Significance
Ephexin family guanine nucleotide exchange factors (GEFs) act downstream of Eph signaling to regulate cytoskeletal dynamics, playing critical roles in diverse cellular processes. To achieve precise regulation of Eph signaling, the GEF activities of Ephexins need to be tightly controlled. We here present the crystal structures of autoinhibited Ephexin4, which reveal that both N- and C-terminal fragments directly contact with and inhibit the DH catalytic domain of Ephexin4. This double inhibition mechanism may be commonly utilized by other Ephexins and SGEF. Phosphorylation of a conserved tyrosine at its N terminus and association of PDZ protein(s) to its C-terminal PDZ-binding motif may relieve the autoinhibited Ephexin4. These results reveal versatile autoinhibitory mechanisms that fine-tune the GEF activities of Ephexin family RhoGEFs.
Keywords: Ephexin, RhoGEF, autoinhibition, crystal structure
Abstract
Ephexin family guanine nucleotide exchange factors (GEFs) transfer signals from Eph tyrosine kinase receptors to Rho GTPases, which play critical roles in diverse cellular processes, as well as cancers and brain disorders. Here, we elucidate the molecular basis underlying inhibition and activation of Ephexin family RhoGEFs. The crystal structures of partially and fully autoinhibited Ephexin4 reveal that the complete autoinhibition requires both N- and C-terminal inhibitory modes, which can operate independently to impede Ras homolog family member G (RhoG) access. This double inhibition mechanism is commonly employed by other Ephexins and SGEF, another RhoGEF for RhoG. Structural, enzymatic, and cell biological analyses show that phosphorylation of a conserved tyrosine residue in its N-terminal inhibitory domain and association of PDZ proteins with its C-terminal PDZ-binding motif may respectively relieve the two autoinhibitory modes in Ephexin4. Our study provides a mechanistic framework for understanding the fine-tuning regulation of Ephexin4 GEF activity and offers possible clues for its pathological dysfunction.
Rho GTPases are master regulators of cytoskeletal dynamics and play pivotal roles in diverse cellular processes, including cell polarity, cell motility, cell division, and synaptic signaling (1–3). Typically, Rho GTPases function as molecular switches that cycle between an active guanosine triphosphate (GTP)-bound form and an inactive guanosine diphosphate (GDP)-bound form. Upon activation, they interact with a wide range of downstream effectors, such as actin cytoskeletal regulators, kinases, and scaffold proteins, to drive essential changes in cytoskeletal architecture necessary for corresponding physiological functions (4, 5). Rho GTPases are activated by guanine nucleotide exchange factors (GEFs) and inactivated by GTPase-activating proteins (GAPs) (6, 7). These regulatory proteins are precisely controlled so that Rho GTPase activities are spatiotemporally initiated or suppressed in response to various upstream signals from cell-surface receptors, such as integrins, growth factors, and tyrosine kinase receptors, among others (4).
Eph-interacting exchange protein (Ephexin) family RhoGEFs activate Rho GTPases, including RhoA, Rac, Cdc42, and RhoG (8). This family consists of five known members in most vertebrate species (Ephexin1 to -5). A common feature of the Ephexin family proteins is that they all associate with and act downstream of Eph receptors, the largest subfamily of tyrosine kinase receptors that are activated by Ephrins and participate in various cellular processes (8–11). Specifically, Ephexin1 (also known as NGEF or ARHGEF27) regulates axon growth cone dynamics and spine morphogenesis via binding to EphA4 and activation of RhoA (9, 12–14). Ephexin4 (also named as ARHGEF16) activates RhoG by interacting with EphA2, which promotes RhoG/ELMO/DOCK/Rac signaling and regulates cell migration (15, 16). Ephexin5 (also known as Vsm-RhoGEF or ARHGEF15) functions together with EphB2 to regulate excitatory synapse development (17). Notably, the biological functions of Ephexin2 and Ephexin3 remain elusive although they are known to activate RhoA (8). Therefore, the Ephexin family RhoGEFs serve as the regulatory hubs that link Ephrin-Eph signaling with cytoskeletal dynamics through spatiotemporal regulation of Rho GTPases. Dysfunctions of Eph-Ephexin–mediated Rho signaling have been associated with a variety of diseases, ranging from cancers to brain disorders (18–22).
Each member of the Ephexin family proteins contains a Dbl homology (DH) domain, responsible for catalyzing guanine nucleotide exchange, and an adjacent regulatory pleckstrin homology (PH) domain. In addition, all members possess an Src homology 3 (SH3) domain C-terminal to the DH–PH domain tandem, except Ephexin5 (Fig. 1A). Ephexin4 contains an additional type-I PDZ-binding motif (PBM) at its very C terminus (Fig. 1A). Previous work has demonstrated that elimination of the C-terminal SH3 domain in Ephexin4 can significantly increase its GEF activity toward RhoG, suggesting that Ephexin4 may adopt an autoinhibited conformation via SH3-mediated intra- or intermolecular interactions (23, 24). Intriguingly, such an SH3-mediated autoinhibition mechanism may be also applicable to Ephexin1 and Ephexin3 (25). In addition to the C-terminal inhibition, several lines of evidence have suggested that an evolutionarily conserved helix preceding the catalytic DH domain (referred to as inhibitory helix [IH]) (Fig. 1A) also contributes to the inhibitory functions in Ephexin1 to -3, by binding directly to DH (9, 25, 26). Phosphorylation of a conserved tyrosine residue within the IH could potentially relieve the autoinhibition and activate Ephexin GEF activity (9, 25). However, the molecular basis underlying these autoinhibition and activation events is not well understood. The topic of whether a common regulatory mechanism is shared by all Ephexins remains an intriguing and potentially informative aspect of Rho GTPase biology.
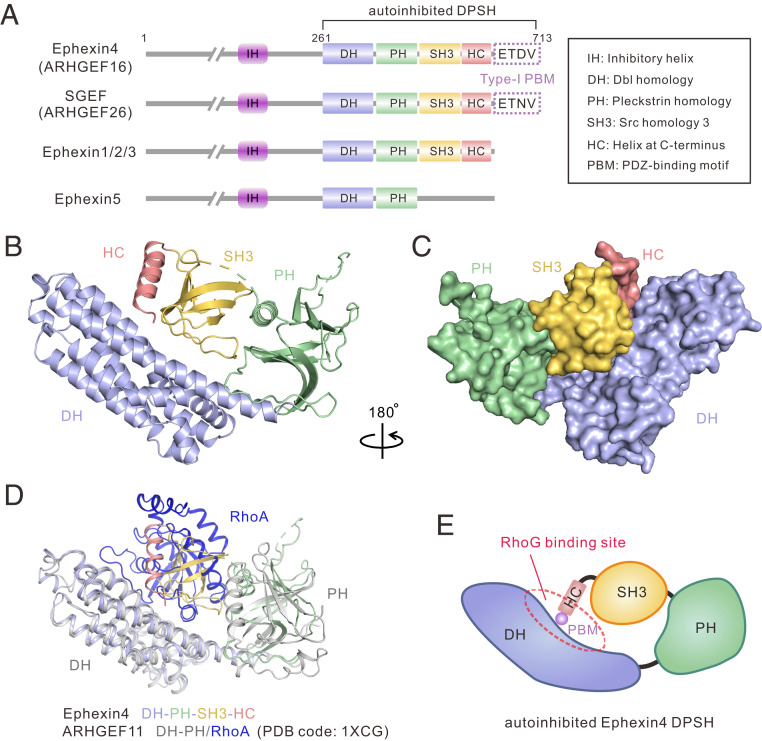
Fig. 1.
