Abstract
Schizophrenia (SZ) is a common and debilitating psychiatric disorder with limited effective treatment options. Although highly heritable, risk for this polygenic disorder depends on the complex interplay of hundreds of common and rare variants. Translating the growing list of genetic loci significantly associated with disease into medically actionable information remains an important challenge. Thus, establishing platforms with which to validate the impact of risk variants in cell-type-specific and donor-dependent contexts is critical. Towards this, we selected and characterized a collection of 12 human induced pluripotent stem cell (hiPSC) lines derived from control donors with extremely low and high SZ polygenic risk scores (PRS). These hiPSC lines are publicly available at the California Institute for Regenerative Medicine (CIRM). The suitability of these extreme PRS hiPSCs for CRISPR-based isogenic comparisons of neurons and glia was evaluated across 3 independent laboratories, identifying 9 out of 12 meeting our criteria. We report a standardized resource of publicly available hiPSCs on which we hope to perform genome engineering and generate diverse kinds of functional data, with comparisons across studies facilitated by the use of a common set of genetic backgrounds.
Keywords: Schizophrenia, Polygenic risk score, Human induced pluripotent stem cell, CRISPR/Cas9
Introduction
Schizophrenia (SZ) is a disorder marked by extremely heterogeneous clinical presentation and an equally complex genetic risk architecture, with contributions from common and rare variants (reviewed [1]). To date, genetic studies have identified 145 loci with common single nucleotide polymorphisms (SNPs) of small effect sizes [2], 8 rare copy number variations (CNVs) of relative high penetrance [3], and 2 genes with rare but high penetrance loss-of-function mutations (SETD1A[4] and RBM12[5]) that are significantly associated with risk for SZ. Although rare variants are typically highly penetrant, common variants individually result in small increases in the odds ratio (OR < 1.3) for SZ [6], accounting for a substantial portion of disease risk only in aggregate. There is a growing consensus that genetic risk converges between psychiatric [7] but not neurodegenerative disorders [8], particularly focused on genes expressed during fetal cortical development [9, 10, 11, 12], and involved in synaptic biology [3, 13, 14, 15, 16] or gene regulation [13, 14, 17].
Although genome-wide association studies (GWAS) [2] and large-scale exome sequencing projects (https://schema.org/) are increasingly dissecting the genetic architecture of risk for psychiatric disease, clinical translation of this knowledge is lagging. Many disease-associated variants are noncoding, suggesting a role in gene regulation [18]; nearly half of the known SZ SNPs likely differentially regulate nearby gene expression [19]. Although the regulatory impact of these SNPs can be predicted by overlapping GWAS with multiomic transcriptomic (expression quantitative trait loci [20, 21]) and epigenetic (histone modification [22] and chromatin accessibility [23] and looping [24, 25]) datasets, functionally demonstrating how these loci act as causal contributors to disease risk remains an intractable problem. There is an increasing need to functionally dissect the causal mechanisms underlying disease risk [26, 27], as many important questions remain unanswered. What are the mechanisms by which this growing list of common SNPs act within the diverse cell types of the brain to contribute to SZ etiology? Is there pathway-level convergence of these disorder-associated genes, and if so, does convergence occur in a cell-type-specific manner? Do these common variants of small effect interact in a strictly additive fashion [26], or through more complex epistatic [28] or omnigenic models [29]? Systematic functional validation of these many risk variants requires a more tractable and amendable model. For instance, one that can perform network-level perturbations to understand GWAS risk variants that may act additively and/or synergistically [30].
Theoretically capable of generating every cell type in the human body, human induced pluripotent stem cells (hiPSCs) are a useful tool with which to generate patient-specific cells for functional genomic studies. hiPSC-derived neurons and glia most resemble their prenatal in vivo counterparts [31, 32, 33, 34, 35, 36] and are particularly well suited to test the neurodevelopmental impact of the many psychiatric risk variants predicted to exert their influence during fetal cortical development [12]. Emerging brain organoids [37] and more complex assembloid [38, 39, 40] models enable studies with extended maturation periods in more physiologically relevant contexts. Although case/control hiPSC-based models demonstrate concordance with postmortem datasets [36], common genetic differences between donors (independent of diagnosis) lead to substantial interindividual heterogeneity between hiPSC lines [41, 42]. Given the limited sample sizes feasible in hiPSC studies, the most rigorous and well-powered approach to study individual variants is through isogenic comparisons on the same donor background [43]. The isogenic approach has been recently made possible by the CRISPR-based genome editing toolbox [44] to empirically evaluate the impact of SZ-associated changes in DNA sequence, endogenous gene expression, DNA methylation, histone modification, and chromatin confirmation (reviewed [45]). We [23, 46, 47] and others [48, 49] have applied genome editing to facilitate the empirical validation of putative causal variants through precise isogenic comparisons on defined genetic backgrounds. However, due to the limited number of hiPSC lines used in each study while the cellular phenotypic expressivity may be influenced by genetic backgrounds, we are so far unable to evaluate and compare the effect. Thus, the field is in need of a set of hiPSC lines with defined genetic backgrounds that can be used by different investigators to produce data that can be cross-replicated and integrated.
Towards the goal of facilitating functional genomic studies of how risk variants interact with donor background in a cell-type-specific manner, here we characterized the genomic integrity, pluripotency, and gene transfection and neural differentiation efficiencies, as well as CRISPR genome edibility of 12 control hiPSCs derived from donors with exceptionally high and low polygenic risk scores (PRS) for SZ (Fig. 1). Through complementary studies conducted at all 3 sites, we collectively identified the 9 hiPSCs most appropriate for neuronal induction and CRISPR editing. These publicly available hiPSC lines with extreme SZ PRS will not only help evaluate the reproducibility of functional genomic studies, but also facilitate the integration of datasets generated by different laboratories, revealing any convergent impacts of independent SZ-GWAS genes/variants.
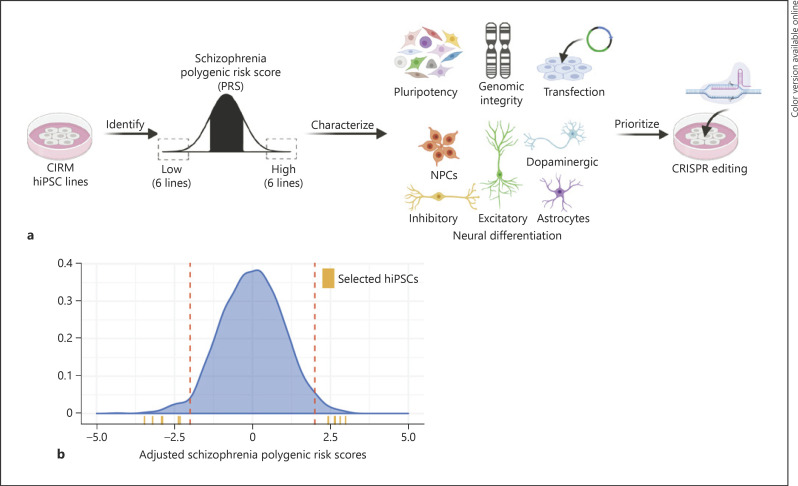
Fig. 1.
Identification, characterization, and prioritization of high and low SZ PRS hiPSC lines. a Schematic summary of analysis pipeline. Figure created with BioRender.com. b Identification of hiPSC lines with >2 SD lower or higher PRS for SZ based on PGC2. The density distribution (blue) depicts the standardized ancestry-adjusted risk scores for ∼1,500 CIRM hiPSC lines (currently available for distribution on the CDI website), the dotted red lines represent >2 SD, and the orange ticks indicate the 6 high and 6 low PRS hiPSC lines of European ancestry selected for this study. SZ, schizophrenia; PRS, polygenic risk scores; hiPSC, human induced pluripotent stem cell; SD, standard deviations; NPCs, neural progenitor cells.
Materials and Methods
Prioritization of High and Low PRS hiPSCs
There are uniform demographic and clinical data for the entire CIRM hiPSC cohort (https://www.cirm.ca.gov/researchers/ipsc-repository). All available lines were validated across the following metrics: (1) chromosomal integrity (Illumina Infinium HumanCore BeadChip), no amplifications >5 MB (resolution of traditional G-banding assay) on SNP arrays with LogRDev score <0.5 that were not pre-existing in the donor; (2) pluripotency (qPCR of 48 mRNAs), a nonprobabilistic binary linear classifier identifies the gene expression of the sample as iPSC based on an appropriate training set; (3) identity confirmation (PCR assay for 48 SNPs), ≤1 mismatch between donor and hiPSC line; (4) loss of reprogramming transgenes (PCR for 2 plasmid EBNA and OriP sequences), detection of ≤1 plasmid copy per 100 cells or a decrease in the number of plasmid copies detected at passage 5; (5) mycoplasma negative (qPCR for 8 species); and (6) sterility (microbiological testing by third-party service provider). We selected only those hiPSCs derived from a common somatic cell source (peripheral blood mononuclear cells), with an absence of acquired structural variants and identity/gender discrepancies (indicative of sample swaps), and from unrelated donors of European ancestry, as SZ PRS was calculated from European subjects [2] and is ancestry dependent [50]. Out of 30 samples with a PRS of at least 2 standard deviations from the mean, 3 female and 3 male subjects were selected per risk group.
hiPSC Culture
hiPSCs were thawed with mTeSR1 (STEMCELL, #85850) and 1 μM H1152 (Millipore, #555550), seeded on Matrigel (Corning, #354230) coated 6-well plates, and subsequently adapted to further culture in StemFlex media (Gibco, #A3349401). hiPSCs at ∼70% confluence (1.5 × 106 cells/well of a 6-well plate) were incubated in EDTA (Life Technologies, #15575-020) for 4 min at room temperature (RT), the EDTA was aspirated, and the cells were dissociated in fresh StemFlex media and seeded onto Matrigel-coated plates at 3–5 × 105 cells/well. The media were replaced every other day for 4 days until the next passage. For freezing and shipping to partners, cells were dissociated with EDTA and subsequently frozen in StemFlex medium with 10% DMSO (Sigma, #D2650) at −80°C and transferred to liquid nitrogen for long-term storage. All hiPSCs were tested and are mycoplasma free.
