Abstract
Background
A number of studies have linked positive Ki-67 expression with the prognosis of osteosarcoma (OS) patients. However, the results have been conflicting. To address this controversy, we conducted an analysis using a meta-analysis and a TCGA dataset to estimate the value of Ki-67 expression in the prognosis of OS.
Methods
A comprehensive search for relevant papers was conducted using NCBI PubMed, Embase, Springer, ISI Web of Knowledge, the Cochrane Library, and CNKI regardless of the publication year. The associations between Ki-67 expression and the clinical features and main prognostic outcomes of OS were measured. The TCGA dataset was also analyzed. The pooled odds ratio (OR) and its 95% confidential intervals (CIs) were utilized for statistical analysis.
Results
Overall, a total of 12 studies with 500 cases were included, and the results indicated that the expression of Ki-67 was significantly associated with Enneking stage (OR = 6.88, 95% CI: 2.92–16.22, p < 0.05), distant metastasis (OR = 3.04, 95% CI: 1.51–6.12, p < 0.05) and overall survival (OR = 8.82, 95% CI: 4.68–16.65, p < 0.05) in OS patients. Additionally, we observed no significant heterogeneity among all retrieved studies. Associations between Ki-67 expression and overall survival and disease-free survival of sarcoma were confirmed using the TCGA and Kaplan-Meier plotter datasets.
Conclusion
The present study strongly suggests that positive Ki-67 expression was associated with Enneking stage, distant metastasis, and overall survival of OS, and it may be used as a potential biomarker to predict prognosis and guide clinical therapy for OS.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-021-07880-y.
Keywords: Ki-67, Meta-analysis, Prognosis, Clinicopathological features, Osteosarcoma, TCGA dataset
Background
Osteosarcoma (OS) is a common malignant bone tumor that mainly originating from the metaphysis of long bones [1–3]. Many factors are responsible for prognosis, including demographics, sensitivity to chemotherapy and tumor size, site, and stage [4, 5]. In all age groups, up to 25% of OS patients have metastatic disease, occurring most frequently in the lung [6, 7]. The 5-year overall survival rate is significantly reduced in patients with metastases [8–10]. The increasing incidence of OS [11] has not only severely affected people’s health year by year but also increased social burden [12]. Despite substantial progress that has been made in the diagnosis and treatment of OS, the outcomes of patients remain unsatisfactory due to incomplete understanding of the mechanisms of the disease [13].
In recent years, although we have made great progress in the surgical treatment of osteosarcoma, the 5-year survival rate of osteosarcoma patients is still only approximately 60–70%, and the 5-year survival rate of osteosarcoma patients with lung metastasis is only approximately 10–20% [14]. In recent years, many prognostic biomarkers of osteosarcoma have been reported; for example, LRRC15 can be used as a prognostic biomarker and is an emerging therapeutic target [15]. Transferrin receptor-1 and VEGF may be potential prognostic factors [16]. Circulating miR-25-3p can be used as a novel diagnostic and prognostic biomarker [17]. Currently, the indicators for prognosis were mainly about location, tumor size, recurrence rate, clinical stage and distant metastasis. To measure these indicator was not precise and efficient. Therefore, it was really essential to identify a more representative biomarker for providing an effective prognosis for OS [4, 5].
The Ki-67 antigen was first identified by Gerdes and colleagues in 1983 with the use of a mouse monoclonal antibody. This name was derived from the German city of Kiel and the clone number on a 96-well plate [18]. The gene is located on chromosome 10q25-ter17, and the Ki-67 antigen is a nonhistone protein comprised of two isoforms that weigh 395 kDa and 345 kDa [19]. The protein is only present in the cells at G1, S, and G2 phases of the cell cycle and mitosis but is absent in resting cells at the G0 phase [20], which suggests its fundamental role in the regulation of cell proliferation. Indeed, overexpression of Ki-67 in cancer cells indicates its predictive potential in various neoplasms [21]. Scotlandi et al. reported that the expression of Ki-67 was related to the level of malignancy in bone tumors [22], while Gail et al. found that positive expression of Ki-67 staining was not significantly associated with the median relapse-free survival in Ewing’s sarcoma [23]. Although numerous clinical studies concerning the relationship between Ki-67 and OS have been published in recent years [24–30], there is still a great degree of inconsistency among studies. Therefore, the role of Ki-67 in the prognosis of osteosarcoma is still uncertain. The main purpose of a meta-analysis is to reflect the results of previous studies more objectively and comprehensively to draw conclusions closer to the truth. In the present study, we performed a meta-analysis to assess the prognostic value of Ki-67 expression in OS patients. We present the following article in accordance with the PRISMA reporting checklist.
Methods
Search strategy and study selection
A systematic literature search of NCBI PubMed, Embase, Springer, ISI Web of Knowledge, the Cochrane Library, and CNKI was conducted to identify all relevant articles without language and publication year limitations. The ending date of literature collection was January 2020. Three search terms, “Ki-67”, “osteosarcoma”, and “prognosis” were combined with the Boolean operator “and”. The search strategies were as follows: (1) marker of proliferation Ki-67 or MKI67 or Ki-67 or MIB-1 or Mindbomb E3 ubiquitin protein ligase 1; (2) osteogenic tumor or osteosarcoma; and (3) prognostic or prognosis or survival. Two authors searched the papers independently and excluded irrelevant papers. The reference part of retrieved articles was screened in case of missing the original search.
