Abstract
Three antifungal macrolides cyphomycin (1), caniferolide C (2) and GT-35 (3) were isolated from Streptomyces sp. ISID311, a bacterial symbiont associated with Cyphomyrmex fungus-growing ants. The planar structures of these compounds were established by 1 and 2D NMR data and MS analysis. The relative configurations of 1–3 were established using Kishís universal NMR database method, NOE/ROE analysis and coupling constants analysis assisted by comparisons with NMR data of related compounds. Detailed bioinformatic analysis of cyphomycin biosynthetic gene cluster confirmed the stereochemical assignments. Compounds 1–3 displayed high antagonism against different strains of Escovopsis sp., pathogen fungi specialized to the fungus-growing ant system. Compounds 1–3 also exhibited potent antiprotozoal activity against intracellular amastigotes of the human parasite Leishmania donovani with IC50 values of 2.32, 0.091 and 0.073 μM, respectively, with high selectivity indexes.
Keywords: Fungus-growing ants, Leishmania donovani, macrolides, polyketides, Streptomyces
Graphical Abstract
1. Introduction
Visceral leishmaniasis is a serious infection caused by two leishmanial species, Leishmania infantum and L. donovani.1 Leishmaniasis is transmitted by the bite of female phlebotomine sandflies. The disease occurs in parts of the tropics, subtropics, and southern Europe. An estimated 50 – 90 thousand new cases of visceral leishmaniasis occur worldwide annually. It is fatal if left untreated in over 95% of cases.2 Treatments of leishmaniasis include pentavalent antimonials, amphotericin B and its liposomal formulation, miltefosine, paromomycin and pentamidine. The last drug approved for treatment of this disease was miltefosine in 2014.3,4 Treatment of this protozoal disease remains costly, complex, and insufficient, therefore new therapeutics to combat leishmaniasis are needed.
Natural products play a major role in drug discovery and development.5 Actinobacteria have been the source of approximately two thirds of all clinically used natural product antibiotics, and of many anticancer, anthelmintic, and antifungal drugs.6 Indeed, the leishmanicidal drugs amphotericin and paromomycin are natural products discovered from Streptomyces species.7 Actinobacteria engage in complex defensive symbiotic interactions providing chemical defenses against pathogens to a number of different hosts, including insects such as fungus-growing ants, fungus-growing termites, stingless bees8,9, beetles and wasps.10–16 The fungus-growing ants (tribe Attini; subtribe Attina) collect plant and other material in the environment to cultivate a fungus for food inside their colonies.17 Symbiotic actinobacteria, usually present in the exoskeleton of the ants, produce small molecules that protect the fungal garden against the specialized pathogenic fungus Escovopsis sp.18 Due to their antifungal ecological role, these natural products can be explored to discover new therapeutic agents against human pathogens.19 Notably, the evolutionary constraints of this system have made it a fruitful discovery resource for antifungal compounds.15,16,18,20–23 As part of an International Cooperative Biodiversity Group (ICBG) project24, we were especially interested in the discovery of new antifungal, antibacterial and antiprotozoal secondary metabolites produced by bacterial symbionts of fungus-growing ants.24,25
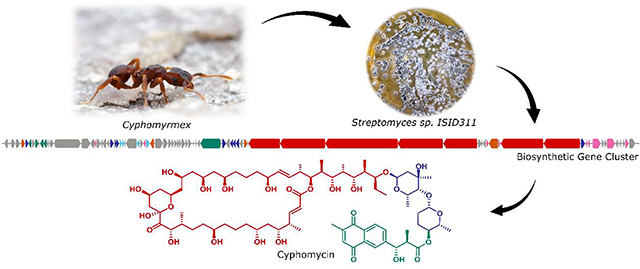
Streptomyces sp. ISID311 was isolated from the exoskeleton of Cyphomyrmex ants and was recently found to produce cyphomycin (1), a new polyketide showing in vivo efficacy in murine models of fungal infections.15 In this initial report, we only established the planar structure of 1 as well as its in vitro and in vivo antifungal activities. Here we describe the complete structural determination of cyphomycin (1), including the relative and absolute configurations using extensive NMR analyses and in silico analysis of the biosynthetic gene cluster. We also report the isolation of two structurally related compounds (2 and 3), as well as the potent antileishmanial activities of the three compounds against intracellular amastigotes and promastigotes.
2. Materials and methods
2.1. General Experimental Procedures
Reversed Phase HPLC was performed using a Shimadzu Prominence HPLC system. The HPLC purification was carried out at 4 mL min−1 with a C18 semipreparative column (Phenomenex Luna, C18(2), 5μm, 250 × 10 mm). The mass spectrometry data of 1 and 3 were acquired with a Bruker MaXis Ultra-High Resolution Quadrupole Time-of-Flight MS coupled to Waters Acquity UPLC system operated by Bruker Hystar software; and for 2, the data were acquired with a UPLC (Shimadzu) coupled to a micrOTOF II mass spectrometer (Bruker Daltonics). The 1D and 2D NMR spectra of 1–3 were recorded in CD3OD with a Bruker Avance 500 MHz spectrometer (500I) with 5 mm a 13C/15N(1H) cryogenic probe, National Magnetic Resonance Facility at Madison, Wisconsin, USA. ROESY, NOESY and NOESY-1D data of compound 1-3 were recorded with 500 MHz (DRX-500) (Bruker UK, Coventry, UK) at the Department of Chemistry, Faculty of Philosophy, Sciences and Letters at Ribeirão Preto, University of São Paulo, Brazil. E.COSY data for 1 were recorded using a Bruker Avance III 600 MHz spectrometer equipped with a 5 mm 13C/15N(1H) TCI cryogenic probehead located at the Department of Chemistry of Federal University of São Carlos, Brazil. Specific optical rotations were recorded using a P-2000 digital polarimeter (Jasco, Tokyo, Japan).
