CsAS2, CsARF3, and CsKAN1), auxin biosynthesis (e.g., CsYUC2/4/6), and auxin transport (e.g., CsPIN1) in shoot tips of CsDFB1 transgenic plants and WT plants. Error bars indicate SD of three biological replicates from different plants. Gene IDs: ASYMMETRIC LEAVES1 (CsAS1), Csa3G264750; ASYMMETRIC LEAVES2 (CsAS2), Csa2G070920; AUXIN RESPONSE FACTOR3 (CsARF3), Csa6G518210; KANADI1 (CsKAN1), Csa3G194380; YUCCA2 (CsYUC2), Csa1G242600; YUCCA4 (CsYUC4), Csa2G379350; YUCCA6 (CsYUC6), Csa2G375750; and PIN-FORMED1 (CsPIN1), Csa4G430820. (C) Immunolocalization of IAA in shoot tips of CsDFB1 transgenic plants and WT plants using fluorescein isothiocyanate (FITC)-conjugated (Upper) and alkaline phosphatase (AP)-conjugated secondary antibody (Lower). Green fluorescence and purple staining indicate the signals. (Scale bar, 200 μm.) (D) IAA level in the shoot tips of CsDFB1 transgenic plants and WT plants. Error bars indicate SD of three biological replicates from different plants. (E) Representative photograph of CsDFB1-OE plants and WT plants with or without treatment with 50 μM NAA. Blue numbers inside white boxes indicate the number of petals. (Scale bar, 1 cm.) Significance analysis was conducted with the two-tailed Student’s t test (**P < 0.01; *P < 0.05). Abbreviations: gfw−1, per gram fresh weight; fm, floral meristem; and NAA, 1-naphthylacetic acid.
Most importantly, we also noted strong enrichment of the GO categories auxin metabolic process, response to auxin, auxin transport, and auxin-activated signaling pathway (Fig. 3A). As auxin plays a crucial role in both floral organ development and vascular differentiation (5), we hypothesized that CsDFB1 might be involved in auxin homeostasis. Indeed, several YUCCA auxin biosynthetic genes (CsYUC2, CsYUC4, and CsYUC6) were expressed at levels two- to fourfold higher in the RNAi lines relative to WT, with slightly lower levels in the OE lines (Fig. 3B). Interestingly, CsDFB1-RNAi lines often developed three cotyledons and formed fasciated shoots, phenotypes that were reminiscent of the auxin transport-deficient transposon insertional mutant pin1::En134 in Arabidopsis (45) (SI Appendix, Fig. S5). We thus turned to auxin efflux carrier genes in cucumber: we discovered that the expression of the cucumber PIN1 ortholog CsPIN1 was reduced in the RNAi lines and increased in the OE plants (Fig. 3B).
Consistent with these results, immunolocalization of IAA on shoot tips demonstrated a strong auxin accumulation in the floral meristems (Fig. 3C) and floral primordia (SI Appendix, Fig. S6) of RNAi lines relative to WT plants. Conversely, OE shoot tips exhibited a weaker IAA signal. These results were corroborated by ultra-performance liquid chromatography (UPLC) followed by tandem mass spectrometry (MS/MS) measurements of IAA levels in shoot tips (Fig. 3D). Moreover, exogenous application of the synthetic auxin 1-naphthaleneacetic acid (NAA) increased the number of petals in WT plants and restored normal floral organ numbers and morphology in CsDFB1-OE plants, including a suppression of floral organ fusion (Fig. 3E and SI Appendix, Fig. S7). These results suggested that CsDFB1 most likely affected floral organogenesis and vascular differentiation via regulating auxin biosynthesis and transport-dependent local auxin distribution.
CsDFB1 Interacts with CsPHB to Inhibit CsPHB-Mediated Activation of CsYUC2 and Repression of CsPIN1.
