Significance
Growth and differentiation factor-15 (GDF-15) acts through its receptor, GFRAL, expressed within the area postrema to reduce food intake and body weight. However, we do not yet understand GFRAL neuron transcriptional profile, regulation, the direct downstream target sites of GFRAL neurons, or the necessary cell types required for GDF-15 activity. Herein, we identify the genetic signature of GFRAL neurons and show that these cells are positively regulated by a number of aversive stimuli, project to both the nucleus of the solitary tract and more densely to the parabrachial nucleus, and that CGRPPBN neurons are required for the aversive and anorectic effects of GDF-15.
Keywords: GFRAL, GDF-15, area postrema, obesity, CGRP
Abstract
The TGFβ cytokine family member, GDF-15, reduces food intake and body weight and represents a potential treatment for obesity. Because the brainstem-restricted expression pattern of its receptor, GDNF Family Receptor α–like (GFRAL), presents an exciting opportunity to understand mechanisms of action for area postrema neurons in food intake; we generated GfralCre and conditional GfralCreERT mice to visualize and manipulate GFRAL neurons. We found infection or pathophysiologic states (rather than meal ingestion) stimulate GFRAL neurons. TRAP-Seq analysis of GFRAL neurons revealed their expression of a wide range of neurotransmitters and neuropeptides. Artificially activating GfralCre-expressing neurons inhibited feeding, decreased gastric emptying, and promoted a conditioned taste aversion (CTA). GFRAL neurons most strongly innervate the parabrachial nucleus (PBN), where they target CGRP-expressing (CGRPPBN) neurons. Silencing CGRPPBN neurons abrogated the aversive and anorexic effects of GDF-15. These findings suggest that GFRAL neurons link non–meal-associated pathophysiologic signals to suppress nutrient uptake and absorption.
Several lines of evidence link high circulating levels of the cytokine, growth and development factor 15 (GDF-15), to the reduced hunger, decreased food intake, and weight loss that occur in a variety of cancers (1). Consequently, GDF-15 and its analogs are candidates that could decrease feeding and promote weight loss in individuals with obesity. Indeed, exogenous GDF-15 treatment leads to substantial weight loss in mice, rats, and nonhuman primates (2–5).
The identification of the receptor that mediates the anorexigenic effects of GDF-15, glial cell line-derived neurotrophic factor (GDNF) family receptor α–like (GFRAL), has provided an opportunity to understand mechanisms of food-intake suppression. While a wide range of tissues synthesize and secrete GDF-15 in response to cellular stress (6–9), GFRAL has a uniquely narrow expression pattern restricted to a small population of central nervous system (CNS) neurons. Importantly, the hypothalamus and other forebrain regions contain no GFRAL, but rather GFRAL-expressing cells reside in the hindbrain, primarily in the area postrema (AP), with a smaller number in the adjacent nucleus tractus solitarius (NTS) (2–5).
To date, the study of GFRAL neurons has largely been conducted through the use of GDF-15; in a range of animal models, exogenous GDF-15 reduces food intake, slows gastric emptying, and produces aversive responses (2, 5, 10–15). High GDF-15 doses also activate neurons in a range of CNS sites, as measured by FBJ murine osteosarcoma viral oncogene homolog (FOS)-immunoreactivity (-IR), including the AP/NTS, parabrachial nucleus (PBN), paraventricular nucleus of the hypothalamus (PVN), central nucleus of the amygdala (CeA), and bed nucleus of the stria terminalis (1, 4). While the PBN is one site of direct communication (13), neither the entirety of downstream projection sites from APGFRAL neurons or the cell types that mediate the potent anorexic and aversive effects of GDF-15 have been identified.
Because the neurons targeted by GDF-15 modulate feeding behavior and body weight, they represent an attractive target for the treatment of obesity. However, knowing that GDF-15 is produced under pathological conditions and is aversive raises the concern that activating this signaling pathway could have adverse side effects. We generated genetic tools that allow direct manipulation of GFRAL-expressing neurons to facilitate a better understanding of how they inhibit appetite.
Results
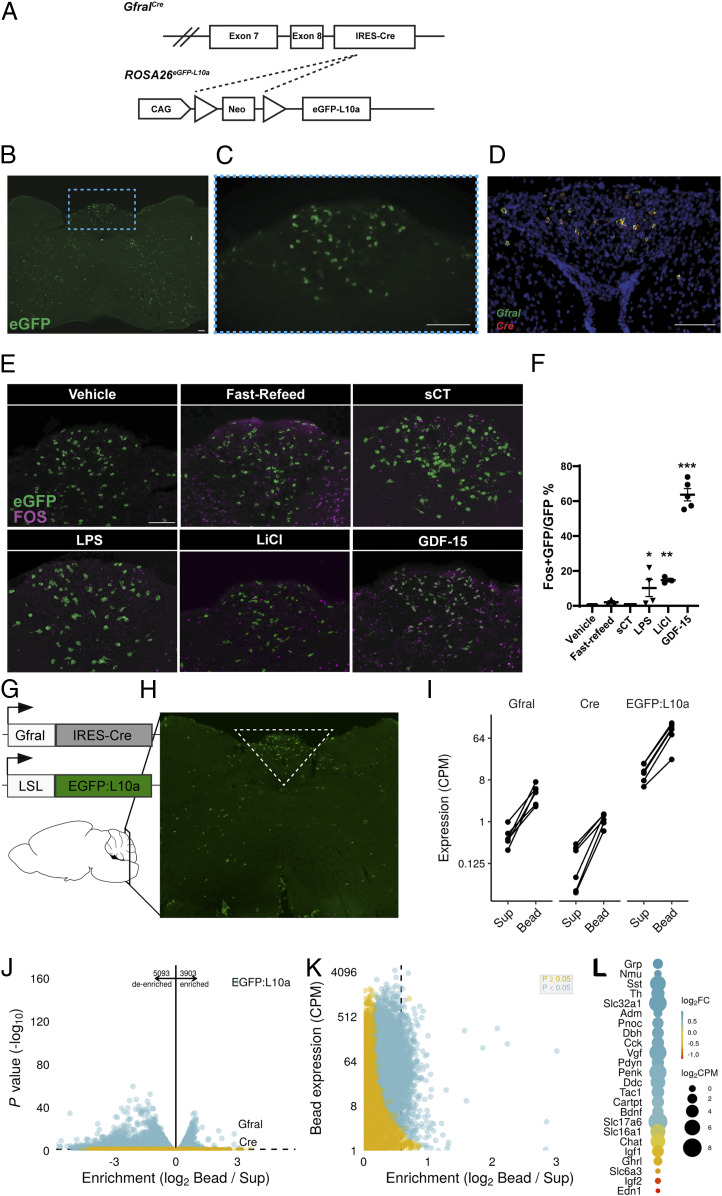
While recent studies have elucidated some aspects of the biology of GDF-15 and its receptor, GFRAL, we know little about the regulation, function, and downstream mediators of GFRAL-expressing neurons. To study GFRAL neurons independently of GDF-15 biology, we targeted Cre recombinase to the Gfral locus. Breeding GfralCre onto the Cre-inducible ROSA26eGFP-L10a reporter background [(16) GFRALeGFP mice, Fig. 1A] revealed the expected presence of many enhanced green florescent protein (eGFP)-expressing neurons within the AP as well as scattered eGFP-containing cells elsewhere within the hindbrain (Fig. 1 B and C). In situ hybridization (ISH) revealed the restriction of Gfral expression to the AP/NTS [in agreement with previous observations (2–5)], where it colocalized with Cre messenger RNA (mRNA) (Fig. 1D). The presence of eGFP reporter expression outside of the AP/NTS presumably indicates some transient developmental expression of GfralCre in these areas (SI Appendix, Fig. S6). Consistent with the postulated crucial role for AP GFRAL neurons in the control of food intake, direct GDF-15 administration to rat AP (but not the NTS) produced a strong anorexic response (SI Appendix, Fig. S1).
