Significance
Cartilage is essential in vertebrate development and adulthood. Cartilage growth plates ensure skeletal growth until closing at puberty, and articular cartilage ensures lifelong structural and functional integrity of joints. Chondrocytes build cartilage in development, governed by the transcription factor SOX9. Using mouse models and transcriptome profiling approaches, we show here that SOX9 also has key roles to maintain growth plates open postnatally and to protect adult articular cartilage from osteoarthritic degradation. In particular, SOX9 safeguards the lineage fate of chondrocytes by preventing their dedifferentiation into skeletogenic mesenchymal progenitors followed by redifferentiation into osteoblasts. These findings provide insights into cellular plasticity and its molecular control in developmental, physiological, and pathological processes within and beyond the skeletal system.
Keywords: cartilage, cell differentiation, lineage determination, SOX9, transcriptional regulation
Abstract
Cartilage is essential throughout vertebrate life. It starts developing in embryos when osteochondroprogenitor cells commit to chondrogenesis, activate a pancartilaginous program to form cartilaginous skeletal primordia, and also embrace a growth-plate program to drive skeletal growth or an articular program to build permanent joint cartilage. Various forms of cartilage malformation and degeneration diseases afflict humans, but underlying mechanisms are still incompletely understood and treatment options suboptimal. The transcription factor SOX9 is required for embryonic chondrogenesis, but its postnatal roles remain unclear, despite evidence that it is down-regulated in osteoarthritis and heterozygously inactivated in campomelic dysplasia, a severe skeletal dysplasia characterized postnatally by small stature and kyphoscoliosis. Using conditional knockout mice and high-throughput sequencing assays, we show here that SOX9 is required postnatally to prevent growth-plate closure and preosteoarthritic deterioration of articular cartilage. Its deficiency prompts growth-plate chondrocytes at all stages to swiftly reach a terminal/dedifferentiated stage marked by expression of chondrocyte-specific (Mgp) and progenitor-specific (Nt5e and Sox4) genes. Up-regulation of osteogenic genes (Runx2, Sp7, and Postn) and overt osteoblastogenesis quickly ensue. SOX9 deficiency does not perturb the articular program, except in load-bearing regions, where it also provokes chondrocyte-to-osteoblast conversion via a progenitor stage. Pathway analyses support roles for SOX9 in controlling TGFβ and BMP signaling activities during this cell lineage transition. Altogether, these findings deepen our current understanding of the cellular and molecular mechanisms that specifically ensure lifelong growth-plate and articular cartilage vigor by identifying osteogenic plasticity of growth-plate and articular chondrocytes and a SOX9-countered chondrocyte dedifferentiation/osteoblast redifferentiation process.
Lineage commitment and differentiation are pivotal decisions that cells make regularly to ensure proper development and homeostasis of multicellular organisms. These decisions are controlled by complex molecular networks, downstream of which dedicated transcription factor sets deploy specific genetic programs. These decisions are often thought to be irreversible, but evidence is mounting that even differentiated cells exhibit plasticity. Their switch to another lineage can occur through transdifferentiation (transition from one to another differentiated cell type) or through dedifferentiation (reversal to a progenitor state) followed by redifferentiation (into a different cell type) (1). While enforceable experimentally in many cells (e.g., induction of pluripotent stem cells), it has been documented to this date in only a discrete number of events in vivo (e.g., epithelial-to-mesenchymal transition, cancer, and heterotopic ossification) (2–4). Thorough evaluation of cellular plasticity prevalence and underpinnings are prerequisites to understanding normal and disease processes and eventually tailor efficient disease treatments.
The skeleton is a model system to study cell fate and differentiation mechanisms. It arises developmentally from multipotent mesenchymal cells, often called osteochondroprogenitors. Guided by spatiotemporal cues, these cells commit to chondrogenesis or osteoblastogenesis to build cartilage or bone, respectively (5–7). Cartilage exists in several forms. Articular cartilage (AC) is a mostly resting tissue that protects opposing bone ends in synovial joints throughout life, whereas growth plates (GPs) are transient, dynamic structures that drive skeletal growth while being gradually replaced by bone (endochondral ossification). Besides expressing a pancartilaginous (PC) program, AC chondrocytes (ACCs) express an articular program and GP chondrocytes (GPCs) a GP program. Through the latter, GPCs proliferate in columns and then undergo growth arrest, prehypertrophy, and hypertrophy. After decade-long debates regarding the ultimate fate of GPCs, recent lineage-tracing studies have provided evidence that many cells escape apoptosis and convert into osteoblasts (OBs) (8, 9). However, many questions remain unanswered (10, 11). For instance, do the same factors originally specify chondrocytes and subsequently maintain their identity? Does osteoblastic plasticity develop during GPC maturation or is it set in progenitors and never lost regardless of differentiation stage, cartilage type, and age? Does the chondrocyte-to-osteoblast switch occur via transdifferentiation or dedifferentiation/redifferentiation? The answers to these questions should provide important insights into fundamental biological and disease processes.
Since SOX9 heterozygous mutations were linked to campomelic dysplasia, a perinatally lethal skeletal malformation disease (12, 13), studies in model systems have revealed that SOX9 is a master chondrogenic transcription factor (14, 15). It may not be a pioneer factor opening chromatin at specific loci, but it potently transactivates PC genes and thereby drives embryonic chondrogenesis (16–19). We also reported that it keeps fetal GPCs alive and prevents osteoblastic conversion of prehypertrophic cells (20). This fate shift involves up-regulation of Runx2, encoding a master transcription factor in GPCs and OBs, and activation of WNT/β-catenin signaling, required for overt osteoblastogenesis (7, 21). SOX9 is thus an essential effector of the PC and GP programs and GPC lineage gatekeeper in utero.
Short stature and severe kyphoscoliosis characterize surviving campomelic dysplasia individuals (22), strongly suggesting important roles for SOX9 in the postnatal skeleton. Sox9 inactivation in chondrocytes in juvenile and adult mice was reported to cause GP shrinking and proteoglycan loss in AC (23). However, it remains unknown if the chondrocyte differentiation programs and their plasticity are still SOX9 dependent. Addressing these questions, we show here that SOX9 prevents GP closure postnatally and protects AC from osteoarthritis. It up-regulates the PC and GP programs, but is dispensable for the AC program, except in load-bearing regions. Both GPCs and ACCs exhibit enduring osteogenic plasticity, with SOX9 preventing their osteoblastogenesis through dedifferentiation/redifferentiation.
Results
SOX9 Expression Lessens as GPs Retire and Articular Cartilage Ages.
