Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can cause acute respiratory disease and multiorgan failure. Finding human host factors that are essential for SARS-CoV-2 infection could facilitate the formulation of treatment strategies. Using a human kidney cell line—HK-2—that is highly susceptible to SARS-CoV-2, we performed a genome-wide RNAi screen and identified virus dependency factors (VDFs), which play regulatory roles in biological pathways linked to clinical manifestations of SARS-CoV-2 infection. We found a role for a secretory form of SARS-CoV-2 receptor, soluble angiotensin converting enzyme 2 (sACE2), in SARS-CoV-2 infection. Further investigation revealed that SARS-CoV-2 exploits receptor-mediated endocytosis through interaction between its spike with sACE2 or sACE2-vasopressin via AT1 or AVPR1B, respectively. Our identification of VDFs and the regulatory effect of sACE2 on SARS-CoV-2 infection shed insight into pathogenesis and cell entry mechanisms of SARS-CoV-2 as well as potential treatment strategies for COVID-19.
Keywords: SARS-CoV-2, RNAi, ACE2, sACE2, COVID-19, virus dependency factor, vasopressin
Graphical abstract
Using an infection-permissive human kidney cell line, Yeung et al. show that a soluble form of ACE2 that is cleaved and liberated from the host cell surface mediates SARS-CoV-2 binding and uptake by receptors involved in renin-angiotensin system signaling.
Introduction
Coronavirus disease 2019 (COVID-19) caused by a novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a pandemic with >109 million confirmed cases and approximately 2.4 million fatalities (https://covid19.who.int/). SARS-CoV-2 can cause substantial pulmonary diseases. Besides, numerous extrapulmonary manifestations such as cardiac and renal complications have been reported to be associated with an increased risk of death in patients with COVID-19 (Gupta et al., 2020; Li et al., 2020).
Host factors required for SARS-CoV-2 infection are largely unknown due to the lack of highly susceptible human cell lines for studying SARS-CoV-2 infection. As of now, a few studies have attempted to identify host factors required for SARS-CoV-2 infection either using non-human cells or modified human cells—that may not truly reflect the authentic infection process. These include studies using the CRISPR screening approach to identify host factors in SARS-CoV-2-infected Vero-E6 cells, which are of non-human origin (Wei et al., 2020), or in modified human cell lines—A549 and Huh7.5.1—transduced with SARS-CoV-2 entry factors to become susceptible to SARS-CoV-2 infection (Daniloski et al., 2020; Heaton et al., 2020; Schneider et al., 2021; Wang et al., 2021). In addition, another study reported the use of a focused CRISPR screening to target a small number of host factors, which were recently identified to be involved in SARS-CoV-2 infection (Hoffmann et al., 2021). Therefore, there remains a pressing need to identify host factors essential for SARS-CoV-2 infection, which may uncover potential therapeutic targets for SARS-CoV-2 and provide insights into COVID-19 pathogenesis.
SARS-CoV-2 utilizes a cellular receptor, angiotensin converting enzyme II (ACE2), for cell entry (Zhou et al., 2020). Besides being a virus receptor, ACE2 functions as an important regulator of the renin-angiotensin system (RAS) (Gheblawi et al., 2020). To serve its regulatory role in the RAS, cellular ACE2 (cACE2) first needs to be transported to the cell surface, where it is cleaved by host proteases such as disintegrin and metalloproteinase 17 (ADAM17), to release an enzymatically active soluble form of ACE2 (sACE2) into the plasma (Lambert et al., 2005). The sACE2 retains an intact SARS-CoV-2 interaction site, suggesting its ability to bind to SARS-CoV-2. Interestingly, a recent study has observed that ACE2 shedding may be induced by regulatory pathways influenced by COVID-19 infection and that the concentrations of sACE2 may correlate with the level of systemic inflammation that occurs (Kornilov et al., 2020). However, most of the current research focus on studying the function of cACE2 in SARS-CoV-2 pathogenesis; the impact of circulating sACE2 on viral entry is largely unknown.
Tissue tropism of SARS-CoV-2 cannot be fully explained only by the expression pattern of cACE2. While a majority of cell susceptibility studies are based solely on the mRNA level of cACE2 (Sungnak et al., 2020), studies on its protein expression pattern is limited (Bertram et al., 2012; Hamming et al., 2004). Recently, Wang et al. (2020) comprehensively investigated the tropism of SARS-CoV-2 in human tissues and discovered discordance between mRNA and protein expression levels of ACE2 in many tissues (Wang et al., 2020). Indeed, studies of cACE2 mRNA and protein expression levels showed that only a small population of the lung cells has a detectable expression level of cACE2—that is in contrast to the common knowledge that the lungs are the primary site of infection (Wang et al., 2020; Zou et al., 2020). Potentially, circulating sACE2, which retains the interaction site for binding to SARS-CoV-2, could interact with SARS-CoV-2 in the extracellular compartment forming complexes, which may affect the infectivity.
In this study, we report the identification of a human renal cell line—HK-2—which is highly susceptible to SARS-CoV-2. Using HK-2 cells, we performed a genome-wide RNAi screening and successfully identified virus dependency factors (VDFs), the expression of which is required for SARS-CoV-2 infection. Detailed analysis of these VDFs suggested that perturbation of their functions could be linked to clinical symptoms and complications of COVID-19. Finally, we elucidated the important but overlooked role of sACE2 in SARS-CoV-2 infection using multiple methods, thus advancing our understanding of pathogenesis and treatment strategies for COVID-19.
Results
Identification of a human cell line highly susceptible to SARS-CoV-2 infection
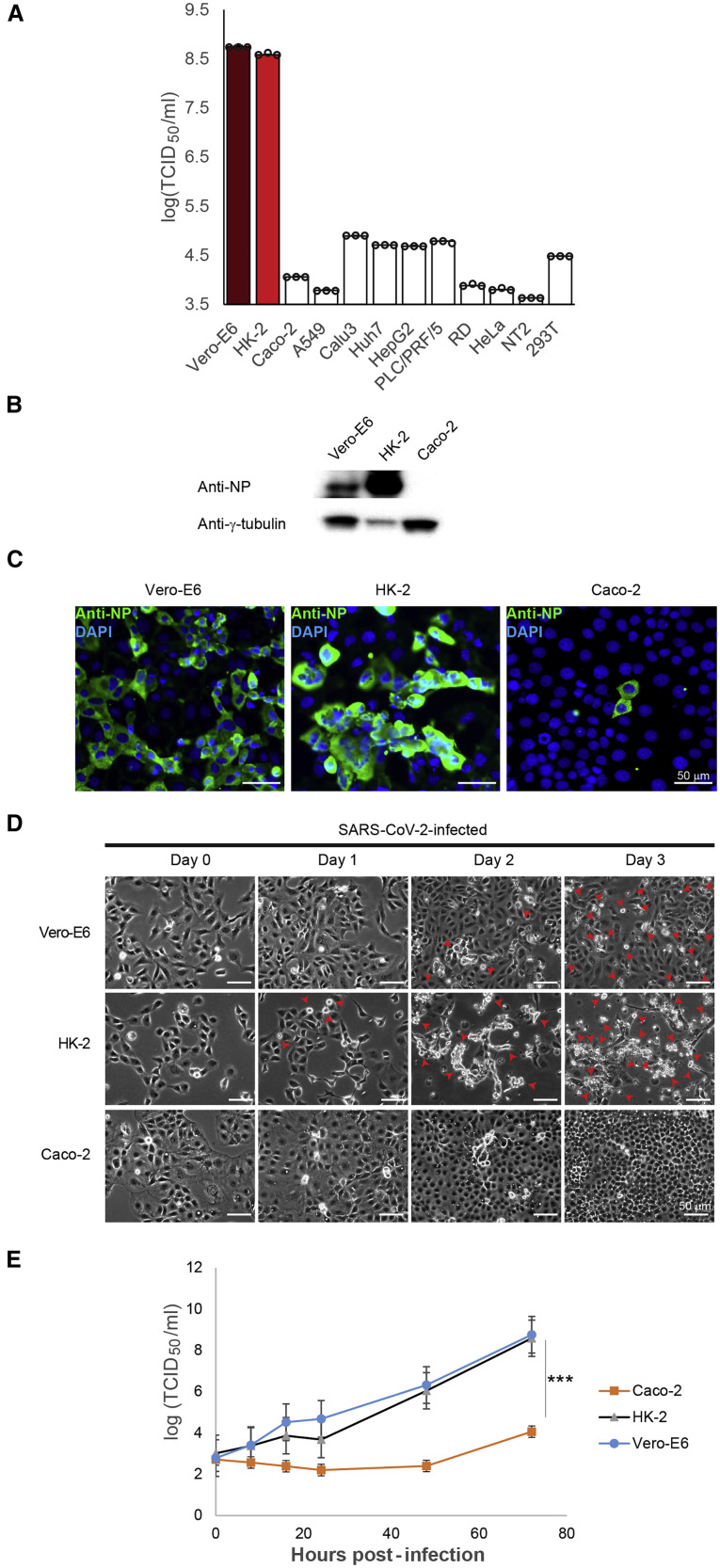
To find an optimal human cell line that is highly susceptible to SARS-CoV-2 infection, we examined 11 human cell lines of different organs, including HK-2, Caco-2, A549, Calu3, Huh7, HepG2, PLC/PRF/5, RD, HeLa, NT2, and 293T, along with Vero-E6 by inoculating the virus at a multiplicity of infection (MOI) of 1. At 72 h post-infection, median tissue culture infective dose (TCID50) results showed that Vero-E6 cells strongly supported SARS-CoV-2 replication with high viral titer of 5.7 × 108 TCID50/mL (Figure 1 A). Much lower viral titers, ranging from 4.2 × 103 to 7.9 × 104 TCID50/mL, were detected in most human cell lines (Figure 1A). Intriguingly, SARS-CoV-2 can replicate efficiently in human renal proximal tubule cells (HK-2) with high viral titers of up to 3.8 × 108 TCID50/mL (Figure 1A). Among these cell lines are Vero-E6 and Caco-2, representing unmodified cell lines of non-human and human origins, respectively, which are known to be susceptible to SARS-CoV-2 (Bojkova et al., 2020). Therefore, effective replication of SARS-CoV-2 in HK-2 and Vero-E6 cells was further confirmed by western blotting (WB) and immunofluorescence assay (IFA), which detected strong viral protein expression in the infected cells (Figures 1B and 1C). In comparison, WB analysis was unable to detect nucleocapsid protein (NP) expression in Caco-2 cells (Figure 1B), which was consistent with the IFA results showing a few positive-stained cells (Figure 1C). Consistent with viral load and the protein expression data (Figures 1A–1C), strong and moderate cytopathic effects (CPEs) were observed in HK-2 and Vero-E6 cells, respectively, but not in Caco-2 cells upon SARS-CoV-2 infection (Figure 1D). SARS-CoV-2 exhibited similar replication kinetics in HK-2 and Vero-E6, whereas in Caco-2 cells, it replicated less efficiently (Figure 1E). Taken together, HK-2 strongly supports the lytic infection of SARS-CoV-2, representing a suitable cell line model of human origin for studying the molecular interactions between SARS-CoV-2 and human host.
Figure 1.
Susceptibilities of human cell lines of different organs to infection by SARS-CoV-2
(A) Human cell lines (HK-2 [red], Caco-2, A549, Calu3, Huh7, HepG2, PLC/PRF/5, RD, HeLa, NT2, and 293T) and Vero-E6 (deep red) were subjected to infection by SARS-CoV-2. Cell lysates were collected and assayed for infectious viral titer.
(B) Western blot (WB) analysis detecting viral antigens using anti-nucleocapsid protein (NP) antibody in SARS-CoV-2-infected Vero-E6, HK-2, and Caco-2 cells.
(C) Immunofluorescence assay (IFA) of Vero-E6, HK-2, and Caco-2 cells infected with SARS-CoV-2 using rabbit antiserum against NP. Scale bars represent 50 μm.
(D) SARS-CoV-2-induced cytopathic effects (CPEs) were monitored in infected Vero-E6, HK-2, and Caco-2 cells. Arrows point to the cells showing CPEs with rounding up of cells progressively detaching from the monolayer.
(E) Virus replication kinetics in infected Vero-E6, HK-2, and Caco-2 were monitored. Cell lysates were harvested from respective time points and viral titers were determined.
The TCID50/mL results from (A) and (E) were derived from three independent experiments. Each data point and error bar depict the mean value and SEM, respectively. Statistical analyses were carried out using Student’s t test. Statistical significance is indicated by the asterisks (∗∗∗p < 0.001). Images from (B–D) are representatives of three independent experiments.
