Significance
In this work, we elucidate a checkpoint dampening mechanism in yeast. Using complementary biochemical and genetic approaches, we show that the Srs2 DNA helicase removes one of the first DNA damage sensors and the associated checkpoint kinase from chromatin, thus preventing hyperactivation of the DNA damage response. We further show that this role of Srs2 is separable from its well-known function as an antirecombinase and mainly accounts for Srs2’s contribution to genotoxin resistance. Our findings also shed light into potential means to regulate the dynamic association of RPA with single-stranded DNA in other cellular contexts and stimulate studies of checkpoint dampening and RPA regulation in other organisms.
Keywords: RPA regulation, Srs2, checkpoint dampening, genotoxic stress, recombinational repair
Abstract
The DNA damage checkpoint induces many cellular changes to cope with genotoxic stress. However, persistent checkpoint signaling can be detrimental to growth partly due to blockage of cell cycle resumption. Checkpoint dampening is essential to counter such harmful effects, but its mechanisms remain to be understood. Here, we show that the DNA helicase Srs2 removes a key checkpoint sensor complex, RPA, from chromatin to down-regulate checkpoint signaling in budding yeast. The Srs2 and RPA antagonism is supported by their numerous suppressive genetic interactions. Importantly, moderate reduction of RPA binding to single-strand DNA (ssDNA) rescues hypercheckpoint signaling caused by the loss of Srs2 or its helicase activity. This rescue correlates with a reduction in the accumulated RPA and the associated checkpoint kinase on chromatin in srs2 mutants. Moreover, our data suggest that Srs2 regulation of RPA is separable from its roles in recombinational repair and critically contributes to genotoxin resistance. We conclude that dampening checkpoint by Srs2-mediated RPA recycling from chromatin aids cellular survival of genotoxic stress and has potential implications in other types of DNA transactions.
Cellular survival of genotoxic stress relies on the highly conserved DNA damage checkpoint (DDC). The DDC can sense genome lesions and induce protection mechanisms such as cell cycle arrest that provides time for genome repair (1). Defects in DDC underlie numerous human genome instability syndromes and influence tumorigenesis (2). A universal DDC sensor in eukaryotes is the RPA complex that has strong affinity to single-strand DNA (ssDNA), a structure commonly generated under genotoxic stress (3). The RPA-ssDNA filament can recruit an apical checkpoint kinase via direct binding to its obligate cofactor. In budding yeast, for example, RPA binding to the Mec1 checkpoint kinase’s cofactor Ddc2 targets the Mec1-Ddc2 complex to DNA lesion sites (4, 5). This is an early and critical event to initiate the Mec1-mediated DDC. Subsequent Mec1 activation of a key downstream effector kinase Rad53 leads to phosphorylation of a myriad of substrates to induce cell cycle arrest and other cellular changes (6).
While turning on the DDC is crucial for cells to cope with genotoxic stress, its timely termination is equally important, partly because cell cycle resumption is required for continued growth (1). Specific phosphatases were found to directly antagonize the DDC kinases (7, 8). More recently, checkpoint dampening factors Slx4 and Sae2 were shown to antagonize the DDC adaptor protein, Rad9, in yeast. Rad9 association with damaged chromatin promotes Rad53 activation, whereas Slx4 and Sae2 favor Rad9 removal from chromatin (9, 10). Slx4 and Sae2 have been classically viewed as DNA repair factors that enable different nucleolytic steps during homologous recombination (HR) (11–13). However, their recently discovered roles in DDC dampening appear to be more critical for genotoxin resistance (9, 10, 14), highlighting the importance of DDC termination and its close relationship with DNA repair factors.
Given the central role of RPA in the DDC pathway, it is conceivable that persistent association of RPA and Mec1-Ddc2 with ssDNA could be a major impediment in DDC termination. How cells cope with this issue and whether active removal of RPA from DNA is needed for DDC termination are not known. Understanding these questions will shed light on DDC control and cellular survival of genotoxic stress. It may also more broadly enhance our understanding of genome maintenance since RPA-ssDNA association affects most DNA transaction processes.
To address the above questions, we have been searching for a potential factor that may act as an RPA antagonist during DDC termination. One candidate is the Srs2 DNA helicase, because it has been implicated in terminating DDC signaling after a single double-strand break (DSB) is generated in cells (15). Srs2 has been predominantly studied as an antirecombinase that can remove the Rad51 recombinase from ssDNA to limit recombinational reactions (16, 17). However, whether this or another role of Srs2 is required in checkpoint regulation has not been clear. In this study, we uncovered numerous suppressive genetic interactions between Srs2 and RPA, revealing their antagonistic relationship. Using RPA mutants generated and biochemically characterized in this work, we elucidate the basis of Srs2 and RPA antagonism, demonstrating that Srs2 can promote DDC dampening by removing RPA and an associated checkpoint kinase from chromatin. Our data further show that this role of Srs2 is critical not only in a single DSB situation but also in genotoxin conditions.
Results
Srs2 Loss Rescues the DNA Damage Sensitivities of rfa1 Mutants.
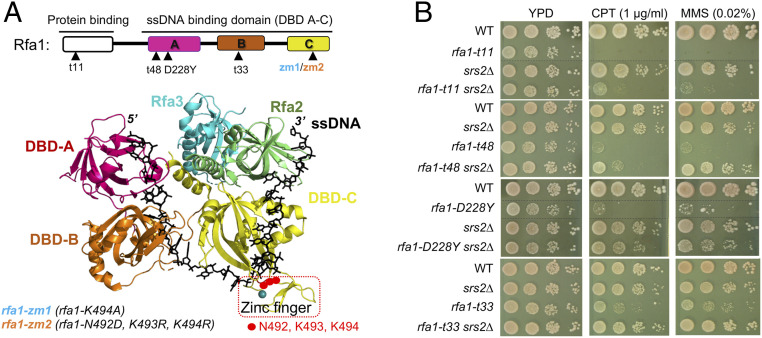
An antagonistic relationship between Srs2 and RPA predicts suppressive interactions among their mutants. We examined four commonly used hypomorphic alleles of RPA (RPA is essential) that affect its large subunit Rfa1. Rfa1 contains three ssDNA binding domains (DBD-A to DBD-C) and a protein binding domain (Fig. 1A). The examined alleles include rfa1-t11 that affects its protein binding domain and reduces interactions with Ddc2 and other proteins, and three DBD mutants that reduce ssDNA binding, namely rfa1-t48, rfa1-D228Y (affecting DBD-A), and rfa1-t33 (affecting DBD-B); all alleles are known to cause pleiotropic defects (Fig. 1A) (5, 18–20).
Fig. 1.
RPA and Srs2 show antagonistic relationship. (A, Upper) Schematic of the Rfa1 protein domains and rfa1 mutant alleles. (Lower) Structure of the Ustilago maydis RPA in complex with ssDNA (black sticks) (23). The Zn2+ finger region is boxed red. (B) rfa1 mutants’ sensitivity toward CPT and MMS is suppressed by srs2Δ. A 10-fold serial dilution of cells of the indicated genotypes were spotted and growth was assessed after incubation at 30 °C for 3 d. Dashed lines indicate removal of superfluous rows.