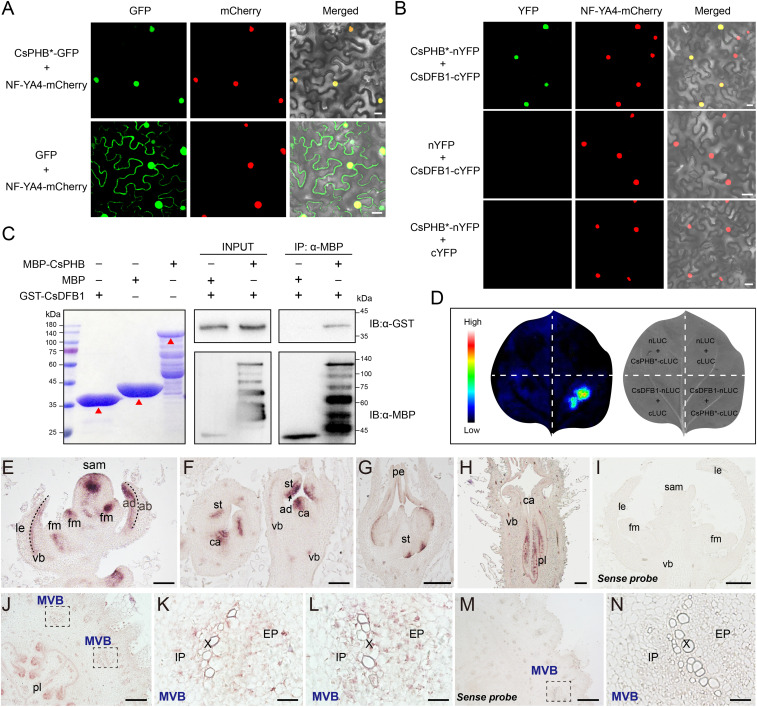
Since we did not identify known DNA-binding domains in CsDFB1, we speculated that the protein might function as a coregulator with other transcription factors. We thus tested the potential for interaction between CsDFB1 and major regulators of auxin-dependent floral development (8, 23–26). Bimolecular fluorescence complementation (BiFC) assays showed that CsDFB1 interacts with the HD-ZIP III transcription factor PHABULOSA (CsPHB), with the YABBY-type transcription factor CsYAB2, and with itself. However, CsDFB1 did not interact with the YABBY-type transcription factor CRABS CLAW (CsCRC) or the C2H2-type zinc finger transcription factor SUPERMAN (CsSUP) (SI Appendix, Fig. S8). Based on the phenotypic similarity (in terms of petal number) between the cucumber CsPHB gain-of-function mutants cul-1 and cul-2 (46) and CsDFB1-RNAi plants, we selected CsPHB (Csa6G525430) for further characterization.
Previous reports had shown that the PHB mRNA is highly unstable due to transcript cleavage mediated by microRNA165 (miR165) and miR166 (46, 47). Therefore, to allow PHB mRNA and PHB protein to accumulate, we generated a miRNA-resistant version of CsPHB (designated CsPHB*) by introducing a synonymous mutation in the miRNA-binding site to prevent miRNA cleavage while retaining the wild-type protein sequence. CsPHB* localized to the nucleus, as determined by transient infiltration of a CsPHB*-GFP construct in N. benthamiana leaves (Fig. 4A). BiFC assays combined with firefly luciferase complementation imaging (LCI) and pull-down assays confirmed that CsDFB1 interacted with CsPHB in vivo and in vitro (Fig. 4 B–D). Furthermore, in situ hybridization localized CsPHB mRNA to the adaxial region of the floral meristems and floral primordia, as well as in the vascular system (Fig. 4 E–N); the CsPHB mRNA accumulation pattern therefore overlapped with that of CsDFB1 (Fig. 1 F–O).
Fig. 4.
CsDFB1 interacts with CsPHB in the floral meristems, floral primordia, and vasculature. (A) Subcellular localization of CsPHB in N. benthamiana leaves. CsPHB*-GFP and GFP were coinfiltrated with the mCherry-labeled nuclear marker NF-YA4-mCherry. miRNA-resistant CsPHB is designated CsPHB*. (Scale bar, 20 μm.) (B) Interaction of CsDFB1 and CsPHB by bimolecular fluorescence complementation assays in N. benthamiana leaves. CsPHB*-nYFP and CsDFB1-cYFP were coinfiltrated with the mCherry-labeled nuclear marker NF-YA4-mCherry. CsDFB1-cYFP and nYFP, or CsPHB*-nYFP and cYFP, were used as negative controls. (Scale bar, 20 μm.) (C) Pull-down assays confirming that CsDFB1 interacts with CsPHB in vitro. Immobilized MBP and MBP-CsPHB were used to pull down GST-CsDFB1, and detected by immunoblot with an anti-GST antibody. The red triangle indicates the respective target protein. (D) Interaction between CsDFB1 and CsPHB by firefly luciferase complementation imaging assays in N. benthamiana leaves. CsPHB*-cLUC and CsDFB1-nLUC were coinfiltrated into N. benthamiana leaves. CsPHB*-cLUC and nLUC, cLUC and CsDFB1-nLUC, or nLUC and cLUC, were used as negative controls. The pseudocolor bar shows luminescence intensity. (E–N) In situ hybridization detection of CsPHB transcripts in the shoot tip (E–I) and ovary/fruit (J–N) from wild-type cucumber. (E) Floral meristem. (F) Floral bud at early stage. (G) Male floral bud. (H) Female floral bud. (J) Cross-section of ovary/fruit. (K and L) Magnified images of MVB indicated by the black dashed boxes in J. (N) Magnified image of MVB indicated by the black dashed box in M. Negative control of the CsPHB sense probe in the shoot tip (I) and ovary (M and N). Purple staining indicates the signal. (Scale bars, 200 μm in E–J and M; 50 μm in K, L, and N.) Abbreviations: GST, glutathione S-transferase; MBP, maltose binding protein; ab, abaxial; ad, adaxial; sam, shoot apical meristem; le, leaf or leaf primordia; fm, floral meristem; vb, vascular bundle; pe, petal primordia; st, stamen primordia; ca, carpel primordia; pl, placenta; MVB, main vascular bundle; EP, external phloem; X, xylem; and IP, internal phloem.