Inclusion and exclusion criteria
The inclusion criteria were as follows: (1) articles were published in Chinese or English; (2) papers contained original research on humans; (3) the full text was available and sufficient information was provided for estimation; (4) pathological results (i.e., the gold standard) were used for the diagnosis of OS; and (5) Ki-67 in OS was measured with immunohistochemistry.
The exclusion criteria were as follows: (1) Repeated researches. (2). Reports without survival outcome.(3) Wrong article types without original data. (4) No cut-off value for Ki-67 indicated in the articles. (5) No biopsy for diagnosis.
Data extraction
Based on the exclusion and inclusion criteria, two investigators independently evaluated the eligibility of all retrieved papers. Discrepancies between the 2 investigators were resolved by discussion with a third investigator to reach consensus. Relevant information was extracted from the included studies, including Ki-67 assessment methods, case number, sex, median age, publication date, research country inclusion period, and first author. We contacted the corresponding author when further information was needed. If we did not receive any replies after three emails, we excluded the study.
Assessment of included studies
The Newcastle-Ottawa Scale (NOS) [31] was used to evaluate the quality of all the published papers. The included studies were divided into three categories according to the score: 0–3, 4–5, and 6–8 were considered low quality, medium quality, and high quality, respectively.
Assessment of prognosis in the TCGA dataset and Kaplan-Meier plotter dataset
Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) was adopted for further evaluation of the differential expression pattern of Ki-67 between normal samples and cancer for various tumors in the TCGA dataset. Additionally, the associations between Ki-67 and overall survival and disease-free survival were plotted as Kaplan-Meier curves using the TCGA dataset and Kaplan-Meier plotter dataset (https://kmplot.com/analysis/index.php?p=service).
Statistical analysis
The OR and its 95% CI were calculated to evaluate the relationship between the incidence of Ki-67 overexpression and the prognosis of OS patients. The chi-square test was conducted to estimate heterogeneity [32]. A random effect model was used when there was significant heterogeneity (p < 0.10 and I-squared > 50%) [33]. Otherwise, (p > 0.10 and I-squared ≤50%), we chose a fixed-effect model [34]. Sensitivity analysis was conducted by sequentially omitting one of the studies to identify the underlying influence of the individual studies and assess the stability of the results. All the data analyses were conducted using STATA 12.0 software (StataCorp LP, College Station, TX, USA). Significance of a two-tailed test was set at p < 0.05.
Results
Search results
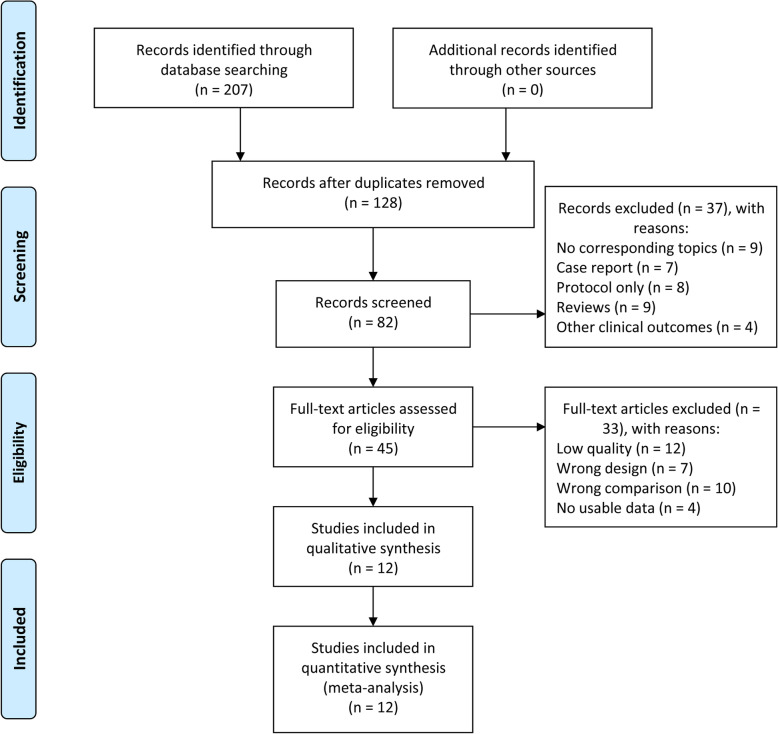
A total of 207 articles were retrieved in the primary search, and 128 reports remained after removing duplicated publications. Then, 46 papers were deleted after further screening, and 82 publications remained. Among them, 37 articles were excluded because they were not related to this topic. After further assessment of the 45 potentially eligible articles, 33 were excluded because of the lack of clinical studies. Finally, 12 relevant articles [24–30, 35–39] published from 1998 to 2018 were adopted in the presented meta-analysis (Fig. 1).
Fig. 1.
Flow diagram of paper selection
Study characteristics
The main features of the 12 remaining studies containing 500 OS patients are listed in Table 1. All the patients involved in the eligible articles were Asian. Immunohistochemistry (IHC) detection methods were used in these studies. Among all these articles, one study lacked data on patient gender, two did not provide the median age, and one paper was missing information on the inclusion period.