Identification of host factors essential for SARS-CoV-2 replication using a genome-wide RNAi screening
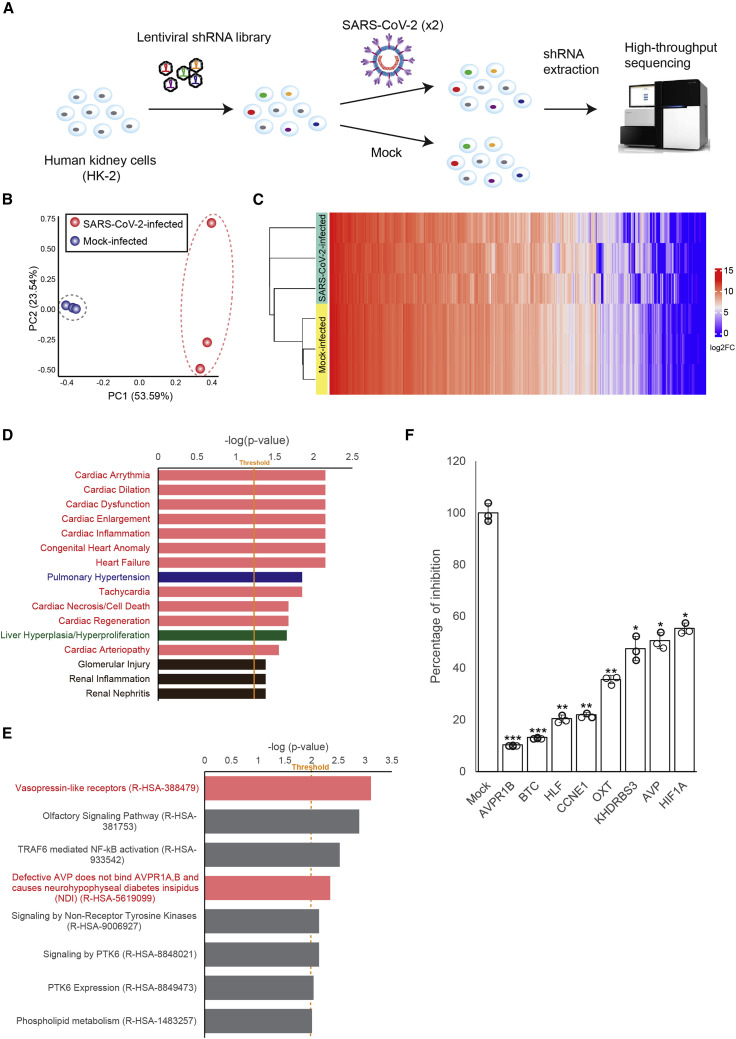
To identify the VDFs essential for SARS-CoV-2 replication, we used a puromycin-marked feline immunodeficiency virus (FIV)-based lenti-vector-short hairpin RNA (shRNA) library to target human transcripts in HK-2 cells followed by the challenge of SARS-CoV-2 (Figures 2 A and S1 ). We anticipated that HK-2 cells that were knocked down for mRNA—which were dispensable for SARS-CoV-2 replication—would succumb rapidly upon infection. In contrast, a shRNA clone silenced an mRNA essential for lytic SARS-CoV-2 replication would survive. The identity of shRNA in the survival cells was determined by high-throughput sequencing (Figure 2A). Principal component analysis (PCA) plot and heatmap based on the abundance of all identified shRNA sequences mapped against the human reference genome revealed good spatial-temporal separation between SARS-CoV-2-infected and mock-infected samples (Figures 2B and 2C). Pathway analysis of VDFs with ≥3-fold enrichment in SARS-CoV-2-infected HK-2 cells revealed 16 significantly affected biological pathways (p < 0.05) related to cardiac (pink), pulmonary (blue), liver (green), and renal (brown) diseases (Figure 2D). Notably, 11 of these 16 biological pathways were related to cardiac diseases (pink; Figure 2D). In addition, PANTHER pathway analysis of the identified VDFs was performed using a cut-off p value of 0.001, and eight major affected molecular pathways were discovered (Figure 2E). Among these, the top-affected molecular pathways “Vasopressin-like receptors (R-HSA-388479),” and “Defective AVP does not bind AVPR1A,B and causes neurohypophyseal diabetes insipidus (NDI) (R-HSA-5619099)” were also involved in the regulation of the cardiovascular system (pink; Figure 2E). Knockdown of individual VDFs involved in the vasopressin-related pathway using corresponding small interfering RNAs (siRNAs) resulted in significant inhibition of SARS-CoV-2 infection (p < 0.01; 45.6%–90.7% of inhibition; Figure 2F). The strong inhibitory effects warranted further study of the VDFs that are functionally related to the vasopressin pathway on SARS-CoV-2 infection.
Figure 2.
Identification of host factors essential for SARS-CoV-2 infection using a genome-wide RNAi screening
(A) Schematic representation of RNAi screening to identify virus dependency factors (VDFs) essential for SARS-CoV-2 infection. The identity of shRNA in the survived and mock-infected control cells were determined via high-throughput sequencing.
(B) Principal component analysis of the shRNA-targeted genes identified in SARS-CoV-2- (red dots; n = 3) and mock-infected (blue dots; n = 3) shRNA-expressing cell clones.
(C) Heatmap analysis of all shRNA-targeted genes identified in SARS-CoV-2- and mock-infected shRNA-expressing cell clones.
(D) Pathway analysis of the identified VDFs essential for SARS-CoV-2 infection. Analysis of VDFs with ≥3-fold enrichment in infected HK-2 cells revealed significantly (p < 0.05) affected biological pathways related to cardiac (pink), pulmonary (blue), liver (green), and renal diseases (brown).
(E) PANTHER pathway analysis of VDFs with ≥3-fold enrichment that are essential for SARS-CoV-2 infection. Significantly affected (p < 0.001) pathways related to the regulation of cardiovascular system (pink) are indicated.
(F) siRNA knockdown of VDFs involved in the vasopressin-related pathway in HK-2 cells leads to an inhibition of SARS-CoV-2 replication as assessed by qRT-PCR. Transfection of non-targeting siRNA was included as the negative control (mock). The results were derived from three independent experiments. Each data point and error bar depict the mean value and SEM, respectively. Statistical analyses were carried out using Student’s t test. Statistical significance is indicated by the asterisks (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
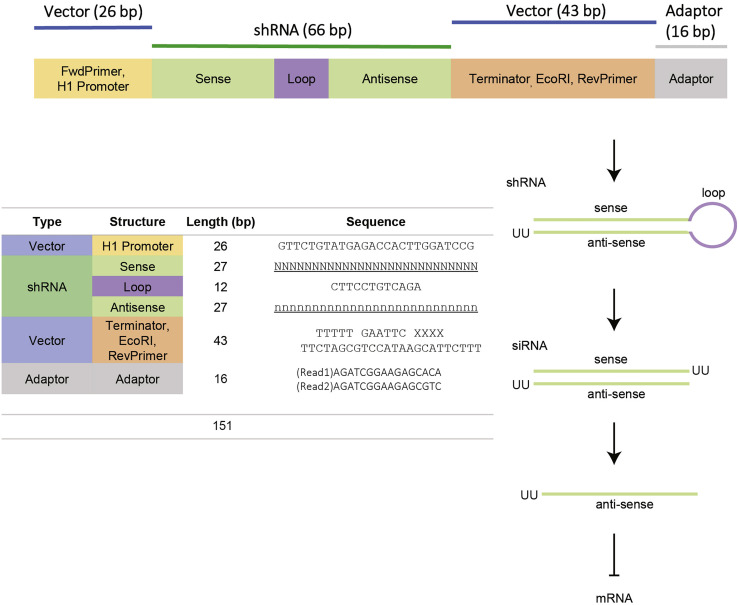
Figure S1.
Design of the shRNA expression cassette, related to Figure 2
VDFs of cytokinesis, vesicle trafficking, and endosomal/lysosomal system play important roles in SARS-CoV-2 infection
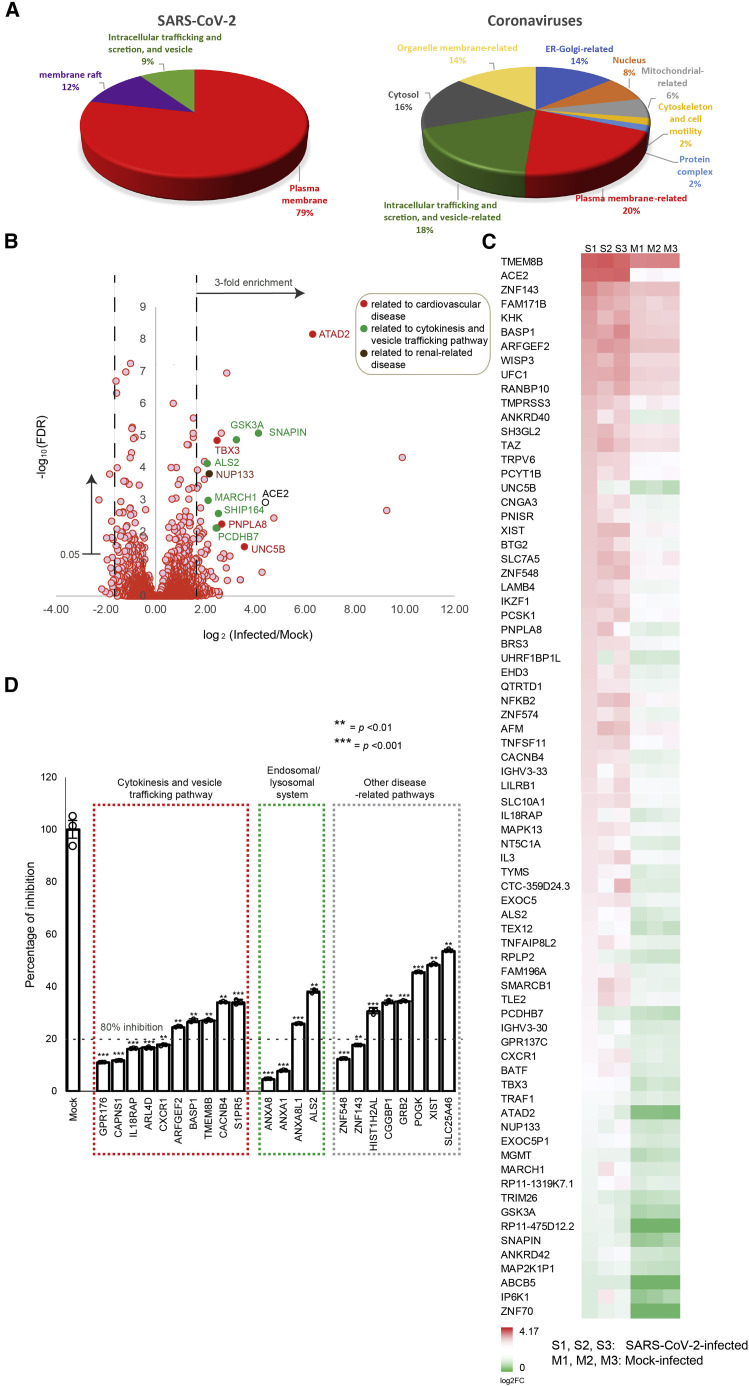
Vasopressin can enter cells via receptor-mediated endocytosis through membrane remodeling and vesicle trafficking (Innamorati et al., 2001). Indeed, our gene ontology (GO) enrichment analysis revealed a close relationship of the VDFs to plasma membrane (79%), membrane raft (12%), as well as intracellular trafficking and secretion, and vesicle (9%), which are known to be important in regulating cytokinesis and vesicle trafficking (Figure 3 A; left). Similarly, we also performed GO enrichment analysis using gene candidates that were previously reported to be important for the replication of other coronaviruses (Ashburner et al., 2000; Bairoch et al., 2005; McKusick, 2007; Oughtred et al., 2019; Pruitt et al., 2005). Consistently, gene candidates related to plasma membrane (20%) as well as intracellular trafficking and secretion, and vesicle (18%) also contributed substantially to the biogenesis of coronaviruses (Figure 3A; right).
Figure 3.
VDFs related to cytokinesis, vesicle trafficking, and endosomal/lysosomal system are important for SARS-CoV-2 infection
(A) Gene ontology (GO) enrichment analysis of VDFs identified in our RNAi screening (left) and that reported by other coronavirus studies (right) using a threshold p value of 0.05. The percentage of each GO term is shown in pie charts.
(B) Volcano plot analysis of VDFs identified in our RNAi screening. Gene annotation revealed that some enriched VDFs ≥3-fold enrichment and false positive rates (FDRs) <5% have functional roles related to the regulation of the cardiovascular system (filled red), cytokinesis and vesicle trafficking pathway (filled green), and renal-related disease (filled brown).
(C) Heatmap analysis of the ≥3-fold enriched VDFs with FDRs < 0.05. S1–3 and M1–3 are biological replicates of SARS-CoV-2- and mock-infected samples, respectively.
(D) siRNA knockdown of selected VDFs in HK-2 cells leads to an inhibition of SARS-CoV-2 replication as assessed by qRT-PCR. Transfection of non-targeting siRNA was included as negative control (mock). Data points below the dotted line indicate siRNA inhibitory effects on SARS-CoV-2 infection to be >80%. The results were derived from three independent experiments. Each data point and error bar depict the mean value and SEM, respectively. Statistical analyses were carried out using Student’s t test. Statistical significance is indicated by the asterisks (∗∗p < 0.01; ∗∗∗p < 0.001).
Next, we performed a volcano plot analysis to identify VDFs that could be involved in SARS-CoV-2 replication (Figure 3B). Manual gene annotation revealed that certain enriched VDFs have functional roles related to the regulation of the cardiovascular system, the cytokinesis and vesicle trafficking pathway, and the renal-related disease (Figure 3B), which was consistent with the results generated from our pathway and GO enrichment analyses (Figures 2D, 2E, and 3A). The differential gene enrichment of the VDFs with a false positive rate (FDR) of <5% is displayed in the heatmap (Figure 3C). Of these, 22 VDFs were selected according to their biological roles for further validation. Consistent with our RNAi screening results, knockdown of individual VDFs using corresponding siRNAs resulted in significant inhibition of SARS-CoV-2 infection (p < 0.01; 46.5%–95.4% of inhibition; Figure 3D). Particularly, potent inhibition (>80%) was observed in some VDFs with biological functions related to the control of cytokinesis and vesicle trafficking pathway (GPR176, CAPNS1, IL18RAP, ARL4D, and CXCR1; red boxed; Figure 3D) and the endosomal/lysosomal system (ANXA8 and ANXA1; green boxed; Figure 3D). Together, our results evidenced that cytokinesis, vesicle trafficking, and the endosomal/lysosomal system are important biological pathways for the replication of SARS-CoV-2.