2.2. Collection and isolation of actinobacteria from fungus-growing ant
Collections of biological samples and research on genetic resources were authorized in Brazil by SISBIO #46555–5 and CNPq #010936/2014–9. ISID311 was isolated from winged males Cyphomyrmex ants collected in campus of the University of São Paulo – Ribeirão Preto (GPS coordinates: S 21°10’04.4” W 47°50’49.5”) on October of 2015. The ants were washed with 500 μL of sterile deionized water, vortexed for 30 s and then plated on chitin medium supplemented with the antifungals nystatin and cycloheximide (per liter: 4 g chitin, 0.7 g K2HPO4, 0.3 g KH2PO4, 0.5 g MgSO4·5H2O, 0.01 g FeSO4·7H2O, 0.01 g ZnSO4·7H2O, 0.01 g MnCl2·4H2O, 20 g of agar, 0.04 g/L nystatin, and 0.05 g/L cycloheximide). After two weeks of growth at 28 °C, bacterial colonies were subcultured onto International Streptomyces Project Medium 2 (ISP-2) agar pH 7.2 with antifungals (0.04 g/L nystatin, and 0.05 g/L cycloheximide).26
2.3. DNA Extraction and Sequencing of Actinobacteria
The DNA extraction procedure was modified from Kumar et al.27, in which the pellet was washed in 500 μL of 10.3% sucrose, centrifuged for 1 min at 10.000 g and the supernatant discarded. Then 450 μL of TSE buffer pH 7.8 supplemented with lysozyme (20 mg/mL) were added and incubated for 20–30 min at 37 °C. After, 13 μL of proteinase K was added and incubated for another 15 min at 55 °C and then 250 μL of 2% SDS, gently mixed until to form a clear solution. Then 300 μL of phenol: chloroform pH 8.0 were added and mixed and centrifuged for 10 min at 4 °C. The supernatant was transferred to another tube and 60 μL of 3 M NaOAc pH 6.0, plus 700 μL of isopropanol were added. The contents were mixed until “white strings” appeared and then centrifuged for 1 min to 10.000 g, and the supernatant discarded. The pellet was washed with 70% ethanol and centrifuged again at 10.000 g for 1 min. After drying the ethanol completely, the DNA was resuspended in 30 μL of deionized H2O.
Some steps were added for sequencing of actinobacterial genomes. The pellets were resuspended in 500 μL TE buffer (10 mM Tris–HCl, 0.1 mM EDTA; pH 8.1) plus 10 μL of RNAse and the tubes were rocked for 15 min at room temperature to dissolve the pellet and then 100 μL 5 M NaCl pH 6.0 were added and mixed. Following, 70 μL of CTAB/NaCl (10 mL 1M Tris (pH 8.4), 5 mL 0.5M EDTA (pH 8.0), 28 mL 5M NaCl, 2 g CTAB, 57 mL of deionized H2O) were added, mixed and incubated for 10 min at 55 °C and then cooled to 37 °C. After this, 500 μL of chloroform were mixed and centrifuged for 10 min at room temperature. The top layers were transferred to a new tube and 300 μL of phenol and 150 μL of chloroform were mixed and centrifuged for 10 min at 4 °C. The top layers were transferred with a wide bore tip to a tube containing 300 μL of CHCl3, mixed and centrifuged briefly for 1 min and then the top layers were transferred to a tube containing 50 μL of NaOAc pH 6.0 plus 400 μL of isopropanol and mixed until “white strings” appear and solution become uniform. The DNA were centrifuged briefly, poured off the supernatant and the remainder pipetted off. And then, the DNA were resuspended in 100–200 μL of ultra-pure H2O. The DNA were quantified and checked for quality with NanoDrop® (Thermo Scientific).
2.4. PCR Amplification
PCR amplification of the 16S rRNA gene of actinobacteria was performed using the primers: 27F and 1492R. The EconoTac® DNA Polymerase Kit (Lucigen, USA) was used and the final reaction volume of 15 μL contained: 8 μL Econotaq, 0.5 μL of each primer 27F and 1492R, 0.5 μL DMSO, 4.5 μL Deionized H2O and 1 μL DNA (10 ng/μL). Amplification followed the following profile: an initial denaturation step at 94 °C for 3 min followed by 32 cycles of amplification of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 2 minutes and a final extension step of 72 °C for 5 min. The PCR product was detected by agarose gel electrophoresis and visualized by ultraviolet (UV) fluorescence after staining with ethidium bromide.
2.5. Sequencing and purification reaction
The primers 27F and 1492R were used again for the sequencing of the 16S rRNA gene. The sequencing reaction of the PCR products contained: 1.5 μL 5X buffer, 1 μL primer (10 μM), 1 μL BigDye 3.1 (Applied Biosystems), 0.5 μL DMSO, 1 μL PCR product DNA and deionized water to make up the total volume of 10 μL. The program used consisted of 95 °C for 3 min, followed by 35 cycles of 96 °C for 10 s, 58 °C for 3 min and a final extent of 72 °C for 7 min. The sequencing reaction was purified with the Axyprep Mag Dyeclean purification kit (Axygen) in which 5 μL of magnetic beads solution and 31 μL of 85% ethanol were added for each reaction. The tubes were placed on a magnetic plate for 3 min and then the liquid was removed. 100 μL of 85% ethanol was added for 30 s and then the liquid was discarded. 100 μL of 85% ethanol was added again for 30 s and after discarded. The liquid was removed as much as possible with a pipette. The DNA was resuspended in 25 μL of deionized H2O.
2.6. Sequencing, editing sequences and bioinformatics analysis
Sequencing was performed at the Center for Genetics and Biotechnology at the University of Wisconsin - Madison (Biotech Center, UW - Madison, WI, USA). The sequences were edited and used for assembly of the contigs in the SecMan Pro Software (DNASTAR). Contigs were used to search for homologous sequences in the NCBI - GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and Eztaxon (http://www.ezbiocloud.net/eztaxon/identify).
