Abstract
Objectives
To determine the impact of the 32 bp deletion (CCR5Δ32) in the coding region of the C-C chemokine receptor 5 (CCR5) on the risk of contracting SARS-CoV-2 and severe COVID-19.
Methods
Cross-sectional study among stem cell donors registered with DKMS in Germany. Genetic information was linked to self-reported COVID-19 outcome data. Multivariable regression models were fitted to determine the risk of contracting SARS-CoV-2, severe respiratory tract infection (RTI) and respiratory hospitalization.
Results
CCR5 information was available for 110 544 donors who were tested at least once for SARS-CoV-2; 5536 reported SARS-CoV-2 infection. For 4758 donors, the COVID-19 disease course was fully evaluable; 498 reported no symptoms, 1227 described symptoms of severe respiratory tract infection, of whom 164 required respiratory hospitalization. The distribution of CCR5Δ32 genotypes (homozygous wild-type vs CCR5Δ32 present) did not differ significantly between individuals with or without SARS-CoV-2 infection (odds ratio (OR) 0.96, 95% CI 0.89-1.03, P = 0.21) nor between individuals with or without symptomatic infection (OR 1.13, 95% CI 0.88-1.45, P = 0.32), severe RTI (OR 1.03, 95% CI 0.88-1.22, P = 0.68) or respiratory hospitalization (OR 1.16, 95% CI 0.79-1.69, P = 0.45).
Conclusions
Our data implicate that CCR5Δ32 mutations do not determine the risk of SARS-CoV-2 infections nor the disease course.
Trial registration
We registered the study with the German Center for Infection Research (https://dzif.clinicalsite.org/de/cat/2099/trial/4361).
Keywords: CCR5Δ32, CCR5, SARS-CoV-2, COVID-19
The 32 bp deletion (CCR5Δ32) in the coding region of the C–C chemokine receptor 5 (CCR5) is a common variant and diminishes CCR5 expression on the cell surface. CCR5Δ32 carriers show resistance to human immunodeficiency virus type-1 (HIV-1) infection due to CCR5 being one of two co-receptors for HIV-1 (Benkirane et al., 1997). In contrast, fatal influenza courses (Falcon et al., 2015) and West Nile virus (Lim et al., 2008) infection have been linked to an impaired effector cell recruitment in CCR5Δ32 carriers.
The role of the receptor in SARS-CoV-2 remains elusive. The CCR5 gene is located at 3p21.31, a gene cluster region associated with severe COVID-19 courses (Severe Covid-19 GWAS Group et al., 2020). Increased risk of infection and mortality correlated with the prevalence of the CCR5Δ32 allele in one population study (Panda et al., 2020). On the contrary, leronlimab, a CCR5 blocking antibody, showed potential clinical benefit in individual patients with severe COVID-19 implying that CCR5 might mediate immune dysregulation caused by the SARS-CoV-2 infection (Patterson et al., 2020). Two ongoing randomized clinical trials evaluate leronlimab in patients with COVID-19 (NCT04343651, NCT04347239).
Here, we present results from a cross-sectional study conducted among stem cell donors aged 18–61 years registered with DKMS in Germany. The study aimed to identify immunogenetic risk factors for severe SARS-CoV-2 infections (Schetelig et al., 2020). All participants provided informed consent for study participation. Medical data on SARS-CoV-2 tests, risk factors, symptoms and treatment were collected with a standardized health questionnaire. The medical information was linked to existing data on CCR5Δ32 deletions determined at donor registration (Solloch et al., 2017). Donors who are homozygous for this mutation are preferably selected for patients with HIV infections (Hütter et al., 2009).