Table 1.
Features of 12 articles included in this meta-analysis
| REF | First author | Year | Cases | Gender (M/F) |
Median age | Inclusion period | Method | ki-67 cut-off | Ethnicity | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Wang et al. | 2018 | 50 | 28/22 | 24.15 | 2015–2017 | IHC | A1*B1 > 3 | Asian | 8 |
| 2 | Fu et al. | 2017 | 20 | 9/11 | 11.27 | 2008–2012 | IHC | A1 ≥ 1 | Asian | 8 |
| 3 | Li et al. | 2017 | 21 | 11/10 | 24.33 | 2011–2013 | IHC | A1 ≥ 1 | Asian | 8 |
| 4 | Lin et al. | 2014 | 55 | 31/24 | 17.42 | 2004–2011 | IHC | A1*B2 > 3 | Asian | 7 |
| 5 | Li et al. | 2014 | 94 | 59/35 | – | 2006–2010 | IHC | B3 ≥ 2 | Asian | 7 |
| 6 | Matsumoto et al. | 2013 | 29 | 19/10 | 19.00 | 1978–2007 | IHC | A1 ≥ 1 | Asian | 8 |
| 7 | Junior et al | 2003 | 25 | 13/12 | 29.00 | 1958–2001 | IHC | A1 ≥ 1 | Asian | 8 |
| 8 | Xu et al. | 2003 | 30 | 16/14 | 19.83 | – | IHC | A3 ≥ 2 | Asian | 7 |
| 9 | Zhang et al. | 2003 | 30 | 16/14 | – | 1994–1999 | IHC | A3 ≥ 2 | Asian | 7 |
| 10 | Peng et al. | 2002 | 62 | – | 17.00 | 1995–2001 | IHC | A1 ≥ 1 | Asian | 8 |
| 11 | Zhang et al. | 2001 | 45 | 28/17 | 17.60 | 1993–1998 | IHC | A1 ≥ 1 | Asian | 8 |
| 12 | Liao et al. | 1998 | 39 | 26/13 | 20.10 | 1992–1995 | IHC | A1 ≥ 1 | Asian | 7 |
A: Positive cell percentage: A1: scored 0 (< 5%), 1 (6–25%), 2 (26–50%), 3 (> 50%); A2: scored 1 (< 25%), 2 (26–50%), 3 (51–75%), 4 (> 75%); A3: scored 0 (< 5%), 1 (5–20%), 2 (> 20%)
B: Staining intensity: B1: scored 0 (light yellow), 1 (brownish yellow), 2 (brown); B2: scored 0 (absence of staining), 1 (weak staining), 2 (middle staining), 3 (strong staining); B3: scored 0 (absence of staining), 1 (light yellow), 2 (brownish yellow), 3 (brown)
Qualitative assessment
The quality of eligible studies was evaluated by NOS. A higher score (0–9) represents better methodology. The NOS scores of these 12 studies ranged from 7 to 8 (average score = 7.58) (Table 1); further information is provided in Supplementary Table 1.
Relationship between Ki-67 and OS
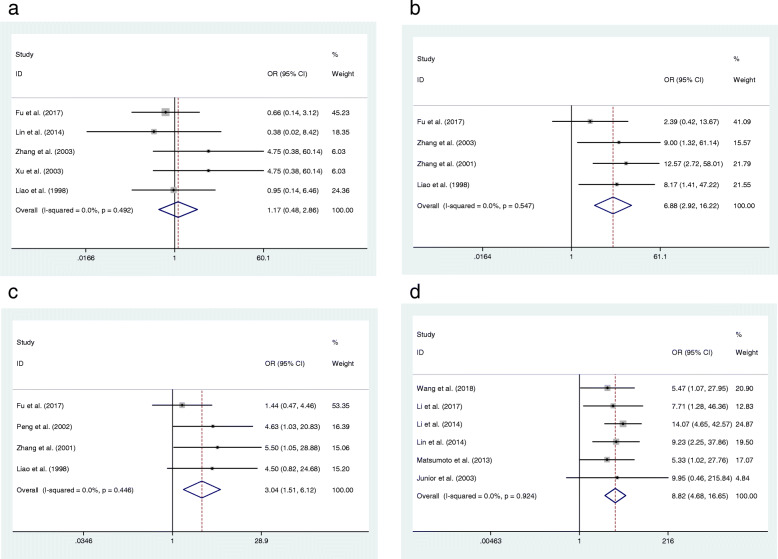
In the present study, we assessed the relationship of Ki-67 expression and clinicopathological features or prognosis of OS. No significant heterogeneity among those eligible studies was found (I2 < 50%), and a fixed-effect model was applied to combine the results of individual studies. The relation was evaluated by the pooled OR with its 95% CI. The results of the meta-analysis indicated that overexpression of Ki-67 in OS was associated with the Enneking stage of tumors (OR = 6.88, 95% CI: 2.92–16.22, p < 0.05) (Fig. 2b). Moreover, Ki-67 was shown to be correlated with distant metastasis (OR = 3.04, 95% CI: 1.51–6.12, p < 0.05) (Fig. 2c). Additionally, six papers (Table 2) were enrolled to explore the association between expression of Ki-67 and over survival of OS and we found that there was an association between the positive expression of Ki-67 and the 5-year overall survival of OS (OR = 8.82, 95% CI: 4.68–16.65, p < 0.05) (Fig. 2d). However, Ki-67-positive expression was confirmed to be irrelevant to OS classification (OR = 1.17, 95% CI: 0.48–2.86, p > 0.05) (Fig. 2a).