The genome was analyzed in AntiSMASH 4.028 to identify all the possible BGC present in the 103 contigs. All the nucleotide and amino acid alignments were performed in Geneious Prime 2020.1.1 using Muscle 3.8.425.29 The AT domains phylogenetic tree was elaborated by the stochastic algorithm IQ-TREE.30 The GenBank assembly accession for Streptomyces sp ISID311 is GCA_007994955.1 (https://www.ncbi.nlm.nih.gov/assembly/GCF_007994955.1/).
2.7. Purification of compounds 1–3
The bacterium Streptomyces sp. ISID311 was grown in broth A-medium (20 g soluble starch, 10 g glucose, 5 g peptone, 5 g yeast extract, 5 g CaCO3 per liter) using Fernbach flasks [5 × (1 L of medium in a flask of 2.8 L + 70 g of HP20)] and placed in shakers for 7 days at 28 °C and 200 rpm. The HP20 was filtered and washed with distilled water and soaked with acetone. The organic solvent was filtered and dried under vacuum. Then, a partition of ethyl acetate/water was performed, and the organic phase was separated and dried to give the crude extract (2.3044 g). It was purified by SPE-C18 (55 μm, 20 g) using the following gradient: 200 mL (20% MeOH-H2O, A1: 72.8 mg); 200 mL (40% MeOH-H2O, A2: 152.5 mg); 200 mL (60% MeOH-H2O, A3: 132.3 mg); and 200 mL (100% MeOH, A4: 1.9349 g).
The fraction A4 was purified by semi-preparative HPLC using the column C18 semi preparative (Phenomenex Luna, C18(2), 5μm, 250 × 10 mm) and the following gradient of MeOH and H2O (containing 0.1% of acetic acid) at 4 mL/min: min 1–20 (linear gradient from 80% MeOH-H2O to 100% MeOH); min 20–22 (isocratic flow of 100% MeOH); min 22–22.5 (linear gradient from 100% MeOH to 80% MeOH-H2O); and min 22.5–27.5 (isocratic flow of 80% MeOH-H2O) to yield 1 (89.0 mg, tR = 11.3 min), 2 (14.2 mg, tR = 11.8 min) and 3 (33.7 mg, tR = 12.3 min).
2.8. Agar-based antifungal bioassay
Fractions or pure compounds were tested against four different Escovopsis strains (ICBG711, ICBG740, ICBG741 and ICBG1251, isolated from Trachymyrmex, Acromyrmex, Apterostigma and Atta colony, respectively), one Trichoderma ICBG1100 (isolated from garden of an Attine colony), and one Cunninghamella ICBG752 (isolated from garden of an Apterostigma colony). Each sample-fungi challenge was replicated two times and done on ISP-2 agar. Bacteria were placed in the center of ISP-2 agar petri dishes and grown alone during 7 days; fungal strains were then point-inoculated near the edge of the culture (3 cm from the center for Escovopsis, Trichoderma and Cunninghamella). Compounds (100 μg) or positive control (100 μg of miconazole) were placed directly in the center of the ISP-2 agar plate. Challenges were monitored during 21 days at 28 °C.19
2.9. In vitro evaluation of natural products on Leishmania donovani.
Leishmania donovani axenic cultures (MHOM/ET/67/HU3) were maintained in M199 medium (pH 7.4) supplemented with 10% heat-inactivated fetal calf serum and grown at 28 °C.31 Stock solutions of compounds were prepared in 100% DMSO at 10 mM. For the assays, compounds were diluted in culture medium and tested at 10 concentrations (0.125 to 64 μM, 2-fold serial dilutions). In the promastigote assay, fluorescence measures were performed in 96-well flat-bottom microtiter plates, with each well containing 100 μL of culture medium with an initial population of 1×105 L. donovani cells obtained from axenic cultures in logarithmic growth. In negative control wells (100% parasite growth) all assay components were added except for the testing compounds. As positive control, the reference drug miltefosine was used (0.5 to 64 μM, 2-fold serial dilutions). After 72 hours of incubation, the plates were inspected using an inverted microscope to assure sterile conditions. Next, 10 μL of Alamar Blue®32 (12.5 mg resazurin dissolved in 100 mL distilled water) was added to each well and the plates were incubated for 3 hours. This indicator of cell viability permeates into viable parasites, where it is reduced to the highly fluorescent compound resorufin.33,34 After this incubation time the plates were read with a microplate fluorometer under an excitation wavelength of 536 nm and an emission wavelength of 588 nm. Growth inhibition was expressed as a percentage of the fluorescence of the negative control wells (100% parasite growth). The IC50 values were determined using SigmaPlot. Dose-response curves were fitted using log inhibitor concentration vs. normalized response (between 0% and 100%) with variable slope, and IC50 was automatically calculated. In the intracellular amastigote assay, THP-1 cells were seeded at 2 × 104/well (RPMI-1640 medium, 100 μL) in 96-well flat-bottom microtiter plates with phorbol 12-myristate 13-acetate (PMA) at 20 ng/mL. After incubation for 72 hours (5% CO2, 37 °C), medium was aspirated and late-stage promastigotes were added (2 × 105/well, 100 μL). After 24 hours of incubation, medium was aspirated to clear extracellular parasites, compounds were added in serial dilutions (100 μL) and the plates were incubated for 120 hours. All plates included negative controls and miltefosine as a positive control. Following incubation, the medium was removed, and the cells were fixed in methanol and stained with Giemsa. The average number of intracellular amastigotes per THP-1 cell was determined using an inverted microscope.35 Growth inhibition was expressed as a percentage of the average number of amastigotes per macrophage in the negative control wells. IC50 values were determined as described for the promastigote assay. For the selectivity assay, THP-1 cells were seeded at 2 × 104/well (RPMI-1640, 100 μL) in 96-well flat-bottom microtiter plates with PMA at 20 ng/mL.36 After incubation for 72 hours (5% CO2, 37 °C), medium was aspirated, compounds were added in serial dilutions (100 μL) and the plates were incubated for 120 h. All plates included negative controls and doxorubicin as a positive control. Following incubation, 10 μL of Alamar Blue® was added to each well and the plates were incubated for 3 hours. Next, the plates were read with a microplate fluorometer under an excitation wavelength of 536 nm and an emission wavelength of 588 nm. Growth inhibition was expressed as a percentage of the fluorescence of the negative control wells. IC50 values were determined as described for the promastigote and amastigote assays.