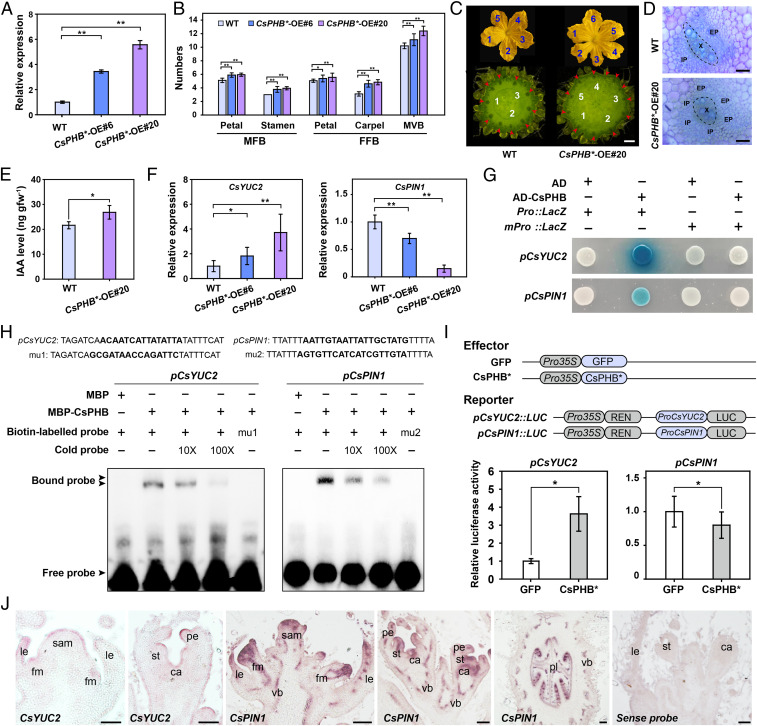
Next, we generated transgenic cucumber lines overexpressing CsPHB* (Fig. 5A); these CsPHB*-OE lines phenotypically mimicked the gain-of-function mutants cul-1 and cul-2 (46). Indeed, CsPHB*-OE lines displayed more floral organs and MVBs (Fig. 5 B and C), in addition to a defective vascular pattern (Fig. 5D), phenotypes that are similar to those of CsDFB1-RNAi plants (Fig. 2). IAA levels were slightly elevated in CsPHB*-OE lines (Fig. 5E). Consistent with this result, CsYUC2 expression rose two- to fourfold, depending on the transgenic line, while CsPIN1 transcript levels decreased (Fig. 5F). Therefore, we hypothesized that CsDFB1 and CsPHB might regulate floral organogenesis and vascular differentiation by controlling auxin distribution in cucumber.
Fig. 5.
CsPHB regulates floral development and vascular patterning by controlling CsYUC2 and CsPIN1 expression. (A) RT-qPCR analysis of CsPHB expression in the floral buds of WT and CsPHB overexpression lines (CsPHB*-OE#6 and CsPHB*-OE#20). Error bars indicate SD of three biological replicates from different plants. (B) Summary of floral organ and MVB numbers in WT and CsPHB overexpression plants. Error bars indicate SD of 20 flowers from around 10 individual plants. (C) Phenotype of male flower and ovary/fruit from WT and the CsPHB*-OE#20 line. Blue and white numbers indicate the number of petals and carpels, respectively; red arrows show the positions of MVBs. (Scale bar, 1 cm.) (D) Hand sections of ovary MVB stained with toluidine blue. (Scale bar, 100 μm.) (E) IAA levels in shoot tips of CsPHB*-OE#20 and WT plants. Error bars indicate SD of three biological replicates. (F) RT-qPCR analysis of CsYUC2 and CsPIN1 expression in shoot tips of CsPHB*-OE plants and WT plants. Error bars indicate SD of three biological replicates. (G) Yeast one-hybrid assays testing the binding of CsPHB to the CsYUC2 and CsPIN1 promoters. (H) EMSAs showing that CsPHB binds to the CsYUC2 and CsPIN1 promoters. Competition for binding was performed using 10× and 100× unlabeled probes; mutant probe (mu1 and mu2), and MBP was used as negative controls. (I) Transcriptional activity of CsPHB* on the CsYUC2 (Left) and CsPIN1 (Right) promoters in cucumber protoplasts. Data are normalized to samples expressing GFP. Error bars indicate SD of three biological replicates. (J) In situ hybridization detection of CsYUC2 and CsPIN1 transcripts in shoot tips of wild-type cucumber. (Scale bar, 100 μm.) Significance analysis was conducted with the two-tailed Student’s t test (**P < 0.01; *P < 0.05). Abbreviations: X, xylem; IP, internal phloem; EP, external phloem; gfw−1, per gram fresh weight; MBP, maltose binding protein; sam, shoot apical meristem; le, leaf or leaf primordia; fm, floral meristem; vb, vascular bundle; pe, petal primordia; st, stamen primordia; ca, carpel primordia; pl, placenta.
