Abstract
Background/Aims
Extracellular vesicles (EVs), including microvesicles and exosomes, deliver bioactive cargo mediating intercellular communication in physiological and pathological conditions. EVs are increasingly investigated as therapeutic agents and targets, but also as disease biomarkers. However, a definite consensus regarding EV isolation methods is lacking, which makes it intricate to standardize research practices and eventually reach a desirable level of data comparability. In our study, we performed an inter-laboratory comparison of EV isolation based on a differential ultracentrifugation protocol carried out in 4 laboratories in 2 independent rounds of isolation.
Methods
Conditioned medium of colorectal cancer cells was prepared and pooled by 1 person and distributed to each of the participating laboratories for isolation according to a pre-defined protocol. After EV isolation in each laboratory, quantification and characterization of isolated EVs was collectively done by 1 person having the highest expertise in the respective test method: Western blot, flow cytometry (fluorescence-activated cell sorting [FACS], nanoparticle tracking analysis (NTA), and transmission electron microscopy (TEM).
Results
EVs were visualized with TEM, presenting similar cup-shaped and spherical morphology and sizes ranging from 30 to 150 nm. NTA results showed similar size ranges of particles in both isolation rounds. EV preparations showed high purity by the expression of EV marker proteins CD9, CD63, CD81, Alix, and TSG101, and the lack of calnexin. FACS analysis of EVs revealed intense staining for CD63 and CD81 but lower levels for CD9 and TSG101. Preparations from 1 laboratory presented significantly lower particle numbers (p < 0.0001), most probably related to increased processing time. However, even when standardizing processing time, particle yields still differed significantly between groups, indicating inter-laboratory differences in the efficiency of EV isolation. Importantly, no relation was observed between centrifugation speed/k-factor and EV yield.
Conclusions
Our findings demonstrate that quantitative differences in EV yield might be due to equipment- and operator-dependent technical variability in ultracentrifugation-based EV isolation. Furthermore, our study emphasizes the need to standardize technical parameters such as the exact run speed and k-factor in order to transfer protocols between different laboratories. This hints at substantial inter-laboratory biases that should be assessed in multi-centric studies.
Keywords: Extracellular vesicles, Ultracentrifugation, Standardization, Inter-laboratory comparison
Introduction
Extracellular vesicles (EVs) are nanometer-sized particles that are enclosed by a lipid bilayer and shed from cells into the extracellular environment [1, 2]. These vesicles, which contain a variety of bioactive molecules, proteins, and lipids, but also nucleic acids such as mRNA and microRNAs, represent a previously unrecognized form of intercellular communication [3]. There has been a tremendous increase in investigations, either developing EVs as therapeutic agents, as targets of therapeutic interventions, or as biomarkers.
For instance, platelet microparticles, at earlier times considered to be plasma membrane fragments and rather “dust,” are involved in haemostasis, thrombotic, vascular, and inflammatory diseases, but also in cancer and metastasis, and are increased in different disease settings and upon platelet activation [4]. Erythrocyte-derived microparticles have been observed to increase in red blood cell concentrates upon storage, but also in certain disease settings (e.g., malaria), and are similarly considered to be involved in haemostasis, thrombosis, and inflammation [5]. EVs are increasingly considered as circulating biomarkers, for example, to diagnose malignancies [6], but they are also considered as therapeutic agents, for example, from mesenchymal stromal cells [7, 8].
EVs can be isolated from cell-conditioned medium or body fluids. EVs are grouped into different classes based on their physicochemical features such as size or density and mechanisms of release. The term exosome is used to define vesicles formed in endosomal multivesicular bodies and secreted when these fuse with the plasma membrane. Microvesicles, on the other hand, are thought to bud directly from the plasma membrane. As a major obstacle to the field, the isolated material used in EV studies is generally a heterogeneous mixture of different EV types and common contaminants (e.g., lipoproteins) [2, 9].
Thus, due to their heterogeneity and small size, isolation and analysis of EVs still present significant challenges [9, 10]. The most common method of EV isolation was described in 1996 [11], which involves several centrifugation and ultracentrifugation steps, where cells, cellular debris, and larger particles are discarded and EVs are sedimented by sequentially increasing centrifugal forces [12].
Starting at its very beginning, EV research faced technical limitations and challenges. Through many rigorous studies and community efforts including review articles, guidelines, and position papers, the field now seems to have a decent grasp of minimal requirements for EV studies [9, 13, 14], the importance of reporting experimental details [15], and the intricacies of various biofluids [16]. Despite these advances, EV isolation itself is still one of the most important and most debated topics in the community. Researchers can select from a wide range of commercial and in-house isolation methods, each with their own advantages and disadvantages [17, 18, 19]. For differential ultracentrifugation, which is still the most commonly used isolation method, there are reports on how parameters such as run time, centrifugation speed, and rotor type impact both EV yield and contamination with non-EV material [20, 21].
In contrast to our growing understanding of the complexities of different isolation methods, one aspect that has largely been neglected is the fact that virtually all EV studies are single-centric, and that reproducibility across laboratories has yet to be systematically assessed. Even if perfect in-house reproducibility was to be assumed, the ability to replicate EV isolates in different laboratories needs to be validated separately. While the past few years have seen a push towards inter-laboratory performance comparisons for analytical methods [22], failing to assess EV isolation itself on a broader basis will contribute to the rampant reproducibility crisis and hamper both basic research and translational applications of EVs [23].
To shed light on the reproducibility of an established isolation protocol in terms of EV yield and characteristics, we performed the first inter-laboratory comparison of EV isolation based on a pre-defined differential centrifugation protocol according to published protocols [24] and investigated the degree of reproducibility between 4 laboratories in 2 independent rounds of isolation. This inter-laboratory comparison aimed to separately analyse the degree of reproducibility between the participating laboratories in each round of isolation rather than between the 2 independent rounds.
Parallel EV preparations were isolated from one pool of conditioned medium of HCT116 colorectal cancer cells in each of the laboratories. After EV isolation, aliquots were distributed to the person with the highest expertise in the respective test method and then analysed as paired analysis by nanoparticle tracking analysis (NTA), transmission electron microscopy (TEM), Western blot (WB), and fluorescence-activated cell sorting (FACS). All laboratories were well able to isolate EVs using the pre-defined protocol, but in spite of similarities in EV morphology and size, there were distinct differences in their quantities and protein profiles. These variabilities were not only due to different equipment used, for example, rotors and ultracentrifuges, but mainly due to the operator's handling. Therefore, reproducibility and standardization of defined protocols and methods should be controlled in multi-centre studies to improve workflows and decrease batch-to-batch variation.
Materials and Methods
Cell Culture
The colorectal cancer cell line HCT116, used as a model of parental EV-secreting cells, was obtained from the European Collection of Authenticated Cell Cultures and cultured in DMEM-F12 medium (Thermo Fisher Scientific, USA) supplemented with 10% heat-inactivated fetal bovine serum (Thermo Fisher Scientific), 2.5 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Thermo Fisher Scientific) in incubators with 5% CO2 atmosphere at 37°C. Cell morphology was constantly monitored by microscopic observation (AxioVert100 Zeiss, Germany). To ensure that the HCT116 cells were free of mycoplasma contaminations, the cultures were frequently analysed using a PCR-based mycoplasma Detection Kit (Minerva Biolabs, Germany) according to the manufacturer's protocol.
