Significance
Sex chromosomes are not only involved in genetic sex determination—they are also important factors in sexual conflict and speciation. Using laboratory experiments and population genetic modeling, we show that the sex chromosomes of Drosophila melanogaster can coevolve antagonistically. We found that swapping sex chromosomes between five D. melanogaster populations increased male fitness, apparently at the cost of reduced offspring survival. After 25 generations, these fitness effects had disappeared, consistent with the resolution of conflict after disrupting antagonistically coevolved X- and Y-linked genes. Our population genetic models show that antagonistic coevolution between sex chromosomes is a biologically plausible explanation for our empirical findings. Together, our empirical and theoretical results provide support for a potential path to speciation through sexual conflict.
Keywords: coevolution, compensatory evolution, interlocus sexual conflict, sex chromosome evolution
Abstract
Antagonistic interactions between the sexes are important drivers of evolutionary divergence. Interlocus sexual conflict is generally described as a conflict between alleles at two interacting loci whose identity and genomic location are arbitrary, but with opposite fitness effects in each sex. We build on previous theory by suggesting that when loci under interlocus sexual conflict are located on the sex chromosomes it can lead to cycles of antagonistic coevolution between them and therefore between the sexes. We tested this hypothesis by performing experimental crosses using Drosophila melanogaster where we reciprocally exchanged the sex chromosomes between five allopatric wild-type populations in a round-robin design. Disrupting putatively coevolved sex chromosome pairs resulted in increased male reproductive success in 16 of 20 experimental populations (10 of which were individually significant), but also resulted in lower offspring egg-to-adult viability that affected both male and female fitness. After 25 generations of experimental evolution these sexually antagonistic fitness effects appeared to be resolved. To formalize our hypothesis, we developed population genetic models of antagonistic coevolution using fitness expressions based on our empirical results. Our model predictions support the conclusion that antagonistic coevolution between the sex chromosomes is plausible under the fitness effects observed in our experiments. Together, our results lend both empirical and theoretical support to the idea that cycles of antagonistic coevolution can occur between sex chromosomes and illustrate how this process, in combination with autosomal coadaptation, may drive genetic and phenotypic divergence between populations.
Sex chromosomes have a number of unique properties that distinguish them from autosomes, and one another, including their mode of inheritance, the selection experienced in the two sexes, and gene content (1). These processes are interdependent and strongly influence the genetic variation harbored on each sex chromosome. The Y chromosome is inherited exclusively through males and is therefore exposed only to selection in males. Because the Y chromosome is male limited, any Y-linked genetic variation that is not male beneficial should experience purifying selection, which is consistent with empirical results (2–4). In contrast, although the X chromosome is not sex limited, it still experiences different selection pressures than both the autosomes and the Y chromosome (1). The X chromosome is largely exposed to selection in females as it spends two-thirds of its time in females (1), but it is also always exposed to selection when present in males since it is hemizygous in males, (i.e., X-linked genes are unsheltered by dominance effects in males) (5). Hence, whether genes on the X chromosome are female or male beneficial can depend on the dominance coefficient (6). Collectively, these unique properties have made sex chromosomes important factors in two major fields within evolutionary biology: speciation (7) and sexual conflict (1, 8).
Here we aimed to investigate whether interactions between sex chromosomes contribute to between-population divergence at the intraspecific level, as has previously been shown in interspecific comparisons (7). To do so, we performed experimental crosses to “swap” either an X or a Y chromosome between five geographically isolated wild-type populations of Drosophila melanogaster, allowing us to investigate the effects of novel interactions between the sex chromosomes on male reproductive fitness. As we outline below, we expected males carrying a novel sex chromosome to experience changes in reproductive success of opposite sign, depending on whether the source populations happen to accumulate incompatibilities that involve the Y or the incompatibilities are driven by sexually antagonistic selection.
When two allopatric populations diverge, each will accumulate unique genetic mutations scattered throughout the genome that may not affect the fitness in either population on their own, but may affect the sterility and viability of hybrid offspring if the two populations come into secondary contact (e.g., the Dobzhansky–Muller model of postzygotic isolation) (9, 10). Because the Y chromosome in Drosophila is inactive in somatic cells (11), it is more likely that Y-linked incompatibilities will cause sterility than reduced viability, which has been documented in multiple Drosophila species crosses (summarized in ref. 12). We therefore expected to find evidence of sterility or decreased fertility in males if mutation accumulation on the Y chromosome were an important factor in population divergence in this species. We found no such evidence (Results) and conclude that accumulation of sex-linked incompatibilities influencing male sterility has not been a major contribution to divergence in our five wild-type populations.
Alternatively, sexually antagonistic coevolution between the sex chromosomes could, in principle, provide a mechanism for the sex chromosomes to contribute to evolutionary divergence between populations (13), as suggested by studies of Y-linked regulation of autosomal gene expression (14). The theory of sexually antagonistic coevolution is based on the Red Queen process (15). In short, when males increase their reproductive success through an adaptation that is simultaneously detrimental to females, it creates selection for a counter-adaptation in females to regain their lost fitness. The process may be repeated in multiple cycles over evolutionary time until a resolution is reached or a palliative adaptation ends the conflict (16, 17). Sexually antagonistic coevolution can be considered a form of interlocus sexual conflict (13) if the traits involved are encoded by different genes in males and females (18). As first proposed by Rice and Holland (13) in 1997, when the loci in question are located on the sex chromosomes, this could lead to cycles of sexual antagonistic coevolution between the sex chromosomes. Depending on the relative sizes of the sex chromosomes and autosomal genome, and the extensive regulatory function of sex-linked genes, these cycles could of course also result in coadaptation with the autosomes.
There are three different ways that a Y-linked male-beneficial allele could harm females and possibly generate sexual conflict: 1) The allele could recombine onto the X and harm female carriers, 2) the allele could cause males to limit females’ mating opportunities (e.g., by harming them), or 3) the allele could harm offspring. Scenario 1 is not relevant for our experimental results because recombination does not occur in D. melanogaster males and there are no pseudoautosomal regions on the sex chromosomes; hence there is no opportunity for a Y-linked allele to recombine onto an X chromosome (19, 20). Scenario 2 appears inconsistent with our empirical findings (Results) but could potentially result in similar coevolutionary cycles caused by conflict over mating opportunities, which we explore further in SI Appendix, section C. Hereafter, we focus on scenario 3. If a Y-linked male-beneficial mutation arises that increases male reproductive success but decreases offspring fitness, it should spread through a population because it is subject to selection in males only. The sexually antagonistic male-beneficial mutation creates selection favoring compensatory mutations that might arise on the X chromosome or autosomes to restore female fitness again (e.g., by increased survival of her offspring). Several mechanisms could potentially generate such sexual conflict over offspring survival in Drosophila. For example, male Drosophila can bias paternity by delivering seminal-fluid proteins that induce accelerated egg laying by females (21), potentially resulting in smaller eggs and therefore reduced offspring viability (22). A Y-linked allele influencing expression of seminal-fluid proteins could therefore create compensatory selection for resistance to accelerated egg laying, which would be disadvantageous alone. Similarly, a Y-linked allele that increases a male’s performance in sperm competition (e.g., by improving sperm displacement) could also cause increased offspring mortality due to polyspermy (23).
