Abstract
Objective
T-cell responses against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are observed in unexposed individuals. We evaluated the impact of this pre-existing cellular response on incident SARS-CoV-2 infections.
Methods
This was a follow-up study of 38 seronegative healthcare workers (HCWs) with previous evaluation of CD8+ and CD4+ T-cell responses after stimulation with SARS-CoV-2 structural proteins. Infection was considered in the presence of a positive RT-PCR test and/or confirmed seroconversion.
Results
Twenty of the 38 HCWs included (53%) had a previous specific CD8+ T-cell response to peptides encompassing the spike protein (S) in 13 (34%), the membrane (M) in 17 (45%), or/and the nucleocapsid (N) in three (8%). During a follow-up of 189 days (interquartile range (IQR) 172–195), 11 HCWs (29%) had an RT-PCR-positive test (n = 9) or seroconverted (n = 2). Median duration of symptoms was 2 days (IQR 0–7), and time to negative RT-PCR was 9 days (IQR 4–10). Notably, six incident infections (55%) occurred in HCWs with a pre-existing T-cell response (30% of those with a cellular response), who showed a significantly lower duration of symptoms (three were asymptomatic). Three of the six HCWs having a previous T-cell response continued to test seronegative. All the infected patients developed a robust T-cell response to different structural SARS-CoV-2 proteins, especially to protein S (91%).
Conclusion
A pre-existing T-cell response does not seem to reduce incident SARS-CoV-2 infections, but it may contribute to asymptomatic or mild disease, rapid viral clearance and differences in seroconversion.
Keywords: COVID-19, Cross-reactivity, Healthcare workers, Immune response, SARS-CoV-2, T-cell response
Graphical abstract
Introduction
Data from other coronavirus infections have demonstrated that cellular immunity is a determinant for long-term protection [1], a crucial fact since antibody levels against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) wane quickly during the follow-up period [2,3]. Notably, recent studies described T-cell responses to viral peptides in patients not exposed to SARS-CoV-2, probably due to cross-reactivity to common coronavirus infections [4]. However, there are no data regarding how this T-cell response can intervene in the evolution of SARS CoV-2 infection. Thus, we investigated the incidence and characteristics of SARS-CoV-2 infections during follow-up in healthcare workers (HCWs) initially evaluated for the presence of T-cell immunity.
Methods
A cohort of 38 uninfected HCWs (asymptomatic and without specific IgG antibodies) underwent blood analysis in May 2020 to evaluate the presence of T-cell immune response against SARS-CoV-2, and were followed to ascertain the incidence of COVID-19. Incident cases were defined as presence of a positive RT-PCR test on nasopharyngeal swab, or/and seroconversion during the follow-up. At the end of November 2020, all the remaining HCWs underwent specific serological testing to evaluate asymptomatic infections. Mild/moderate disease was defined as the absence/presence of radiological infiltrates and lack of hypoxaemia (oxygen saturation ≥95% on room air). No severe disease was observed [5].
The study was approved by our ethic committee (EC162/20; NCT04402827). Written informed consent was obtained from all the participants.
Both at inclusion and at the end of follow-up, the presence of antibodies was assessed by SARS-CoV-2 ELISA (COVID-19-SARS-CoV-2 IgG ELISA, Demeditec, Germany).
The presence of a cellular immune response was assessed at the same time points. SARS-CoV-2-specific CD4+ and CD8+ T cells were measured using in vitro stimulation with SARS-CoV-2 peptide pools of viral proteins encompassing the spike (S), membrane (M), and nucleocapsid (N), followed by quantification of CD4+ and CD8+ T-cell-specific interferon (IFN)-γ in live cell flow cytometry, using peripheral blood mononuclear cell (PBMC) samples from all subjects. It was considered significantly reactive if the proportion of positive cells in stimulated wells was at least 2-fold higher in comparison with the negative control wells (unstimulated). A detailed description is included as a supplementary file, including the flow cytometry gating strategy (see web-only Supplementary Material).
