Significance
The boom of single-cell sequencing technologies in the past decade has profoundly expanded our understanding of fundamental biology. Today, tens of thousands of cells can be measured by single-cell RNA-seq in one experiment. However, single-cell DNA-sequencing studies have been limited by false positives and cost. Here we report META-CS, a single-cell whole-genome amplification method that takes advantage of the complementary strands of double-stranded DNA to filter out false positives and reduce sequencing cost. META-CS achieved the highest accuracy in terms of detecting single-nucleotide variations, and provided potential solutions for the identification of other genomic variants, such as insertions, deletions, and structural variations in single cells.
Keywords: single-cell sequencing, mutations, false positives, Tn5 transposition, complementary DNA strands
Abstract
Single-nucleotide variants (SNVs), pertinent to aging and disease, occur sporadically in the human genome, hence necessitating single-cell measurements. However, detection of single-cell SNVs suffers from false positives (FPs) due to intracellular single-stranded DNA damage and the process of whole-genome amplification (WGA). Here, we report a single-cell WGA method termed multiplexed end-tagging amplification of complementary strands (META-CS), which eliminates nearly all FPs by virtue of DNA complementarity, and achieved the highest accuracy thus far. We validated META-CS by sequencing kindred cells and human sperm, and applied it to other human tissues. Investigation of mature single human neurons revealed increasing SNVs with age and potentially unrepaired strand-specific oxidative guanine damage. We determined SNV frequencies along the genome in differentiated single human blood cells, and identified cell type-dependent mutational patterns for major types of lymphocytes.
Genome-wide determination of single-nucleotide variants (SNVs) in single cells has been challenging due to false positives (FPs) from two sources. First, polymerases used for amplification make errors. The error rates of base substitution during in vitro DNA synthesis for most DNA polymerases range from 10−4 to 10−6 (1), indicating that thousands of FPs can be generated in the first cycle of amplification for a human genome of 6 billion bp. Second, the harsh conditions of cell lysis and amplification cause DNA damage, which, together with damage that occurred naturally in the live cell (e.g., deamination and oxidative damage), can be misrecognized by DNA polymerases and turned into errors. For example, it has been reported that deamination of cytosine resulted in the artifact of C>T transitional mutation in single-cell multiple displacement amplification (MDA) (2). These FPs usually significantly outnumbered naturally occurring SNVs, posing a limitation on single-cell genomics.
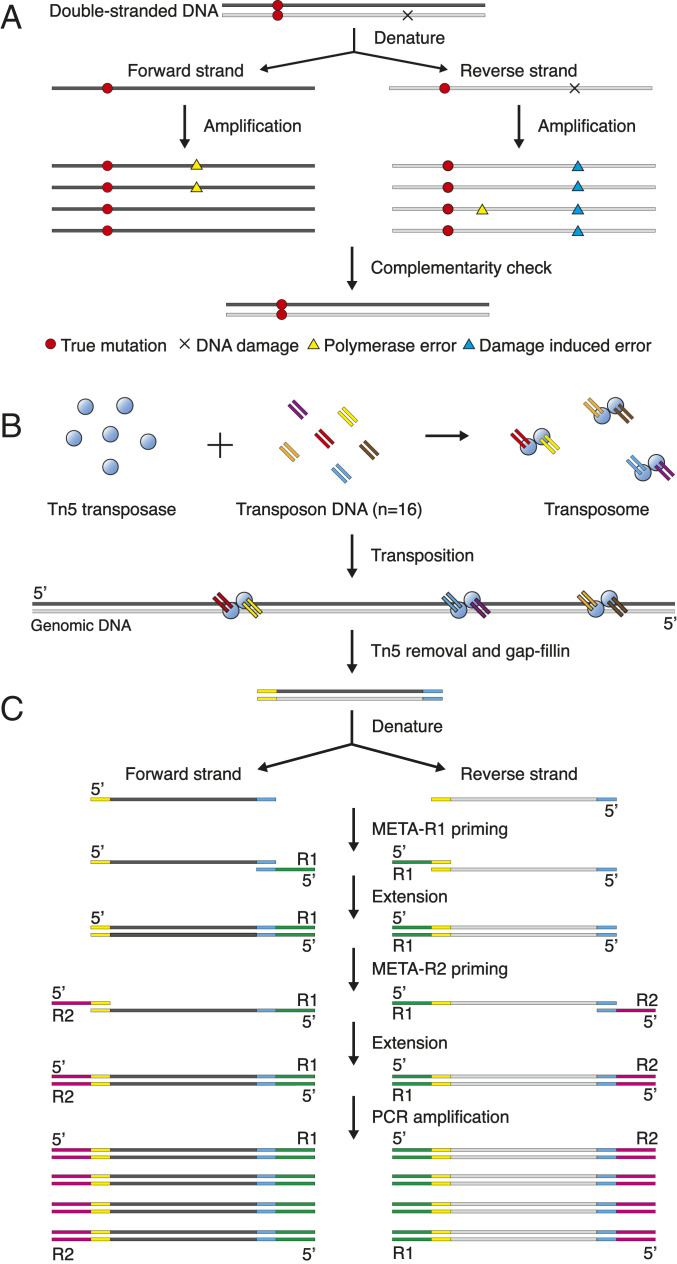
While a true SNV has to be on the same position of both strands, both polymerase errors and DNA damage occur only on one strand of DNA. Therefore, FPs can be filtered out through checking the complementarity of the two strands after sequencing (Fig. 1A). Current methods to achieve this, involving ligation of strand-specific adapters (3, 4) or physical separation of the two strands (5), are labor-intensive and suffer from significant sample loss of single cells. In silico SNV-calling algorithms, on the other hand, rely on heterozygous single-nucleotide polymorphisms and the assumption that single strands are amplified uniformly, which still leads to FPs when one of the strands fails to be amplified (6, 7). Here, we report a whole-genome amplification (WGA) method termed multiplexed end-tagging amplification of complementary strands (META-CS), which is able to separately label and amplify the two strands of DNA in a one-tube reaction and accurately identify de novo SNVs from a single cell.
Fig. 1.