Extracellular Vesicle Isolation
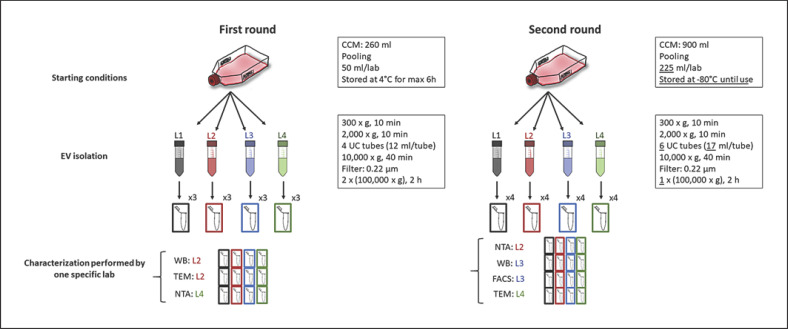
1.7 × 105 (first round) and 1.8 × 105 (second round) cells/cm2 were seeded in T175 flasks (first round: 13 × T175; second round: 45 × T175). After overnight culture, the cells were washed twice with phosphate-buffered saline (PBS, Thermo Fisher Scientific) and cultured for another 24 h in 20 mL/T175 flask DMEM-F12 medium containing 1% EV-depleted FBS (depleted from 20% FBS via overnight centrifugation at 100,000 g at 4°C). Cell-conditioned medium (CCM) was collected, pooled, split into 50-mL tubes, and distributed to the participating laboratories. In the first round, the CCM aliquots were stored at 4°C and processed within 6 h in each laboratory. In the second isolation, the medium was immediately frozen at −80°C and thawed just prior to EV isolation. Each laboratory processed 1 aliquot of the CCM pool independently according to the predefined protocol. EV isolation was performed by differential centrifugation analogous to published protocols [24] (Fig. 1): first, at 300 g for 10 min at 4°C, second at 2,000 g for 10 min at 4°C, third at 10,000 g for 40 min at 4°C, and fourth at 100,000 g for 2 h at 4°C to pellet the EVs. Before the fourth centrifugation step, CCM was filtered using a 0.22-μm pore size filter. This ultracentrifugation step was performed twice in the first and once in the second round (Fig. 1). All laboratories used swing out rotors and available equipment (Table 1). Fresh tubes were used for each centrifugation. In the first round, EV pellets obtained from 12 mL CCM (1 tube) each were resuspended in 30- and 50-µL PBS for TEM and NTA, respectively. For WB analysis, EV pellets obtained from 24 mL CCM (2 tubes) were resuspended in a total volume of 50 µL radioimmunoprecipitation assay (RIPA) buffer. In the second round, EV pellets from 34 mL CCM (2 tubes) were resuspended in 100 µL PBS, where 45 µL were subjected to NTA, 25 µL to TEM, and 30 µL to FACS. For WB analysis, EV pellets from the 170 mL CCM (10 tubes), were resuspended in 100 µL RIPA buffer. EV data were submitted to the EV‑TRACK knowledgebase (http://evtrack.org/; EV track ID: EV190090) [15]. Paired analysis of the 4 EV samples was performed by 1 person with the highest expertise in the respective test method to assure comparability of data.
Fig. 1.
Schematic workflow of EV preparation and characterization. The experimental workflow of the first (left) and second (left) isolation round is depicted and technical differences between the 2 rounds are highlighted in the boxes (underlined).
Table 1.
Description of equipment parameters of ultracentrifugation-based EV isolation across individual laboratories
| Laboratory | Rotor | Ultracentrifuge |
|---|---|---|
| First round | ||
| 1.1 | TH-641 (Thermo Fisher Scientific) | Optima L-90K (Beckman Coulter, USA) |
| 1.2 | SW 28.1 (Beckman Coulter) | Optima LE-80K (Beckman Coulter) |
| 1.3 | Sorvall SureSpin 630 (Thermo Fisher Scientific) | Sorvall Discovery 100 (Thermo Fisher Scientific) |
| 1.4 | Sorvall SureSpin 630 (Thermo Fisher Scientific) | Sorvall Discovery 100 (Thermo Fisher Scientific) |
| Second round | ||
| 2.1 | Sorvall SureSpin 630 (Thermo Fisher Scientific) | Sorvall WX Ultra 100 (Thermo Fisher Scientific) |
| 2.2 Sorvall SureSpin 630 (Thermo Fisher Scientific) | Sorvall WX Ultra 100 (Thermo Fisher Scientific) | |
| 2.3 Sorvall SureSpin 630 (Thermo Fisher Scientific) | Sorvall WX Ultra 100 (Thermo Fisher Scientific) | |
| 2.4 | SW28.1 (Beckman Coulter) | Optima LE-80K (Beckman Coulter) |
All laboratories used swing out rotors in their respective ultracentrifuge.
Transmission Electron Microscopy
In detail, 5 μL of EV suspensions were left to settle on 100 mesh formvar-coated copper grids (Plano, Germany), contrasted with 2% aqueous uranyl acetate (negative stain), air-dried, and visualized using a JEM-1400 transmission microscope (JEOL, USA) equipped with a Tietz 2 K digital camera (TVIPS, Germany) at 80 kV.
Nanoparticle Tracking Analysis
In the first round, 2 µL of concentrated EV suspensions were diluted in sterile-filtered PBS 1:100 and visualized using the NanoSight LM10 NTA device (Malvern Instruments, UK). Each sample was measured 5 times for 45 s (Screen Gain 1.0, camera level 12) with at least 200 valid tracks per video to obtain particle concentration and size distribution. In the second round, 1 μL of concentrated EVs was diluted in sterile-filtered PBS in a dilution range between 1:2,000 and 1:4,000 and visualized using the ZetaView (sensitivity 80%, shutter 100, 11 positions, 2 cycles; Particle Metrix, Germany).