Statistical analysis
Comparisons between groups were performed using χ2 or Fisher's exact tests for categorical variables, and the Mann–Whitney test or one-way analysis of variance (Kruskal–Wallis test) with Dunn's correction for multiple comparisons, as appropriate. Analysis of paired observations during follow-up was performed using the Wilcoxon rank t-test. Statistical significance was defined as two-sided p values < 0.05. Statistics were done with IBM SPSS Statistics, version 23.0.
Results
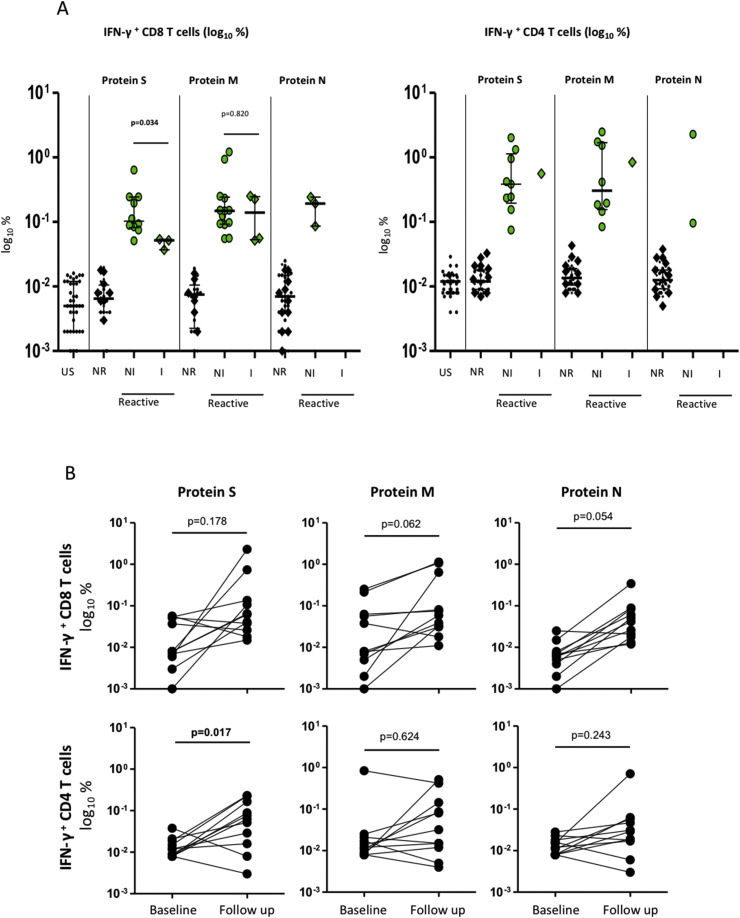
Initially, 20 out of the 38 serologically negative HCWs (53%) presented T-cell responses against structural proteins of SARS-CoV-2, based mainly in a reactive CD8+ response toward peptides of protein S (13, 34%), M (17, 45%), or/and N (3, 8%) (Fig. 1 A). Of note, seven participants had a CD8+ T-cell response only to protein M, whereas three had an exclusive response to protein S.
Fig. 1.
IFN-γ producing CD8+ and CD4+ T-cell (log%) in both cases responding to peptides spanning the immunogenic domains of the SARS-CoV-2 spike (S), membrane (M), and nucleocapsid proteins (N) in HCWs at inclusion in May 2020, and according to subsequent SARS-CoV-2 infection during follow up. Fig. 1A: detection of IFN-γ-producing CD8+ T-cells and CD4+ T-cells (%): US, unstimulated or negative controls; NR, non-reactive response, below 2-fold increase in stimulated well compared to unstimulated well; R, reactive CD8+ and CD4+ T-cells, both in patients not infected (NI) or infected (I) during follow up. Diamonds indicate incident infections. Lines indicate median values. There were significant differences for IFN- γ producing CD8 T cells in response to protein S among NI and I patients. Fig. 1B showed individual changes of IFN-γ producing CD8+ and CD4+ T-cells in response to stimulation with structural viral peptides at inclusion and after incident SARS-CoV-2 infections during the follow up (N = 11). Lines represent the change of response for each individual.