Identification of SNVs by META-CS. (A) FPs on single strands can be filtered out through sequencing the two complementary strands of double-stranded DNA (dsDNA). Complementary strands are shown in dark and light gray. DNA damage and polymerase errors occur randomly on one of the two strands, while true mutations are detected at the same position on both strands. (B) Transposition of the META-CS transposome to single-cell DNA. A mixture of 16 unique transposon sequences (only 6 are shown for simplicity) are mixed with Tn5 transposase with an equal molar ratio to form transposome complexes, which cut the single-cell DNA and tag each fragment with two random transposon sequences. (C) Single-cell WGA of the forward and reverse strand by META-CS.
Results
META-CS for SNV Identification.
META-CS is based on our previously developed WGA method, multiplexed end-tagging amplification (8). Briefly, genomic DNA from single-cell lysates was fragmented by Tn5 transposition, and each fragment was randomly tagged with 2 out of 16 unique transposon sequences which served as priming sites in the following reactions (Fig. 1B). The use of multiple transposon sequences provided fragment-specific barcodes and reduced the amplification loss associated with the intramolecular hairpin structure that may form when a fragment is tagged by the same sequence on both ends (SI Appendix, Fig. S1). Next, DNA fragments were melted by heating, and the two single strands were preamplified by two sequential polymerase extension reactions to obtain strand-specific labeling (Fig. 1C). Excess primers were removed by exonuclease I after each round of polymerase extension reaction. The resulting products, amplified separately from the forward and reverse strands of the original DNA, could be distinguished by the way they mapped to the reference genome. SNVs were then determined as variants agreed by both strands (SI Appendix, Fig. S2). The complete META-CS procedures were performed in one single tube and could easily scale up to multiwell plates.
To determine the specificity of our strand-labeling strategy, we used a synthetic single-stranded DNA as template and sequenced the amplification product. We observed four discordant strands out of 173,635 reads, corresponding to a strand conversion rate of 2.3 × 10−5. This suggested a high specificity of the strand-labeling strategy, although different templates and other combinations of META primers might result in different strand conversion rates.
Characterization of Detection Accuracy.
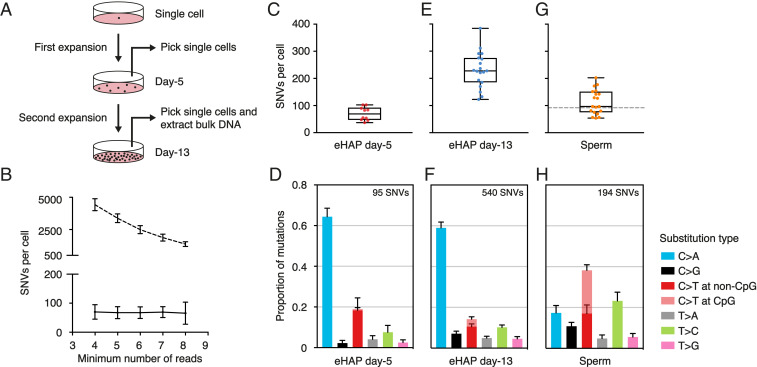
To evaluate the accuracy of META-CS, we clonally expanded a single ancestor cell from a human haploid cell line (eHAP) for 5 d to a few hundred cells (Fig. 2A). Ten single cells were picked by mouth pipetting and successfully amplified by META-CS (SI Appendix, Fig. S3). The rest of the cells were used for a second expansion of another 8 d to millions of cells for bulk DNA extraction to represent the ancestor cell’s genome. Both single-cell and bulk samples were mapped to the human reference genome and de novo SNVs were identified in single cells compared with the ancestor cell.
Fig. 2.
Validation of META-CS in clonally expanded single cells and human sperm. (A) Experimental design for clonal expansion of an eHAP single cell. (B) Number of SNVs per cell with regard to the changing of calling threshold. The horizontal axis represents the minimum number of reads required to call a mutation. For callings with the strand filter (solid line), thresholds are set as a4s2, a5s2, a6s3, a7s3, and a8s4. For calling without the strand filter (dashed line), thresholds are set as a4s0, a5s0, a6s0, a7s0, and a8s0. Error bars represent SD. (C, E, and G) Box and whisker plot of the number of SNVs determined in day-5 eHAP (C), day-13 eHAP (E), and human sperm single cells (G). Each dot represents a single cell. The dashed line in G indicates the number of germline mutations at age 56 by fitting a linear regression model to data obtained from family trios (16). (D, F, and H) Relative contribution of mutation types for day-5 eHAP (D), day-13 eHAP (F), and sperm (H). Data are represented as the mean contribution of each mutation type from all single cells. Error bars represent SE. The total number of mutations is indicated (Top).
We first determined the minimum number of sequencing reads required to confidently identify a SNV. We used the letter “a” (for allelic depth) to represent the minimum number of total reads, and the letter “s” (for stranded allelic depth) to represent the minimum number of strand-specific reads. To filter out sequencing errors, we started with a total number of no less than four reads, requiring at least two reads from each strand (a4s2). We then examined the robustness of our calling with respect to the thresholds. Compared with FPs that occur on only one strand of DNA, true positives existing on both strands of DNA generally have higher allelic frequencies after amplification, and are thus less sensitive to the changing of threshold. Increasing the threshold of the strand filter from a4s2 to a8s4, we found that the number of SNVs detected from the eHAP single cells only decreased ∼2-fold (95/48) (SI Appendix, Fig. S4). In comparison, for callings without the strand filter, increasing the threshold from a4s0 to a8s0 led to an ∼6.4-fold decrease (11,153/1,754). Notably, after adjustment for detection efficiency, the number of SNVs remained almost unchanged for callings with the strand filter but decreased continuously for callings without the strand filter (Fig. 2B). This suggested that META-CS was able to measure mutation rates with as few as four sequencing reads.