We tested the above rfa1 mutants with srs2∆ in genotoxic conditions caused by the Top1 trapping compound camptothecin (CPT) or by the DNA methylation agent methylmethane sulfonate (MMS). Under low drug concentrations wherein srs2∆ cells showed proficient growth, all tested rfa1 mutants showed strong sensitivity as seen previously (Fig. 1B) (20). Strikingly, CPT and MMS sensitivities of all tested rfa1 mutants were rescued by srs2Δ (Fig. 1B). Compared with rfa1-t11, rfa1 mutants affecting DBD-A and DBD-B showed stronger improvement upon Srs2 loss, suggesting that srs2∆ suppression is greater toward DNA binding-defective rfa1 alleles (Fig. 1B). As cells were spotted in 10-fold serial dilutions, srs2∆ suppression of rfa1-t48, rfa1-D228Y, and rfa1-t33 was estimated to be ∼10–1,000-fold. The positive genetic interactions seen here are unique, since rfa1 mutants show extensive negative interactions with other mutants (21). To our knowledge, srs2∆ is the first suppressor of four rfa1 alleles, thus providing strong evidence for the antagonistic relationship between Srs2 and RPA.
rfa1 Mutants Affecting DBD-C Moderately Reduce ssDNA Binding.
The examined rfa1 mutants cause pleiotropic defects and reduced Rfa1 protein levels (22), thus were not well suited for determining the basis of the RPA and Srs2 antagonism. We, therefore, attempted to generate mild rfa1 mutants that slightly impair ssDNA binding but maintain protein levels and overall functions. We focused on DBD-C as it contacts more nucleotides on ssDNA than the other DBDs, thus may better tolerate DNA binding mutations (23). DBD-C also uniquely harbors a peripherally located Zn-finger domain that contacts the ssDNA backbone (Fig. 1A and SI Appendix, Fig. S1A) (23). We reasoned that small perturbations in the Zn-finger domain may specifically reduce RPA-ssDNA binding without affecting Rfa1 folding. We thus mutated three DNA-contacting residues (N492, K493, K494) located at the N-terminal boundary of the Zn finger domain to generate two alleles, referred to as rfa1-zm1 (K494A) and rfa1-zm2 (N492D, K493R, K494R), or collectively as rfa1-zm hereafter (Fig. 1A and SI Appendix, Fig. S1A).
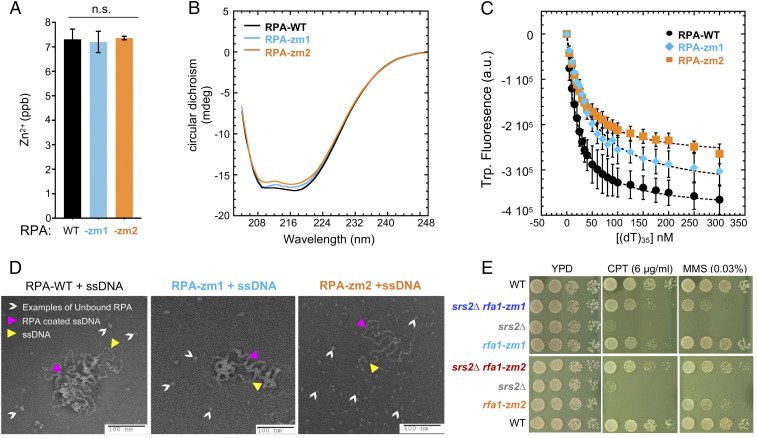
We examined how the newly generated mutants affect RPA complex behavior in vitro. The RPA complex containing the rfa1-zm1 or rfa1-zm2 (RPA-zm1 or -zm2) formed heterotrimers and behaved similar to the WT complex during purification (SI Appendix, Fig. S1B). Moreover, mutant RPA complexes contained WT levels of Zn2+ as assayed by inductively coupled plasma mass spectrometry, suggesting that the Zn-finger structure was intact (Fig. 2A). Further, secondary structure analyses using circular dichroism showed that RPA-zm1 or RPA-zm2 exhibited a WT profile (Fig. 2B). These data suggest that RPA-zm1 and RPA-zm2 do not affect the overall structure of the complex.
Fig. 2.
rfa1-zm mutants moderately reduce ssDNA binding and suppress srs2∆ genotoxin sensitivity. (A) ICP-MS results show that the concentration of Zn2+ per mole of RPA is similar between WT and mutant RPA complexes. (B) WT and mutant RPA complexes have similar CD spectra. (C) Intrinsic tryptophan fluorescence signals upon ssDNA binding by RPA complexes. (D) Negative stain electron microscopy images of RPA filaments on M13 ssDNA. RPA bound to ssDNA as nucleoprotein complexes are denoted by pink arrowheads. Examples of DNA-free RPA is denoted by white arrowheads. Examples of ssDNA region is marked with yellow arrowheads. More free RPA molecules were observed for RPA-zm1 and even more for RPA-zm2 compared to WT RPA. (E) rfa1-zm mutants suppress srs2Δ sensitivity toward CPT and MMS. Experiments were done as described in Fig. 1B.
We next assayed the affinity of RPA complexes toward (dT)35 ssDNA using tryptophan quenching. RPA-zm1 or RPA-zm2 exhibited a moderate reduction in overall ssDNA binding: while WT RPA bound to (dT)35 with a KD of 20.6 ± 7 nM, RPA-zm1 and RPA-zm2 exhibited a KD of 29.8 ± 5 nM and 45 ± 9 nM, respectively (Fig. 2C). A stronger defect seen for RPA-zm2 is consistent with three DNA-contacting residues being mutated compared to a single residue being mutated in RPA-zm1 (SI Appendix, Fig. S1A).
Altered Density of the RPA-zm-ssDNA Nucleoprotein Filament.
To gain a deeper understanding of the reduced ability of RPA-zm to associate with ssDNA, we used electron microscopy to visualize the nucleoprotein filaments formed by the mutant proteins. The 6.4-kb M13 ssDNA was used for imaging longer RPA nucleoprotein filaments. We observed that WT RPA formed highly compacted nucleoprotein filaments with almost no free ssDNA visible (Fig. 2D). In contrast, lower DNA compaction was observed for RPA-zm1 and more so for RPA-zm2 (Fig. 2D). This finding supports the reduced ssDNA binding ability of RPA-zm and further suggests increased access to ssDNA by other proteins that can remodel or displace mutant RPA from DNA. Collectively, our in vitro data demonstrate that RPA-zm1 and RPA-zm2 maintain overall WT attributes but display reduced ssDNA binding properties.
rfa1-zm Alleles Suppress DNA Damage Sensitivity of srs2Δ Cells.
We subjected rfa1-zm1 and rfa1-zm2 mutants to a series of in vivo analyses. Unlike commonly used rfa1 alleles, rfa1-zm mutants maintained Rfa1 protein levels with or without CPT and MMS treatment (SI Appendix, Fig. S1C) and supported growth at 24 °C, 30 °C, and 37 °C on either normal media or media containing low dose of CPT and MMS (SI Appendix, Fig. S1 D and E). In addition, chromosome replication in rfa1-zm cells was similar to that of wild type (SI Appendix, Fig. S1 F and G). These data differentiate rfa1-zm from commonly used rfa1 mutants and are consistent with in vitro WT-like attributes of RPA-zm complexes described above.