During a median follow-up of 189 days (interquartile range (IQR) 172–195, 18.5 person-years of follow up), 11 of these 38 HCWs (29%) were finally considered as incident cases of infection; seven were studied because of symptoms, two due to contact with a partner, and two were asymptomatic seroconversions. Characteristics of these 11 infected HCWs are detailed in Table 1 .
Table 1.
Clinical characteristics of 38 seronegative heath care workers according to incident SARS-CoV-2 infection during the follow up
| Overall N = 38 |
Infected N = 11 |
Not infected N = 27 |
p-value | |
|---|---|---|---|---|
| Age (years) | 38 [22-60] | 41 [25-60] | 36 [22-57] | 0.975 |
| Sex (female) | 21 (55%) | 7 (64%) | 14 (52%) | 0.721 |
| Body Mass Index (Kg/m2) | 23.1 (20.3-25.5) | 23.1 (20-23.6) | 23 (20-26) | 0.612 |
| HCWs1: | 0.582 |
|||
| Physicians | 26 (68%) | 9 (82%) | 17 (63%) | |
| Nurses | 12 (32%) | 2 (18%) | 10 (37%) | |
| Concomitant comorbidities | 0.987 | |||
| Hypertension | 1 (3%) | — | 1 (4%) | |
| Diabetes | 1 (3%) | — | 1 (4%) | |
| Working at COVID-19 ward | 14 (37%) | 4 (36%) | 10 (37%) | 0.287 |
| Exposure to aerosol generating procedures2 | 12 (32%) | 2 (18%) | 10 (37%) | 0.456 |
| Time to evaluation (days)3 | 189 [172-195] | 159 [147-170] | 190 [188-196] | <0.001 |
| Previous T-cell response | ||||
| CD8+ reactive | 20 (53%) | 6 (55%) | 14 (52%) | 0.880 |
| CD4+ reactive | 13 (34%) | 2 (18%) | 11 (41%) | 0.268 |
| SARS-CoV-2 RT-PCR | ||||
| Positive | 9 (24%) | 9 (82%) | — | |
| Negative | 8 (21%) | — | 8 (30%) | |
Data are expressed as median and interquartile range, and percentage. Mann-Whitney U test for statistical differences between variables. HCW, health care workers; RT-PCR, reverse transcriptase- PCR; 1chi-square test; 2aerosol-generating procedures included airway suction, application of a high-flow O2 instrument, bronchoscopy, endotracheal intubation, tracheostomy, nebulizer treatment, sputum induction, positive pressure ventilation, manual ventilation, and cardiopulmonary resuscitation; 3Time from study inclusion to positive RT-PCR testing or final serologic testing, depending of the group.
Deidentified clinical data with the characteristic of all the infected HCWs are available in the Supplementary Material Table S1. Three HCWs were asymptomatic and eight had a mild disease, with resolution of symptoms in a median time of 2 days (IQR 0–7), and median time until a negative RT-PCR was 9 days (IQR 7–10).
Six out of the 11 HCWs (55%) with incident infection had a previous CD8+ T-cell response to SARS-CoV-2 proteins, representing 30% of those with a T-cell response (six out of 20). These six infected HCWs showed a limited CD8+ T-cell response to SARS-CoV-2 epitopes (three to protein M, two to protein S, one to proteins S and M, Fig. 1A). Strikingly, a significantly weaker CD8+ T-cell response to protein S was observed in infected patients (p 0.034). In any case, a pre-existing T-cell response was associated with short duration of symptoms (1.5 versus 7; p 0.029), and the three infected individuals with a cellular response to protein S were asymptomatic.
Of interest, HCW#6 had a previous CD8+ T-cell response to protein M and became infected after contact with an undiagnosed patient. Before developing symptoms, he attended a dinner with co-workers, spreading the infection to HCWs#7, #8 and #10, who developed symptomatic SARS-CoV-2 infection.
After infection, antibodies against SARS-CoV-2 were not detected in three HCWs with pre-existing T cells (one with a response to S). Nevertheless, T-cell response was observed in these 11 HCWs after incident infection, with emergence of a robust CD8+ T-cell response involving the three studied structural proteins in seven cases (64%), and with response to protein S in ten cases (91%) (Fig. 1B; see also web-only Supplementary Material Table S1 and Fig. S1).