The specific mutations causing sexual conflict will likely differ between allopatric populations, and we would therefore expect to find an increase in male reproductive fitness when a Y chromosome with male beneficial mutations is paired with an X chromosome without the corresponding compensatory mutation(s). According to the sexually antagonistic coevolutionary model, we also predict a decrease in female fitness when mating with males harboring a Y chromosome paired to a novel X chromosome.
Another corollary of the antagonistic coevolutionary model is that the effect of disrupting coevolved sex chromosomes on male and female fitness should decay over subsequent generations as new compensatory mutations arise or novel combinations of segregating alleles achieve a similar compensatory effect. We tested this prediction by examining male and offspring fitness in our experimental populations after 25 generations of experimental evolution.
To complement our empirical results and help formalize the hypothesis of sexually antagonistic coevolution between the sex chromosomes, we developed population genetic models describing the evolution of two interacting loci located in different genomic regions (i.e., unlinked): a Y-linked locus influencing both adult male fertilization success (i.e., sperm competition) and subsequent offspring survival and a compensatory locus affecting only offspring survival located on either an autosome or the X chromosome.
Results
Empirical Evidence for Sexually Antagonistic Coevolution.
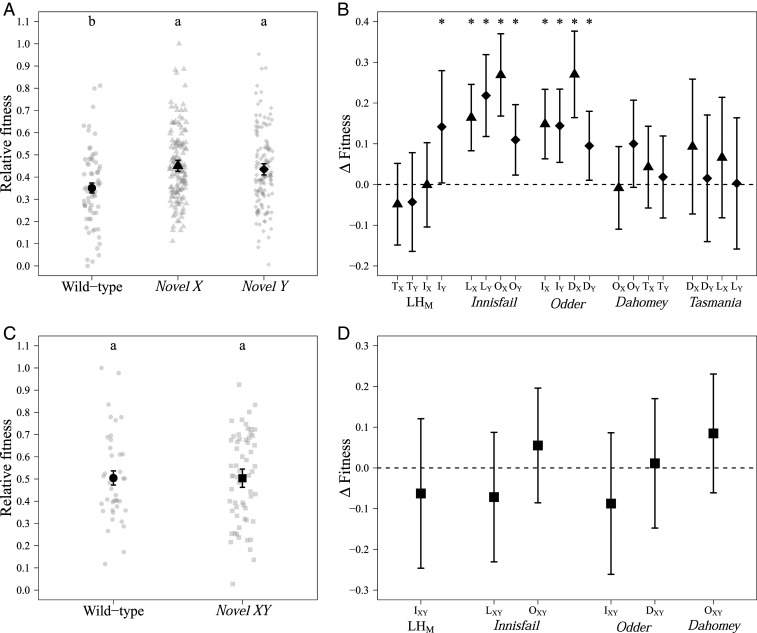
To empirically test for evidence of population-specific interactions between the sex chromosomes in D. melanogaster, we crossed five outbred laboratory-adapted wild-type populations derived from four continents and three climatic zones in a round-robin crossing design (Fig. 1). We created 20 novel populations where either the X chromosome (novel X treatment) or the Y chromosome (novel Y treatment) from one wild-type population (wt) was incorporated into another population (SI Appendix, section A, Fig. S1). We found a significant effect of treatment on male relative reproductive fitness (SI Appendix, section A, Table S1), with males from the novel X or novel Y treatments having a significantly higher relative fitness than wt males (Fig. 2A and SI Appendix, section A, Table S2).
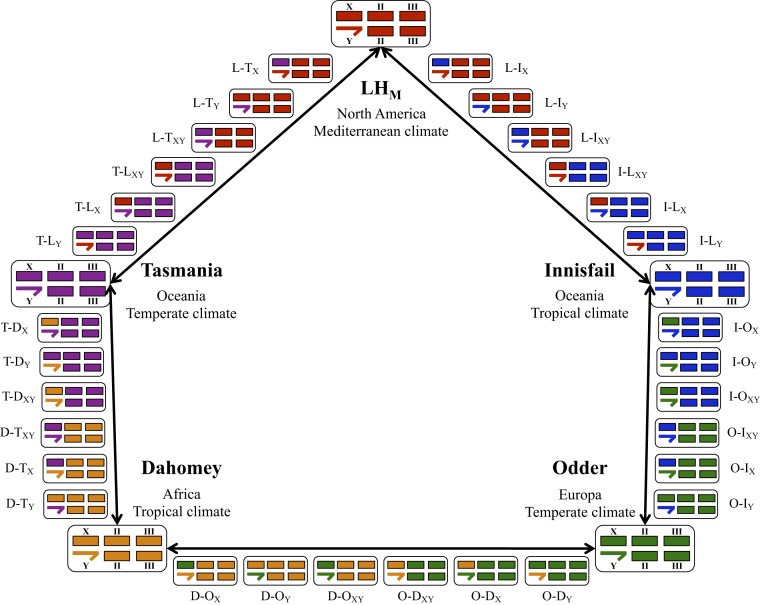
Fig. 1.
Schematic of the round-robin experimental cross design. Each of the five wild-type populations is color coded: LHM (L, red), Innisfail (I, blue), Odder (O, green), Dahomey (D, orange), and Tasmania (T, purple). We crossed each wild-type population (large boxes) with two others from either a different continent or a different climate. This procedure generated four novel genotypes from each cross, so that every combination of the novel sex chromosome pairs experienced a different autosomal genetic background. In total, 10 Novel X and 10 Novel Y genotypes were generated (small boxes). We also generated two Novel XY genotypes from each population cross, where a sex chromosome pair were paired with a novel set of autosomes. Colored bars represent the two major chromosomes (II and III) and chromosome X, with chromosome Y depicted by a half arrow. Novel genotypes are annotated as “genetic background–origin of sex chromosome,” with the novel sex chromosome indicated by an X or Y subscript.
Fig. 2.
Male reproductive fitness assays at generation 0. (A) Relative reproductive fitness for wt (circles), novel X (triangle), and novel Y (diamonds) males. We detected statistically significant (P = 2.45e−04) increases in reproductive fitness for both novel X and novel Y males compared to the wild-type populations (Tukey HSD; P < 0.05). Shown is mean (±SE) of fitted values from the linear model. The raw data are plotted as gray points. (B) Change in relative fitness between the wt and the novel populations. ∆Fitness = ωnovel population − ωwild type with bars indicating bootstrap 95% confidence (Materials and Methods). Ten of 20 novel populations (novel X, triangle; novel Y, diamond) had significantly higher fitness than their wt counterpart (indicate by asterisks), and 16 populations had changed in a positive direction, which was more than expected by chance (P = 0.01). Each of the five wild types is indicated underneath its four novel populations. Uppercase letters indicate which population the novel sex chromosome originates from (D, Dahomey; I, Innisfail; L, LHM; O, Odder; and T, Tasmania) and the novel sex chromosome is denoted by X or Y subscripts. (C) Relative reproductive fitness for wt (circles) and novel XY (squares) treatment groups. There was no significant difference between wt and novel XY. Shown is mean (±SE) of fitted values from the linear model. The raw data are plotted as gray points. (D) Change in relative fitness between the wt and the novel XY population (∆Fitness = ωnovel XY population − ωwild type) with bars indicating bootstrap 95% confidence. Novel XY: squares.