Vasopressin interacts with sACE2 and spike of SARS-CoV-2
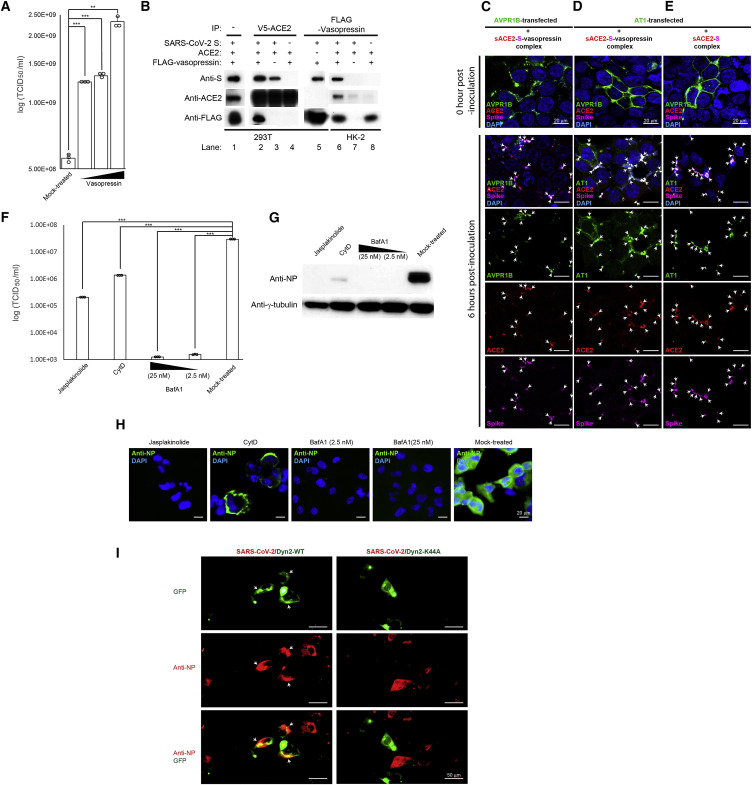
In view of the importance of vasopressin-related VDFs in SARS-CoV-2 infection (Figure 2F), we further tested if SARS-CoV-2 infection depends on vasopressin. Compared to the mock-treated control, HK-2 cells pretreated with increasing doses of vasopressin showed a significant increase in the SARS-CoV-2 infectivity as determined by the TCID50 assays (Figure 4 A). It is known that circulating sACE2 is an important regulator of the RAS, while vasopressin production could also be regulated by the RAS (Reid et al., 1983). Given that sACE2 retains an intact interaction site for binding to SARS-CoV-2, we hypothesized that there may be interaction between vasopressin and sACE2, which could modulate the SARS-CoV-2 infectivity. To examine their interactions, we spiked recombinant spike (S) of SARS-CoV-2 into culture medium of 293T cells, which were doubly transfected with FLAG-tagged vasopressin and V5-tagged ACE2. Secretions of sACE2 and vasopressin were confirmed by WB analyses of the conditioned medium of transfected cells (Figure 4B; lane 1). As expected, pull-down of sACE2 could effectively co-immunoprecipitate the S (Figure 4B; lane 2). Intriguingly, vasopressin was detected in the same co-immunoprecipitation, suggesting that sACE2 could form a complex with S and vasopressin (Figure 4B; lane 2). To further examine if there was any interaction between S and vasopressin, we removed vasopressin from the co-immunoprecipitation experiment. Pull-down of sACE2 was able to co-immunoprecipitate the S (Figure 4B; lane 3), but the amount was lower compared to the reaction containing vasopressin (Figure 4B; lane 1). Remarkably, when S was removed from the co-immunoprecipitation reaction, vasopressin was no longer able to be co-immunoprecipitated by sACE2, suggesting that the S was required for the formation of sACE2-S-vasopressin complex (Figure 4B; lane 4). The interaction between vasopressin and S was supported by successful co-immunoprecipitation of S by pulling down of vasopressin in the absence of ACE2 (Figure 4B; lane 5). The interactions between vasopressin, sACE2, and S of SARS-CoV-2 were further confirmed by reciprocal co-immunoprecipitation using HK-2 cells. It is expected that the HK-2 cells, which are highly susceptible to SARS-CoV-2, could have sufficient endogenous sACE2 for the formation of the sACE2-S-vasopressin complex. Consistent with the co-immunoprecipitation results using 293T cells, pull-down of the vasopressin using supernatants of FLAG-tagged vasopressin transfected HK-2 cells—in the presence of S—was able to co-immunoprecipitate endogenous sACE2 and S (Figure 4B; lane 6). In contrast, sACE2 was unable to be co-immunoprecipitated by vasopressin in the absence of S (Figure 4B; lane 8). Overall, our results supported the possible formation of sACE2-S-vasopressin complex, in which the strength of the sACE2-S interaction could be enhanced in the presence of vasopressin.
Figure 4.
Formation of a protein complex containing sACE2, S of SARS-CoV-2, and/or vasopressin facilitates SARS-CoV-2 cell entry via Dyn2-dependent endocytosis
(A) Effect of vasopressin on SARS-CoV-2 infectivity. The HK-2 cells were pretreated with increasing doses of vasopressin followed by SARS-CoV-2 infection. Mock-treated HK-2 cells were included as a control.
(B) Co-immunoprecipitation of sACE2, vasopressin, and S. WB analysis of conditioned supernatants of FLAG-tagged vasopressin- and V5-tagged ACE2-doubly transfected 293T cells with spike-in recombinant S (lane 1). Co-immunoprecipitation of V5-tagged ACE2 (lanes 2–4) and FLAG-tagged vasopressin (lane 5) from conditioned supernatants of transfected 293T cells using anti-V5 and anti-FLAG antibodies, respectively. Co-immunoprecipitation of FLAG-tagged vasopressin from conditioned supernatants of transfected HK-2 cells using anti-FLAG antibodies (lane 6–8). Detection of S, ACE2, and vasopressin was performed using specific antibodies.
(C) IFA of SARS-CoV-2 S, sACE2, and AVPR1B-transfected 293T cells. The 293T cells were transfected with a plasmid encoding AVPR1B prior to inoculation with HK-2 conditioned supernatant containing sACE2, recombinant S, and vasopressin. The cells were fixed and immunostained with anti-S (magenta), anti-ACE2 (red), and anti-AVPR1B (green) antibodies. Scale bars represent 20 μm.
(D and E) IFA of SARS-CoV-2 S, sACE2, and AT1 receptor-transfected 293T cells using specific antibodies. The 293T cells were transfected with a plasmid encoding YFP-tagged AT1 receptor prior to inoculation with HK-2 conditioned supernatant containing sACE2, recombinant S of SARS-CoV-2, and/or vasopressin. The cells were fixed and immunostained with respective anti-S (magenta) and anti-ACE2 (red) antibodies. Scale bars represent 20 μm.
(F) HK-2 cells were treated with Jasplakinolide, Cytochalasin D (CytD), or Bafilomycin A1 (BafA1) prior to SARS-CoV-2 infection. Cell lysates were assayed for viral titer. Mock-treated HK-2 cells were included as control.
(G) WB analysis detecting viral antigens via anti-NP antibodies in drug-pretreated HK-2 cells as described in (F).
(H) IFA of SARS-CoV-2 NP (green) in drug-pretreated HK-2 cells described in (F) using specific antibodies. Cells were counterstained with DAPI to label nuclei (blue). Scale bars represent 20 μm.
(I) IFA of SARS-CoV-2 NP (red) in infected HK-2 cells using specific antibodies. The HK-2 cells were transfected with a plasmid encoding GFP-tagged wild-type dynamin 2 (Dyn2-WT) or GFP-tagged dominant-negative Dyn2 (Dyn2-K44A) prior to SARS-CoV-2 infection. Scale bars represent 50 μm.
The results from (A) and (F) were derived from three independent experiments. Each data point and error bar depict the mean value and SEM, respectively. Statistical analyses were carried out using Student’s t test. Statistical significance is indicated by the asterisks (∗∗p < 0.01; ∗∗∗p < 0.001). Images from (B–E) and (G–I) are representatives of three independent experiments. Arrows point to co-localized fluorescent signals.
Vasopressin functions by binding to its membrane receptors, which could trigger a process called receptor-mediated endocytosis. Consistent with our interaction assay results, which showed that vasopressin could form a complex with sACE2 and S (Figure 4B), knockdown of a vasopressin receptor—AVPR1B—potently inhibited SARS-CoV-2 infection (89.66% ± 0.14 inhibition; Figures 2F and S2 ). Therefore, we hypothesized that SARS-CoV-2 may enter cells via the receptor-mediated endocytosis through AVPR1B. To test this hypothesis, non-permissive 293T cells were first transfected with a plasmid overexpressing AVPR1B, followed by inoculating the conditioned supernatants, which contained the sACE2-S-vasopressin complex prepared in Figure 4B. IFA showed that the AVPR1B was primarily localized on the plasma membrane of the transfected 293T cells (Figure 4C). At 6 h post-inoculation, the AVPR1B-transfected cells showed decreased immunofluorescent intensity on the plasma membrane but increased puncta-staining pattern in the cytosol (Figure 4C). IFA revealed a strong co-localization signal between S and AVPR1B at the puncta (Figure 4C), suggesting internalization of sACE2-S-vasopressin complex via AVPR1B.
Figure S2.
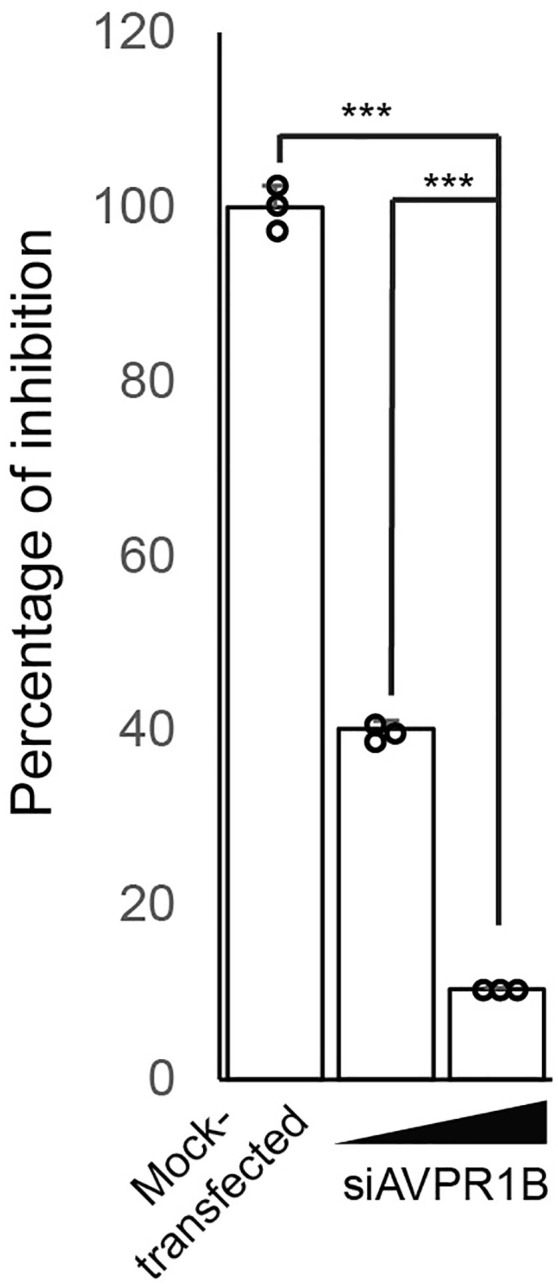
Knockdown effect of AVPR1B on SARS-CoV-2 infectivity, related to Figure 2
The HK-2 cells were transfected with different doses of siAVPR1B 24 h before SARS-CoV-2 infection. Transfection of non-targeting siRNA was included as negative control (Mock-transfected). Statistical analyses were carried out using Student’s t test. Statistical significance is indicated by the asterisks (∗∗∗p < 0.001).
Interaction between ACE2 and cellular receptor AT1 has been reported (Deshotels et al., 2014). To test if the sACE2-S-vasopressin complex—like AVPR1B—could also be internalized into cells via AT1, we performed overexpression experiments using the same experimental conditions as described above. Over the infection time course, IFA results detecting the YFP-tagged AT1 and recombinant S showed that 293T cells transfected with the AT1 also displayed increasing puncta staining pattern in the cytosol after inoculation with conditioned supernatant containing sACE2-S-vasopressin complex (Figure 4D). As our interaction assay showed that the sACE2 could interact with S in the absence of vasopressin (Figure 4B), we tested if the sACE2-S complex could also enter cells by utilizing the same cell entry mechanism. Under the same experimental conditions, similar puncta staining pattern could be observed in the YFP-tagged AT1-transfected 293T cells following inoculation of the sACE2-S complex (Figure 4E). Together, our results suggested that the sACE2-S complex and sACE2-S-vasopressin complex could enter cells via receptor-mediated endocytosis through AVPR1B and/or AT1 receptors.
Dynamin 2-dependent endocytosis is important for SARS-CoV-2 to establish an effective infection
Receptor-mediated endocytosis is known to be regulated by cytokinesis, vesicle trafficking, and the endosomal/lysosomal system (Simonetti and Cullen, 2019). To further examine the role of cytokinesis and vesicle trafficking in SARS-CoV-2 infection, we treated the HK-2 cells with two cytoskeletal drugs—Jasplakinolide and Cytochalasin D (CytD)—prior to SARS-CoV-2 infection. Compared to the mock-treated control cells, HK-2 cells pretreated with Jasplakinolide and CytD showed 2.15-log (p < 0.001) and 1.33-log (p < 0.001) reductions in viral load, respectively (Figure 4F). WB and IFA results confirmed undetectable and minimal SARS-CoV-2 NP expression in pretreated cells (Figures 4G and 4H). Besides a substantial reduction in the number of positively stained cells in the CytD-treated HK-2 cells, we observed an altered staining pattern with the viral antigen expression primarily restricted to the plasma membrane forming a “ring-like” structure (Figure 4H). These results suggested that perturbation of the cytoskeletal organization may hamper the entry of SARS-CoV-2 into the cytosol.
Besides, CytD is known to be able to modulate the function of the endosomal-lysosomal system. Indeed, our RNAi screening also identified several VDFs that are related to this system, of which four (ANXA8, ANXA1, ANXA8L1, and ALS2) have been validated (Figure 3D). Given that endosomal acidification plays important roles in receptor-mediated cell entry in some viruses (Glebov, 2020), we examined if the change of endosomal pH would affect SARS-CoV-2 replication by impairing the function of the proton pump—vacuolar H+-ATPase (V-ATPase)—which is responsible for controlling the endosomal pH via a specific inhibitor, Bafilomycin A1 (BafA1) (Cotter et al., 2015). Pretreatment of HK-2 cells using the BafA1 at a concentration of 25 ηM showed a significant inhibition (>4-log) of SARS-CoV-2 infection when compared with the mock-treated control cells (p < 0.001; Figure 4F). Pretreatment with a 10-fold lower dose (2.5 ηM) of BafA1 also achieved a similar level of inhibition (>4-log) (p < 0.001; Figure 4F). Consistently, viral protein expression was not detected by WB and IFA in the BafA1-treated cells (Figures 4G and 4H).
Thus far, our results supported a potential cell entry mechanism by which SARS-CoV-2 exploits the endocytosis pathway (Figure 4C–4H). Receptor-mediated endocytosis is known to involve membrane scission of nascent vesicles from plasma membrane by dynamin 2 (Dyn2) (González-Jamett et al., 2013). To further test the requirement of Dyn2, we targeted its function by transfecting a plasmid overexpressing the dominant-negative Dyn2 (Dyn2-K44A) prior to SARS-CoV-2 infection. In contrast to the GFP-wild-type Dyn2-transfected cells showing strong co-localization between the GFP-tagged Dyn2 and SARS-CoV-2 NP, no co-localization was detected in the GFP-Dyn2-K44A-transfected cells (Figure 4I), suggesting entry of SARS-CoV-2 into the cells through the Dyn2-dependent endocytosis pathway.