3. Results and discussion
The bacterium Streptomyces sp. ISID311 showed strong antagonism against different Escovopsis strains (Fig. S1–S4). Further, the crude extract obtained from bacterial culture in A-medium strongly inhibited the growth of L. donovani (94% at 20 μg/mL). Bioassay-guided fractionation led to the isolation of three antifungal structurally related macrolides cyphomycin (1), caniferolide C (2) and GT-35 (3) (Fig. 1).
Fig. 1.
Compounds isolated from bacterium Streptomyces sp. ISID311
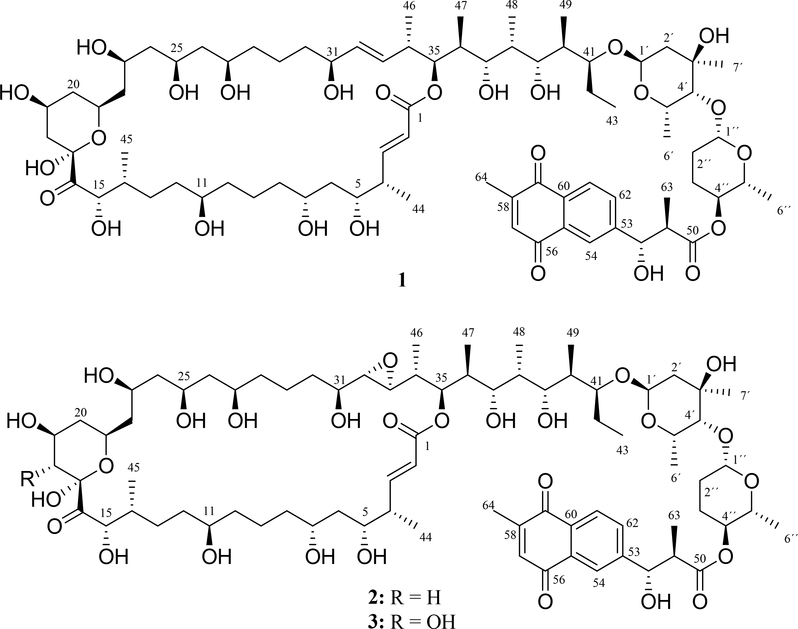
The molecular formula of 1 was determined to be C77H122O26 based on positive ion HRESIMS ([M + H]+ m/z 1463.8282, err 1.0 ppm) (Fig. S5). The 1H and 13C NMR spectral data of 1 (Fig. S6–S14) are shown in Table 1. The 13C NMR spectrum showed 77 signals assigned to 12 methyl, 18 methylene, 36 methine, 5 carbonyl carbons, 4 tertiary carbons sp2 and 2 quaternary carbon groups by multiplicity-edited gHSQC experiment (Fig. S15). The gCOSY and HSQC-TOCSY spectra revealed connectivity from H-2 to H-15 and H-18 to H-43 (Fig. S16–S19). The connection of these two spin systems was revealed with the HMBC correlation of H-18 (δH 2.09 and 1.33) to C-16 and C-17; H-15 (δH 4.69) to C-16; and H-35 (δH 5.14) to C-1 (Fig. S20–S22). The presence of two sugar units was evidenced by 1H NMR and 13C NMR data, mainly signals of two anomeric protons and carbons at δH 5.01 (H-1´) and δH 4.58 (H-1´´); and δC 96.4 (C-1´) and δC 104.0 (C-1´´), respectively (Figure S7, S8 and S13). The first sugar unit was established as α-axenose by HMBC, COSY, ROESY correlations (Figure 2A and Figure S16, S23–S28) and NMR data comparison of sugar units of caniferolide A-D37. There are two 1H spin systems from H-1´ to H-2´ and from H-4´ to H-6´; and HMBC correlations from methyl H-7´ to C-2´, C-3´ and C4´. The connection of this sugar to side chain of macrolactone was observed with HMBC correlations of H-1´ to C-41 (Figure S23). The small coupling constant of H-1´ and H-4śuggested they should be in equatorial orientation. The ROE correlation of H-41 to H-5´ supported the axial orientation of H-5´. The second sugar unit was established as β-amicetose. The chemical shifts of β-amicetose were compared to the natural products caniferolides A-D37. This sugar was assigned by the 1H spin system from H-1´´ to H-6´´ by HMBC and COSY spectra (Figure S16, S23–S25, S27 and S28). The large coupling constants 3JH-1´´,H-2´áx (7.6 Hz) and 3JH-4´´,H-5´´ (9.3 Hz); and ROESY/NOESY correlation between H-1´´ and H-5´´ (Figure S28 and S29) suggested the axial orientation of H-1´´, H-4´´ and H-5´´. The connection of both sugars was observed by the HMBC correlation of H-1´´ to C-4´ (Figure S23). The HMBC correlation of H-4´´ to C-50 connected the β-amicetose with naphthoquinone moiety (Figure S21). The naphthoquinone derivative moiety in 1 has identical NMR data (Table 1 and Fig. S30–S32) as observed for caniferolides A-D37. So, configuration of C-51 and C-52 is also anti (Figure 2B). Extensive NMR and MS analyses suggested that compound 1 was the macrolide natural product cyphomycin15. The key COSY, HSQC-TOCSY and HMBC correlations of 1 are shown in Fig. S33. Other similar macrolides also have been described, such as compound axenomycin B38, GT-3539, PM100117 and PM10011840, deplelides A and B41, astolides A and B42, and recently caniferolides A-D37 (Fig. S34). We reported previously the specific rotation value for cyphomycin15 as −96.4. We obtained a new value (+25.5) in other measurement after a carefully calibration of polarimeter with a reference, which is more acceptable based in the literature37,40.