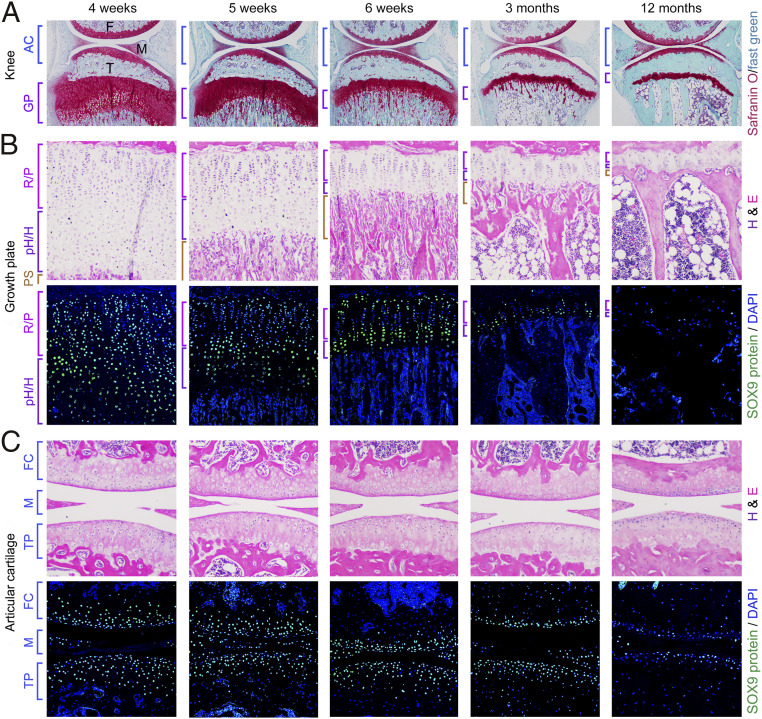
We first asked if SOX9 expression in chondrocytes changes as mice reach adulthood and start aging. We focused on the tibia proximal GP and the knee, a complex osteoarthritis-prone joint. The GP was large in 4-wk-old juveniles, lost two-thirds of its height as mice attained adulthood 2 wk later, and further shrank as mice reached 1 y (Fig. 1 A and B). The primary spongiosa dwindled in adulthood, proving cessation of endochondral ossification. Thus, although GPs remain open in adult mice, they are vestigial. SOX9 protein and RNA expression were high in most GPCs of young mice, but weak and limited to a few chondrocytes in aging mice (Fig. 1A and SI Appendix, Fig. S1A). AC had similar depth and organization in juvenile and aging mice but its SOX9 expression strongly decreased with aging (Fig. 1 A and C and SI Appendix, Fig. S1A). This SOX9 expression lessening as GPs retired and AC aged suggested key roles for SOX9 to maintain vigorous GPs and AC in youth.
Fig. 1.
SOX9 expression declines as GPs retire and AC ages. (A) Sections through the knee of 4-wk- to 12-mo-old mice. Staining, Safranin O (cartilage) and Fast Green. F, femur; M, meniscus; T, tibia. (B) Tibia proximal GPs. (Top) Hematoxylin and eosin (H&E) staining. (Bottom) SOX9 antibody (green) and DAPI (blue, cell nuclei) staining. R/P, relesting and proliferative zones; pH/H, prehypertrophic and hypertrophic zones. PS, primary spongiosa. (C) Knee joint. FC, femoral condyle AC; M, menisci; TP, tibial plateau AC.
SOX9 Ensures Lifelong GP and Articular Cartilage Integrity.
To test the importance of SOX9 in cartilage postnatally, we generated mice carrying Sox9fl/fl (conditional null), AcanCreERT2 (chondrocyte expression of tamoxifen-activatable Cre recombinase), and R26tdT (tdTomato expression following Cre-mediated recombination) alleles. Sox9 inactivation in 4-wk-old mice prompted GP closure within 2 wk (SI Appendix, Fig. S1B), starting with loss of proteoglycans and the hypertrophic zone. Mutant mice were consequently smaller than control littermates, but looked otherwise healthy (SI Appendix, Fig. S1C). Nonmineralized AC also lost proteoglycans, but remained otherwise intact (Fig. 2A). Similar results were obtained when Sox9 was inactivated in 3-mo-old mice (Fig. 2A and SI Appendix, Fig. S1D). While losing SOX9, mutant GPCs and ACCs became tdTomato+ (SI Appendix, Fig. S1 E and F). Steady cell density corroborated that SOX9-deprived ACCs remained alive. Intrigued by the different fates of mutant GPs and AC, we asked whether SOX9 depletion affected AC susceptibility to posttraumatic osteoarthritis (OA). We gave tamoxifen to 3-mo-old mice and performed DMM surgery (destabilization of the medial meniscus) (24) 1 mo later. Two months later, sham-operated controls had pristine AC, with an OA score close to 0 on the six-grade Osteoarthritis Research Society International (OARSI) scale (25), whereas mutants scored at 0.5 due to proteoglycan loss (Fig. 2B). DMM-subjected controls showed AC proteoglycan loss and superficial erosion, scoring of 2, whereas mutants massively eroded AC, scoring at 4. SOX9 is thus required to keep GPs open and AC integrity, including resistance to osteoarthritis.
Fig. 2.
SOX9 is required to keep GPs open and AC healthy. (A) Tibia proximal GPs and knee sections from control and SOX9 mutant mice treated and analyzed at various time points as indicated. Staining, Safranin O/Fast Green. Note that, in this and all other figures showing several time points, control pictures are displayed at one time point only, but these pictures are representative of the data obtained at all time points. (B, Left) Sections through medial femoral condyles (MFCs) and tibial plateaus (MTPs) of control and mutant mice treated as indicated. Staining, Safranin O/Fast Green. (B, Right) OARSI scores. Dots, scores for individual mice. Bars and brackets, means ± SD for six to eight animals. P values are indicated for statistical significance of differences between controls and mutants calculated using Student’s t test.
SOX9 Ensures Survival and Proliferation of GPs but Not Articular Chondrocytes.
GPCs proliferate actively at the columnar stage, growth arrest upon prehypertrophy, and many ultimately die, whereas most ACCs are growth arrested and long lived. EdU (5-ethynyl-2′-deoxyuridine) incorporation assays showed that GPCs quickly stopped proliferating following Sox9 inactivation (SI Appendix, Fig. S2A). TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) assays detected an increased death rate of mutant GPCs, starting with mature cells and then spreading to others (SI Appendix, Fig. S2B). MMP13 (matrix metalloproteinase 13) immunostaining showed that cell death coincided with endochondral ossification. In contrast, cell proliferation, cell death, and MMP13 signals remained quasi absent in mutant AC (SI Appendix, Fig. S2 C and D). SOX9 is thus needed to keep GPCs, but not ACCs, proliferative and alive.
SOX9 Is Required for High Expression of Pancartilaginous and GP Genes, and Its Loss Is Not Compensated for by SOX8.
To gauge the SOX9 dependency of the GPC and ACC transcriptomes, we performed RNA sequencing (RNA-seq) assays on laser capture-microdissected tissues from Sox9 control and mutant mice 7 and 14 d after tamoxifen treatment and from Sox8/Sox9 control and mutant mice 14 d after tamoxifen treatment (SI Appendix, Fig. S3 A and B). We included Sox8/9 mutants because initial analysis of Sox9 mutant data revealed that Sox8, which acts redundantly with Sox9 in other lineages (26, 27), was expressed weakly in control GPs and AC and still detectable in Sox9 mutant tissues (Dataset S1 and SI Appendix, Fig. S4 A–C). However, since AC and GPs were indistinguishable in control and Sox8fl/flAcanCreERT2/+ mice and in Sox9fl/flAcanCreERT2/+ and Sox8fl/flSox9fl/flAcanCreERT2/+ mice (SI Appendix, Fig. S4D), we concluded that SOX8 has no essential functions in chondrocytes and we coanalyzed RNA-seq data from Sox9 and Sox8/Sox9 mutants.