ACE2 shedding modulates the SARS-CoV-2 infectivity
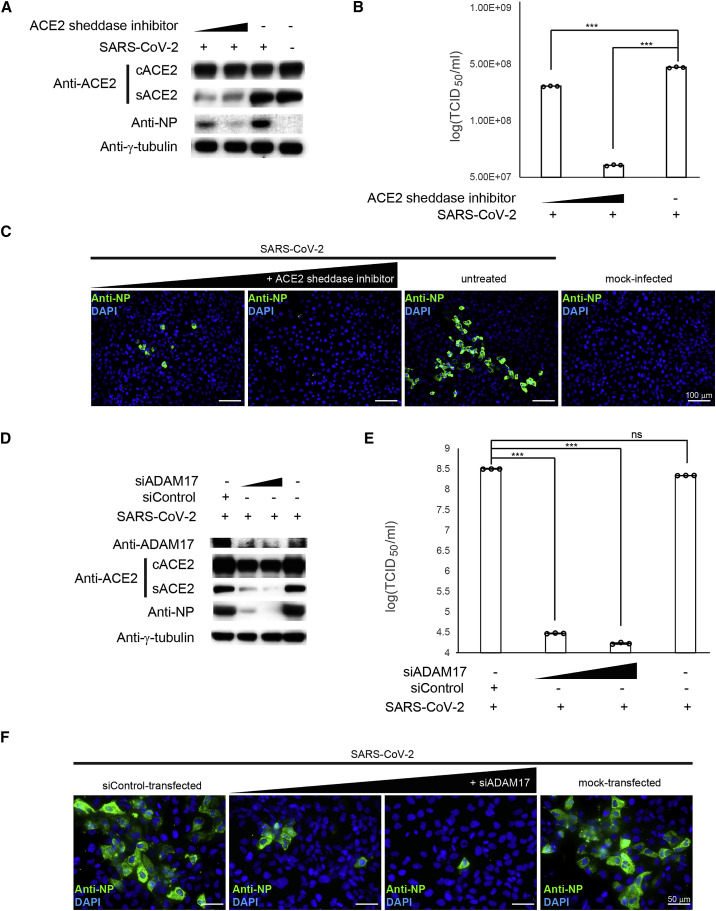
Dyn2-dependent endocytosis involves a vesicular transport event to facilitate the internalization and recycling of receptors (Cullen and Steinberg, 2018). In addition to being a receptor for SARS-CoV-2, ACE2 controls blood pressure homeostasis, which requires complex regulatory events involving protein trafficking as well as post-translational modifications. First, surface trafficking and sorting are required for plasma membrane expression of the cACE2. On the surface, cACE2 requires proteolytic cleavage by proteases such as disintegrin and ADAM17 to release circulating sACE2 to the extracellular space. It is conceivable that the shedding of cACE2 could modulate SARS-CoV-2 infectivity. To test this possibility, prior to SARS-CoV-2 infection, we pretreated HK-2 cells with GW280264X, which inhibits the enzymatic activity of ADAM17. Following the treatment, successful inhibition of sACE2 but not cACE2 expression was confirmed by WB (Figure 5 A). Notably, pretreatment of GW280264X potently inhibited SARS-CoV-2 infection in a dose-dependent manner as shown by WB, TCID50, and IFA (Figures 5A–5C).
Figure 5.
ACE2 shedding modulates the SARS-CoV-2 infectivity
(A) WB analysis showing expression levels of cellular ACE2 (cACE2) and secretory ACE2 (sACE2) in HK-2 cells pretreated with different doses of ACE2 sheddase inhibitor prior to SARS-CoV-2 infection (lanes 1 and 2). Untreated (lane 3) and mock-infected (lane 4) controls were included. SARS-CoV-2 NP was detected using specific antibodies.
(B) Cell lysates of samples described in (A) were assayed for viral titer. Untreated and mock-infected cells were included as controls.
(C) IFA of SARS-CoV-2 NP (green) in pretreated HK-2 cells described in (A) using specific antibodies. Cells were counterstained with DAPI to label nuclei (blue). Scale bars represent 100 μm.
(D) WB analysis showing expression levels of cACE2 and sACE2 in HK-2 cells transfected with different amount of siADAM17 (lanes 2 and 3). siControl-transfected (lane 1) and mock-transfected (lane 4) controls were included. The SARS-CoV-2 NP was detected using specific antibodies.
(E) Cell lysates of samples described in (D) were assayed for viral titer.
(F) IFA of SARS-CoV-2 NP (green) in pretreated HK-2 cells as described in (D) using specific antibodies. Cells were counterstained with DAPI to label nuclei (blue). Scale bars represent 50 μm.
TCID50/mL results from (B) and (E) were derived from three independent experiments. Each data point and error bar depict the mean value and SEM, respectively. Statistical analyses were carried out using Student’s t test. Statistical significance is indicated by the asterisks (∗∗∗p < 0.001; ns: p > 0.05). Images from (A), (C), (D), and (F) are representatives of three independent experiments.
In addition, we independently performed knockdown experiments to confirm the regulatory effect of ADAM17 on SARS-CoV-2 infectivity. Consistent with the results of the GW280264X treatment experiment, a reduction of sACE2 was also observed in the siADAM17-transfected cells (Figure 5D). Dose-dependent reductions in viral loads (p < 0.001) and NP expression were observed in siADAM17-transfected cells (Figure 5D–5F). Overall, our results suggested that targeting ACE2 shedding could modulate SARS-CoV-2 infectivity.
SARS-CoV-2 infection depends on sACE2
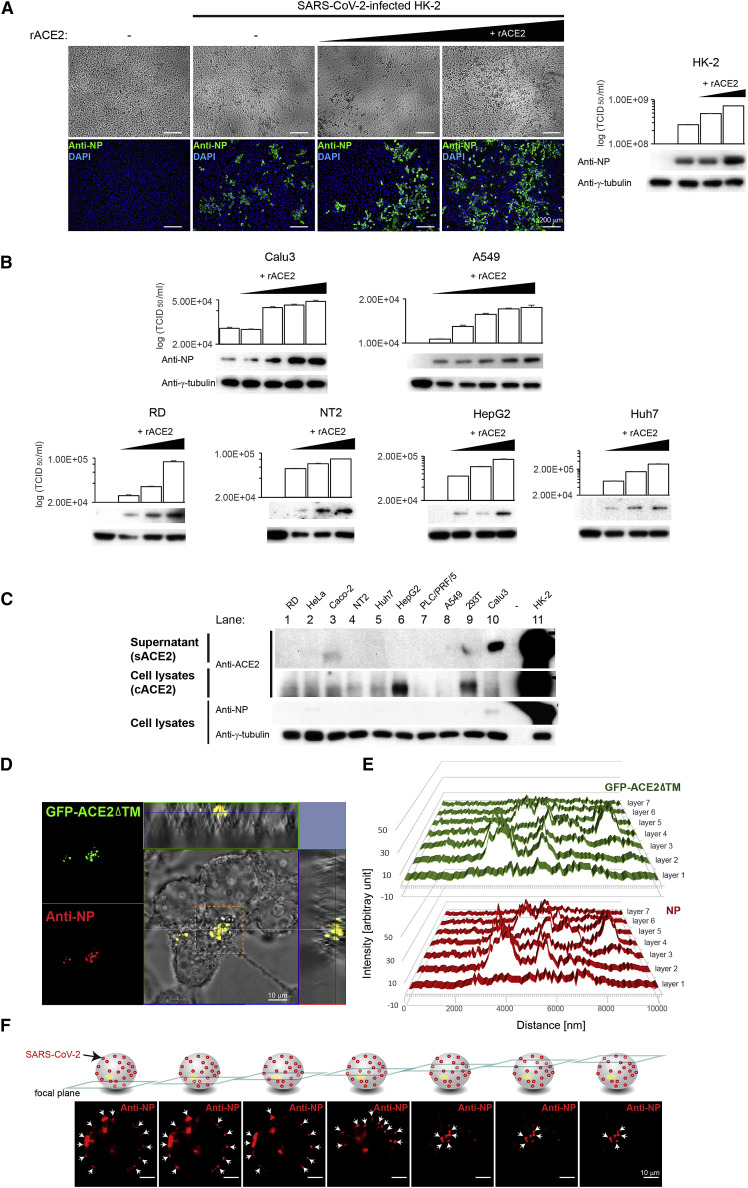
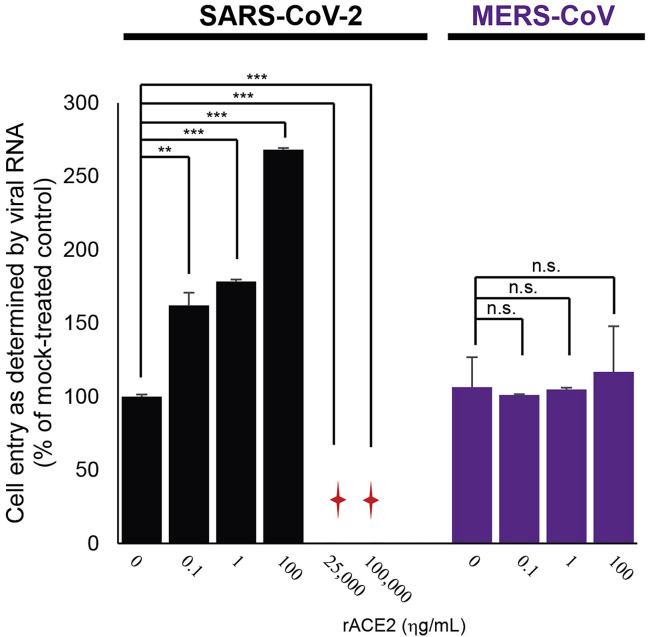
Our results showed that sACE2 could facilitate virus cell entry through receptor-mediated endocytosis, suggesting its potentially important role on SARS-CoV-2 infection. To further substantiate its role, we tested the effect of sACE2 on SARS-CoV-2 infectivity in HK-2 cells at a low MOI of 0.01. Under this infection condition, ∼10% of cells could be infected (Figure 6 A). With increasing doses of recombinant ACE2 (rACE2) on HK-2 cells, SARS-CoV-2 infectivity increased to 30% and 50%, respectively, as evidenced by stronger CPEs and higher expression of SARS-CoV-2 NP observed in the infected cells (Figure 6A). The results suggested that the addition of rACE2, mimicking the sACE2, could enhance SARS-CoV-2 infectivity. This effect, however, was not observed when performing the experiment using Middle East respiratory syndrome (MERS)-CoV under the same conditions (Figure S3 ). To further confirm the role of sACE2 in SARS-CoV-2 infection, we repeated the same experiment using two human lung cell lines—Calu3 and A549. Consistently, WB and TCID50 results showed a dose-dependent augmentation of SARS-CoV-2 infectivity in both cell lines treated with an increasing dose of rACE2 (Figure 6B). Intriguingly, cell lines—including RD, NT2, HepG2, and Huh7—derived from organs other than the lung also showed increase in SARS-CoV-2 susceptibility when treated with an increasing dose of rACE2 (Figure 6B).
Figure 6.
SARS-CoV-2 infection depends on sACE2
(A) Left: IFA of SARS-CoV-2 NP (green) in HK-2 cells pretreated with different doses of recombinant ACE2 (rACE2) prior to infection by SARS-CoV-2. Untreated and mock-infected controls were included. Cells were counterstained with DAPI to label nuclei (blue). Corresponding CPE were also shown. Scale bars represent 200 μm. Right: WB analysis detecting viral antigens using anti-NP antibodies in HK-2 cells described in (A). Corresponding TCID50 results were presented above the blot images.
(B) WB analysis detecting viral antigens using anti-NP antibodies in SARS-CoV-2-infected cells pretreated with increasing doses of rACE2. Corresponding TCID50 results were presented above the blot images.
(C) WB analysis detecting expression levels of cACE2 and sACE2 in human cell lines of different organs that were infected with SARS-CoV-2. SARS-CoV-2 NP was detected using specific antibodies.
(D) Confocal image with orthogonal projections of SARS-CoV-2 infected 293T cells transfected with mutant GFP-ACE2ΔTM. Co-localization signal (yellow) was observed between SARS-CoV-2 NP (red) and mutant ACE2 (GFP-ACE2ΔTM; green). Scale bar represents 10 μm. The 3D reconstruction of the confocal images is available in Video S1.
(E) Profiles of fluorescent intensity of NP (red) and GFP-ACE2ΔTM (green) measured at different layers of the confocal images described in (D; boxed area in orange).
(F) Optical sectioning of the SARS-CoV-2-infected 293T cell expressing the GFP-ACE2ΔTM. Arrows indicate NP (red), which showed a punctate pattern predominately localized on the cell surface. Scale bars represent 10 μm. Images from (A–D) and (F) are representatives of three independent experiments. Each data point and error bar depict the mean value and SEM, respectively.
Figure S3.
Effect of ACE2 on SARS-CoV-2 and MERS-CoV cell entry, related to Figure 6
The HK-2 cells were treated with different doses of recombinant ACE2 (rACE2) protein prior to SARS-CoV-2 (black) or MERS-CoV (purple) infection separately. Cell entry of the SARS-CoV-2- or MERS-CoV-inoculated HK-2 cells was evaluated by measuring viral RNAs using qRT-PCR. Sample with undetectable viral RNA is marked with a red cross.
To investigate the role of sACE2 under physiologically relevant conditions, we selected a panel of human cell lines from different organs and examined their relationship between the susceptibility to SARS-CoV-2 and sACE2 and cACE2 expressions. Consistent with previous studies on the rare expression of ACE2 in many cell types (Lukassen et al., 2020; Wang et al., 2020), WB results demonstrated that the protein expression of ACE2 could only be detected in a few cell lines (Figure 6C). Expectedly, high expression of sACE2 and cACE2 was found in the conditioned supernatants and cell lysates of HK-2 cells (Figure 6C; lane 11), respectively, and the results were in line with the high susceptibility of HK-2 to SARS-CoV-2 (Figure 1). As shown by the detection of NP in the infected cell lysates, relatively high expression level of sACE2 but undetectable level of cACE2 in Calu3 cells seemed to support SARS-CoV-2 infection (Figure 6C; lane 10). We also noted the expression of sACE2 but not cACE2 in Caco-2 cells (Figure 6C; lane 3). Although WB analysis was unable to detect the NP expression (Figures 1B and 6C; lane 3), IFA and TCID50 assays consistently detected viral antigens and RNAs, though at a much lower levels compared to HK-2 cells, in the infected Caco-2 cells (Figures 1C and 1E). Notably, HepG2 cells were unable to support SARS-CoV-2 infection despite a strong expression of cACE2 (but not sACE2) detected in this cell line (Figure 6C; lane 6).