Protein Extraction and Western Blot
EV pellets were resuspended in RIPA buffer (50 mM Tris-HCl, pH7.4, 150 mM NaCl, 1% Triton X-100, 1% Na-deoxycholate, 0.1% SDS, 0.1 mM CaCl2, and 0.01 mM MgCl2 supplemented with protease inhibitor cocktail (Thermo Fisher Scientific). Proteins extracted from HeLa and HCT116 cells were used as cellular controls. Protein concentration was determined using the BCA assay process with Pierce BCA Protein Assay Kit (Thermo Fisher Scientific), following the manufacturer's instructions. Equal amounts of protein (20 µg) were loaded and separated on 4–15% Mini-PROTEAN TGX Precast Gels (Bio-Rad, USA). Protein and cell lysates were treated with protein loading dye (Laemmli sample buffer; Bio-Rad) with freshly added β-mercaptoethanol (10%; v/v; Sigma, Germany) and boiled for 5 min at 95°C before SDS-PAGE. Proteins were subsequently transferred to nitrocellulose blotting membrane (0.2 µm; 1060000; GE Healthcare, USA). Membranes were blocked in 5% BSA (Carl Roth, Germany) in 0.1% Tween in TBS (TBS-T). After blocking, blots were probed with the following primary antibodies diluted in 5% milk/TBS-T: TSG-101 (1:500 dilution, clone 4A10;Thermo Fisher Scientific), Alix (1:500 dilution, clone 3A9;, Biolegend, USA), CD81 (1:300 dilution, clone 5A6; Biolegend), CD63 (assessed under non-reducing conditions, 1:300 dilution, clone MX-49.129.5; Santa Cruz, Germany), CD9 (1:300 dilution, clone C-4; Santa Cruz), and calnexin (1:500 dilution, clone E10; Santa Cruz). After overnight incubation at 4°C, membranes were washed 3 times with TBS-T and subsequently incubated with the secondary antibody dilution: anti-mouse IgG HRP (1:2000 dilution; NA931V; GE Healthcare) for 1 h at room temperature followed by washing. Blots were then developed using WesternBright ECL (541004; Biozym Scientific, Germany) and protein bands were detected using the FusionCapt Advanced Solo 4 (Vilber, Germany).
Fluorescence-Activated Cell Sorting Analysis
FACS measurement of HCT116-derived EVs and their parental cells was performed with BD FACS Canto II, using BD FACSDiva software (BD Biosciences). HCT116-derived EVs were captured on anti-human CD9 beads for flow detection (Exosome-Human CD9 beads; Thermo Fisher Scientific). CD9 beads (20 µL) were washed with 1 mL of assay buffer (PBS + 0.1% BSA sterile-filtered 0.22 µm; BSA, Carl Roth). EVs were added 1:10 v/v to the assay buffer and incubated overnight at 4°C, end-over-end mixing. On the next day, bead-bound EVs were isolated with a magnetic separator and washed several times prior to EV staining. To assess antigen expression, samples were incubated in the dark with the following titrated monoclonal antibodies for 45 min under agitation at 1,000 rpm: CD9 PerCP-Cy 5.5 (BD Biosciences), CD63 Brilliant violet 421 (Biolegend), CD81 PE/Cy7 (Biolegend), TSG101 and calnexin, both Alexa fluor 647 and Alix PE (all Santa Cruz). After washing, beads were retrieved through magnetic separation. EV-bead samples diluted in PBS were measured acquiring a minimum of 50,000 events at low speed. Sterile 0.22-µm-filtered PBS and EVs not labelled with any antibody served as controls. An extracellular staining of cells incubated for 20 min with CD9, CD63, and CD81 and an intracellular staining − 30 min fixation in IC fixation buffer (eBioscience, USA), wash with 1× permeabilization buffer (Invitrogen, USA) and stain for 30 min in permeabilization buffer for TSG101, Alix, and calnexin − was performed on cells. All data were analysed using FlowJo software version 10.0.0.8.
Statistical Analysis
Results of the NTA were analysed using one-way ANOVA and Tukey's post hoc test with p < 0.05 considered statistically significant. Results depicted as box-whisker plots show interquartile range; whiskers: 10th and 90th percentile; line: median. GraphPad Prism (version 7.00; GraphPad Software, USA) was used for statistical analysis and visualization of results.
Results
Comparative EV Isolation: First Round
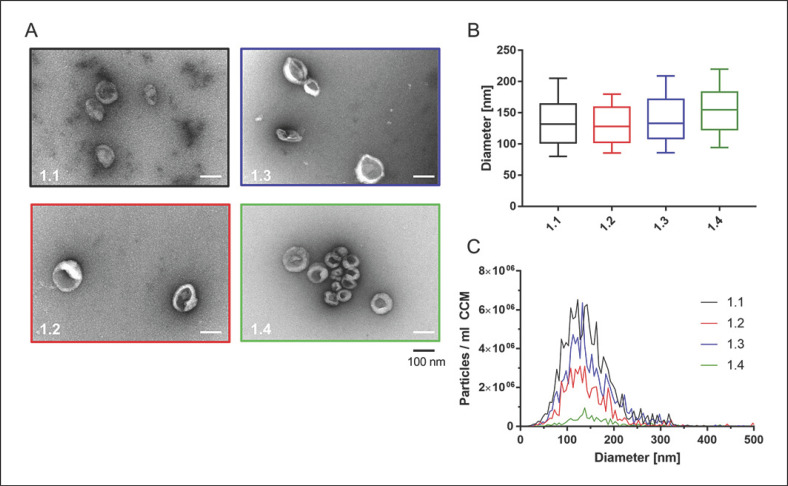
The aim of the first round of our inter-laboratory study was to qualitatively and quantitatively evaluate EV preparations, isolated by ultracentrifugation in the 4 different laboratories using a pre-defined protocol. The human epithelial colorectal cancer cell line HCT116 was chosen as a model of parental EV-secreting cells. CCM was collected and distributed to the 4 participating laboratories for EV isolation by ultracentrifugation, using a pre-defined protocol and their laboratory-specific technical equipment (Fig. 1; Table 1). For the subsequent EV quantification and characterization, the 4 EV preparations were analysed pairwise by 1 person with the highest expertise in the respective test method (Fig. 1). TEM analysis showed spherical and cup-shaped EVs in all 4 preparations (Fig. 2A). EV sizes ranged from around 30 to 150 nm in diameter.
Fig. 2.
Characterization of EVs (first round). A TEM pictures from all EV preparations are depicted. Scale bar, 100 nm. B NTA data of isolated EV sizes. C Full-size profiles are shown for each EV preparation (replicate measurements for each EV isolate). Box: interquartile range; whiskers: 10th and 90th percentile; line: median. Particle concentrations were significantly different with p < 0.01 (1.2 vs. 1.3), p < 0.001 (1.1 vs. 1.3, 1.2 vs. 1.4), and p < 0.0001 (1.1 vs. 1.2, 1.1 vs. 1.4, 1.3 vs. 1.4).
NTA showed that median sizes of isolated particles differed slightly between groups, ranging from 128.3 ± 14.5 nm (laboratory 1.2) to 154.9 ± 42.7 nm (laboratory 1.4) per group (Fig. 2B; Table 2), with a coefficient of variation (Cv) of 8.80% across the groups. Calculating the particle concentrations by NTA revealed significantly different particle yields per ml of medium (Table 2). Laboratory 1.1 isolated most particles, and significantly more than 1.2 (p < 0.0001), 1.3 (p = 0.0035), and 1.4 (p < 0.0001), which is reflected in an overall Cv of 70.88%. The EV preparation with the lowest yield (1.4) was isolated after storing CCM at 4°C for 6 h, whereas the other isolations were initiated within 2 h after CCM preparation. Removing data obtained from L4 from the analysis, inter-group variation was significantly reduced for both particle yield per mL of medium (Cv: 40.93%) and median particle diameter (Cv: 1.96%).
Table 2.
