Abstract
SARS-CoV-2 is a global challenge due to its ability to spread much faster than the SARS-CoV, which was attributed to the mutations in the receptor binding domain (RBD). These mutations enhanced the electrostatic interactions. Recently, a new strain is reported in the UK that includes a mutation (N501Y) in the RBD, that is possibly increasing the infection rate. Here, using Molecular Dynamics simulations (MD) and Monte Carlo (MC) sampling, we show that the N501 mutation enhanced the electrostatic interactions due to the formation of a strong hydrogen bond between SARS-CoV-2-T500 and ACE2-D355 near the mutation site. In addition, we observed that the electrostatic interactions between the SARS-CoV-2 and ACE2 in the wild type and the mutant are dominated by salt-bridges formed between SARS-CoV-2-K417 and ACE2-D30, SARS-CoV-2-K458, ACE2-E23, and SARS-CoV-2-R403 and ACE2-E37. These interactions contributed more than 40% of the total binding energies.
Keywords: SARS-CoV, SARS-CoV-2, Binding domains, Electrostatic interactions, Molecular dynamics, Monte Carlo
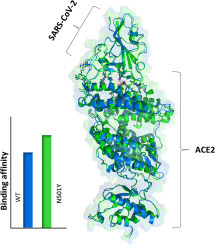
Graphical abstract
SARS-CoV-2 is the infectious agent of the highly spreading coronavirus disease 2019. Since the beginning of 2020, the number of infections is spreading numerously in almost everywhere around the world [[1], [2], [3], [4]]. Currently, the total number of confirmed cases reached more than 80 million cases and nearly 2 million deaths in more than 190 countries. Previous studies reported that the angiotensin converting enzyme 2 (ACE2) is the receptor, which facilitates its binding and entry to the host cells [5,6]. Recently, it was shown that the spread rate of the virus has become much faster due to different genetic changes in the receptor-binding domain and the FURIN cleavage site [7].These changes include the mutation of Asparagine at position 501 to Tyrosine (N501Y), which is one of the residues in the RBD-ACE2 contact area. Experimental findings showed that the N501Y could enhance the binding affinity of SARS-CoV2 spike protein to ACE2 [8,9].
Binding affinity of viruses to the host receptors is mainly affected by different protein–protein electrostatic interactions [10,11]. A change in different pivotal residues at the binding site could affect viral-host cells fusion and hence the infectivity of the virus [5,[12], [13], [14]]. Therefore, we herein study how the N501Y mutation in the RBD of the SARS-CoV2 can alter the binding of the virus to ACE2. We focus on the role of the electrostatic interactions on the binding energy, where it is known to be dominant among different protein-protein interactions [15]. In this study, we used a combined Molecular dynamic (MD) and Monte Carlo (MC) simulations to assess the molecular interactions between RBD of S-protein and ACE2 for the N501Y mutant and compared our results with the wild type. The crystal structure (PDB ID: 6M17) [12] was optimized using openMM [16]. Then, several rotamers were built by MCCE to by rotating each rotatable bond by 60o to appropriately sample the sidechains conformations. In order to build the N501Y mutant, the sidechain of N501 is replaced by aromatic.sidechain of tyrosine using MCCE.
The electrostatic interactions are calculated for the optimized most occupied conformer of the proteins by solving Poisson Boltzmann equation.
The electrostatic interactions between the different conformers were calculated using DELPHI [17]. Then, MCCE is used to generate Boltzmann distribution for all conformer using MC sampling for the wild type and the N501Y mutant at pH 7. The most occupied conformers were subjected to MD minimization again using openMM [16]. The resulting structures were used to calculate the electrostatic energies between the RBD of SARS-CoV-2 and the ACE2 using DELPHI. In WT, the maximum vdW interaction of −1.889 kcal/mol was reported between residue RBD-N501 and ACE2-K353. While For the mutated structure, a maximum vdW interactions of −2.441 Kcal/mol was between RBD-Q498 and ACE2-Y41. The Maximum electrostatic interactions of −9.55 kcal/mol was observed between RBD-K417 and ACE2-D30 in wild type, and of −8.39 kcal/mol between RBD-K458 and ACE2-E23 in mutated structure. The electrostatic interactions between the SARS-CoV-2 and ACE2 in the wild type and the mutant are dominated by salt-bridges formed between SARS-CoV-2-K417 and ACE2-D30, SARS-CoV-2-K458, ACE2-E23, and SARS-CoV-2-R403 and ACE2-E37 (Fig. 1 ).
Fig. 1.