Fig. 1.
GDF-15 activates GFRAL neurons. (A) Schematic of the GfralCre-mediated excision of the Lox-Stop-Lox cassette from the ROSA26eGFP-L10a allele to generate GFRALeGFP mice. (B) Representative image of coronal hindbrain (Bregma -7.5) eGFP-IR (green) in GFRALeGFP mice. (C) High magnification of boxed AP region in B. (D) Representative image of ISH for Gfral (green) and Cre (red) transcripts in AP/NTS of GfralCre mice. (E and F) Representative image and quantification of FOS-IR (magenta) in GFRAL (eGFP+; green) neurons from GFRALeGFP mice in response to VEH, Refeeding, sCT, LPS, LiCl, and GDF-15 (n = 3–6). (G and H) Schematic of TRAP-Seq dissection of AP/NTS from GfralCre/+;Rosa26eGFP-L10a mice. (I) Expression of Gfral, Cre, and eGFP-L10a in Sup and immunoprecipitated actively translating ribosomes (bead). (J and K) Analysis of de-enriched and enriched transcripts within GFRAL neurons. (L) Assessment of neurotransmitter enrichment (Log2 Fold change gene expression in Bead compared with Sup) and expression levels in GFRAL neurons. Data are shown as mean ± SEM and analyzed with one-way ANOVA with Dunnet’s post hoc test (F), *P < 0.05, **P < 0.01, and ***P < 0.001. (Scale bar, 100 μm.)
We first sought to define the types of stimuli that activate GFRAL neurons. Unlike other hindbrain cells known to modulate food intake (17–19), refeeding following an overnight fast did not promote the accumulation of FOS-IR in eGFP-labeled AP GFRAL neurons in GFRALeGFP mice and neither did treatment with the appetite-suppressing calcitonin receptor (CALCR) agonist salmon calcitonin (sCT). In contrast, lipopolysaccharide (LPS), LiCl (which causes GI distress), and GDF-15 promoted FOS-IR in many AP GFRAL cells (Fig. 1 E and F). These data suggest that GFRAL neurons do not respond to meal-related signals and that GFRAL cells are distinct from CALCR neurons which respond to sCT and meal-related cues (18). Instead, GFRAL neurons respond to signals associated with pathophysiology, including GDF-15, GI distress, and bacterial infection. These data agree with other recent reports (11, 13).
To better understand the gene signature of brainstem GFRAL neurons, we performed translating ribosomal affinity purification (TRAP)-based RNA sequencing on AP/NTS GFRAL neurons (Fig. 1 G and H). As expected, Gfral, Cre, and eGFP-L10a transcripts were significantly enriched within our pull down (Fig. 1I), validating the approach. In addition, we also found ∼5,000 transcripts that were significantly de-enriched and 4,000 transcripts significantly enriched within GFRAL neurons (Fig. 1 J and K). Specifically examining transcripts involved in the synthesis or trafficking of neurotransmitters or neuropeptides reveals that GFRAL neurons have a complex neurotransmitter complement, including GABA (γ-aminobutyric acid)-ergic markers (Slc32a1), glutamatergic markers (Slc17a6), several neuropeptides (gastrin-releasing peptide [Grp], Neuromedin U [Nmu], and somatostatin [Sst]), and enzymes required for epinephrine synthesis (Fig. 1L).