Particle sizes and concentrations determined by NTA (first round)
| L1.1 | L1.2 | L1.3 | L1.4 Cv, % | p value | ||
|---|---|---|---|---|---|---|
| Median diameter, nm | 131.8±9.7 | 128.3±15.5 | 133.3±12.8 | 154.9±42.7 | 8.80 0.3170 | |
| Particles per mL CCM | 1.28×108±1.18×107 | 5.39×107±2.32×107 | 9.17×107±1.04×107 | 1.09×107±2.28×106 | 70.88 | <0.0001 |
All data are presented as arithmetic mean ± SD of measurements within one group. CCM, cell-conditioned medium.
The results of our first isolation round showed that despite a pre-defined protocol, isolation yielded EVs of differing sizes, significantly reduced EV yield in 1 group, and an insufficient amount of material for EV protein characterization. From the data, we assumed that increased storage time could have a detrimental effect on the final yield. For the second round, settings were adapted as illustrated in Figure 1, mainly increasing the starting volume, omitting ultracentrifugation-based washing at 100,000 g, and cryopreserving the CCM before isolation to overcome extended storage times at ambient temperature (Fig. 1).
Comparative EV Isolation: Second Round
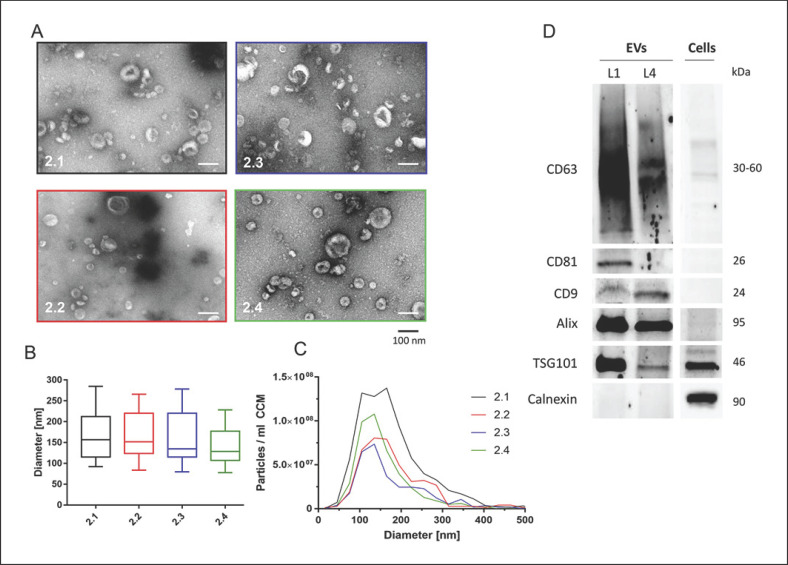
Due to the increased starting volume, EVs isolated within the second round could be subjected to a more comprehensive characterization. TEM revealed the EV-typical spherical and cup-shaped morphology with sizes ranging from 20 to 180 nm (Fig. 3A).
Fig. 3.
Characterization of EVs (second round). A TEM analysis showed spherical and cup-shaped EVs isolated by all laboratories. Scale bar, 100 nm. B NTA of size profiles of isolated EVs. C EV concentrations and size distributions are shown. Particle concentrations were significantly different with p < 0.05 (2.1 vs. 2.2, 2.1 vs. 2.4) and p < 0.01 (2.1 vs. 2.3) or failed to reach statistical significance (2.2 vs. 2.4, 2.3 vs. 2.4). D WB analysis confirmed EV-enriched and EV-depleted protein marker expression in EV and cellular lysates. Protein sizes are indicated.
Next, NTA confirmed sizes ranging from 128.7 ± 17.7 nm (laboratory 2.4) to 156.9 ± 8.6 nm (laboratory 2.1) between groups (Fig. 3B; Table 3). As in the first round, particle sizes across laboratories differed significantly (p = 0.0215), and the Cv of median diameters was 9.38%. Particle yield was significantly different and highest in laboratory 2.1 (compared to 2.2 p = 0.0182, 2.3 p = 0.0016, and 2.4 p = 0.0168; Fig. 3C; Table 3). Inter-group variations in particle yield were still high, but lower than in the first round (Cv: 40.49%).
Table 3.
Particle sizes and concentrations determined by NTA (second round)
| L2.1 | L2.2 | L2.3 | L2.4 Cv, % | p value | ||
|---|---|---|---|---|---|---|
| Median diameter, nm | 156.9±8.6 | 151.5±23.7 | 134.6±18.3 | 128.7±17.7 | 9.38 | 0.0215 |
| Particles per mL CCM | 7.50×108±8.05×107 | 4.15×108±2.68×108 | 3.12×108±2.21×108 | 4.12×108±1.22×108 | 40.49 | 0.0023 |
All data are mean ± SD of replicate measurement within one laboratory. CCM, cell-conditioned medium.
For the characterization of isolated EVs, WB and FACS analyses were performed. Due to lower yields in samples 2.2 and 2.3, WB analysis was only performed on samples from 2.1 to 2.4. Overall, common EV marker proteins like CD63, CD81, CD9, Alix, and TSG101 were found to be expressed in EVs. The endoplasmic reticulum-associated protein calnexin was only observed in cellular protein lysates but not in the EV protein extracts, confirming the absence of cellular protein contamination in the EV protein samples (Fig. 3D). Despite equal protein loading, 2.1 samples appeared to contain larger numbers of EVs as indicated by higher band intensities for CD63, CD81, Alix, and TSG101. Interestingly, CD9 prevailed with a stronger band in 2.4 than in 2.1.
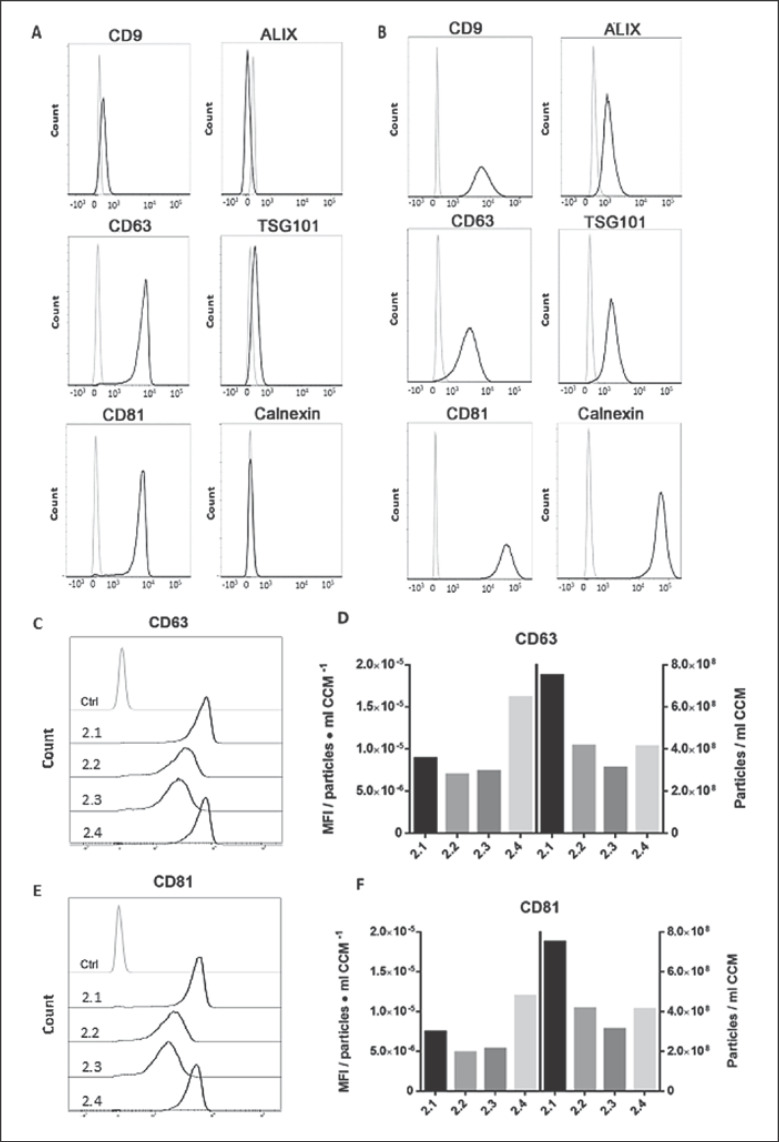
FACS analysis was used as a complementary approach to EV marker characterization, given that a much lower amount of EV material was required. EVs bound to CD9 beads showed positive staining for the typical EV surface markers CD9 (27.3%), CD63 (99.2%), and CD81 (98.6%), as well as TSG101 (25.4%) − all values for EV sample from laboratory 2.1. EV samples from laboratories 2.2–2.4 were also tested for these EV surface markers: CD9 (0.77, 0.69, and 9.82%, respectively), CD63 (97.8, 98 and 99.1%), CD81 (97.3, 97.2, and 98.5%), and TSG101 (3.69, 1.99, and 16.8%; data not shown).