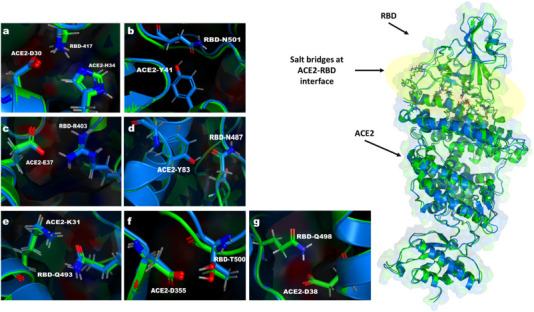
Salt-bridges between RBD and ACE2 in both of WT and N501Y mutated structure. The WT RBD and ACE2 are shown in Blue while the N501Y mutated RBD and ACE2 are shown in Green. (a-g) Different salt-bridges interactions (interaction energies are shown in Table.S1). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
These residues contribute by ~20 Kcal/mol of the total electrostatic interactions between the SARS-CoV-2 and ACE2 (Table S1). However, the binding between the SARS-CoV-2 and ACE2 is favored by ~4 Kcal/mol in the mutant (Table 1 ) due to a stronger hydrogen bond between SARS-CoV-2-T500 and ACE2-D355 (Fig. 2a, Fig. 3A).
Table 1.
The interaction energies between SARS-CoV-2-RBD and ACE2 in both WT and N501Y mutated structures.
| Coulomb (Kcal/mol) | Van der Waals (Kcal/mol) | Total (Kcal/mol) | |
|---|---|---|---|
| Wt | −18.38 | −31.56 | −49.94 |
| N501Y | −21.91 | −32.59 | −53.91 |
Fig. 2.
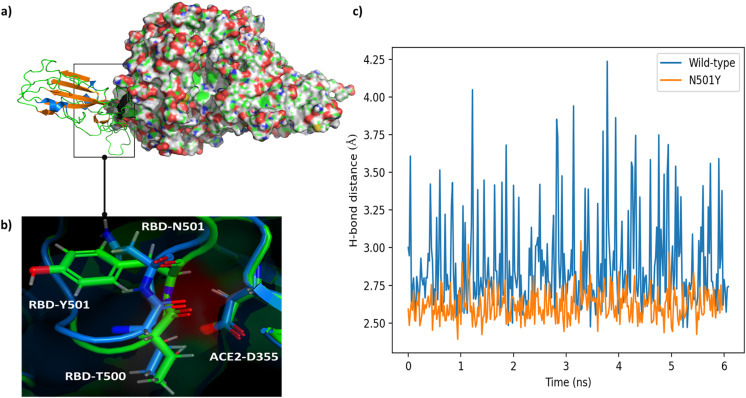
(a) [PDB code 6M17]: SARS-CoV2 RBD and human ACE2 complex. RBD is shown in cartoon view. (b) Residues at position 501 for both of WT and N501Y mutated structures and interacting residue in the vicinity of position 501.The WT RBD and ACE2 are shown in Blue while the N501Y mutated RBD and ACE2 are shown in Green. (c) The distance in Å between OG1 of SARS-CoV-2 and ACE2-T500 and OD2 of D355 through 6 ns MD trajectory.
Fig. 3.
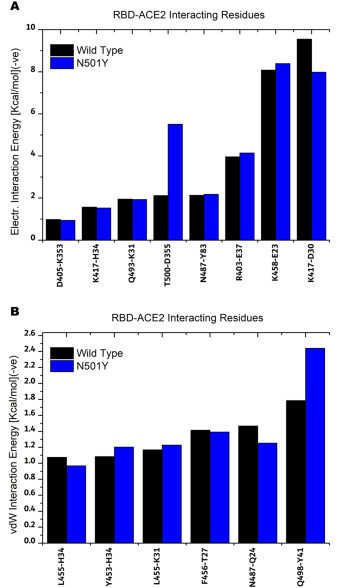
Representation of a selected favorable Electrostatic and vdW interactions in both of WT and N501Y complexes. (Table S1).
Upon N501Y mutation, RBD-K417 and ACE2-D30 exhibit less attraction than that for WT by ~1.57 Kcal/mol (Fig. 3A). While this mutation has a noticeable effect on the vdW interaction between RBD-Q498 and ACE2-Y41, by which interaction increased by about 0.66 Kcal/mol (Fig. 3B). Other interactions are shown to exhibit minor changes after mutation.
In agreement with other studies [18,19], our data demonstrated a short hydrogen bond (~1.7 Å) between OD1 of RBD-N501 and OH of ACE2-Y41 at the WT SARS-CoV-2 RBD-ACE2 contact region (Fig. 4 ), with electrostatic energy of ~ −2.15 Kcal/mol S1 (Table.S1). While in case of mutated complex this hydrogen bond was replaced by a longer one (~2.1 Å) between OH of ACE2-Y41 and backbone nitrogen of RBD—Y501.
Fig. 4.