To investigate the interaction between the sex chromosomes in more detail, we calculated the difference in fitness between the novel populations and their wt counterparts (∆Fitness = ωnovel population − ωwild type). We used bootstrap methods to model the data and found that 10 of the 20 novel populations were significantly different from zero in a positive direction (Fig. 2B and SI Appendix, section A, Table S3). Sixteen of the 20 point estimates were also positive, which is significantly different from what would be expected under random deviations (two-sided binomial test, P = 0.01).
We attempted to disentangle possible interactions between the novel sex chromosomes and the autosomes in two different ways. First, we tested the difference in relative male fitness between populations with the same novel sex chromosome combination but different autosomal background (e.g., I-OY and O-IX; SI Appendix, section A, Table S4). Second, we created a novel XY treatment where a pair of sex chromosomes from one wt population were introduced into the autosomal background from another wt population (SI Appendix, section A, Fig. S2). We found no significant differences between the novel populations (SI Appendix, section A, Table S4) and no effect of treatment on relative fitness in the assay with novel XY treatments (Fig. 2C and SI Appendix, section A, Table S1). Neither were any of the differences in relative fitness between novel XY populations and their wt populations significantly different from zero (Fig. 2D). When comparing the novel populations within the round-robin cross, we also found no evidence of autosomal interaction effects between lines (SI Appendix, section A, Table S5). It therefore seems that introducing an individual novel sex chromosome caused the fitness differences rather than interactions with the autosomal background.
Male Beneficial Traits.
To tease apart which fitness components were driving the overall pattern of increased relative fitness in the novel populations, we looked at a number of phenotypic traits that are correlated with male fitness. Interestingly, we found that the increase in fitness was related to different traits in the two novel sex chromosome treatments. For novel X males, we found that the increase in fitness was correlated with an increase in size, as novel X males were significantly larger than both wt and novel Y males (SI Appendix, section A, Fig. S3 and Tables S1 and S2). For novel Y males, we found that the increase in fitness was correlated with an increased ability to displace other males’ sperm (SI Appendix, section A, Fig. S4 and Table S1), as novel Y males were significantly better at displacing sperm compared to wt males (SI Appendix, section A, Table S2). These differences were generally consistent across lines (SI Appendix, section A). Unlike the results for male reproductive fitness, where we did not find evidence of autosomal involvement, it is clear that interactions with the autosomes do play a part in the abovementioned phenotypic traits. Males with the same sex chromosome combination but different autosomal backgrounds do not have identical phenotypes for thorax size and sperm competition (e.g., I-OY and O-IX; SI Appendix, section A, Figs. S3 and S4). This suggests that the autosomal background modifies the effect of the mismatched sex chromosomes.
Female Harmful Traits.
If the sex chromosomes coevolved antagonistically, the increase in male fitness should come at a cost to female fitness. We found no difference on female lifespan when continually exposed to males between the three treatments (SI Appendix, section A, Fig. S5 and Table S1). We did find a significant effect of treatment on total offspring number (SI Appendix, section A, Fig. S6 and Table S1) with novel X males siring a lower number of live offspring compared to wt males (SI Appendix, Table S2). However, we did not find any difference in number of eggs laid by females mated with males from the different treatments (SI Appendix, section A, Fig. S7 and Table S1). The discrepancy between number of eggs and live offspring suggests a trade-off between offspring number and offspring quality, which was confirmed by a significant difference in egg-to-adult viability (SI Appendix, section A, Fig. S8 and Table S1). Novel X males sired significantly lower numbers of viable offspring than the other two treatments (SI Appendix, section A, Table S2). So, mating with novel X males decreases a female’s fitness by reducing the overall number of live offspring she produces. A decrease in offspring survival could be caused by meiotic drives seen in a departure from 50:50 sex ratio (24). We did not find a significant difference in sex ratio between the different treatments (SI Appendix, section A, Fig. S9 and Table S1).
We were unable to establish whether novel Y males had any harmful effects on female fitness through the assays performed in this experiment, and differences between lines for these measures were not very consistent except for total offspring number (SI Appendix, section A). But we were able to exclude costs to females associated with increased male harassment (e.g., due to larger size), males inducing higher fecundity in females, and a reduction in offspring viability.
Counter-Adaptation.
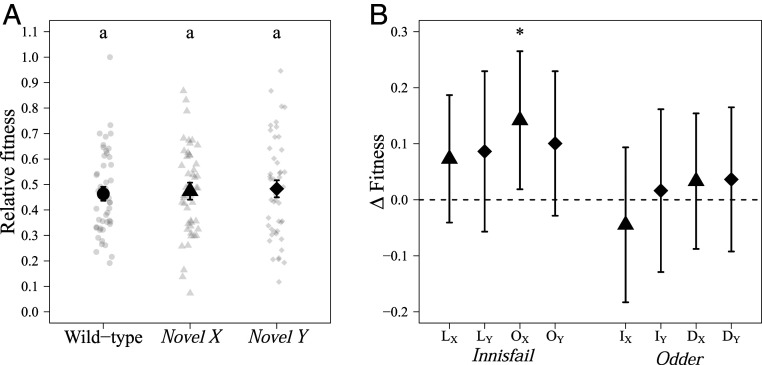
Any shift toward increased fitness in one sex at the cost of the other sex should lead to counter-adaptation, which we expected to see as a reduction in male fitness over microevolutionary time. After 25 generations of experimental evolution (SI Appendix, section A, Fig. S10), we repeated both the male reproductive fitness assay and two of the significant phenotype assays to test whether the interactions between the novel pairs of sex chromosomes had changed. As predicted, we no longer found a significant treatment effect on male fitness (Fig. 3A and SI Appendix, section A, Fig. S11 and Table S6). Indeed, we found that ∆Fitness (∆Fitness = ωnovel population − ωwild type) between novel populations and their wild-type counterparts had diminished in magnitude and were no longer significantly different from 0 (except for one population, I-OX) (Fig. 3B and SI Appendix, section A, Table S7). We also no longer found an effect of the treatments on sperm competition (SI Appendix, section A, Fig. S12 and Table S6). We did find a significant effect of treatment on offspring egg-to-adult survival (SI Appendix, section A, Fig. S13 and Table S6). However, unlike at the start of the experiment, this was not due to a decrease in offspring viability, but instead both novel treatments had higher offspring viability than the wt treatment, with novel Y males being significantly higher than wt. There was also a significant effect of the treatments on sex ratio (SI Appendix, section A, Fig. S14 and Table S6) as novel Y males produced significantly more male offspring than wt males (SI Appendix, section A, Table S8). However, this significant difference was not due to a change in sex ratio in the novel Y populations; rather it was due to a (putatively stochastic) change in the sex ratio in the wt populations.
Fig. 3.