Three Germ-Layer Differentiation
After thawing, cells were passaged 1–2 times in mTeSR1 media before differentiation into the 3 germ layers using STEMdiffTM Definitive Endoderm Kit (STEMCELL, #05110), STEMdiffTM Mesoderm Induction Medium (STEMCELL, #05220), and dual-SMAD [51] inhibition for ectoderm differentiation. hiPSCs were dissociated with Accutase Cell Detachment Solution (Innovative Cell Technologies, #AT-104) for 10 min at 37°C and seeded on Matrigel-coated 24-well plates in mTeSR1 medium with 1 μM H1152 at a density of 2.1 × 105 cells/cm2 for endoderm and ectoderm differentiation and 5 × 104 cells/cm2 for mesoderm differentiation. Endoderm and mesoderm differentiations were performed according to the manufacturer's protocol, whereas ectoderm was induced with neural induction medium: DMEM/F12 (Thermo Fisher, #10565018), 1% N-2 supplement (Thermo Fisher, #17502048), 10 µM SB431542 (Tocris, #1614), and 200 nM LDN193189 (Tocris, #6053) with daily medium changes. Cells were harvested on day 5, and RNA was prepared using Trizol isolation.
Neuron and Glia Generation
NPC Differentiation from hiPSCs
Our neural progenitor cells (NPCs) were generated using PSC Neural Induction Medium (Thermo Fisher) and passaged using Rosette selection reagent (STEMCELL, #05832). To estimate purity, we stained PAX6 (Biolegend, #PRB-278P) and NESTIN (Sigma, #MAB5326).
NGN2-Glutamatergic Neuron Induction from hiPSCs [52]
On day 1, a 6-well plate was coated with Matrigel. On day 0, hiPSCs were dissociated with Accutase Cell Detachment Solution (Innovative Cell Technologies, #AT-104), counted, and transduced with rtTA (Addgene 20342) and NGN2 (Addgene 99378) lentiviruses in StemFlex media containing 10 μM Thiazovivin (Millipore, #S1459). They were subsequently seeded at 1 × 106 cells/well in the prepared 6-well plate. On day 1, the medium was switched to nonviral induction medium (DMEM/F12 [Thermo Fisher, #10565018], 1% N-2 [Thermo Fisher, #17502048], and 2% B27-RA [Thermo Fisher, #12587010]), and doxycycline (dox) was added to each well at a final concentration of 1 μg/mL. At day 2, transduced hiPSCs were treated with 500 μg/mL G418 (Thermo Fisher, #10131035). At day 4, the medium was replaced including 1 μg/mL dox and 4 μM cytosine arabinoside (Ara-C) to reduce the proliferation of nonneuronal cells. On day 5, young neurons were dissociated with Accutase Cell Detachment Solution (Innovative Cell Technologies, #AT-104), counted, and seeded at a density of 1 × 106 per well of a Matrigel-coated 12-well plate. The medium was switched to Brainphys neuron medium (Brainphys [STEMCELL, #05790], 1% N2, 2% B27-RA, 1 μg/mL Natural Mouse Laminin [Thermo Fisher, #23017015], 20 ng/mL BDNF [R&D, #248], 20 ng/mL GDNF [R&D, #212], 250 μg/mL dibutyryl cyclic-AMP [Sigma, #D0627], and 200 mML-ascorbic acid [Sigma, #A4403]). For seeding, 10 μM Thiazovivin (Millipore, #S1459), 500 μg/mL G418, and 4 μM Ara-C and 1 μg/mL dox were added. At day 6, the medium was replaced with Brainphys neuron medium with 4 μM Ara-C and 1 μg/mL dox. Subsequently, 50% of the medium was replaced with fresh neuronal medium (lacking dox and Ara-C) once every other day until the neurons were fixed or harvested at day 21.
ASCL1/DLX2-GABAergic Neuron Induction from hiPSCs [53] and NPCs [54]
GABAergic neurons were generated from NPCs by overexpression of Dlx2 and Ascl1. NPCs were seeded at a density of 5.0 × 105 cells per well of a 24-well tissue plate. At day 1, NPCs were transduced with CMV-rtTA (Addgene ID: 19780), TetO-Ascl1-T2A-Puro (Addgene ID: 97329), and TetO-Dlx2-IRES-Hygro (Addgene ID: 97330), incubated for 15 min at 37°C, and spinfected at 1,000 g for 1 h at RT. At day 0, the media were replaced with NPC media containing 1 μg/mL dox (Sigma, #D9891). At day 1, the NPC media were replaced with NPC media containing 1 μg/mL dox, 1 μg/mL puromycin (Sigma, #P7255), and 250 μg/mL hygromycin (Thermo Fisher, 10687010). On day 7, NPCs were introduced to neuronal media via half-media changes. Cells received 1 μg/mL of dox until DIV 14. To prevent the proliferation of dividing mitotic progenitors, 50 nM cytosineβ-D-arabinofuranoside, also known as Ara-C (Sigma, #C6645), was supplemented in the neuronal media. Neurons were harvested at day 32 for analysis.
ASCL1/NURR1/LMX1a-Dopaminergic Neuron Induction from hiPSCs, Adapted and Modified from [55, 56, 57]
On day 1, a 6-well plate was coated with Matrigel. On day 0, hiPSCs were dissociated with Accutase Cell Detachment Solution (Innovative Cell Technologies, #AT-104), counted, and transduced with rtTA (Addgene 20342), ASCL1, NURR1, and LMX1 (Addgene, #97329) lentiviruses. hiPSCs were mixed in a conical tube in low-volume StemFlex media containing 10 μM Thiazovivin (Millipore, #S1459) and subsequently seeded at 1 × 106 cells/well in the prepared 6-well plate. At day 1, the medium was switched to nonviral induction medium (DMEM/F12 [Thermo Fisher, #10565018], 1% N-2 [Thermo Fisher, #17502048], and 2% B27-RA [Thermo Fisher, #12587010]) and 1 μg/mL dox. On day 2, the medium was replaced with induction media with1 μg/mL dox and 1 μg/mL puromycin. At day 3, the medium was replaced with induction media with1 μg/mL dox and 2 μg/mL puromycin. On day 5, the medium was replaced with induction media with 1 μg/mL dox and 1 μg/mL puromycin and 2 μM Ara-C. On day 6, antibiotic selection was withdrawn. On day 7, young neurons were dissociated with Accutase Cell Detachment Solution (Innovative Cell Technologies, #AT-104) and seeded at a density of 0.57 mio/cm2 in Brainphys neuron medium (Brainphys [STEMCELL, #05790], 1% N2, 2% B27-RA, 1 μg/mL Natural Mouse Laminin [Thermo Fisher, #23017015], 20 ng/mL BDNF [R&D, #248], 20 ng/mL GDNF [R&D, #212], 250 μg/mL dibutyryl cyclic-AMP [Sigma, #D0627], and 200 mML-ascorbic acid [Sigma, #A4403]). For accelerated maturation, part of the neurons were seeded on a layer of human astrocytes. In this case, the medium was supplemented with 2% FBS (Thermo Fisher, #10082147). Subsequently, 50% of the medium was replaced with fresh neuronal medium once every other day until the neurons were fixed or harvested on day 35. Dox and Ara-C were withdrawn on day 14.