Next, we attempted to decipher how CsDFB1 influences CsPHB-mediated transcriptional regulation in cucumber. Sequence analysis identified typical binding sites for HD-ZIP III transcription factors [GTAAT(G/C)ATTAC] in the CsYUC2 and CsPIN1 promoters (SI Appendix, Fig. S9) (23, 48). Accordingly, yeast one-hybrid assays indicated that CsPHB binds to these promoters (Fig. 5G). Electrophoretic mobility shift assays (EMSAs) confirmed the physical interaction of CsPHB and the CsYUC2 and CsPIN1 promoters (Fig. 5H). This binding was specific, as CsPHB failed to bind to mutant probes for the CsYUC2 and CsPIN1 promoters (Fig. 5H). To validate this interaction in vivo, we next performed luciferase (LUC) transactivation assays in cucumber protoplasts. LUC activity derived from the CsYUC2 promoter was significantly enhanced upon cotransformation with CsPHB*; by contrast, LUC activity from the CsPIN1 promoter was repressed upon cotransformation with CsPHB* (Fig. 5I). We also established that the accumulation patterns of CsYUC2 and CsPIN1 transcripts (Fig. 5J) coincided with those of CsPHB (Fig. 4 E–I) based on in situ hybridization assays, as would be expected from genes involved in a regulatory network. These results indicated that CsPHB is a direct activator of CsYUC2 and a repressor of CsPIN1 transcription in cucumber.
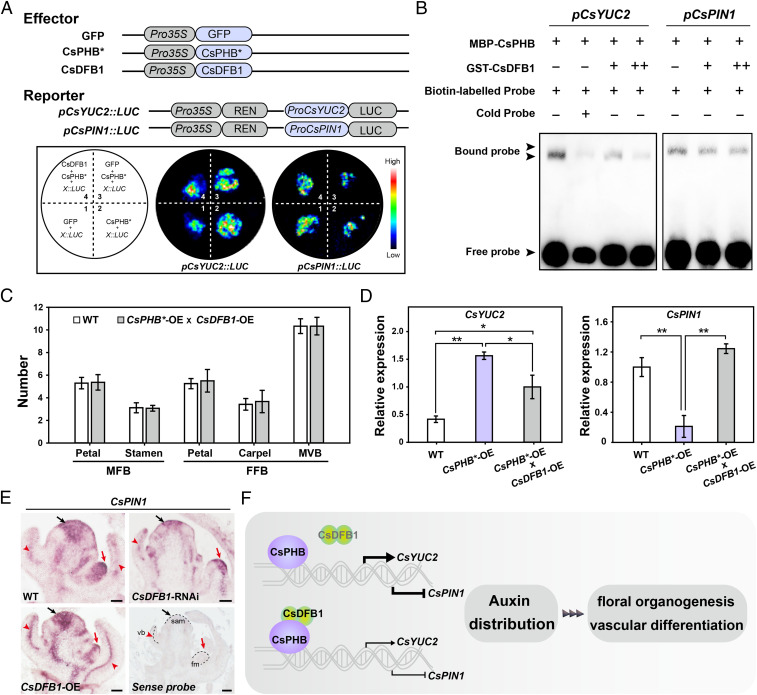
Building upon the LUC transactivation assays and EMSAs described above in which we tested CsPHB alone, we next determined what influence, if any, CsDFB1 might have on CsPHB binding to the CsYUC2 and CsPIN1 promoters (Fig. 6 A and B). Accordingly, we transiently coinfiltrated N. benthamiana leaves with the LUC reporters and the CsPHB and CsDFB1 effectors. CsDFB1 impaired the CsPHB-dependent activation of CsYUC2 and the repression of CsPIN1 (Fig. 6A) by preventing CsPHB binding to its cognate DNA-binding site (Fig. 6B). We also generated CsPHB*-OE (♀) × CsDFB1-OE (♂) hybrid plants. Notably, overexpression of CsDFB1 rescued the floral organ and vascular bundle defects in CsPHB*-OE lines, as well as the abnormal expression of CsYUC2 and CsPIN1 (Fig. 6 C and D). Finally, we tested the accumulation pattern of CsPIN1 transcripts in WT, CsDFB1-RNAi, and CsDFB1-OE transgenic plants (Fig. 6E). Compared to WT plants, CsPIN1 transcript levels were greatly reduced in the shoot apical meristems, floral meristems, and VBs of CsDFB1-RNAi plants, but higher in CsDFB1-OE plants, especially in the vasculature (Fig. 6E).