In day-5 eHAP single cells, META-CS achieved an average (±SD) of 50.9 ± 12.2% (n = 10) genome coverage and detected 9.5 ± 5.9 autosomal de novo SNVs per cell, which corresponded to 70 ± 25 SNVs after correction with a strand-specific detection efficiency of 15.9 ± 6.1% (Fig. 2C, SI Appendix, Fig. S5, and Dataset S2). Among the 95 detected SNVs, 63 were C>A transversions (Fig. 2D). The transition-to-transversion (Ti/Tv) ratio was 0.27, which was consistent with the number previously reported from a similar single-cell clonal expansion experiment, where FPs were filtered out through sequencing kindred cells due to the lack of reliable technique at the time (9). Furthermore, the mutational spectrum of the 95 SNVs was different from the error spectrum of the Q5 DNA polymerase (Ti/Tv = 0.89) used for amplification and spectra of DNA damage, which were mainly composed of C>T transitions (10). As a comparison, mutations identified without applying the strand filter (a4s0) showed a very different spectrum that was predominated by transition mutations (Ti/Tv = 1.73) (SI Appendix, Fig. S6).
To validate this C>A-dominated mutational spectrum, we further amplified and sequenced eHAP single cells picked on day 13 (after the second expansion). If the true mutational spectrum of eHAP kindred cells was different from the spectrum of FPs, the observed spectrum would shift as the cells obtained more true mutations with cell division. With the same threshold of a4s2, we identified an average of 231 ± 65 (n = 19) de novo SNVs per cell, suggesting that cells from day 13 indeed accumulated more mutations compared with cells from day 5 (Fig. 2E). Nevertheless, the mutational spectrum of day-13 cells (540 mutations) remained almost identical to day-5 cells (Pearson correlation coefficient r = 0.995) with a Ti/Tv ratio of 0.31 (Fig. 2F).
Validation of META-CS with Single Human Sperm.
We then asked if META-CS can distinguish mutational spectra in different cell types. We amplified and sequenced sperm cells from a healthy male donor in his late 50s. There are three advantages of using single sperm as a technical control. First, the germline mutation rate of sperm is very low, making characterization of FPs more accurate. Second, the mutation rates and spectra of sperm across a wide range of ages have been characterized in both population and single-cell studies (11–14). Third, it has been shown that using single cells in G2/M phase can significantly increase amplification efficiency and accuracy (15), thus complicating method evaluation. In contrast, sperm do not undergo cell cycles, which rules out G2/M-phase cells.
With META-CS, we identified an average of 114 ± 45 (n = 19) germline SNVs per cell (Fig. 2G). This number is close to the typical number of mutations (∼92 SNVs, 95% CI 80 to 105) at the donor’s age, as determined from a previously published dataset of large-scale family trios (16). Instead of the C>A transversions that predominated the mutational spectrum of eHAP kindred cells, the spectrum of sperm was mainly composed of C>T and T>C transitions (Ti/Tv = 1.46) (Fig. 2H). Moreover, the spectrum identified from single sperm cells by META-CS was largely consistent with the spectrum derived from the data of family trios in ref. 16 (r = 0.93) (SI Appendix, Fig. S7). A previous study of single human sperm reported a much higher Ti/Tv ratio of 5.6 with a C>T predominant spectrum (14), which was possibly due to FPs generated during WGA.
Taken together, these data indicated that most of the SNVs identified by META-CS in single eHAP and sperm cells were true positives. We estimated the upper bound of the false positive rate (FPR) of META-CS to be ∼2.4 × 10−8 (70/2.9 Gb) for day-5 eHAP kindred cells. However, the true FPR of META-CS was very likely much lower, masked by de novo mutations generated during in vitro clonal expansion.
De Novo SNVs in Human Neurons.
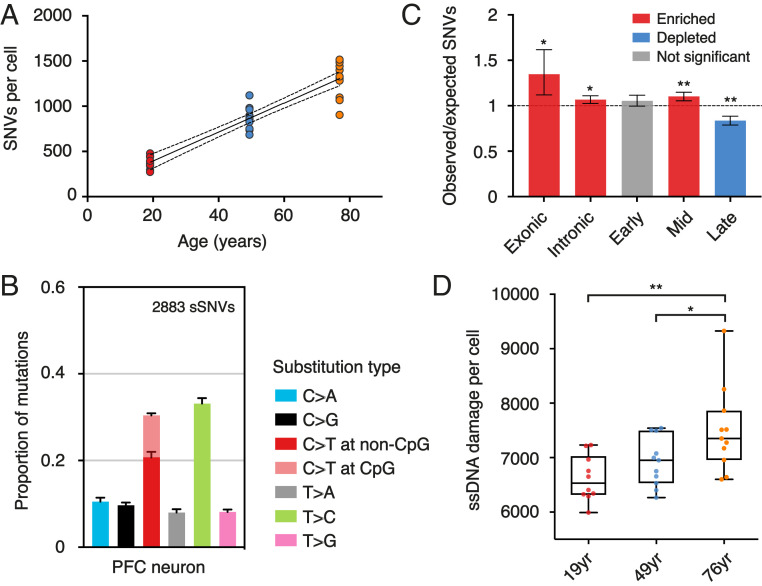
Somatic SNVs that accumulate in mature neurons have long been speculated to play a role in aging and neural degeneration. It was not until recently, however, that studies of single neurons had been performed by single-cell MDA (17). With the aim of characterizing the mutational patterns in neurons with a higher accuracy, we amplified single prefrontal cortex (PFC) neurons from postmortem brain tissues of three individuals. We identified an average of 379 ± 66 (n = 10, 19-y-old), 871 ± 123 (n = 11, 49-y-old), and 1,304 ± 202 (n = 11, 76-y-old) SNVs, corresponding to an increase of ∼16 SNVs per year (Fig. 3A). Our results supported the observation that SNVs in neurons accumulate with age (18). However, the number of SNVs identified by META-CS was generally lower compared with previous studies (17, 18), indicating that unrecognized FPs still existed with other methods.
Fig. 3.
SNVs and DNA damage identified in single human neurons. (A) SNVs in single neurons identified from three individuals (red for 19 y old, blue for 49 y old, and orange for 76 y old). Each dot represents a single cell. The dataset is fitted by a linear regression line with 95% CI. (B) Relative contribution of mutation types in PFC neurons. Data are represented as the mean contribution of each mutation type from all 32 single cells. Error bars represent SE. The total number of mutations is indicated (Top). (C) Enrichment and depletion of SNVs from single neurons in exons, introns, and replication timing domains (early, mid, and late). The expected values are calculated assuming that SNVs distribute randomly along the genome. Error bars denote 95% CI. *P < 0.005, **P < 0.0001, two-tailed binomial test. (D) ssDNA damage in single neurons from three individuals. Each dot represents a single neuron. Data are adjusted by detection efficiency and shown as a box and whisker plot. *P < 0.05, **P < 0.01, two-tailed t test.