NFIB/SOX9-Astrocyte Induction from hiPSCs [58]
On day 0, hiPSCs were dissociated with Accutase Cell Detachment Solution (Innovative Cell Technologies, #AT-104), counted, and transduced with rtTA (Addgene 20342) and inducible Nfib (Addgene, #117271) and Sox9 (Addgene, #117269) lentiviruses. hiPSCs and viruses were mixed in a conical tube in low-volume StemFlex media containing 10 μM Thiazovivin (Millipore, #S1459) and subsequently seeded at 0.75 × 106 cells/well in the prepared 6-well plate. Expression was induced on day 1 in StemFlex media with 2.5 μg/mL dox (continued throughout). On day 2, the medium was switched to expansion medium (DMEM/F-12, 10% FBS, and 1% N2 supplement) with 1 μg/mL puromycin and 400 μg/mL hygromycin for selection. On days 4–6, expansion medium was gradually switched to FGF medium (Neurobasal, 2% B27 supplement, 1% NEAA, 1% Glutamax, and 1% FBS, 8 ng/mL FGF2, 5 ng/mL CNTF, and 10 ng/mL BMP4, from Peprotech). On day 7, cells were dissociated with Accutase Cell Detachment Solution and replated in Matrigel-coated wells. On days 8 and 9, FGF medium was gradually switched to maturation medium (1:1 DMEM/F-12 and Neurobasal, 1% N2, 1% sodium pyruvate, and 0.5% Glutamax; 5 mg/mL N-acetyl-cysteine, 250 mg/mL dbcAMP; 5 ng/mL heparin-binding EGF-like growth factor, 10 ng/mL CNTF [Peprotech, #450-13], and 10 ng/mL BMP4 [Peprotech, #120-05]). Subsequently, 50% of the medium was replaced with maturation medium every 2–3 days.
hiPSC CRISPR Transfection and Validation
hiPSC Transfection JHU
Firstly, hiPSCs were seeded at a concentration of 250,000 cells/well of a 6-well plate and when 30% confluent, typically on the third day, subjected to lipofectamine transfection. The transfection medium was prepared by mixing 2 solutions. The first consisted of 2 μL/well pmaxGFP DNA (LONZA, 1 μg/μL) diluted in 250 μL/well Opti-MEM® medium (Thermo Fisher Scientific) to obtain 500 ng DNA/well concentration. The second is 10 μL/well LipofectamineTM STEM Transfection Reagent (Thermo Fisher Scientific) diluted in 250 μL/well Opti-MEM® medium. The transfection medium consists of a 1:1 ratio of the first (pmaxGFP DNA and Opti-MEM®) and second (LipofectamineTM and Opti-MEM®) solutions that were incubated for 10 min before addition to the cells. Meanwhile, the cell medium was removed and replaced with 2 mL/well mixture of ROCK inhibitor and StemFlexTM medium (Thermo Fisher Scientific) at 1:1,000 ratio. After incubation, 500 μL/well transfection medium was added to the cells with the ROCK inhibitor and StemFlexTM and then incubated at 37°C for 4 h. Afterward, 2 mL/well of StemFlexTM medium was added to the mix and incubated at 37°C overnight. After about 24 h, the media were changed and replaced with 2 mL/well of StemFlexTM medium and then incubated at 37°C overnight. After about 48 h, the cells can be washed, lifted, and analyzed by fluorescence-activated cell sorting (FACS). Note that controls were prepared following a similar procedure, but instead of pmaxGFP DNA, the same volume of water was added.
hiPSC Transfection UC
All iPSC lines were passaged according to manufacturer's protocols. In brief, 50,000 cells were plated on Matrigel (Corning, #354234) coated 24-well plates with mTeSR1 (STEMCELL, #85850) media. Twenty-four hours after plating cells, each well was treated with 1.2 μL Lipofectamine STEM reagent (Thermo Fisher, #STEM00001) and 500 ng pmaxGFP (Lonza, PBP3-00675) combined into 50 µL Opti-MEM media following Lipofectamine STEM reagent protocol. The cells were overlaid with fresh mTeSR1 the following day. Forty-eight hours after transfection, the cells were washed with 1× PBS and treated with Accutase (STEMCELL, cat#07922) for 10 min followed by 200 g centrifugation. The supernatant was removed and cells resuspended in fresh mTeSR media. The cell suspension was pipetted through Falcon tubes with cell-strainer caps and then analyzed by MACSQuant Analyzer 10 Flow Cytometer (Miltenyi Biotec) for 10,000 events.
hiPSC Nucleofection ISMMS
hiPSCs were transfected using the Lonza P3 Primary Cell 4D-Nucleofector Kit (V4XP-3024) according to the manufacturer's instructions. In brief, cells were dissociated following 15-min Accutase Cell Detachment Solution (Innovative Cell Technologies, #AT-104) incubation at 37°C, and 1 × 106 cells were centrifuged at 800 g for 5 min. Pellet was resuspended in 100 μL nucleofector solution containing 800 ng GFP plasmid. The suspension was quickly transferred to a nucleofection cuvette, transfected in the Lonza 4D nucleofector program “CA-137,” and seeded onto a Matrigel-covered 6-well plate in StemFlex containing 1 μM H1152 (Millipore, #555550). On the next day, the medium was replaced. Two days after nucleofection, cells were dissociated using Accutase, and stained with LIVE/DEADTM Fixable Far Red Dead Cell Stain Kit (Thermo Fisher, #L34973). The cells were washed with PBS, filtered using FACS tubes with cell-strainer caps, and subsequently analyzed using the BD FACSCantoTM II counting 10,000 events.
hiPSC SNP rs4702 Editing Using Cas9 Protein
hiPSCs were transfected with 10 μg TrueCutTM Cas9 Protein v2 (Thermo Fisher, #A36498), 8 μg synthetic gRNAs (Synthego): FURIN rs4702 A-G conversion: GGCTGGTTTTGTAAGATACT; FURIN rs4702 G-A conversion: GGCTGGTTTTGTAAGATGCT, and 100 pmol of the respective repair ssODNs (Thermo Fisher): rs4702 G->A: TZFFATAGAACCAGCAATGCTGGGCCTGTTTAAATTACAAGAAAAAAATCACTGTGCACCAACCCAGZATCTTACAAAACCAGCCGGGCTGGCCAOEA; rs4702 A->G: TZFFATAGAACCAGCAATGCTGGGCCTGTTTAAATTACAAGAAAAAAATCACTGTGCACCAACCCAGOATCTTACAAAACCAGCCGGGCTGGCCAOEA using the Lonza P3 Primary Cell 4D-Nucleofector Kit (V4XP-3024). In brief, cells were dissociated following 15-min Accutase Cell Detachment Solution (Innovative Cell Technologies, #AT-104) incubation at 37°C, and 1 × 106 cells were centrifuged at 800 g for 5 min. Pellet was resuspended in 100 μL nucleofector solution and subsequently mixed with the assembled RNP (ribonucleoprotein complex) and the repair Oligo. The suspension was quickly transferred to a nucleofection cuvette, transfected in the Lonza 4D nucleofector program “CA-137,” and seeded onto a Matrigel-covered 6-well plate in 3 mL StemFlex containing 10 μM Thiazovivin (Millipore, #S1459). The cells were subjected to a 48-h cold shock at 32°C to enhance homology directed repair. One day after nucleofection, the medium was replaced by fresh StemFlex medium without Thiazovivin. The medium was changed every other day, until 70–80% confluence. Cells were dissociated with Accutase, the majority of cells were frozen, ∼1/5 was lysed for analysis, and 1,500 cells were seeded onto a 10-cm dish containing 1 million mouse embryonic fibroblasts and cultured in StemFlex media for clonal expansion. The media were replaced every other day for 7–10 days until colonies were well visible. Colonies were then picked, split in several pieces, and half was transferred into a well of 2 matched Matrigel-coated 96-well plates for maintenance and analysis, respectively.
Analysis of rs4702 Genomic Locus
The rs4702 region was amplified using AmpliTaq GoldTM 360 Master Mix (Thermo Fisher, #4398901) and primers against the rs4702 region (f: GGAATAGTTGAGCCCCAAGTCC, r: TGACTTGGGCCCACATCCAG). PCR conditions were as follows: 95°C for 10 min followed by 35 cycles (95°C for 30 s, 60°C for 30 s, and 72°C for 60 s) and 72°C for 7 min. The restriction enzymes SfaN1 (NEB; cuts specifically rs4702 G) and Bsr1 (NEB; cuts specifically rs4702 A) were used for further analysis of the PCR product of the bulk edit and the picked clones. After identification of potential successfully edited clones, the sequence was confirmed by Sanger using the forward primer.
hiPSC SNP Editing Efficiency Evaluation by ABEmax System
For testing bulk DNA base editing efficiency, we used a customized pβactin-ABEmax-puro vector system for which we cloned the dCas9-adenine base editor part from the ABEmax vector (Addgene, #112095) [59] into the pSpCas9(BB)-2A-Puro (PX459) V2.0 vector (Addgene, #62988) [60]. We edited a common SNP rs7148456 that showed allele-specific open chromatin in BAG5[61] in 2 hiPSC lines: CW70372 (low PRS) and CW30525 (high PRS). Cells for editing were plated on 4-well dishes (Thermo Scientific) coated with Matrigel (Corning). iPSCs were cultured with mTeSR plus (STEMCELL) and passaged with ReLeSR (STEMCELL). All cells were incubated, maintained, and cultured at 37°C with 5% CO2. Once hiPSCs reached ∼70% confluency, they were treated with 1.5 μL Lipofectamine STEM (Thermo Fisher Scientific) according to manufacturer's protocols and 750 ng ABEmax BAG5 targeting plasmid. Twenty-four hours after transfection, cells were treated with 0.5 μg/mL puromycin (InvivoGen). Forty-eight hours after transfection, cells were treated with 0.25 μg/mL puromycin. Seventy-two hours after transfection, cells were washed with DPBS (Thermo Fisher Scientific) and detached from plates with Accutase (STEMCELL). QuickEx (Lucigen) was used for rapidly extracting genomic DNAs from the transfected cells according to manufacturer's protocol. The extracted DNAs were used as a template for PCR, followed by Sanger sequencing. Primers used for genomic DNA amplification were as follows: BAG5_rs7148456_onTar_L: TTCCCCTCCCCACCCTTTTA and BAG5_rs7148456_onTar_R: CGCTCAGACCTAGTCGGGA.