Fig. 6.
CsDFB1 attenuates CsPHB-mediated regulation of CsYUC2 and CsPIN1. (A) The pCsYUC2::LUC and pCsPIN1::LUC reporter constructs were transiently infiltrated in N. benthamiana leaves together with the CsPHB* or CsPHB* and CsDFB1 effectors. The pseudocolor bar shows luminescence intensity. GFP effector was used as negative control. “X” in the diagram represents pCsYUC2 or pCsPIN1. (B) EMSA showing that CsDFB1 attenuates the binding of CsPHB to the CsYUC2 and CsPIN1 promoters. A gradient concentration of GST-CsDFB1 was applied (+, 1.0 μg; ++, 2.0 μg). (C) Summary of floral organ and vascular numbers in WT and F1 hybrid plants derived from the cross between CsPHB*-OE#20 line (♀) × CsDFB1-OE#3 line (♂). Data represent means ± SD (n = 10). (D) RT-qPCR analysis of CsYUC2 and CsPIN1 expression in shoot tips of CsPHB*-OE plants, F1 hybrid plants between CsPHB*-OE#20 line and CsDFB1-OE#3 line, and WT plants. Error bars indicate SD of three biological replicates from different plants. (E) In situ hybridization detection of CsPIN1 transcripts in shoot tips of the WT and CsDFB1 transgenic plants. Black arrows indicate shoot apical meristem, red arrows indicate floral meristem, and red arrowheads indicate vascular bundle. (Scale bar, 100 μm.) (F) A proposed working model for CsDFB1. CsDFB1 can dimerize and modulates auxin distribution through interacting with CsPHB to attenuate its DNA-binding activity toward the promoters of the auxin biosynthetic gene CsYUC2 and the auxin efflux carrier CsPIN1, and thus regulates floral organogenesis and vascular differentiation in cucumber. Significance analysis was conducted with the two-tailed Student’s t test (**P < 0.01; *P < 0.05).
Taken together, our findings reveal that CsDFB1 functions as a repressor of CsPHB to attenuate CsPHB-mediated activation of CsYUC2 and repression of CsPIN1 expression, and thus participates in controlling local auxin distribution and plant development in cucumber (Fig. 6F).
Discussion
Local auxin biosynthesis and polar auxin transport are essential to establishing local auxin gradients throughout plant development (7, 10). Local auxin biosynthesis is controlled by various environmental (high temperature, shading, and sugars) as well as endogenous signals (phytohormones and developmental processes) (25, 26, 49–53). Transcriptional regulation of YUC genes, encoding the rate-limiting step of auxin biosynthesis (5), is crucial to controlling local auxin biosynthesis and thus is indispensable for proper plant development. In Arabidopsis, the transcription factor PLETHORA (PLT) controls spiral phyllotaxis by ensuring sufficient production of auxin at the shoot apex, probably by regulating YUC1 and YUC4 expression (15). The master regulator of flower determinacy AGAMOUS (AG) and the YABBY-type transcription factor CRABS CLAW (CRC) also regulate auxin biosynthesis by physically binding to the YUC4 promoter (25). However, auxin levels are not by themselves sufficient to initiate auxin-dependent biological processes: establishing an accurate auxin gradient is also essential. PIN proteins are major auxin efflux carriers that are required to establish auxin gradients and are associated with various important developmental events (10). Although many studies have focused on PIN regulation at the posttranscriptional level, transcriptional regulation of PIN is also vital for plant development. For instance, the MADS-box transcription factor AGL14/XAANTAL2 (XAL2) regulates auxin transport and auxin gradient in roots by acting on PIN transcription in Arabidopsis (54). Similarly, cucumber CsFUL1 functions as a transcriptional repressor that inhibits CsPIN1 and CsPIN7 expression and hence regulates fruit length (8).