Crystal structure of Ephexin4DPSH. (A) Schematic diagrams showing the conserved domain organizations of Ephexins and SGEF. The PBM sequences of Ephexin4 and SGEF are shown. The domain color coding is consistent throughout this paper. The domain keys are also shown here. (B) Ribbon diagram representation of the Ephexin4DPSH structure. (C) Surface representation showing the overall architecture of the Ephexin4DPSH. (D) Superposition of ARHGEF11 DH-PH/RhoA (PDB ID code: 1XCG) and the Ephexin4DPSH (this study) structures. (E) Schematic representation of autoinhibited Ephexin4DPSH. The circle indicates the DH active site.
In the present study, we report crystal structures of Ephexin4 in its partially and fully autoinhibited states. Structural analyses show that the complete inhibition of Ephexin4 involves both N- and C-terminal inhibitory modes. We further demonstrate that this double inhibitory mechanism is conserved among the Ephexin family proteins. Interestingly, association of PDZ proteins with Ephexin4 relieves its C-terminal inhibition while N-terminal inhibition may be regulated by phosphorylation of a conserved tyrosine residue preceding its DH–PH catalytic domain tandem. Moreover, we predict and verify that another RhoGEF, SGEF (also known as ARHGEF26), also adopts a similar autoinhibited architecture and can be activated by binding to PDZ protein. In addition, our study provides a mechanistic clue for how a cancer-associated variant may lead to aberrant Ephexin4 GEF activity.
Results
Crystal Structure of the Autoinhibited Ephexin4DPSH.
To understand how SH3 domain-mediated interdomain interactions suppress the GEF activity of Ephexin4, we first sought to crystallize the Ephexin4DH-end fragment (amino acids 261 to 713) (Fig. 1A). Ephexin4DH-end was crystallized in the P3112 space group with four molecules in the asymmetric unit. Single-wavelength anomalous dispersion (SAD) data were collected using selenomethionine-substituted protein crystals (Table 1). The structure was determined and refined at 2.39 Å resolution (Table 1 and Fig. 1 B and C).
Table 1.
Data collection and refinement statistics
| Crystal | ||||
| Ephexin4DPSH (SeMet) | Ephexin4DPSH (Native) | Ephexin4IDPSH | ||
| Data collection and processing | ||||
| Source | SSRF-BL19U1 | SSRF-BL19U1 | SSRF-BL18U1 | |
| Wavelength, Å | 0.97775 | 0.97775 | 0.97915 | |
| Space group | P3112 | P3112 | P65 | |
| Unit cell (a,b,c), Å | 144.5, 144.5, 290.7 | 144.4, 144.4, 290.5 | 143.1, 143.1, 138.5 | |
| Unit cell (α,β,γ), ° | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | |
| Resolution range, Å | 50–2.59 (2.75–2.59) | 50.00–2.39 (2.48–2.39) | 50.00–3.00 (3.11–3.00) | |
| No. of unique reflections | 108,283 (20,188) | 136,390 (13,383) | 31,720 (3,161) | |
| Redundancy | 20.1 (20.7) | 13.7 (13.7) | 7.0 (6.6) | |
| I/σ(I) | 9.0 (2.4) | 9.7 (2.7) | 14.2 (1.5) | |
| Completeness, % | 99.6 (97.5) | 99.8 (98.4) | 96.5(90.2) | |
| Rmerge, %* | 18.2 (85.6) | 18.8 (85.4) | 8.9 (95.5) | |
| CC1/2 | 99.1 (85.9) | 99.2 (88.7) | 99.8 (53.9) | |
| Wilson B | 46.0 | 43.5 | 82.6 | |
| Phase determination | ||||
| Anomalous scatterer | Selenium (23 of 28 possible sites) | |||
| Mean FOM | 0.3027 | |||
| Structure refinement | ||||
| Resolution, Å | 48.42–2.39 | 46.83–3.00 | ||
| Rcryst†/Rfree‡, % | 17.95/20.24 | 18.50/23.10 | ||
| rmsd bonds, Å/angles, ° | 0.009/1.150 | 0.013/1.660 | ||
| No. of protein atoms | 13,980 | 7,349 | ||
| No. of solvent atoms | 613 | 0 | ||
| Average B factor, Å2 | 53.7 | 88.3 | ||
| Ramachandran plot, % | ||||
| Most favored regions | 98.2 | 97.8 | ||
| Additionally allowed | 1.8 | 2.2 | ||
| Generously allowed | 0 | 0 | ||
Numbers in parentheses represent the value for the highest resolution shell. FOM, figures of merit; rmsd, root-mean-square deviation.
Rmerge = ∑|Ii − Im|/∑Ii, where Ii is the intensity of the measured reflection, and Im is the mean intensity of all symmetry related reflections.
Rcryst = Σ||Fobs| − |Fcalc||/Σ|Fobs|, where Fobs and Fcalc are observed and calculated structure factors.
Rfree = ΣT||Fobs| − |Fcalc||/ΣT|Fobs|, where T is a test dataset of about 5% of the total reflections randomly chosen and set aside prior to refinement.
In the structure, Ephexin4DH-end consists of four major structural elements: DH, PH, SH3, and a previously undefined α-helix C-terminal to the SH3 domain (amino acids 693 to 707; referred to as HC; this four-element fragment is hereafter abbreviated as Ephexin4DPSH) (Fig. 1B). HC binds tightly to SH3, forming the SH3–HC domain tandem, which then engages in contact with the DH–PH (Fig. 1 B and C). Thus, a steric hindrance is generated by SH3–HC-mediated intramolecular interactions, which prevents RhoG from binding to the Ephexin4 DH, as reflected in the superposition of the ARHGEF11–RhoA complex structure (PDB ID code: 1XCG) with the Ephexin4DPSH structure (Fig. 1 D and E). The overall architecture of Ephexin4DPSH is similar to the closed conformation of the SH3–DH–PH domain tandem in Collybistin (also known as ARHGEF9) (PDB ID code: 4MT6) and Asef (also named as ARHGEF4) (PDB ID code: 2PZ1) (SI Appendix, Fig. S1). However, the relative orientation of the SH3, DH, and PH domains is strikingly different among these structures (SI Appendix, Fig. S1). Notably, the type-I PBM of Ephexin4 is not observed in the structure, probably due to its flexible conformation in the crystal.
Details of Interfaces Required for Ephexin4DPSH Interdomain Interactions.
In general, the Ephexin4DPSH interfaces between domains can be divided into four regions and involve both polar and hydrophobic interactions (Fig. 2A). Specifically, at the DH–HC interface, R706HC forms an interdomain salt bridge with D439DH (Fig. 2B and SI Appendix, Fig. S2). By contrast, at the DH–SH3 interface, E679SH3 forms hydrogen bonds with N470DH and H474DH. In addition, the aliphatic chain of R676SH3 contacts with the hydrophobic surface formed by P435, L436, L459, S463, and V466 from DH, and L707 from HC (Fig. 2C). The PH–SH3 interface is stabilized by polar interactions. For example, R614PH forms an electrostatic interaction with E651SH3 (SI Appendix, Fig. S3). Several charge–charge and hydrogen-bonding interactions further reinforce the formation of the SH3–HC domain tandem, such as the K632SH3–E673SH3–R709HC interaction network (SI Appendix, Fig. S3).
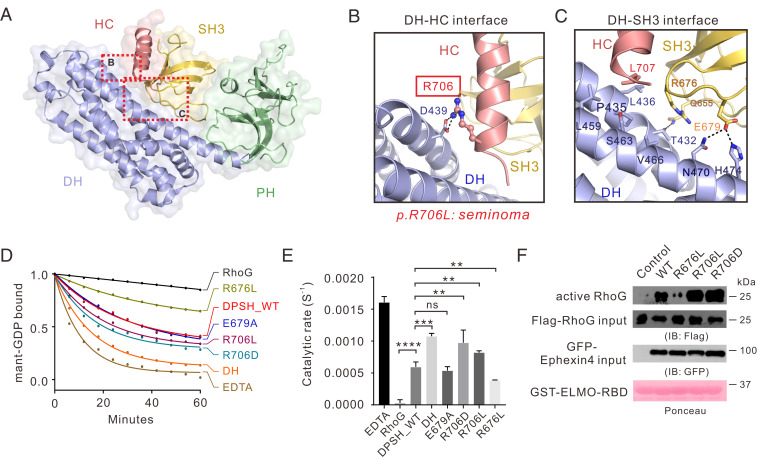
Fig. 2.