Molecular Analysis
Real-Time Quantitative PCR
Cells were harvested with Trizol, and total RNA extraction was carried out following the manufacturer's instructions. Quantitative transcript analysis was performed using a QuantStudio 7 Flex Real-Time PCR System with the Power SYBR Green RNA-to-Ct Real-Time qPCR Kit (all Thermo Fisher). Total RNA template (25 ng per reaction) was added to the PCR mix, including primers. qPCR conditions were as follows: 48°C for 15 min and 95°C for 10 min followed by 45 cycles (95°C for 15 s and 60°C for 60 s). All qPCR data are collected from at least 3 independent biological replicates of one experiment. Data analyses were performed using GraphPad PRISM 8 software.
Primers used were as follows: 18S (f: ACACGGACAGGATTGACAGA, r: NANOG [f: CAGCTGTGTGTACTCAATGATAGATTTC, r: GGACATCTAAGGGCATCACAG]; GGAGAATTTGGCTGGAACTGCATG); PAX6 (f: GGAGTGAATCAGCTCGGTGG, r: GGTCTGCCCGTTCAACATCC); LMX1A (f: TCCAGGTGTGGTTCCAAAAC, r: GGTTCATGATTCCTTCCATCCC); HAND1 (f: AAAGGCTCAGGACCCAAGAAG, r: TGATCTTGGAGAGCTTGGTGTC); HOPX (f: CGAGGAGGAGACCCAGAAATG, r: GACGGATCTGCACTCTGAGG); FOXP2 (f: CAGTCACCCCGATTACCCAG, r: GGGGCAATTTCTGATGACATGG); SOX17 (f: GAACGCTTTCATGGTGTGGG, r: CACGACTTGCCCAGCATCT); GAD2 (Thermo Fisher, Hs00609534_m1); and vGAT (Thermo Fisher, Hs00369773_m1).
Immunostaining and Microscopy
Cells were washed with ice-cold PBS and fixed with 4% PFA/sucrose PBS solution at pH 7.4 for 20 min, RT. Then, fixative solution was replaced with permeabilizing solution (0.1% Triton-X PBS) for 20 min. After washed with PBS, cells were incubated with blocking solution (5% donkey serum in PBS) for 1 h, at RT. The blocking solution was aspirated and replaced with the same solution with primary antibodies (MAP2-Ck, Abcam ab5392, 1:500; Nanog-Gt, R&D AF1997, 1:200; TRA-1-60 IgM-Ms, Millipore MAB4360, 1:100; GFAP-Ck Aves AB_2313547, 1:1,000; MAP2AB-Ms, Sigma M1406, 1:500; synapsin1-Ms Synaptic Systems, 1:500; TH-Rb, Pel-Freez P40101, 1:1,000; S100beta-Ms, Sigma S2532, 1:1,000; HuNu, Sigma MAB1281B, 1:200; MAP2, Synaptic Systems 188 003, 188 011, 1:700; and GABA, 1:1,000, Sigma A2052) overnight at 4°C. Cells were then incubated with secondary antibodies (Alexa 488 anti-Rabbit, Dk, Jackson ImmunoResearch, 711-545-152, 1:500; Alexa 568 anti-Chicken, Gt, Thermo Fisher, A-11041, 1:500; and Alexa 647 anti-mouse, Dk, Jackson ImmunoResearch, 715-605-150, 1:500), prepared in blocking solution, for 2 h at RT, followed by DAPI staining for 3–4 min and PBS washing 3 times.
Cells were imaged with a Zeiss LSM 780, Nikon C2 confocal microscope, or a Thermo Fisher HCS CX7 microscope. The images were analyzed using ImageJ, except astrocytes, which were quantified using the CellProfiler software. Data points represent 10–33 images from 2 biological replicates.
Results
Identification of hiPSCs Derived from Donors with Extreme High and Low PRS for SZ
The California Institute of Regenerative Medicine (CIRM) hiPSC collection is one of the largest US-based single-deriver collections of genotyped hiPSCs (1,618 donors), generated by a standardized, nonintegrating episomal reprogramming approach in a single production facility (http://catalog.coriell.org/CIRM). Genome-wide SNP genotype data (from SNP microarray Illumina Infinium HumanCore BeadChip) were used to derive the SZ PRS using Psychiatric Genomics Consortium (PGC) 2 GWAS summary statistics [2] for all the non-SZ donors. We identified high and low PRS hiPSC lines with an adjusted PRS >2 standard deviations and therefore with an expected >10-fold relative risk for disease [2]. After excluding hiPSCs with potential sample swaps, gender discrepancies, and structural variants, this totaled 28 hiPSCs derived from peripheral blood mononuclear cells from unrelated donors of European descent. From these, we prioritized hiPSCs from 12 unrelated high (3 males and 3 females) and low (3 males and 3 females) PRS donors (shaded, Table 1) for a collective trisite evaluation (Icahn School of Medicine at Mount Sinai, University of Chicago, and Johns Hopkins University) to assess their suitability for neural induction and CRISPR editing.
Table 1.
Selection of high and low SZ PRS hiPSC lines (bold) from extreme genotypes
| Donor ID | Sex | Description | Age (at biopsy) | PRS |
|---|---|---|---|---|
| CW30274 | M | No psychiatric diagnosis | 63 | 2.98 |
| CW30525 | F | No psychiatrie diagnosis | 56 | 2.81 |
| CW20184 | M | ASD | 24 | 2.65 |
| CW30421 | F | No psychiatric diagnosis | 66 | 2.63 |
| CW40201 | F | No psychiatric diagnosis | 76 | 2.43 |
| CW30454 | M | No psychiatric diagnosis | 67 | 2.42 |
| CW20041 | F | No psychiatric diagnosis | 9 | 2.27 |
| CW30265 | F | No psychiatrie diagnosis | 55 | 2.26 |
| CW20195 | F | ASD | 24 | 2.12 |
| CW30350 | F | No psychiatric diagnosis | 12 | 2.09 |
| CW60130 | F | No psychiatric diagnosis | 6 | 2.09 |
| CW70016 | F | No psychiatric diagnosis | 49 | 2.06 |
| CW70191 | F | No psychiatric diagnosis | 61 | 2.05 |
| CW50094 | M | No psychiatric diagnosis | 70 | −2.06 |
| CW50106 | F | No psychiatric diagnosis | 67 | −2.11 |
| CW40187 | F | No psychiatric diagnosis | 71 | −2.14 |
| CW30484 | F | No psychiatric diagnosis | 23 | −2.14 |
| CW70004 | M | No psychiatric diagnosis | 65 | −2.21 |
| CW70280 | M | No psychiatric diagnosis | 59 | −2.23 |
| CW70164 | F | No psychiatric diagnosis | 83 | −2.32 |
| CW30108 | M | No psychiatric diagnosis | 53 | −2.32 |
| CW70372 | M | No psychiatric diagnosis | 54 | −2.37 |
| CW50101 | F | No psychiatric diagnosis | 78 | −2.39 |
| CW40067 | F | No psychiatric diagnosis | 76 | −2.70 |
| CW70179 | F | No psychiatric diagnosis | 86 | −2.88 |
| CW30190 | M | No psychiatric diagnosis | 63 | −2.91 |
| CW70142 | F | No psychiatric diagnosis | 82 | −3.20 |
| CW30154 | F | No psychiatric diagnosis | 66 | −3.46 |
SZ, schizophrenia; PRS, polygenic risk scores; hiPSC, human induced pluripotent stem cell; ASD, autism spectrum disorder.
Validation of Pluripotency and Genomic Integrity of Extreme PRS hiPSCs
hiPSCs were rigorously validated by the CIRM IPSC repository for genomic integrity, pluripotency using a 48-gene classifier, and loss of the reprogramming transgenes (see the section Materials and Methods). All 12 extreme PRS hiPSC lines were subsequently re-evaluated by our 3 laboratories to confirm pluripotency and genomic integrity.
Two hiPSC lines demonstrated abnormal growth at 2 or more sites: low_PRS_4 showed low postthaw viability and attachment and high_PRS_3 showed low adhesion and spontaneous differentiation (Table 2). We confirmed pluripotency marker expression by immunohistochemistry (NANOG and TRA-1-60, Fig. 2a). The differentiation potential was evaluated by rapid 5-day trilineage-directed differentiation [62], followed by qPCR against early lineage-specific transcripts (ectoderm: PAX6 and LMX1A; mesoderm: HOPX; ectoderm: FOXP2 and SOX17; and pluripotency: NANOG) (Fig. 2b).
Table 2.