SNVs in PFC neurons were mainly T>C and C>T transitions (Fig. 3B), which were likely results of adenine and cytosine deamination (19). It has been previously shown that human de novo mutations were enriched in late-replicating domains, possibly due to an accumulation of single-stranded DNA (ssDNA) during the late stages of DNA replication (20). Mutations in nondividing neurons, in contrast, were depleted from late-replicating domains but enriched in transcribed regions (Fig. 3C), supporting the hypothesis that SNVs in mature neurons partially resulted from transcription-associated DNA damage (17).
Since ssDNA damage can be turned into strand-specific “mutations” during amplification, they can be detected by META-CS. Now, having defined somatic SNVs as those found on both strands, we further defined ssDNA damage as having the mutational allele on only one strand, while having the reference allele on the other (Fig. 1A and SI Appendix, Fig. S8). Note that damage called under this definition consists of not only biological damage that occurred in live cells but also artifacts generated during WGA. For example, a relatively high temperature (70 °C for 15 min) is required during cell lysis to inactivate the Qiagen protease. Such heat may cause additional DNA damage. Nevertheless, we reasoned that WGA-associated artifacts were constant for samples treated under the same procedures and, therefore, variations observed across samples should reflect real sample differences. We found that there was a significant increase in ssDNA damage in the 76-y-old brain compared with the younger ones (Fig. 3D). Strand-specific spectrum analysis revealed that the increase mainly came from G>T transversions (SI Appendix, Fig. S9), which were reported to be a result of guanine oxidation to 8-hydroxyguanine (21, 22). However, there might be other types of DNA damage which were not detected by META-CS because certain bases, such as uracil and inosine, were not preferred by the Q5 DNA polymerase. Future work may use an alternative polymerase, such as the New England BioLabs (NEB) Q5U DNA polymerase, to address this issue.
De Novo SNVs in Human Peripheral Blood Cells.
Somatic mutations in hematopoietic stem and progenitor cells (HSPCs) have been studied and associated with blood cancers (23, 24). SNVs in single HSPCs can be characterized through sequencing the bulk DNA of a single-cell clone expanded in vitro (25, 26). However, matured peripheral blood cells cannot be expanded from a single cell and hence have not been widely investigated. With the rapid development of cancer immunotherapy, it is necessary to closely examine the genomic variations of immune cells.
With META-CS, we amplified and sequenced a total of 53 single peripheral blood mononuclear cells (PBMCs) from a healthy male donor (same as the sperm donor). An average of 1,494 ± 721 SNVs per cell were identified. This number was more than one order of magnitude higher than the germline mutations of the same individual, but comparable with other tissues as measured by clonal organoid cultures derived from multipotent cells (27), which indicated a higher mutational burden in somatic cells than in germ cells (28).
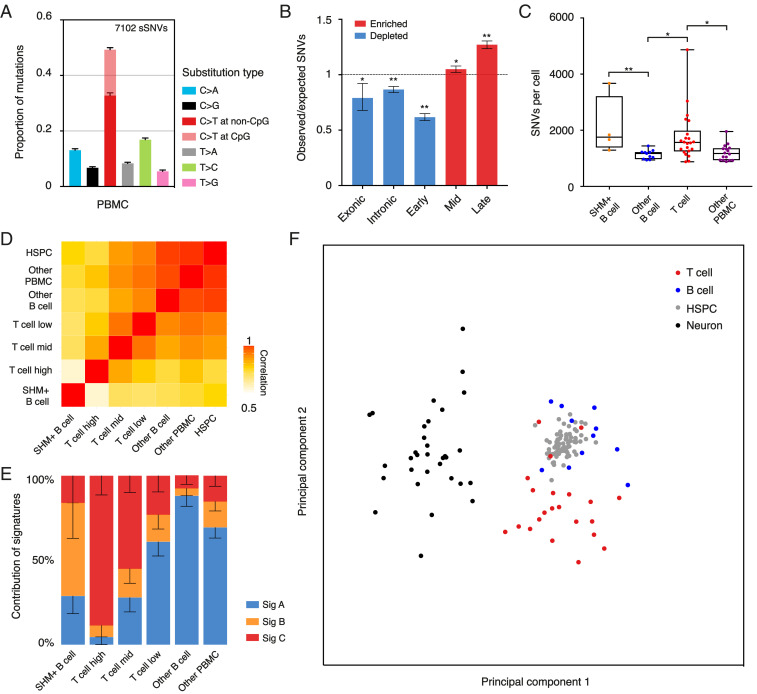
The mutational spectrum of PBMCs, which mainly consisted of C>T transitions (Fig. 4A), showed a strong correlation with the spectrum of hematopoietic stem cells (r = 0.991) (SI Appendix, Fig. S10). In addition, the genomic distribution of SNVs was depleted from transcribed regions but enriched in late-replicating domains (Fig. 4B), contrary to the neurons we studied but similar to mutations found in multipotent cells (27), suggesting that SNVs in PBMCs were mainly from HSPCs.
Fig. 4.
Mutational frequency and spectrum of SNVs vary across different types of cells. (A) Relative contribution of mutation types for PBMCs. Data are represented as the mean contribution of each mutation type from all single cells (53 PBMCs). Error bars represent SE. The total number of SNVs is indicated (Top). (B) Enrichment and depletion of SNVs detected in PBMCs in exons, introns, and replication timing domains (early, mid, and late). The expected values are calculated by assuming that SNVs distribute randomly along the genome. Errors bars denote 95% CI. *P < 0.005, **P < 0.0001, two-tailed binomial test. (C) Box and whisker plot of the number of SNVs identified in PBMCs. Each dot represents a single cell. *P < 0.05, **P < 0.01, two-tailed t test. (D) Correlation matrix of the mutational spectrum in a trinucleotide context for PBMCs and HSPCs. Data of HSPCs were obtained from ref. 25. (E) Proportion of the total number of detected SNVs in PBMCs as contributed by each mutational signature. Error bars represent SE. (F) Principal-component analysis of the mutational spectrum in a trinucleotide context for a T cell, B cell (SHM+ B cells were excluded), HSPC, and neuron. Each dot represents a single cell.