Reproductive fitness assays at generation 25. We selected 8 of the 10 significant populations for the evolution experiment. (A) Relative reproductive fitness for wt (black circle), novel X (triangle), and novel Y (diamonds) males. Novel treatment males no longer had a higher fitness relative to wt males after 25 generations of experimental evolution. Mean (±SE) of fitted values from the linear model. The raw data are plotted as gray points. (B) Change in relative fitness between wt and novel X (triangles) and novel Y (diamonds) populations (∆Fitness = ωnovel population − ωwild type), with bootstrap 95% confidence intervals (Materials and Methods). The asterisk indicates whether ∆Fitness was significantly greater than zero (Tukey HSD; P < 0.05). Each of the wt populations is indicated underneath its four novel populations. Uppercase letters indicate which population the novel sex chromosome originates from (D, Dahomey; I, Innisfail; L, LHM; O, Odder) and the novel sex chromosome is denoted by X or Y subscripts.
Population Genetic Models.
Consider two alternative genetic systems involving two unlinked loci: a Y-linked locus (Y, with alleles Y and y) and a compensatory locus located on either an autosome (A, with alleles A and a) or the X chromosome (X, with alleles X and x) (the autosomal and X-linked models, respectively), in a large population with discrete generations. In both models, a mutant y chromosome increases male fertilization success by a rate of 1 + sm relative to the wild type (Y), but also reduces viability of offspring resulting from matings with females carrying the wild-type allele at the compensatory locus, A (or X). Specifically, the fitnesses of offspring resulting from matings between mutant y males and females with genotypes AA, Aa, and aa (or XX, Xx, and xx) at the compensatory locus are 1 – so, 1 – ho so, and 1, respectively. At the same time, females carrying the mutant a (or x) compensatory allele may incur a “cost of compensation” in terms of offspring viability when mating with wild-type (Y) males, such that the fitnesses of offspring resulting from these matings are 1, 1 – hc sc, and 1 – sc, respectively (see Tables 1 and 2 and Materials and Methods for a summary and description of fitness expressions). These fitness expressions create a scenario of antagonistic coevolution between a potentially male-beneficial Y-linked mutation and the compensatory locus. Reminiscent of standard theories of compensatory evolution (e.g., refs. 25 and 26), each of the mutant alleles (y and either a or x) reduces offspring survival in isolation. However, unlike previous theory, the two loci are located on different chromosomes that do not recombine with one another; if a mutant y chromosome spreads to fixation, compensation at the population level requires that the compensatory mutation also fixes [see also recent models of “father’s curse” for similar scenarios, but without compensatory evolution or parental effects on offspring viability (27); a full description of the models is presented in Materials and Methods and in SI Appendix, section B]. Our theoretical analyses focus on 1) the evolutionary invasion of rare mutants at each locus individually and 2) single bouts of coevolution, beginning with invasion of a single-copy mutant y chromosome in a population initially fixed for the wild-type Y chromosome and for the wild-type A (or X) allele at the compensatory locus and completing when both mutant alleles y and a (or x) become fixed (see ref. 28 for a similar approach in the context of mito-nuclear coevolution).
Table 1.
Fitness expressions for the Y-linked locus (Y) influencing male siring success
| Male Y genotype | Mating success |
| Y | 1 |
| y | 1 + sm |
Table 2.
Fitness expressions for the viability of offspring resulting from all possible combinations of parental genotypes at Y and the compensatory locus A (or X)
| Father’s Y genotype | Mother’s genotype | ||
| AA, XX | Aa, Xx | aa, xx | |
| Y | 1 | 1 − hcsc | 1 − sc |
| y | 1 − so | 1 − hoso | 1 |
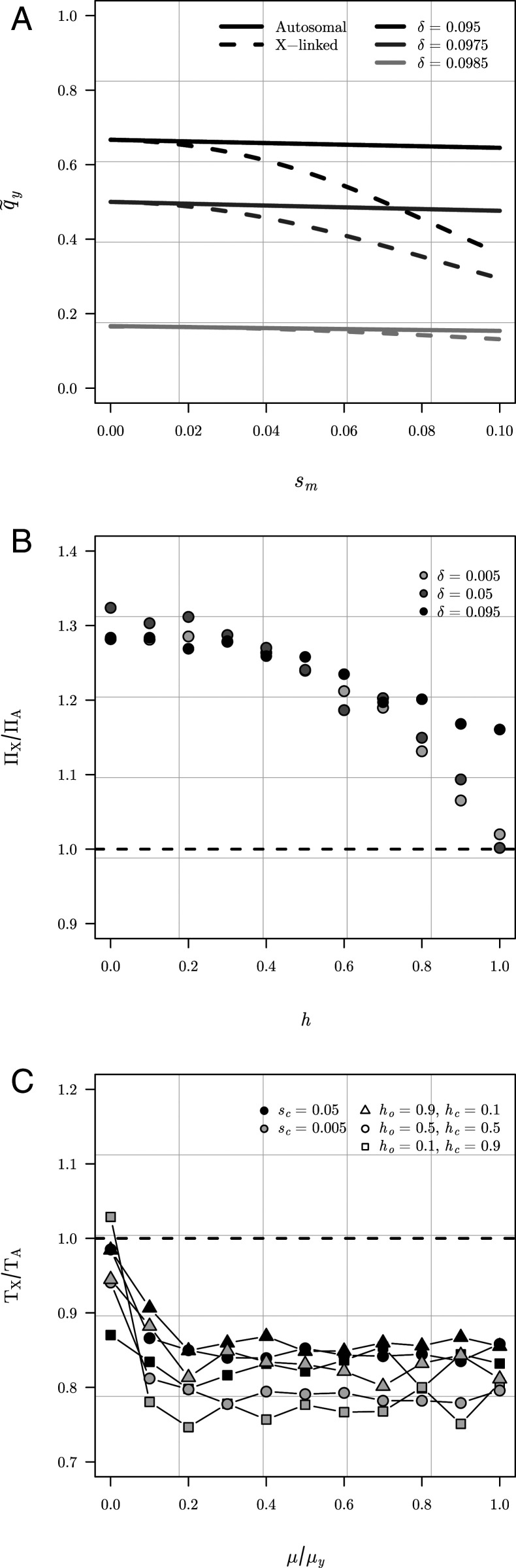
Evolutionary invasion analyses of the two models reveal two key results. First, intuitively, invasion of a rare mutant y chromosome into a population initially fixed for the wild-type alleles at both loci requires that the increase in fertilization success for mutant males is greater than the accompanying reduction in offspring survival. Neglecting second-order terms, a mutant y chromosome can spread when δ = sm − so > 0. Second, whether the genomic location of the compensatory locus (either autosomal or X linked) influences the invasion conditions for the compensatory mutation depends on the relative rate of mutation at the compensatory and Y-linked loci. If compensatory evolution is slow relative to the evolution of the Y-linked locus (e.g., the compensatory mutation rate is much smaller than the mutation rate to a mutant y), the mutant y chromosome is most likely to fix before a new compensatory mutation occurs. In this case, the genomic location of the compensatory locus does not influence the invasion conditions for a compensatory mutation. All males carry the mutant y, and so compensatory mutations will spread if there is any selection against the wild-type allele at the compensatory locus (i.e., 0 < ho,so < 1 for both models). Differences between the models emerge when new compensatory mutations can arise while the mutant y chromosome is still segregating in the population. If there is any cost of compensation for females (i.e., 0 < sc), compensatory mutations will experience purifying selection while qy is small because most males carry the wild-type Y chromosome. As qy increases, more matings involve mutant y males and females homozygous for the wild-type allele at the compensatory locus. When qy reaches a threshold frequency, ( for the X-linked model), compensatory mutations will be favored by selection (derivations provided in SI Appendix, section B). As illustrated in Fig. 4A for the case of additive compensatory fitness effects (h = ho = hc = 1/2), is always less than or equal to and the difference between the two thresholds is greatest when Overall, these results suggest that the genomic location of compensatory mutations (autosomal or X linked) will be most important when 1) the mutant y chromosome is strongly beneficial for males and there is little or no cost of compensation for females (i.e., when ) and 2) compensatory evolution is not limited by mutational variation (i.e., when mutant y chromosomes and compensatory mutations are likely to cosegregate in the population).