We then tested how rfa1-zm mutants alone or in combination with srs2∆ affected genotoxic resistance. At a high dose of CPT, srs2∆ cells showed a stronger growth defect than rfa1-zm1 cells (Fig. 2E). Strikingly, rfa1-zm1 rescued srs2∆ growth to the level of rfa1-zm1 cells (Fig. 2E). A mutual suppression was seen between rfa1-zm2 and srs2∆: While either single mutant exhibited strong CPT sensitivity, their combined mutations supported close to WT levels of resistance (Fig. 2E). We estimated rfa1-zm suppression of srs2∆ CPT sensitivity to be 100–1,000-fold. Similar results were seen in MMS conditions: rfa1-zm1 suppressed srs2∆ sensitivity and a mutual suppression was seen for rfa1-zm2 and srs2∆ (Fig. 2E). These results using mildly defective rfa1 alleles further support the antagonistic relationship between RPA and Srs2.
rfa1-zm Mutants Reduce Hyper-Checkpoint of srs2Δ Cells in CPT and MMS Conditions.
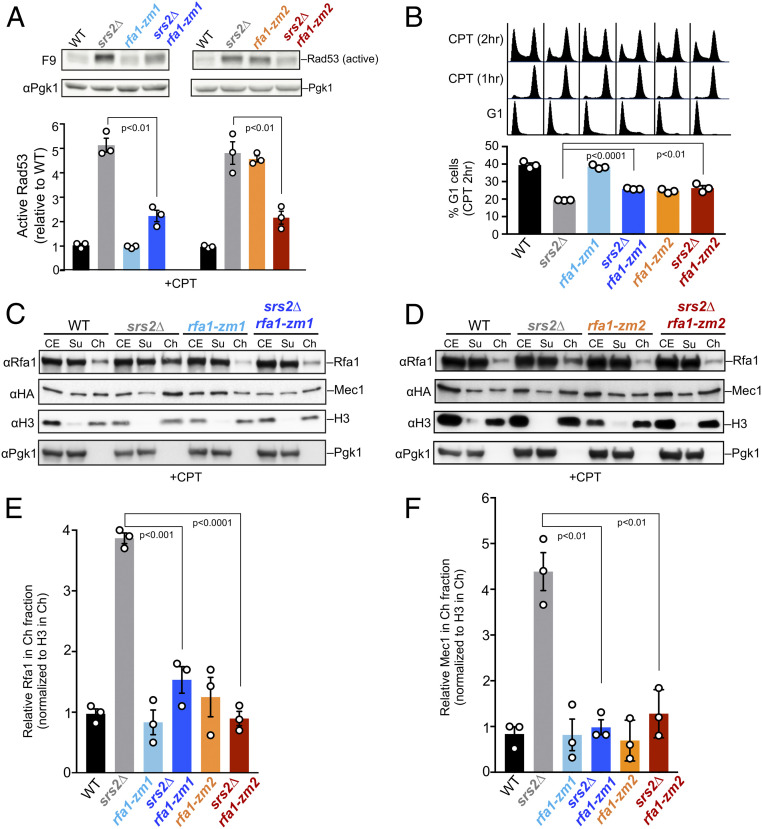
The overall proficiency of rfa1-zm mutants enabled us to determine the mechanisms by which they suppress srs2 sensitivities to genotoxic stress. To this end, we first queried DDC levels by monitoring Rad53 activation, which is detected by the F9 antibody (24). Compared with WT cells, srs2∆ cells exhibited ∼fivefold increase in active Rad53 level after CPT treatment, suggesting DDC hyperactivation (Fig. 3A). Both rfa1-zm mutants reduced this increase by ∼50% (Fig. 3A). While active Rad53 levels in rfa1-zm1 cells were similar to wild type, rfa1-zm2 cells exhibited a similar increase as srs2Δ cells, thus rfa1-zm2 and srs2∆ were mutually suppressive in this assay as seen in drug resistance tests (Fig. 2E). Similar results were seen in MMS conditions: Increased levels of active Rad53 in srs2∆ cells were suppressed by rfa1-zm1 or rfa1-zm2 (SI Appendix, Fig. S2A).
Fig. 3.
rfa1-zm mutants reduce hyperactivation of Rad53 and hyperchromatin association of RPA and Mec1 in srs2Δ cells. (A) rfa1-zm mutants reduce the levels of active Rad53 in srs2∆ cells. G1-arrested cells were released into cycling in the presence of CPT for 2 h. Activated Rad53 was detected by the F9 antibody by immunoblotting. Active Rad53 signals were compared to the Pgk1 loading control and normalized to wild type. (B) rfa1-zm mutants allow better G1 entry of srs2∆ cells. Experiment was performed as in A, except that FACS of samples of indicated time point is shown at the top and a graph for percentage of G1 cells after 2 h of CPT treatment (CPT 2 h) is at the bottom. (C and D) rfa1-zm mutants reduce chromatin-bound Rfa1 and Mec1 in srs2∆ cells upon CPT treatment. Rfa1 and HA-tagged Mec1 in cell extract (CE), chromatin-bound (Ch), and chromatin-unbound (Su) fractions were examined. H3 and Pgk1 are markers for Ch and Su fractions, respectively. (E and F) Quantification of the levels of chromatin-bound Rfa1 (E) and Mec1 (F) after being normalized to H3. For A, B, E, and F, mean of three biological isolates per genotype is graphed with error bars representing SEM. Statistically significant difference by Student’s t test are indicated by P values.
We next queried exit from G2/M arrest as another DDC readout. WT and rfa1-zm cells were arrested in G2/M phase after 1 h of CPT treatment (Fig. 3B). Another hour later, while ∼40% WT cells exited this arrest and moved on to G1, only ∼19% srs2Δ cells behaved this way (Fig. 3B). The ∼twofold reduction of G1 cells in srs2∆ background is consistent with the hyperactivation of Rad53 described above (Fig. 3A). In line with their abilities to reduce active-Rad53 levels in srs2∆ cells, rfa1-zm1 and rfa1-zm2 allowed more srs2∆ cells to transition to G1 (Fig. 3B). Similar results were obtained in MMS treatment. In brief, when cells were released from MMS treatment, ∼42% WT cells progressed into G1, while only ∼16% srs2Δ cells did so (SI Appendix, Fig. S2B). Again, rfa1-zm1 and rfa1-zm2 increased G1 percentages in srs2Δ cells, and the increase was about 40% of the level seen for srs2∆ cells (SI Appendix, Fig. S2B). Thus, the above data suggests that a common mechanism of rfa1-zm suppression of srs2∆ sensitivity toward genotoxins is via DDR down-regulation.
rfa1-zm Mutants Reduce Hyper-Checkpoint of srs2Δ Cells upon a Single DSB Generation.