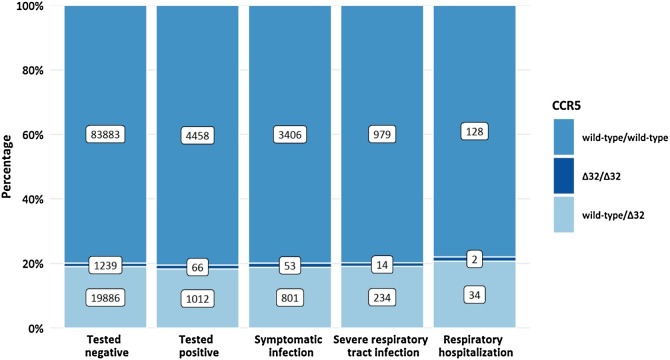
CCR5 information was available for 110 544 study participants who were tested at least once for SARS-CoV-2. Of these, 22 203 (20.1%) had a CCR5Δ32 deletion (Figure 1 ). The prevalence of CCR5Δ32 was analyzed in 4 nested groups of participants based on the reported medical data: individuals with SARS-CoV-2 infection, symptomatic infection, severe respiratory tract infection (RTI), and respiratory hospitalization. Severe RTI was defined by a combination of at least cough and fever, cough and dyspnea, fever and dyspnea, or myalgia and dyspnea and included the subgroup of patients who needed in-patient care with or without supplemental oxygen or mechanical ventilation. The impact of the CCR5Δ32 variant on the risk of individuals to contract SARS-CoV-2 and to experience a more severe course of COVID-19 was tested in multivariable logistic regression models adjusted for sex, age, diabetes mellitus, arterial hypertension, smoking status and for the month of testing in case of risk of symptomatic infection.
Figure 1.
CCR5 genotype distribution among participants grouped according to the clinical course of COVID-19.
SARS-CoV-2 infection was reported by 5536 participants with CCR5 information. The severity of the clinical course of COVID-19 was fully evaluable for 4758 individuals. Of those, 498 reported an asymptomatic COVID-19 course and 1227 described symptoms compatible with severe RTI, with 164 requiring respiratory hospitalization (Figure 1). This distribution of asymptomatic and symptomatic cases was in line with national surveillance data (Robert Koch Institute, 2020).
The genotype distribution of CCR5Δ32 did not differ significantly between infected and non-infected individuals (homozygous wild-type vs CCR5Δ32 present, odds ratio (OR) 0.96, 95% CI 0.89-1.03, P = 0.21). No significant differences for the prevalence of CCR5Δ32 were found among patients with different clinical courses: symptomatic infections, OR 1.13, 95% CI 0.88-1.45, P = 0.32; severe RTI, OR 1.03, 95% CI 0.88-1.22, P = 0.68; respiratory hospitalizations, OR 1.16, 95% CI 0.79-1.69, P = 0.45 (Table 1 ). Since the distribution of asymptomatic vs symptomatic cases largely depends on the testing strategy, symptomatic cases might be over-represented in our data collected during a period when individuals tested were mainly those who showed symptoms suggesting SARS-CoV-2 infection. However, our analysis did not show a trend for a different distribution of CCR5Δ32 among asymptomatic and symptomatic cases (Figure 1); it is therefore highly unlikely that under-representation of asymptomatic cases biased our results.
Table 1.
Impact of CCR5Δ32 on SARS-CoV-2 infection and the course of the disease.
| CCR5 | SARS-CoV-2 infection |
Evaluable infection* | Symptomatic infection |
Severe RTI |
Respiratory hospitalization |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | (95%-CI) | p | n | OR | (95%-CI) | p | OR | (95%-CI) | p | OR | (95%-CI) | p | |
| Wild-type homozygous | 1 | 3816 | 1 | 1 | 1 | ||||||||
| CCR5Δ32 homozygous | 1.05 | (0.81–1.36) | 0.70 | 59 | 1.12 | (0.47–2.67) | 0.80 | 0.92 | (0.50–1.70) | 0.80 | 1.21 | (0.29–5.13) | 0.79 |
| CCR5Δ32 + wild-type | 0.95 | (0.88–1.02) | 0.16 | 883 | 1.13 | (0.88–1.46) | 0.34 | 1.04 | (0.88–1.23) | 0.63 | 1.15 | (0.78–1.7) | 0.47 |
| Wild-type homozygous | 1 | 3816 | 1 | 1 | 1 | ||||||||
| CCR5Δ32 present | 0.96 | (0.89–1.03) | 0.21 | 942 | 1.13 | (0.88–1.45) | 0.32 | 1.03 | (0.88–1.22) | 0.68 | 1.16 | (0.79–1.69) | 0.45 |
Legend: CI, confidence interval; n, number of cases; OR, odds ratio; p, p-value; RTI, respiratory tract infection. Odds ratios were calculated in a multivariable logistic regression model containing information on age, sex, BMI, diabetes mellitus, arterial hypertension, smoking status and month of testing.
Infections reported for January to July 2020.
We did not find a statistically significant difference, although the study was powered sufficiently. For example, to detect a change to the risk for respiratory hospitalization by factor 2 for individuals with a heterozygous CCR5Δ32 deletion, the study had a power of 85%. The power was close to 100% for a comparable change to the risk of infection, symptomatic infection, or severe RTI, with a 5% significance level.