Here, we discovered a transcription regulatory module, consisting of CsDFB1 and CsPHB, acting in local auxin biosynthesis and transport in cucumber. CsDFB1 encodes a cystatin-like protein from the plant cystatin superfamily (also known as the phytocystatins), which are generally considered to be involved in defense responses and regulation of protein turnover (28–37). Unexpectedly, silencing or overexpression of CsDFB1 resulted in defects in floral organogenesis (leading to malformed flowers) and vascular differentiation, which are classic aspects of plant development governed by auxin (4–6). Auxin content was higher in CsDFB1-RNAi lines but lower in CsDFB1-OE lines. More importantly, the floral defects of CsDFB1-OE lines were rescued by the application of exogenous auxin, further supporting the idea that CsDFB1 participates in modulating local auxin distribution.
CsDFB1 physically interacted with CsPHB, an HD-ZIP III transcription factor with critical roles in meristem formation, polarity establishment, and vascular differentiation (46, 55–60). In Arabidopsis, the gain-of-function mutation phb-1d produces rod-like leaves because of a transformation from abaxial to adaxial cell fate (60). The loss of function of PHB and the related factor REVOLUTA (REV) results in a reduction in flower meristem activity and floral organ number (57, 59). In cucumber, gain-of-function mutations in CsPHB result in curly leaves and increased floral organ number (46). Several HD-ZIP III transcription factors are also involved in regulating auxin homeostasis. In poplar (Populus trichocarpa), PtrHB7 acts with the Aux/IAA9-ARF5 module as a key regulator to control secondary xylem and phloem differentiation (55, 56). REV directly binds to the YUC5 and TAA1 promoters and induces their expression in Arabidopsis (23). In this study, overexpression of CsPHB* caused an increased number of floral organs and abnormal vascular differentiation concomitant with auxin accumulation. We demonstrated that CsPHB is a direct regulator that acts upstream of CsYUC2 and CsPIN1 but with opposite outcomes, as CsPHB induced CsYUC2 expression but repressed CsPIN1 expression. Accordingly, auxin content was elevated in CsPHB*-OE plants. Notably, in situ hybridization showed that CsDFB1 shared an expression pattern with CsPHB both spatially and temporally, indicating that CsDFB1 and CsPHB may indeed cooperate to fine tune the dynamics of local auxin homeostasis. In addition, CsDFB1 appeared to act as an attenuator of CsPHB function in the context of the activation of CsYUC2 expression and the repression of CsPIN1 expression. Adding CsDFB1 to the EMSAs reduced the binding of CsPHB to the two promoters. Moreover, overexpression of CsDFB1 rescued the abnormal expression of CsYUC2 and CsPIN1 in CsPHB*-OE plants. Thus, changes in auxin dynamics, based on the modulation of local auxin biosynthesis and polar transport, may explain how CsDFB1 affects floral and vascular development in cucumber.
Interestingly, in Arabidopsis sup mutants, the up-regulation of YUC1 and YUC4 and the down-regulation of PIN3 and PIN4 expression result in elevated auxin levels at the boundary between whorls 3 and 4, leading to increased number of reproductive organs (26). By contrast, the numbers of petals, stamens, and carpels all increased in CsDFB1-RNAi lines and CsPHB*-OE lines, indicating that the CsDFB1-CsPHB module influences auxin biosynthesis and auxin transport at whorls 2, 3, and 4. Whether the CsDFB1-CsPHB module functions redundantly with or in parallel to SUP in controlling the development of whorls 3 and 4 needs to be clarified.
Besides CsPHB, CsDFB1 also interacted with the YABBY-type transcription factor CsYAB2 as well. HD-ZIP III and YABBY-type transcription factors have opposite effects on polarity establishment (58, 61). Recently, the YABBY-type transcription factor CRC was reported to directly bind to the YUC4 promoter and activate its expression in flower primordia in Arabidopsis (25). It would be interesting to explore whether and how CsDFB1 may influence CRC-mediated auxin biosynthesis in cucumber.
CsDFB1 was expressed in the phloem, like its homologs AtCYS1 in Arabidopsis (31) and CmPP1 in pumpkin (35), suggesting that CsDFB1 might have similar functions to these cystatin family members in vascular tissues. However, CsDFB1 did not inhibit the activity of the cysteine proteinase papain in vitro. A previous study has reported that the expression of rice cystatins is suppressed by auxin treatment during callus differentiation (62). More interestingly, an aspartic protease in rice (OsAsp1) was recently shown to interact with a zinc finger transcription factor OsTIF1 to dismiss OsTIF1-mediated transcriptional inhibition of auxin biosynthesis gene OsTAA1 and thus increase IAA content (63). It will therefore be worth examining whether CsDFB1 function in auxin homeostasis evolved from an ancestor that originally possessed cystatin function.
Collectively, our findings revealed a regulatory pathway of local auxin distribution during floral organogenesis and vascular differentiation in cucumber. Whether this pathway is conserved in other species, and how it interacts with other known regulatory pathways, will be fruitful avenues of future inquiry.