We also examined the effects of rfa1-zm mutants in srs2Δ cells after a single DSB was generated by the HO endonuclease, analogous to the initial study implicating Srs2 in DDC (15). In this system, upon galactose-induced HO expression, a DSB is generated on chromosome III. Repairing this break can proceed after extensive resection over a 30-kb region, which exposes a homologous sequence to support single-strand annealing (SSA) (SI Appendix, Fig. S3A), although the repair can also occur by break-induced replication (15, 25). In this system, srs2Δ cells are sensitive to the generation of a single DSB due to persistent DDC signaling (15). Interestingly, the two rfa1-zm mutants suppressed this sensitivity by ∼100-fold (SI Appendix, Fig. S3B). Moreover, while srs2∆ cells showed increased levels of active Rad53 as seen previously (15), both rfa1-zm1 and rfa1-zm2 were able to reduce this (SI Appendix, Fig. S3C). In conjunction with our data in CPT and MMS conditions, these results suggest that rfa1-zm is a general suppressor of hyper-checkpoint levels in srs2∆ cells under different types of DNA damage conditions. We focused on the CPT and MMS conditions hereafter.
Increased Chromatin Association of RPA and Mec1 in srs2Δ Cells Is Rescued by rfa1-zm.
We moved on to test whether CPT and MMS sensitivity and hyper-checkpoint seen in srs2∆ cells are due to increased chromatin association of RPA and Mec1. Chromatin fractionation tests performed in CPT conditions showed that srs2Δ led to ∼fourfold increase of Mec1 and RPA on chromatin compared with WT cells (Fig. 3 C–F). Importantly, both rfa1-zm1 and rfa1-zm2 reversed this effect (Fig. 3 C–F). Similar results were obtained in MMS conditions (SI Appendix, Fig. S4). These data suggest that checkpoint hyperactivation associated with srs2∆ can be explained by increased chromatin association of RPA and the Mec1 kinase. Given the reduced ability of RPA-zm to associate with ssDNA, its mutants may bypass the need for Srs2 in releasing RPA and Mec1-Ddc2 from DNA. Our data support the notion that Srs2 can displace RPA and the associated Mec1-Ddc2 from chromatin in vivo, thus dampening the checkpoint.
rfa1-zm Mutants Are Proficient for HR and Do Not Rescue Hyper-Recombination Phenotype of srs2∆ Cells.
Srs2 is well studied as an antirecombinase that removes the Rad51 recombinase from ssDNA (16, 17). Indeed, Srs2 loss increases HR rates (hyper-rec) and HR intermediate levels, which cause lethality when HR intermediate removing enzymes, such as the Sgs1 helicase, are also absent (26, 27). Both types of defects are suppressible by reducing Rad51 functions (28, 29). As RPA acts upstream of Rad51 during HR, it is formally possible that rfa1-zm may mimic a rad51 mutant. We addressed this possibility by asking if rfa1-zm behave like rad51 mutants in reducing HR in srs2∆ cells or rescuing sgs1∆ srs2∆ lethality.
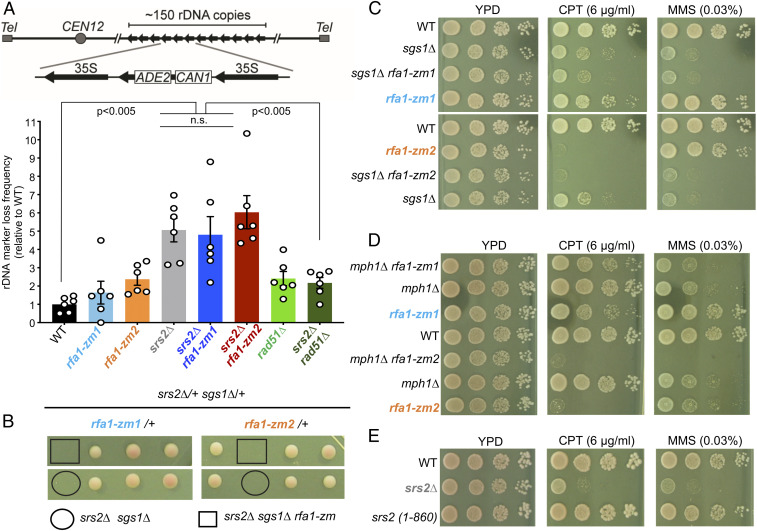
Using a HR assay in the ribosomal DNA (rDNA) locus, wherein HR can lead to the loss of an internal ADE2-CAN1 cassette, we confirmed that srs2∆ led to ∼fivefold increase in recombination frequency (30) (Fig. 4A). As expected, rad51∆ suppressed this increase (Fig. 4A). In contrast, rfa1-zm mutants did not rescue the hyper-rec phenotype seen in srs2∆ cells (Fig. 4A). In addition, rfa1-zm mutants did not affect HR frequency themselves; their moderately increased frequency from that of WT cells was not statistically significant (Fig. 4A).
Fig. 4.
rfa1-zm mutants rescue of srs2∆ genotoxic sensitivity is separable from HR regulation. (A) rDNA marker loss rate measurement. (Upper) Schematic of the assay. (Lower) Averages of marker loss rates with error bars representing SEM and significant difference of P < 0.005 (Student’s t test) is indicated. n = 6, three colonies from two biological isolates were used. (B) rfa1-zm mutants do not rescue srs2∆ sgs1∆ synthetic lethality. Representative tetrads of diploids heterozygous for indicated mutations are shown. Spore clones were grown at 30 °C for 2 d. (C and D) rfa1-zm mutants do not rescue DNA damage sensitivity of sgs1∆ and mph1∆. Cells of the indicated genotypes were spotted, and experiment was done as described in Fig. 1B. (E) Srs2 C-terminal domain is largely dispensable for coping with DNA damage. Cells of the indicated genotypes were spotted, and experiment was done as described in Fig. 1B.
We also tested HR frequency using an assay that produces recombinants by gene conversion or deletion outside the rDNA locus (SI Appendix, Fig. S5A) (31). In this system, two copies of defective leu2 genes containing different mutations can be used to restore the WT LEU2 genes using Rad51-mediated gene conversion or Rad51-independent SSA. While both events generate Leu+ cells, only gene conversion retains the URA3 marker inserted between the two copies of leu2 genes. As shown previously, srs2∆ increased gene conversion and LEU2 recombination rates (SI Appendix, Fig. S5A). Again, rfa1-zm mutants did not reduce either rate or rescue the hyper-rec phenotype of srs2∆ (SI Appendix, Fig. S5A), These results distinguish rfa1-zm from rad51 mutants. We note that rfa1-zm mutants did not change gene conversion rates but showed moderate increases in overall LEU2 recombination rate, suggesting possible increase in SSA events.
rfa1-zm Suppression of srs2∆ Drug Sensitivity Is Separable from HR Regulation.
Additional tests described below further demonstrate that rfa1-zm suppression of srs2∆ genotoxin sensitivity is separable from HR regulation. First, rfa1-zm mutants did not rescue the srs2∆ sgs1∆ synthetic lethality (Fig. 4B). Second, we examined rfa1-zm interaction with Sgs1 and Mph1, two DNA helicases that disfavor HR intermediate formation similar to Srs2 (32). Distinct from the previously known rad51 suppression of sgs1 or mph1 mutant defects, rfa1-zm mutants did not rescue the CPT or MMS sensitivities of either helicase mutant (Fig. 4 C and D) (33–35). Thus, rfa1-zm suppression of srs2∆ drug sensitivity is specific and not phenocopied when helicases with roles similar to Srs2 in HR are mutated. These data argue that rfa1-zm bypasses the need of Srs2 via a mechanism separable from modulating HR.