Hundreds of genes were significantly up- and down-regulated in Sox9 mutant AC and GPs (SI Appendix, Fig. S3C, and Dataset S2), and pathway analyses indicated their primary relevance to cartilage and bone (Datasets S3 and S4). Among them, PC and GP markers, including Col2a1 (collagen-II), Acan, Hapln1 (link protein), Fgfr3 (fibroblast growth factor receptor-3, columnar GPC marker), Ihh (Indian hedgehog, prehypertrophic marker), and Col10a1 (collagen-X, hypertrophic marker) were sizably down-regulated (Dataset S1 and Fig. 3A). RNA in situ hybridization confirmed the Acan, Ihh, and Col10a1 down-regulation and quick turnover of the GP hypertrophic zone following Sox9 and Sox8/9 inactivation (Fig. 3B and SI Appendix, Fig. S4 E and F). A few columnar cells became weakly positive for Ihh and Col10a1 in mutant GPs, and for Ihh in mutant AC, and these markers were still detectable at RNA and protein levels in AC months after Sox9 inactivation (SI Appendix, Fig. S4 E and G). Sox5 and Sox6, which encode transcription factors enhancing SOX9 activity in chondrocytes, had their expression unchanged in mutant GPs and AC (SI Appendix, Fig. S4 H and I). Lacking transactivation capability, their proteins, however, likely did not suffice to maintain PC and GP genes on (18, 28).
Fig. 3.
SOX9 ensures high-level expression of PC and GP markers. (A) RNA-seq assay. Three pairs of control and mutant mice received tamoxifen at 3 mo and were analyzed 7 or 14 d later, as indicated. Averages of NRPKM values obtained for control and mutant samples, and statistically significant (P < 0.05) differences obtained in paired t tests for control and mutant data are indicated. (B) In situ detection of Acan, Ihh, and Col10a1 RNA (magenta) in tibia GP and knee AC sections. Sox9 control and mutant mice were given tamoxifen at 3 mo and analyzed at indicated times. Ihh RNA signals were amplified with Adobe Photoshop. Arrows, cells positive for Ihh RNA. Counterstaining, hematoxylin. See SI Appendix, Fig. S4E, for entire-knee pictures.
We concluded that SOX9 is dispensable, as is SOX8, to keep the PC and GP programs on, but is required for robust expression of these programs in fully differentiated ACCs and GPCs.
SOX9 Backs the Articular Program in Load-Bearing Regions and Inhibits Early-Osteoarthritis Signature Genes.
The AC program contains ACC-specific and ACC/FLS-specific (shared by ACC and synovial fibroblasts) genes, whose SOX9 dependence remains unknown. Prg4 (lubricin), an ACC/FLS marker, was partially down-regulated, but still strongly expressed in mutant AC (Fig. 4A and Dataset S1). This finding was not unexpected since FLSs do not express Sox9 (SI Appendix, Fig. S4J). The ACC-specific markers Cytl1 (cytokine-like protein-1), Cilp (cartilage intermediate-layer protein), Smoc2 (SPARC-related modular calcium-binding protein-2), and Clu (clusterin) had their expression slightly altered in mutant AC (Fig. 4A and Dataset S1). Noticeably, weeks and even months after Sox9 or Sox8/Sox9 inactivation, Prg4, Smoc2, and Clu expression was virtually lost in the center of femoral condyle and tibial plateau AC, regions subject to high mechanical load, but was largely intact elsewhere (Fig. 4 B and C and SI Appendix, Fig. S4 K and L). The AC program thus requires SOX9’s support only in load-bearing regions. Interestingly too, Smoc2 and Clu were weakly expressed in the first and last layers of control GPs, presumably in stem cells and terminal chondrocytes, respectively, and were expressed in most remaining GPCs 2 wk after Sox9 inactivation. Expression of these genes is thus likely SOX9 independent in AC and GP.
Fig. 4.
SOX9 is dispensable for AC marker expression, except in load-bearing regions. (A) RNA-seq assay of AC markers. Data were generated and plotted as in Fig. 3A. (B) In situ detection of Prg4, Smoc2, and Clu RNAs (magenta). Sox9 control and mutant mice were given tamoxifen at 3 mo and analyzed 15 d later. Counterstaining, hematoxylin. Green arrows, Smoc2- and Clu-expressing GPCs. Black arrows, load-bearing AC regions down-regulating AC markers. (C) Same experiment as in B, except that mice were analyzed 5 mo after tamoxifen treatment. (D) RNA-seq assay of early-osteoarthritis and inflammation markers. Data were generated and plotted as in Fig. 3A. P values are indicated for differences between control and mutant samples reaching (P < 0.05) and approaching (P < 0.25) statistical significance.
Early-OA signs in SOX9-deficient AC and rapid tissue degeneration upon trauma could be due to cartilage matrix deficiency only, or also to wider gene expression changes. OA was identified as a top canonical pathway altered in mutant AC (Datasets S3 and S4). Down-regulated genes included Timp3 (inhibitor of matrix metalloproteinases) and Frzb (WNT signaling inhibitor), and up-regulated genes included Adamts5 (metalloproteinase targeting aggrecan) (Fig. 4D). Other genes up-regulated in human and canine OA and SOX9-null AC included genes for matrix components (e.g., Vcan, versican, and Tnc, tenascin-C) and catabolic factors (e.g., Ctsb, cathepsin-B) (29–31). Inflammation contributes to OA (32), but typical inflammation markers were not detected (e.g., IL6, interleukin-6) or altered (e.g., S100a8, alarmin) in early-OA human knees (30) and SOX9-deficient AC. SOX9 expression is thus required to maintain lifelong AC integrity.
SOX9 Inhibits Osteoblastogenesis of GP and Articular Chondrocytes.
We next asked if SOX9-deprived ACCs and GPCs converted in OBs. Expression of Runx2 and Sp7, encoding essential osteogenic transcription factors, were globally unchanged in mutant GPs and AC (Fig. 5A). Runx2 RNA was easily detected in control hypertrophic GPCs and OBs, but hardly in ACCs and resting/columnar GPCs, and Sp7 RNA was reliably seen only in OBs (Fig. 5B and SI Appendix, Fig. S5A). Their RNA levels increased in remaining GPCs by day 4, and even more by day 7 of Sox9 loss. They also became readily detectable in mutant AC, but only in the femur load-bearing region. Col1a1 (collagen-I, early OB marker) and Bglap/Bglap2 (osteocalcin, mature OB marker) were globally up-regulated in mutant AC and GPs (Fig. 5A) and detected in the same regions as Runx2 and Sp7 (Fig. 5B and SI Appendix, Fig. S5B). The RUNX2 and osteocalcin proteins became visible in mutant GPCs and ACCs 7 to 10 d after Sox9 inactivation (Fig. 5C). Both proteins could be seen throughout AC by day 14, but most strongly in load-bearing regions. AC still expressed RUNX2, osteocalcin, and collagen-I months after Sox9 inactivation (SI Appendix, Fig. S5C), but remained unmineralized (SI Appendix, Fig. S5D), possibly because of incomplete matrix turnover.
Fig. 5.
Sox9 inactivation prompts chondrocyte osteoblastogenesis. (A) RNA-seq assay of OB markers. Data were generated and plotted as in Figs. 3A and 4D. (B) In situ detection of Runx2, Sp7, and Bglap/Bglap2 RNAs (magenta) in tibia proximal GP and knee AC. Sox9 control and mutant mice received tamoxifen at 3 mo and were analyzed at indicated times. Counterstaining, hematoxylin. RNA signals for Runx2 and Sp7 were amplified using Adobe Photoshop. See SI Appendix, Fig. S5A, for entire-knee sections. (C) In situ detection of tdTomato (red) and RUNX2 and osteocalcin immunostaining (green). Sox9 control and mutant mice received tamoxifen at 3 mo and were analyzed at indicated times. Cell nuclei are stained with DAPI. Blue brackets, GP. Note the progressive disappearance of RUNX2 signal in the mutant GP 2 and 4 d after tamoxifen treatment, reflecting the loss of hypertrophic GPCs and the fact that nonhypertrophic GPCs have not up-regulated RUNX2 expression yet.