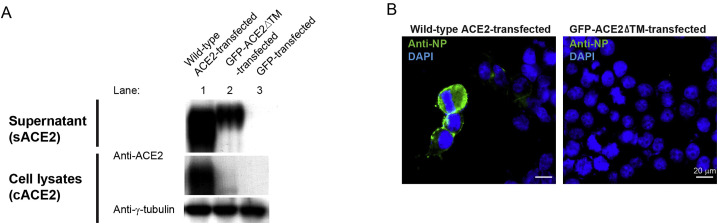
To monitor sACE2 during the SARS-CoV-2 infection process, we utilized a GFP-tagged ACE2 mutant lacking the transmembrane domain—GFP-ACE2ΔTM—to mimic sACE2 (Procko, 2020) for infection. We first validated the subcellular localization of GFP-ACE2ΔTM by transfecting it into 293T cells. In contrast to the cells transfected with the wild-type ACE2, where the ACE2 proteins were readily detected in both cell lysates and conditioned supernatants (Figure S4 A, lane 1), cells transfected with the GFP-ACE2ΔTM could only be detected in the conditioned supernatants (Figure S4A, lane 2), suggesting that most ACE2 (i.e., GFP-ACE2ΔTM) was exported out into the extracellular space and not retained inside the cells. These results were consistent with the IFA results where the GFP signal could be detected in cells transfected with the wild-type ACE2 but not the GFP-ACE2ΔTM (Figure S4B). We then performed confocal microscopy analysis of the GFP-ACE2ΔTM-transfected cells upon SARS-CoV-2 infection. Remarkably, orthogonal projections of confocal sections revealed strong co-localization of SARS-CoV-2 NP and GFP-ACE2ΔTM (Figure 6D). Three-dimensional (3D) reconstruction of the confocal images is available in Video S1. Profiles of fluorescent intensity measured at different focal planes also revealed a high degree of overlapping signals between the SARS-CoV-2 NP and GFP-ACE2ΔTM (Figure 6E; Video S1). Further optical sectioning of the infected cells revealed that similar to the formation of vesicles during the endocytosis, a punctate pattern of viral antigen predominantly localized on the cell surface (Figure 6F). Together, these results support that sACE2 could interact with SARS-CoV-2 during the infection process and that sACE2 has a potentially determining role in SARS-CoV-2 infection.
Figure S4.
Characterization of subcellular localization of the wild-type and mutant ACE2, related to Figure 6
(A) WB analysis showing expression levels of cACE2 and sACE2 in 293T cells transfected with a plasmid encoding wild-type ACE2 (lane 1), GFP-tagged mutant ACE2 lacking the transmembrane domain (GFP-ACE2ΔTM) (lane 2), or GFP (lane 3) prior to SARS-CoV-2 infection.
(B) IFA of SARS-CoV-2 NP (green) in 293T cells transfected with wild-type ACE2 (left) and mutant GFP-ACE2ΔTM (right). Cells were counterstained with DAPI to label nuclei (blue). Scale bars represent 20 μm.
Discussion
We have successfully identified VDFs that are important for SARS-CoV-2 to establish a productive virus replication (Figure 7 ), and their annotated functions were correlated with clinical manifestation and complications of SARS-CoV-2 infection. Increasing evidence support the association between COVID-19 and cardiovascular and renal-related diseases (Camm and Camm, 2020; Yang et al., 2020). These observations were also in line with the RNAi screening results, in which the functional annotation of some enriched VDFs and their biological pathways were also linked to the regulation of cardiac and renal functions. In fact, pathway analysis of VDFs showed ∼70% of significantly affected biological pathways are related to cardiac diseases (pink; Figure 2D). The most affected biological pathway is related to cardiac arrhythmias, which is often observed in SARS-CoV-2-infected patients, especially in critically ill cases (Figure 2D) (Babapoor-Farrokhran et al., 2020; Karamchandani et al., 2020; Rav-Acha et al., 2020). Besides, we also identified three significantly affected renal disease-related pathways (brown; Figure 2D), which is in line with the high incidence of acute kidney injury reported in SARS-CoV-2-infected cases (Yang et al., 2020). A previous study also reported the presence of coronavirus-like particles in >75% kidney biopsies of deceased SARS-CoV-2 patients (Su et al., 2020). Furthermore, PANTHER pathway analysis of the enriched VDFs identified vasopressin-related pathways as the top-affected molecular pathways (Figure 2E). The important roles of these vasopressin-related VDFs in SARS-CoV-2 infection were further validated by performing knockdown experiment (Figure 2F). Previous studies showed that vasopressin can induce differentiation of stem cells into cardiomyocytes and promote heart muscle homeostasis (Yasin et al., 1994). It can also control tonicity of body fluids by converting to arginine vasopressin (AVP) that can further regulate the arterial blood pressure (Demiselle et al., 2020). Consistently, due to their regulatory roles in maintaining homeostasis of kidney and heart, dysregulated expressions of vasopressin and AVP were also directly linked to the development of renal failure and cardiovascular system diseases (Czarzasta et al., 2018). Therefore, further investigation of the identified VDFs involved in cardiac and renal disease development may provide mechanistic insights into SARS-CoV-2-induced extrapulmonary diseases.
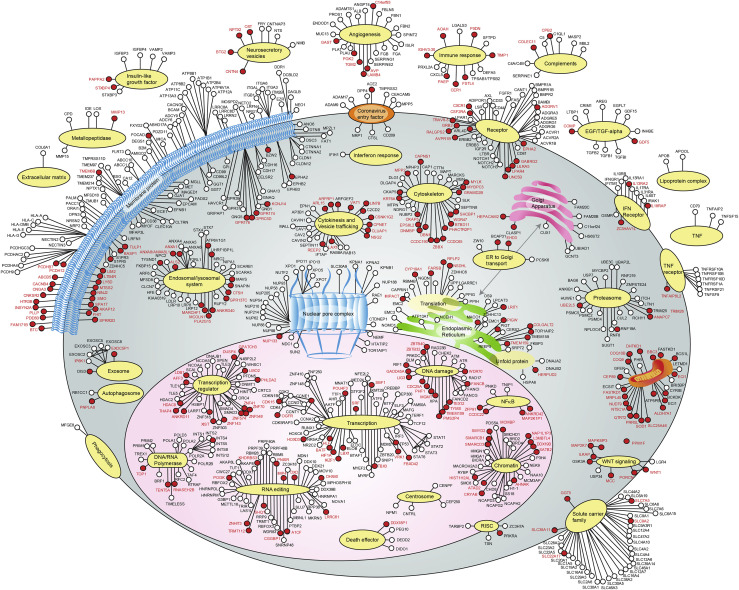
Figure 7.
A coronavirus life cycle map
The function and subcellular localization of VDFs were determined based on the use of multiple databases (please see STAR Methods) and the literature. VDFs that were discovered only in this study are indicated by red-filled dots, and the gene symbols are written in red.VDFs discovered in this study and previously reported in other studies are indicated by red-filled dots, and the gene symbols are written in black. VDFs identified in other studies are indicated by open dots, and the gene symbols are written in black. The information of the VDFs identified in this study is listed in Table S1.
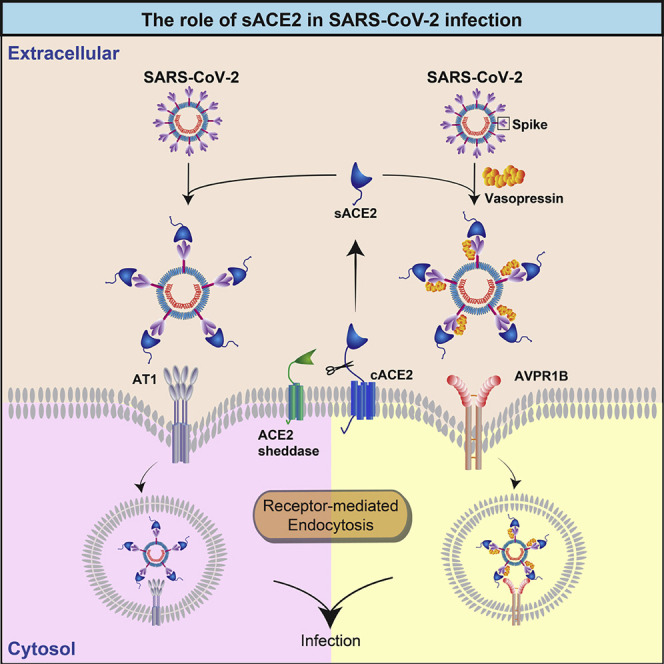
sACE2 and vasopressin play important roles in SARS-CoV-2 infection. ACE2, being a receptor for SARS-CoV-2 (Wan et al., 2020), is also known to serve as one of the key regulators controlling the release of vasopressin into the plasma for the maintenance of blood pressure homeostasis via the RAS (Matsukawa and Miyamoto, 2011; Reid et al., 1983). To serve its regulatory role in the RAS, the tissue-bound form of ACE2 (i.e., cACE2) is shed by proteases to produce the sACE2, which can then enter the circulatory system. As sACE2 preserves the binding site for SARS-CoV-2, it is possible that sequestration of SARS-CoV-2 by sACE2 may enable cell entry of tissues where cACE2 is poorly expressed. Indeed, our in vitro data showed that endogenous sACE2 could interact with the S of SARS-CoV-2 in the extracellular compartment (Figure 4B). The resulting sACE2-S complex could then enter cells through receptor-mediated endocytosis via the AT1 surface receptor (Figures 4D and 4E). Additionally, we found that the S of SARS-CoV-2 could interact with vasopressin forming an sACE2-S-vasopressin complex, which facilitated cell entry via another vasopressin receptor, AVPR1B (Figures 4B and 4C). This new cell entry mechanism may explain our data showing that cells from various organs could be sensitized to SARS-CoV-2 upon administration of rACE2 (Figures 6A and 6B).
sACE2 expression contributes to the cell line susceptibility to SARS-CoV-2. Little or low infectivity of SARS-CoV-2 was detected in all tested human cell lines, except for the HK-2 cells (Figure 6C). In contrast to HK-2 cells, SARS-CoV-2 is unable to replicate efficiently in 293T, although both cell lines were derived from human kidney. We speculate that the differential susceptibility may be linked to their differences in sACE2 level. We also noted that while highly susceptible HK-2 cells exhibited very strong expressions of both cACE2 and sACE2 (Figure 6C; lane 11), expression of cACE2 alone does not render the cells susceptible to SARS-CoV-2 as exemplified in HepG2 and 293T cells where cACE2, but not sACE2, was detected (Figure 6C; lanes 6 and 9). In contrast, expression of sACE2 alone in Caco-2 and Calu3 cells was able to support SARS-CoV-2 infection ( Figure 6C; lanes 3 and 10). Although WB results failed to detect SARS-CoV-2 NP expression in Caco-2 cells, our qRT-PCR and IFA results confirmed the presence of the SARS-CoV-2 RNA and protein in the infected Caco-2 cells (Figures 1A, 1C and 1E). The low expression level of sACE2 in Caco-2 cells may weakly support SARS-CoV-2 infection. This observation coincides with IFA and WB results, which showed a dose-dependent augmentation of SARS-CoV-2 infectivity in cells administered with an increasing dose of rACE2 (Figures 6A and 6B). Together, our in vitro infection data using human cell lines that originated from different organs support the important role of sACE2 in SARS-CoV-2 infection.
We discovered the dual role of sACE2 in SARS-CoV-2 infection. Modulating the SARS-CoV-2 infectivity using recombinant sACE2 has been previously suggested as a treatment strategy for COVID-19. Attempts have been made to utilize recombinant soluble human ACE2 to inhibit SARS-CoV-2 infection using in vitro model (Cocozza et al., 2020; Monteil et al., 2020). In these studies, very high concentrations of rACE2 [∼10–200 μg/mL of ACE2, concentrations are much higher than its physiological range in plasma, i.e., ηg/mL; Ridwan et al., 2019; Sama et al., 2020) were required to achieve inhibitory effects. Indeed, our results were also in line with their findings, where 25 and 100 μg/mL of rACE2 could inhibit SARS-CoV-2 infection (Figure S3). We speculate that the addition of excessive amounts (i.e., μg/mL level) of recombinant ACE2 may saturate endocytic recycling of the ACE2 receptor, competing with the SARS-CoV-2-ACE2 complex for cell entry, therefore resulting in the reduction of SARS-CoV-2 infectivity. In contrast, rACE2 concentrations close to physiological range (i.e., ηg/mL level) could enhance SARS-CoV-2 infection (Figures 6A, 6B, and S3). Interestingly, a similar phenomenon was recently reported in a study that provided an important clue on the in vivo effect of the rACE2 in SARS-CoV-2-infected patient (Zoufaly et al., 2020). The study showed that substantial and steady increases in viral loads were detected in tracheal aspirates (from ∼103 copies/mL at day 0 to ∼105 copies/mL at day 2) and nasopharyngeal swabs (from ∼104 copies/mL at day 0 to ∼105 copies/mL at day 5), respectively, after the administration of rACE2. Although the patient eventually recovered after the appearance of neutralizing antibodies, this in vivo data and our findings suggested that treatment that may alter sACE2 level in patients with COVID-19 should be carefully considered.
The discovery of a highly permissive human renal cell line—HK-2—to SARS-CoV-2 is useful for studying the biogenesis of SARS-CoV-2 and the possible therapeutic options for COVID-19. The physiological and pathological relevance of using this HK-2 cell line in studying SARS-CoV-2 biogenesis is supported by recent ultrastructural studies that detected SARS-CoV-2 in renal autopsy of patients with COVID-19 (Farkash et al., 2020). The versatility of HK-2 is further supported by our RNAi screening results, in which many successfully identified VDFs were not only linked to renal-related diseases but also other clinical manifestations and complications of COVID-19 (Figures 2D and 2E). Furthermore, most current studies on SARS-CoV-2 infection were based on the use of pseudovirus-expressing viral surface S protein or non-human cell lines, which may not truly reflect an authentic infection process (Heaton et al., 2020; Shang et al., 2020). Our findings using authentic virus to infect physiologically relevant HK-2 cells revealed that SARS-CoV-2 entered the cells mainly through Dyn2-dependent endocytosis (Figures 4F–4I). The involvement of cytokinesis, vesicle trafficking, and the endosomal/lysosomal system in SARS-CoV-2 cell entry was further confirmed through the use of inhibitors (Jasplakinolide, CytD, and BafA1) and a dominant-negative Dyn2 construct (Figures 4F–4I). Of note, our results showed that a potent inhibition (>4-log) of SARS-CoV-2 replication could be achieved even when cells were pretreated with a low dose of BafA1 (i.e., 2.5 ηM) (Figures 4F–4H), which could inhibit receptor-mediated endocytosis (Harada et al., 1996). Further investigation of the BafA1 and its analogs as potential therapeutic agents for COVID-19 is warranted.