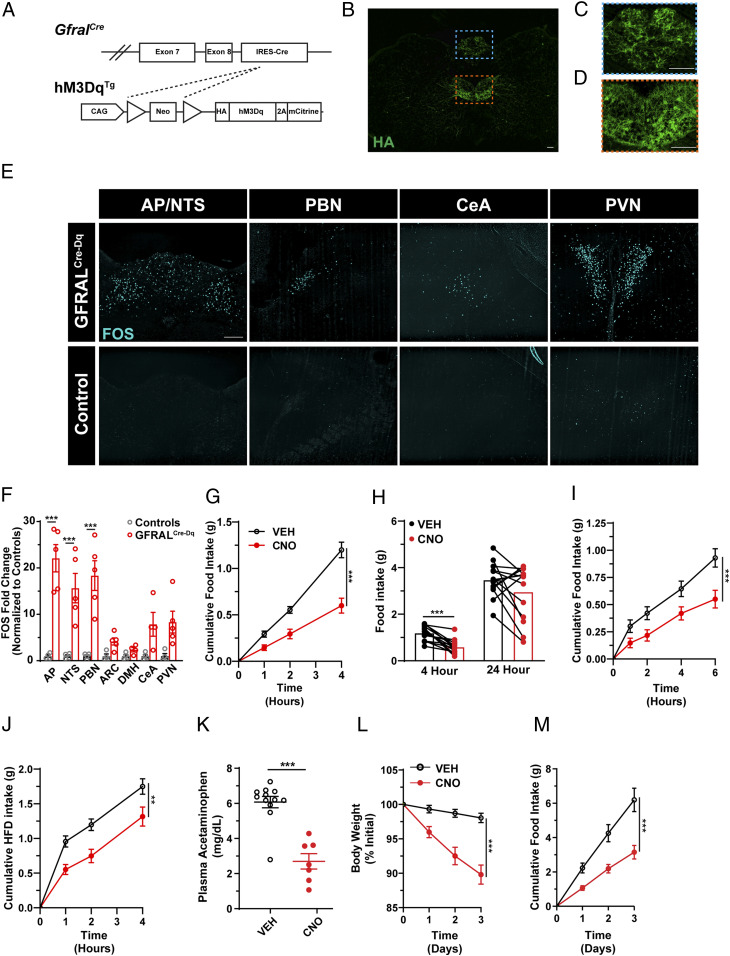
Because the manipulation of neurons in the mouse AP by the stereotaxic injection of viral vectors has proven difficult, we took advantage of the restricted Gfral expression pattern and utilized genetic systems to study AP GFRAL neurons. To study the functional capacity of AP GFRAL neurons, we crossed GfralCre mice onto the Cre-inducible hM3DqTg background (20) to express the activating (hM3Dq) designer receptor exclusively activated by designer drugs (DREADD) in GFRAL neurons (GFRALCre-Dq mice), permitting the activation of these cells by CNO administration (Fig. 2A). While we observed strong DREADD expression within the AP of GFRALCre-Dq mice, we also noted DREADD expression outside of the AP—most prominently in the hypoglossal and facial nuclei (Fig. 2 B–D and SI Appendix, Fig. S2). CNO injection in GFRALCre-Dq mice increased FOS-IR within the AP, consistent with the expected activation of hM3Dq-expressing GFRAL neurons by CNO. Notably, a number of other regions also displayed increased FOS-IR, including the NTS, PBN, CeA, and the PVN (Fig. 2 E and F), which likely constitute regions downstream of GFRAL neurons, as exogenous GDF-15 also promotes FOS-IR in these areas (1, 4).
Fig. 2.
DREADD-mediated activation of Gfralcre neurons suppresses food intake and reduces body weight. (A) Schematic of GfralCre-mediated excision of the Lox-Stop-Lox cassette from the hM3DqTg line to produce GFRALCre-Dq mice. (B and C) Representative image of HA-IR (green) in coronal hindbrain sections (Bregma -7.5) from GFRALCre-Dq mice and higher magnification of AP (C, region in blue dashed box) and hypoglossal nucleus (D, region in orange dashed box). (E and F) Representative image (E) and quantification (F) of FOS-IR (cyan) in the AP/NTS, PBN, CeA, and PVN from CNO-treated (1 mg/kg; ip) GFRALCre-Dq and control animals (n = 4). (G) Food intake over the first 4 h of the dark phase in GFRALCre-Dq mice following either VEH (black lines) or CNO (red lines) injection 30 min prior to the onset of the dark cycle (n = 14). (H) Paired analysis of 4- and 24-h food intake in GFRALCre-Dq animals administered CNO or VEH. (I) Light phase food intake levels following a 10-h overnight fast in GFRALCre-Dq mice administered VEH or CNO (n = 18). (J) Dark phase food intake of a palatable diet (60% fat) in lean GFRALCre-Dq animals injected with either VEH or CNO (n = 14). (K) Plasma acetaminophen concentrations 30 min after oral gavage in controls and GFRALCre-Dq animals administered CNO (n = 7–12). (L and M) Body weight and food intake of DIO GFRALCre-Dq animals injected twice daily with either VEH or CNO (n = 11). Data are shown as mean ± SEM. One-way ANOVA with Dunnet’s post hoc test (F) Two-way ANOVA (G, I, J, L, and M), paired t test (H) or unpaired t test (K) was performed, **P < 0.01, ***P < 0.001. (Scale bar, 100 μm.)
To determine the ability of GFRAL neuron activation to modulate feeding behavior, we administered CNO 30 min prior to the onset of the dark cycle and monitored food intake. In GFRALCre-Dq mice, CNO treatment dramatically suppressed food intake compared to saline injection, although this effect was attenuated by 24 h (Fig. 2 G and H). Importantly, CNO did not alter food intake in control animals (lacking either Cre or DREADD expression, SI Appendix, Fig. S2). We also examined the ability of GFRAL-neuron activation to suppress food intake in the face of an elevated drive to feed. Following a 10-h fast or acute exposure to palatable food (60% fat diet), activation of GFRAL neurons by CNO treatment decreased food intake by ∼50% (Fig. 2 I and J). We also found that CNO treatment of GFRALCre-Dq mice decreased gastric emptying (Fig. 2K).
To determine whether chronic activation of GFRAL neurons could suppress food intake and decrease body weight over longer periods of time, we treated GFRALCre-Dq mice with CNO over 3 d. This CNO-dosing scheme resulted in the continued suppression of food intake and loss of body weight for the duration of the experiment (Fig. 2 L and M).
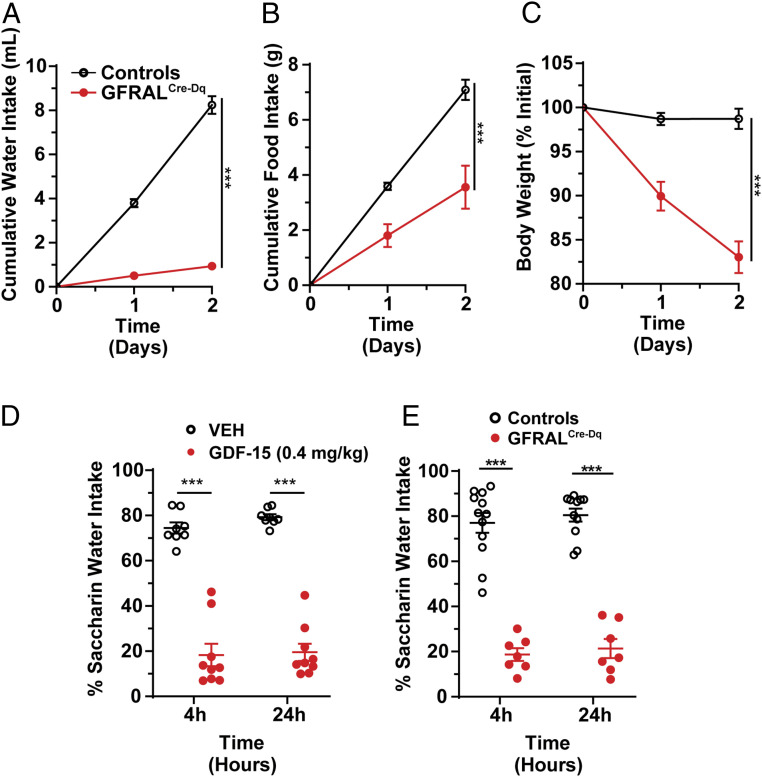
In a separate experiment initially designed as a chronic CNO-dosing paradigm, we provided mice with CNO- and glucose-containing water as their only source of drinking water. To our surprise, GFRALCre-Dq mice suppressed their liquid consumption compared to control mice (which received the same CNO- and glucose-containing drinking water) to a greater extent than they decreased food intake. CNO also reduced body weight in GFRALCre-Dq mice (Fig. 3 A–C). Injection of CNO (intraperitoneal injection [ip]) did not decrease normal water intake, however (SI Appendix, Fig. S5), and thus GFRAL neuron activation does not simply suppress thirst but rather, these experiments suggest activating GFRAL neurons results in aversive responses.