Similar to META, META-CS exhibited high amplification uniformity and was able to detect V(D)J recombination (SI Appendix, Fig. S11) (8). By V(D)J recombination patterns, 15 cells were inferred to be B lymphocytes and 22 cells were inferred to be T lymphocytes (SI Appendix, Fig. S12). B cells are known to undergo somatic hypermutation (SHM) during maturation, which introduces a high rate of mutation in immunoglobulin genes to fine-tune the antibody response. Out of the 15 B cells, we identified 4 SHM+ B cells defined as having at least three mutations clustering within 2-kb regions of an immunoglobulin gene (SI Appendix, Fig. S13). Mutations detected from immunoglobulin gene regions in SHM+ B cells were enriched in the predefined SHM hotspot motif (SI Appendix, Fig. S14) (29). In contrast, none of the T cells or other PBMCs had such hypermutations within the same regions. Similar to a recent study, we found that SHM+ B cells had a higher SNV frequency compared with other B cells (Fig. 4C) (30). It has been shown that targets of SHM are not limited to immunoglobulin genes, and aberrant SHM has been linked to certain types of B cell lymphomas (31, 32). However, it is unclear how SHM contributed to a higher rate of mutation across the whole genome.
Interestingly, after excluding the four SHM+ B cells, other B cells and PBMCs showed a lower number of SNVs than T cells (Fig. 4C). To further investigate the mutational mechanism, we divided the T cells into three subgroups based on the number of SNVs, corresponding to 3,038 ± 1,061 (high, n = 5), 1,645 ± 113 (mid, n = 8), and 1,170 ± 186 (low, n = 9) SNVs (SI Appendix, Fig. S15). A correlation matrix of the mutational spectrum in a trinucleotide context revealed that the spectrum of SHM+ B cells was most distinct from other blood cells (Fig. 4D and SI Appendix, Fig. S16). Excluding the SHM+ B cells, the spectra of other B cells and other PBMCs were closer to HSPCs than T cells. Within the T cell subgroups, the higher the SNV number, the further the spectrum diverged from other blood cells. By using nonnegative matrix factorization (33), we decomposed the mutational spectra of single blood cells into three mutational signatures (Fig. 4E and SI Appendix, Fig. S17). Signature A showed a high correlation with the mutational spectrum of HSPCs (r = 0.976) and contributed to most of the mutations detected in other B cells and other PBMCs. Signature B was mostly found in SHM+ B cells. Signature C mainly contributed to T cells and its contribution correlated with the number of SNVs in T cell subgroups. This observation implied that, compared with other B cells and PBMCs whose mutations were mostly from HSPCs, one or multiple additional mutational processes that generated mutations at a higher rate and caused a different mutational spectrum must exist in T cells. There are many potential mutational processes during the maturation and maintenance of T cells. An illustration of this is the reported long-lasting immunological memory maintained by a rapid turnover of memory T cells (34), which likely results in an accumulation of additional mutations, especially for immunological responses occurring at early stages of life.
Discussion
We present META-CS, a single-cell whole-genome amplification method that leverages the complementarity of the two strands from double-stranded DNA to achieve accurate SNV identification. META-CS exhibited four major advantages compared with previous methods. First, META-CS is not limited to diploid cells with heterozygous single-nucleotide polymorphisms (SNPs), which would be useful for haploid cells or cancer cells with aneuploidy. Second, the method is very robust. The success rate of single-cell amplification is around 90%. For example, 32 samples were successfully amplified from 36 single human neurons. Third, the whole reaction is completed in a single tube, which simplifies the experiment procedure and reduces sample loss. Fourth, with META-CS, a mutation can be identified with as few as four reads, which significantly reduces sequencing cost. In contrast to the 30 to 60× sequencing depth commonly used for single-cell SNV identification, most cells were sequenced between 3 and 8× in this work (Dataset S2).
We validated the accuracy of META-CS with clonally expanded kindred cells and human sperm and further applied the method to various human primary tissues. The mutational pattern of a single cell reflects the history of the cell’s and its precursor cells’ DNA damage and repair processes, which are closely related to cell function-specific features, such as transcription, DNA methylation, chromatin structure, and DNA replication. With highly accurate SNVs identified by META-CS, we were able to distinguish cell types based on single-cell mutational spectra. Principal-component analysis of single-cell mutational spectra in a trinucleotide context showed that neurons and blood cells were clearly separated into two clusters (Fig. 4F). Within the cluster of blood cells, despite being derived from common ancestral stem cells, the majority of T cells were well-separated from B cells. The clustering of B cells and HSPCs indicated the stem cell origin of most mutations found in B cells. Although the underlying mechanism causing the T cell-specific mutational spectrum needs further investigation, it is evident that, with the capability to detect single-cell SNVs with high accuracy, META-CS will expand our understanding of fundamental biology including development, aging, and generation of diseases such as cancer.
Materials and Methods
Human Subjects.
The peripheral blood and sperm cells were collected from a deidentified male donor with written consent. The procedure was approved by the Institutional Review Board at Harvard.
Fresh-frozen postmortem brain samples from the prefrontal cortex were obtained from the NIH NeuroBioBank at the University of Maryland. All three samples were Caucasians and tested negative for HIV and hepatitis B surface antigen. The youngest sample, UMBN4916, was a 19-y-and-47-d-old male with a postmortem interval (PMI) of 5 h, and died of drowning in a car accident. The intermediate sample, UMBN4915, was a 49-y-and-160-d-old male with a PMI of 5 h, and died of atherosclerotic cardiovascular disease. The oldest sample, UMBN5219, was a 76-y-and-348-d-old female with a PMI of 3 h, and died of complications of cancer.
Published Data.
Germline de novo SNVs from family trios were taken from table S4 of ref. 16. Parental SNVs were extracted with the “Phase_combined” column annotated as “father” and the “Phase_source” column annotated as “three_generation” or “both_approaches”.
Exonic and transcribed regions were downloaded from the UCSC Table Browser (35).