Modulating the shedding of sACE2 may provide insights into treatment strategies for COVID-19. A previous study suggested that the ADAM17 activity could be correlated with infectivity of SARS-CoV, which also utilized ACE2 as a cellular receptor (Haga et al., 2008). The authors reported that the S of SARS-CoV induced the ADAM17 activity causing an enhanced ACE2 shedding, which was positively correlated with SARS-CoV infectivity. It is possible that cells could be sensitized to both SARS-CoV-2 and SARS-CoV by enhancing the production of sACE2 through the induction of ADAM17 activity. On the flip side, controlling the activity/expression of ADAM17 could be a potential treatment strategy for COVID-19. Indeed, we demonstrated that inhibition of ADAM17 by a sheddase inhibitor, GW280264X, or by a specific siRNA potently suppressed SARS-CoV-2 infection (Figures 5A–5F). The therapeutic potential of targeting the ADAM17 has also been widely investigated in cancer research with minimal side effects (Bandsma et al., 2015; Blaydon et al., 2011), suggesting that it may represent a safe target in controlling SARS-CoV-2 infection. Overall, our identification of inhibitors, including the sheddase inhibitor, the cytoskeletal drugs and/or siRNAs/drugs targeting the VDFs, could provide insights into future development of drug combination therapy, which can be considered as an effective strategy in preventing the generation of drug-resistant mutants, particularly in the case of RNA viruses (Presloid and Novella, 2015), for the treatment of COVID-19.
Limitations of the study
This study has several limitations as the findings are based on HK-2 cells, which are of renal origin. As the primary infection site of SARS-CoV-2 is lung, further examinations will be needed to study the involvement of these factors during pulmonary infection. Besides, during the preparation of our manuscript, several CRISPR/Cas9 screening studies have been published, in which the VDFs identified are largely different in these studies. One reason may be due to the key difference between CRISPR and RNAi, where CRISPR results in true loss-of-function effects, which is in contrast to RNAi that may create hypomorphic phenotypes, thus allowing us to study essential genes whose knockout may otherwise be cell lethal. While both technologies are considered powerful tools to identify VDFs for replication, perhaps combining results from each approach could help fully to understand the underlying biology of SARS-CoV-2.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ACE-2 | R&D | Cat#AF933 |
| ADAM17 | Abcam | Cat#ab2051 |
| γ-Tubulin | Sigma-Aldrich | Cat#T6557 |
| AVPR1B | Abcam | Cat#ab116246 |
| SARS-CoV-2 NP (Rabbit polyclonal) | This paper | N/A |
| SARS-CoV-2 Spike (Mouse monoclonal) | GeneTex | Cat#GTX632604 |
| FLAG | Sigma-Aldrich | Cat# F7425 |
| EZview™ Red ANTI-FLAG M2 Affinity Gel | Sigma-Aldrich | Cat# F2426 |
| ANTI-V5 agarose affinity gel | Sigma-Aldrich | Cat# A7345 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific (Invitrogen) | Cat#A-11055 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific (Invitrogen) | Cat#A-11008 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Thermo Fisher Scientific (Invitrogen) | Cat#A-11012 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific (Invitrogen) | Cat#A-21235 |
| Goat anti-Mouse IgG (H+L) Secondary Antibody, HRP | Thermo Fisher Scientific (Invitrogen) | Cat#62-6520 |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, HRP | Thermo Fisher Scientific (Invitrogen) | Cat#31460 |
| Rabbit anti-Goat IgG (H+L) Secondary Antibody, HRP | Thermo Fisher Scientific (Invitrogen) | Cat#81-1620 |
| Bacterial and Virus Strains | ||
| MAX Efficiency™ DH5α Competent Cells | Thermo Fisher Scientific | Cat# 18258012 |
| SARS-CoV-2, isolate HKU-001a | Laboratory of Microbiology, HKU | N/A |
| MERS-CoV, EMC/2012 strain | A gift from Ron Fouchier (Erasmus Medical Center) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Pierce Protease and Phosphatase Inhibitor Mini Tablets, EDTA-free | Thermo Fisher Scientific | Cat# A32961 |
| Fetal bovine serum (FBS) | Thermo Fisher Scientific (GIBCO™) | Cat#10082 |
| PBS, pH 7.4 | Thermo Fisher Scientific (GIBCO™) | Cat#10010023 |
| Trypsin-EDTA (0.25%), phenol red | Thermo Fisher Scientific (GIBCO™) | Cat#25200056 |
| Dulbecco’s Modified Eagle Medium (DMEM) | Thermo Fisher Scientific (GIBCO™) | Cat#11965-092 |
| Minimal Essential Media (MEM) | Thermo Fisher Scientific (GIBCO™) | Cat#11095-080 |
| DMEM/F12 (1:1) (1X) | Thermo Fisher Scientific (GIBCO™) | Cat#11320-033 |
| Paraformaldehyde Aqueous Solution-16% | Electron Microscopy Sciences | Cat#30525-89-4 |
| Triton X-100 | Sigma-Aldrich | Cat# 93443 |
| Immun-Blot PVDF Membrane | Bio-Rad | Cat#1620177 |
| Lipofectamine 2000 | Invitrogen | Cat# 11668027 |
| Lipofectamine™ Transfection Reagent | Thermo Fisher Scientific (Invitrogen) | Cat#18324020 |
| PLUS™ Reagent | Thermo Fisher Scientific (Invitrogen) | Cat#11514015 |
| VECTASHIELD® Mounting Medium with DAPI | Vector Laboratories | Cat#H-1200 |
| Jasplakinolide, Actin polymerization and stabilization inducer | Abcam | Cat#ab141409 |
| Cytochalasin D | Sigma-Aldrich | Cat#C8273 |
| Bafilomycin A1 | Sigma-Aldrich | Cat#SML1661 |
| Angiotensin Converting Enzyme-2, ACE2 (Human recombinant, expressed in HEK293 cells) | Sigma-Aldrich | Cat#SAE0064 |
| SARS-CoV-2 (2019-nCoV) Spike Protein (S1+S2 ECD, His tag) | Sino Biological | Cat#40589-V08B1 |
| Vasopressin | Origene | Cat#TP316816 |
| Deposited Data | ||
| RNA-seq data | This paper | GEO accession number: GSE159272 |
| Experimental Models: Cell Lines | ||
| RD | ATCC | Cat#CCL-136 |
| H1HeLa | ATCC | Cat#CRL-1958 |
| Caco-2 | ATCC | Cat#HTB-37 |
| NT2 | ATCC | Cat#CRL1973 |
| Huh-7 | JCRB cell bank of Okayama University | Cat#JCRB0403 |
| HepG2 | ATCC | Cat#HB-8065 |
| PLC/PRF/5 | ATCC | Cat#CRL-8024 |
| A549 | ATCC | Cat#CL-185 |
| Calu3 | ATCC | Cat#HTB-55 |
| 293T | ATCC | Cat#CRL-3216 |
| HK-2 | ATCC | Cat#CRL-2190 |
| Vero-E6 | ATCC | Cat#CRL-1586 |
| Oligonucleotides | ||
| SARS-CoV-2 E-gene, Forward: ACAGGTACGTTAATAGTTAATAGCGT | This paper | N/A |
| SARS-CoV-2 E-gene, Reverse: ATATTGCAGCAGTACGCACACA | This paper | N/A |
| Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH), Forward: TCACCACCATGGAGAAGGC | This paper | N/A |
| Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH), Reverse: GCTAAGCAGTTGGTGGTGCA | This paper | N/A |
| GNF primer ATTTATTGTATCTGTGGGAGCCTC | This paper | N/A |
| Fwd GNF primer TGCATGTCGCTATGTGTTCTGGGA | This paper | N/A |
| Rev GNF primer ACAAAGCACTGGAAGCTATCGAA | This paper | N/A |
| H1 Forward primer GTTCTGTATGAGACCACTTGGATCC | This paper | N/A |
| RevPrimer AAAGAATGCTTATGGACGCTAGAA | This paper | N/A |
| Novel Coronavirus 2012Real-Time RT-PCR assay | CDC | Cat#KT0136 |
| ON-TARGETplus Human ADAM17, siRNA - SMARTpool, 5nmol | Horizon Discovery/Dharmacon | Cat#L-003453-00-0005 |
| ON-TARGETplus Human TMEM8B, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-017716-02 |
| ON-TARGETplus Human BASP1, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-019008-00 |
| ON-TARGETplus Human ZNF143, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-013965-00 |
| ON-TARGETplus Human XIST, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#R-188298-00 |
| ON-TARGETplus Human HIST1H2AL, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-011434-01 |
| ON-TARGETplus Human SLC25A46, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-007353-02 |
| ON-TARGETplus Human CGGBP1, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-015703-00 |
| ON-TARGETplus Human ARFGEF2, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-012208-02 |
| ON-TARGETplus Human ZNF548, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-016400-02 |
| ON-TARGETplus Human POGK, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-017451-00 |
| ON-TARGETplus Human ALS2, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-014168-00 |
| ON-TARGETplus Human IL18RAP, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-007950-00 |
| ON-TARGETplus Human CXCR1, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-005646-00 |
| ON-TARGETplus Human CACNB4, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-009062-00 |
| ON-TARGETplus Human CAPNS1, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-009979-00 |
| ON-TARGETplus Human ARL4D, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-011583-00 |
| ON-TARGETplus Human GPR176, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-005522-00 |
| ON-TARGETplus Human S1PR5, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-004791-00 |
| ON-TARGETplus Human ANXA8L2, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-011570-01 |
| ON-TARGETplus Human ANXA8, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-183964-00 |
| ON-TARGETplus Human GRB2, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-019220-00 |
| ON-TARGETplus Human ANXA1, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-011161-00 |
| ON-TARGETplus Human AVP, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-010997-00 |
| ON-TARGETplus Human OXT, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-011185-00 |
| ON-TARGETplus Human AVPR1B, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-005431-00 |
| ON-TARGETplus Human HLF, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-010514-02 |
| ON-TARGETplus Human BTC, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-011597-01 |
| ON-TARGETplus Human CCNE1, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-003213-00 |
| ON-TARGETplus Human HIF1A, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-004018-00 |
| ON-TARGETplus Human KHDRBS3, siRNA- SMARTpool, 0.1nmol | Horizon Discovery/Dharmacon | Cat#L-012748-01 |
| Recombinant DNA | ||
| K44A dynamin 2-pEGFP | A gift from Sandra Schmid | Addgene #34687 |
| Wt dynamin 2-pEGFP | A gift from Sandra Schmid | Addgene #34686 |
| pcDNA3.1 | Thermo Fisher Scientific (Invitrogen) | Cat#V79020 |
| pcDNA3.1-ACE2-GFP | A gift from Utpal Pajvani | Addgene #154962 |
| pcDNA3.1-ACE2ΔTM-GFP | Chan et al., 2020 | Addgene #145171 |
| Vasopressin V1b receptor (AVPR1B) (NM_000707) Human Tagged ORF Clone | OriGene | Cat# RC215919 |
| pCXN2-HA-AT1R-YFP | Inuzuka et al., 2016 | Addgene #101659 |
| Software and Algorithms | ||
| DAVID Bioinformatics Resources | Huang et al., 2007 | https://david.ncifcrf.gov/tools.jsp |
| PANTHER (Protein Analysis Through Evolutionary Relationships) | Mi et al., 2019 | http://www.pantherdb.org/ |
| Cutadapt | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| HTSeq | Anders et al., 2015 | https://htseq.readthedocs.io/en/master/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| R | Gu et al., 2016; Ito and Murphy, 2013 | https://www.r-project.org/ |
| UniProt | Bairoch et al., 2005 | https://www.uniprot.org/ |
| Gene Ontology | Ashburner et al., 2000 | http://geneontology.org/ |
| NCBI Reference Sequence | Pruitt et al., 2005 | https://www.ncbi.nlm.nih.gov/refseq/ |
| NCBI OMIM database | McKusick, 2007 | https://www.ncbi.nlm.nih.gov/omim |
| BioGRID | Oughtred et al., 2019 | https://thebiogrid.org/ |
| PubMed | NCBI | https://pubmed.ncbi.nlm.nih.gov/ |
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Man Lung Yeung (pmlyeung@hku.hk).
Materials availability
All requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact author. Materials will be made publicly available either through publicly available repositories or via the authors upon execution of a Material Transfer Agreement.
Data and code availability
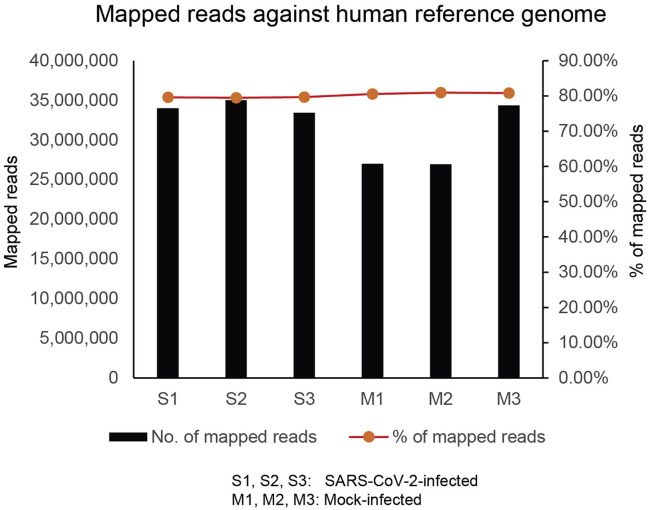
Additional Supplemental Items, including supplementary figures, tables and video, are available from Mendeley Data: https://data.mendeley.com/datasets/gcdpsxd6d5/2. All sequencing data associated with this study have been deposited to NCBI with GEO accession number: GSE159272. The sequencing throughput and a summary of the read counts are available in Table S2 (see also Figure S5 ).
Figure S5.
Mapping of high-throughput sequencing reads to human genome, related to Figures 2, 3, and 7
Experimental model and subject details
Cells
Eleven human cell lines (HK-2, Caco-2, A549, Calu-3, Huh7, HepG2, PLC/PRF/5, RD, HeLa, NT2, and 293T) derived from different organs were maintained in a humidified atmosphere at 37°C with 5% CO2, in Dulbecco’s modified Eagle’s medium (DMEM) or minimum essential medium (MEM; GIBCO) or DMEM/F12 (GIBCO) containing 10% (v/v) fetal bovine serum (FBS, GIBCO). Similarly, a type of African green monkey kidney epithelial cells—Vero-E6—were maintained in DMEM containing 10% (v/v) FBS. All cell lines have been authenticated by short tandem repeat (STR) profiling and tested negative for mycoplasma contamination (Pangenia Lifesciences Limited).