Interdomain interactions in Ephexin4DPSH. (A) The combined surface and ribbon representations of the Ephexin4DPSH structure showing that the DH, PH, and SH3–HC units couple tightly with each other. The DH–HC and DH–SH3 interfaces are indicated as dotted boxes. (B and C) Zoomed-in view of the DH–HC (B) and DH–SH3 (C) interface. The key residue R706 associated with cancer are indicated by the red box and the side chain of R706 is shown in the stick mode. (D) Representative in vitro GEF assays showing that the R706D mutation significantly increased the Ephexin4DPSH GEF activity for RhoG compared to WT. (E) Quantification of GEF assays testing the role of residues at DH–HC and DH–SH3 interfaces in regulation of Ephexin4 activity shown in D. Values are expressed as mean ± SD. ns, not significant, ****P < 0.0001, ***P < 0.001, **P < 0.01. All GEF assays in this study were performed using three independent protein preparations with at least duplicate measurements. (F) Either WT or mutant variants of Ephexin4 were cotransfected with Flag-tagged RhoG in HEK 293T cells. Active RhoG was coprecipitated with GST-ELMO2-RBD and detected by immunoblotting.
We then purified various mutants of DPSH (SI Appendix, Fig. S4) and performed in vitro GEF assays to evaluate the role of several key residues at these interdomain interfaces in regulation of the GEF activity of Ephexin4. As expected, the DH domain displayed much stronger GEF activity than wild-type (WT) DPSH (DPSHWT) (Fig. 2 D and E). Compared with DPSHWT, DPSHR706D (the mutant at the DH–HC interface), but not DPSHR614A, DPSHT617A, and DPSHR614A/T617A (the mutants at the PH–SH3 interface) or DPSHE679A (the mutant at the DH–SH3 interface), exhibited a significant increase in GEF activity toward RhoG (Fig. 2 D and E and SI Appendix, Fig. S3), thus suggesting that the DH–HC interaction plays a more critical role than DH–SH3 and PH–SH3 interactions in suppression of GEF activity. Interestingly, substitution of R676SH3 with Leu (R676L) led to a further decreased GEF activity than DPSHWT (Fig. 2 D and E), most likely due to the increased hydrophobicity essential for a more closed conformation of DPSH.
Disease-Causing Mutation in Autoinhibited Ephexin4DPSH.
Given that EphA2 is frequently overexpressed and mutated in a variety of cancers, it would not be surprising that its downstream RhoGEF Ephexin4 has been associated with tumorigenesis and cancer metastasis (15, 27, 28). Several somatic mutations in Ephexin4 have been found in patients with various cancers (29). Specifically, a missense mutation (p.R706L) located at the DH–HC interface was identified in patients with seminoma (30). This variant appears likely to impair the DH–HC coupling, which would thus interfere with autoinhibition. In line with our prediction, DPSHR706L displayed enhanced GEF activity compared with DPSHWT in vitro GEF assays (Fig. 2 D and E).
We next sought to verify the above data through cell-based GEF assays. To this end, we used glutathione S-transferase (GST)-ELMO-RBD to pull down the active form of RhoG from cell lysates when various Ephexin4 constructs were expressed. We previously showed that the Ras-binding domain of ELMO family proteins is sufficient to bind active RhoG (31). As expected, full-length Ephexin4WT showed effective GEF activity toward RhoG in cells (Fig. 2F). Consistent with in vitro GEF assays, both Ephexin4R706D and Ephexin4R706L displayed much stronger GEF activity, compared with that of Ephexin4WT, while the GEF activity of the Ephexin4R676L variant toward RhoG decreased in comparison with Ephexin4WT (Fig. 2F).
Binding of the PDZ Domain Relieves Autoinhibition of Ephexin4DPSH.
How is the autoinhibited Ephexin4DPSH activated? As reported by earlier studies, we noticed that SGEF, another RhoGEF for RhoG, could be activated by the binding of its type-I PBM (-ETNV) to DLG1 PDZ1-2 (32) (Fig. 1A). SGEF and Ephexin4 share highly conserved domain organization and sequence properties (Fig. 1A and SI Appendix, Fig. S5). In particular, the residues required for autoinhibition of Ephexin4 are highly conserved in SGEF (SI Appendix, Fig. S5), suggesting that SGEF may also adopt a similar autoinhibited conformation as Ephexin4. We first verified this hypothesis by showing that substitution of R868 (corresponding to R706 in Ephexin4 at the HC–DH interface) with Asp (i.e., SGEFR868D) resulted in a significant increased GEF activity (SI Appendix, Fig. S6). Interestingly, addition of DLG1 PDZ1-2 significantly enhanced the GEF activity of SGEF (SI Appendix, Fig. S6). These results led us to hypothesize that Ephexin4 may also be activated by PDZ proteins such as DLG1.
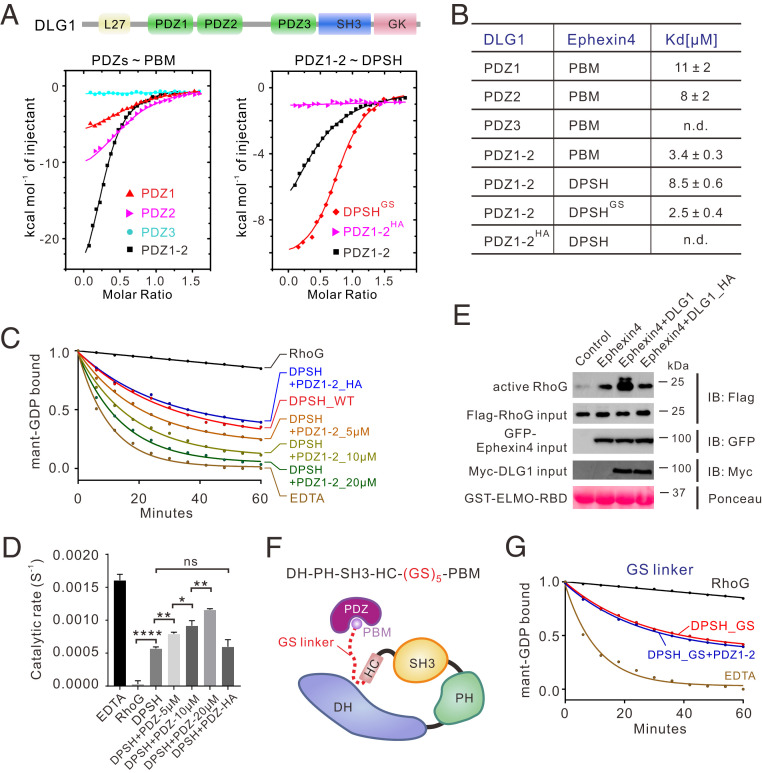
We first examined the interactions between Ephexin4 PBM and DLG1 PDZs. Isothermal titration calorimetry (ITC) data demonstrated that both the PDZ1 and PDZ2 domains of DLG1, but not PDZ3, bound to Ephexin4 PBM with a dissociation constant [Kd] of ∼10 µM. PDZ1-2 displayed a stronger binding affinity (Kd: ∼3 µM) than PDZ1 or PDZ2 (Fig. 3 A and B). Based on the known type-I PDZ-PBM structures, we designed a DLG1 PDZ1-2 double mutant (H256A/H351A; hereafter referred to as HA mutant) which we predicted to disrupt the PDZ–PBM interaction. Indeed, PDZ1-2HA showed no detectable binding affinity toward the Ephexin4 PBM (Fig. 3 A and B). Satisfyingly, we found that addition of PDZ1-2 would significantly enhance the GEF activity of Ephexin4DPSH while the Ephexin4-binding deficient mutant PDZ1-2HA did not (Fig. 3 C and D). Similar results were observed in cell-based GEF assays (Fig. 3E).