We then generated Sox9+/+R26tdT/+AcanCreERT2/+ and Sox9fl/flR26tdT/+AcanCreERT2/+ mice to ask whether mutant GPC-derived OBs survived in endochondral bone. Of all endochondral bone cells present below control GPs, 30 to 40% were tdTomato+ 2 and 9 mo after tamoxifen treatment, and the same was true for bone cells occupying the former mutant GPs (SI Appendix, Fig. S5E). This result suggested that Sox9 deletion did not affect the fate of GPC-derived OBs.
By showing OB marker expression in cells still embedded in original cartilage matrix, these data convincingly showed that both GPCs and ACCs exhibit osteogenic plasticity throughout life and that SOX9 prevents their premature or ectopic osteoblastogenesis.
Chondrocytes Transit through Skeletal Progenitor Stages as They Convert into Osteoblasts.
To test whether chondrocytes convert into OBs via transdifferentiation or dedifferentiation/redifferentiation in wild-type and SOX9-deficient conditions, we treated control and mutant mice with tamoxifen at postnatal day 8 (P8) or P14, i.e., when GPs are very active and primary and secondary ossification centers fully developing. Since major changes could already be seen on GP and AC histological sections 5 d later (SI Appendix, Fig. S6A), we chose this time point for single-cell RNA-seq (scRNA-seq) analysis. Uniform manifold approximation and projection (UMAP) of gene expression profiles performed with mesenchymal and skeletal cells identified 35 subpopulations, including progenitor cells, ACCs, and GPCs and OBs (Fig. 6 A and B and SI Appendix, Fig. S7 A and B and Datasets S5 and S6). Pseudotemporal cell trajectory analysis projected that control GPCs became OBs through sequentially undergoing hypertrophic and terminal differentiation (clusters 29 and 30), and reaching mesenchymal (cluster 31), OB progenitor (cluster 32), preosteoblast (cluster 33), and osteoblast states (cluster 34) (Fig. 6C). Similarly, scRNA-seq data generated for endochondral cells from 1- to 1.5-mo-old mice linked GPCs to OBs via equivalent terminal GPCs, mesenchymal, and osteoprogenitor stages (33) (SI Appendix, Fig. S7 C–G and Dataset S7). Interestingly, most GPCs in mutant pups were projected to skip late proliferative stages, prehypertrophy, and hypertrophy, and to directly reach terminal GPC, mesenchymal, and osteoblast progenitor stages on their path toward osteoblastogenesis (Fig. 6 A–C).
Fig. 6.
SOX9 prevents chondrocyte dedifferentiation/osteoblastic redifferentiation. (A) UMAP plot of skeletal cell populations extracted from the femoral and tibial epiphyses of two pairs of control (Sox9fl/fl) and mutant (Sox9fl/flAcanCreERT2/+) mouse littermates. Mice were treated with tamoxifen 4 and 5 d before analysis at P13 and P19. Cells were segregated into 35 clusters, as indicated. (B) Dot plot showing the relative expression of marker genes across clusters. The darkness and size of the dots reflect the average RNA level in expressing cells and the proportion of expressing cells, respectively. (C) Monocle pseudotemporal ordering of cell clusters. (D) RNA in situ detection of terminal GPC and skeletal progenitor markers (magenta) in Sox9 control and mutant mice treated with tamoxifen at 3 mo and analyzed 4 to 14 d later. The Nt5e and Sox4 RNA signals were amplified using Adobe Photoshop. See SI Appendix, Fig. S8A, for entire-knee sections.
Validating these predictions in real time and real space, RNA hybridization assays showed that Sox9 inactivation prompted proliferating GPCs to activate markers of terminal GPCs, including Col10a1 and Mgp (matrix GLA protein); terminal GPCs and skeletal progenitors, including Nt5e (ecto-5′-nucleotidase or CD73) (34, 35), Sox4 (36–38), Mmp13 and Ibsp (integrin-binding sialoprotein); skeletal progenitors, such as Postn (periostin), Col3a1, and Mmp2; and pre-OBs and OBs, including Sp7, Col1a1, and Bglap (Fig. 6D and SI Appendix, Figs. S6 B and C and S8 A and B). Several markers, including Mgp, Nt5e, Mmp13, Ibsp, Col3a1, and Sp7 were also up-regulated or ectopically activated in subsets of mutant ACCs. Importantly, the detection of all these RNAs in cells that were still embedded in original GPs and AC tissue matrix gave assurance that chondrocytes themselves, and not invading endochondral progenitor cells, were undergoing osteoblastogenesis.
Of note, Sox2, Nanog, and Oct4 expression, which marks pluripotent stem cells and was detected in chondrocytes becoming OBs in mouse bone fracture callus (39), was undetectable in our mutant samples (Datasets S1, S5, and S6). Gdf5 (growth-and-differentiation factor-5) expression, marking embryonic joint progenitors (40, 41), was also undetected. Pthlh (parathyroid hormone-related protein) expression, marking GP stem cells (42), remained confined to top-layer mutant GPCs (SI Appendix, Fig. S8C). Thus, SOX9-deficient chondrocytes unlikely reverted to a pluripotent, joint progenitor or GP stem-cell stage.
Together, these data indicated that GPCs and ACCs attain osteoblastogenesis upon physiological or forced loss of Sox9 expression by transiting through skeletal progenitor states, and thus through dedifferentiation/redifferentiation.
SOX9 May Involve Skeletogenic Pathways in the Control of Chondrocyte Fate.
Looking for mechanisms whereby SOX9 may inhibit chondrocyte-to-osteoblast conversion, we focused on the TGFβ (transforming growth factor-β) and BMP (bone morphogenetic protein) pathways, based on their key roles in skeletal development, adult homeostasis, and cell fate determination. TGFβ keeps ACCs and GPCs at immature stages at least in part by lowering RUNX2 activity and stability (43–46) and also promotes such cell fate changes as epithelial-to-mesenchymal transition (4). Tgfbr3, the TGFβ receptor gene most expressed in control AC, was down-regulated in mutant AC, whereas other TGFβ ligand and receptor genes were unaffected in mutant AC and GPs (Dataset S1). Accordingly, TGFβ signaling, assessed by pSMAD2/3 immunostaining, lost partial strength in mutant AC a week after Sox9 inactivation and continued to underperform for months (Fig. 7A). BMP signaling promotes GPC maturation and OB differentiation, namely by up-regulating RUNX2 RNA and protein levels, and also permits chondrogenesis and osteogenesis from progenitor cells (47–50). While BMP ligand and receptor genes were globally unaffected by Sox9 inactivation, Chrdl2, Grem1, Nog, and Bambi, encoding secreted inhibitors, were down-regulated in mutant AC, and Grem1 and Nog were also down-regulated in mutant GPs (Dataset S1). Accordingly, BMP signaling, assessed by pSMAD1/5/9 immunostaining, increased in mutant AC and GPs by day 7 after Sox9 loss and remained elevated for months in AC (Fig. 7B).
Fig. 7.