Fig. 3.
Activation of GFRAL neurons, like GDF-15, promotes aversive responses. (A–C) Water intake (A), food intake (B), and body weight (C) in control and GFRALCre-Dq mice over 2 d during which time the only source of drinking water contained CNO (0.025 mg/mL; n = 6). (D) CTA assay in wild-type mice injected with either VEH or GDF-15 during conditioning (n = 8–9). (E) CTA in GFRALCre-Dq mice and controls injected with CNO in conditioning phase (n = 7–12). Data are shown as mean ± SEM. Two-way ANOVA (A–E) was performed, ***P < 0.001.
To test whether GDF-15 signaling can provoke a conditioned taste aversion (CTA), we gave mice saccharin paired with either GDF-15 or vehicle (VEH) and then used a two-bottle test to assess their preference for saccharin versus water. We found GDF-15 produced a strong CTA (Fig. 3D), consistent with the findings of others (9–11). Furthermore, providing mice with saccharin along with artificial activation of GFRALCre-Dq mice with CNO also produced a strong CTA to saccharin (Fig. 3E). These studies show that GDF-15 and the artificial activation of GFRAL neurons mimics gastrointestinal malaise in the CTA assay.
To define the brain regions that GFRAL neurons innervate, we crossed GfralCre mice onto the Cre-inducible ROSA26Syn-TdTomato reporter background. We observed high rates of recombination outside of the AP. This made it impossible to decipher which terminals resulted from AP GFRAL neurons. Because we observed GfralCre-mediated recombination in regions inconsistent with adult Gfral expression patterns, we hypothesized this recombination was due to the transient developmental expression of GfralCre, as has been previously described (21).
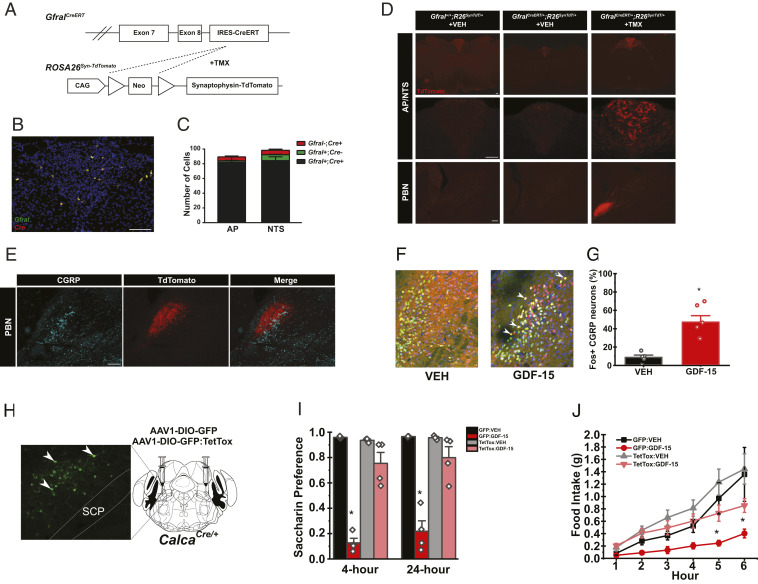
To overcome this limitation, we generated a second Gfral knock-in allele to express tamoxifen-inducible CreERT (GfralCreERT; Fig. 4A), permitting the manipulation of Gfral-expressing neurons in adulthood. To define the distribution of GfralCreERT neurons and reveal the circuits by which GFRAL neurons act, we crossed GfralCreERT onto the Cre-inducible ROSA26Syn-TdTomato background to express the synaptophysin-TdTomato fusion protein in GFRAL neurons (Fig. 4A). Following tamoxifen (TMX) administration, we observed reporter expression only in mice containing GfralCreERT and only within the AP (Fig. 4D). While the GfralCreERT reporter revealed fewer GFRAL neurons than the GfralCre model (SI Appendix, Figs. S3, S4, and S6), ISH revealed that the reporter expression in the GfralCreERT model correctly identified GFRAL neurons (Fig. 4 B and C). Interestingly, activating GfralCreERT neurons reconstituted the aversion and gastric emptying, but not anorectic, effects observed in the GfralCre mice (SI Appendix, Figs. S4 and S5). This seems likely because of reduced labeling of GFRAL expressing cells in the conditional mouse model. Examining the entire brain for TdTomato-labeled terminals revealed that GFRAL neurons project only to the NTS and the external lateral PBN (Fig. 4D).
Fig. 4.
AP GFRAL neurons project to the PBN and CGRPPBN neurons are required for GDF-15-mediated feeding suppression and aversion (A) Schematic of GfralCreERT– and TMX-mediated recombination of the Lox-Stop-Lox cassette from the ROSA26Syn-TdTomato line to generate GFRALCreERT-TdTomato mice. (B and C) Representative image and quantification of ISHs for Cre (red) and Gfral (green) transcripts in the AP/NTS of GfralCreERT/CreERT mice (n = 3). (D) Representative image of TdTomato-IR (red) in AP/NTS and PBN of Gfral+/+ mice, GFRALCreERT-SynTdT mice, and GFRALCreERT-SynTdT mice administered TMX or VEH, as indicated. (E) Representative image of TdTomato- (red) and CGRP-IR (cyan) within the PBN from TMX-treated GFRALCreERT-SynTdT mice. (F and G) Representative images (F) and quantification (G) of PBN FOS-IR 4 h following VEH or GDF-15 (400 μg/kg) administration (DNA: blue, CGRP: green, and FOS: red; n = 5). (H) Verification of GFP:TetTox expression within the PBN of CalcaCre mice (Green: GFP). (I) CTA in AAV-DIO-GFP or AAV-DIO-GFP:TetTox injected CalcaCre:GFP/+ mice administered with either VEH or GDF-15 during conditioning (n = 4). (J) Dark phase food intake of in AAV-DIO-GFP or AAV-DIO-GFP:TetTox injected CalcaCre:GFP mice injected with either VEH or GDF-15 1 h prior to the onset of the dark cycle (n = 5). SCP= superior cerebellar peduncle. Data are shown as mean ± SEM and analyzed by Two-way ANOVA *P < 0.05. (Scale bar, 100 μm.)