Third, we examined a Srs2 mutant (srs2-1-860) that is devoid of the Rad51 binding regions (26, 36). The growth of srs2-1-860 cells was similar to wild type on both CPT and MMS media (Fig. 4E). Thus, while the loss of Srs2 antirecombinase function causes hyper-rec, its contribution toward DNA damage sensitivity is limited. Along this line, while rad51 mutants suppress the HR-related phenotype of srs2∆ (28, 29), rad51∆ did not reduce active-Rad53 levels in srs2Δ cells in CPT conditions (SI Appendix, Fig. S5B). In summary, multiple lines of evidence support the conclusion that srs2∆ genotoxin sensitivity and rfa1-zm suppression of this sensitivity are separable from HR regulation.
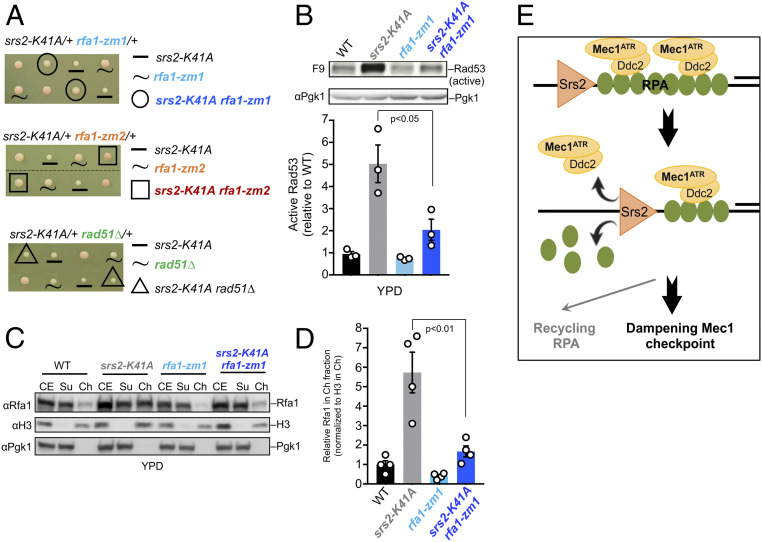
rfa1-zm Mutants Rescue Multiple Defects Caused by Srs2 Helicase Inactivation.
Thus far, our data suggest that Srs2 contributes to checkpoint dampening by counteracting RPA association with chromatin. Given Srs2’s helicase activity, we asked whether this function is required for RPA removal from chromatin. To this end, we examined if impairing Srs2 helicase activity by using the srs2-K41A allele (37) could increase RPA chromatin association and checkpoint signaling and if these defects could be relieved by rfa1-zm mutants. We found that srs2-K41A was more deleterious than srs2∆ by causing slow growth. Interestingly, rfa1-zm1 or rfa1-zm2 rescued this growth defect, while rad51Δ did not (Fig. 5A). In addition, rfa1-zm1 or rfa1-zm2, but not rad51∆, suppressed CPT and MMS sensitivities of srs2-K41A (SI Appendix, Fig. S6A). Moreover, srs2-K41A led to increased levels of active Rad53 and chromatin-associated RPA even during normal growth (Fig. 5 B–D) and both defects were rescued by rfa1-zm1 and rfa1-zm2 (Fig. 5 B–D and SI Appendix, Fig. S6 B–D). Collectively, these findings suggest that Srs2 helicase activity is required to remove RPA from chromatin, and such a role is important for growth likely by countering checkpoint hyperactivation.
Fig. 5.
rfa1-zm1 rescues multiple defects of srs2-K41A cells. (A) Slow growth of srs2-K41A is rescued by rfa1-zm and not by rad51∆. Representative tetrads of diploids heterozygous for indicated mutations are shown. Spore clones were grown at 30 °C for 2 d. (B) rfa1-zm1 reduces the level of active Rad53 in srs2-K41A cells. Protein extracts prepared from cultures growing in YPD were examined by immunoblotting, and quantification is presented as in Fig. 3A. (C and D) rfa1-zm1 reduces chromatin-bound Rfa1 in srs2-K41A cells. Experiments were done as in Fig. 3C, except cells were grown in YPD media, and data are presented and analyzed as in Fig. 3E, except n = 4. (E) Working model: Srs2 removes RPA and associated Mec1–Ddc2 complexes from chromatin to reduce the Mec1-mediated DDC signaling and promote RPA recycling.
Discussion
While DNA damage checkpoint activation is well studied, its termination is less understood. Pioneering work in yeast has demonstrated the importance of DDC termination and identified factors such as phosphatases that counter DDC kinases. More recent studies discovered additional DDC dampening factors (1). Here, we present multiple lines of evidence to support a mechanism by which the DNA helicase Srs2 removes RPA and associated Mec1 kinase from chromatin, thus reducing DDC signaling (Fig. 5E). We show that this role is important for cells to cope with different genotoxins and is separable from HR regulation.
We demonstrate the Srs2 and RPA antagonism by uncovering strong and suppressive interactions between srs2 and two types of rfa1 mutants. While srs2∆ rescued CPT and MMS sensitivities of commonly used rfa1 mutants, its own sensitivities were rescued by newly generated rfa1-zm mutants. The mutually suppressive relationship seen here is specific, as srs2 and rfa1 mutants show negative interactions with many other mutants (21). Indeed, rfa1-zm mutants did not rescue drug sensitivity of cells lacking Sgs1 and Mph1, which play similar roles to Srs2 in HR, pointing to the specificity of rfa1-zm suppression of srs2∆ drug sensitivity.
We further determined the suppression mechanisms by employing rfa1-zm1 and rfa1-zm2. Both mutants moderately reduce ssDNA binding without grossly affecting overall RPA structure, RPA expression levels, RPA complex formation, cell growth, or DNA replication. Significantly, in both CPT and MMS conditions, rfa1-zm mutants down-regulated the increased levels of active Rad53 in srs2∆ cells and promoted srs2∆ cells to exit G2/M arrest. Thus, better survival of rfa1-zm srs2∆ mutants in both drug conditions is likely due to reducing DDC hyperactivation caused by Srs2 loss.
We went on to show that loss of Srs2 or its helicase activity leads to accumulation of RPA and Mec1 on chromatin. Importantly, both defects were suppressed by rfa1-zm mutants. This data suggests that Srs2 helicase activity is required to remove RPA and RPA-bound Mec1 from chromatin, and this role can be bypassed by moderate perturbation of RPA–ssDNA interaction. Given that Srs2 was recently shown to remove RPA from ssDNA in vitro, Srs2 likely directly displaces RPA from chromatin to down-regulate DDC (38, 39).
Finally, we showed that the antirecombinase role of Srs2 is not required for DDC dampening and that the observed rfa1-zm suppression is not due to reducing HR. Unlike previous findings for rad51 mutants, rfa1-zm did not suppress the srs2∆ hyper-rec phenotype nor sgs1∆ srs2∆ lethality and did not reduce HR levels. Moreover, a srs2 mutant lacking the entire Srs2 C-terminal domain involved in binding Rad51 and other factors, such as PCNA and SUMO, is proficient for CPT and MMS resistance (40, 41). These findings support the notion that the importance of Srs2 helicase activity in the face of genotoxins lies in removing RPA from DNA. Collectively, our data suggest a cellular role of Srs2 in RPA regulation that promotes DDR dampening and drug resistance. This role could provide a molecular explanation to the earlier findings that Srs2 null mutant could not turn off the Mec1 checkpoint upon generation of a single DSB (15). We note that our model does not rule out other additional effects of Srs2 in regulating checkpoint.