The regions of replication timing domains were downloaded from ref. 27 with the link https://wgs11.op.umcutrecht.nl/mutational_patterns_ASCs/data/genomic_regions/.
SNVs in human hematopoietic stem cells were downloaded from Mendeley Data (“Population dynamics of normal human blood inferred from spontaneous somatic mutations”: https://data.mendeley.com/datasets/yzjw2stk7f/1) in ref. 25. SNVs from clones labeled as “BMH” were used in the mutational spectrum and principal-component analysis.
Cell Culture.
The human haploid cell line, eHAP, was purchased from Horizon Discovery (C669). Prior to clonal expansion, the cells were first seeded in a 10-cm cell-culture dish supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin in Iscove’s modified Dulbecco’s media (IMDM) (Thermo Fisher; 12440053) and maintained at a confluency of <70% according to the cell-culture protocol from Horizon Discovery.
On day 0, spent medium from the culture plate was collected and centrifuged at 300 × g for 5 min; the supernatant was transferred to ∼96 wells on a 384-well cell-culture plate with 50 microliters per well. Upon medium removal, the cells on the plate were trypsinized using TrypLE (Gibco; 12605036) at 37 °C for 3 to 5 min, followed by addition of 10 mL complete medium (IMDM with 10% FBS and 1% penicillin-streptomycin) to quench TrypLE. The cells were centrifuged at 300 × g for 5 min and resuspended and diluted in complete medium to one cell per 10 microliters. Ten microliters of such a cell suspension was added to each well with medium on the 384-well plate (so that on average there was one cell in 60 μL of medium per well). After 3 h, each well was visually inspected under a microscope to check which wells contained one cell per well.
On day 5, cells in one well (which initially had one cell) were trypsinized using the same trypsinization procedure above (with TrypLE at 37 °C for 3 to 5 min, followed by addition of complete medium); 12 of the single cells were picked by mouth pipetting for the assay, while the rest of the cells were reseeded into one well on a 12-well plate with 2 mL of fresh complete medium.
The reseeded cells were cultured and maintained at a confluency of <70% for 8 more days, and on day 13 they were harvested for fluorescence-activated cell sorting (FACS) (using a BD FACSJazz) and bulk genomic DNA extraction.
Bulk DNA Extraction and Library Preparation.
Bulk DNA was extracted following the manufacturer’s protocol with the Qiagen DNeasy Blood & Tissue Kit (69504). Libraries were prepared with the Illumina TruSeq DNA PCR-Free Library Prep Kit (20015962).
Isolation of Single Cells.
Blood was drawn into K2EDTA-coated tubes (BD) and placed on ice immediately. Peripheral blood lymphocytes were isolated with Ficoll-Paque PLUS (GE) with 1× phosphate-buffered saline + 2 mM ethylenediaminetetraacetic acid (EDTA) as the salt solution (8).
Sperm were isolated from freshly ejaculated semen after swim-up in G-IVF PLUS (Vitrolife).
Neuronal nuclei were isolated as previously described (36). Both density gradient centrifugation and immunostaining (anti-NeuN, Alexa Fluor 488-conjugated; Millipore; MAB377X) were included. RNase inhibitor was omitted.
Single cells were mouth pipetted or sorted (FACSJazz flow cytometer; BD) into 0.2-mL ultraviolet-irradiated DNA low-bind tubes (MAXYMum Recovery; Axygen) containing lysis buffer.
Whole-Genome Amplification and Library Preparation by META-CS.
Tn5 transposase was expressed from the pTXB1-Tn5 plasmid (Addgene; 60240) and purified as previously described (37). DNA oligos were ordered from IDT with polyacrylamide-gel electrophoresis purification.
META-CS transposon DNA was prepared as previously described (8). We used 16 META tags in this work (Dataset S1).
Each of the 16 META transposons was annealed at a final concentration of 5 μM in annealing buffer (10 mM Tris, pH 7.5, 50 mM NaCl, 1 mM EDTA) and then pooled with equal volumes. Tn5 transposase and the pooled META transposons were mixed at an equal molar ratio at room temperature for 30 min to form transposomes, which were diluted to a final concentration of 125 nM and stored at −80 °C.
Single cells were lysed in 2 μL META lysis buffer (20 mM Tris, pH 8.0, 20 mM NaCl, 0.15% Triton X-100, 25 mM dithiothreitol, 1 mM EDTA, 1.5 mg/mL Qiagen protease [19155], 500 nM carrier ssDNA) at 50 °C for 1 h, 70 °C for 15 min. The sequence of the carrier ssDNA is the same as in LIANTI (5′-TCAGGTTTTCCTGAA-3′) (2). Lysed cells could be stored at −80 °C if not immediately amplified.
Single-cell lysate was transposed by the addition of 8 μL transposition mix (leading to a final concentration of 10 mM TAPS, pH 8.5, 5 mM MgCl2, 8% polyethylene glycol 8000, and 0.38 nM META transposome) and incubation at 55 °C for 10 min. Transposases were removed by the addition of 2 μL stop buffer containing 300 nM NaCl, 45 mM EDTA, 0.01% Triton X-100, and 1 mg/mL Qiagen protease and incubation at 50 °C for 30 min, 70 °C for 15 min.
First-strand tagging was performed by the addition of 13 μL Strand Tagging Mix 1 containing 5 μL Q5 reaction buffer, 5 μL Q5 high GC enhancer, 0.8 μL 50 μM (total) Adp1 primer mix (Dataset S1), 0.6 μL 100 mM MgCl2, 0.6 μL water, 0.5 μL 10 mM (each) dNTP mix, 0.25 μL of 20 mg/mL bovine serum albumin (NEB), and 0.25 μL Q5 DNA polymerase (NEB; M0491) and incubation at 72 °C for 3 min, 98 °C for 30 s, 62 °C for 5 min, 72 °C for 1 min. Adp1 primers were removed by the addition of 1 μL exonuclease I (NEB; M0293) and incubation at 37 °C for 30 min, 80 °C for 20 min.