Summary of pluripotency, identity confirmation (>99% concordance with the original CIRM data), genomic aberrations, neural induction, transfection efficiency, and CRISPR editing across 12 high and low PRS hiPSCs
| Study ID | CIRM donor ID | Cell growth | Pluripotency | Identity confirmation | Genome integrity | Neural induction | Transfection efficiency, % | Suitable for editing | CRISPR edited |
|---|---|---|---|---|---|---|---|---|---|
| high_PRS_1 | CW30274 | ✓ | ✓ | ✓ | ✓ | ✓ | 27 | ✓ | ✓ |
| high_PRS_2 | CW30525 | ✓ | ✓ | ✓ | ✓ | ✓ | 32 | ✓ | ✓ |
| high_PRS_3 | CW20184 | Abnormal | ✓ | nt | nt | Low GABA yield | 16 | No | |
| high_PRS_4 | CW30421 | ✓ | ✓ | ✓ | ✓ | ✓ | 38 | ✓ | ✓ |
| high_PRS_5 | CW40201 | ✓ | ✓ | ✓ | ✓ | ✓ | 26 | ✓ | ✓ |
| high_PRS_6 | CW30454 | ✓ | ✓ | ✓ | ✓ | ✓ | 40 | ✓ | |
| low_PRS_1 | CW30108 | ✓ | Low mesoderm | ✓ | >1 ×107 bp del | ✓ | 33 | No | |
| low_PRS_2 | CW70372 | ✓ | ✓ | ✓ | ✓ | ✓ | 30 | ✓ | ✓ |
| low_PRS_3 | CW70179 | ✓ | ✓ | ✓ | ✓ | ✓ | 28 | ✓ | ✓ |
| low_PRS_4 | CW30190 | Abnormal | ✓ | ✓ | >1 ×107 bp del | Low GABA yield | 25 | No | |
| low_PRS_5 | CW70142 | ✓ | ✓ | ✓ | ✓ | ✓ | 26 | ✓ | ✓ |
| low_PRS_6 | CW30154 | ✓ | ✓ | ✓ | ✓ | ✓ | 27 | ✓ |
PRS, polygenic risk scores; hiPSC, human induced pluripotent stem cell; nt, not tested: high_PRS_3 was not fully evaluated owing to growth abnormalities.
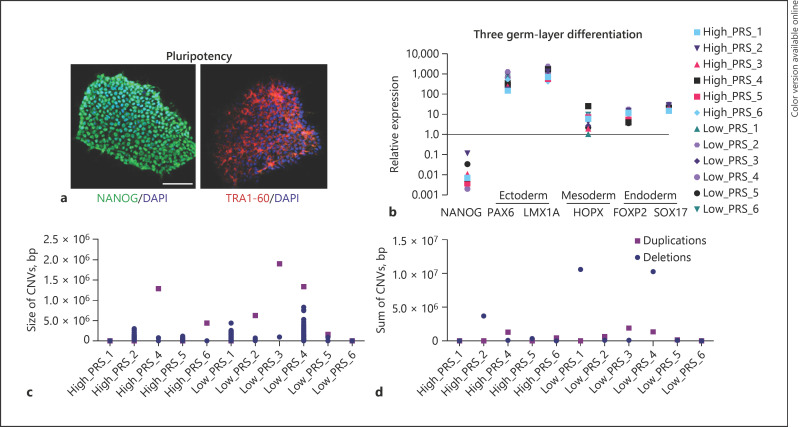
Fig. 2.
Validation, identity confirmation, and CNV analysis of extreme PRS hiPSC lines. a Representative immunofluorescence staining against the pluripotent markers NANOG and TRA-1-60. Scale bar, 100 µm. b Directed 3 germ layer differentiation of extreme PRS hiPSC lines for 5 days with subsequent qPCR analysis against typical lineage markers, normalized to hiPSC control and 18S. c Identification of CNVs depicted by size. d Sum of deletions and duplications present in the selected hiPSC lines after expansion. CNV, copy number variation; PRS, polygenic risk scores; hiPSC, human induced pluripotent stem cell.
hiPSCs were subjected to genotyping on the Multi-EthnicGlobal_D1 chip (Illumina Inc.) to confirm donor identity against genotype data provided by the CIRM repository. The GenomeStudio CNV Region Report Plug-in v2.1.2 evaluated genome integrity and confirmed the absence of gross karyotypic abnormalities: high_PRS_4, low_PRS_3, and low_PRS_4 showed duplications >1.0 × 106 bp, but more critically, high_PRS_4 exhibited an additional number of deletions >5.0 × 105 bp, adding up to >1.0 × 107 bp (Fig. 2c–e). Overall, 10 of 12 extreme PRS hiPSCs showed robust proliferation and demonstrated pluripotency and genome integrity indicating overall suitability for functional genomic studies (Table 2).
Demonstration of Neuronal and Glial Generation
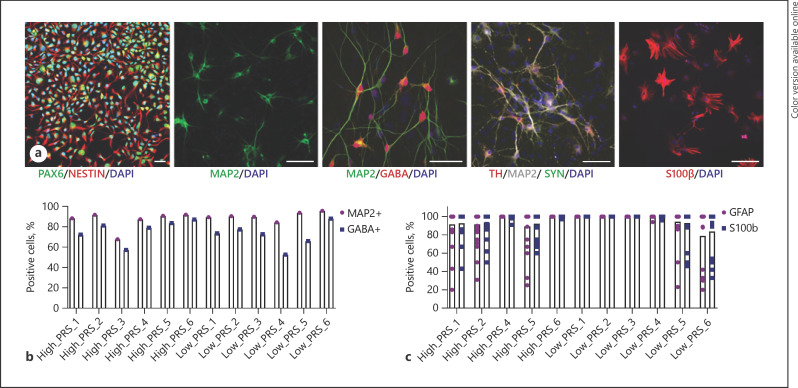
To ensure that the hiPSCs can robustly generate the neural subtypes most commonly linked to SZ (glutamatergic neurons [63], GABAergic neurons [63], dopaminergic neurons [64], and astrocytes [65]), each laboratory independently evaluated neural differentiation and induction using different methodologies already well established at each site [23, 30, 54, 66, 67]. Forebrain NPCs were generated from all 12 lines [68]; immunofluorescence staining of PAX6 and NESTIN exceeded 95% in 10 out of 12 lines, with NPCs from 2 donors showing impurities (low_PRS_1 and low_PRS_4) (Fig. 3a). Glutamatergic neuron induction by overexpression of NGN2 in hiPSCs [52] resulted in robust expression of glutamatergic genes by 21 days after induction (Fig. 3a). GABAergic neuronal induction via overexpression of DLX2 and ASCL1 was efficient from both hiPSCs [53] and NPCs [54] (Fig. 3a). Thirty-two days after induction, NPC-induced GABAergic neurons expressed GABAergic neuronal genes glutamate decarboxylase 2 (GAD2) and vesicular GABA transporter (vGAT) by qPCR (online suppl. Fig. 1; see www.karger.com/doi/10.1159/000512716 for all online suppl. material). GABAergic neurons were stained with HuNu, MAP2, and GABA, demonstrating ∼80% yield for 9 out of 12 lines (exceptions included 65% for low_PRS_5 and ∼50% for high_PRS_3 and low_PRS_4) (Fig. 3a, b). Those hiPSCs with reduced propensity for neuronal induction also had abnormalities in hiPSC culture and CNV analysis, suggesting that hiPSCs with irregular growth may be more likely to show defective differential potential as well. Dopaminergic neurons, a third neuronal cell type implicated in SZ, were achieved by a novel lentiviral induction strategy: overexpression of ASCL1, NURR1, and LMX1A from hiPSCs [55, 56, 57]. We derived dopaminergic neurons coexpressing tyrosine hydroxylase and synapsin after 5 weeks of induction when cocultured with astrocytes for 6 of 12 hiPSC lines (Fig. 3a); the variability between hiPSC lines and induction batches was high, so we recommend that dopaminergic neuron differentiation be individually optimized for the remaining hiPSC lines. We queried expression of the 67 non-MHC SZ risk genes associated by prediXcan [69] with the SZ-PGC2 loci [2, 70], finding that 62.6, 79.1, and 76.1% are expressed at >0.5 log2RPKM, respectively, in our existing NGN2-glutamatergic, ASCl1/DLX2-GABAergic, and ASCL1, NURR1, and LMX1A-dopaminergic neuron RNAseq datasets [54].
Fig. 3.
Demonstration of robust neuronal and glial induction. a Representative images of astrocyte and neuron (glutamatergic, GABAergic, and dopaminergic) populations. Scale bar, 50 µm. b Quantification of MAP2- and GABA-positive cells after 32 days of GABAergic neuron maturation. c Quantification of GFAP- and S100β-positive cells after 21 days of astrocyte maturation.
Lentiviral cotransduction of dox-inducible SOX9 and NFIB yields astrocytes [58]. We have demonstrated high efficiency induction of astrocytes from the PRS hiPSCs, with 10 out of 11 hiPSCs yielding population of >90% GFAP- and S100β-positive cells by day 21, with only low_PRS_6 showing a slightly lower efficiency (∼80%) (Fig. 3a, c). Overall, all 9 of 10 extreme hiPSCs with robust proliferation demonstrated pluripotency and genome integrity and showed strong capacity to generate neurons and glia through standard methods (Table 2).