Fig. 3.
Binding of the PDZ domain relieves Ephexin4DPSH autoinhibition. (A) ITC curves of the bindings between WT or mutants of DLG PDZs and Ephexin4. (B) Summary of ITC-based measurements of binding affinities between WT or mutants of DLG PDZs and Ephexin4. n.d., not detectable. (C) Representative in vitro GEF assays showing that binding of DLG1 PDZ1-2, but not the PBM-binding deficient mutant, PDZ1-2HA, significantly enhanced GEF activity. (D) Quantification of GEF assays testing the role of the DLG1 PDZ domains in regulation of Ephexin4 activity shown in C. All results are expressed as mean ± SD. ns, not significant, ****P < 0.0001, **P < 0.01, *P < 0.1. (E) HEK 293T cells were transfected with the indicated plasmids. Active RhoG was coprecipitated with GST-ELMO2-RBD and detected by immunoblotting (IB). (F) Schematic diagram showing the engineering of a GS linker (GSGSGSGSGS) between HC and PBM in Ephexin4DPSH. (G) Representative in vitro GEF assays of DPSHGS with or without DLG1 PDZ1-2. Note that all GEF assays were performed using three independent protein preparations with at least duplicate measurements.
We then investigated the mechanism by which PDZ binding relieved the autoinhibited conformation of Ephexin4DPSH. Since the PBM is physically close to the SH3–HC inhibitory module (the linker between the PBM and HC contains only two residues) (SI Appendix, Fig. S5), we hypothesized that binding between PDZ and Ephexin4 could result in a steric clash between PDZ and the Ephexin4 DH domain. More specifically, we speculated that the DH domain would be pushed away from the inhibitory module (i.e., SH3–HC–PBM) when DLG1 was bound, thus allowing RhoG access. We reasoned that, if the linker between PBM and SH3–HC was long enough, the effects of the PDZ–PBM interaction on Ephexin4 autoinhibition would be limited. To test this hypothesis, we inserted a flexible loop (GSGSGSGSGS) between HC and PBM (referred to as DPSHGS) (Fig. 3F). We found that, although DPSHGS bound to PDZ1-2 with a similar affinity as PBM did (Fig. 3 A and B), the binding of PDZ1-2 no longer activated DPSHGS (Fig. 3G), thereby substantiating our hypothesis.
The further question remained as to whether this mechanism of Ephexin4 activation was conserved among other PDZ protein(s). Previously, the PDZ-containing protein Tip1 was reported to bind to the Ephexin4 PBM and subsequently activate Ephexin4 in human papillomavirus (HPV) viral carcinogenesis (33). We verified this interaction by showing that Tip1 PDZ bound to Ephexin4DPSH with a comparable affinity to that of DLG1 PDZ1-2 (Fig. 3B and SI Appendix, Fig. S7). The effective increase in the GEF activity of Ephexin4DPSH by Tip1 PDZ indicated that Tip1 employs a similar mechanism to that utilized by DLG1 PDZ1-2 in the activation of Ephexin4 (SI Appendix, Fig. S7). Collectively, these results indicated that C-terminal inhibition of Ephexin4 could be relieved by binding of type-I PDZ, which results in synergistic promotion of its GEF activity toward RhoG.
Characterization of the N-Terminal Inhibition in Ephexin4.
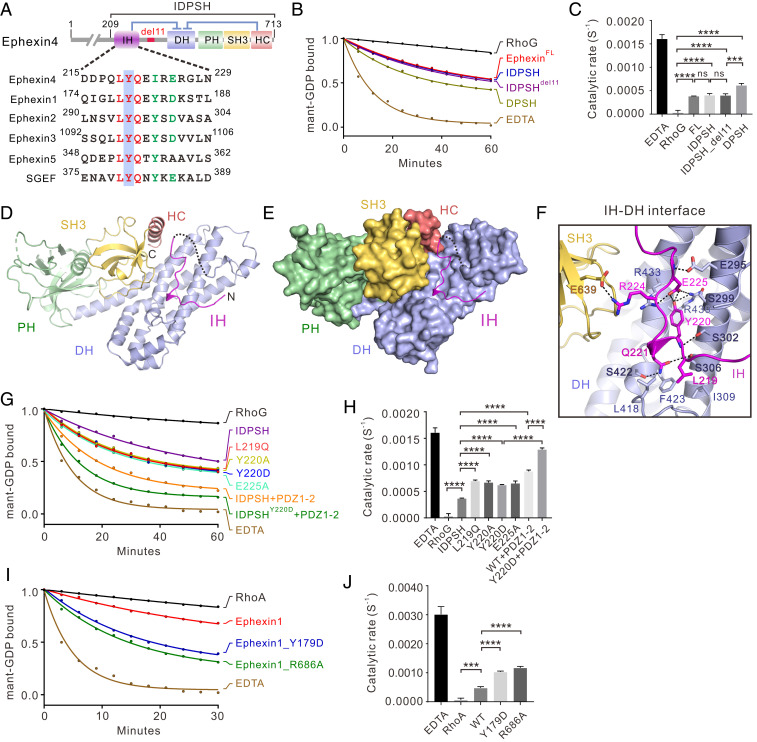
Several lines of evidence have shown that the IH domain preceding the catalytic DH domain plays an inhibitory role in Ephexin1-3, but whether it is present in Ephexin4 and inhibits its GEF activity has not yet been established. Careful sequence analysis revealed that the IH domain, especially the “LYQ” motif, is conserved among all Ephexin family proteins, as well as SGEF (Fig. 4A). To evaluate the potentially inhibitory function of the IH domain in Ephexin4, we compared the GEF activity between Ephexin4DPSH and Ephexin4209-713 which includes both the IH and DPSH domains (Ephexin4IDPSH hereafter) (Fig. 4A). We found that Ephexin4IDPSH exhibited a reduction of GEF activity toward RhoG, compared with Ephexin4DPSH (Fig. 4 B and C), suggesting that the IH domain also plays an inhibitory role in Ephexin4. Notably, Ephexin4IDPSH displayed a similar GEF activity to that of full-length Ephexin4 (Fig. 4 B and C), indicating that Ephexin4IDPSH likely represents the complete autoinhibited form of Ephexin4.
Fig. 4.
Double inhibition mechanism in Ephexin4. (A) Schematic diagram of the domain organization of Ephexin4. The amino acid sequence alignment of the N-terminal inhibitory domain of Ephexins and SGEF is also shown, with identical and conserved residues indicated by red and green, respectively. The tyrosine residue which may be phosphorylated in vivo is highlighted with a blue box. (B) Representative in vitro GEF activity assays of full-length and truncated variants of Ephexin4 (IDPSH, DPSH, IDPSHdel11, and DPSH). (C) Quantification of GEF experiments using various forms of Ephexin4 shown in B. Results are expressed as mean ± SD. ns, not significant, ****P < 0.0001, ***P < 0.001. (D) Ribbon diagram representation of the Ephexin4IDPSH structure. (E) The combined surface and ribbon representations of the structure of the Ephexin4IDPSH. (F) Close-up view of the interface between IH and DH. (G) Representative in vitro GEF assays showing that fully autoinhibited Ephexin4 requires both N- and C-terminal inhibition and can be activated independently. (H) Quantification of GEF assays testing the role of the two inhibitory elements in regulation of Ephexin4 activity shown in G. All results are expressed as mean ± SD. ****P < 0.0001. (I) Mutation of functionally conserved residues in Ephexin1 that correspond to residues in the Ephexin4 inhibition interfaces led to significantly increased Ephexin1 GEF activity toward RhoA in vitro. (J) Quantification of GEF assays testing the role of residues at the two inhibitory interfaces in regulation of Ephexin1 GEF activity shown in I. All results are expressed as mean ± SD. ****P < 0.0001, ***P < 0.001. Note that all GEF experiments were performed using three independent protein preparations with at least duplicate measurements.