Fig. 2.
Forest plot of the association between Ki-67 expression and OS. a: classification, b: enneking stage, c: metastasis, d: overall survival
Table 2.
Features of papers for survival of ki-67 on osteosarcoma
| No. | Trial | Year | High expression | Low expression | Outcomes | Follow-up (month) | ||
|---|---|---|---|---|---|---|---|---|
| Death | 5-year survival | Death | 5-year survival | |||||
| 1 | Wang et al. | 2018 | 13 | 19 | 2 | 16 | over survival | 60 |
| 2 | Li et al. | 2017 | 12 | 7 | 2 | 9 | over survival | 60 |
| 3 | Li et al. | 2014 | 26 | 17 | 5 | 46 | over survival | 60 |
| 4 | Lin et al. | 2014 | 24 | 13 | 3 | 15 | over survival | 60 |
| 5 | Matsumoto et al. | 2013 | 12 | 3 | 6 | 8 | over survival | 60 |
| 6 | Junior et al. | 2003 | 13 | 9 | 0 | 3 | over survival | 60 |
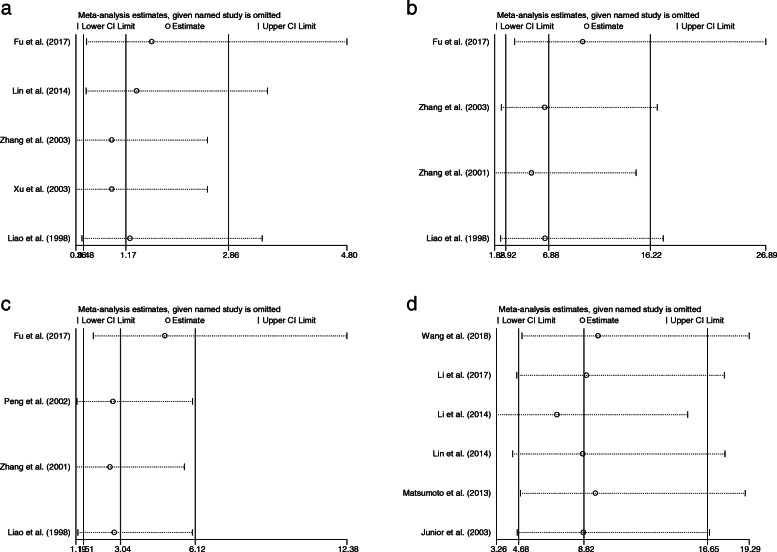
Sensitivity analysis
A sensitivity analysis was conducted to assess the stability of the results of the meta-analysis. The heterogeneity did not change significantly when omitting one of the combined papers. Therefore, we could conclude that the analysis results did not rely on individual studies, and the conclusion was credible (Fig. 3).
Fig. 3.
Sensitivity analysis indicating the association between Ki-67 expression and OS. a: classification, b: enneking stage, c: metastasis, d: overall survival
Association between Ki-67 and OS prognostic features in the TCGA dataset
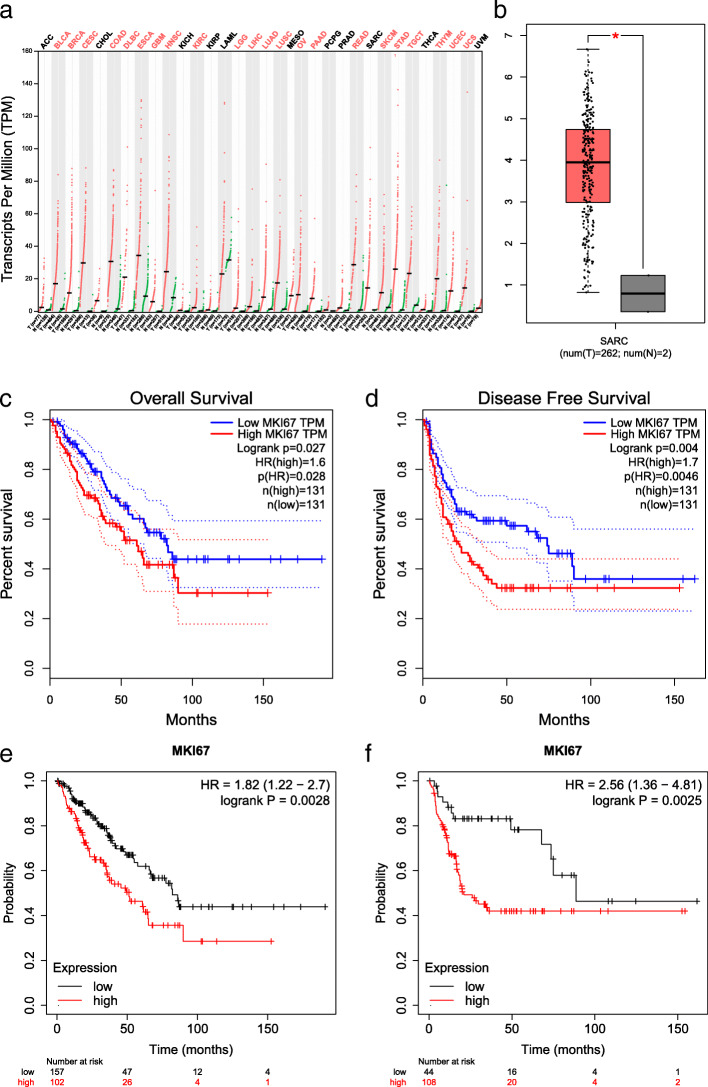
The TCGA pan-cancer dataset and Kaplan-Meier plotter dataset were used to further validate the relationship between Ki-67 positive expression and prognostic features of OS. The results indicated that Ki-67 was significantly upregulated in many cancers including sarcoma (SARC), stomach adenocarcinoma (STAD), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), colon adenocarcinoma (COAD) and liver hepatocellular carcinoma (LIHC) (Fig. 4a). In addition, we found that Ki-67 was significantly upregulated in sarcoma (SARC) (Fig. 4b). Furthermore, the relationship between Ki-67 expression and OS and DFS was shown by Kaplan-Meier curves. The results of the TCGA dataset indicated that Ki-67 positive expression was significantly associated with overall survival (p < 0.05, HR = 1.6) (Fig. 4c) and disease-free survival (p < 0.05, HR = 1.7) (Fig. 4d). The results of the Kaplan-Meier plotter showed that Ki-67 positive expression was significantly related to overall survival (p < 0.05, HR = 1.8) (Fig. 4e) and recurrence-free survival (p < 0.05, HR = 2.5) (Fig. 4f) in sarcoma.
Fig. 4.
Analysis for the expression and prognosis of Ki-67 in cancers. a: differential expression of Ki-67 in various cancers, b: differential expression of Ki-67 in sarcoma, c-f: association between Ki-67 and over survival (c and e), disease free survival (d) and recurrence free survival (f) in sarcoma
Discussion