Transfection Efficiency
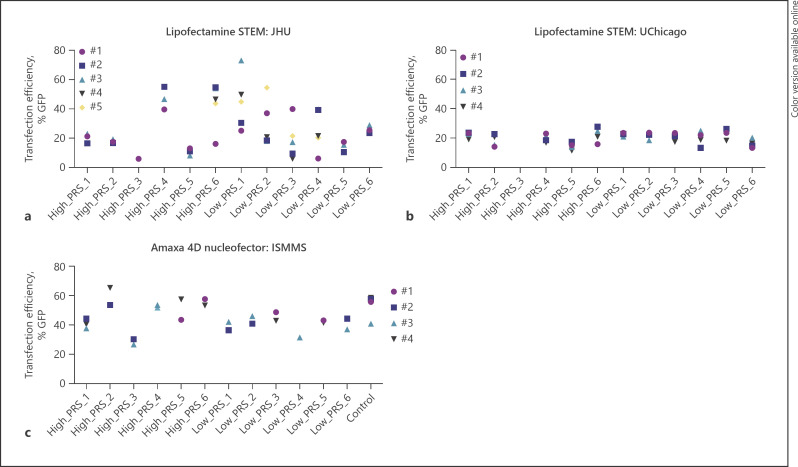
An important perquisite for CRISPR editing is the ability to introduce Cas9, template DNA, and gRNAs. hiPSC lines are commonly known to be difficult to genetically manipulate; optimization of transfection conditions and enriching for transfected cells are critical for efficiently recovering modified clones [71]. To assess the likely efficiency of each hiPSC line for subsequent CRISPR engineering, we tested the efficiency of 2 commonly used methods to introduce DNA across our laboratories: transfection and nucleofection. We evaluated Lipofectamine STEM transfection and Amaxa nucleofection approaches with a GFP reporter followed by FACS analysis (Fig. 4). Lipofectamine methods yielded highly variable efficiencies that differed between sites (JHU: 6–73% [Fig. 4a] and UChicago: 16–23% [Fig. 4b]). Nucleofection was generally more efficient: 36–65% (Fig. 4c). Across all methods and sites, the 2 hiPSCs without lowest transfection efficiencies (high_PRS_3 and low_PRS_4) consistently showed the poorest culture and neuronal generation as well. Overall, from 9 of 9 extreme hiPSCs with robust proliferation, demonstrating pluripotency, genome integrity, and neuronal/glia induction, we observed transfection efficiencies sufficient to achieve efficient CRISPR/Cas9 SNP editing (Table 2).
Fig. 4.
Evaluation of transfection efficiency. Percentage GFP-positive cells using Lipofectamine STEM and the Lonza 4D nucleofector across 3 different sites: a Transfection with Lipofectamine STEM at JHU, b Transfection with Lipofectamine STEM at UChicago, c Transfection with Amaxa 4D nucleofector at ISMMS.
Appropriateness for CRISPR-Editing
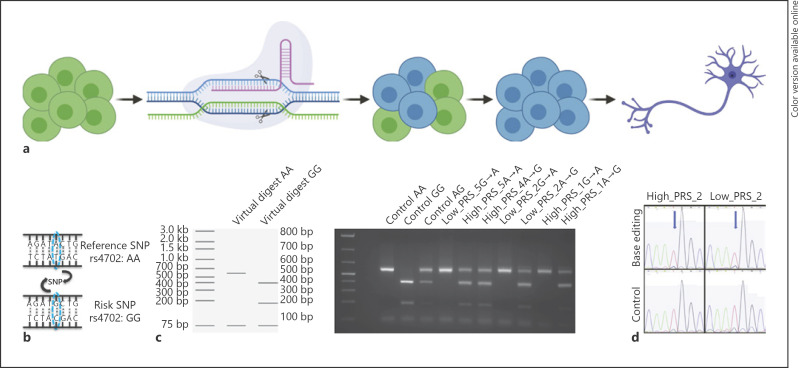
To evaluate the suitability for CRISPR/Cas9 editing, we applied 2 independent methodologies (Cas9-based editing [60] and dCas9-based DNA base editing [72]) against 2 independent noncoding SNPs already edited in our laboratories. First, we retargeted the SZ risk SNP rs4702 within the 3′ region of the gene FURIN[46]. hiPSC lines were nucleofected with a Cas9 protein/gRNA complex and a respective repair oligonucleotide for either the wild-type allele A or the risk allele G (Fig. 5a, b). Bulk analysis of editing efficiency was performed by restriction enzyme digest, revealing efficient editing in both directions (whether starting from either heterozygous alleles (low_PRS_5, low_PRS_2, and high_PRS_1) or homozygous AA alleles (high_PRS_4 and high_PRS_5)). Correctly edited clones were identified from 5 of 5 hiPSC lines evaluated, with efficiencies of 21.7, 25, 52.2, 50, 27.8, 26, and 37.5%, respectively (Fig. 5c).
Fig. 5.
Demonstration of CRISPR editing of SZ risk SNPs. a Overview of editing pipeline, using CRISPR/Cas9 and a repair oligonucleotide to achieve homologous recombination (HDR). Figure created with BioRender.com. b Example of SZ GWAS-SNP rs4702, with reference allele A and risk allele G. c Bulk editing analysis of FURIN rs4702 using the restriction enzyme SfANI, cutting the A allele once and the G allele twice. d Sanger sequencing results from base editing of 2 extreme PRS hiPSC lines. Top row: BAG5 rs7148456 edited (T to C), with blue arrow pointing to SNP; bottom row: unedited homozygous controls. Note the small peak of the edited C allele that was absent in the unedited controls and a much lower peak of T allele in the edited sample versus unedited controls. SZ, schizophrenia; SNPs, single nucleotide polymorphisms; GWAS, genome-wide association studies; HDR, homology directed repair.
Second, we applied dCas9 to retarget the SZ risk SNP rs7148456 associated with allele-specific open chromatin at the BAG5 locus [61]. In 2 hiPSC lines, we transfected a customized ABEmax vector system [72] to express gRNA, dCas9, and ABEmax in a single vector. This DNA base editor is expected to have higher editing efficiency than homologous-directed repair in CRISPR/cas9 SNP editing, with minimal DNA off-target editing because the deactivated Cas9 does not create double-strand DNA breaks [72]. hiPSCs were transfected using Lipofectamine STEM reagent. After puromycin screening, bulk DNAs were extracted approximately 72 h after transfection and sequenced. We approximated the editing efficiencies (A to G) based on A to G (or T to C on the minus strand) percentage by Sanger sequencing, which showed an 11.2 and 13.8% editing efficiency for high_PRS_2 and low_PRS_2, respectively (Fig. 5d). Overall, while we believe that all 9 validated extreme PRS hiPSC (5 high and 4 low) should be suitable for CRISPR-editing, for 7, we have empirically demonstrated efficient allelic conversion of a single noncoding SNP (Table 2).
Discussion
A major challenge for the field of psychiatric genetics is translating highly complex genetic insights into medically actionable information. The integration of hiPSCs and CRISPR editing is becoming the preferred means to test the function of common and rare disease-associated variants. Many studies now follow a design whereby CRISPR-edited isogenic lines are contrasted; however, there is tremendous variability in the efficiency with which different hiPSC lines respond to CRISPR editing and neuron differentiation [73]. Towards this, we conducted a collaborative and systematic trisite validation of the pluripotency, genomic integrity, propensity for neuronal differentiation, and CRISPR-editing capabilities of 6 high and 6 low PRS control hiPSCs, prioritizing 9 lines suitable for functional validation of SZ variants. All hiPSC lines were grown, induced to neural fates, and transfected at all 3 sites; moreover, for CRISPR editing, the editability and efficiency were tested for multiple lines by different labs using different SNP editing approaches (Table 3). Thus, we expect that other laboratories will find them straightforward to adapt for CRISPR-based studies of psychiatric disease.
Table 3.
Summary of site at which pluripotency, identity confirmation, genomic aberrations, neural induction, transfection efficiency, and CRISPR editing were evaluated across 12 high and low PRS hiPSCs
| Study ID high_PRS_1 | CIRM donor ID CW30274 | Cell growth ISMMS, JHU, UC | Pluripotency ISMMS | Identity confirmation JHU | Genome integrity JHU | Neural induction ISMMS, JHU, UC | Transfection efficiency ISMMS, JHU, UC | CRISPR edited ISMMS |
|---|---|---|---|---|---|---|---|---|
| high_PRS_2 | CW30525 | ISMMS, JHU, UC | ISMMS | JHU | JHU | ISMMS, JHU, UC | ISMMS, JHU, UC | UC |
| high_PRS_3 | CW20184 | ISMMS, JHU, UC | ISMMS | nt | nt | UC | ISMMS, UC | |
| high_PRS_4 | CW30421 | ISMMS, JHU, UC | ISMMS | JHU | JHU | ISMMS, JHU, UC | ISMMS, JHU, UC | ISMMS |
| high_PRS_5 | CW40201 | ISMMS, JHU, UC | ISMMS | JHU | JHU | ISMMS, JHU, UC | ISMMS, JHU, UC | ISMMS |
| high_PRS_6 | CW30454 | ISMMS, JHU, UC | ISMMS | JHU | JHU | ISMMS, JHU, UC | ISMMS, JHU, UC | |
| low_PRS_1 | CW30108 | ISMMS, JHU, UC | ISMMS | JHU | JHU | ISMMS, JHU, UC | ISMMS, JHU, UC | |
| low_PRS_2 | CW70372 | ISMMS, JHU, UC | ISMMS | JHU | JHU | ISMMS, JHU, UC | ISMMS, JHU, UC | ISMMS, UC |
| low_PRS_3 | CW70179 | ISMMS, JHU, UC | ISMMS | JHU | JHU | ISMMS, JHU, UC | ISMMS, JHU, UC | ISMMS |
| low_PRS_4 | CW30190 | ISMMS, JHU, UC | ISMMS | JHU | JHU | ISMMS, JHU, UC | ISMMS, JHU, UC | |
| low_PRS_5 | CW70142 | ISMMS, JHU, UC | ISMMS | JHU | JHU | ISMMS, JHU, UC | ISMMS, JHU, UC | ISMMS |
| low_PRS_6 | CW30154 | ISMMS, JHU, UC | ISMMS | JHU | JHU | ISMMS, JHU, UC | ISMMS, JHU, UC | ISMMS |
PRS, polygenic risk scores; hiPSC, human induced pluripotent stem cell; nt, not tested.