TGFβ and BMP signaling likely contributes to chondrocyte fate changes upon Sox9 inactivation. (A) pSMAD2/3 immunostaining in Sox9 control and mutant mice given tamoxifen at 3 mo and analyzed at indicated times. Cell nuclei are stained with DAPI. Blue brackets, GPs. Graphs, pSMAD2/3 signal intensities. Dots, values obtained for individual mice. Bars and brackets, means ± SD for four animals per time point. P values obtained in t tests of differences between controls and mutants in the first week (days 2 to 7 combined), second and third week (days 10 to 21 combined), and 2 and 9 mo after Sox9 inactivation are shown in purple. P values for differences between early (2 to 7 d) and late (10 to 21 d) times are shown in blue for controls and in red for mutants. (B) pSMAD1/5/9 immunostaining conducted and presented as in A, except that differences between controls and mutants were assessed at 2 to 4 d and 7 to 21 d after Sox9 inactivation. (C) RNA levels of various genes in Sox9fl/fl primary chondrocytes transduced with Ad-GFP (controls) or Ad-CRE (mutant) adenovirus, and treated with 5 ng/mL TGFβ1 or 200 ng/mL BMP2 for 24 h. RNA values were measured by qRT-PCR and calculated relative to those for Hprt and control cells (Ad-GFP, no additive). Dots, individual values obtained in four independent experiments, each with one pair of control and mutant cells. Average values and SDs are shown. P values of ≤0.05 in paired t tests are indicated for conditions testing the effects of Sox9 inactivation and/or the effects of growth factors. (D) Western blots of β-actin, SOX9, RUNX2, and SP7 present in extracts from cells treated as in C. The RUNX2/β-actin and SP7/β-actin ratios are presented relative to that obtained for cells treated with Ad-GFP, and with Ad-GFP/BMP2, respectively. Data are representative of multiple similar experiments.
To test how TGFβ and BMP signaling could contribute to SOX9-deficient chondrocyte fate changes, we transduced Sox9fl/fl primary chondrocytes with GFP or CRE-encoding adenovirus, and treated them with TGFβ1 or BMP2. TGFβ1 partially down-regulated Sox9 expression, but did not change the expression of the hypertrophic GPC marker Col10a1, terminal GPC marker, Sox4, and OB-determining genes Runx2 and Sp7 (Fig. 7C). Interestingly, it up-regulated the expression of the progenitor marker Col3a1 in synergy with Sox9 inactivation, suggesting that, since TGFβ signaling was still active in Sox9 mutant GPs in vivo, it could have contributed to the reversal of mutant chondrocytes to progenitor states (Fig. 7C). Sox9 inactivation and BMP2 similarly down-regulated Col10a1 expression, but did not synergize, suggesting that Sox9 inactivation in vivo may have inhibited the GP program at least in part through increased BMP signaling (Fig. 7C). BMP2 showed a trend toward down-regulating Sox4 and Col3a1 expression, indicating that it unlikely promoted chondrocyte terminal maturation and dedifferentiation. Interestingly, although it did not change the Runx2 RNA level, it greatly increased the Sp7 RNA and RUNX2 and SP7 protein levels, especially in the absence of Sox9, strongly suggesting that the increase in BMP signaling observed in mutant ACCs and GPCs in vivo contributed to converting chondrocytes into osteoblasts (Fig. 7 C and D).
We concluded that SOX9 could control the fate of GPCs and ACCs at least in part by modulating skeletogenic pathways, including progenitor-inductive and osteoinductive activities of TGFβ and BMP signaling, respectively.
Discussion
Keeping GPs open throughout development and AC intact throughout adulthood is essential for vertebrate health. The present study filled critical knowledge gaps regarding the mechanisms underlying these processes by adding key roles to the list of SOX9’s chondrogenic activities in GPCs and ACCs (Fig. 8A) and by uncovering a unique progenitor state progression that chondrocytes transit through as they convert into osteoblasts (Fig. 8B).
Fig. 8.
Roles of SOX9 in established GPs and AC. (A) SOX9 is required to keep GPs open and prevent AC from preosteoarthritic degeneration, namely by preventing osteoblastogenesis of GPCs and load-bearing ACCs. (B) SOX9 ensures and paces the multistep differentiation of GPCs and helps maintain the ACC permanent phenotype, at least in part by preventing GPCs and ACCs from reaching terminal and progenitor stages leading to osteoblast differentiation.
To this date, the mechanisms that control postnatal GPs remain poorly understood, despite evidence that genetic, traumatic, and other conditions can cause serious skeletal abnormalities by untimely inducing GP closure (51). Partly explaining this gap is the fact that rodents are deemed unsuitable models to fill it because they never close their GPs. We here largely overturned this assumption by showing that GPs of sexually mature mice are vestigial. Further, we showed that Sox9 down-regulation is critical to cause GP retirement or closure. The factors that down-regulate SOX9 at puberty likely include estrogen signaling, as this pathway is well known to cause pubertal growth arrest and was also shown to control Sox9 expression in chondrocytes in vitro (52–55). Our findings should thus motivate further research to fully decode the mechanisms driving GP maintenance and closure.
Adult AC is unique by means of its composition and functions, but also by its vulnerability. Built in development for life, it lacks regenerative ability, despite undergoing high mechanical and physiological stress. It is thus prone to osteoarthritic degeneration. Therapies remain scarce for OA, mainly due to limited understanding of ACC identity and regulation. SOX9 expression declines in human OA, but only in late disease stages (56), raising a question on whether this event is a cause or consequence of OA. We showed that SOX9-depleted AC promptly eroded under posttraumatic OA conditions. SOX9 may help maintain a hard-wearing cartilage matrix and direct AC-saving responses of chondrocytes to trauma. Supporting these assumptions, Sox9 inactivation resulted in a pre-OA transcriptomic profile; cartilage matrix deficiencies predispose to OA (57); and SOX9 mitigates the chondrocyte response to proinflammatory cytokines (58).
GPs and AC overlap in composition, but have distinct roles and fates. Our transcriptome profiling data for GPCs and ACCs from the same mice are invaluable resources to compare the two chondrocyte types and assess their SOX9 dependence. They showed that the PC program was not completely turned off upon Sox9 or even Sox9 and Sox8 inactivation. Together with our previous observation in Sox9-null limb buds (17), this finding infers that, despite being essential to develop and maintain cartilage, SOX9 is unlikely to be a chondrogenic pioneer factor. The GP program was found to require SOX9 in both fetal (20) and adult GPCs, but since many GPCs die upon losing Sox9, a question arises as whether SOX9 directly activates this program or only keeps the cells alive. Our finding that ACCs down-regulated GP markers upon Sox9 inactivation, but remained alive, asserts that SOX9 may directly control this program. In contrast, SOX9 was found to be dispensable for the AC program, except in load-bearing regions. An indirect role in AC is supported by the expression of several AC genes, but not Sox9, in FLS. Thus, SOX9 may counteract chondrocyte fate-changing signals triggered by loading conditions.