Structure of the Complete Autoinhibited Ephexin4.
To uncover the molecular basis underlying the fully autoinhibited state of Ephexin4, we first attempted to purify and crystallize a series of constructs, including the native full-length Ephexin4 and Ephexin4IDPSH. However, efforts to crystallize these proteins were unsuccessful, and we found instead that a truncated form of IDPSH carrying a deletion of an 11-residue flexible loop between the IH and DH domains (amino acids 242 to 252) (i.e., IDPSHdel11) could be successfully crystallized. It is noteworthy that IDPSHdel11 displayed similar GEF activity as that of IDPSH_WT (Fig. 4 B and C), which implied that this deletion variant represents the complete autoinhibited Ephexin4. The structure of IDPSHdel11 was solved using the molecular replacement method, with the DPSH structure as the search model (Table 1).
No significant conformational differences between DPSH and IDPSH were observed in the DH–PH–SH3–HC interdomain interaction network. In the structure, the IH domain adopts an extended loop conformation instead of a previously predicted helix to occupy the pocket created by the DH and SH3–HC (Fig. 4 D and E). Direct contact between the IH and DH obstructs RhoG binding (SI Appendix, Fig. S8), which reasonably explains why the IH further limited Ephexin4 GEF activity. The electron densities of most residues of IH were well defined (SI Appendix, Fig. S2). Specifically, structural analysis showed that L219IH forms hydrophobic contacts with L418, F423, and I309 from DH while Y220IH forms polar interaction networks with R430DH, R433DH, and E225IH. In addition, the main chain of Y220IH forms an additional hydrogen bond with S302DH. Q221IH forms two hydrogen bonds with S422DH and S306DH (Fig. 4F). Moreover, R224IH binds to E639SH3 via an electrostatic interaction (Fig. 4F).
Consistent with our structural analysis, replacement of the L219IH hydrophobic residue with a polar residue Gln (L219Q) significantly relieved the autoinhibition (Fig. 4 G and H and SI Appendix, Fig. S4). Substitution of Y220IH with either Ala or Asp resulted in an obvious increased GEF activity (Fig. 4 G and H and SI Appendix, Fig. S4). In addition, the E225A substitution could also activate the GEF activity, most likely due to disruption of the aforementioned polar interaction networks (Fig. 4 G and H and SI Appendix, Fig. S4). These data suggested that the key residues at the IH–DH interface are essential for IH-mediated suppression of Ephexin4 GEF activity. Notably, these key residues are highly conserved in Ephexin4 from different species (Fig. 4A and SI Appendix, Fig. S5), which indicates that IH-mediated inhibition is conserved during evolution.
A Double Inhibitory Mechanism Is Conserved among Ephexins and SGEF.
Our above findings illustrated the respective mechanisms of autoinhibition mediated by SH3–HC (C-terminal inhibition) and by IH (N-terminal inhibition), both of which are required for repression of Ephexin4 GEF activity. In line with structural analysis, our biochemical data showed that the addition of PDZ1-2 further increased the GEF activity of IDPSHY220D (i.e., in this construct, IH-mediated inhibition is relieved) (Fig. 4 G and H). Moreover, the IDPSHY220D/PDZ1-2 group exhibited much greater levels of guanine nucleotide exchange than did the IDPSHWT/PDZ1-2 group (Fig. 4 G and H). These data indicated that the two inhibitory modes may work independently. To further verify this point, we compared the GEF activity of IDPSH, IDPSH_Y220D, and DPSH. We reasoned that, if the two inhibitory modes operate independently, loss of one inhibitory element would not destabilize the other. Satisfyingly, IDPSHY220D displayed a similar GEF activity as DPSH (SI Appendix, Fig. S9). These results collectively support the conclusion that Ephexin4 adopts a double inhibitory conformation in which N- and C-terminal inhibition may operate independently.
The question then remained as to whether this double inhibitory mechanism is shared in common with other Ephexins and SGEF. Taking advantage of our solved crystal structures, we performed a detailed, structure-based amino acid sequence alignment of all reported Ephexins, as well as SGEF (SI Appendix, Fig. S5). This analysis revealed that the residues involved in the IH–DH and HC–DH interfaces are highly conserved among Ephexin1-4 and in SGEF (Fig. 4A and SI Appendix, Fig. S5). Therefore, we reasonably hypothesized that Ephexin1-3 and SGEF should share the same or highly similar inhibitory mechanisms. We chose Ephexin1 to verify our prediction. In in vitro GEF assays, Ephexin1 showed effective GEF activity toward RhoA (Fig. 4I), which was in agreement with previous reports (13, 25). Substitution of Y179 (corresponding to Y220 in Ephexin4IH) (Fig. 4A) with Asp dramatically enhanced the capacity of Ephexin1 to catalyze nucleotide exchange on RhoA (Fig. 4 I and J). Replacement of R686 (corresponding to R706 in Ephexin4HC) (SI Appendix, Fig. S5) with Ala also substantially increased the GEF activity of Ephexin1 (Fig. 4 I and J). Taken together, these results indicated that the double inhibitory mechanism is highly conserved among other Ephexins and SGEF.
Relief of Ephexin4 Autoinhibition Promotes Cell Migration.
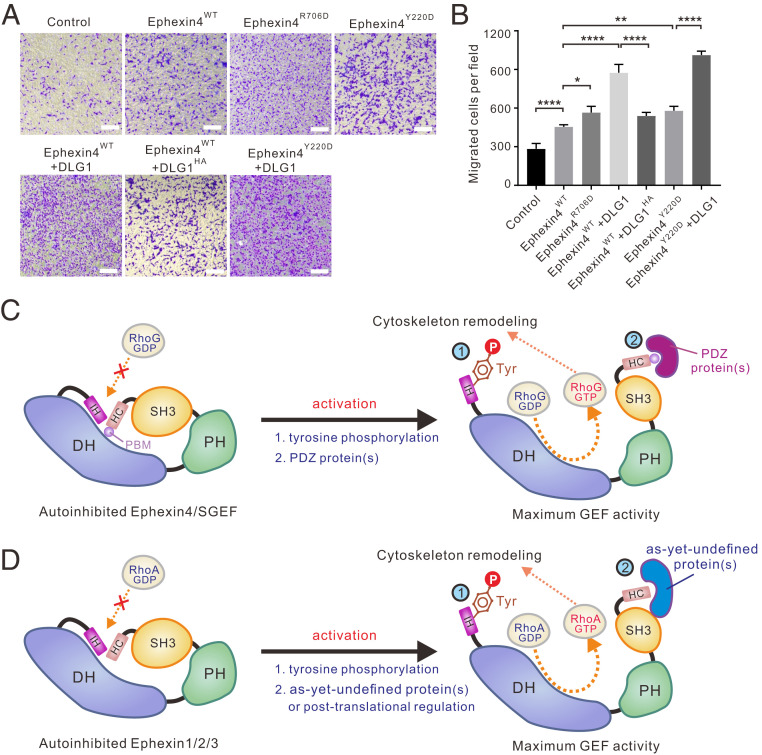
Since Ephexin4 acts downstream of EphA2 to promote cell migration through activation of RhoG, we next used cell migration assays to examine the biological relevance of the double inhibition and activation mechanisms of Ephexin4 in vivo. In transwell migration assays, we found that expression of WT Ephexin4 significantly promoted cell migration (Fig. 5 A and B). We therefore predicted that mutations which relieved autoinhibition of Ephexin4 in our in vitro experiments would further enhance cell migration. In support of this hypothesis, expression of either EphexinR706D (with relieved C-terminal inhibition) or EphexinY220D (with relieved N-terminal inhibition) led to enhanced cell migration (Fig. 5 A and B). Moreover, coexpression of the PDZ protein DLG1 with Ephexin4 remarkably increased the invasive capability of cells. The increased migration rate was most likely caused by DLG1-mediated relief of C-terminal inhibition of Ephexin4 since the Ephexin4-binding deficient mutant of DLG1 (i.e., DLG1HA) did not promote cell migration as effectively as WT DLG1 (Fig. 5 A and B). As expected, coexpression of DLG1 with Ephexin4Y220D caused the highest cell migration rate because Ephexin4 was present in the fully activated state in this scenario (Fig. 5 A and B).