Discussion
In this study we investigated and compared incident SARS-CoV-2 infections in a cohort of HCWs according to a pre-existing T-cell immune response, a fact observed in 30–81% of unexposed individuals in recent studies [[6], [7], [8]]. We found that 53% of seronegative HCWs had a T-cell response to structural viral proteins. Several studies suggest that these pre-existing T cells were mainly cross-reactive, with comparable affinity to SARS-CoV-2 and common cold coronaviruses [4,9]. This fact could also be suggested by the predominance of a response to protein M in our study, which showed 90% structural identity with that of other coronaviruses [10].
Nonetheless, no studies have addressed the incidence and clinical importance of this pre-existing T-cell response. In our study, six of the 11 incident infections (55%) were observed in HCWs with pre-existing T-cell immunity. As expected, cellular response does not provide sterilizing immunity, and indeed it could be associated with transmission, as we showed with one HCW in our cohort. However, as demonstrated with influenza, some degree of pre-existing cellular immunity correlate with less severe disease [11]. This could explain the short duration of symptoms and rapid viral clearance, although we cannot exclude the beneficial effect of the young age and the absence of comorbidities.
We offer data about the functional capacity and evolution of pre-existing T-cell responses. HCWs who acquired COVID-19 (3/7 and 3/13 with response to proteins M and S, respectively) previously had a weaker IFN-γ-producing CD8+ T-cell response. It is not known which is the most protective profile of cellular response, although T-cell reactivities in convalescent patients covered multiple SARS-CoV-2 proteins [3,12], and we observed a rapid and extensive SARS-CoV-2-specific CD8+ T-cell response affecting multiple epitopes after infection.
Our study has several limitations, including the small number of HCWs that limits statistical associations. First, the possibility of previous asymptomatic infections and misclassification was possible but unlikely because of the high sensitivity of the serological test, and the differences in T-cell responses with convalescent patients. Second, initial T-cell evaluation was performed 5–6 months before infection, and we cannot preclude a slight decrease in T-cell immunity during this period. Finally, we did not include T-cell response to other structural proteins, such as ORF1a, which have been showed to be immunodominant in some studies [13].
In conclusion, we found that 30% of HCWs with a pre-existing T-cell response acquired a SARS-CoV-2 infection during a 6-month follow-up, confirming that symptomatic, transmissible SARS-CoV-2 infection is possible in the presence of previous cellular immunity. However, our data suggest that this T-cell background, although weak, could modify seroconversion rates and could help to attenuate the clinical course, explaining differences in duration and severity of the disease.
Author contributions
JLC and AV contributed equally to this work. JC and AV conceived of and designed the study, analysed the results, and wrote the manuscript. All other authors conducted the study and collected data, revised the manuscript, and approved the final version.
Transparency declaration
All the authors declare no conflicts of interest. All research was conducted within the guidelines of ethical principles and local legislation. No external funding was received for this work, and it was supported by departmental discretionary funds available to the first corresponding author.
Acknowledgments
We thank Ana Abad for database management.
Editor: L. Leibovici
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.02.020.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Channappanavar R., Zhao J., Perlman S. T cell-mediated immune response to respiratory coronaviruses. Immunol Res. 2014;59:118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Self W.H., Tenforde M.W., Stubblefield W.B., Feldstein L.R., Steingrub J.S., Shapiro N.I. Decline in SARS-CoV-2 antibodies after mild infection among frontline health care personnel in a multistate hospital network—12 States, April–August 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1762–1766. doi: 10.15585/mmwr.mm6947a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.-B., Olsson A. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168 e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501 e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 8.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 9.Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 10.Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilkinson T.M., Li C.K., Chui C.S., Huang A.K.Y., Perkins M., Liebner J.C. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 12.Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 13.Kared H., Redd A.D., Bloch E.M., Bonny T.S., Sumatoh H., Kairi F. CD8+ T cell responses in convalescent COVID-19 individuals target epitopes from the entire SARS-CoV-2 proteome and show kinetics of early differentiation. bioRxiv. 2020 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.