Lineage-tracing studies have supported the concept that osteoblastogenesis is an alternate fate to apoptosis for terminal GPCs in fetal and neonatal mice. Our findings firmly secured this concept by showing activation of exclusive OB markers by SOX9-depleted cells whose chondrocyte origin was unequivocal since the cells were still located in their original cartilage matrix. Further, we expanded this concept to adult, resting/proliferating GPCs and permanent ACCs, and showed that SOX9 is a gatekeeper of the lineage fate of all these cells. This same role was previously proposed for SOX9, but based on transgenic overexpression of SOX9 in hypertrophic chondrocytes in young mice, leaving open the question of whether endogenous SOX9, whose level is low in adult GPCs and ACCs, fulfills this role (59). Chondrocyte-to-osteoblast transition was proposed in bone fracture healing to occur through reversal to a pluripotent stage and to be facilitated by the invading vasculature (39). We did not find evidence of reversal to a pluripotent state, but provided evidence of a transition through terminal GPC, osteoprogenitor, and preosteoblast states. Transition through these states, but not through prehypertrophy and hypertrophy, is likely required for chondrocyte reprogramming, as SOX9-deficient chondrocytes virtually skipped prehypertrophy and hypertrophy, and underwent osteoblastogenesis soon after activating progenitor markers. The idea that the vasculature may foster chondrocyte osteoblastogenesis is intriguing as it could explain why GPCs switched fate more readily than ACCs. While forced inactivation of Sox9 may only precipitate the normal lineage change of GPCs, OB conversion of ACCs is definitely not normal. It could, however, be pathological, as proposed in temporomandibular joint OA (60), and leading to osteophyte formation in OA, but this possibility remains to be investigated.
A pivotal mechanism whereby SOX9 safeguards the immature GPC and ACC fate is undoubtedly by restricting RUNX2 expression. Overexpression studies in nonskeletal cells in vitro have shown that SOX9 can physically interact with RUNX2, inhibit its activity, and induce its degradation (61, 62). We cannot rule out that such a physical battle takes place between SOX9 and RUNX2 in immature GPCs and ACCs. However, the fact that it took a week for RUNX2 to become detectable following Sox9 inactivation suggests an indirect mechanism. Our data and previous studies concur that several signaling pathways may be involved, including PTHrP, TGFβ, WNT, and BMP signaling, as they are modulated by SOX9 and their roles include cellular plasticity control (4). Interestingly, we found that BMP signaling was not only increased but was also more osteoinductive upon SOX9 loss. We speculate that the mechanisms may be distinct in GPs and AC. On one hand, MGP was shown at low level to inhibit this BMP activity in marrow stromal cells in vitro and at a high level to potentiate it (63). Thus, the low level of MGP in ACCs and immature GPCs may help maintain the cells’ fate, whereas substantial MGP up-regulation in terminal GPCs in wild-type and Sox9-null conditions may help BMP convert chondrocytes into OBs. On the other hand, OB conversion and BMP signaling up-regulation in SOX9-depleted AC load-bearing regions show an intriguing parallelism with a mechanism proposed for fibrodysplasia ossificans progressiva (FOP). This heterotopic ossification disease is caused by ACVR1 gain-of-function mutations (BMP receptor type I) (64). FOP cells display mechanotransduction responses on soft substrates typical of wild-type cells on bone-like stiff substrates (65). Combined, abnormal mechanical signals and increased BMP signaling led to ectopic osteoblastogenesis. Thus, SOX9 loss may cause ACC osteoblastogenesis through increasing BMP signaling and mechanotransduction, the latter possibly resulting from stiffening of the mutant cartilage matrix.
In conclusion, this study has demonstrated that GPCs and ACCs rely throughout life on their developmental master SOX9 to restrain plasticity toward dedifferentiation to unique progenitor states and switching to the fate of their closest ontogenic and anatomical relatives, osteoblasts. SOX9 is thereby pivotal to keep vigorous GPs and enduring AC. These findings should help elucidate mechanisms underlying various types of inherited and acquired skeletal diseases and answer questions on cellular plasticity underpinnings within and beyond the skeletal system.
Materials and Methods
Animals.
Mice were used as approved by the institutional animal care and use committees. They carried Sox9 conditional null (66), AcanCreERT2 (44), and R26tdT (67) alleles. A Sox8 conditional null allele was generated using an MGI:98370/tm1a(EUCOMM)Wtsi targeting vector from the European Conditional Mouse Mutagenesis Program. Genotyping was done by PCR using specific primers (SI Appendix, Table S1). Cre activity was induced in AcanCreERT2/+ mice by intraperitoneal administration of tamoxifen (1 mg/10 g body weight; Sigma-Aldrich) for 2 to 5 consecutive days. The first day of injection was used to report time elapsed between treatment and analyses. DMM and sham surgeries were performed as described (24). OARSI scores were calculated in sections generated at four levels of knee medial portions (70 µm apart) and stained with Safranin O/Fast Green (25).
Histology Analysis and In Situ Assays.
For histology analysis and immunostaining, knees were fixed in 4% paraformaldehyde for 48 h at 4 °C and demineralized in 15% (wt/vol) EDTA (ethylenediaminetetraacetic acid) (pH 7.4) for 10 d at 4 °C. Paraffin and frozen sections were made at 7-µm thickness in the sagittal plane. Staining with Safranin O/Fast Green and hematoxylin/eosin was done following standard protocols. tdTomato was visualized in frozen sections. Tissue mineralization was assessed on undecalcified frozen sections stained with Alizarin Red. Immunostaining was performed using sections, antibodies, and antigen retrieval methods as listed (SI Appendix, Tables S2 and S3). Fluorescence signals were quantified by measuring integrated density and mean fluorescence on selected tissue areas using ImageJ 1.5a software (NIH). RNA in situ hybridization was performed using RNAscope 2.5 HD detection reagent kit-RED (Advanced Cell Diagnostics) on paraffin sections of tissues fixed in 4% paraformaldehyde for 48 h and demineralized in Morse’s solution for 48 h at room temperature. Probes were as listed (SI Appendix, Table S4). Cell proliferation was assessed by EdU (Invitrogen) incorporation (68). EdU was injected intraperitoneally at 5 mg/10 g body weight 4 h before being killed. Cell death was assessed using the ApopTag Fluorescein In Situ Apoptosis Detection kit (EMD-Millipore). Slides were mounted using DAPI-containing Vectashield (Vector Laboratories). Images were acquired using a Leica TCS SP8 confocal microscope, ZEISS Axio Scan.Z1 slide scanner microscope, or a Nikon TE2000 inverted microscope, and were processed using AdobePhotoshop CC 19.1.6 software.
Laser Capture Microdissection and RNA-Seq Assays.
Knee sections from control and mutant mice were prepared using the CryoJane tape method (69). RNA was extracted from 30 7-µm sections per sample using Qiagen RNeasy Plus Microkit. RNA quality and quantity were assessed on a 2200 Tape Station (Agilent). All samples had an RNA integrity number ≥6. Libraries, prepared using 10 ng RNA and the SMARTer Stranded Total RNA-seq v2–Pico Input Mammalian kit (Takara Bio), were sequenced on a NovaSeq sequencer (Illumina) in paired-end dual-indexed format (2 × 100 bp). Sequencing data (≥50 million reads per sample) were demultiplexed into FASTQ files using the DRAGEN genome pipeline (Illumina). The Strand NGS integrated platform was then used for the analysis, management, and visualization of the data, as previously described (18). In brief, reads were mapped to the mouse genome (mm10), and unmapped and duplicate reads were filtered out. DESeq software was used to normalize RNA levels by computing them as numbers of reads per kilobase of exons per million total reads (NRPKM). The statistical significance of fold differences in RNA levels between control and mutant samples was determined using a two-tailed paired t test, with a P value of ≤0.05 being considered significant. Genes were selected for downstream analyses based on an average NRPKM value of ≥3 in at least one sample type.
Pathway Analyses.
Differentially expressed genes were uploaded into the IPA System (Ingenuity Systems, Qiagen) and overlaid with the global molecular network in the Ingenuity Pathway Knowledge Base (IPKB) to identify canonical pathways, diseases, and biofunctions. Gene Ontology (GO) analysis (http://geneontology.org/) was similarly performed and statistically evaluated using the PANTHER overrepresentation test. Data significance was further assessed using Fisher exact tests with a false discovery rate (FDR) correction.