Table 1.
1H and 13C NMR (MeOH-d4, 500/125 MHz) data of cyphomycin (1), caniferolide C (2) and GT-35 (3)
| No. | 1 δC | 1 δH, mult. (J in Hz) | 2 δC | 2 δH, mult. (J in Hz) | 3 δC | 3 δH, mult. (J in Hz) |
|---|---|---|---|---|---|---|
| 1 | 168.7 | --- | 168.6 | --- | 168.6 | --- |
| 2 | 122.1 | 5.86, br d (15.8) | 122.3 | 5.92, d (15.8) | 122.3 | 5.93, d (15.8) |
| 3 | 153.5 | 7.06, dd (15.8, 6.8) | 154.0 | 7.10, dd (15.8, 6.7) | 154.0 | 7.09, dd (15.8, 6.9) |
| 4 | 43.5 | 2.47, m | 43.4 | 2.49, m | 43.3 | 2.50, m |
| 5 | 74.2 | 3.78, m | 74.2 | 3.82, m | 74.1 | 3.84, m |
| 6 | 41.7 | 1.60, m | 41.7 | 1.50–1.65 | 41.6 | 1.53–1.67, m |
| 7 | 71.8 | 3.78, m | 71.9 | 3.79, m | 71.8 | 3.81, m |
| 8 | 38.9 | 1.32–1.62, m | 39.0 | 1.36–1.54, m | 38.8 | 1.33–1.60, m |
| 9 | 23.0 | 1.45–1.68, m | 23.1 | 1.46–1.56, m | 22.9 | 1.46–1.56, m |
| 10 | 38.5 | 1.32–1.62, m | 38.5 | 1.35–1.56 | 38.4 | 1.31–1.65, m |
| 11 | 72.9 | 3.51, m | 72.4 | 3.52, m | 72.8 | 3.51, m |
| 12 | 36.4 | 1.38–1.47, m | 36.4 | 1.40–1.46, m | 36.2 | 1.35–1.45, m |
| 1.53–1.63 m | 1.52–1.61, m | 1.52–1.69, m | ||||
| 13 | 31.9 | 1.69, m | 32.1 | 1.31, m | 31.6 | 1.19–1.27, m |
| 1.30, m | 1.69, m | 1.45–1.76, m | ||||
| 14 | 36.2 | 2.25, m | 36.3 | 2.23, m | 36.7 | 2.17, m |
| 15 | 75.8 | 4.69, d (2.2) | 75.8 | 4.67, d (2.1) | 75.4 | 4.59, d (3.9) |
| 16 | 210.6 | --- | 210.6 | --- | 208.7 | --- |
| 17 | 99.4 | --- | 99.4 | --- | 99.7 | --- |
| 18 | 41.1 | 2.09, m | 41.2 | 2.08, m | 75.2 | 3.49, d (9.2) |
| 1.33, m | 1.33, m | |||||
| 19 | 64.9 | 4.08, m | 64.9 | 4.07, m | 69.6 | 3.87, m |
| 20 | 42.0 | 1.96, m | 42.0 | 1.94, m | 40.9 | 1.98, m |
| 1.21, m | 1.22, m | 1.43, m | ||||
| 21 | 67.7 | 4.20, m | 67.8 | 4.20, m | 67.4 | 4.24, m |
| 22 | 45.6 | 1.45–1.74, m | 45.6 | 1.51–1.68, m | 45.1 | 1.47–1.72, m |
| 23 | 65.4 | 4.08, m | 65.4 | 4.07, m | 65.5 | 4.00, m |
| 24 | 46.3 | 1.43–1.67, m | 46.3 | 1.53, m | 46.2 | 1.40–1.69, m |
| 25 | 68.2 | 4.04, m | 68.4 | 4.06, m | 68.5 | 4.05, m |
| 26 | 46.0 | 1.40–1.76, m | 45.7 | 1.58, m | 45.6 | 1.39–1.75, m |
| 27 | 71.3 | 3.79, m | 71.6 | 3.79, m | 71.7 | 3.76, m |
| 28 | 38.4 | 1.32–1.62, m | 38.5 | 1.35–1.56 | 38.5 | 1.31–1.66, m |
| 29 | 22.7 | 1.51, m | 22.4 | 1.34–1.40, m | 22.5 | 1.30–1.39, m |
| 1.60–1.66, m | 1.61–1.70, m | |||||
| 30 | 38.4 | 1.32–1.62, m | 33.8 | 1.28–1.39, m | 33.8 | 1.28–1.38, m |
| 1.40–1.44, m | 1.40–1.49, m | |||||
| 31 | 72.4 | 3.94, m | 70.9 | 3.43, m | 71.0 | 3.42, m |
| 32 | 135.2 | 5.53, dd (15.6, 4.5) | 63.7 | 2.78, m | 63.7 | 2.78, dd (4.1, 2.0) |
| 33 | 133.1 | 5.49, dd (15.6, 7.2) | 60.6 | 2.73, br d (8.2) | 60.6 | 2.73, dd (8.4, 2.0) |
| 34 | 40.4 | 2.54, m | 40.5 | 1.58, m | 40.5 | 1.59, m |
| 35 | 77.3 | 5.14, br d (9.5) | 75.1 | 5.33, br d (10.4) | 75.1 | 5.33, br d (10.4) |
| 36 | 38.7 | 1.97, m | 38.5 | 1.94, m | 38.6 | 1.94, m |
| 37 | 78.5 | 3.39, br d (9.5) | 78.0 | 3.37, m | 78.0 | 3.38, br d (9.5) |
| 38 | 36.0 | 1.81, m | 36.1 | 1.81, m | 36.1 | 1.81, m |