Fig. 5.
Relief of Ephexin4 autoinhibition promotes cell migration. (A) Transwell migration assays were performed to measure the cell migration activities of HeLa cells transfected with the indicated plasmids. (Scale bar: 20 μm.) (B) Quantification of cell migration abilities of the indicated plasmids described in A. Data are expressed as mean ± SD for each group from three independent experiments. ****P < 0.0001, **P < 0.01, *P < 0.1. (C and D) Depiction of the double inhibition and activation mechanisms in Ephexins and SGEF. The double inhibition involves both N-terminal (IH-mediated) and C-terminal (SH3–HC-mediated) inhibitory modes which prevents the small GTPases (e.g., RhoG and RhoA) from binding to the catalytic DH domain. Phosphorylation of a conserved tyrosine residue in IH relieves the N-terminal inhibition in Ephexins and SGEF. Binding of PDZ-containing protein(s) relieves the C-terminal inhibition in Ephexin4 and SGEF, which possess a PBM at the very C terminus (C) while, in other Ephexins (D), association with as-yet-undefined protein(s) or posttranslational modifications may contribute to the activation process.
Discussion
Ephexin family RhoGEFs function downstream of Eph family tyrosine kinase receptors, playing essential roles in both physiological and pathological conditions. To achieve precise, spatiotemporal control of Rho signaling in response to diverse upstream signals, the nucleotide exchange activity of the Ephexin family requires tight regulation. Like many other GEFs, Ephexins were reported to be autoregulated through intramolecular interactions; several autoinhibitory mechanisms have been proposed. It remains uncertain, however, whether there is a general inhibitory mechanism applicable to all members of the Ephexin family. In this study, we systematically investigated the autoinhibition and activation mechanisms of Ephexin4, which activates RhoG in response to EphA2 signaling, through a combination of biochemical, biophysical, and cell biological approaches. We demonstrate that complete autoinhibition of Ephexin4 involves both N-terminal IH-mediated and C-terminal SH3–HC-mediated inhibitory modes. Structural investigation provides insights into the molecular basis for the abovementioned double inhibition mode. We then prove that such a double inhibition mode is employed by all members of the Ephexin family, and that SGEF, another RhoGEF for RhoG, also adopts a similar autoinhibitory conformation.
What is the mechanism for activation of Ephexins? Previous reports have shown that the C-terminal proline-rich region of ELMO family proteins can bind to the SH3 domain of Ephexin4. Moreover, this interaction eliminated the steric hindrance caused by the SH3 domain, thus activating Ephexin4 (23). However, purified full-length ELMO2 protein was not able to activate Ephexin4 in our in vitro GEF assays (SI Appendix, Fig. S10), suggesting that the molecular basis by which ELMO activates Ephexin4 is potentially more complex in vivo. In this study, our study reveals that binding of PDZ proteins (e.g., DLG1 or Tip1) to Ephexin4 released its C-terminal inhibition. A steric clash is most likely created by PDZ binding, which results in a conformational change in Ephexin4 that allows RhoG access and consequently stimulates GEF activity (Fig. 5C). A similar activation mechanism may be utilized by SGEF since it also contains a type-I PBM in the C-terminal flank of its SH3–HC domain tandem. Indeed, human DLG was reported to interact with SGEF to activate its GEF activity toward RhoG through PDZ–PBM interaction, which can contribute to HPV-induced malignancy (32). Nevertheless, it may be reasonable to speculate that C-terminal–mediated inhibition of Ephexins lacking the PBM (i.e., Ephexin1-3) cannot be relieved by binding to PDZ protein(s). Alternatively, binding of as-yet-undefined protein(s) to the SH3 or HC may also generate a conformational change in these Ephexins and thereby activate their GEF activities for RhoA (Fig. 5D). Notably, activation of SH3-mediated inhibition by binding to their cognate activator(s) has been observed in other RhoGEFs, such as Asef and Collybistin. During activation of Asef, binding of the armadillo repeats of adenomatous polyposis coli (APC) to the APC-binding region (ABR) adjacent to Asef SH3 results in a steric clash between DH and APC, leading to a conformational change in Asef and subsequent stimulation of its GEF activity (34). In Collybistin, SH3-mediated inhibition is relieved by binding of a proline-rich region in synaptic adhesion molecule Neuroligin-2 to the SH3 domain of Collybistin, thus activating its GEF activity (35). It cannot be ruled out that posttranslational modification may also participate in the regulation of SH3-mediated inhibition. It has been shown that bivalent association of the C-terminal SH3 domain with its DH and PH domains inhibited the GEF activity of Vav (19, 36). Phosphorylation of multiple tyrosine residues in the SH3 domain relieved Vav autoinhibition, thus suggesting another regulatory mechanism that potentially contributes to autoinhibition and activation (36).
The conserved tyrosine residue in the IH domain of Ephexin1-3 has been shown to be phosphorylated by Src in vitro and in vivo in response to Eph signaling (9, 25). Given the close connection between Ephexins and Eph signaling pathways, it would not be surprising that the corresponding conserved tyrosine residue in Ephexin4 may also be phosphorylated by Eph kinases and/or Src family members. Mechanistically, structural analyses conducted here indicate that phosphorylation of this Tyr (Y220 in Ephexin4) would disrupt the polar interaction network at the DH–IH interface in Ephexins (Fig. 4F). In accordance with this analysis, a phosphomimetic mutant of either Ephexin4 or Ephexin1 greatly enhanced their corresponding GEF activities (Fig. 4). Therefore, we propose that phosphorylation of the conserved tyrosine residue in the IH domain of Ephexins is a general activation mechanism for relieving the N-terminal inhibitory mode (Fig. 5 C and D).
In summary, the results presented here not only provide mechanistic details that broaden our understanding of the allosteric regulation of Ephexin family RhoGEFs, but also may offer possible insights for the pathogenesis of diseases related to mutations in Ephexins. The double inhibition and activation mechanisms provide a multilayered regulatory system for spatiotemporal control of enzymatic activities of Ephexins in response to distinct external stimuli. Moreover, since the two inhibitory elements operate independently, multiple upstream signals might also independently relieve each inhibitory mode to fine-tune the GEF activity of Ephexins during cell signaling.
Methods
Constructs.
Mouse Arhgef16 (encoding Ephexin4; GenBank: NP_001106215.1), RhoG (GenBank: NM_019566.3), RhoA (GenBank: NM_016802.5), and TAX1BP3 (encoding TIP1; GenBank: NM_029564.2) were amplified from mouse brain complementary DNA (cDNA) library. Rat DLG1 (GenBank: AM710296.2) was a gift from Mingjie Zhang, Hong Kong University of Science and Technology, Hong Kong, China. Human NGEF (encoding Ephexin1) and Arhgef26 (encoding SGEF) were in a friendly way provided by Jiahuai Han, Xiamen University, Xiamen, China. Various constructs of these genes were amplified by a standard PCR method and cloned into pGEX-4T-1, pET-32M3C (a modified pET-15b vector with an N-terminal Trx-His6 tag), pET-M3C (with an N-terminal His6 tag), pEGFP-C1, pCDNA3.1-Flag, or pCMV-Myc vector. Mutations were created using the site-directed mutagenesis method and confirmed by DNA sequencing.