Single-Cell RNA-Seq and Data Analysis.
Distal femur and proximal tibial epiphyses from mouse pups were digested with Liberase (1 mg/mL; Sigma Aldrich) for 1 h at 37 °C to remove soft tissues. They were then cut into 1- to 2-mm3 pieces and digested with Liberase (2 mg/mL) for 2 additional hours. Single cells were passed through a 70-μm cell strainer in PBS (phosphate buffered saline) containing 0.04% BSA (bovine serum albumin). Their numbers and viability were quantified before encapsulation into emulsion droplets by Chromium Controller (10× Genomics). Libraries were constructed using a Chromium Single Cell 3′ v3 reagent kit (10× Genomics). cDNA libraries were profiled in a NovaSeq sequencer using 100-cycle paired-end reads, generating ∼200 million reads per sample. Data were processed using the 10× Genomics workflow. Cell Ranger (10× Genomics) was used for demultiplexing, barcode assignment, and unique molecular identifier (UMI) quantification. Downstream analyses were performed using Seurat v3 and Monocle 3. Cells with >6,000 and <200 expressed genes and >10% mitochondrial transcripts were excluded. Data were normalized using the SCTransform normalization method and integrated as described (70) before principal component analysis and UMAP were performed. Skeletal lineage cells were reclustered and subjected to trajectory modeling and pseudotemporal ordering in Monocle 3 (71). Clusters were ordered using OB/osteocytes as an initial root node. The pseudotime scale was inverted in graphs. Bone and marrow scRNA-seq data (GEO accession no. GSE145477) (33) were likewise analyzed.
Primary Chondrocytes, Western Blotting, and qRT-PCR.
Costal chondrocytes isolated from Sox9fl/fl pups (72) were seeded at 106/10 cm2 and cultured in standard conditions. They were infected the next day with 500 pfu/cell Ad-CMV-iCRE or eGFP adenovirus (Vector Biolabs). Four days later, they were serum starved for 16 h and then treated for 24 h with or without BMP2 (200 ng/mL; R&D Systems) or TGFβ1 (5 ng/mL; R&D Systems). Whole-cell extracts (25 µg protein) were made in RIPA (radio-immunoprecipitation assay) buffer and subjected to Western blotting with listed antibodies (SI Appendix, Table S5). Signals were detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) on a ChemiDoc Imaging System (Bio-Rad Laboratories) and quantified using Image Lab software (Bio-Rad Laboratories). Total RNA was prepared using the RNeasy kit (Qiagen). cDNA was synthesized using the High-Capacity cDNA Reverse Transcription kit, and qPCR was performed using the QuantStudio 3 Real-Time PCR system, SYBR Green PCR Master Mix (Thermo Fisher Scientific), and custom-designed primers (SI Appendix, Table S6). mRNA levels were calculated relative to those of Hprt according to the ΔΔCt method.
Supplementary Material
Acknowledgments
We thank all members of the Translational Research Program in Pediatric Orthopedics at the Children’s Hospital of Philadelphia, members of the Biostatistics and Data Management Core of the Children’s Hospital of Philadelphia Research Institute, and K. S. Zaret for invaluable advice. This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (AR072649 and AR068308 to V.L.; AR062908 to M.P.; AR066098 to L.Q.; and AR069619 to the Penn Center for Musculoskeletal Disorders).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019152118/-/DCSupplemental.
Data Availability
Bulk and single-cell RNA-seq data are available in the GEO (Gene Expression Omnibus) database (GSE154381 and GSE162033, respectively) (73, 74). Bone and marrow scRNA-seq data were accessed from GEO database (GSE145477) (75).
References
- 1.Merrell A. J., Stanger B. Z., Adult cell plasticity in vivo: De-differentiation and transdifferentiation are back in style. Nat. Rev. Mol. Cell Biol. 17, 413–425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta P. B., Pastushenko I., Skibinski A., Blanpain C., Kuperwasser C., Phenotypic plasticity: Driver of cancer initiation, progression, and therapy resistance. Cell Stem Cell 24, 65–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaji D. A., et al., Cellular plasticity in musculoskeletal development, regeneration, and disease. J. Orthop. Res. 38, 708–718 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piera-Velazquez S., Jimenez S. A., Endothelial to mesenchymal transition: Role in physiology and in the pathogenesis of human diseases. Physiol. Rev. 99, 1281–1324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hallett S. A., Ono W., Ono N., Growth plate chondrocytes: Skeletal development, growth and beyond. Int. J. Mol. Sci. 20, E6009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozhemyakina E., Lassar A. B., Zelzer E., A pathway to bone: Signaling molecules and transcription factors involved in chondrocyte development and maturation. Development 142, 817–831 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long F., Ornitz D. M., Development of the endochondral skeleton. Cold Spring Harb. Perspect. Biol. 5, a008334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L., Tsang K. Y., Tang H. C., Chan D., Cheah K. S., Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc. Natl. Acad. Sci. U.S.A. 111, 12097–12102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X., et al., Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 10, e1004820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jing Y., Wang Z., Li H., Ma C., Feng J., Chondrogenesis defines future skeletal patterns via cell transdifferentiation from chondrocytes to bone cells. Curr. Osteoporos. Rep. 18, 199–209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong S. A., et al., Microenvironmental regulation of chondrocyte plasticity in endochondral repair-A new frontier for developmental engineering. Front. Bioeng. Biotechnol. 6, 58 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster J. W., et al., Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372, 525–530 (1994). [DOI] [PubMed] [Google Scholar]
- 13.Wagner T., et al., Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79, 1111–1120 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Hojo H., Ohba S., Insights into gene regulatory networks in chondrocytes. Int. J. Mol. Sci. 20, E6324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefebvre V., Angelozzi M., Haseeb A., SOX9 in cartilage development and disease. Curr. Opin. Cell Biol. 61, 39–47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akiyama H., Chaboissier M. C., Martin J. F., Schedl A., de Crombrugghe B., The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16, 2813–2828 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C. F., Angelozzi M., Haseeb A., Lefebvre V., SOX9 is dispensable for the initiation of epigenetic remodeling and the activation of marker genes at the onset of chondrogenesis. Development 145, dev164459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C. F., Lefebvre V., The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 43, 8183–8203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohba S., He X., Hojo H., McMahon A. P., Distinct transcriptional programs underlie Sox9 regulation of the mammalian chondrocyte. Cell Rep. 12, 229–243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dy P., et al., Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev. Cell 22, 597–609 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komori T., Runx2, an inducer of osteoblast and chondrocyte differentiation. Histochem. Cell Biol. 149, 313–323 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Mansour S., et al., The phenotype of survivors of campomelic dysplasia. J. Med. Genet. 39, 597–602 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry S. P., Liang S., Akdemir K. C., de Crombrugghe B., The postnatal role of Sox9 in cartilage. J. Bone Miner. Res. 27, 2511–2525 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glasson S. S., Blanchet T. J., Morris E. A., The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage 15, 1061–1069 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Glasson S. S., Chambers M. G., Van Den Berg W. B., Little C. B., The OARSI histopathology initiative–Recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthritis Cartilage 18 (suppl. 3), S17–S23 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Barrionuevo F., Scherer G., SOX E genes: SOX9 and SOX8 in mammalian testis development. Int. J. Biochem. Cell Biol. 42, 433–436 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Wegner M., Stolt C. C., From stem cells to neurons and glia: A Soxist’s view of neural development. Trends Neurosci. 