Fig. 4.
Summary of theoretical results. (A) Threshold frequencies of the mutant y chromosome at which a new compensatory mutation will experience positive selection for the autosomal ( solid lines) and X-linked ( dashed lines) models, respectively. As the mutant y chromosome sweeps to fixation, X-linked compensatory mutations will become selectively favored earlier than autosomal ones. Results are shown for the case of additive fitness (ho = hc = 1/2), sm = 0.1, and sc = 0.005. (B) The relative probability of invasion for new autosomal and X-linked compensatory mutations into populations initially fixed for the mutant y chromosome is always greater than or equal to 1, suggesting that when compensatory evolution is limited by mutational variation, the probability of invasion will be higher for X-linked compensatory mutations except under complete dominance. For simplicity we assumed equivalent dominance for compensatory and cost of compensation fitness effects (i.e., hc = 1 – ho). Results are shown for three different values of δ (recall that δ = sm − so), where n = 1,000, sm = 0.1, and sc = 0.005. Each point indicates the mean of 106 replicate Wright–Fisher simulations for δ = 0.005 and δ = 0.05, but 5.0 × 106 for δ = 0.095. (C) The relative time to complete a coevolutionary cycle for the autosomal vs. X-linked models plotted as a function of the relative mutation rates at the compensatory and Y-linked loci. To explore the consequences of slow vs. fast compensatory dynamics, μY was held constant at 10−3, while μi (where ) was incremented from 10−7 to μY. At low values of μi/μY, the rate of compensatory evolution is limited by mutational variation, but at intermediate to high values, the mutant y chromosome and compensatory mutations are more likely to segregate simultaneously. Results are shown for two different costs of compensation (sc) and three different dominance scenarios (we again assume equivalent dominance, hc = 1 – ho), and population size n = 1,000.
To complement our analytic results, we performed Wright–Fisher simulations to estimate two other important properties of coevolutionary cycles predicted by our models: 1) the probability of invasion of single-copy autosomal (ΠA) and X-linked (ΠX) compensatory mutations into populations of N individuals that are initially fixed for the mutant y chromosome and 2) the total time to complete a single bout of coevolution between the Y-linked and compensatory loci ( and respectively) under recurrent mutation, selection, and drift. Although the invasion conditions for mutant compensatory mutations are the same for both models when the mutant y chromosome is initially fixed, the probability of eventual fixation for a single-copy mutation is larger for an X-linked compensatory locus (ΠX) than an autosomal one (ΠA) except for the special case of complete dominance for both the compensatory and cost of compensation fitness effects (i.e., when ho = 1 and hc = 1 – ho) (Fig. 4B). The average time to complete a coevolutionary cycle is also smaller for an X-linked than an autosomal compensatory locus, provided that compensatory evolution is strongly limited by mutational variation (i.e., μa or μx is not dramatically lower than μy) (Fig. 4C).
Overall, our theoretical models support the plausibility of antagonistic coevolution between the sex chromosomes as an explanation for our empirical findings: They predict that X-linked compensatory mutations will be favored over more parameter space, have a higher fixation probability, and result in more rapid coevolutionary cycles than autosomal ones. However, the models do not predict that antagonistic coevolution is more likely, on average, to involve X-linked compensatory mutations. All else being equal, compensatory mutations are still more likely to occur and fix on an autosome than the X chromosome in D. melanogaster because they have a larger mutational target size (the autosomal genome is roughly twice the size of the X chromosome and there are three-quarters as many X chromosomes as each autosome) (29). Nevertheless, it is worth noting that our empirical results at least suggest that there is more segregating variation for compensatory traits on the X than on autosomes (our experiment was too short for mutational variance to have had much effect, so any adaptation to novel X or Y chromosomes was most likely via standing genetic variation).
Discussion
We investigated whether interactions between the sex chromosomes could contribute to between-population divergence at the intraspecific level. Taken together, our empirical results demonstrated that exchanging a sex chromosome between populations of D. melanogaster had an overall positive effect on male fitness. However, we found that different phenotypic traits were associated with the increase in male reproductive fitness depending on whether it was a novel X or a novel Y chromosome that was introduced, suggesting that the autosomal background also plays a role.
For the novel Y populations, we found that males gained a fitness advantage through improved sperm displacement. This improvement in male sperm competitive ability in the novel Y populations is consistent with the expectation that male fitness should increase if coevolved male-beneficial/female-detrimental and female compensatory genes have been disrupted in the experimental cross. However, with our data we cannot definitively say whether the increase in novel Y male fitness occurs because they carry a unique Y-linked male-beneficial mutation or because the local population harbors less effective X-linked or autosomal compensatory mutations.
For the novel X populations, we found that the males gained a mating advantage from an overall increase in body size, which is an important factor in male mating success and fitness (30, 31). Whether the increase in size is a result of direct effects of the novel X chromosome or because the local X had some suppressive function on Y–autosome interactions, which are released in its absence, is unclear. Nevertheless, our findings are again consistent with a scenario in which the local population lacks coevolved X-linked compensatory mutations, resulting in increased novel X male fitness.
Thus, our results indicate that different phenotypic and genetic mechanisms may be responsible for the increased male fitness observed in the two novel sex chromosome treatments, presumably in conjunction with coadaptation on the autosomes. That different mechanisms are operating in the two novel treatments is not altogether unexpected. For example, it is well known that the D. melanogaster ejaculate contains various proteins that manipulate female reproductive biology (32). Y-linked mutations could affect the expression level or binding efficiency of these manipulative compounds, resulting in increased receptor activation in females. Ancestral activation levels could then be restored in females by compensatory mutations, which reduce receptor number or sensitivity. Unless the receptor is completely sex limited, this compensatory mutation will likely also result in correlated changes in receptor activity in males, potentially causing downstream phenotypic changes.
Taken together with the absence of change in male reproductive fitness in the novel XY populations, the observed patterns of male reproductive fitness are most consistent with the expected signature of antagonistic coevolution between the sex chromosomes and not with that of accumulated sex-linked incompatibilities influencing male sterility.