Consequently, our data do not support the previously suggested association between CCR5Δ32 and the risk of contracting SARS-CoV-2 or the risk of severe COVID-19 courses (Panda et al., 2020).
It is unclear to what extent the effects of CCR5Δ32 deletion and a transient drug-induced CCR5 blockade as described by Patterson (Patterson et al., 2020) are comparable; therefore, our results should not be interpreted as an indicator that leronlimab will not be effective. Our data may trigger further research to better understand the impact of heterozygous or homozygous CCR5Δ32 compared to CCR5 blockade in the course of SARS-CoV-2 infection.
Funding statement
DKMS initiated and conducted this study. The Federal Ministry of Education and Research(BMBF) supported the study by a research grant (COVID-19 call (202), reference number 01KI20177).
Conflict of interest statement
The authors declare that there is no conflict of interest.
Ethical approval
The responsible Institutional Review Board of the Technische Universität Dresden (IRB00001473) approved the study. The study was conducted in compliance with the principles of the Declaration of Helsinki. All participants provided informed consent.
Acknowledgements
We are very grateful to all registered DKMS donors who participated in this study. In addition, we would like to acknowledge the dedicated support of many of our colleagues in different departments. Further, we would like to acknowledge a research grant from BMBF (reference number 01KI20177) that partially facilitated this study.
References
- Benkirane M., Jin D.Y., Chun R.F., Koup R.A., Jeang K.T. Mechanism of transdominant inhibition of CCR5-mediated HIV-1 infection by ccr5delta32. J Biol Chem. 1997;272(49):30603–30606. doi: 10.1074/jbc.272.49.30603. [DOI] [PubMed] [Google Scholar]
- Falcon A., Cuevas M.T., Rodriguez-Frandsen A., Reyes N., Pozo F., Moreno S. CCR5 deficiency predisposes to fatal outcome in influenza virus infection. J Gen Virol. 2015;96(8):2074–2078. doi: 10.1099/vir.0.000165. [DOI] [PubMed] [Google Scholar]
- Hütter G., Nowak D., Mossner M., Ganepola S., Müssig A., Allers K. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360(7):692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- Lim J.K., Louie C.Y., Glaser C., Jean C., Johnson B., Johnson H. Genetic deficiency of chemokine receptor CCR5 is a strong risk factor for symptomatic West Nile virus infection: a meta-analysis of 4 cohorts in the US epidemic. J Infect Dis. 2008;197(2):262–265. doi: 10.1086/524691. [DOI] [PubMed] [Google Scholar]
- Panda A.K., Padhi A., Prusty B.A.K. CCR5 Δ32 minor allele is associated with susceptibility to SARS-CoV-2 infection and death: an epidemiological investigation. Clin Chim Acta. 2020;510:60–61. doi: 10.1016/j.cca.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson B.K., Seethamraju H., Dhody K., Corley M.J., Kazempour K., Lalezari J. CCR5 inhibition in critical COVID-19 patients decreases inflammatory cytokines, increases CD8 T-cells, and decreases SARS-CoV2 RNA in plasma by day 14 [published online ahead of print, 2020 Nov 10] Int J Infect Dis. 2020;103:25–32. doi: 10.1016/j.ijid.2020.10.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert Koch Institute . 2020. Situation report of the Robert Koch Institute COVID-19, 13.10.2020.https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/Situationsberichte/Okt_2020/2020-10-13-en.pdf?__blob=publicationFile [Google Scholar]
- Schetelig J., Baldauf H., Wendler S., Heidenreich F., Real R., Kolditz M. Severity of respiratory infections due to SARS-CoV-2 in working population: age and body mass index outweigh ABO Blood group. medRxiv. 2020 doi: 10.1101/2020.11.05.20226100. 11.05.20226100. [DOI] [Google Scholar]
- Severe Covid-19 GWAS Group, Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A. Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. 2020;383(16):1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solloch U.V., Lang K., Lange V., Böhme I., Schmidt A.H., Sauter J. Frequencies of gene variant CCR5-Δ32 in 87 countries based on next-generation sequencing of 1.3 million individuals sampled from 3 national DKMS donor centers. Hum Immunol. 2017;78(11–12):710–717. doi: 10.1016/j.humimm.2017.10.001. [DOI] [PubMed] [Google Scholar]