Exogenous GDF-15 induces FOS-IR in the external lateral PBN, with a partial overlap with calcitonin gene-related peptide (CGRP)-expressing neurons (4). As CGRPPBN neurons have previously been described to mediate aversive and anorectic effects (22, 23), we hypothesized that this cell population might be required for GDF-15 action. Indeed, direct projections from AP GFRAL neurons reside in proximity and largely overlap with CGRP-IR within the PBN (Fig. 4E). Furthermore, activation of AP GFRAL neurons in the GfralCreERT Dq model promoted FOS-IR in CGRPPBN neurons (SI Appendix, Fig. S4), and exogenous GDF-15 promoted FOS-IR in ∼50% of CGRPPBN cells (Fig. 4 F and G). To test the requirement for CGRPPBN neurons in GDF-15 action, we injected the Cre-dependent adeno-associated virus (AAV) double-floxed gene with inverted orientation (DIO)-green florescent protein (GFP): tetanus toxin (TetTox) into the PBN of CalcaCre:GFP animals (CGRPPBN-TetTox mice) to express the light chain of TetTox in CGRPPBN cells, silencing them (Fig. 4H). We found GDF-15 failed to promote a CTA to saccharine-laced water in CGRPPBN-TetTox mice and that CGRPPBN-TetTox mice exhibited an attenuated anorectic response to GDF-15 (Fig. 4 I and J), indicating CGRPPBN neurons mediate the aversive and anorectic responses to GDF-15.
Discussion
The important function and therapeutic potential of GDF-15, the hindbrain-restricted expression of GFRAL, and the dearth of information about roles for AP neural populations in the control of food intake and body weight prompted us to study GFRAL neurons. In addition to using genetic approaches to confirm that GFRAL neurons reside predominantly in the AP, we demonstrate that AP GFRAL neurons respond to anorectic doses of GDF-15, validating our genetic model. We also found that signals associated with systemic infection or illness stimulate GFRAL cells, while meal-related signals do not activate GFRAL neurons.
Assessing the neurotransmitter profile of GFRAL neurons through TRAP-Seq revealed a wide range of genes required for glutamatergic (Slc17a6), GABA-ergic (Slc32a1), and noradrenergic transmission and a range of neuropeptides including Grp, Nmu, and SSt, suggesting the potential existence of multiple subgroups of GFRAL cells and providing for a rich set of possibilities for how GFRAL neurons impact the activity of downstream neurons.
Acutely activating GfralCre neurons potently reduces food intake in a variety of feeding conditions (as well as promoting a CTA and decreasing gastric emptying), and chronic activation of GfralCre neurons results in substantial reductions in food intake and body weight. Interestingly, utilizing the GfralCreERT allele in conjunction with the hM3Dq DREADD resulted in substantially fewer cells expressing the DREADD, but was still sufficient to produce a CTA and decrease gastric emptying rates (all independent of changes in food intake). The difference in phenotypes between the GfralCre and GfralCreERT models aligns with dose-dependent behavioral responses to GDF-15, wherein low doses of GDF-15 induce aversion while considerably higher concentrations are required to suppress food (9–11). Our data support the observation that it takes considerably less GFRAL neuron activation to produce an aversive response than to produce anorexia.
Exogenous GDF-15 administration promotes a sense of visceral malaise as assessed by CTA and other measures (9–11) (and our data). This aversion is not limited to simply activating GFRAL, rather, our data show these neurons are inherently aversive and very likely any intervention designed to activate GFRAL-expressing neurons will result in visceral malaise. This presents a potential challenge to the use of GDF-15 or its analogs for the pharmacological treatment of obesity, since GDF-15 would be much less attractive to use if its weight loss effects result primarily from a feeling of visceral illness. This represents the challenge of employing a target such as the GDF-15/GFRAL system that is not part of the normal regulation of body weight but rather modulates food intake under pathophysiological conditions. There exist other examples of pharmacological agents (such as long-acting glucagon-like peptide-1 receptor agonists) that produce potent CTAs in rodents and cause initial visceral illness in humans (24, 25). However, most patients who continue on these drugs become tolerant to the nausea-inducing effects but continue to lose weight (26). Hence, the effects of GDF-15 on nausea may ultimately be separable from its effects on food intake and weight.
Indeed, several conditions that result in chronically elevated circulating GDF-15 do not result in persistent nausea. Metformin treatment increases circulating GDF-15, which plays a major role in the ability of metformin to cause weight loss (6), although most metformin-treated patients do not report persistent nausea. GDF-15 levels are also elevated in the second trimester of pregnancy (27). Nevertheless, the second trimester is actually associated with reduced visceral illness and emesis as compared to the first trimester for the majority of women (28).
We sought to use our genetic tools to identify the downstream systems that mediate the potent effects of GDF-15. Given the aversive responses to GDF-15 and activation of GFRAL neurons, we hypothesized that critical populations of cells in the hindbrain linked to pathophysiological responses would be important targets for GFRAL neurons. Indeed, GFRAL neurons project strongly to the PBN but not to hypothalamic sites that are critical for the homeostatic control of food intake and body weight. Within the PBN, CGRP-expressing cells are critical for transmitting negative-valence signals in response to a variety of pathophysiologic stimuli, including cancer anorexia (23, 29) which is associated with elevated circulating levels of GDF-15. Interestingly, strong projections from GFRAL-expressing neurons colocalize with CGRPPBN cells and activating a small number of GfralCreERT neurons results in FOS-IR in many CGRP-expressing PBN neurons. Importantly, silencing CGRPPBN cells abrogates the ability of GDF-15 to reduce food intake or produce a conditioned taste aversion. These data make a very strong case that CGRP-expressing cells represent critical downstream target for GFRAL-expressing neurons in the AP.