| 39 | 79.4 | 3.52, m | 79.5 | 3.51, br d (7.5) | 79.5 | 3.52, dd (9.2, 2.3) |
| 40 | 38.5 | 2.00, m | 38.5 | 2.00, m | 38.5 | 1.99, m |
| 41 | 80.5 | 3.88, m | 80.5 | 3.87, m | 80.5 | 3.87, m |
| 42 | 22.9 | 1.64, m | 23.0 | 1.62, m | 22.9 | 1.63, m |
| 1.36, m | 1.40, m | 1.40, m | ||||
| 43 | 11.5 | 0.98, t (7.2) | 11.5 | 0.99, t (7.5) | 11.5 | 0.99, t (7.4) |
| 44 | 14.1 | 1.09, d (6.7) | 13.6 | 1.11, d (6.5) | 13.5 | 1.11, d (6.8) |
| 45 | 13.6 | 0.79, d (6.4) | 13.5 | 0.79, d (6.5) | 13.8 | 0.84, d (6.8) |
| 46 | 17.3 | 1.03, d (6.5) | 14.5 | 1.07, d (6.5) | 14.4 | 1.07, d (6.8) |
| 47 | 9.7 | 0.90, d (6.8) | 9.4 | 0.86, d (6.7) | 9.4 | 0.86, d (6.9) |
| 48 | 5.0 | 0.90, d (6.8) | 4.9 | 0.89, d (6.7) | 4.9 | 0.89, d (6.9) |
| 49 | 10.6 | 0.78, d (6.4) | 10.6 | 0.78, d (6.5) | 10.6 | 0.78, d (6.9) |
| 1´ | 96.4 | 5.01, br s | 96.4 | 5.00, br s | 96.4 | 5.01, br s |
| 2´ | 37.9 | 1.93, dd (3.70, 14.0) | 38.0 | 1.92, br d (13.8) | 38.0 | 1.91, dd (3.0, 14.0) |
| 1.55, d (14.0) | 1.55, br d (13.8) | 1.56, d (14.0) | ||||
| 3´ | 71.2 | --- | 71.2 | --- | 71.2 | --- |
| 4´ | 81.7 | 3.31, m | 81.7 | 3.31, br s | 81.7 | 3.31, br s |
| 5´ | 64.7 | 4.44, m | 64.7 | 4.44, m | 64.7 | 4.44, m |
| 6´ | 17.3 | 1.21, d (6.6) | 17.3 | 1.21, d (5.7) | 17.3 | 1.21, d (5.7) |
| 7´ | 27.5 | 1.24, s | 27.5 | 1.24, s | 27.5 | 1.24, s |
| 1´´ | 104.0 | 4.58, br d (7.6) | 104.0 | 4.58, br d (7.7) | 104.0 | 4.58, br d (7.0) |
| 2´´ | 31.3 | 2.01, m | 31.4 | 2.01, m | 31.4 | 2.02, m |
| 1.56, m | 1.56, m | 1.57, m | ||||
| 3´´ | 28.6 | 2.10, m | 28.6 | 2.10, m | 28.6 | 2.10, m |
| 1.56, m | 1.55, m | 1.56, m | ||||
| 4´´ | 74.4 | 4.45, m | 74.4 | 4.44, m | 74.4 | 4.44, m |
| 5´´ | 74.5 | 3.54, dq (6.1, 9.3) | 74.5 | 3.54, m | 74.5 | 3.52, dq (6.3, 9.6) |
| 6´´ | 18.5 | 1.19, d (6.1) | 18.5 | 1.19, d (5.7) | 18.5 | 1.19, d (6.3) |
| 50 | 175.5 | --- | 175.6 | --- | 175.6 | --- |
| 51 | 48.9 | 2.78, dq (8.5, 7.1) | 49.0 | 2.78, dq (7.0, 8.6) | 49.0 | 2.79, dq (8.6, 7.0) |
| 52 | 76.4 | 4.86, d (8.5) | 76.5 | 4.85, d (8.6) | 76.5 | 4.85, d (8.6) |
| 53 | 150.3 | --- | 150.4 | ---- | 150.4 | --- |
| 54 | 125.3 | 8.01, br s | 125.3 | 8.02, br s | 125.3 | 8.02, d (1.5) |
| 55 | 133.5 | --- | 133.6 | --- | 133.6 | --- |
| 56 | 186.2 | --- | 186.3 | --- | 186.2 | --- |
| 57 | 136.5 | 6.88, br d (1.4) | 136.5 | 6.89, d (1.1) | 136.5 | 6.89, br d (1.3) |
| 58 | 149.8 | --- | 149.9 | --- | 149.9 | --- |
| 59 | 186.3 | --- | 186.3 | --- | 186.3 | --- |
| 60 | 132.9 | --- | 133.0 | --- | 133.0 | --- |
| 61 | 127.6 | 8.07, d (8.0) | 127.6 | 8.08, d (8.0) | 127.6 | 8.08, d (8.0) |
| 62 | 133.2 | 7.79, dd (8.0, 1.3) | 133.2 | 7.79, br d (8.0) | 133.2 | 7.79, dd (8.1, 1.2) |
| 63 | 14.3 | 0.94, d (7.1) | 14.3 | 0.94, d (7.0) | 14.3 | 0.94, d (7.0) |
| 64 | 16.4 | 2.17, s | 16.4 | 2.17, br s | 16.4 | 2.17, d (1.3) |
Fig. 2.
Relative configuration proposed for fragment: A) disaccharide and B) naphthoquinone units.