Protein Expression and Purification.
Recombinant RhoG protein was expressed in Escherichia coli BL21 (DE3) cells in the presence of a chaperone expression plasmid pG-Tf2. Expression of the Tf2 chaperone protein was induced with 10 ng/mL tetracycline at 37 °C (optical density at 600 nm [OD600]: 0.4 to 0.6) for 4 h. Cells were then cooled to 16 °C and induced with isopropyl-β-d-thiogalactoside (IPTG) at a final concentration of 0.2 mM for 18 h. Other recombinant proteins were expressed at 16 °C for 18 h in E. coli BL21 (DE3) cells, induced by 0.2 mM IPTG. The His6-tagged proteins were purified by Ni2+-nitrilotriacetic acid (NTA) agarose affinity chromatography followed by Superdex-200 26/60 size-exclusion chromatography (SEC). GST-tagged proteins were purified by GSH-Sepharose affinity chromatography follower by SEC purification. For the crystallography, various Trx-tagged Ephexin4 proteins were cleaved by HRV-3C protease at 4 °C overnight, and the Trx-His6 was then removed by another step of SEC purification in a buffer containing 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM (ethylenedinitrilo)tetraacetic acid (EDTA) and 1 mM dithiothreitol (DTT). For selenomethionine-labeled Ephexin4DPSH (Se-Met Ephexin4DPSH), pET-32M3C-Ephexin4DPSH was expressed in E. coli B834 (DE3) cells using M9 medium supplemented with 4 g/L glucose, 5 mg/L VB1, 2 mM MgSO4, 0.1 mM CaCl2, 100 mg/L lysine, 100 mg/L threonine, 100 mg/L phenylalanine, 100 mg/L seleomethionine, 50 mg/L leucine, 50 mg/L isoleucine, and 50 mg/L valine, at 16 °C for 18 h, induced by 0.2 mM IPTG. Se-Met Ephexin4DPSH was purified by Ni2+-NTA agarose affinity chromatography followed by a Superdex-200 26/60 SEC. Trx-His6 was cleaved by HRV-3C protease at 4 °C overnight and then removed by another step of SEC purification in a buffer containing 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA and 1 mM DTT.
ITC Assay.
ITC assays were carried out on a MicroCal iTC200 system (Malvern Panalytical) at 25 °C. Various PDZ and Ephexin4 fragments were dissolved in the buffer containing 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 1mM DTT. The PDZ proteins (∼500 μM) were loaded into syringe. In each titration, 2 μL aliquot of PDZ proteins were injected into the cell placed with the various Ephexin4 fragments (∼50 μM). The time interval was 120 s in each titration to ensure the titration peak returned to the baseline. Data were analyzed with a one-site binding model by Origin 7.0.
Cell-Based GEF Activity Assay.
Flag-tagged RhoG, WT, and mutation forms of GFP-tagged Ephexin4, and Myc-tagged DLG1 and its mutants were cotransfected into HEK293T cells as indicated. Cells were harvested after 24 h and lysed by an ice-cold cell lysis buffer [50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), pH 7.4, 150 mM NaCl, 10% glycerol, 2 mM MgCl2, 1% Triton X-100, and protease inhibitor mixture]. After centrifugation, the supernatants were incubated with GST-tagged ELMO-RBD protein preloaded to 30 μL of GSH Sepharose 4B slurry beads in the lysis buffer at 4 °C for 1 h. After extensive wash, the captured proteins (e.g., active RhoG or active RhoA) were eluted by 20 µL of 2× sodium dodecyl sulfate polyacrylamide gel electrophoresis loading dye and detected by Western blot using the anti-Flag antibody (Sigma), GFP antibody (Santa Cruz), and Myc-antibody (Santa Cruz), respectively.
Crystallization, Data Collection, and Structure Determination.
All crystals were yielded by the sitting-drop vapor diffusion method with drops consisting of 0.5 μL of protein (∼10 mg/mL) and 0.5 μL of reservoir solution at 16 °C. The best crystals of both native and Se-Met Ephexin4DPSH were grown in 1.5 M ammonium sulfate, 100 mM Tris, pH 7.5, 12% glycerol. The Ephexin4IDPSH crystals were grown in 2.8 M sodium acetate, pH 7.0. Crystals were cryoprotected in the corresponding reservoir solution with 25% glycerol. Diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) and processed by XDS (37).
The structure of Ephexin4DPSH was determined by SAD phasing by CRANK2 (38) in the CCP4 suite. Twenty-three of the 28 expected Set-Met sites were identified using the 3.2-Å dataset. The model was built and refined using the native 2.39-Å dataset. The structure of Ephexin4IDPSH was solved by the molecular replacement method using the native DPSH structure as searching model by PHASER (39). Rigid-body refinement was performed using REFMAC in CCP4 (40), and the density maps were examined in Coot (41). Further refinement was performed using PHENIX (42) and Coot (41) iteratively. For PHENIX refinement, the translation, rotation, and screw-rotation parameters were used. The final models were validated by MolProbity (43) to make sure the Rwork/Rfree values, Molprobity scores, geometry, and stereochemistry were favorable. The final refinement statistics of these structures are listed in Table 1. Structural diagrams were prepared by PyMOL (https://pymol.org/2/).
In Vitro GEF Assay.
All GEF assays were carried out on a Microplate Reader (Bio Tek) at 25 °C using a buffer containing 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM DTT, and 5 mM MgCl2. N-methylanthraniloyl (MANT)-GDP loading was performed as previously described (44). Briefly, 100 μM small GTPase (RhoG or RhoA) was incubated with a threefold molar excess of MANT-GDP (Invitrogen) on ice in the buffer of 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM DTT, 5 mM EDTA. After incubation on ice for 30 min, MgCl2 (final concentration of 15 mM) was added to stop the MANT-GDP loading. The mixture was then purified through a PD-10 column to remove the excess MANT-GDP. In each GEF assay, MANT-GDP–bound RhoG or RhoA (2 μM) was incubated with their corresponding GEF proteins (Ephexin4: 5 μM, SGEF: 3 μM, Ephexin1: 0.5 μM) with or without DLG1 proteins (5 to 20 μM), in the presence of excess (20 μM) Gpp(NH)p, a nonhydrolyzable analog of GTP. The EDTA (20 mM) group served as a positive control in which EDTA was added to deplete the Mg2+ from the nucleotide binding site of the GTPases, which led to a complete dissociation of the bound MANT-GDP from the GTPase. The fluorescence intensity of 20 mM EDTA at 0 min (the beginning of reaction) and 60 min (the end of reaction) were set as 100% and 0%, respectively. The observed decrease of the fluorescence signal (excitation at 360 nm and emission 450 nm) due to the release of MANT-GDP from RhoG or RhoA was fitted as a single-exponential decay mode using GraphPad Prism to determine the observed pseudo first-order exchange rate constant (Kobs) using the equation
where It is the emission intensity at time t, I0 the initial emission intensity, and I∞ the final emission intensity. All nucleotide exchange experiments were performed using three independent protein preparations with at least duplicate measurements.
Cell Migration Assay.