This publicly available resource of well-characterized extreme PRS hiPSC lines will facilitate the study of SZ risk in a well-defined cohort that is comparable across different sites. High and low PRS lines can be used to address the question of penetrance when modeling rare or common variants, to examine the potential impact of genetic background on phenotypic expressivity in hiPSC-derived cells. Using this well-characterized cohort will reduce the time and resources spent validating CRISPR-editing conditions across hiPSCs of lesser quality or with less available genetic information. Eventually, if adopted across additional psychiatric genetics laboratories, we will improve the reproducibility of hiPSC-based studies between research groups and increase the power of isogenic CRISPR-based experiments by facilitating meta-analyses with an ever-growing dataset of isogenic hiPSCs. Most importantly, this resource will facilitate the future integration of datasets generated across laboratories. Such meta-analyses identify convergent effects between SZ-GWAS risk variants.
We note 3 limitations to this resource. First, while we expect that the concept of shared extreme PRS hiPSCs will prove to be of enduring value, this specific hiPSC cohort might have to be redefined as PRS calculations improve (by expanded PGC3-GWAS cohort size [74] and/or incorporation of pathway-specific information [75]), which could necessitate the inclusion of additional extreme PRS hiPSCs. Second, being derived entirely of donors of European descent, this cohort is not well suited to test transancestry effects. Third, and most importantly, this collection is unlikely to prove to be adequately powered to resolve sex-specific and/or high versus low PRS effects and is mostly intended for the validation of isogenic functional genomic studies across a diverse collection of donor backgrounds. Of course, experimentally deconvolving the extent to which individual risk factors exert variable effects across extreme PRS donor backgrounds will help to resolve the extent to which polygenic risk reflects additive [26, 76] or more complex epistatic [28] or omnigenic models [29] of inheritance and will in turn shape future refinements of PRS.
In a broader context, the limitation of using only European ancestry-based PRS in our study reflects an inherent limitation for the genetic research field in the usefulness of PRS in diverse populations. There are important scientific, ethical, social, and legal imperatives to ensure genetic findings have the broadest relevance to patients worldwide. The historical lack of diversity in medical genetic research, particularly the overrepresentation of European ancestry samples, limits our ability to fine-map disease-associated variants and accurately estimate PRS across diverse populations [77]. The predictive performance of European ancestry-derived PRS is lower in non-European ancestry samples [78]. Scores constructed using a transancestry meta-analysis markedly improved predictive value for participants of both European and African descent [79]. These findings highlight the need for considering population-specific linkage disequilibrium architecture and variant frequencies when applying PRS to cohorts of non-European ancestry and bolster the rationale for future effort of expanding our resource to include more highly and low SZ PRS samples of diverse ethnicity.
In summary, we describe a publicly available hiPSC collection from donors with extreme PRS for SZ and characterize the suitability to test functional effects of SZ risk variants in a context-dependent manner. As a shared resource, these hiPSC lines will enable the cross-lab reproducibility evaluation and data integration. However, given the polygenic nature of SZ and other neuropsychiatric disorders, we acknowledge that a larger number of iPSC lines of extreme SZ PRS would be needed for delineating the effect of polygenic risk background on disease modeling. Nonetheless, we anticipate that a similar strategy will prove broadly useful across many complex genetic disorders, although of course, each disorder will require identification and validation of a unique set of extreme PRS hiPSCs.
Statement of Ethics
All hiPSC lines were obtained from the CIRM hiPSC Repository funded by the California Institute of Regenerative Medicine (CIRM). Ethical approval was not required because the hiPSC lines, having been obtained from a public repository, are thus not considered to be human subject research.
Conflict of Interest Statement
The authors declare no conflicts of interest.
Funding Sources
This work was supported by a supplement to the National Institute of Health (NIH) grant R01MH113215 (D.A.), R56 MH101454 (K.J.B.), R01MH106575 and R01MH116281 (J.D.), R01MH106056 (K.J.B. and S.A.), and U01 MH115727 (S.M. and K.E.).
Author Contributions
K.J.B., J.D., and D.A. conceptualized this collaborative approach. K.J.B., J.D., D.A., K.R., H.Z., and S.A. contributed to experimental design and wrote the manuscript. S.G., K.E., and S.M. identified the high and low PRS hiPSC lines in the CIRM hiPSC repository. K.R., H.Z., T.P., S.W., D.D., and S.A. conducted all pluripotency experiments, transfections, neuron production, neuronal differentiation, and CRISPR-editing experiments. D.A. conducted all genomic and CNV analyses on the hiPSC lines.
Availability of Data and Material
All source donor hiPSCs are available from the CIRM repository (https://www.cirm.ca.gov/researchers/ipsc-repository). Genotype data will be made publicly available prior to publication.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgements
Special thanks are due to Dr. David Panchision at NIMH for program guidance on this collaborative project. Portions of Figures 1 and 5 were created with BioRender.com.
References
- 1.Sullivan PF, Geschwind DH. Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell. 2019;177((1)):162–83. doi: 10.1016/j.cell.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardinas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50((3)):381–89. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh T, Kurki MI, Curtis D, Purcell SM, Crooks L, McRae J, et al. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci. 2016;19((4)):571–7. doi: 10.1038/nn.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinberg S, Gudmundsdottir S, Sveinbjornsson G, Suvisaari J, Paunio T, Torniainen-Holm M, et al. Truncating mutations in RBM12 are associated with psychosis. Nat Genet. 2017;49((8)):1251–4. doi: 10.1038/ng.3894. [DOI] [PubMed] [Google Scholar]
- 6.Schizophrenia Working Group of the Psychiatric Genomics C. Ripke S, Neale BM, Corvin A, Walters JTR, Farh KH, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511((7510)):421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee PH, Anttila V, Won H, Feng Y-CA, Rosenthal J, Zhu Z, et al. Genome wide meta-analysis identifies genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. bioRxiv. 2019:528117. doi: 10.1016/j.cell.2019.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brainstorm C, Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018:360. doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149((3)):525–37. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loohuis LM, Vorstman JA, Ori AP, Staats KA, Wang T, Richards AL, et al. Genome-wide burden of deleterious coding variants increased in schizophrenia. Nat Commun. 2015;6:7501. doi: 10.1038/ncomms8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154((3)):518–29. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schork AJ, Won H, Appadurai V, Nudel R, Gandal M, Delaneau O, et al. A genome-wide association study of shared risk across psychiatric disorders implicates gene regulation during fetal neurodevelopment. Nat Neurosci. 2019;22((3)):353–61. doi: 10.1038/s41593-018-0320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180((3)):568–e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87((6)):1215–33. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506((7487)):185–90. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506((7487)):179–84. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Network Pathway Analysis Subgroup of the Psychiatric Genomics C, International Inflammatory Bowel Disease Genetics C, International Inflammatory Bowel Disease Genetics Consortium I Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337((6099)):1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaffe AE, Straub RE, Shin JH, Tao R, Gao Y, Collado-Torres L, et al. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat Neurosci. 2018;21((8)):1117–25. doi: 10.1038/s41593-018-0197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19((11)):1442–53. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobbyn A, Huckins LM, Boocock J, Sloofman LG, Glicksberg BS, Giambartolomei C, et al. Landscape of conditional eQTL in dorsolateral prefrontal cortex and co-localization with schizophrenia GWAS. Am J Hum Genet. 2018;102((6)):1169–84. doi: 10.1016/j.ajhg.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Girdhar K, Hoffman GE, Jiang Y, Brown L, Kundakovic M, Hauberg ME, et al. Cell-specific histone modification maps in the human frontal lobe link schizophrenia risk to the neuronal epigenome. Nat Neurosci. 2018;21((8)):1126–36. doi: 10.1038/s41593-018-0187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forrest MP, Zhang H, Moy W, McGowan H, Leites C, Dionisio LE, et al. Open chromatin profiling in hiPSC-derived neurons prioritizes functional noncoding psychiatric risk variants and highlights neurodevelopmental loci. Cell Stem Cell. 2017;21((3)):305. doi: 10.1016/j.stem.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sey NYA, Hu B, Mah W, Fauni H, McAfee JC, Rajarajan P, et al. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nature Neuro. 2020;23((4)):583–93. doi: 10.1038/s41593-020-0603-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajarajan P, Borrman T, Liao W, Schrode N, Flaherty E, Casiño C, et al. Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science. 2018;362((6420)):362. doi: 10.1126/science.aat4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wray NR, Wijmenga C, Sullivan PF, Yang J, Visscher PM. Common disease is more complex than implied by the core gene omnigenic model. Cell. 2018;173((7)):1573–80. doi: 10.1016/j.cell.2018.05.051. [DOI] [PubMed] [Google Scholar]