GDF-15 levels are not regulated in a manner consistent with an endogenous regulator of body weight (9) and the bulk of the data suggests that a lack of GDF-15 or GFRAL minimally impacts body weight under normal conditions (2, 4, 5, 15, 30). Similarly, CGRPPBN neurons contribute to the decreased feeding and body weight associated with pathophysiologic states such as cancer, chemotherapy, or infection (31) but are not required for normal body weight regulation (32).
Thus, our findings suggest that AP GFRAL neurons detect pathophysiologic signals for relay to CGRPPBN cells as part of the system that promotes aversion and attenuates nutrient uptake during pathophysiologic states. AP GFRAL neurons appear to have no direct communication with critical hypothalamic sites associated with the long-term regulation of body weight. Nevertheless, chronic activation of GFRAL neurons via exogenous GDF-15 (2–5) or via CNO-mediated DREADD activation results in profound and sustained weight loss. The key question is whether such a system can be safely targeted as a new treatment to promote weight loss. Understanding the neural circuits that are downstream from AP GFRAL neurons will not only be important in answering this question but also will identify future therapeutic strategies that would target these brainstem systems.
Materials and Methods
Stereotaxic Surgeries and Infusion Site Verification in Rats.
Male Long Evans rats were fed Tso’s 40% butter-fat diet for up to 8 wk. Two weeks prior to surgery they were singly housed and handled daily. On the day of surgery, each rat was anesthetized using 5% isoflurane and maintained on 2% isoflurane. Totals of 0.03 mg/kg buprenex and 0.5 mg/kg meloxicam were provided as analgesia. Each rat was placed in the stereotaxic frame and an incision was made through the skin across the skull. The bregma and the lambda were both identified and the skull was adjusted to be horizontal between these. For AP cannulation a hole was drilled on the ridge at the caudal end of the skull (∼15 mm caudal to bregma and 0 mm lateral to the midline; the bregma distance varied between rats depending on the location of the skull ridge). For NTS cannulation, a hole was drilled 0.5 mm rostral to the skull ridge and 0.9 mm lateral to the midline. Cannulation depth for both AP and NTS, respectively, varied between 9.4 and 10.2 mm ventral to bregma. The exact depth was tested in three to five rats in each cohort prior to the actual surgery as skull thickness and size varied between high-fat fed cohorts. For all cannulation surgeries, the cannula was held in place by dental acrylic (fujicem) and the rostral part of the incision was closed with wound clips. Animals received analgesia for 2 d after surgery: 0.03 mg/kg buprenex twice a day and 0.5 mg/kg ostilox once daily. Wound clips were removed 7 d postsurgery.
At the end of each experiment the rats were euthanized using CO2 and decapitation. A total of 0.5 μl blue dye was then slowly infused over 2 min and the brain was extracted and frozen in tissue tech on dry ice. Each brain was sectioned on a cryostat and injection site was verified by the presence of the blue dye using a rat brain atlas (Paxinos and Watson, 2013) for reference.
Neuropile Infusions and Food Intake in Rats.
After surgery, the rats were handled daily with the dummy cannula being pulled out and inserted to habituate rats to cannula manipulations. Infusions started 2 wk after surgery, when all rats were fully healed. On each infusion day, food was removed 2 h prior to lights out. After 1 h, infusions were initiated with 1 μL of VEH, GDF-15 (doses of 0.33, 1, or 2 μg), or salmon calcitonin (0.4 μg) being slowly infused over 1 min with an additional 1-min wait before the infusion needle was removed. Food was then given back exactly 1 h after the infusion. Food intake was measured 1, 2, and 23 h after infusion. Each rat had three infusions with doses randomized for infusion day and were allowed a minimum of 1 wk of recovery between each infusion.
Stereotaxic Surgery in Mice.
A mix of male and female mice (CalcaCre:GFP/+) were anesthetized with isoflurane (1 to 4%) and mounted in a stereotaxic frame (Kopf). Using standard surgical techniques, virus (AAV-DIO-GFP:TetTox or AAV-DIO-GFP) was injected bilaterally via a glass micropipette attached to a microinjector (Nanoject II) targeting the PBN (Anterior/Posterior -4.8 mm; Medial/Lateral ±1.3 mm, Dorsal/Ventral −3.3 mm, relative to bregma).
High-Fat Diet Feeding.
GFRALCre-Dq mice were fed a high-fat diet (60% fat by kCal; research diets D12492) from weaning to 8 wk of age prior to study. For GFRALCreERT-Dq studies, mice were fed high-fat diet from weaning, administered tamoxifen at 5 wk of age and given an additional 5 wk of HFD feeding prior to studies.
Tamoxifen Dosing.
Animals were dosed at 8 wk of age (except for the case of high fat diet fed animals, which were administered tamoxifen at 5 wk of age) with 150 mg/kg tamoxifen dissolved in corn oil, delivered ip once per day for 5 d. Controls were VEH treated. All experiments using GfralCreERT mice were performed for a minimum of 3 wk posttamoxifen administration.
Conditioned Taste Aversion.
Mice (both male and females were used for all studies) were housed in custom cages with two ports on the front of the cage that accepted drinking bottles constructed of glass test tubes fitted with a rubber stopper and spout. On days 1 to 5 mice were injected with saline (0.01 mL/kg, subcutaneous) once daily. On the afternoon of day five the standard home cage water bottle was removed and replaced by custom drinking bottles filled with water. On day eight water bottles were removed 1 h before lights out. On day nine a single preweighted bottle filled with 0.15% saccharin was offered 2 h prior to lights out. At lights out on day nine, saccharin was removed and animals were injected with either VEH or compound (GDF-15 [0.4 mg/kg, delivered sc; Novo Nordisk] or CNO 1 mg/kg ip; Tocris Bioscience) and water was returned. On day 10, water was removed from the cage 1 h before lights out. On day 11, 1 h before lights out two preweighed bottles were placed in the cage ports, one containing water and the other 0.15% saccharin. Bottles were weighed after 4 and 24 h.
Food-Intake Studies.
Mice (both male and females were used for all studies) were singly housed a minimum of 1 wk prior to studies. For acute studies, on the day of the experiment, food was removed at 4 PM and mice were randomized between CNO (1 mg/kg) or VEH (sterile saline 0.9%) treatment. CNO or VEH was administered via ip injection 30 min prior to the onset of darkness and the food hopper was returned with a preweighed amount of food (generally ∼10 g) at the onset of lights-out. Food was weighed 1, 2, 4, 16, and 24 h following the onset of the dark phase. Mice were given 1 wk of rest, prior to the repetition of the experiment under inversed treatment conditions. In a subset of experiments, water intake was measured by the same experimental paradigm. Controls were either mice that did not express either Cre or hM3Dq. For chronic CNO dosing, singly housed mice were administered CNO once at 9 AM and once at 5 PM for up to 4 d. Food and body weight were monitored daily.