Supporting the WB results, almost no staining was observed for calnexin (0.94%), indicating high purity of EVs. Nevertheless, Alix was not detectable on EVs by FACS, contrary to the WB analysis (Fig. 4A). All markers were validated to be expressed in cells by either extra- or intracellular staining (Fig. 4B). Expression intensities varied between groups, which prompted us to assess whether expression intensities correlated with particle counts. To this end, mean fluorescence intensity (MFI) values for CD63 (Fig. 4C) and CD81 (Fig. 4E) were calculated and expressed in relation to the particle concentrations in preparations from each laboratory (Fig. 4D, F). Higher expression intensities were found for laboratory 2.4, followed by 2.1, whereas 2.2 and 2.3 depicted lower but similar intensities amongst them.
Fig. 4.
FACS-based characterization of isolated EVs (second round). A FACS histograms depicting the relative fluorescence/marker intensity of EV preparation 2.1 (black line) against unstained EV particle control (grey line). B Corresponding marker expression in HCT116 cells (extracellular staining for CD9, CD63, and CD81 and intracellular staining for Alix, TSG101, and calnexin). C, E Mean fluorescence intensity (MFI) raw values of CD63 (C) and CD81 (E) marker expression from laboratories 2.1–2.4. D, F MFI values per particle concentration of CD63 (D) and CD81 (F; left y axis) against the respective particle concentration per ml CCM (right y axis).
The results from our second isolation round also demonstrated a strong variation in EV sizes and yield, discerning however, lower inter-group variation compared to the first round. We confirmed that higher EV yield generated stronger band intensities despite equal protein loading. Additionally, FACS analysis not only revealed the presence of typical EV surface markers, but indeed indicated no direct correlation between MFI values and particle concentration in our preparations.
Calculation of Actual Centrifugation Forces and k-Factors
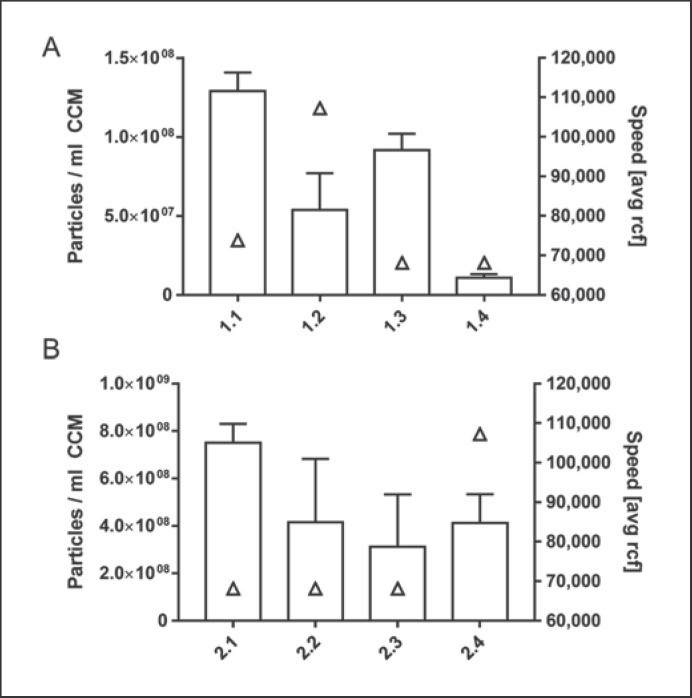
Especially the laboratory-specific differences in EV yields prompted us to compare rotor details and actual centrifugation forces between the first and second round of EV preparations (Table 4). It became obvious that the protocol-based instruction of “using 100,000 g” was interpreted in different laboratories in 2 different ways: either as maximum or as average speed. Accordingly, the respective k-factors as indicators of the relative pelleting efficiency turned out to be different (Table 4; Fig. 5). However, EV yields could not be correlated to speed. In the first and second round of isolation, 2 and 3 laboratories, respectively, used the same centrifuge and rotor (Table 4). Comparing these data still showed apparent differences in EV yield (Fig. 5), leading to a Cv of 46.51%.
Table 4.
Rotor type and technical specifications
| Laboratory | Rotor | Actual speed, rpm | rcf avg | k-factor |
|---|---|---|---|---|
| First round | ||||
| 1.1 | TH-64.1 | 24,200 | 73,823 | 327.2 |
| 1.2 | SW 28.1 | 28,000 | 107,215 | 276.0 |
| 1.3 | SureSpin 630 | 23,000 | 68,135 | 456.4 |
| 1.4 | SureSpin 630 | 23,000 | 68,135 | 456.4 |
| Second round | ||||
| 2.1 | SureSpin 630 | 23,000 | 68,135 | 456.4 |
| 2.2 | SureSpin 630 | 23,000 | 68,135 | 456.4 |
| 2.3 | SureSpin 630 | 23,000 | 68,135 | 456.4 |
| 2.4 | SW 28.1 | 28,000 | 107,215 | 276.0 |
Fig. 5.
Correlation of centrifugation speed and particle yield in the first (A) and second (B) round of EV isolation. Particle counts (bars, left yaxis, replicate measurements of each EV isolate) vs. speed (avg rcf; triangles, right y axis) obtained in different laboratories are depicted.
Surprisingly, we found that even when comparing laboratories with the same protocol interpretation and technical equipment, no direct correlation between total EV particle yield and centrifugation speed in either isolation round was seen.