Viruses
SARS-CoV-2 (HKU-001a) was isolated from a nasopharyngeal aspirate of a laboratory-confirmed COVID-19 patient in Hong Kong (To et al., 2020). MERS-CoV (EMC/2012 strain) and was a gift from Ron Fouchier (Erasmus Medical Center)(Zaki et al., 2012). The isolates were propagated through Vero-E6 cells (ATCC) in DMEM (GIBCO) supplemented with 10% FCS (GIBCO) and 100 units/mL penicillin plus 100 μg/mL streptomycin (1% PS). All experiments entailing live SARS-CoV-2 followed the approved standard operating procedures of our Biosafety Level 3 (BSL-3) facility (Yeung et al., 2016).
Method details
Production of shRNA library and stable clones
FIV-based shRNA library was produced as described previously (Yeung et al., 2009; Yeung et al., 2018). Briefly, 2 μg of the shRNA library in lentiviral constructs (SBI) were co-transfected with 10 μg of the pPACK packaging plasmid mix into HEK293T cells using Lipofectamine and Plus reagents according to the manufacturer’s protocol (ThermoFisher). At 48 h after transfection, conditioned culture media were collected to harvest the packaged viruses. In parallel, GFP-packaged viruses were produced. The infectivity of the viruses was established based on the percentage of the GFP-transduced HK-2 cells. Four days after transduction, the cells were selected for 3 weeks in a growth medium containing 3 μg/mL of puromycin. The shRNA-transduced HK-2 cells were challenged with SARS-CoV-2 (MOI = 1). Three days after infection when CPE of the SARS-CoV-2-infected GFP-transduced HK-2 cells reached over 90%, the shRNA-transduced HK-2 cells were recovered by replenishing with DMEM/F12 (GIBCO) containing 10% (v/v) of FBS (GIBCO). After 24 h, the survived shRNA-transduced HK-2 cells were re-challenged with SARS-CoV-2 using the same infection conditions. At day 3 post-infection, total RNA of the shRNA-transduced HK-2 cells was harvested using TRIzol (ThermoFisher). Seven micrograms of total RNA was reverse transcribed into cDNAs in the presence of 10 μM of GNF primer (5′-ATTTATTGTATCTGTGGGAGCCTC-3′), 100 mM of dithiothreitol, 10 mM of each dNTP, 1 μM reverse transcriptase buffer, and 200 U of SuperScript® III Reverse Transcriptase (Invitrogen). The reaction mixtures were incubated at 50°C for 1 h. The reaction was stopped by heat inactivation at 72°C for 5 min. Half of the reaction mixture was then transferred to a tube containing 1 × PCR reaction buffer, 20 mM of dNTP, 20 μM of Fwd GNF primer (5′-TGCATGTCGCTATGTGTTCTGGGA-3′), 20 μM of Rev GNF primer (5′-ACAAAGCACTGGAAGCTATCGAA-3′), and iProof (Bio-rad). PCR amplification of the shRNA target region was performed under the following conditions: Step 1:94°C for 4 min; Step 2: 94°C for 30 s and then 68°C for 1 min; Step 3: Repeat Step 2 for 20 cycles; Step 4: 68°C for 3 min. Nested PCR reactions using 1 μL of the 1st round PCR products were performed using 100 μM of H1 Forward primer (5′-GTTCTGTATGAGACCACTTGGATCC-3′), 100 μM of RevPrimer (5′-AAAGAATGCTTATGGACGCTAGAA-3′), and iProof (Bio-Rad). PCR amplification of the shRNA target region was performed under the following conditions: Step 1:94°C for 2 min, 50°C for 2 min and then 68°C for 1 min; Step 2: 94°C for 30 s and then 68°C for 30 s; Step 3: Repeat Step 2 for 18 cycles; Step 4: 68°C for 3 min. The PCR products were then gel purified using QIAquick PCR purification kit (QIAGEN) as described in the manufacturer’s protocol. The purified PCR products were then submitted to Centre for PanorOmic Sciences at HKU for high-throughput sequencing.
Knockdown of gene expressions by siRNAs
For independent validation, siRNAs (Human ON-TARGETplus) were purchased from ThermoFisher. These siRNAs were transfected into the HK-2 cells using Lipofectamine 2000 (ThermoFisher) according to the manufacturer’s protocol. Forty-eight h after transfection, the cells were challenged with SARS-CoV-2 (MOI = 1). At day 3 post-infection, total RNA of the shRNA-transduced HK-2 cells were harvested as described above.
High-throughput sequencing
All samples for high-throughput sequencing were processed by the Centre for PanorOmic Sciences at HKU. Briefly, libraries of each sample were prepared by KAPA Hyper Prep Kit (KAPA Biosystems) according to the manufacturer’s protocol. Fifty nanograms of each library was then first purified by AMPure XP (Beckman Coulter) before end-repairing. Next, the 3′ end of the libraries were ligated to adaptors using xGen Dual Index UMI Adapters (IDT) according to the manufacturer’s protocol. The adaptor-ligated libraries were then validated by Agilent Bioanalzyer, Qubit, and qPCR for quality control analysis. Then, the adaptor-ligated libraries were sequenced using NovaSeq 6000 (2 × 150 bp paired-end reads) System (Illumina). The dataset of the shRNA screening results has been deposited in NCBI with GEO accession number: GSE159272. The sequencing throughput and a summary of the read counts are available in Table S2 (see also Figure S5).
Virus titration by TCID50 assay
The median tissue culture infective dose (TCID50) per mL was determined for SARS-CoV-2 in Vero-E6 cells as previously described (Yeung et al., 2018). Briefly, cells were seeded in 96-well plates at a density of 5 × 104 cells/well in 150 μL of DMEM. The virus was serially diluted by half-log from 103 to 1014 in DMEM. One hundred microliters of each dilution were added per well; and the plates were observed daily for CPE for five consecutive days.
Confocal and immunofluorescence microscopic analysis
Vero-E6, HK-2, 293T, and Caco-2 cells were fixed in 4% paraformaldehyde solution in 1 × PBS containing 0.1% Triton X-100. After 1 h blocking with 3% bovine serum albumin (BSA) at room temperature, the cells were stained with anti-SARS-CoV-2 antibodies (developed by our group), anti-FLAG (Sigma) and/or with anti-ACE2 antibodies (R&D systems) for 1 h at room temperature as we previously described (Yeung et al., 2008). Unbound antibodies were washed away six times with 1 × PBS. Positively stained cells were detected by secondary IgG (H+L) antibodies conjugated either with Alexa Fluor 488 or Alexa Fluor 594 (Life Technology) for 30 min at room temperature. Following six 1 × PBS washes, the stained cells were mounted onto glass slides with VECTASHIELD mounting medium with 4′,6′-diamidino-2-phenylindole (DAPI) (Vector Lab) and examined with a Leica TCS-NT microscope (Leica Microsystem) or a LSM700 confocal microscope (Zeiss).
Reverse transcription and quantitative real-time PCR (qRT-PCR)
Total RNAs and viral RNAs were isolated using TRIzol (ThermoFisher) and Viral RNA Mini kit (QIAGEN), respectively, as described in our previous literature (Yeung et al., 2016)[REMOVED HYPERLINK FIELD].[REMOVED HYPERLINK FIELD] Following RNA quantification, one microgram of RNA was reverse transcribed into cDNA using random hexamers. Detection of SARS-CoV-2 and MERS-CoV was performed using primers 5′-ACAGGTACGTTAATAGTTAATAGCGT-3′ and 5′-ATATTGCAGCAGTACGCACACA-3′ targeting E gene of SARS-CoV-2 as described by WHO and by Novel Coronavirus 2012 Real-Time RT-PCR assay (CDC; Catalog # KT0136), respectively. Sample normalization was done by quantifying their respective housekeeping gene expression using primers 5′-TCACCACCATGGAGAAGGC-3′ and 5′-GCTAAGCAGTTGGTGGTGCA-3′ (GAPDH). For each reaction, equal amounts of cDNA were mixed with FS Universal SYBR Green Master Rox (Roche) plus 5pmol each of forward and reverse primers. Amplification was carried out at 95°C for 15 s and 60°C for 1 min for 55 cycles in a 7900 real-time PCR detection system (ABI).
Western blot (WB) analysis
Cell lysates or tissue extracts were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Proteins were transferred to polyvinylidene difluoride (PVDF) membrane by electroblotting (Hoefer) at constant current of 150mA overnight. Detection of antigens was performed using anti-SARS-CoV-2 antibodies (developed by our group) or anti-hACE2 antibodies (R&D systems) as previously described (Yeung et al., 2016). As loading controls, the membranes were stripped with Restore western blot stripping buffer (Pierce) before reprobing with anti-γ-tubulin (Sigma).
Co-immunoprecipitation assay
Recombinant SARS-CoV-2 S proteins (Sino Biological) were incubated with protein lysates expressing V5-tagged or untagged ACE2 and FLAG-tagged vasopressin under previously described conditions (Yeung et al., 2018) with the following modifications: both recombinant S and the protein lysates were mixed in the presence of 0.1% Tween 20 at 4°C overnight with shaking. The next day, the anti-FLAG® M2 affinity agarose gel (Sigma) or anti-V5 agarose affinity gel (Sigma) was added into the mixture and allowed to incubate for an additional 2.5 h. Following six washes with a buffer containing 50 mM of NaH2PO4, 300 mM of NaCl, and 20 mM of imidazole pH 8.0, agarose-bound proteins were fractionated by SDS-PAGE and detected by WB analyses using anti-S (Abcam), anti-FLAG (Sigma), and anti-ACE2 (Abcam) antibodies.
Cytopathic effect (CPE) measurement
The CPEs of unfixed, unstained, infected cells were measured using an optical microscope, with the condenser down and the iris diaphragm partly closed as described in “Cytopathic Effects of Viruses Protocols,” by Sushman E and Blair C (ASM Microbe Library. American Society for Microbiology. Archived from the original on June 2, 2012. Retrieved 20 November 2014). We consider total detachment of the monolayer cell as 100%. To determine the percentage of CPE, Vero-E6, HK-2, and Caco-2 cells were inoculated with SARS-CoV-2 at a virus titer of MOI = 1. The virus induced CPEs were monitored daily.
Identification of virus-dependency factors (VDFs)
High-throughput sequencing raw reads were processed with Cutadapt (Martin, 2011) removing constant lentivirus vector and hair-pin loop nucleotide sequences. Resulting sequences were then filtered to contain only the anti-sense shRNA arms ranging from 15 to 40 bp, which were representative of the shRNA sequences. The shRNA sequences were then aligned to human reference genome (GENCODE GRCh38) using Bowtie2 (Langmead and Salzberg, 2012). Subsequently, the number of reads mapping to shRNA probes and shRNA target genes were counted using HTSeq (Anders et al., 2015). The shRNA probes and shRNA target genes were generated by retrieving unique and properly paired alignments to non-overlapping genomic locations with a mapping quality over 15, following by target gene annotation (GENCODE GRCh38 annotation v24). Finally, shRNA counts were median-normalized and analyzed with DESeq2 (Love et al., 2014) to infer changes between samples and to evaluate statistical significance. Next, we adjusted the gene counts of raw reads within each treatment group by median normalization. For each treatment group, the gene counts with missing values from individual replicates were removed. The resulting datasets were subjected to clustering analyses, including principal components analysis (PCA), volcano plot, and heatmap hierarchical clustering analysis, using the stat function in R, ggplot2 function in R, and Complex Heatmap, respectively (Gu et al., 2016; Ito and Murphy, 2013).
Pathway analysis
The average of the adjusted gene counts from SARS-CoV-2-infected samples was compared with that of the mock-infected samples. Fold changes of gene counts between the SARS-CoV-2-infected samples and the mock-infected samples were generated with counts from replicates higher than 100. GO enrichment analysis was performed on 3-fold enriched genes using DAVID GO Cellular Component using default parameters with p < 0.05 (Huang et al., 2007). Similarly, molecular pathway analysis was conducted by analyzing the enriched VDFs into PANTHER (Protein Analysis Through Evolutionary Relationships) using the FISHER test type (Mi et al., 2019).
The coronavirus life cycle map
To generate the coronavirus life cycle map, relevant genes reported from different database: UniProt (Bairoch et al., 2005), Gene Ontology (Ashburner et al., 2000), NCBI Reference Sequence (Pruitt et al., 2005), NCBI OMIM database (McKusick, 2007) and BioGRID (Oughtred et al., 2019) were obtained. Text mining of the genes properties, including biological processes, molecular functions, and cellular components, were analyzed using PubMed query via iHop (Hoffmann and Valencia, 2004). The information was summarized and integrated into a database framework to form a schematized coronavirus viral life cycle. Newly discovered VDFs were added manually into this framework according to the cellular functions inferred from literature curation and database review. All SARS-CoV-2 VDFs identified in this study are available in Table S1.
Quantification and statistical analysis
Statistical significance was determined as p < 0.05 using GraphPad Prism 8 unless otherwise indicated. Experiments were analyzed by unpaired two-tailed t tests.
Acknowledgments
This study was partly supported by funding from the Sanming Project of Medicine in Shenzhen, China (SZSM201911014); the Innovation and Technology Fund (ITF) and its commissioned research program of the Food and Health Bureau and Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance of the Department of Health, The Government of the Hong Kong Special Administrative Region, China; the Theme-based Research Scheme, General Research Fund and the Collaborative Research Fund (C5012-15E) of the University Grant Committee; and donations of Michael Seak-Kan Tong, Richard Yu, and Carol Yu, the Shaw Foundation of Hong Kong, May Tam Mak Mei Yin, Lee Wan Keung Charity Foundation Limited, Hong Kong Sanatorium & Hospital, Hui Ming, Hui Hoy and Chow Sin Lan Charity Fund Limited, Chan Yin Chuen Memorial Charitable Foundation, Marina Man-Wai Lee, the Hong Kong Hainan Commercial Association South China Microbiology Research Fund, the Jessie & George Ho Charitable Foundation, Perfect Shape Medical Limited, Kai Chong Tong, Foo Oi Foundation Limited, Tse Kam Ming Laurence, Betty Hing-Chu Lee, Ping Cham So, and the Lo Ying Shek Chi Wai Foundation. We also thank the support from the Centre for PanorOmic Sciences, Genomics, and Bioinformatics Core, at the University of Hong Kong. The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the paper.