CNO in Drinking Water.
CNO was dissolved in water (supplemented with 1% glucose) at a concentration of 2.5 mg/100mL H2O. Mice were singly housed a minimum of 1 wk prior to experiments. Regularly supplied water was removed and replaced with water bottles containing CNO-laced water as the only source of drinking water. Water, food consumption, and mouse body weight were monitored daily. Water was replaced daily.
Gastric Emptying.
Mice were fasted 4 h prior to CNO injections (1 mg/kg ip; Tocris Bioscience). Thirty minutes following CNO injection, mice were gavaged with 100 mg/kg acetaminophen dissolved in water and tail blood samples were collected for acetaminophen assay (Sekisui Diagnostics 506-30 Acetaminophen L3k Assay) 15 min following gavage.
FOS Studies.
Liraglutide (400 μg/kg; delivered ip; Novo Nordisk), salmon calcitonin (400 μg/kg; delivered ip; Bachem), LiCl (84 mg/kg delivered ip; Sigma), and LPS (50 μg/kg delivered ip; Sigma) were injected into GfralCre/+;Rosa26eGFP-L10a/+ mice 2 h prior to sacrifice. Additionally, a separate cohort was fasted 16 h (5 PM to 9 AM) and permitted 2 h of food access prior to sacrifice, controls for refed mice were mice euthanized under fasting conditions. Mice were given GDF-15 (400 μg/kg; delivered sc; Novo Nordisk) and euthanized and perfused after 4 h. Following fixation and staining, FOS+ cells were quantified in ImageJ within the AP and normalized to the number of GFP+ cells.
In Situ Hybridization.
Mice were anesthetized with isoflurane and euthanized by decapitation. Brains were dissected, flash frozen in isopentane, chilled on dry ice, and stored at -80 °C. Sections were sliced at 16 mm thickness using a cryostat (Leica) and every fourth section was thaw-mounted onto slides, allowed to dry for 1 h at −20 °C, and then further stored at −80 °C. Slides were then processed for RNAScope ISH per the manufacturer’s protocol (Advanced Cell Diagnostics). The multiplex fluorescent assay (320850) was used to visualize Gfral (439141-C2) and Cre (312281-C3) probes using Amp 4 Alt-A. At each of eight coronal planes for each mouse, four images comprising the entire NTS/AP complex were obtained with a QImaging Retiga 6000 monochrome camera attached to an Olympus BX53 fluorescent microscope under 20× objective. The four images were then stitched together using Photoshop (Adobe). CellProfiler was used to process all images identically to remove nonspecific background and outline-specific cells using the DAPI nuclear signal and analyze the presence or absence of a signal for both probes. For each mouse, the total number of cells identified as positive for either or both probes were added from all eight coronal planes (for NTS) or for the subset of the six planes where AP was present. Subsequently, the sums from the three mice were averaged for each region.
Antibody Staining.
Mice were euthanized with isoflurane and then perfused by gravity flow with phosphate buffered saline for 5 min followed by an additional 5 min of 10% formalin fixation. Brains were then removed and postfixed in 10% formalin for 24 h at room temperature, before being moved to 30% sucrose for a minimum of 24 h at room temperature. Brains were then sectioned 30-μm thick, free-floating sections and stained. Sections were treated sequentially with 1% hydrogen peroxide/0.5% sodium hydroxide, 0.3% glycine, 0.03% sodium dodecyl sulfate, and blocking solution (phosphate-buffered saline with 0.1% triton, 3% normal donkey serum; Fisher Scientific). The sections were incubated overnight at room temperature in rabbit anti-FOS (FOS, #2250, Cell Signaling Technology, 1:1,000; HA #C29F4, Cell Signaling Technology 1:1,000; CGRP ab81887, Abcam, 1:2,000). The following day, sections were washed and incubated with either biotinylated (1:200 followed by avidin-biotin complex [ABC] amplification and 3,3′-diaminobenzidine [Thermo Fisher Scientific]) or fluorescent secondary antibody to visualize proteins. Immunofluorescent staining was performed using primary antibodies (GFP, GFP1020, Aves Laboratories; DSRed, 632392, Clontech); antibodies were reacted with species-specific Alexa Fluor-488, 568 conjugated secondary antibodies (Invitrogen, Thermo Fisher,1:300). Images were collected on an Olympus BX51 microscope. Images were pseudocolored and cell counts performed using ImageJ (NIH).
TRAP.
Mice (GfralCreROSA26eGFP-L10a) were treated with VEH or 400 μg/kg GDF-15 and euthanized 6 h postinjection with isolfuorane and decapitated. The brain was subsequently removed from the skull the AP/NTS dissected from the brainstem and homogenized in lysis buffer. Between 15 to 20 mice were used for TRAP experiments. GFP-tagged ribosomes were immunoprecipitated and RNA isolated as previously described (33). RNA was subject to ribodepletion and the resultant mRNA was fragmented and copied into first strand complementary DNA (cDNA). The products were purified and enriched by PCR to create the final cDNA library. Samples were sequenced on a 50-cycle single end run on a HiSeq 2500 (Illumina) according to manufacturer’s protocols.
FASTQ files were filtered using fastq_quality_filter from fastx_toolkit to remove reads with a phred score < 20. Then reads were mapped using Spliced Transcripts Alignment to a Reference (STAR) with a custom genome containing the Ensembl reference and sequences and annotation for Cre and eGFP:L10a. Count tables were generated using the STAR–quantMode GeneCounts flag.
Count tables were analyzed in R 3.6.3 and were subject to quality control to ensure read adequate library size (20 to 30 million reads), enrichment of positive control genes (e.g., EGFP:L10a, Cre, and Gfral), and appropriate sample similarity in both hierarchical clustering of Euclidean distance and t-distributed stochastic neighbor embedding/uniform manifold approximation and projection space (e.g., bead samples are more similar to one another than to any supernatant [Sup] sample). All samples passed quality control. Enriched genes were determined using DESeq2, including an effect of sample pair in the model to account for pairing of the bead–Sup samples (∼pair + cells).
Quantification and Statistical Analysis.