Discussion/Conclusion
This pilot study aimed at examining the transferability of basic ultracentrifugation protocols for EV isolation and the technical variation induced by equipment and operator that remains even for standardized protocols. Even though all participating laboratories were able to enrich EVs from pooled HCT116 CCM, differences in results were obvious in both rounds of isolation. As expected, these differences seemed to be predominantly quantitative as demonstrated by significant variation in NTA-based particle counts but similar EV morphology. Unexpectedly, the amount of protein pelleted in some of the laboratories was insufficient for analysis by immunoblotting, which could be mitigated in future studies by increasing starting volumes or including more sensitive methods such as flow cytometry/FACS. Indeed, we were able to detect the tetraspanins CD63 and CD81 in preparations from all laboratories by FACS even though their concentration in some lysates was too low to be detected on WB. Additionally, while inter-laboratory evaluations of EV yield and composition are a valuable starting point, more advanced studies might also assess their functionality or levels of disease-related markers associated with EVs from each study site.
At first glance, these quantitative differences might seem obvious since centrifugation speed and k-factors were not matched across all rotors used in this study. Despite similar rpm, for instance, centrifugation time would have had to be substantially increased for the SureSpin rotor to match the performance of the high-rcf SW 28.1 rotor. It is therefore surprising that particle yield did not seem to correlate with centrifugation speed and that the highest particle yields were not generated by the laboratory using the SW 28.1 rotor. Even using identical centrifuges, rotors and run parameters did not produce highly similar results (Fig. 1, laboratories 1.3 and 1.4; Fig. 2, laboratories 2.1–2.3). The variation in particle yield observed under these conditions (second round, Cv: 46.51%) seems to be attributable to operator effects, although it is impossible to estimate true inter-operator variability without having a good estimate of intra-operator variability across multiple isolations. A follow-up multi-centre study, thus, should also record intra-operator, intra-laboratory, and inter-laboratory variability.
We suspect that among the multitude of isolation methods, ultracentrifugation is particularly prone to operator biases due to subtle handling differences such as removing supernatant by decanting versus pipetting and resuspending pellets. As such, our study should be considered as a starting point to initiate more detailed intra- and inter-laboratory comparisons. It would be highly interesting to extend these to additional EV isolation methods, particularly those relevant to manufacturing therapeutic EVs, for which standardized workflows and minimal batch-to-batch variation in yield, purity, and potency are imperative [7, 25].
In addition to the well-established impact of storage conditions and freeze-thaw cycles on isolated vesicles [26, 27, 28], our data seem to indicate that storage time impacts EV yield. In the first round of isolation, particle yield for laboratory 1.4, in which CCM was stored for a longer period of time prior to EV separation, was remarkably reduced despite using identical UC parameters to those for laboratory 1.3. Of course, to verify this assumption, additional experiments are required permuting storage times and temperatures. However, the reduced overall variability of particle concentrations in the second round already supports our notion. Furthermore, previous reports already linked shorter processing time to increased particle concentrations [29]. For plasma and serum, a direct impact of time between blood draw and initial centrifugation on EV yield has already been reported [30]. Reduced concentrations after longer storage could be mediated by EV adhesion to plastic surfaces or disruption of vesicles. Since particles isolated by laboratory 1.4 trended towards larger diameters, it is tempting to speculate that EV loss might also be induced by aggregation of vesicles.
Necessitated by these challenges and the limitations of current assays, reference materials tailor-made for EV experiments have been highly anticipated. Appropriate EV mimetics such as the trackable recombinant EVs, recently presented by the Hendrix and Giebel laboratories [31, 32], as well as monodisperse and bimodal spherical reference material [33] will be helpful to calibrate instruments, benchmark methods, and normalize results. Using reference material/particles as spike-ins in multi-centric comparative studies will facilitate the assessment of equipment- and operator-dependent variability. By comparing the sizes of silica or polystyrene-based nanoparticle references in an inter-laboratory study, it was suggested that determining/measuring particle sizes of heterogeneous and bimodal samples is more difficult compared to monodisperse and homogeneous samples [33]. The study performed by Nicolet et al. [33] emphasized the technical benefits of reference material for the calibration of EV measurements. Since EVs are typically isolated and characterized as heterogeneous and polydisperse populations, evidence suggested that EV size determination requires careful data interpretation [33].
Admittedly, the conclusions that can be derived from an inter-laboratory study involving only 4 laboratories, only 2 rounds of isolations, and a basic characterization of EVs are limited; yet our findings prompt to perform larger studies across multiple institutions. Further, while we demonstrated inter-operator variability and the need for thoroughly standardized protocols, we cannot accurately quantify variability between laboratories due to not having assessed intra-operator variability, which might have accounted for a non-significant proportion of overall variability. However, all contributing partners were experienced in EV isolation and characterization [6, 17, 24, 34, 35, 36, 37, 38, 39, 40, 41].
Given its widespread utilization in the community, we chose to assess reproducibility of EV isolation based on ultracentrifugation, but similar studies clearly need to include additional methods such as those based on precipitation, size-exclusion chromatography, or filtration. Future studies that benchmark methods and compare their robustness, reproducibility, and efficiency should be performed for additional biofluids besides cell culture supernatant. Using exogenous reference material and more sophisticated methodology will additionally improve conclusions drawn from follow-up studies.
Which conclusions regarding experimental standardization and variability can thus be drawn from this study?
Unambiguous and Extensive Reporting
Many publications, for instance, report centrifugation speed without explicitly specifying if it relates to the average centrifugal force in the middle of the tube (rcf avg) or the maximal centrifugal force at the bottom of the tube (rcf max). In our study, the protocol-based instruction of “using 100,000 g” was interpreted differently (as maximum or average speed) between the laboratories, resulting in different applied centrifugal forces (Table 4). Although we did not observe a relation, we agree with the MISEV2018 guidelines to report g-forces and k-factors in addition to rotor types (fixed angle or swinging bucket) to transfer a given isolation protocol across rotors [13]. In addition, our data underline the need to report sample storage and handling, including processing time. We advise strict adherence to the published protocols by Théry et al. [42] and the MISEV guidelines [13, 14] to facilitate reproducibility.
Alternative Methods, Easy to Standardize or Automate
Given the intrinsic intra-and inter-laboratory variability in UC-based methods, alternative methods that are faster, facile, efficacious, and easier to standardize or even automate, appear to be desired especially when considering clinical utility.
Intra- and Inter-Laboratory Studies for Harmonization and Standardization
Given the EV community's learning curve in dealing with previous and ongoing challenges, which led to the development of valuable tools such as EV-TRACK and the MISEV guidelines [13, 14, 15], we are confident that a better understanding of inter-laboratory variability will lead to improved harmonization and standardization. Indeed, comprehensive efforts to increase rigor and standardization were already initiated by the International Society for Extracellular Vesicles (ISEV), paving the way for the development of community-wide conventions and standardized methods. In detail, the planning should consider existing knowledge, already identified challenges/limitations, and strategies for improvement, especially regarding (a) starting material and pre-processing, (b) isolation protocols, (c) sample and assay-specific pre-analytics, (d) defined and validated analytical methods for EV quantification and characterization, and finally (e) a pre-defined statistical analysis plan [9, 13, 14, 15, 16, 27, 43, 44, 45, 46, 47, 48, 49, 50, 51].