OS is a primary malignant bone tumor among young adults [40]. OS incidence has an age-specific bimodal pattern: the highest incidence occurs in adolescence and among those older than age 60 [41]. The metaphyses of long bones are the most common sites of OS in young patients. OS incidence is similar in childhood and adolescence and varies little by sex and race worldwide [42–44]. OS is characterized by easy metastasis and recurrence. Individuals with metastatic disease tend to have much poorer outcomes and lower 5-year survival. Although chemotherapy has improved the overall survival rate, the fatality rate is still high. The 5-year survival was approximately 60% for OS patients without visible metastases at the time of diagnosis but was reduced to 15% if tumor metastasis occurred in patients. Therapies for OS have not changed significantly over the past 3 decades, and this bottleneck needs to be overcome as soon as possible. Incremental progress is possible in OS therapies if novel prognostic biomarkers are included in clinical trials [45–47].
Ki-67, also called MKI-67, is expressed only in actively proliferating cells and is a proliferation-related nuclear antigen. Due to the overexpression of Ki-67 in cancer cells, it has been proposed as a prognostic biomarker of cancer [21]. Numerous retrospective studies have reported on the relationship between Ki-67 expression and the prognosis of prostate cancer [48], renal cell carcinoma [49] and several other cancers [50, 51].
In this study, we focused on the predictive effect of Ki-67 positive expression on the prognosis of OS. Li and Zhang suggested that the level of Ki-67 was related to the prognosis of patients with OS [25], but Junior and colleagues were not able to find a correlation between the marker and the prognosis, possibly because of the small number of cases [37]. Although many studies have suggested that Ki-67 is useful for predicting tumor grade [25, 26, 29], there are still other investigators who have drawn the opposite conclusion [26]. The relationship between Ki-67 and metastasis is also controversial [28, 29, 39, 52]. Considering the conflicting results, we investigated the correlation of Ki-67 expression with the clinicopathologic features and prognosis of OS using meta-analysis. The results identified Ki-67 as a predictive marker for reduced 5-year overall survival (OR = 8.82, 95% CI: 4.68–16.65, p < 0.05) in patients with OS. It can also be used as an independent risk factor for distant metastasis (OR = 3.04, 95% CI: 1.51–6.12, p < 0.05). Furthermore, the Ki-67 index indicated surgical Enneking staging of OS (OR = 6.88, 95% CI: 2.92–16.22, p < 0.05), while positive expression of Ki-67 was not related to OS classification (OR = 1.17, 95% CI: 0.48–2.86, p > 0.05). Additionally, the relationship between Ki-67 and worse survival outcomes in sarcoma was further confirmed using the TCGA dataset and Kaplan-Meier plotter dataset. In summary, the present study revealed that Ki-67 was a valuable marker of OS clinicopathological features and prognosis.