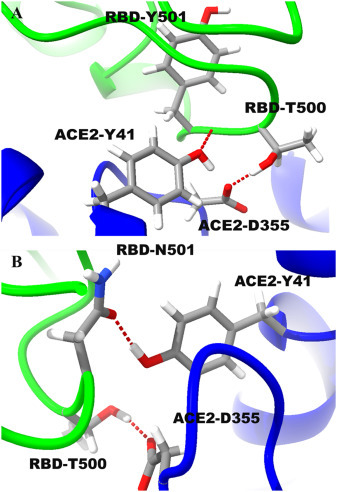
The interactions between the residue at position 501 in SARS-CoV2 RBD and nearby residues. (A) Shows interaction between mutated site Y501 in RBD and human ACE2 complex. (B), shows interaction between N501 in the native RBD and human ACE2 complex. The RBD and ACE2 are shown in Green and Blue, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The N501Y mutant is shown to decrease the repulsion between carboxylate of ACE2-D355 and backbone oxygen of RBD-T500 RBD-T500 as the distance decreased from 3.81 to 3.42 Å.
In addition, the hydrogen bond between the carboxylate of ACE2-D355 and HG1 of RBD-T500 is shortened from 1.65 to 1.57 Å in the mutant. To further asses the stability of this hydrogen bond, we ran MD trajectories for 6 ns Fig. 1(c). The trajectories show that there is a significant variation in the bond length in the wild type, while in the mutant, the variation is much less due to the strong interactions. However, the average distance in between OG1 of SARS-CoV-2 and ACE2-T500 and OD2 of D355 is shorter for the mutant (2.61 Å) compared to the wild type (2.85 Å).
In summary, we showed that the binding affinity of SARS-CoV-2 to human ACE2 is higher in the N501Y mutated structure than that in WT because of the significant change in the electrostatic interactions [[18], [19], [20]]. Upon mutation, the salt-bridge electrostatic interaction increased between T500 and D355 in the RBD and ACE2, respectively, to be ~3.39 kcal/mol more negative than that in the WT. This is shown to be due to the shorter atomic distances between ACE2-D355 and SARS-CoV2-T500.
Credit author statement
Fedaa Ali, Formal analysis, simulations, writing, Amal Kasry, supervision, review, Funding acquisition and editing, Muhamed Amin, Conceptualization, Simulations, Writing – review & editing.
Declaration of Competing Interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.medidd.2021.100086.
Appendix A. Supplementary data
Supplementary material
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan J.F.-W., Yuan S., Kok K.-H., To K.K.-W., Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.052. 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14(8) doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A. 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. [Google Scholar]
- 8.Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.-Q. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369(6511):1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5) doi: 10.1016/j.cell.2020.08.012. 1295–1310.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norel R., Sheinerman F., Petrey D., Honig B. Electrostatic contributions to protein-protein interactions: fast energetic filters for docking and their physical basis. Protein Sci. 2008;10(11):2147–2161. doi: 10.1110/ps.12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheinerman F. Electrostatic aspects of protein–protein interactions. Curr Opin Struct Biol. 2000;10(2):153–159. doi: 10.1016/S0959-440X(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 12.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ali F., Elserafy M., Alkordi M.H., Amin M. ACE2 coding variants in different populations and their potential impact on SARS-CoV-2 binding affinity. Biochem Biophys Reports. 2020;24:100798. doi: 10.1016/j.bbrep.2020.100798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amin M., Sorour M.K., Kasry A. Vol. 11. The Journal of Physical Chemistry Letters; 2020. Comparing the Binding Interactions in the Receptor Binding Domains of SARS-CoV-2 and SARS-CoV. pp. 4897–4900. [DOI] [PubMed] [Google Scholar]
- 15.Nicholls A., Honig B. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 16.Eastman P., Swails J., Chodera J.D., McGibbon R.T., Zhao Y., Beauchamp K.A. OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLoS Comput Biol. 2017;13(7) doi: 10.1371/journal.pcbi.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L., Li C., Sarkar S., Zhang J., Witham S., Zhang Z. DelPhi: a comprehensive suite for DelPhi software and associated resources. BMC Biophys. 2012;5(1):9. doi: 10.1186/2046-1682-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos C., Passos G.A. bioRxiv; 2021. The High Infectivity of SARS-CoV-2 B.1.1.7 Is Associated with Increased Interaction Force between Spike-ACE2 Caused by the Viral N501Y Mutation. [Google Scholar]
- 19.Luan B., Wang H., Huynh T. Molecular mechanism of the N501Y mutation for enhanced binding between SARS-CoV-2’s spike protein and human ACE2 receptor. bioRxiv. 2021 doi: 10.1101/2021.01.04.425316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fratev F. The SARS-CoV-2 S1 spike protein mutation N501Y alters the protein interactions with Both HACE2 and human derived antibody: a free energy of perturbation study. bioRxiv. 2020 doi: 10.1101/2020.12.23.424283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material