Mitochondrial–nuclear interactions have previously been shown to affect male fertility in D. melanogaster (33), and both novel X and novel Y chromosomes were expressed with novel mitochondria. However, the novel XY chromosomes also were expressed with novel mitochondria and we found no effect on male fitness in these populations, suggesting that the magnitude of any mito-nuclear interactions was minor in these populations compared to the effect of interactions between the sex chromosomes.
Since the reproductive fitness results were consistent with antagonistic coevolution, we expected to find a corresponding decrease in female fitness. It has previously been shown that mating with males has a negative effect on female lifespan (34–36), and though our results also showed a reduction in female lifespan when continuously exposed to males, we found no difference between the three treatments. We instead found that offspring sired by novel X males had a significantly lower egg-to-adult survival rate. We did not find the decrease in offspring survival to be caused by a distortion in sex ratio, which suggests the effect is not due to meiotic drive or sexually antagonistic zygotic drive (i.e., mortality resulting from competition between opposite-sex siblings) (37). In addition, there are no known sex ratio meiotic drivers in D. melanogaster. Thus, the decrease in offspring survival was most likely caused by the father’s genotype. Reproduction is costly for females (35), so any eggs which do not produce live offspring are an expense and will over time reduce the reproductive fitness of the females. We were not able to identify any traits in females that were directly negatively affected when mating with novel Y males. A possible cause to investigate in the future is the reduction of female receptivity after mating with males, which could lead to a reduction in female lifetime reproductive fitness.
As a final test of the hypothesis of antagonistic coevolution, we carried out an evolution experiment for 25 generations to see whether we would observe any counter-adaptation. After 25 generations, we found a reduction in fitness indicating strong selection pressure for compensatory evolution, which is supported by the simulation results. In principle, this subsequent reduction in male fitness could be due to new X-linked compensatory mutations; however, given the timescale of the experiment it is more likely that any compensatory evolution by females must utilize standing genetic variation in the stock populations, with novel allelic combinations achieving a compensatory effect.
Collectively, our empirical results are consistent with predictions for antagonistic coevolution between the sex chromosomes. Interestingly, although the increase in male reproductive fitness is caused by the change of a sex chromosome, the specific fitness-related traits, which seem to drive the increase, are affected by an interaction between the novel sex chromosome combinations and the autosomal background. Thus, the antagonistic coevolution between the sex chromosomes seems to have happened concurrently with adaptation in the rest of the genome.
To formally establish the plausibility of antagonistic coevolution as an explanation for our empirical findings, we developed theoretical population genetic models based loosely on our experiments. Our theoretical predictions support the conclusion that antagonistic coevolution 1) is plausible under the fitness effects observed in our empirical experiments and 2) will result in more rapid coevolutionary cycling when it involves both sex chromosomes than the Y and an autosome, especially when compensatory dynamics are not slow relative to those of male-beneficial Y-linked mutations. An interesting corollary to these results is that antagonistic coevolution can potentially drive more rapid among-population divergence on the X chromosome than on the autosomes.
In conclusion, we empirically examined the role of the sex chromosomes in evolutionary divergence among allopatric populations of D. melanogaster. We found evidence of intragenomic conflict between the sex chromosomes that was not completely independent of interactions with the autosomes, but was independent of interaction with the mitochondria. Disrupting coevolved sex chromosomes resulted in an overall increase in reproductive fitness for novel males, increased body size (for novel X males) and sperm displacement (for novel Y males), but also decreased offspring egg-to-adult viability. The accompanying reduction in offspring viability created indirect selection on females to mitigate the loss in fitness resulting from mating with a novel male. After 25 generations of experimental evolution novel males no longer enjoyed higher reproductive fitness indicating that the sexually antagonistic effects of disrupting coevolved sex chromosomes had been resolved, probably by standing genetic variation for compensatory alleles in the experimental populations. Overall, although the particular costs and benefits associated with introgression of a novel sex chromosome must be considered preliminary at this stage, our empirical results were consistent with a sexually antagonistic coevolutionary model of sex chromosome evolution in allopatric populations. Analysis of our theoretical models supports the plausibility of antagonistic coevolution between the sex chromosomes under the fitness effects observed in the experiments and predicts that such coadaptation is more likely to involve sex chromosomes than autosomes provided that compensatory evolution is not limited by mutational variation. These insights into the interactions between the sex chromosomes can help further our understanding of early speciation events. Antagonistic coevolutionary cycles between the sex chromosomes will most likely follow different trajectories in different populations due to random mutations and the interaction between the environment and sexual conflict (38), resulting in genetic and phenotypic divergence between allopatric populations. Thus, over evolutionary time, antagonistic coevolutionary cycles could lead to hybrid incompatibility and thereby contribute to speciation.
Materials and Methods
Empirical Methods.
Drosophila husbandry and fly stocks.
We used five outbred laboratory-adapted wild-type populations: 1) Dahomey, Africa, tropical (30); 2) Innisfail, Oceania, tropical; 3) LHM, North America, Mediterranean (39); 4) Odder, Europa, temperate (40); and 5) Tasmania, Oceania, temperate. We maintained all wild-type populations under the standard LHM culturing protocol (25 °C, 12- to 12-h light-to-dark cycle, 60% relative humidity, cornmeal-molasses-yeast medium; SI Appendix, section A) (39) for at least two generations before the cross. For the fitness assays we used an outbred LHM population homozygous for the visible brown eye (bw) genetic marker (LHM-bw).
Male reproductive fitness.
We estimated male reproductive fitness as the proportion of live offspring sired by males of the target treatment. Five adult target males were placed in a vial for 2 d with 10 competitor LHM-bw males and 15 virgin LHM-bw females. The females were transferred into a single test tube to oviposit for 18 h after which the females were discarded, and the test tubes were left under standard LHM conditions for 12 d, after which we counted the adult offspring and recorded their eye color to assess paternity. The bw genetic marker is recessive to the wild-type red eye-color allele, so all red-eyed offspring can be assigned to red-eyed target males. We calculated relative fitness of the target males by dividing the fitness for each replicate by the maximum fitness across all replicates. We measured male fitness for three different assays: at generation 0 (n = 2 blocks × 7 experimental replicates × 25 populations; fitness estimates for 70 individuals per population), at generation 25 (n = 2 blocks × 3 experimental replicates × 2 replicate populations × 14 populations; fitness estimates for 60 individuals per population), and for the sex chromosome–autosome interactions (n = 10 experimental replicates × 10 populations; fitness estimates for 50 individuals per population).
A detailed description of the crosses to create the novel populations (Fig. 1) can be found in SI Appendix, section A. Also in SI Appendix, section A are descriptions of the evolution experiment, male thorax size, sperm competition assay, male effect on female fecundity, and offspring egg-to-adult viability assay.
Statistical procedures.