There are several limitations of our study into consideration. First, potential publication bias may exist as articles with positive results are easier to publish, which may influence the overall results. Second, the language of the included documents was limited to English and Chinese, which may have also had an impact on the accuracy of the results. Third, the results from these dataset were about sarcoma rather than osteosarcoma, which may affect the validation for this meta-analysis. Fourth, all included patients were from Asian, so the ethnicity may also attribute to potential bias. Last but not least, although all of the patients included were diagnosed with the gold standard (the pathological result), the pathological stage of each patient may also have had an effect on the outcome to some extent. Further multicenter studies with larger sample sizes are needed to reveal the internal correlation of Ki-67 and its predictive role in clinical work; this will decrease sample biases and minimize unavoidable random errors in the meta-analysis process.
Conclusion
In the present study, a meta-analysis was performed to evaluate the relationship between Ki-67 expression and the clinicopathological features and prognosis of OS. Our study showed that Ki-67 positivity was related to the OS Enneking stage and distant metastasis. The results of the meta-analysis and TCGA dataset also indicated a dismal 5-year overall survival for OS patients with Ki-67 expression. Ki-67 may be a valuable biomarker for OS prognosis.
Supplementary Information
Additional file 1 Table S1. Qualitative assessment of included study.
Acknowledgments
The authors thank Dr. Ayub Abdulle nur, Dr. Yanshan Zhu and Dr. Ming Zeng for English language support in preparing revised manuscript.
Abbreviations
- OS
osteosarcoma
- PIP3
phosphatidylinositol 3,4,5-trisphosphate
- CDK2
cyclin-dependent kinases 2
- CNKI
China National Knowledge Internet database
- CBM
Chinese Biological Medical Database
- NOS
Newcastle-Ottawa quality assessment scale
- OR
odds ratio
- 95% CI
95% confidence interval
- TCGA
The Cancer Genome Atlas.
Authors’ contributions
JZ, MZ, LW, YZ, and WW conceived and designed the study, and also critically revised the manuscript. MZ, JZ, and WW conducted the experiments and drafted the manuscript. MZ, YL, JZ, and WW contributed to the revision of the manuscript. All of the authors have read and approved the final manuscript.
Funding
This work was supported by the Mittal Innovation Project of Central South University (Grant No. GCX20190879Y), the Fundamental Research Funds for the Central Universities of Central South University (Grant No. 2018zzts930).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Consent for publication
All studies included in this study got informed consent from each study participant and that each study was approved by ethics committee or institutional review board.
Competing interests
The authors declare that they do not have any competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ming Zeng, Email: zengmingfu2@csu.edu.cn.
Jian Zhou, Email: jian.zhou@csu.edu.cn.
Lifang Wen, Email: 462243236@qq.com.
Yanshan Zhu, Email: 916723699@qq.com.
Yingquan Luo, Email: luoyingquan@csu.edu.cn.
Wanchun Wang, Email: wanchun.wang@csu.edu.cn.
References
- 1.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and end results program. CANCER-AM CANCER SOC. 2009;115(7):1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirabello L, Troisi RJ, Savage SA. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int J Cancer. 2009;125(1):229–234. doi: 10.1002/ijc.24320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anfinsen KP, Devesa SS, Bray F, Troisi R, Jonasdottir TJ, Bruland OS, Grotmol T. Age-period-cohort analysis of primary bone cancer incidence rates in the United States (1976-2005) Cancer Epidemiol Biomark Prev. 2011;20(8):1770–1777. doi: 10.1158/1055-9965.EPI-11-0136. [DOI] [PubMed] [Google Scholar]
- 4.Gorlick R. Osteosarcoma: clinical practice and the expanding role of biology. J Musculoskelet Neuronal Interact. 2002;2(6):549–551. [PubMed] [Google Scholar]
- 5.Durnali A, Alkis N, Cangur S, Yukruk FA, Inal A, Tokluoglu S, Seker MM, Bal O, Akman T, Inanc M, et al. Prognostic factors for teenage and adult patients with high-grade osteosarcoma: an analysis of 240 patients. Med Oncol. 2013;30(3):624. doi: 10.1007/s12032-013-0624-6. [DOI] [PubMed] [Google Scholar]
- 6.Marko TA, Diessner BJ, Spector LG. Prevalence of metastasis at diagnosis of osteosarcoma: an international comparison. Pediatr Blood Cancer. 2016;63(6):1006–1011. doi: 10.1002/pbc.25963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duchman KR, Gao Y, Miller BJ. Prognostic factors for survival in patients with high-grade osteosarcoma using the surveillance, epidemiology, and end results (SEER) program database. Cancer Epidemiol. 2015;39(4):593–599. doi: 10.1016/j.canep.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Meyers PA, Heller G, Healey J, Huvos A, Lane J, Marcove R, Applewhite A, Vlamis V, Rosen G. Chemotherapy for nonmetastatic osteogenic sarcoma: the memorial Sloan-Kettering experience. J Clin Oncol. 1992;10(1):5–15. doi: 10.1200/JCO.1992.10.1.5. [DOI] [PubMed] [Google Scholar]