- 27.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169((7)):1177–86. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang G, Wang W, Paunic V, Heydari H, Costanzo M, Liu X, et al. Discovering genetic interactions bridging pathways in genome-wide association studies. Nat Commun. 2019;10((1)):4274. doi: 10.1038/s41467-019-12131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Li YI, Pritchard JK. Trans effects on gene expression can drive omnigenic inheritance. Cell. 2019;177((4)):1022–e6. doi: 10.1016/j.cell.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schrode N, Ho SM, Yamamuro K, Dobbyn A, Huckins L, Matos MR, et al. Synergistic effects of common schizophrenia risk variants. Nat Genet. 2019;51((10)):1475–85. doi: 10.1038/s41588-019-0497-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry. 2015;20((3)):361–8. doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109((31)):12770–5. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015;12:671–8. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165((5)):1238–54. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, Vogt D, et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013;12((5)):573–86. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffman GE, Hartley BJ, Flaherty E, Ladran I, Gochman P, Ruderfer DM, et al. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat Commun. 2017;8((1)):2225. doi: 10.1038/s41467-017-02330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570((7762)):523–7. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545((7652)):54–9. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim KY, Sun P, et al. hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell. 2019;24((3)):487–e7. doi: 10.1016/j.stem.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat Methods. 2017;14((7)):743–51. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carcamo-Orive I, Hoffman GE, Cundiff P, Beckmann ND, D'Souza SL, Knowles JW, et al. Analysis of transcriptional variability in a large human iPSC library reveals genetic and non-genetic determinants of heterogeneity. Cell Stem Cell. 2017;20((4)):518–e9. doi: 10.1016/j.stem.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546((7658)):370–5. doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffman GE, Schrode N, Flaherty E, Brennand KJ. New considerations for hiPSC-based models of neuropsychiatric disorders. Mol Psychiatry. 2019;24((1)):49–66. doi: 10.1038/s41380-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16((5)):299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajarajan P, Flaherty E, Akbarian S, Brennand KJ. CRISPR-based functional evaluation of schizophrenia risk variants. Schizophr Res. 2020;217:26–36. doi: 10.1016/j.schres.2019.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schrode N, Ho SM, Yamamuro K, Dobbyn A, Huckins L, Matos MR, et al. Synergistic effects of common schizophrenia risk variants. Nat Genet. 2019;51((10)):1475–85. doi: 10.1038/s41588-019-0497-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pham X, Song G, Lao S, Goff L, Zhu H, Valle D, et al. The DPYSL2 gene connects mTOR and schizophrenia. Transl Psychiatry. 2016;6((11)):e933. doi: 10.1038/tp.2016.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515((7527)):414–8. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tai DJ, Ragavendran A, Manavalan P, Stortchevoi A, Seabra CM, Erdin S, et al. Engineering microdeletions and microduplications by targeting segmental duplications with CRISPR. Nat Neurosci. 2016;19((3)):517–22. doi: 10.1038/nn.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim MS, Patel KP, Teng AK, Berens AJ, Lachance J. Genetic disease risks can be misestimated across global populations. Genome Biol. 2018;19((1)):179. doi: 10.1186/s13059-018-1561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chambers SM, Mica Y, Lee G, Studer L, Tomishima MJ. Dual-SMAD inhibition/WNT activation-based methods to induce neural crest and derivatives from human pluripotent stem cells. Methods Mol Biol. 2016;1307:329–43. doi: 10.1007/7651_2013_59. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78((5)):785–98. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang N, Chanda S, Marro S, Ng YH, Janas JA, Haag D, et al. Generation of pure GABAergic neurons by transcription factor programming. Nat Methods. 2017;14((6)):621. doi: 10.1038/nmeth.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barretto N, Zhang H, Powell SK, Fernando MB, Zhang S, Flaherty EK, et al. ASCL1- and DLX2-induced GABAergic neurons from hiPSC-derived NPCs. J Neurosci Methods. 2020;334:108548. doi: 10.1016/j.jneumeth.2019.108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caiazzo M, Dell'Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476((7359)):224–7. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 56.Kim J, Su SC, Wang H, Cheng AW, Cassady JP, Lodato MA, et al. Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell. 2011;9((5)):413. doi: 10.1016/j.stem.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Addis RC, Hsu FC, Wright RL, Dichter MA, Coulter DA, Gearhart JD. Efficient conversion of astrocytes to functional midbrain dopaminergic neurons using a single polycistronic vector. PLoS One. 2011;6((12)):e28719. doi: 10.1371/journal.pone.0028719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Canals I, Ginisty A, Quist E, Timmerman R, Fritze J, Miskinyte G, et al. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat Methods. 2018;15((9)):693–6. doi: 10.1038/s41592-018-0103-2. [DOI] [PubMed] [Google Scholar]
- 59.Koblan LW, Doman JL, Wilson C, Levy JM, Tay T, Newby GA, et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol. 2018;36((9)):843–6. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8((11)):2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang S, Zhang H, Zhou Y, Qiao M, Zhao S, Kozlova A, et al. Allele-specific open chromatin in human iPSC neurons elucidates functional disease variants. Science. 2020;369((6503)):561–5. doi: 10.1126/science.aay3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rehbach K, Kesavan J, Hauser S, Ritzenhofen S, Jungverdorben J, Schüle R, et al. Multiparametric rapid screening of neuronal process pathology for drug target identification in HSP patient-specific neurons. Sci Rep. 2019;9((1)):9615. doi: 10.1038/s41598-019-45246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skene NG, Bryois J, Bakken TE, Breen G, Crowley JJ, Gaspar H, et al. Genetic identification of brain cell types underlying schizophrenia. bioRxiv. 2017;50((6)):825–33. doi: 10.1038/s41588-018-0129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dong X, Liao Z, Gritsch D, Hadzhiev Y, Bai Y, Locascio JJ, et al. Enhancers active in dopamine neurons are a primary link between genetic variation and neuropsychiatric disease. Nat Neurosci. 2018;21((10)):1482–92. doi: 10.1038/s41593-018-0223-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Birnbaum R, Jaffe AE, Chen Q, Shin JH, BrainSeq C, Kleinman JE, et al. Investigating the neuroimmunogenic architecture of schizophrenia. Mol Psychiatry. 2018;23((5)):1251–60. doi: 10.1038/mp.2017.89. [DOI] [PubMed] [Google Scholar]
- 66.Zhang S, Zhang H, Qiao M, Zhou Y, Zhao S, Kozlova A, et al. Allele-specific open chromatin in human iPSC neurons elucidates functional non-coding disease variants. bioRxiv. 2019:827048. doi: 10.1126/science.aay3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doostparast Torshizi A, Armoskus C, Zhang H, Forrest MP, Zhang S, Souaiaia T, et al. Deconvolution of transcriptional networks identifies TCF4 as a master regulator in schizophrenia. Sci Adv. 2019;5((9)):eaau4139. doi: 10.1126/sciadv.aau4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan Y, Shin S, Jha BS, Liu Q, Sheng J, Li F, et al. Efficient and rapid derivation of primitive neural stem cells and generation of brain subtype neurons from human pluripotent stem cells. Stem Cells Transl Med. 2013;2((11)):862–70. doi: 10.5966/sctm.2013-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huckins LM, Dobbyn A, Ruderfer DM, Hoffman G, Wang W, Pardiñas AF, et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat Genet. 2019;51((4)):659–74. doi: 10.1038/s41588-019-0364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schizophrenia Working Group of the Psychiatric Genomics C Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511((7510)):421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li K, Wang G, Andersen T, Zhou P, Pu WT. Optimization of genome engineering approaches with the CRISPR/Cas9 system. PLoS One. 2014;9((8)):e105779. doi: 10.1371/journal.pone.0105779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang TP, Zhao KT, Miller SM, Gaudelli NM, Oakes BL, Fellmann C, et al. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat Biotechnol. 2019;37((6)):626–31. doi: 10.1038/s41587-019-0134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Volpato V, Smith J, Sandor C, Ried JS, Baud A, Handel A, et al. Reproducibility of molecular phenotypes after long-term differentiation to human iPSC-derived neurons: a multi-site omics study. Stem Cell Reports. 2018;11((4)):897–911. doi: 10.1016/j.stemcr.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Borglum AD, Breen G, et al. Psychiatric genomics C: psychiatric genomics: an update and an agenda. Am J Psychiatry. 2018;175:15–27. doi: 10.1176/appi.ajp.2017.17030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi SW, O'Reilly PF. PRSice-2: polygenic risk score software for biobank-scale data. Gigascience. 2019;8((7)):giz082. doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wainschtein P, Jain DP, Yengo L, Zheng Z, Cupples LA, Shadyab AH, et al. Recovery of trait heritability from whole genome sequence data. bioRxiv. 2019:588020. [Google Scholar]
- 77.Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100((4)):635–49. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duncan L, Shen H, Gelaye B, Meijsen J, Ressler K, Feldman M, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun. 2019;10((1)):3328. doi: 10.1038/s41467-019-11112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bigdeli TB, Genovese G, Georgakopoulos P, Meyers JL, Peterson RE, Iyegbe CO, et al. Contributions of common genetic variants to risk of schizophrenia among individuals of African and Latino ancestry. Mol Psychiatry. 2020;25((10)):2455–67. doi: 10.1038/s41380-019-0517-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Data Availability Statement
All source donor hiPSCs are available from the CIRM repository (https://www.cirm.ca.gov/researchers/ipsc-repository). Genotype data will be made publicly available prior to publication.