RPA regulation by Srs2 adds a means to reduce Mec1-checkpoint signaling. The multitude of mechanisms down-regulating DDC speak for the importance of this event. It will be interesting to determine the coordination of different checkpoint dampening mechanisms in the future. In addition, understanding how different checkpoint dampening pathways are regulated will be informative. Since Mec1 phosphorylation of Slx4 and Sae2 helps to down-regulate checkpoint (9, 42), we envision that Mec1 activation itself triggers checkpoint down-regulation, similar to the role of CDK kinase in cell cycle control (43). Our model predicts that Mec1 targeting Srs2 and/or RPA could favor Srs2-mediated RPA removal from chromatin. Mec1-mediated phosphorylation of Rfa1 and Rfa2 has been shown, and potential Mec1 phosphorylation sites on Srs2 as well as the RPA chaperone Rtt105 were recently reported (44–46). Examining these phosphorylation events individually or in combination in the context of checkpoint dampening will be conducted to gain further insight into Srs2-mediated RPA regulation.
We note that like the Slx4 and Sae2 DDC dampening factors, Srs2’s DDC dampening role has a more prominent effect on genotoxin resistance than its known function in HR. It is thus worthwhile to reinterpret phenotype of Srs2 loss (and Sae2 and Slx4 loss) by considering both DDC-dampening and DNA repair roles. As homologs of all three factors exist in mammals, it is tempting to speculate that similar DDC dampening functions may also help cope with genotoxic or oncogenic stress in other organisms.
RPA is a central player in DDC and most DNA transaction processes. Although it is an abundant complex, its levels are limited and RPA exhaustion can cause lethality in mammalian cells (47). It is thus conceivable that regulating RPA-ssDNA dynamics can be important in broader contexts besides DDC control. We speculate that Srs2-like proteins, such as RTEL1, in higher eukaryotes (48) may provide a means to control RPA dynamics or prevent its exhaustion. In addition, given that the RPA-ssDNA filament is a major platform to recruit many DNA processing proteins, recycling RPA from DNA may provide a means to reset the RPA-ssDNA platform, allowing the removal of unproductive or erroneous intermediates for new repair attempts. This may complement the RPA degradation mechanisms discovered recently to facilitate RPA removal from ssDNA, which can be difficult to achieve otherwise due to its strong affinity to ssDNA (49, 50). It is also interesting to note that an RPA chaperone, Rtt105, can aid a subset of RPA functions in yeast (51). The opposite roles played by Rtt105 and Srs2 in RPA regulation is reminiscent to that of histone chaperone and remodelers in histone regulation. Since RPA and histones are the main protectors of ssDNA and dsDNA, respectively, and both are platforms for protein recruitment, it is conceivable that as seen for histones, RPA regulation has a much broader influence on genome functions than so-far documented.
Materials and Methods
Yeast Strains and Genetic Techniques.
Standard procedures were used for cell growth and media preparation. Strains used are provided in SI Appendix, Table S1 and are isogenic to W1588-4C, a RAD5 derivative of W303 (MATa ade2-1 can1-100 ura3-1 his3-11,15, leu2-3, 112 trp1-1 rad5-535) (52), except G1052, G1053, and their derivatives. rfa1-zm mutant alleles were generated following standard CRISPR-Cas9 method (53) to produce markerless allele replacement at the endogenous locus. All alleles were verified by sequencing. Standard yeast genetic procedures were used for tetrad analyses and spotting assays, and at least two biological duplicates were used for each genotype.
Detection of Rfa1 Protein Level in Cells.
Cells were treated as indicated in the text before collection. Cells were then lysed by bead beating in the presence of 20% trichloroacetic acid. The pellets were recovered by centrifugation and incubated with 1× Laemmli buffer at 95 °C for 5 min to recover proteins. Subsequently, proteins were separated on 3–8% Tris-acetate gels (Life Technologies) followed by Western blotting with anti-Rfa1 antibody (a kind gift from Steven J. Brill, Department of Molecular Biology and Biochemistry, Rutgers University, Piscataway, NJ). Pgk1 was used as a loading control and was detected by using anti-Pgk1 antibody (22C5D8, Invitrogen).
Cell Synchronization and Detection of the Active Form of Rad53.
Log-phase cultures were arrested in G1 by treatment with 5 μg/mL α-factor for 1.5 h. G1 cells were then released into yeast extract–peptone–dextrose (YPD) media containing 100 μg/mL Protease (Sigma) and 16 μg/mL CPT at 30 °C for 2 h. To analyze the effect of MMS, asynchronous cultures were treated with MMS (0.01% or 0.02%) for 1 h, after which MMS was washed off and cultures were released in fresh YPD media for 4 h. Protein extracts were prepared and active Rad53 form was detected as described previously (54). Briefly, 2 × 108 cells were collected and protein extract was prepared by standard TCA method. Proteins were separated on gradient gels (Bio-Rad) followed by Western blotting with the F9 antibody (a kind gift from Marco Foiani and Daniele Piccini, The FIRC Institute of Molecular Oncology, Milan, Italy) to detect active Rad53 levels. Pgk1 was used as a loading control and was detected by anti-Pgk1 antibody (22C5D8, Invitrogen). Accurate quantification of protein bands was achieved by scanning the Western blots using a LAS-3000 luminescent image analyzer (Fujifilm) with a linear dynamic range of 104. The signal intensities of nonsaturated bands were measured using ImageJ software. For graphs, data are shown as mean and SEM except in SI Appendix, Fig. S5A. Statistical differences were determined using Student’s t tests.
Cell Cycle Analyses.
Cells were either synchronized in G1 and treated with CPT or grown asynchronously and treated with MMS as described above. Samples for flow cytometry were collected at the indicated time points, and cell cycle progression was monitored as described previously (55).
Chromatin Fractionation.
Chromatin fractionation was performed as described previously (56). Briefly, spheroplasts from log-phase cells were lysed using extraction buffer (20 mM pH 6.6 PIPES-KOH, 150 mM KOAc, 2 mM Mg(OAc)2, 1 mM NaF, 0.5 mM Na3VO4, 1× Sigma protease inhibitors, 1% Triton X-100) for 5 min on ice. Lysates were centrifuged at 16,000 × g for 15 min on a sucrose cushion. Chromatin pellets were washed and resuspended with extraction buffer. Protein loading buffer was added to all fractions and boiled for 5 min followed by SDS-PAGE and Western blotting. Rfa1 was detected by an anti-Rfa1 antibody (a kind gift from Steven J. Brill); HA-tagged Mec1 was detected by an anti-HA antibody (3F10, Santa Cruz Biotechnology). Histone H3 was used as the marker for chromatin-associated proteins and was detected by an anti-H3 antibody (ab46765, Abcam). Pgk1 was used as a marker for nonchromatin-associated proteins and was detected by an anti-Pgk1 antibody (22C5D8, Invitrogen).
rDNA Marker Loss Frequency.