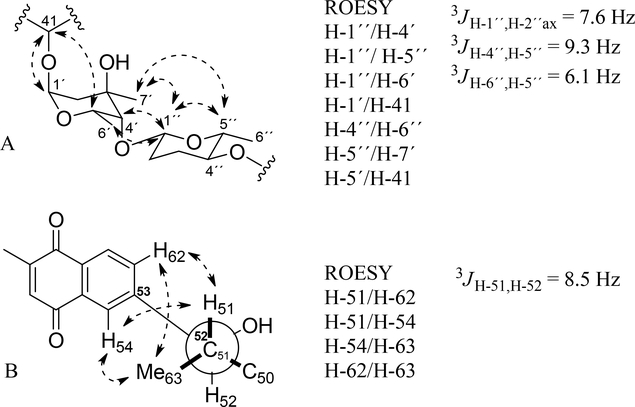
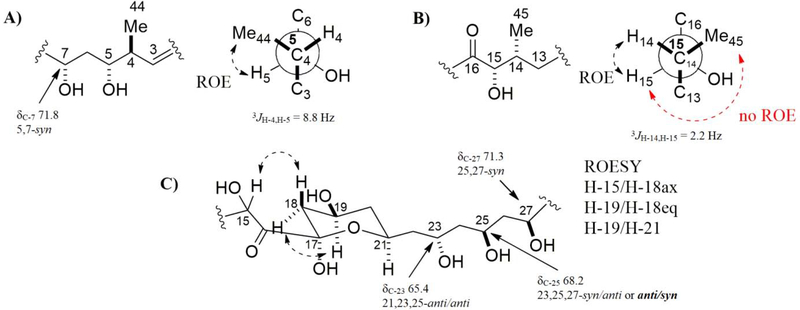
The partial relative configuration of the macrolactone moiety of 1 was determined using ROESY, NOESY-1D, E.COSY, J-based configuration analysis and Kishís universal NMR database method.43,44 The coupling constants were measured by 1H NMR and E.COSY spectra (Figure S35–S38). The 1H-1H ROESY correlation of H-5/H-44 (Figure S39), large coupling constant 3JH-4,H-5 (8.8 Hz) (Figure S36) and NMR data comparison with caniferolide A-D37 indicated a 5,44-syn configuration (Figure 3A). The configuration of C-5 as syn with C-7 was proposed based in shift of C-7 (δC-7 71.8) and data set I of Kishís universal NMR data base (Figure S40). The 1H-1H ROESY correlation of H-14/H-15; the no ROE correlation of H-15/H-45; and small coupling constant 3JH-14,H-15 (2.2 Hz) suggested the syn configuration of H-14 with H-15 (Figure 3B and Figure S41). The 1H-1H ROESY correlation of H-18ax/H-15, H-18eq/H-19, H-19/H-21 and H-21/H-23 inferred a chair conformation of the tetrahydropyran moiety with equatorial substituents, C-16, 19-OH and C-22, and axial hydroxyl group at C-17 (Figure 3C, Figure S42 and S43). The relative configurations of oxymethine carbons C-23, C-25 and C-27 were assigned as anti/syn using 13C NMR signals (δC 65.4, 68.2 and 71.3, respectively) and data set I and II of Kishís universal NMR data base (Figure S40). The NMR data of C-2 to C-27 of 2 and caniferolide C37 is identical, suggesting the same relative configuration.
Fig. 3.
Relative configuration proposed for fragments: A) C-4 to C-7, B) C-14 to C-15, and C) C-17 to C-27.
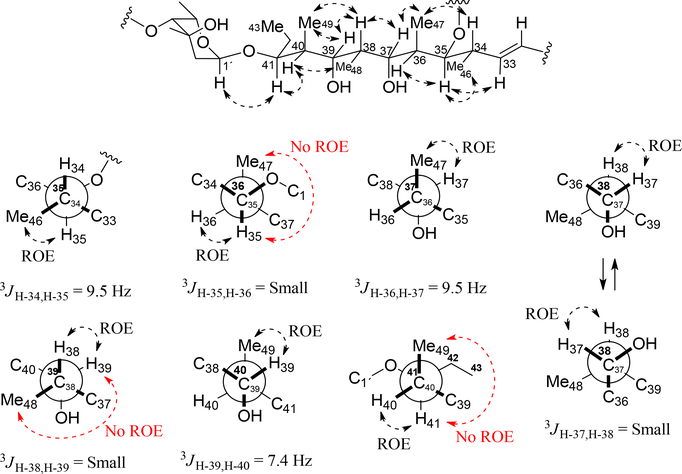
The 1H-1H ROESY correlation of H-33/H-35, H-35/H-46, H-35/H-36, H-34/H-47, H-37/H-47, H-37/H-38, H-38/H-39, H-38/H-49, H-39/H-49, H-40/H-48, H-40/H41 and H-41/H-1´ (Figure S39, S41, S44–S46 and Figure 4); the absence of 1H-1H ROESY correlation of H-35/H-47, H-39/H-48 and H-41/H-49; the large coupling constants 3JH-34,H-35 (9.5 Hz), 3JH-36,H-37 (9.5 Hz) and 3JH-39,H-40 (7.4 Hz); small coupling constants 3JH-35,H-36, 3JH-37,H-38 and 3JH-38,H-39; data set III of Kishís universal NMR data base for configuration of C-37 to C-39 [δC-48 5.0, 37,38,39-syn/syn] (Figure S40); and NMR data comparison with caniferolide A-D37 suggested that relative configuration of C-34 to C-41 in 1 is identical. The difference between 1 and caniferolide C is in C-32/C-33. Compound 1 has a double bond in C-32 (δC 135.2) and C-33 (δC 133.1); while caniferolide C has an epoxide in C-32 (δC 63.7) and C-33 (δC 60.6).
Fig. 4.
Relative configuration proposed for fragment of C-32 to C-43.