Materials and Methods
A detailed description of plant materials, plant growth conditions, bioinformatic sequence analysis, vector construction and plant transformation, gene expression analysis (in situ hybridization, RT-qPCR, and RNA-seq), histochemical staining, quantification of endogenous auxins, immunolocalization of IAA, phytohormone treatments, subcellular localization, Western blot analysis, the protein–protein interaction assays (BiFC, LCI, and pull-down), the DNA–protein interaction assays (Y1H, EMSA, and dual-LUC), and any associated references are available in SI Appendix, Materials and Methods. Accession numbers used for the phylogenic analysis are listed in SI Appendix, Table S1. Primers used in this study are listed in SI Appendix, Table S2.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31972398), the National Key Research and Development Program of China (2019YFD1000300), the Beijing Innovation Consortium of Agriculture Research System (BAIC01), the earmarked fund for China Agriculture Research System (CARS-23), and the 111 Project of the Ministry of Education of P.R.C. (B17043). We thank Prof. Nan Ma (China Agricultural University) for critically reading and improving the manuscript. We thank PlantEditors (www.planteditors.com) for editing this manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023942118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Robert H. S., et al., Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat. Plants 4, 548–553 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prigge M. J., et al., Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. eLife 9, e54740 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su Y. H., Liu Y. B., Zhang X. S., Auxin-cytokinin interaction regulates meristem development. Mol. Plant 4, 616–625 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fàbregas N., et al., Auxin influx carriers control vascular patterning and xylem differentiation in Arabidopsis thaliana. PLoS Genet. 11, e1005183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng Y., Dai X., Zhao Y., Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang K., et al., AUXIN RESPONSE FACTOR3 regulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling. Plant Cell 30, 324–346 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brumos J., et al., Local auxin biosynthesis is a key regulator of plant development. Dev. Cell 47, 306–318.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Zhao J., et al., A functional allele of CsFUL1 regulates fruit length through repressing CsSUP and inhibiting auxin transport in cucumber. Plant Cell 31, 1289–1307 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandolfini T., Molesini B., Spena A., Molecular dissection of the role of auxin in fruit initiation. Trends Plant Sci. 12, 327–329 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Friml J., et al., Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Teale W. D., Paponov I. A., Palme K., Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7, 847–859 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Casanova-Sáez R., Voß U., Auxin metabolism controls developmental decisions in land plants. Trends Plant Sci. 24, 741–754 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Zheng Z., et al., Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat. Plants 2, 16025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mähönen A. P., et al., PLETHORA gradient formation mechanism separates auxin responses. Nature 515, 125–129 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinon V., Prasad K., Grigg S. P., Sanchez-Perez G. F., Scheres B., Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 1107–1112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y., Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu. Rev. Plant Biol. 69, 417–435 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Brumos J., Alonso J. M., Stepanova A. N., Genetic aspects of auxin biosynthesis and its regulation. Physiol. Plant. 151, 3–12 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Tao Y., et al., Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stepanova A. N., et al., TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Swarup R., Péret B., AUX/LAX family of auxin influx carriers-an overview. Front. Plant Sci. 3, 225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adamowski M., Friml J., PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 27, 20–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanneste S., Friml J., Auxin: A trigger for change in plant development. Cell 136, 1005–1016 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Brandt R., et al., Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 72, 31–42 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Zhong R., Ye Z. H., Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiol. 126, 549–563 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaguchi N., et al., Chromatin-mediated feed-forward auxin biosynthesis in floral meristem determinacy. Nat. Commun. 9, 5290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y., et al., SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. EMBO J. 37, e97499 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margis R., Reis E. M., Villeret V., Structural and phylogenetic relationships among plant and animal cystatins. Arch. Biochem. Biophys. 359, 24–30 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Abe K., Emori Y., Kondo H., Suzuki K., Arai S., Molecular cloning of a cysteine proteinase inhibitor of rice (oryzacystatin). Homology with animal cystatins and transient expression in the ripening process of rice seeds. J. Biol. Chem. 262, 16793–16797 (1987). [PubMed] [Google Scholar]