Second-strand tagging was performed by the addition of 4 μL Strand Tagging Mix 2 containing 1 μL Q5 reaction buffer, 1 μL Q5 high GC enhancer, 1 μL 50 μM (total) Adp2 primer mix (Dataset S1), 0.855 μL water, 0.1 μL 10 mM (each) dNTP mix, and 0.05 μL Q5 DNA polymerase and incubation at 98 °C for 30 s, 62 °C for 5 min, 72 °C for 1 min. Adp2 primers were removed by the addition of 1 μL exonuclease I (NEB; M0293) and incubation at 37 °C for 30 min, 80 °C for 20 min.
Strand tagging products were then amplified by the addition of 19 μL PCR mix containing 5 μL NEBNext Multiplex Oligos Universal Primer, 5 μL NEB Index Primers (NEB; E7335S, E7500S, E7710S, E7730S), 4 μL Q5 reaction buffer, 4 μL Q5 high GC enhancer, 0.4 μL 10 mM (each) dNTP mix, 0.4 μL water, and 0.2 μL Q5 DNA polymerase and incubation at 98 °C for 20 s, 10 cycles (or 11 cycles for haploid cells) of [98 °C for 10 s, 72 °C for 2 min], 72 °C for 2 min.
Libraries were purified by DNA Clean and Concentrator-5 columns (Zymo; D4013) and amplification efficiency was checked by Bioanalyzer (SI Appendix, Fig. S3). Single-cell libraries were pooled together, and size selection was performed with Ampure XP beads (Beckman Coulter). To maximize fragment recovery, the pooled library was divided into three groups based on fragment size. Long-size fragments were selected by the addition of 0.6× beads. The supernatant was transferred to a new tube and midsize fragments were selected by further addition of 0.15× beads (final 0.75×). The supernatant was transferred to a new tube and short-size fragments were selected by addition of another 0.15× beads.
Sequencing.
Long-size libraries were sequenced with paired-end 2 × 250 bp on an Illumina HiSeq 2500. Midsize and short-size libraries were sequenced with paired-end 2 × 150 bp on an Illumina HiSeq 4000. Long-size and midsize libraries were sequenced for all single cells; short-size libraries were only sequenced for samples 340-2, 340-3, 340-4, 340-5, 340-13, 340-14, and 340-15. Twenty percent PhiX (Illumina; FC-110-3001) was added to avoid the low-complexity issue at the 19-bp transposase recognition site. A list of sequencing information is shown in Dataset S2.
Data Preprocessing.
We used premeta from https://github.com/lh3/pre-pe to preprocess raw single-cell paired-end reads. This tool identifies transposon insertion sites, merges overlapping ends, and trims Illumina sequencing adapters. We aligned preprocessed single-cell reads with two mappers, BWA-MEM v0.7.17 (38) and Minimap2 v2.12 (39), both with their default settings for short reads. The two-mapper strategy reduces false positive SNV calls caused by false read mapping.
Identification of Single-Nucleotide Variants.
We used lianti pileup from https://github.com/lh3/lianti to generate the initial variant call set, including germline variants and somatic SNVs, with the following command line: “lianti pileup -nLXXX -P20 -b um75-hs37d5.bed -ycf hs37d5.fa bulk.bam sample1.bwa.bam sample1.mm2.bam sample2.bwa.bam sample2.mm2.bam …”, where XXX is the number of single-cell binary alignment map (BAM) files, -P20 ignores alignments with a clipping 20 bp or longer, file “um75-hs37d5.bed” indicates high-quality regions in the hs37d5 version of the human reference genome, and -n considers fragment strand (i.e., the mapping strand of the first read in a read pair) for single-cell BAMs. The output of this command line is a multisample variant call format (VCF) file containing the number of forward and reverse reads at each potential variant site.
We called somatic SNVs by processing the initial call set with the plp-joint.js script from https://github.com/lh3/lianti. For brain samples, we used the command line “plp-joint.js -a4 -s2 -r all.rep -v gnomad-01.snp.txt.gz -u sample.vcf.gz,” where “gnomad-01.snp.txt.gz” gives the list of gnomAD calls having ≥1% population frequency and “all.rep” indicates which BAMs are generated from the same sample/cell. We call an ALT (i.e., nonreference) allele if the allele is supported by at least four reads in total (-a4) and two reads from each strand (-s2). As we are using two mappers, we always choose the smaller read count between the two mappers. The plp-joint.js script also filters somatic calls if the ALT allele balance (the fraction of ALT reads) is below 20%, drops calls overlapping with gnomAD ≥1% calls, removes somatic calls within 100 bp from each other (this filter was removed with option -w0 for calling the somatic hypermutations in B cells), and filters calls within 10 bp from the 5′ or 3′ end of reads. The procedure above calls ALT alleles, including both germline SNPs and somatic SNVs. We classify an ALT allele to be somatic if no bulk reads support the ALT allele. We estimate the false negative rate of SNV by comparing ALT calls in single cells and germline heterozygous SNPs in the bulk.
Sperm and blood cells were sequenced from the same individual. We called somatic SNVs by jointly considering both sperm and blood bulks with command line “plp-joint.js -r all.rep -v gnomad-01.snp.txt.gz -u -a4 -s2 -D40 -A15 -b2 sample.vcf.gz.” With two bulks, we can further call a somatic SNV if only one of the bulks has no reads supporting the SNV allele. The false negative rate in sperm samples is estimated by excluding all heterozygous calls by the plp-joint.js command with -h all.hap option, where “all.hap” indicates which cells are haploid. The rest of the treatment is identical.
SNVs in eHAP cells are called by the command line “plp-joint.js -r all.rep -v gnomad-01.snp.txt.gz -u -a4 -s2 -D40 -A15 -P sample.vcf.gz.” Similar to sperm, the false negative rates are estimated by excluding all heterozygous calls. The calling thresholds are examined with -a and -s options. For example, -a4 -s0 indicates that a call needs to be supported by at least four reads in total without requirement of strand-specific reads.
A full description of SNVs detected in 133 single cells is given in Dataset S3.
Identification of ssDNA Damage.
Potential ssDNA damage-associated mutations and amplification artifacts (including damage generated during WGA and polymerase errors), which are indistinguishable in the experiment, are called with a similar procedure except that we require a call to be supported by at least four reads from the mutational allele and four reads from the reference allele (SI Appendix, Fig. S8 and Dataset S3).
Genomic Distribution Analysis.