- 9.Goorin AM, Schwartzentruber DJ, Devidas M, Gebhardt MC, Ayala AG, Harris MB, Helman LJ, Grier HE, Link MP. Presurgical chemotherapy compared with immediate surgery and adjuvant chemotherapy for nonmetastatic osteosarcoma: pediatric oncology group study POG-8651. J Clin Oncol. 2003;21(8):1574–1580. doi: 10.1200/JCO.2003.08.165. [DOI] [PubMed] [Google Scholar]
- 10.Kempf-Bielack B, Bielack SS, Jurgens H, Branscheid D, Berdel WE, Exner GU, Gobel U, Helmke K, Jundt G, Kabisch H, et al. Osteosarcoma relapse after combined modality therapy: an analysis of unselected patients in the cooperative osteosarcoma study group (COSS) J Clin Oncol. 2005;23(3):559–568. doi: 10.1200/JCO.2005.04.063. [DOI] [PubMed] [Google Scholar]
- 11.Morrow JJ, Khanna C. Osteosarcoma genetics and epigenetics: emerging biology and candidate therapies. Crit Rev Oncog. 2015;20(3–4):173–197. doi: 10.1615/CritRevOncog.2015013713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moriarity BS, Otto GM, Rahrmann EP, Rathe SK, Wolf NK, Weg MT, Manlove LA, LaRue RS, Temiz NA, Molyneux SD, et al. A sleeping beauty forward genetic screen identifies new genes and pathways driving osteosarcoma development and metastasis. Nat Genet. 2015;47(6):615–624. doi: 10.1038/ng.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. NAT REV ENDOCRINOL. 2017;13(8):480–491. doi: 10.1038/nrendo.2017.16. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Zhang W. New molecular insights into osteosarcoma targeted therapy. Curr Opin Oncol. 2013;25(4):398–406. doi: 10.1097/CCO.0b013e3283622c1b. [DOI] [PubMed] [Google Scholar]
- 15.Cui J, Dean D, Wei R, Hornicek FJ, Ulmert D, Duan Z. Expression and clinical implications of leucine-rich repeat containing 15 (LRRC15) in osteosarcoma. J Orthop Res. 2020. [DOI] [PubMed]
- 16.Wu H, Zhang J, Dai R, Xu J, Feng H. Transferrin receptor-1 and VEGF are prognostic factors for osteosarcoma. J Orthop Surg Res. 2019;14(1):296. doi: 10.1186/s13018-019-1301-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara T, Uotani K, Yoshida A, Morita T, Nezu Y, Kobayashi E, Yoshida A, Uehara T, Omori T, Sugiu K, et al. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget. 2017;8(20):33375–33392. doi: 10.18632/oncotarget.16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iqbal A, Tamgadge S, Tamgadge A, Pereira T, Kumar S, Acharya S, Jadhav A. Evaluation of Ki-67 expression in oral submucous fibrosis and its correlation with clinical and histopathological features. J Microsc Ultrastruct. 2020:8(1):20–24. [DOI] [PMC free article] [PubMed]
- 19.Duchrow M, Schluter C, Wohlenberg C, Flad HD, Gerdes J. Molecular characterization of the gene locus of the human cell proliferation-associated nuclear protein defined by monoclonal antibody Ki-67. Cell Prolif. 1996;29(1):1–12. doi: 10.1111/j.1365-2184.1996.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 20.Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133(4):1710–1715. [PubMed] [Google Scholar]
- 21.Li LT, Jiang G, Chen Q, Zheng JN. Ki67 is a promising molecular target in the diagnosis of cancer (review) Mol Med Rep. 2015;11(3):1566–1572. doi: 10.3892/mmr.2014.2914. [DOI] [PubMed] [Google Scholar]
- 22.Scotlandi K, Serra M, Manara MC, Maurici D, Benini S, Nini G, Campanacci M, Baldini N. Clinical relevance of Ki-67 expression in bone tumors. CANCER-AM CANCER SOC. 1995;75(3):806–814. doi: 10.1002/1097-0142(19950201)75:3<806::aid-cncr2820750310>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 23.Amir G, Issakov J, Meller I, Sucher E, Peyser A, Cohen IJ, Yaniv I, Ben AM, Tavori U, Kollender Y, et al. Expression of p53 gene product and cell proliferation marker Ki-67 in Ewing's sarcoma: correlation with clinical outcome. Hum Pathol. 2002;33(2):170–174. doi: 10.1053/hupa.2002.31475. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y. Expression of P27kip1,CyclinE and Ki-67 In human osteosarcoma: Zheng zhou University, 2003.
- 25.Zhang C, Liao W, Li F, Hu R, Luo F, Han S. Expressions of P21WAF1 protein, PCNA and Ki-67 and their significance in osteosarcoma. China Oncology. 2001;11:109–112. [Google Scholar]
- 26.Xu J, Zhang Y, Wang Y, Yin L, Lu Z, Hu X, Wang X. Expression of Ki-67 protein in human osteosarcoma. Henan Journal of Surgery. 2003;9:1–2. [Google Scholar]
- 27.Wang W, Li H, Liu T, Wang X, Yang F, Xie J. Expression of livin, LRH1 and Ki-67 in osteosarcoma and its relationship with prognosis. Oncology Progress. 016: 1914–6,21.