The loss frequency of the ADE2-CAN1 cassette inside the rDNA array was measured as described previously (57). Cells were grown for equal doublings to stationary phase and then plated on synthetic complete (SC) media for total cell counts. Cells were also plated on canavanine-containing media (SC+Can) and incubated at 30 °C for 2 d after which colonies were counted. The frequency of marker loss (57) was calculated as described previously (30) using the formula FR = NCan/NC, where NCan = number of colonies on SC+Can plates and NC = number of cells plated on SC plates. The frequency of marker loss was normalized to wild type.
Pulse Field Gel Electrophoresis.
Cells arrested in G1 were released into S phase for 60 min and embedded into agarose plugs for pulse field gel electrophoresis (PFGE) as previously described (58). Briefly, plugs were treated with zymolyase (20T, MP Biomedicals), proteinase K, and lauroylsarcosine to permeabilize cells. Chromosomes were separated on 1% agarose (Bio-Rad) gels in 0.5× Tris–borate–ethylenediaminetetraacetic acid (EDTA) buffer using the Bio-Rad CHEF-DR III PFGE system. The conditions for gel running were 70–160 s switch time, 5.5 V/cm voltage gradient, and 106° angle for 15 h at 12 °C. The agarose gel was then stained with ethidium bromide. The percentage of gel entry for each chromosome was calculated by dividing the chromosomal band signal by the sum total of the chromosomal band signal and well signal. The positions of each chromosome were indicated as in ref. 59.
Measurement of Recombination Rates at a Non-rDNA Locus.
Recombination rates were measured using the leu2-ri::URA3::leu2-bsteii recombination assay as described previously (54), and the rates were calculated using fluctuation analysis based on the Lea–Coulson Method of the Median. Briefly, cells were grown in YPD to midlog phase and the appropriate number of cells were then plated on SC-LEU, SC-LEU-URA (for gene conversion events), and SC plates. Colonies were counted after incubation at 30 °C for 2 d. Each test was performed with 12 colonies obtained from two spore clones for each genotype and was repeated twice.
RPA Purification.
Saccharomyces cerevisiae RPA was purified as described (39). Briefly, WT and mutant RPA complexes were overexpressed in BL21Ai cells containing plasmid p11d-tscRPA, or the plasmids carrying the respective mutations. The mutations were generated by using the Q5 site-directed mutagenesis kit from New England Biolabs. Four-liter Luria-broth cultures were grown for each protein preparation. Cells were induced with 0.4 mM isopropyl β-ᴅ-thiogalactopyranoside and 0.05% (wt/vol) l-arabinose when they reached OD600 = 0.6 and grown for an additional 3 h at 37 °C. Harvested cells were resuspended in 120-mL cell resuspension buffer (30 mM HEPES, pH 7.8, 300 mM KCl, 0.1 mM EDTA, protease inhibitor mixture, 1 mM phenylmethylsulfonyl fluoride, 10% [vol/vol] glycerol and 10 mM imidazole). Cells were lysed using 400 μg/mL lysozyme followed by sonication. Clarified lysates were fractionated on a Ni2+-NTA agarose column. Protein was eluted using cell resuspension buffer containing 400 mM imidazole. Fractions containing RPA were pooled and diluted threefold with buffer H0 (30 mM HEPES, pH 7.8, 0.1 mM EDTA, 1 mM dithiothreitol [DTT] and 10% [vol/vol] glycerol). The diluted protein sample was then fractionated over a Q-Sepharose column equilibrated with buffer H100 (buffer H0 with 100 mM KCl). Protein was eluted with a linear gradient H100–H400 (superscript denotes final KCl concentration in the buffer). Fractions containing RPA were pooled and diluted with H0 buffer to match the conductivity of buffer H100, and further fractionated over a Heparin column. Protein was eluted using a linear gradient H100–H1000, and fractions containing RPA were pooled and concentrated using an Amicon spin concentrator (30 kDa cutoff). RPA was dialyzed into storage buffer (30 mM HEPES, pH 7.8, 30 mM KCl, 2 mM DTT and 10% [vol/vol] glycerol), flash frozen using liquid nitrogen, and stored at –80 °C. RPA concentration was measured spectroscopically using ε280 = 98,500 M−1cm−1.
Zn2+ Content Using Inductively Coupled Plasma Mass Spectrometry.
Inductively coupled plasma mass spectrometry (ICP-MS) analysis was performed to quantitate the concentration of Zn2+ in the proteins. One hundred microliters of 20 µM RPA-WT, RPA-zm1, or RPA-zm2 was digested for 15 min in concentrated nitric acid using a microwave digestion system (CEM Corporation). The digested samples were then diluted 1:10 using 1% nitric acid and subjected to ICP-MS analysis for Zn2+ content measurements. The concentration of Zn was calculated using a multielement calibration standard (PerkinElmer), specifically calibrated for Zn.
Secondary Structure Determination Using Circular Dichroism.
Circular dichroism (CD) measurements were performed using a Chirascan spectrometer (Applied Photophysics Inc.). A nitrogen fused set up with a cell path of 1 mm was used to perform the experiments at 20 °C. All CD traces were obtained between 200 and 260 nm, and traces were background corrected using CD reaction buffer (5 mM Tris-Cl, pH 7.8, 100 mM KCl, 5 mM MgCl2, 6% glycerol). Two hundred nanomolar RPA-WT, RPA-zm1, or RPA-zm2 was used, and five scans were collected and averaged per sample using 1-nm step size and 1-nm bandwidth.
DNA Binding Using Tryptophan Fluorescence Quenching.
Intrinsic tryptophan fluorescence quenching data were collected using a PTI-QM40 instrument (Horiba Scientific). Fifty nanomolar RPA (WT or mutant) in a 1.9-mL reaction with reaction buffer (30 mM Hepes, pH 7.8, 100 mM KCl, 5 mM MgCl2, 1 mM β-mercaptoethanol, and 6% [vol/vol] glycerol) was equilibrated to 25 °C in a 2-mL quartz cuvette, and fluorescence scans were collected at a 1-min interval after adding increasing concentrations of (dT)35 oligonucleotide. Changes in intrinsic tryptophan fluorescence were monitored by exciting the sample at 290 nm and capturing emission at 325 nm. Data were fit to a Michaelis–Menten-hyperbola to obtain KD values.
Negative Stain Electron Microscopy.
Samples for negative stain electron microscopy were prepared by mixing 505 nM RPA with 0.25 μg of M13mp18 circular ssDNA in RPA reaction buffer (30 mM HEPES pH 7.8, 6% [vol/vol] glycerol, 100 mM KCl, 1 mM βME, and 5 mM MgCl2). The protein-nucleic acid mixture was incubated at room temperature for 10 min. Freshly glow-discharged grids (carbon film on 200 mesh copper grid, Ted Pella Inc.) were incubated for 2 min over 4 μL of protein DNA drops. Excess sample was removed with three water washes followed by staining twice with uranyl formate (2% solution) for 30 s. Samples were air dried, and micrographs were acquired with a JEOL JEM-1400 120kV transmission electron microscope at 80,000× magnification.