Overlapping regions of the downloaded browser extensible data (BED) files of exonic and transcribed regions were merged with BEDTools (40). Intronic regions were determined by the subtraction of exonic regions from transcribed regions. The number of mutations observed was determined by intersecting with surveyed genomic regions (for autosomes only). The expected number of mutations was calculated by multiplying the genome-wide mutation frequency by the length of surveyed genomic regions. Two-tailed binomial tests were performed with P < 0.05 considered significant.
Mutational Signature Analysis.
The mutational frequency of each single cell (or single-cell clone derived from stem cells) was calculated in a context of 96 trinucleotide substitution, with 5′ and 3′ sequences acquired from GRCh37. Three mutational signatures were extracted using SigProfiler (41) from 53 single peripheral blood mononuclear cells. The contribution of three mutational signatures to each cell type or group was calculated by the mean contribution for each single cell within the group.
Principal-Component Analysis.
Principal-component analysis was performed using MATLAB. The mutational frequency in a context of 96 trinucleotide substitutions of 32 PFC neurons, 22 T cells, 11 B cells, and 73 hematopoietic stem cell clones was used for the analysis shown in Fig. 4F.
Data Availability
Raw sequencing data reported in this article have been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. PRJNA533595). Code is available at GitHub (https://github.com/lh3/lianti and https://github.com/lh3/pre-pe).
Supplementary Material
Acknowledgments
We thank S. Mulepati (Harvard University, currently Intellia Therapeutics) and C. Chen (Harvard University, currently NIH) for their advice at the early stage of the experiment design. This work was performed during the transition period when X.S.X. relocated to Peking University and was supported by the Beijing Advanced Innovation Center for Genomics at Peking University and by a generous gift grant from Dr. Xianhong Wu to Harvard University.
Footnotes
Competing interest statement: D.X., L.T., C.-H.C., and X.S.X. are investors on Patent WO2018217912A1 filed by the president and Fellows of Harvard College that covers META-CS.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013106118/-/DCSupplemental.
References
- 1.Tindall K. R., Kunkel T. A., Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry 27, 6008–6013 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Chen C., et al., Single-cell whole-genome analyses by linear amplification via transposon insertion (LIANTI). Science 356, 189–194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitt M. W., et al., Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, 14508–14513 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoang M. L., et al., Genome-wide quantification of rare somatic mutations in normal human tissues using massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 113, 9846–9851 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu W. K., et al., Ultraaccurate genome sequencing and haplotyping of single human cells. Proc. Natl. Acad. Sci. U.S.A. 114, 12512–12517 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong X., et al., Accurate identification of single-nucleotide variants in whole-genome-amplified single cells. Nat. Methods 14, 491–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohrson C. L., et al., Linked-read analysis identifies mutations in single-cell DNA-sequencing data. Nat. Genet. 51, 749–754 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan L., Xing D., Chang C. H., Li H., Xie X. S., Three-dimensional genome structures of single diploid human cells. Science 361, 924–928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zong C., Lu S., Chapman A. R., Xie X. S., Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 338, 1622–1626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potapov V., Ong J. L., Examining sources of error in PCR by single-molecule sequencing. PLoS One 12, e0169774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong A., et al., Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488, 471–475 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahbari R.et al.; UK10K Consortium , Timing, rates and spectra of human germline mutation. Nat. Genet. 48, 126–133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldmann J. M., et al., Parent-of-origin-specific signatures of de novo mutations. Nat. Genet. 48, 935–939 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Wang J., Fan H. C., Behr B., Quake S. R., Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150, 402–412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., et al., Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155–160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jónsson H., et al., Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature 549, 519–522 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Lodato M. A., et al., Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 350, 94–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lodato M. A., et al., Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359, 555–559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strachan T., Goodship J., Chinnery P. F., Genetics and Genomics in Medicine (Garland Science/Taylor & Francis Group, New York, 2015). [Google Scholar]
- 20.Stamatoyannopoulos J. A., et al., Human mutation rate associated with DNA replication timing. Nat. Genet. 41, 393–395 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng K. C., Cahill D. S., Kasai H., Nishimura S., Loeb L. A., 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G----T and A—C substitutions. J. Biol. Chem. 267, 166–172 (1992). [PubMed] [Google Scholar]
- 22.Lu T., et al., Gene regulation and DNA damage in the ageing human brain. Nature 429, 883–891 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Mardis E. R., et al., Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361, 1058–1066 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welch J. S., et al., The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–278 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee-Six H., et al., Population dynamics of normal human blood inferred from somatic mutations. Nature 561, 473–478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osorio F. G., et al., Somatic mutations reveal lineage relationships and age-related mutagenesis in human hematopoiesis. Cell Rep. 25, 2308–2316.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blokzijl F., et al., Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milholland B., et al., Differences between germline and somatic mutation rates in humans and mice. Nat. Commun. 8, 15183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogozin I. B., Diaz M., Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol. 172, 3382–3384 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Zhang L., et al., Single-cell whole-genome sequencing reveals the functional landscape of somatic mutations in B lymphocytes across the human lifespan. Proc. Natl. Acad. Sci. U.S.A. 116, 9014–9019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen H. M., Peters A., Baron B., Zhu X., Storb U., Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science 280, 1750–1752 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Pasqualucci L., et al., Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 412, 341–346 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Alexandrov L. B., Nik-Zainal S., Wedge D. C., Campbell P. J., Stratton M. R., Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 3, 246–259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michie C. A., McLean A., Alcock C., Beverley P. C., Lifespan of human lymphocyte subsets defined by CD45 isoforms. Nature 360, 264–265 (1992). [DOI] [PubMed] [Google Scholar]
- 35.Karolchik D., et al., The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krishnaswami S. R., et al., Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat. Protoc. 11, 499–524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Picelli S., et al., Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 24, 2033–2040 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint] (2013). https://arxiv.org/abs/1303.3997 (Accessed 20 March 2019).
- 39.Li H., Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexandrov L., SigProfiler (MATLAB Central File Exchange) (2019). https://www.mathworks.com/matlabcentral/fileexchange/38724-sigprofiler. Accessed 28 April 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data reported in this article have been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. PRJNA533595). Code is available at GitHub (https://github.com/lh3/lianti and https://github.com/lh3/pre-pe).