- 28.Peng T, Qiu J, Wu H, Liang H, Luo C. Expressions of CD44s,MMP-9,and Ki-67: Possible Association with Invasion, Metastasis, and Recurrence of Osteosarcoma. Chinese Journal of Cancer. 2002;21:745–750. [PubMed] [Google Scholar]
- 29.Liao W, Zhou Z, Qu G, Han S, Li F, Qiu J. Expression of nm23 protein and Ki-67 in association with early metastasis in osteosarcomas. Chinese Journal of Cancer. 1998;17:334–337. [Google Scholar]
- 30.Li Y. Prognostic implication of the expression of Ezrin, Ki-67, Rb and TGF-β1 in soft tissue sarcomas: Xinjiang Medical University, 2014..
- 31.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 32.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 34.MANTEL N, HAENSZEL W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 35.Li J, Jin S. Expression of Ki-67 and prognosis in patients with osteosarcoma. Chinese Journal of the Frontiers of Medical Science (Electronic Version). 2018; 9: 121–4.
- 36.Fu Y, Zheng C, Zhang S, Guan H, Li W, Tan J. Expression and clinical significance of thymidine kinase 1 and Ki67 in osteosarcoma. Chinese Journal of Bone and Joint. 2017;6:128–133. [Google Scholar]
- 37.Takahama Junior A, de Abreu AF, Lopes Pinto CA, Lopes Carvalho A, Kowalski LP, Lopes MA. Clinicopathological and immunohistochemical analysis of twenty-five head and neck osteosarcomas. Oral Oncol. 2003;39:521–530. doi: 10.1016/S1368-8375(03)00017-4. [DOI] [PubMed] [Google Scholar]
- 38.Matsumoto I, Oda M, Yachi T, Tsuchiya H, Zen Y, Watanabe G. Outcome prediction of pulmonary metastasectomy can be evaluated using metastatic lesion in osteosarcoma patients. World J Surg. 2013;37:1973–1980. doi: 10.1007/s00268-013-2022-9. [DOI] [PubMed] [Google Scholar]
- 39.Jinluan L, Jianhua L, Zhaoyang W, Wenbin L, Xiang L, Weinan L, Jinyi FENGFW. Expression of CD133, CD117, and Ki-67 in human osteosarcoma and their clinical significance. Chin J Clin Oncol. 2014;41:305–310. [Google Scholar]
- 40.Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):i320–i325. doi: 10.1093/annonc/mdq276. [DOI] [PubMed] [Google Scholar]
- 41.Xu N, Kang Y, Wang W, Zhou J. The prognostic role of CD133 expression in patients with osteosarcoma. Clin Exp Med. 2020;20(2):261–267. doi: 10.1007/s10238-020-00607-6. [DOI] [PubMed] [Google Scholar]
- 42.Biazzo A, De Paolis M. Multidisciplinary approach to osteosarcoma. Acta Orthop Belg. 2016;82(4):690–698. [PubMed] [Google Scholar]
- 43.Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14(11):722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 44.Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92. doi: 10.1007/978-3-319-07323-1_4. [DOI] [PubMed] [Google Scholar]
- 45.Hmada YA, Bernieh A, Morris RW, Lewin J, Allen T. Chondroblastoma-like Osteosarcoma. ARCH PATHOL LAB MED. 2020;144(1):15–17. doi: 10.5858/arpa.2019-0191-RA. [DOI] [PubMed] [Google Scholar]
- 46.Prater S, McKeon B: Osteosarcoma. 2020. [PubMed]
- 47.Whelan JS, Davis LE. Osteosarcoma, Chondrosarcoma, and Chordoma. J Clin Oncol. 2018;36(2):188–193. doi: 10.1200/JCO.2017.75.1743. [DOI] [PubMed] [Google Scholar]
- 48.Nagao K, Yamamoto Y, Hara T, Komatsu H, Inoue R, Matsuda K, Matsumoto H, Hara T, Sakano S, Baba Y, et al. Ki67 and BUBR1 may discriminate clinically insignificant prostate cancer in the PSA range <4 ng/ml. Jpn J Clin Oncol. 2011;41(4):555–564. doi: 10.1093/jjco/hyq233. [DOI] [PubMed] [Google Scholar]
- 49.Zheng K, Zhu W, Tan J, Wu W, Yang S, Zhang J. Retrospective analysis of a large patient sample to determine p53 and Ki67 expressions in renal cell carcinoma. J BUON. 2014;19(2):512–516. [PubMed] [Google Scholar]
- 50.Kloppel G, La Rosa S. Ki67 labeling index: assessment and prognostic role in gastroenteropancreatic neuroendocrine neoplasms. Virchows Arch. 2018;472(3):341–349. doi: 10.1007/s00428-017-2258-0. [DOI] [PubMed] [Google Scholar]
- 51.He Y, Wang N, Zhou X, Wang J, Ding Z, Chen X, Deng Y. Prognostic value of ki67 in BCG-treated non-muscle invasive bladder cancer: a meta-analysis and systematic review. BMJ Open. 2018;8(4):e19635. doi: 10.1136/bmjopen-2017-019635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mardanpour K, Rahbar M, Mardanpour S. Coexistence of HER2, Ki67, and p53 in osteosarcoma: a strong prognostic factor. N Am J Med Sci. 2016;8(5):210–214. doi: 10.4103/1947-2714.183013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Table S1. Qualitative assessment of included study.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.