- 29.Massonneau A., Condamine P., Wisniewski J. P., Zivy M., Rogowsky P. M., Maize cystatins respond to developmental cues, cold stress and drought. Biochim. Biophys. Acta 1729, 186–199 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Martinez M., Cambra I., Carrillo L., Diaz-Mendoza M., Diaz I., Characterization of the entire cystatin gene family in barley and their target cathepsin L-like cysteine-proteases, partners in the hordein mobilization during seed germination. Plant Physiol. 151, 1531–1545 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang J. E., et al., Distinct expression patterns of two Arabidopsis phytocystatin genes, AtCYS1 and AtCYS2, during development and abiotic stresses. Plant Cell Rep. 29, 905–915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belenghi B., et al., AtCYS1, a cystatin from Arabidopsis thaliana, suppresses hypersensitive cell death. Eur. J. Biochem. 270, 2593–2604 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Zhao P., et al., A bipartite molecular module controls cell death activation in the Basal cell lineage of plant embryos. PLoS Biol. 11, e1001655 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Wyk S. G., Du Plessis M., Cullis C. A., Kunert K. J., Vorster B. J., Cysteine protease and cystatin expression and activity during soybean nodule development and senescence. BMC Plant Biol. 14, 294–306 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark A. M., et al., Molecular characterization of a phloem-specific gene encoding the filament protein, phloem protein 1 (PP1), from Cucurbita maxima. Plant J. 12, 49–61 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Liang J., et al., Biotic stress-induced expression of mulberry cystatins and identification of cystatin exhibiting stability to silkworm gut proteinases. Planta 242, 1139–1151 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Kiyosaki T., et al., Gliadain, a gibberellin-inducible cysteine proteinase occurring in germinating seeds of wheat, Triticum aestivum L., specifically digests gliadin and is regulated by intrinsic cystatins. FEBS J. 274, 1908–1917 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Zhao W., et al., CsLFY is required for shoot meristem maintenance via interaction with WUSCHEL in cucumber (Cucumis sativus). New Phytol. 218, 344–356 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Bowman J. L., Smyth D. R., Meyerowitz E. M., The ABC model of flower development: Then and now. Development 139, 4095–4098 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Ditta G., Pinyopich A., Robles P., Pelaz S., Yanofsky M. F., The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935–1940 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Sui X., et al., Transcriptomic and functional analysis of cucumber (Cucumis sativus L.) fruit phloem during early development. Plant J. 96, 982–996 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Lopez-Cobollo R. M., Filippis I., Bennett M. H., Turnbull C. G. N., Comparative proteomics of cucurbit phloem indicates both unique and shared sets of proteins. Plant J. 88, 633–647 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Kelley D. R., Arreola A., Gallagher T. L., Gasser C. S., ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 139, 1105–1109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu B., et al., Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary-specifying genes to promote sepal and petal development. Plant Physiol. 146, 566–575 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gälweiler L., et al., Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Rong F., et al., A mutation in class III homeodomain-leucine zipper (HD-ZIP III) transcription factor results in curly leaf (cul) in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 132, 113–123 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Carlsbecker A., et al., Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sessa G., Steindler C., Morelli G., Ruberti I., The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol. Biol. 38, 609–622 (1998). [DOI] [PubMed] [Google Scholar]
- 49.van der Woude L. C., et al., HISTONE DEACETYLASE 9 stimulates auxin-dependent thermomorphogenesis in Arabidopsis thaliana by mediating H2A.Z depletion. Proc. Natl. Acad. Sci. U.S.A. 116, 25343–25354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sairanen I., et al., Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell 24, 4907–4916 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Z. Y., et al., Functional characterization of the CKRC1/TAA1 gene and dissection of hormonal actions in the Arabidopsis root. Plant J. 66, 516–527 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Franklin K. A., et al., Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. U.S.A. 108, 20231–20235 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ikeda Y., et al., Local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis. Nat. Cell Biol. 11, 731–738 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Garay-Arroyo A., et al., The MADS transcription factor XAL2/AGL14 modulates auxin transport during Arabidopsis root development by regulating PIN expression. EMBO J. 32, 2884–2895 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu C., et al., Auxin-mediated Aux/IAA-ARF-HB signaling cascade regulates secondary xylem development in Populus. New Phytol. 222, 752–767 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Zhu Y., Song D., Sun J., Wang X., Li L., PtrHB7, a class III HD-Zip gene, plays a critical role in regulation of vascular cambium differentiation in Populus. Mol. Plant 6, 1331–1343 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Prigge M. J., et al., Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17, 61–76 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emery J. F., et al., Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Otsuga D., DeGuzman B., Prigge M. J., Drews G. N., Clark S. E., REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25, 223–236 (2001). [DOI] [PubMed] [Google Scholar]
- 60.McConnell J. R., Barton M. K., Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Eshed Y., Baum S. F., Perea J. V., Bowman J. L., Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251–1260 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Yin L., Lan Y., Zhu L., Analysis of the protein expression profiling during rice callus differentiation under different plant hormone conditions. Plant Mol. Biol. 68, 597–617 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Chang S., et al., Auxin apical dominance governed by the OsAsp1-OsTIF1 complex determines distinctive rice caryopses development on different branches. PLoS Genet. 16, e1009157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.