28, 583–588 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Smits P., et al., The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell 1, 277–290 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Clements D. N., Carter S. D., Innes J. F., Ollier W. E., Day P. J., Analysis of normal and osteoarthritic canine cartilage mRNA expression by quantitative polymerase chain reaction. Arthritis Res. Ther. 8, R158 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soul J., et al., Stratification of knee osteoarthritis: Two major patient subgroups identified by genome-wide expression analysis of articular cartilage. Ann. Rheum. Dis. 77, 423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunn S. L., et al., Gene expression changes in damaged osteoarthritic cartilage identify a signature of non-chondrogenic and mechanical responses. Osteoarthritis Cartilage 24, 1431–1440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van den Bosch M. H. J., Inflammation in osteoarthritis: Is it time to dampen the alarm(in) in this debilitating disease? Clin. Exp. Immunol. 195, 153–166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong L., et al., Single cell transcriptomics identifies a unique adipose lineage cell population that regulates bone marrow environment. eLife 9, e54695 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suto E. G., et al., Prospectively isolated mesenchymal stem/stromal cells are enriched in the CD73+ population and exhibit efficacy after transplantation. Sci. Rep. 7, 4838 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ullah I., Subbarao R. B., Rho G. J., Human mesenchymal stem cells–Current trends and future prospective. Biosci. Rep. 35, e00191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattaram P., et al., Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat. Commun. 1, 9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dy P., et al., The three SoxC proteins–Sox4, Sox11 and Sox12–Exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 36, 3101–3117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhattaram P., et al., SOXC proteins amplify canonical WNT signaling to secure nonchondrocytic fates in skeletogenesis. J. Cell Biol. 207, 657–671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu D. P., et al., Cartilage to bone transformation during fracture healing is coordinated by the invading vasculature and induction of the core pluripotency genes. Development 144, 221–234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koyama E., et al., A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev. Biol. 316, 62–73 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rountree R. B., et al., BMP receptor signaling is required for postnatal maintenance of articular cartilage. PLoS Biol. 2, e355 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizuhashi K., et al., Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature 563, 254–258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thielen N. G. M., van der Kraan P. M., van Caam A. P. M., TGFβ/BMP signaling pathway in cartilage homeostasis. Cells 8, E969 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henry S. P., et al., Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage. Genesis 47, 805–814 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang J. S., Alliston T., Delston R., Derynck R., Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 24, 2543–2555 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen R., et al., Cyclin D1-cdk4 induce runx2 ubiquitination and degradation. J. Biol. Chem. 281, 16347–16353 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jun J. H., et al., BMP2-activated Erk/MAP kinase stabilizes Runx2 by increasing p300 levels and histone acetyltransferase activity. J. Biol. Chem. 285, 36410–36419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lyons K. M., Rosen V., BMPs, TGFβ, and border security at the interzone. Curr. Top. Dev. Biol. 133, 153–170 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Shu B., et al., BMP2, but not BMP4, is crucial for chondrocyte proliferation and maturation during endochondral bone development. J. Cell Sci. 124, 3428–3440 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu M., Chen G., Li Y. P., TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 4, 16009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krishnan Y., Grodzinsky A. J., Cartilage diseases. Matrix Biol. 71-72, 51–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonnelye E., Zirngibl R. A., Jurdic P., Aubin J. E., The orphan nuclear estrogen receptor-related receptor-alpha regulates cartilage formation in vitro: Implication of Sox9. Endocrinology 148, 1195–1205 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Chen J., et al., Estrogen via estrogen receptor beta partially inhibits mandibular condylar cartilage growth. Osteoarthritis Cartilage 22, 1861–1868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frank G. R., Role of estrogen and androgen in pubertal skeletal physiology. Med. Pediatr. Oncol. 41, 217–221 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Zeng K., Zhang H. Q., Chen Y., Gao Q., Estradiol via estrogen receptor beta inhibits chondrogenesis of mouse vertebral growth plate in vitro. Childs Nerv. Syst. 32, 461–465 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Zhong L., Huang X., Karperien M., Post J. N., Correlation between gene expression and osteoarthritis progression in human. Int. J. Mol. Sci. 17, E1126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hardingham T., Extracellular matrix and pathogenic mechanisms in osteoarthritis. Curr. Rheumatol. Rep. 10, 30–36 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Ouyang Y., et al., Overexpression of SOX9 alleviates the progression of human osteoarthritis in vitro and in vivo. Drug Des. Devel. Ther. 13, 2833–2842 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lui J. C., et al., Persistent Sox9 expression in hypertrophic chondrocytes suppresses transdifferentiation into osteoblasts. Bone 125, 169–177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruscitto A., et al., Evidence of vasculature and chondrocyte to osteoblast transdifferentiation in craniofacial synovial joints: Implications for osteoarthritis diagnosis and therapy. FASEB J. 34, 4445–4461 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng A., Genever P. G., SOX9 determines RUNX2 transactivity by directing intracellular degradation. J. Bone Miner. Res. 25, 2680–2689 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Zhou G., et al., Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 19004–19009 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zebboudj A. F., Imura M., Boström K., Matrix GLA protein, a regulatory protein for bone morphogenetic protein-2. J. Biol. Chem. 277, 4388–4394 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Pacifici M., Shore E. M., Common mutations in ALK2/ACVR1, a multi-faceted receptor, have roles in distinct pediatric musculoskeletal and neural orphan disorders. Cytokine Growth Factor Rev. 27, 93–104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stanley A., Heo S. J., Mauck R. L., Mourkioti F., Shore E. M., Elevated BMP and mechanical signaling through YAP1/RhoA poises FOP mesenchymal progenitors for osteogenesis. J. Bone Miner. Res. 34, 1894–1909 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kist R., Schrewe H., Balling R., Scherer G., Conditional inactivation of Sox9: A mouse model for campomelic dysplasia. Genesis 32, 121–123 (2002). [DOI] [PubMed] [Google Scholar]
- 67.Madisen L., et al., A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mead T. J., Lefebvre V., Proliferation assays (BrdU and EdU) on skeletal tissue sections. Methods Mol. Biol. 1130, 233–243 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tong W., et al., Periarticular mesenchymal progenitors initiate and contribute to secondary ossification center formation during mouse long bone development. Stem Cells 37, 677–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stuart T., et al., Comprehensive integration of single-cell data. Cell 177, 1888–1902 e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trapnell C., et al., The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lefebvre V., et al., Characterization of primary cultures of chondrocytes from type II collagen/beta-galactosidase transgenic mice. Matrix Biol. 14, 329–335 (1994). [DOI] [PubMed] [Google Scholar]
- 73.Lefebvre V., R. Kc R, A. Haseeb, M. Angelozzi, Bulk RNA-seq of articular and growth plate chondrocytes from control and Sox9 mutant mice. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE154381. Deposited 14 July 2020.
- 74.Lefebvre V., R. Kc R, A. Haseeb, M. Angelozzi, Single-cell RNA-seq of long bone epiphyses from control and Sox9 mutant mice. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE162033. Deposited 23 November 2020.
- 75.Zhong L., et al., Single cell transcriptomics analysis of bone marrow mesenchymal lineage cells. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145477. Accessed 13 August 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk and single-cell RNA-seq data are available in the GEO (Gene Expression Omnibus) database (GSE154381 and GSE162033, respectively) (73, 74). Bone and marrow scRNA-seq data were accessed from GEO database (GSE145477) (75).