All the statistical analyses were conducted in R version 3.4.4 (41). We fitted linear models with treatment as a fixed factor to test each of the dependent variables: male reproductive fitness, thorax size, sperm competition, male effect on female fecundity, egg-to-adult offspring viability, sex ratio, and total offspring number. For assays done in experimental blocks, we added experimental block as a fixed factor, tested for significant effects using ANOVA, and performed post hoc Tukey honestly significant differences (HSD) comparisons for all analyses with a significant treatment effect. The survival curves were calculated using the Kaplan–Meier method (42), and significance was obtained using the Gρ family of tests. We calculated bootstrap 95% confidence intervals around ∆Fitness (∆Fitness = ωnovel population − ωwild type), randomly resampling the data with replacement, and thus recalculating ∆Fitness 10,000 times. We used a two-sided exact binomial test to test whether there were more positive ∆Fitness estimates than expected by chance. For statistical analysis after the 25 generations of experimental evolution we fitted linear mixed models (lme4) (43) with treatment nested within replicate populations to test the dependent variables: male reproductive fitness, egg-to-adult offspring survival, sperm competition, and sex ratio. For assays done in experimental blocks, we added experimental block as a fixed factor, tested for significant effects using ANOVA, and performed post hoc Tukey HSD tests (multcomp) (44) for analyses with significant treatment effect. We calculated bootstrap 95% confidence intervals around treatment means for ∆Fitness by randomly resampling the data with replacement and recalculating ∆Fitness 10,000 times.
Theoretical Models.
We developed two population genetic models, identified by the location of the compensatory locus: the autosomal and X-linked models, respectively. We assumed discrete generations and a life cycle that proceeds as follows: 1) birth, 2) selection on offspring survival, 3) meiosis and mutation, and 4) selection on male mating success. Both loci in each model are biallelic. The Y-linked locus, Y, has wild-type Y and mutant y alleles with frequencies qY and qy = 1 − qY. The compensatory locus is denoted A (with alleles A and a and frequencies qA and qa = 1 − qA) for the autosomal model and X (with alleles X and x and frequencies qX and qx = 1 − qX) for the X-linked model (wild-type alleles are indicated by uppercase letters and mutant alleles by lowercase). Following standard population genetics theory, Y is effectively haploid with strictly paternal inheritance, while the autosomal or X-linked compensatory loci (A or X) are diploid with biparental inheritance.
We based the fitness expressions used in our models loosely on our experimental results. The mutant y chromosome increases male mating success by a factor of 1 + sm relative to the ancestral Y chromosome. Offspring survival depends on the paternal genotype at Y and the maternal genotype at the compensatory locus (A or X), as sires effects on offspring viability (45), and any such negative effects should create selection for females to compensate by maternal effects. Offspring sired by mutant y males may experience reduced survival, depending on their mother’s genotype at the compensatory locus: Matings involving parental genotypes [y: AA], [y: Aa], and [y: aa] result in offspring relative fitness of 1 − so, 1 − hoso, and 1, respectively. To make the models more general, and analytically tractable, we allowed females carrying the mutant a allele to incur a cost of compensation when mating with wild-type (Y) males: Matings involving parental genotypes [Y: AA], [Y: Aa], and [Y: aa] result in relative offspring fitnesses of 1, 1 − hcsc, and 1 − sc, respectively (Tables 1 and 2). Similar to standard theories of compensatory evolution (e.g., refs. 25 and 26) each of the mutant alleles (y and either a or x) is deleterious in isolation. In our models, however, compensation requires that both parents have the appropriate mutant genotype at the other locus.
We model evolutionary invasion and single coevolutionary cycles between the male-beneficial Y-linked locus and the compensatory locus (see ref. 28 for a similar approach in the context of mito-nuclear coevolution). A bout of coevolution begins with the invasion of a single-copy mutant y chromosome in a population initially fixed for the wild-type A (or X) allele at the compensatory locus. The mutant y evolves under net positive selection if the increase in male mating success outweighs the reduction in offspring survival (i.e., if δ = (sm − so) > 0) until it becomes fixed (qy = 1) or is lost from the population (qy = 0). The compensatory locus (A or X) evolves under recurrent mutation and selection, with the population initially fixed for the wild-type A allele (qa = 1 for the autosomal model) or X (qx = 1 for the X-linked model). For simplicity we assume one-way mutation from A → a at a rate ua and X → x at a rate ux per meiosis. If females experience a cost of compensation (i.e., when sc > 0), the mutant compensatory locus will evolve under purifying selection until the mutant y chromosome reaches a threshold frequency, denoted and at which y becomes selectively favored. A coevolutionary cycle completes when the mutant allele becomes fixed at both loci (i.e., qy = qa = 1 or qy = qx = 1).
Our analytic results all assume large population sizes (negligible drift), and an equal sex ratio. To identify the conditions under which rare mutant alleles at each locus can spread during key points of a coevolutionary cycle, we performed a linear stability analysis for each model under three different scenarios: 1) invasion of a mutant y chromosome into populations initially fixed for the wild-type allele at both loci (initial frequencies of qy = 0 and qi = 0, where ), 2) invasion of a rare mutant compensatory allele into a population fixed for y (qy = 1 and qi = 0), and 3) invasion of a mutant compensatory allele into a population with an arbitrary initial frequency of y (qy = qy, qi = 0). Mutant alleles can invade (i.e., the initial equilibrium is unstable) when the leading eigenvalue of the Jacobian of the system of recursions is greater than one (λL > 1) (46). To simplify the analysis for scenario 3, we artificially hold qy constant when performing the invasion analysis. When the initial frequency of y is arbitrary, solving the expression λL > 1 for qy yields the threshold frequency at which compensatory mutations become selectively favored, A full derivation of all models and analytic results is provided in SI Appendix, section B.
We complemented our analytic results with stochastic Wright–Fisher simulations for the autosomal and X-linked models with population size N and an equal sex ratio. We estimated two important properties of coevolutionary cycles from the simulations: 1) the probability of invasion of single-copy autosomal (ΠA) and X-linked (ΠX) compensatory mutations into populations initially fixed for the mutant y chromosome and 2) the total time required to complete a single bout of coevolution between the Y-linked and compensatory loci ( and for the autosomal and X-linked models, respectively) under recurrent mutation, selection, and drift. Additional details and R code for the simulations can be found in SI Appendix, section B.
Supplementary Material
Acknowledgments
We thank Ary Hoffmann, Mads Fristrup Schou, and Stuart Wigby for the wild-type fly stocks and Claire Webster and Qinyang Lee for technical support. Part of the abstract and the empirical methods and results were developed from the PhD thesis of K.K.L.-H. Financial support came from Carl Tryggers Stiftelse (CTS 17:1) (to K.K.L.-H.), the European Research Council (ERC-2015-StG-678148 [to J.K.A.] and ERC-2011-StG-280632 [to E.H.M.]), a Royal Society University Research Fellowship and a Swedish Research Council grant (2011-3701) (to E.H.M.), and a Wenner-Gren International Postdoctoral Fellowship (to C.O.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003359118/-/DCSupplemental.
Data Availability
All data related to the empirical experiment are available at https://github.com/KKLund-Hansen/SexChromCoAdapt and data related to the theoretical model are available at https://github.com/colin-olito/sexChromCoAdapt.