Author contributions
Conceptualization, M.L.Y. and J.L.L.T.; infection experiments, M.L.Y., L.J., and C.Z.; sample coordination and preparation, C.H., J.-P.C., R.Z., Z.C., H.Z., K.-H.C., K.K.-W.T., J.F.-W.C., L.Z., S.Y., T.M.C., Z.C., S.K.P.L., S.-Y.F., and D.-Y.J.; literature review, M.L.Y., J.L.L.T., and K.-Y.Y.; manuscript preparation, M.L.Y., J.L.L.T., D.-Y.J., and K.-Y.Y.; bioinformatics analysis, M.L.Y. and J.L.L.T.; annotation and interactive map preparation, M.L.Y. and J.L.L.T; work supervision, M.L.Y., J.L.L.T., P.C.Y.W., and K.-Y.Y.
Declaration of interests
J.F.-W.C. has received travel grants from Pfizer Corporation Hong Kong and Astellas Pharma Hong Kong Corporation Limited and was an invited speaker for Gilead Sciences Hong Kong Limited and Luminex Corporation. The other authors declare no competing interests.
Published: March 2, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.02.053.
Supplemental information
References
- Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babapoor-Farrokhran S., Rasekhi R.T., Gill D., Babapoor S., Amanullah A. Arrhythmia in COVID-19. SN Compr. Clin. Med. 2020 doi: 10.1007/s42399-020-00454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandsma R.H., van Goor H., Yourshaw M., Horlings R.K., Jonkman M.F., Schölvinck E.H., Karrenbeld A., Scheenstra R., Kömhoff M., Rump P., et al. Loss of ADAM17 is associated with severe multiorgan dysfunction. Hum. Pathol. 2015;46:923–928. doi: 10.1016/j.humpath.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Heurich A., Lavender H., Gierer S., Danisch S., Perin P., Lucas J.M., Nelson P.S., Pöhlmann S., Soilleux E.J. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS ONE. 2012;7:e35876. doi: 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaydon D.C., Biancheri P., Di W.L., Plagnol V., Cabral R.M., Brooke M.A., van Heel D.A., Ruschendorf F., Toynbee M., Walne A., et al. Inflammatory skin and bowel disease linked to ADAM17 deletion. N. Engl. J. Med. 2011;365:1502–1508. doi: 10.1056/NEJMoa1100721. [DOI] [PubMed] [Google Scholar]
- Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Munch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camm C.F., Camm A.J. The cardiac effects of SARS-CoV2: COVID-19 special issue. Eur. Heart J. Case Rep. 2020;4(FI1):1–2. doi: 10.1093/ehjcr/ytaa351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369:1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocozza F., Névo N., Piovesana E., Lahaye X., Buchrieser J., Schwartz O., Manel N., Tkach M., Théry C., Martin-Jaular L. Extracellular vesicles containing ACE2 efficiently prevent infection by SARS-CoV-2 Spike protein-containing virus. J. Extracell. Vesicles. 2020;10:e12050. doi: 10.1002/jev2.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter K., Stransky L., McGuire C., Forgac M. Recent Insights into the Structure, Regulation, and Function of the V-ATPases. Trends Biochem. Sci. 2015;40:611–622. doi: 10.1016/j.tibs.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P.J., Steinberg F. To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 2018;19:679–696. doi: 10.1038/s41580-018-0053-7. [DOI] [PubMed] [Google Scholar]
- Czarzasta K., Cudnoch-Jedrzejewska A., Niemczyk L., Wrzesien R., Tkaczyk M., Puchalska L., Saracyn M., Zmudzki W., Niemczyk S. Effect of Chronic Kidney Disease on Changes in Vasopressin System Expression in the Kidney Cortex in Rats with Nephrectomy. BioMed Res. Int. 2018;2018:2607928. doi: 10.1155/2018/2607928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Jordan T.X., Wessels H.-H., Hoagland D.A., Kasela S., Legut M., Maniatis S., Mimitou E.P., Lu L., Geller E., et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell. 2020;184:92–105.e16. doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demiselle J., Fage N., Radermacher P., Asfar P. Vasopressin and its analogues in shock states: a review. Ann. Intensive Care. 2020;10:9. doi: 10.1186/s13613-020-0628-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshotels M.R., Xia H., Sriramula S., Lazartigues E., Filipeanu C.M. Angiotensin II mediates angiotensin converting enzyme type 2 internalization and degradation through an angiotensin II type I receptor-dependent mechanism. Hypertension. 2014;64:1368–1375. doi: 10.1161/HYPERTENSIONAHA.114.03743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkash E.A., Wilson A.M., Jentzen J.M. Ultrastructural Evidence for Direct Renal Infection with SARS-CoV-2. J. Am. Soc. Nephrol. 2020;31:1683–1687. doi: 10.1681/ASN.2020040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glebov O.O. Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing. FEBS J. 2020;287:3664–3671. doi: 10.1111/febs.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Jamett A.M., Momboisse F., Haro-Acuña V., Bevilacqua J.A., Caviedes P., Cárdenas A.M. Dynamin-2 function and dysfunction along the secretory pathway. Front. Endocrinol. (Lausanne) 2013;4:126. doi: 10.3389/fendo.2013.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Gupta A., Madhavan M.V., Sehgal K., Nair N., Mahajan S., Sehrawat T.S., Bikdeli B., Ahluwalia N., Ausiello J.C., Wan E.Y., et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga S., Yamamoto N., Nakai-Murakami C., Osawa Y., Tokunaga K., Sata T., Yamamoto N., Sasazuki T., Ishizaka Y. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc. Natl. Acad. Sci. USA. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M., Sakisaka S., Yoshitake M., Kin M., Ohishi M., Shakado S., Mimura Y., Noguchi K., Sata M., Tanikawa K. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPases, inhibits the receptor-mediated endocytosis of asialoglycoproteins in isolated rat hepatocytes. J. Hepatol. 1996;24:594–603. doi: 10.1016/s0168-8278(96)80146-2. [DOI] [PubMed] [Google Scholar]
- Heaton B.E., Trimarco J.D., Hamele C.E., Harding A.T., Tata A., Zhu X., Tata P.R., Smith C.M., Heaton N.S. SRSF protein kinases 1 and 2 are essential host factors for human coronaviruses including SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.08.14.251207. [DOI] [Google Scholar]
- Hoffmann R., Valencia A. A gene network for navigating the literature. Nat. Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- Hoffmann H.H., Sanchez-Rivera F.J., Schneider W.M., Luna J.M., Soto-Feliciano Y.M., Ashbrook A.W., Le Pen J., Leal A.A., Ricardo-Lax I., Michailidis E., et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. 2021;29:267–280.e5. doi: 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innamorati G., Le Gouill C., Balamotis M., Birnbaumer M. The long and the short cycle. Alternative intracellular routes for trafficking of G-protein-coupled receptors. J. Biol. Chem. 2001;276:13096–13103. doi: 10.1074/jbc.M009780200. [DOI] [PubMed] [Google Scholar]
- Inuzuka T., Fujioka Y., Tsuda M., Fujioka M., Satoh A.O., Horiuchi K., Nishide S., Nanbo A., Tanaka S., Ohba Y. Attenuation of ligand-induced activation of angiotensin II type 1 receptor signaling by the type 2 receptor via protein kinase C. Sci. Rep. 2016;6:21613. doi: 10.1038/srep21613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Murphy D. Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacometrics Syst. Pharmacol. 2013;2:e79. doi: 10.1038/psp.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamchandani K., Quintili A., Landis T., Bose S. Cardiac Arrhythmias in Critically Ill Patients With COVID-19: A Brief Review. J. of Cardiothorac. and Vasc. Anesth. 2020 doi: 10.1053/j.jvca.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornilov S.A., Lucas I., Jade K., Dai C.L., Lovejoy J.C., Magis A.T. Plasma levels of soluble ACE2are associated with sex, Metabolic Syndrome, and its biomarkers in a large cohort, pointing to a possible mechanism for increased severity in COVID-19. Crit. Care. 2020;24:452. doi: 10.1186/s13054-020-03141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert D.W., Yarski M., Warner F.J., Thornhill P., Parkin E.T., Smith A.I., Hooper N.M., Turner A.J. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2) J. Biol. Chem. 2005;280:30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W., et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal. 2011;17:10–12. [Google Scholar]
- Matsukawa T., Miyamoto T. Angiotensin II-stimulated secretion of arginine vasopressin is inhibited by atrial natriuretic peptide in humans. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011;300:R624–R629. doi: 10.1152/ajpregu.00324.2010. [DOI] [PubMed] [Google Scholar]
- McKusick V.A. Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkruys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oughtred R., Stark C., Breitkreutz B.J., Rust J., Boucher L., Chang C., Kolas N., O’Donnell L., Leung G., McAdam R., et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47(D1):D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presloid J.B., Novella I.S. RNA Viruses and RNAi: Quasispecies Implications for Viral Escape. Viruses. 2015;7:3226–3240. doi: 10.3390/v7062768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko E. The sequence of human ACE2 is suboptimal for binding the S spike protein of SARS coronavirus 2. bioRxiv. 2020 doi: 10.1101/2020.03.16.994236. [DOI] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rav-Acha M., Orlev A., Itzhaki I., Zimmerman S.F., Fteiha B., Bohm D., Kurd R., Samuel T.Y., Asher E., Helviz I., et al. Cardiac arrhythmias among hospitalized Coronavirus 2019 (COVID-19) patients: prevalence, characterization, and clinical algorithm to classify arrhythmic risk. Int. J. Clin. Pract. 2020 doi: 10.1111/ijcp.13788. [DOI] [PubMed] [Google Scholar]
- Reid I.A., Schwartz J., Ben L., Maselli J., Keil L.C. Interactions between vasopressin and the renin--angiotensin system. Prog. Brain Res. 1983;60:475–491. doi: 10.1016/S0079-6123(08)64414-3. [DOI] [PubMed] [Google Scholar]
- Ridwan R., Natzir R., Rasyid H., Patellongi I., Hatta M., Linggi E.B., Bukhari A., Bahrun U. Decreased Renal Function Induced by High-Fat Diet in Wistar Rat: The Role of Plasma Angiotensin Converting Enzyme 2 (ACE2) Biomed. Pharmacol. J. 2019;12:1279–1287. [Google Scholar]
- Sama I.E., Ravera A., Santema B.T., van Goor H., Ter Maaten J.M., Cleland J.G.F., Rienstra M., Friedrich A.W., Samani N.J., Ng L.L., et al. Circulating plasma concentrations of angiotensin-converting enzyme 2 in men and women with heart failure and effects of renin-angiotensin-aldosterone inhibitors. Eur. Heart J. 2020;41:1810–1817. doi: 10.1093/eurheartj/ehaa373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Luna J.M., Hoffmann H.H., Sanchez-Rivera F.J., Leal A.A., Ashbrook A.W., Le Pen J., Ricardo-Lax I., Michailidis E., Peace A., et al. Genome-Scale Identification of SARS-CoV-2 and Pan-coronavirus Host Factor Networks. Cell. 2021;184:120–132.e14. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti B., Cullen P.J. Actin-dependent endosomal receptor recycling. Curr. Opin. Cell Biol. 2019;56:22–33. doi: 10.1016/j.ceb.2018.08.006. [DOI] [PubMed] [Google Scholar]
- Su H., Yang M., Wan C., Yi L.X., Tang F., Zhu H.Y., Yi F., Yang H.C., Fogo A.B., Nie X., Zhang C. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98:219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., et al. HCA Lung Biological Network SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K., Tsang O.T., Yip C.C., Chan K.H., Wu T.C., Chan J.M., Leung W.S., Chik T.S., Choi C.Y., Kandamby D.H., et al. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clin. Infect. Dis. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020;94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Luo W., Huang L., Xiao J., Li F., Qin S., Song X., Wu Y., Zeng Q., et al. A comprehensive investigation of the mRNA and protein level of ACE2, the putative receptor of SARS-CoV-2, in human tissues and blood cells. Int. J. Med. Sci. 2020;17:1522–1531. doi: 10.7150/ijms.46695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K.A., Hayashi J.M., Carlson-Stevermer J., Zengel J.R., Richards C.M., Fozouni P., et al. Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses. Cell. 2021;184:106–119.e14. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Alfajaro M.M., DeWeirdt P.C., Hanna R.E., Lu-Culligan W.J., Cai W.L., Strine M.S., Zhang S.-M., Graziano V.R., Schmitz C.O., et al. Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell. 2020;184:76–91.e13. doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasin S., Costa A., Navarra P., Pozzoli G., Kostoglou-Athanassiou I., Forsling M., Grossman A. Endothelin-1 stimulates the in vitro release of neurohypophyseal hormones, but not corticotropin-releasing hormone, via ETA receptors. Neuroendocrinology. 1994;60:553–558. doi: 10.1159/000126796. [DOI] [PubMed] [Google Scholar]
- Yeung M.L., Yasunaga J., Bennasser Y., Dusetti N., Harris D., Ahmad N., Matsuoka M., Jeang K.T. Roles for microRNAs, miR-93 and miR-130b, and tumor protein 53-induced nuclear protein 1 tumor suppressor in cell growth dysregulation by human T-cell lymphotrophic virus 1. Cancer Res. 2008;68:8976–8985. doi: 10.1158/0008-5472.CAN-08-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung M.L., Houzet L., Yedavalli V.S., Jeang K.T. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J. Biol. Chem. 2009;284:19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung M.-L., Yao Y., Jia L., Chan J.F.W., Chan K.-H., Cheung K.-F., Chen H., Poon V.K.M., Tsang A.K.L., To K.K.W., et al. MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat. Microbiol. 2016;1:16004. doi: 10.1038/nmicrobiol.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung M.L., Jia L., Yip C.C.Y., Chan J.F.W., Teng J.L.L., Chan K.H., Cai J.P., Zhang C., Zhang A.J., Wong W.M., et al. Human tryptophanyl-tRNA synthetase is an IFN-γ-inducible entry factor for Enterovirus. J. Clin. Invest. 2018;128:5163–5177. doi: 10.1172/JCI99411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoufaly A., Poglitsch M., Aberle J.H., Hoepler W., Seitz T., Traugott M., Grieb A., Pawelka E., Laferl H., Wenisch C., et al. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir. Med. 2020;8:1154–1158. doi: 10.1016/S2213-2600(20)30418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional Supplemental Items, including supplementary figures, tables and video, are available from Mendeley Data: https://data.mendeley.com/datasets/gcdpsxd6d5/2. All sequencing data associated with this study have been deposited to NCBI with GEO accession number: GSE159272. The sequencing throughput and a summary of the read counts are available in Table S2 (see also Figure S5 ).
Figure S5.
Mapping of high-throughput sequencing reads to human genome, related to Figures 2, 3, and 7