All data are displayed as mean ± SEM. Statistical analysis was performed in either Graphpad Prism 8 or R 3.6.3 using either t tests or ANOVAs with Dunnet’s post hoc test when appropriate. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of the Molecular Genetics Core of the University of Michigan Diabetes Research Center. P.V.S. was supported by American Diabetes Association Grant 1-19-PDF-099. J.W. was supported by the China Scholarship Council. J.M.A. was supported by NIH Grant T32-DK-101357. This research was also supported in part by NIH Grants R01- DA-24908 (to R.D.P.); P30-DK089503, P01-DK117821, and R01-DK119188 (to R.J.S.); and P30-DK020572 (to M.G.M.).
Footnotes
Competing interest statement: S.B.J. is an employee of Novo Nordisk. D.P.O., M.G.M., and R.J.S. receive research support from Novo Nordisk. R.J.S. and M.G.M. receive research support from AstraZeneca. R.J.S. receives research support from Pfizer, Kintai, and Ionis. R.J.S. also serves as a paid consultant for Novo Nordisk, Kintai, Ionis, and Scohia. R.J.S. has equity positions in Zafgen and ReDesign Health. All other authors report no conflicts of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021357118/-/DCSupplemental.
Data Availability
TRAP-based sequencing data have been deposited in GenBank (accession number GSE160257) (34). All other study data are included in the article and/or supporting information.
References
- 1.Johnen H., et al., Tumor-induced anorexia and weight loss are mediated by the TGF-β superfamily cytokine MIC-1. Nat. Med. 13, 1333–1340 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Mullican S. E., et al., GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and nonhuman primates. Nat. Med. 23, 1150–1157 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Emmerson P. J., et al., The metabolic effects of GDF15 are mediated by the orphan receptor GFRAL. Nat. Med. 23, 1215–1219 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Hsu J.-Y., et al., Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15. Nature 550, 255–259 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Yang L., et al., GFRAL is the receptor for GDF15 and is required for the anti-obesity effects of the ligand. Nat. Med. 23, 1158–1166 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Coll A. P., et al., GDF15 mediates the effects of metformin on body weight and energy balance. Nature 578, 444–448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montero R., et al., GDF-15 is elevated in children with mitochondrial diseases and is induced by mitochondrial dysfunction. PLoS One 11, e0148709 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakayasu E. S., et al., Comprehensive proteomics analysis of stressed human islets identifies GDF15 as a target for type 1 diabetes intervention. Cell Metab. 31, 363–374.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel S., et al., GDF15 provides an endocrine signal of nutritional stress in mice and humans. Cell Metab. 29, 707–718.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borner T., et al., GDF15 induces an aversive visceral malaise state that drives anorexia and weight loss. Cell Rep. 31, 107543 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borner T., et al., GDF15 induces anorexia through nausea and emesis. Cell Metab. 31, 351–362.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frikke-Schmidt H., et al., GDF15 acts synergistically with liraglutide but is not necessary for the weight loss induced by bariatric surgery in mice. Mol. Metab. 21, 13–21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Worth A. A., et al., The cytokine GDF15 signals through a population of brainstem cholecystokinin neurons to mediate anorectic signalling. eLife 9, e55164 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong Y.et al., Long-acting MIC-1/GDF15 molecules to treat obesity: Evidence from mice to monkeys. Sci. Transl. Med. 9, eaan8732 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Tsai V. W.-W.et al., TGF-b superfamily cytokine MIC-1/GDF15 is a physiological appetite and body weight regulator. PLoS One 8, e55174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krashes M. J., et al., An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature 507, 238–242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng W.et al., Leptin receptor-expressing nucleus tractus solitarius neurons suppress food intake independently of GLP1 in mice. JCI Insight 5, e134359 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng W., et al., Calcitonin receptor neurons in the mouse nucleus tractus solitarius control energy balance via the non-aversive suppression of feeding. Cell Metab. 31, 301–312.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roman C. W., Derkach V. A., Palmiter R. D., Genetically and functionally defined NTS to PBN brain circuits mediating anorexia. Nat. Commun. 7, 11905 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu H., et al., Cre-dependent DREADD (designer receptors exclusively activated by designer drugs) mice. Genesis 54, 439–446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z., et al., Identification, expression and functional characterization of the GRAL gene. J. Neurochem. 95, 361–376 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Carter M. E., Soden M. E., Zweifel L. S., Palmiter R. D., Genetic identification of a neural circuit that suppresses appetite. Nature 503, 111–114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter M. E., Han S., Palmiter R. D., Parabrachial calcitonin gene-related peptide neurons mediate conditioned taste aversion. J. Neurosci. 35, 4582–4586 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams J. M., et al., Liraglutide modulates appetite and body weight through glucagon-like peptide 1 receptor-expressing glutamatergic neurons. Diabetes 67, 1538–1548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanoski S. E., Rupprecht L. E., Fortin S. M., De Jonghe B. C., Hayes M. R., The role of nausea in food intake and body weight suppression by peripheral GLP-1 receptor agonists, exendin-4 and liraglutide. Neuropharmacology 62, 1916–1927 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lean M. E. J.et al.; NN8022-1807 Investigators , Tolerability of nausea and vomiting and associations with weight loss in a randomized trial of liraglutide in obese, non-diabetic adults. Int. J. Obes. 38, 689–697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petry C. J., et al., Associations of vomiting and antiemetic use in pregnancy with levels of circulating GDF15 early in the second trimester: A nested case-control study. Wellcome Open Res. 3, 123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eliakim R., Abulafia O., Sherer D. M., Hyperemesis gravidarum: A current review. Am. J. Perinatol. 17, 207–218 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Campos C. A., et al., Cancer-induced anorexia and malaise are mediated by CGRP neurons in the parabrachial nucleus. Nat. Neurosci. 20, 934–942 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran T., Yang J., Gardner J., Xiong Y., GDF15 deficiency promotes high fat diet-induced obesity in mice. PLoS One 13, e0201584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmiter R. D., The parabrachial nucleus: CGRP neurons function as a general alarm. Trends Neurosci. 41, 280–293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campos C. A., Bowen A. J., Schwartz M. W., Palmiter R. D., Parabrachial CGRP neurons control meal termination. Cell Metab. 23, 811–820 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allison M. B., et al., TRAP-seq defines markers for novel populations of hypothalamic and brainstem LepRb neurons. Mol. Metab. 4, 299–309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabatini P. V., Rupp A. C., Myers M. G., Seeley R. J., TRAP-Sequencing analysis of Gfral neurons. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160257. Deposited 27 October 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TRAP-based sequencing data have been deposited in GenBank (accession number GSE160257) (34). All other study data are included in the article and/or supporting information.