References
- 1.Mank J. E., Small but mighty: The evolutionary dynamics of W and Y sex chromosomes. Chromosome Res. 20, 21–33 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chippindale A. K., Rice W. R., Y chromosome polymorphism is a strong determinant of male fitness in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 98, 5677–5682 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rohmer C., David J. R., Moreteau B., Joly D., Heat induced male sterility in Drosophila melanogaster: Adaptive genetic variations among geographic populations and role of the Y chromosome. J. Exp. Biol. 207, 2735–2743 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Griffin R. M., Le Gall D., Schielzeth H., Friberg U., Within-population Y-linked genetic variation for lifespan in Drosophila melanogaster. J. Evol. Biol. 28, 1940–1947 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Vicoso B., Charlesworth B., Evolution on the X chromosome: Unusual patterns and processes. Nat. Rev. Genet. 7, 645–653 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Frank S. A., Patten M. M., Sexual antagonism leads to a mosaic of X-autosome conflict. Evolution 74, 495–498 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Payseur B. A., Presgraves D. C., Filatov D. A., Introduction: Sex chromosomes and speciation. Mol. Ecol. 27, 3745–3748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rice W. R., Sex chromosomes and the evolution of sexual dimorphism. Evolution 38, 735–742 (1984). [DOI] [PubMed] [Google Scholar]
- 9.Dobzhansky T., Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21, 113–135 (1936). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller H. J., Isolating mechanisms, evolution, and temperature. Biol. Symp. 6, 71–125 (1942). [Google Scholar]
- 11.Ashburner M., Golic K. G., Hawley R. S., Drosophila: A Laboratory Handbook (Cold Spring Harbor Laboratory Press, 2005). [Google Scholar]
- 12.Turelli M., Orr H. A., Dominance, epistasis and the genetics of postzygotic isolation. Genetics 154, 1663–1679 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rice W. R., Holland B., The enemies within: Intergenomic conflict, interlocus contest evolution (ICE), and the intraspecific red queen. Behav. Ecol. Sociobiol. 41, 1–10 (1997). [Google Scholar]
- 14.Lemos B., Araripe L. O., Hartl D. L., Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science 319, 91–93 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Arnqvist G., Rowe L., Sexual Conflict (Princeton University Press, 2005). [Google Scholar]
- 16.Van Valen L., The red queen. Am. Nat. 111, 809–810 (1977). [Google Scholar]
- 17.Lessells C. M., The evolutionary outcome of sexual conflict. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 301–317 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker G. A., “Sexual selection and sexual conflict” in Sexual Selection and Reproductive Competition in Insects, M. S. Blum, N. A. Blum, Eds. (Academic Press, 1979), pp. 123–166. [Google Scholar]
- 19.Patterson J. T., Suche M. L., Crossing over induced by X-rays in Drosophila males. Genetics 19, 223–236 (1934). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson Sayres M. A., “Pseudoautosomal linkage, region” in Reference Module in Life Sciences, S. Maloy, K. Hughes, Eds. (Elsevier, 2017), pp. 514–516. [Google Scholar]
- 21.Hollis B., et al., Sexual conflict drives male manipulation of female postmating responses in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 116, 8437–8444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azevedo R. B. R., French V., Partridge L., Life-history consequences of egg size in Drosophila melanogaster. Am. Nat. 150, 250–282 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Eberhard W. G., Female Control: Sexual Selection by Cryptic Female Choice (Princeton University Press, 1996). [Google Scholar]
- 24.Presgraves D. C., Sex chromosomes and speciation in Drosophila. Trends Genet. 24, 336–343 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura M., The role of compensatory neutral mutations in molecular evolution. J. Genet. 64, 7 (1985). [Google Scholar]
- 26.Weinreich D. M., Chao L., Rapid evolutionary escape by large populations from local fitness peaks is likely in nature. Evolution 59, 1175–1182 (2005). [PubMed] [Google Scholar]
- 27.Ågren J. A., Munasinghe M., Clark A. G., Sexual conflict through mother’s curse and father’s curse. Theor. Popul. Biol. 129, 9–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connallon T., Camus M. F., Morrow E. H., Dowling D. K., Coadaptation of mitochondrial and nuclear genes, and the cost of mother’s curse. Proc. Biol. Sci. 285, 20172257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams M. D., et al., The genome sequence of Drosophila melanogaster. Science 287, 2185–2195 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Partridge L., Farquhar M., Lifetime mating success of male fruitflies (Drosophila melanogaster) is related to their size. Anim. Behav. 31, 871–877 (1983). [Google Scholar]
- 31.Pitnick S., Male size influences mate fecundity and remating interval in Drosophila melanogaster. Anim. Behav. 41, 735–745 (1991). [Google Scholar]
- 32.Wolfner M. F., The gifts that keep on giving: Physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity 88, 85–93 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Camus M. F., Wolf J. B. W., Morrow E. H., Dowling D. K., Single nucleotides in the mtDNA sequence modify mitochondrial molecular function and are associated with sex-specific effects on fertility and aging. Curr. Biol. 25, 2717–2722 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Partridge L., Fowler K., Trevitt S., Sharp W., An examination of the effects of males on the survival and egg-production rates of female Drosophila melanogaster. J. Insect Physiol. 32, 925–929 (1986). [Google Scholar]
- 35.Fowler K., Partridge L., A cost of mating in female fruitflies. Nature 338, 760–761 (1989). [Google Scholar]
- 36.Friberg U., Arnqvist G., Fitness effects of female mate choice: Preferred males are detrimental for Drosophila melanogaster females. J. Evol. Biol. 16, 797–811 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Rice W. R., Gavrilets S., Friberg U., Sexually antagonistic “zygotic drive” of the sex chromosomes. PLoS Genet. 4, e1000313 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arbuthnott D., Dutton E. M., Agrawal A. F., Rundle H. D., The ecology of sexual conflict: Ecologically dependent parallel evolution of male harm and female resistance in Drosophila melanogaster. Ecol. Lett. 17, 221–228 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Rice W. R., et al., Inter-locus antagonistic coevolution as an engine of speciation: Assessment with hemiclonal analysis. Proc. Natl. Acad. Sci. U.S.A. 102 (suppl. 1), 6527–6534 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jensen P., et al., Inbreeding effects on standard metabolic rate investigated at cold, benign and hot temperatures in Drosophila melanogaster. J. Insect Physiol. 62, 11–20 (2014). [DOI] [PubMed] [Google Scholar]
- 41.R Core Team , R: A Language and Environment for Statistical Computing (R version 3.4.4, R Foundation for Statistical Computing, Vienna, Austria, 2018). [Google Scholar]
- 42.Therneau T. M., Lumley T., Survival: Survival analysis. https://cran.r-project.org/web/packages/survival/index.html. Accessed 12 February 2021.
- 43.Bates D., Maechler M., Bolker B., Walker S., Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015). [Google Scholar]
- 44.Hothorn T., et al., multcomp: Simultaneous Inference in General Parametric Models, Version 1.4-15. https://cran.r-project.org/web/packages/multcomp/index.html. Accessed 12 February 2021.
- 45.García-González F., Simmons L. W., The evolution of polyandry: Intrinsic sire effects contribute to embryo viability. J. Evol. Biol. 18, 1097–1103 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Otto S. P., Day T., A Biologist’s Guide to Mathematical Modeling in Ecology and Evolution (Princeton University Press, 2007). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data related to the empirical experiment are available at https://github.com/KKLund-Hansen/SexChromCoAdapt and data related to the theoretical model are available at https://github.com/colin-olito/sexChromCoAdapt.