To summarize, our proof-of-concept inter-laboratory study indicates significant equipment- and operator-dependent technical variability in ultracentrifugation-based EV isolation. Reproducing isolation protocols based on centrifugation speed and run time alone is clearly insufficient, and even if the k-factor is appropriately considered, variability is reduced but not eliminated. These findings prompt several questions including how reproducibility is best quantified, which criteria should be assessed, and how sensible thresholds for inter-user variability might be established. Recent advances in dedicated EV technologies will certainly help to answer these questions. Using increasingly sensitive analytical assays and incorporating appropriate reference material allows the detection and quantification of technical biases, which lays the foundation for any effort to reduce variability and increase standardization.
Statement of Ethics
No ethical approval was required for this study.
Disclosure Statement
The authors have no conflicts of interest to declare.
Funding Sources
We acknowledge the financial contribution of The Ministry of Science, Research and the Arts of the State of Baden-Württemberg for the Collaborative Research Training Group “Tissue Analytics for Stem Cell-based Diagnostics and Therapy (TASCDT)” (A.T.C. and K.B.).
Author Contributions
A.T.C., F.F., K.N., T.W., U.E., D.B., C.T., M.S., J.G. and K.B.: conceptualization, methodology, resources, and writing. A.T.C., F.F., K.N., T.W., U.E., S.E.-H., C.T., and I.H.: investigation and formal analysis. M.K., M.S., J.G., and K.B.: funding acquisition, project administration, supervision, and writing: review and editing.
Acknowledgments
We gratefully acknowledge Ulrike Ganserer, IPH Heidelberg, and Stefanie Uhlig from the Mannheim FlowCore for their excellent support in transmission electron microscopy and flow cytometry, respectively.
References
- 1.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013 Feb;200((4)):373–83. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30((1)):255–89. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 3.Tkach M, Théry C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016 Mar;164((6)):1226–32. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 4.E Kehrel B, F Brodde M. State of the art in platelet function testing. Transfus Med Hemother. 2013 Apr;40((2)):73–86. doi: 10.1159/000350469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubin O, Canellini G, Delobel J, Lion N, Tissot JD. Red blood cell microparticles: clinical relevance. Transfus Med Hemother. 2012 Oct;39((5)):342–7. doi: 10.1159/000342228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Worst TS, Previti C, Nitschke K, Diessl N, Gross JC, Hoffmann L, et al. miR-10a-5p and miR-29b-3p as Extracellular Vesicle-Associated Prostate Cancer Detection Markers. Cancers (Basel) 2019 Dec;12((1)):E43. doi: 10.3390/cancers12010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witwer KW, Van Balkom BW, Bruno S, Choo A, Dominici M, Gimona M, et al. Defining mesenchymal stromal cell (MSC)-derived small extracellular vesicles for therapeutic applications. J Extracell Vesicles. 2019 Apr;8((1)):1609206. doi: 10.1080/20013078.2019.1609206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reiner AT, Witwer KW, van Balkom BW, de Beer J, Brodie C, Corteling RL, et al. Concise Review: Developing Best-Practice Models for the Therapeutic Use of Extracellular Vesicles. Stem Cells Transl Med. 2017 Aug;6((8)):1730–9. doi: 10.1002/sctm.17-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014 Dec;3((1)):26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva J, Garcia V, Rodriguez M, Compte M, Cisneros E, Veguillas P, et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer. 2012 Apr;51((4)):409–18. doi: 10.1002/gcc.21926. [DOI] [PubMed] [Google Scholar]
- 11.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996 Mar;183((3)):1161–72. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Théry C, Amigorena S, Raposo G, Clayton A. Curr Protoc Cell Biol. 2006. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Chapter 3: Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 13.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018 Nov;7((1)):1535750–50. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Witwer KW, Soekmadji C, Hill AF, Wauben MH, Buzás EI, Di Vizio D, et al. Updating the MISEV minimal requirements for extracellular vesicle studies: building bridges to reproducibility. J Extracell Vesicles. 2017 Nov;6((1)):1396823. doi: 10.1080/20013078.2017.1396823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Deun J, Mestdagh P, Agostinis P, Akay Ö, Anand S, Anckaert J, et al. EV-TRACK Consortium EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods. 2017 Feb;14((3)):228–32. doi: 10.1038/nmeth.4185. [DOI] [PubMed] [Google Scholar]
- 16.Yuana Y, Böing AN, Grootemaat AE, van der Pol E, Hau CM, Cizmar P, et al. Handling and storage of human body fluids for analysis of extracellular vesicles. J Extracell Vesicles. 2015 Nov;4((1)):29260. doi: 10.3402/jev.v4.29260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buschmann D, Kirchner B, Hermann S, Märte M, Wurmser C, Brandes F, et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J Extracell Vesicles. 2018 Jun;7((1)):1481321–21. doi: 10.1080/20013078.2018.1481321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel GK, Khan MA, Zubair H, Srivastava SK, Khushman M, Singh S, et al. Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci Rep. Mar 29;9:5335. doi: 10.1038/s41598-019-41800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mussack V, Wittmann G, Pfaffl MW. Comparing small urinary extracellular vesicle purification methods with a view to RNA sequencing-Enabling robust and non-invasive biomarker research. Biomol Detect Quantif. 2019 Jun;17:100089. doi: 10.1016/j.bdq.2019.100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cvjetkovic A, Lötvall J, Lässer C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J Extracell Vesicles. 2014 Mar;3((1)):3. doi: 10.3402/jev.v3.23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livshits MA, Khomyakova E, Evtushenko EG, Lazarev VN, Kulemin NA, Semina SE, et al. Isolation of exosomes by differential centrifugation: theoretical analysis of a commonly used protocol. Sci Rep. 2015 Nov;5((1)):17319. doi: 10.1038/srep17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Pol E, Sturk A, van Leeuwen T, Nieuwland R, Coumans F, Mobarrez F, et al. ISTH-SSC-VB Working group Standardization of extracellular vesicle measurements by flow cytometry through vesicle diameter approximation. J Thromb Haemost. 2018 Jun;16((6)):1236–45. doi: 10.1111/jth.14009. [DOI] [PubMed] [Google Scholar]