Supplementary Material
Acknowledgments
We thank Steven J. Brill for providing the anti-Rfa1 antibody; Marco Foiani and Daniele Piccini for providing the F9 antibody; Daniel Durocher, Hannah L. Klein, Maria P. Longhese, Rodney Rothstein and Lorraine S. Symington for sharing strains; and Qing Li for discussion. This work was supported by National Institute of General Medical Sciences Grants R01GM080670 and R01GM131058 (to X.Z.) and R01GM130746 and R01GM133967 (to E.A.) of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020185118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Waterman D. P., Haber J. E., Smolka M. B., Checkpoint responses to DNA double-strand breaks. Annu. Rev. Biochem. 89, 103–133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson S. P., Bartek J., The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maréchal A., Zou L., RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 25, 9–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball H. L., et al., Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol. Cell. Biol. 27, 3367–3377 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou L., Elledge S. J., Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Lanz M. C., Dibitetto D., Smolka M. B., DNA damage kinase signaling: Checkpoint and repair at 30 years. EMBO J. 38, e101801 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leroy C., et al., PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol. Cell 11, 827–835 (2003). [DOI] [PubMed] [Google Scholar]
- 8.O’Neill B. M., et al., Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc. Natl. Acad. Sci. U.S.A. 104, 9290–9295 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohouo P. Y., Bastos de Oliveira F. M., Liu Y., Ma C. J., Smolka M. B., DNA-repair scaffolds dampen checkpoint signalling by counteracting the adaptor Rad9. Nature 493, 120–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu T. Y., Kimble M. T., Symington L. S., Sae2 antagonizes Rad9 accumulation at DNA double-strand breaks to attenuate checkpoint signaling and facilitate end resection. Proc. Natl. Acad. Sci. U.S.A. 115, E11961–E11969 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mimitou E. P., Symington L. S., Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G., Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fricke W. M., Brill S. J., Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 17, 1768–1778 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gobbini E., et al., Sae2 function at DNA double-strand breaks is bypassed by dampening Tel1 or Rad53 activity. PLoS Genet. 11, e1005685 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaze M. B., et al., Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10, 373–385 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Krejci L., et al., DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Veaute X., et al., The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309–312 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Seeber A., et al., RPA mediates recruitment of MRX to forks and double-strand breaks to hold sister chromatids together. Mol. Cell 64, 951–966 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Smith J., Rothstein R., A mutation in the gene encoding the Saccharomyces cerevisiae single-stranded DNA-binding protein Rfa1 stimulates a RAD52-independent pathway for direct-repeat recombination. Mol. Cell. Biol. 15, 1632–1641 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umezu K., Sugawara N., Chen C., Haber J. E., Kolodner R. D., Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics 148, 989–1005 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oughtred R., et al., The BioGRID interaction database: 2019 update. Nucleic Acids Res. 47, D529–D541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng S. K., Gibb B., de Almeida M. J., Greene E. C., Symington L. S., RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat. Struct. Mol. Biol. 21, 405–412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan J., Pavletich N. P., Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 26, 2337–2347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bermejo R., et al., Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 21, 1921–1936 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elango R., et al., Break-induced replication promotes formation of lethal joint molecules dissolved by Srs2. Nat. Commun. 8, 1790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colavito S., et al., Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic Acids Res. 37, 6754–6764 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gangloff S., Soustelle C., Fabre F., Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25, 192–194 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Klein H. L., Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Delta with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157, 557–565 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McVey M., Kaeberlein M., Tissenbaum H. A., Guarente L., The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination. Genetics 157, 1531–1542 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernstein K. A., et al., The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase. Mol. Biol. Cell 22, 1599–1607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S., SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–433 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Daley J. M., Niu H., Sung P., Roles of DNA helicases in the mediation and regulation of homologous recombination. Adv. Exp. Med. Biol. 767, 185–202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liberi G., et al., Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 19, 339–350 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panico E. R., Ede C., Schildmann M., Schürer K. A., Kramer W., Genetic evidence for a role of Saccharomyces cerevisiae Mph1 in recombinational DNA repair under replicative stress. Yeast 27, 11–27 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Prakash R., et al., Yeast Mph1 helicase dissociates Rad51-made D-loops: Implications for crossover control in mitotic recombination. Genes Dev. 23, 67–79 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antony E., et al., Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell 35, 105–115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krejci L., et al., Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J. Biol. Chem. 279, 23193–23199 (2004). [DOI] [PubMed] [Google Scholar]
- 38.De Tullio L., et al., Yeast Srs2 helicase promotes redistribution of single-stranded DNA-bound RPA and Rad52 in homologous recombination regulation. Cell Rep. 21, 570–577 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pokhrel N., et al., Monitoring Replication Protein A (RPA) dynamics in homologous recombination through site-specific incorporation of non-canonical amino acids. Nucleic Acids Res. 45, 9413–9426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolesar P., Sarangi P., Altmannova V., Zhao X., Krejci L., Dual roles of the SUMO-interacting motif in the regulation of Srs2 sumoylation. Nucleic Acids Res. 40, 7831–7843 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Armstrong A. A., Mohideen F., Lima C. D., Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 483, 59–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu T. Y., Garcia V. E., Symington L. S., CDK and Mec1/Tel1-catalyzed phosphorylation of Sae2 regulate different responses to DNA damage. Nucleic Acids Res. 47, 11238–11249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enserink J. M., Kolodner R. D., An overview of Cdk1-controlled targets and processes. Cell Div. 5, 11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartrand A. J., Iyasu D., Marinco S. M., Brush G. S., Evidence of meiotic crossover control in Saccharomyces cerevisiae through Mec1-mediated phosphorylation of replication protein A. Genetics 172, 27–39 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faca V. M., et al., Maximized quantitative phosphoproteomics allows high confidence dissection of the DNA damage signaling network. Sci. Rep. 10, 18056 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim H. S., Brill S. J., MEC1-dependent phosphorylation of yeast RPA1 in vitro. DNA Repair (Amst.) 2, 1321–1335 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Toledo L. I., et al., ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Barber L. J., et al., RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 135, 261–271 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feeney L., et al., RPA-mediated recruitment of the E3 ligase RFWD3 is vital for interstrand crosslink repair and human health. Mol. Cell 66, 610–621.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inano S., et al., RFWD3-mediated ubiquitination promotes timely removal of both RPA and RAD51 from DNA damage sites to facilitate homologous recombination. Mol. Cell 66, 622–634.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Li S., et al., Rtt105 functions as a chaperone for replication protein A to preserve genome stability. EMBO J. 37, e99154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao X., Blobel G., A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. U.S.A. 102, 4777–4782 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DiCarlo J. E., et al., Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dhingra N., Wei L., Zhao X., Replication protein A (RPA) sumoylation positively influences the DNA damage checkpoint response in yeast. J. Biol. Chem. 294, 2690–2699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao X., Rothstein R., The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. U.S.A. 99, 3746–3751 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chung I., Zhao X., DNA break-induced sumoylation is enabled by collaboration between a SUMO ligase and the ssDNA-binding complex RPA. Genes Dev. 29, 1593–1598 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fritze C. E., Verschueren K., Strich R., Easton Esposito R., Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16, 6495–6509 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cremona C. A., et al., Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol. Cell 45, 422–432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng X. P., et al., Acute Smc5/6 depletion reveals its primary role in rDNA replication by restraining recombination at fork pausing sites. PLoS Genet. 14, e1007129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.