Cell migration experiments were carried out using transwell membrane filter inserts (8-μm pore size; Corning Costar). Then, 1 × 105 HeLa cells were seeded into the upper chamber with serum-free medium (Dulbecco’s modified Eagle’s medium [DMEM]). DMEM containing 1% fetal bovine serum (FBS) was placed into the bottom chamber. After incubation for 16 to 18 h at 37 °C, cells in the upper chamber were wiped by cotton buds, and cells adhering to the bottom surface of the membrane were washed once with phosphate-buffered saline (PBS) and fixed by 100% methanol for 10 min. Cells were washed once again with PBS and then stained with Crystal Violet Staining Solution (Beyotime Biotechnology) for 10 min. The migrated cells were counted under a light microscope from five random fields of each well. Data are expressed as mean ± SD for each group from three independent experiments (****P < 0.0001, **P < 0.01, *P < 0.1).
Quantification and Statistical Analysis.
For in vitro GEF assays in this study, all assays were performed using three independent protein preparations with at least duplicate measurements. Values are expressed as mean ± SD (ns, not significant, ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.1). For transwell migration assays, all results were expressed as mean ± SD for each group from three independent migration assays (ns, not significant, ****P < 0.0001, **P < 0.01, *P < 0.1).
Supplementary Material
Acknowledgments
We thank the SSRF BL18U1 and BL19U1 for X-ray beam time; the staff members of the Large-Scale Protein Preparation System and Molecular Imaging System at the National Facility for Protein Science in Shanghai; the Zhangjiang laboratory for providing technical support and assistance in data collection and analysis; and Prof. Mingjie Zhang for valuable discussion and critical comments on the manuscript. This work was supported by grants from the National Key R&D Program of China: 2018YFA0507900 (to J.Z.) and 2019YFA0508402 (to C.W.); grants from the National Natural Science Foundation of China: U2032122 and 31770779 (to J.Z.) and 31670734 and 91953110 (to C.W.); a grant from Science and Technology Commission of Shanghai Municipality: 20S11900200 (to J.Z.); a grant from University of Science and Technology of China Research Funds of Double First-Class Initiative: YD9100002006 (to C.W.); and a grant from the Fundamental Research Funds for the Central Universities: WK9100000029 (to C.W.). C.W. is supported by the Chinese Academy of Sciences Pioneer Hundred Talents Program.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. A.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024465118/-/DCSupplemental.
Data Availability
The atomic coordinates of the Ephexin4DPSH and Ephexin4IDPSH have been deposited in the Protein Data Bank under the accession codes 7CSO and 7CSP, respectively. All other study data are included in the article and/or SI Appendix.
References
- 1.Jaffe A. B., Hall A., Rho GTPases: Biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21, 247–269 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Lawson C. D., Ridley A. J., Rho GTPase signaling complexes in cell migration and invasion. J. Cell Biol. 217, 447–457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spence E. F., Soderling S. H., Actin out: Regulation of the synaptic cytoskeleton. J. Biol. Chem. 290, 28613–28622 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodge R. G., Ridley A. J., Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 17, 496–510 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Schaks M., Giannone G., Rottner K., Actin dynamics in cell migration. Essays Biochem. 63, 483–495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos J. L., Rehmann H., Wittinghofer A., GEFs and GAPs: Critical elements in the control of small G proteins. Cell 129, 865–877 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Fritz R. D., Pertz O., The dynamics of spatio-temporal Rho GTPase signaling: Formation of signaling patterns. F1000 Res. 5, 749 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim K., Lee S. A., Park D., Emerging roles of Ephexins in physiology and disease. Cells 8, E87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahin M., et al., Eph-dependent tyrosine phosphorylation of ephexin1 modulates growth cone collapse. Neuron 46, 191–204 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Kania A., Klein R., Mechanisms of ephrin-Eph signalling in development, physiology and disease. Nat. Rev. Mol. Cell Biol. 17, 240–256 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Pasquale E. B., Eph receptor signalling casts a wide net on cell behaviour. Nat. Rev. Mol. Cell Biol. 6, 462–475 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Shi L., et al., Ephexin1 is required for structural maturation and neurotransmission at the neuromuscular junction. Neuron 65, 204–216 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shamah S. M., et al., EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell 105, 233–244 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Fu W. Y., et al., Cdk5 regulates EphA4-mediated dendritic spine retraction through an ephexin1-dependent mechanism. Nat. Neurosci. 10, 67–76 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Hiramoto-Yamaki N., et al., Ephexin4 and EphA2 mediate cell migration through a RhoG-dependent mechanism. J. Cell Biol. 190, 461–477 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawai H., et al., Ephexin4-mediated promotion of cell migration and anoikis resistance is regulated by serine 897 phosphorylation of EphA2. FEBS Open Bio 3, 78–82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margolis S. S., et al., EphB-mediated degradation of the RhoA GEF Ephexin5 relieves a developmental brake on excitatory synapse formation. Cell 143, 442–455 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porter A. P., Papaioannou A., Malliri A., Deregulation of Rho GTPases in cancer. Small GTPases 7, 123–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasquale E. B., Eph receptors and ephrins in cancer: Bidirectional signalling and beyond. Nat. Rev. Cancer 10, 165–180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sell G. L., Schaffer T. B., Margolis S. S., Reducing expression of synapse-restricting protein Ephexin5 ameliorates Alzheimer’s-like impairment in mice. J. Clin. Invest. 127, 1646–1650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J. C., et al., Increased EphA4-ephexin1 signaling in the medial prefrontal cortex plays a role in depression-like phenotype. Sci. Rep. 7, 7133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debily M. A., et al., Expression and molecular characterization of alternative transcripts of the ARHGEF5/TIM oncogene specific for human breast cancer. Hum. Mol. Genet. 13, 323–334 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Kim K., et al., Intermolecular steric inhibition of Ephexin4 is relieved by Elmo1. Sci. Rep. 7, 4404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim K., et al., The intermolecular interaction of Ephexin4 leads to autoinhibition by impeding binding of RhoG. Cells 7, E211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yohe M. E., Rossman K., Sondek J., Role of the C-terminal SH3 domain and N-terminal tyrosine phosphorylation in regulation of Tim and related Dbl-family proteins. Biochemistry 47, 6827–6839 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yohe M. E., et al., Auto-inhibition of the Dbl family protein Tim by an N-terminal helical motif. J. Biol. Chem. 282, 13813–13823 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Huang D., et al., GLI2 promotes cell proliferation and migration through transcriptional activation of ARHGEF16 in human glioma cells. J. Exp. Clin. Cancer Res. 37, 247 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bralten L. B., et al., Absence of common somatic alterations in genes on 1p and 19q in oligodendrogliomas. PLoS One 6, e22000 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J., et al., Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerami E., et al., The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng Z., et al., Structure of BAI1/ELMO2 complex reveals an action mechanism of adhesion GPCRs via ELMO family scaffolds. Nat. Commun. 10, 51 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishna Subbaiah V., et al., The invasive capacity of HPV transformed cells requires the hDlg-dependent enhancement of SGEF/RhoG activity. PLoS Pathog. 8, e1002543 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliver A. W., et al., The HPV16 E6 binding protein Tip-1 interacts with ARHGEF16, which activates Cdc42. Br. J. Cancer 104, 324–331 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z., et al., Structural basis for the recognition of Asef by adenomatous polyposis coli. Cell Res. 22, 372–386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soykan T., et al., A conformational switch in collybistin determines the differentiation of inhibitory postsynapses. EMBO J. 33, 2113–2133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barreira M., et al., The C-terminal SH3 domain contributes to the intramolecular inhibition of Vav family proteins. Sci. Signal. 7, ra35 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skubák P., Pannu N. S., Automatic protein structure solution from weak X-ray data. Nat. Commun. 4, 2777 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy A. J., et al., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winn M. D., et al., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Adams P. D., et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen V. B., et al., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanie T., Jackson P. K., Guanine nucleotide exchange assay using fluorescent MANT-GDP. Bio Protoc. 8, e2795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates of the Ephexin4DPSH and Ephexin4IDPSH have been deposited in the Protein Data Bank under the accession codes 7CSO and 7CSP, respectively. All other study data are included in the article and/or SI Appendix.