- 23.De Wever O, Hendrix A. A supporting ecosystem to mature extracellular vesicles into clinical application. EMBO J. 2019 May;38((9)):e101412. doi: 10.15252/embj.2018101412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tucher C, Bode K, Schiller P, Claßen L, Birr C, Souto-Carneiro MM, et al. Extracellular Vesicle Subtypes Released From Activated or Apoptotic T-Lymphocytes Carry a Specific and Stimulus-Dependent Protein Cargo. Front Immunol. 2018 Mar;9:534. doi: 10.3389/fimmu.2018.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohde E, Pachler K, Gimona M. Manufacturing and characterization of extracellular vesicles from umbilical cord-derived mesenchymal stromal cells for clinical testing. Cytotherapy. 2019 Jun;21((6)):581–92. doi: 10.1016/j.jcyt.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 26.Cheng Y, Zeng Q, Han Q, Xia W. Effect of pH, temperature and freezing-thawing on quantity changes and cellular uptake of exosomes. Protein Cell. 2019 Apr;10((4)):295–9. doi: 10.1007/s13238-018-0529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeyaram A, Jay SM. Preservation and Storage Stability of Extracellular Vesicles for Therapeutic Applications. AAPS J. 2017 Nov;20((1)):1. doi: 10.1208/s12248-017-0160-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lőrincz AM, Timár CI, Marosvári KA, Veres DS, Otrokocsi L, Kittel Á, et al. Effect of storage on physical and functional properties of extracellular vesicles derived from neutrophilic granulocytes. J Extracell Vesicles. 2014 Dec;3((1)):25465. doi: 10.3402/jev.v3.25465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. 2015 Jul;4((1)):27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bæk R, Søndergaard EK, Varming K, Jørgensen MM. The impact of various preanalytical treatments on the phenotype of small extracellular vesicles in blood analyzed by protein microarray. J Immunol Methods. 2016 Nov;438:11–20. doi: 10.1016/j.jim.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Geeurickx E, Tulkens J, Dhondt B, Van Deun J, Lippens L, Vergauwen G, et al. The generation and use of recombinant extracellular vesicles as biological reference material. Nat Commun. 2019 Jul;10((1)):3288. doi: 10.1038/s41467-019-11182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Görgens A, Bremer M, Ferrer-Tur R, Murke F, Tertel T, Horn PA, et al. Optimisation of imaging flow cytometry for the analysis of single extracellular vesicles by using fluorescence-tagged vesicles as biological reference material. J Extracell Vesicles. 2019 Mar;8((1)):1587567. doi: 10.1080/20013078.2019.1587567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicolet A, Meli F, van der Pol E, Yuana Y, Gollwitzer C, Krumrey M, et al. Inter-laboratory comparison on the size and stability of monodisperse and bimodal synthetic reference particles for standardization of extracellular vesicle measurements. Meas Sci Technol. 2016;27((3)):035701. [Google Scholar]
- 34.Claßen L, Tykocinski LO, Wiedmann F, Birr C, Schiller P, Tucher C, et al. Extracellular vesicles mediate intercellular communication: transfer of functionally active microRNAs by microvesicles into phagocytes. Eur J Immunol. 2017 Sep;47((9)):1535–49. doi: 10.1002/eji.201646595. [DOI] [PubMed] [Google Scholar]
- 35.Fricke F, Mussack V, Buschmann D, Hausser I, Pfaffl MW, Kopitz J, et al. TGFBR2‑dependent alterations of microRNA profiles in extracellular vesicles and parental colorectal cancer cells. Int J Oncol. 2019 Oct;55((4)):925–37. doi: 10.3892/ijo.2019.4859. [DOI] [PubMed] [Google Scholar]
- 36.Fricke F, Michalak M, Warnken U, Hausser I, Schnölzer M, Kopitz J, et al. SILAC-Based Quantification of TGFBR2-Regulated Protein Expression in Extracellular Vesicles of Microsatellite Unstable Colorectal Cancers. Int J Mol Sci. 2019 Aug;20((17)):4162. doi: 10.3390/ijms20174162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yue S, Mu W, Erb U, Zöller M. The tetraspanins CD151 and Tspan8 are essential exosome components for the crosstalk between cancer initiating cells and their surrounding. Oncotarget. 2015 Feb;6((4)):2366–84. doi: 10.18632/oncotarget.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erb U, Zhao K, Wang Z, Xiao L, Zöller M. Murine and human pancreatic tumor exosome recovery in mouse serum: diagnostic and prognostic potential and target cell delivery. Cancer Lett. 2017 Sep;403:1–12. doi: 10.1016/j.canlet.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Erb U, Zöller M. Progress and potential of exosome analysis for early pancreatic cancer detection. Expert Rev Mol Diagn. 2016 Jul;16((7)):757–67. doi: 10.1080/14737159.2016.1187563. [DOI] [PubMed] [Google Scholar]
- 40.Worst TS, von Hardenberg J, Gross JC, Erben P, Schnölzer M, Hausser I, et al. Database-augmented Mass Spectrometry Analysis of Exosomes Identifies Claudin 3 as a Putative Prostate Cancer Biomarker. Mol Cell Proteomics. 2017 Jun;16((6)):998–1008. doi: 10.1074/mcp.M117.068577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gouveia de Andrade AV, Bertolino G, Riewaldt J, Bieback K, Karbanová J, Odendahl M, et al. Extracellular vesicles secreted by bone marrow- and adipose tissue-derived mesenchymal stromal cells fail to suppress lymphocyte proliferation. Stem Cells Dev. 2015 Jun;24((11)):1374–6. doi: 10.1089/scd.2014.0563. [DOI] [PubMed] [Google Scholar]
- 42.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 Apr;Chapter 3((1)):22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 43.Coumans FA, Brisson AR, Buzas EI, Dignat-George F, Drees EE, El-Andaloussi S, et al. Methodological Guidelines to Study Extracellular Vesicles. Circ Res. 2017 May;120((10)):1632–48. doi: 10.1161/CIRCRESAHA.117.309417. [DOI] [PubMed] [Google Scholar]
- 44.Witwer KW, Buzás EI, Bemis LT, Bora A, Lässer C, Lötvall J, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013 May;2((1)):2. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clayton A, Boilard E, Buzas EI, Cheng L, Falcón-Perez JM, Gardiner C, et al. Considerations towards a roadmap for collection, handling and storage of blood extracellular vesicles. J Extracell Vesicles. 2019 Aug;8((1)):1647027. doi: 10.1080/20013078.2019.1647027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soekmadji C, Hill AF, Wauben MH, Buzás EI, Di Vizio D, Gardiner C, et al. Towards mechanisms and standardization in extracellular vesicle and extracellular RNA studies: results of a worldwide survey. J Extracell Vesicles. 2018 Oct;7((1)):1535745. doi: 10.1080/20013078.2018.1535745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maas SL, de Vrij J, van der Vlist EJ, Geragousian B, van Bloois L, Mastrobattista E, et al. Possibilities and limitations of current technologies for quantification of biological extracellular vesicles and synthetic mimics. J Control Release. 2015 Feb;200:87–96. doi: 10.1016/j.jconrel.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Pol E, Coumans FA, Grootemaat AE, Gardiner C, Sargent IL, Harrison P, et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost. 2014 Jul;12((7)):1182–92. doi: 10.1111/jth.12602. [DOI] [PubMed] [Google Scholar]
- 49.Momen-Heravi F, Balaj L, Alian S, Mantel PY, Halleck AE, Trachtenberg AJ, et al. Current methods for the isolation of extracellular vesicles. Biol Chem. 2013 Oct;394((10)):1253–62. doi: 10.1515/hsz-2013-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rocke DM. Robust Statistical-Analysis of Interlaboratory Studies. Biometrika. 1983;70((2)):421–31. [Google Scholar]
- 51.Molenaar J, Cofino WP, Torfs PJ. Efficient and robust analysis of interlaboratory studies. Chemometr Intell Lab. 2018 Apr;175:65–73. [Google Scholar]