The molecular formula of 2 was determined to be C77H122O27 based on positive ion HRESIMS ([M + H]+ m/z 1479.8212, err 2.3 ppm) (Fig. S47). Compound 2 has one additional oxygen atom than compound 1. The differences between compound 1 and 2 are in carbons C-32 and C-33, based on the 13C NMR, gCOSY, gHSQC, gHMBC, gROESY and NOESY-1D (Table 1 and Fig. S48–S61), suggesting that compound 2 is the antifungal macrolide caniferolide C37. The molecular formula of 3 was determined to be C77H122O28 based on positive ion HRESIMS ([M + H]+ m/z 1495.8189, err 0.4 ppm) (Fig. S62). Compound 3 has one additional oxygen atom than 2. The difference between compound 2 and 3 is in carbon C-18, based in the 1 and 2 D NMR data (Table 1 and Fig. S63–S76), suggesting that compound 3 is the patented antifungal macrolide GT-35 (Fig. S34), isolated from Streptomyces sp.39, and recently published as caniferolide B37.
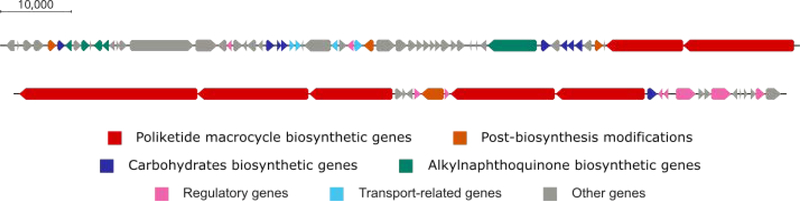
The absolute configuration of caniferolides A-D was recently established by combination of NMR with bioinformatics analysis of the biosynthetic gene cluster.37 With these findings the absolute configuration of some related macrolides, such as PM100117/8, deplelide A and B, and astolide A and B were deduced. The cyphomycin (1) type 1 polyketide synthase BGC predicted by antiSMASH is 194 kb length.28 However, analysis of genes near the predicted cluster boundaries showed 16 additional genes downstream that appear to be related to the biosynthesis of cyphomycin. Therefore, the proposed BGC is 220 kb in length and contains 83 genes (Fig. 5), 50 of them have a high similarity with genes present in the caniferolides A-D BGC37 (Table S1).
Fig. 5.
Cyphomycin biosynthetic gene cluster.
The polyketide macrocycle biosynthesis can be performed by seven type I PKS encoded by cph58-cph62, cph69, and cph70. The seven multimodular PKS is composed of 21 modules, one load (Load M), and 20 extension modules (M1-M20) (Fig. S77). Comparative analysis showed that the amino acid sequence from all the seven PKSs have more than 85% of similarity with the PKSs encoded by scaP1-scaP7, present on S. caniferus CA-271066 BGC. Moreover, the domains organizations in the 21 modules remains the same.37 The substrate specificity of the AT domains was predicted by phylogenetic analysis with the amino acid sequences of the 21 AT domains from cyphomycin PKSs, 47 AT domains known as malonyl-CoA specific, and 44 AT domains known as methylmalonyl-CoA specific.30,45 Seven domains (Load M, M1-M4, M14, and M19) clustered with the 44 methylmalonyl-CoA selective, and the remaining 14 clustered with the 47 malonyl-CoA selective (Fig. S78). The sequences for the DH and ER domains were also aligned to confirm their activity based on conserved motifs (Fig. S79 and S80).46
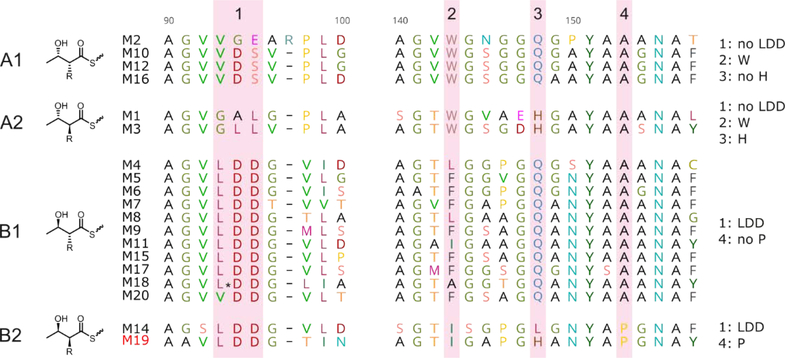
Four conserved motifs were analyzed in the alignment of the 19 KR domains present in the cyphomycin PKSs. Specific residues present in these motifs are responsible to lead the stereochemistry of the chiral centers of the polyketide. The presence of LDD residues in motif 1 characterizes the type B KR domain, which produces a S β-hydroxyl group. In contrast, type A KR domain does not have the LDD motif and produces a R β-hydroxyl group. Residues present in motifs 1, 2, and 3 are responsible for the stereochemistry of α-alkyl groups (Fig. 6).47 The amino acid sequence of all the 19 KR domains of cyphomycin PKSs have more than 80% similarity with the KR domains of caniferolides A-D, and all the residues in the motifs are conserved (Fig. S81).
Fig. 6.
Alignment of the catalytic region of the 19 KR domains. The pattern of resides in the four highlighted motifs, specified in the last column, is important to predict the stereochemistry of the β-hydroxyl and the α-alkyl groups.
Some modifications are performed post PKS biosynthesis. Cytochrome P450, such the ones encoded by genes cph05, cph35, cph56, and cph67, can be responsible for the epoxidation in the double bond predicted at C32 and C33, and oxidations at C16, C25 and C18 (Fig. S82). The hydroxyl group in C25 also could be explained by a domain skipping mechanism in the DH and ER domains present in module M09. Alignments of all cph BGC DHs and the erythromycin DH448 show that cph DH9 has an intact active site and no major gaps, suggesting it is functional and that the M09 hydroxyl is likely installed post-assembly line by one of the BGC’s cytochrome P450s. Additionally, the S configuration predicted for the α-methyl at C4 generated in Module 19 does not match with the NMR data. Caniferolides A-D biosynthesis presented the same discrepancy, which is probably a result of additional methyl-epimerizing activity.37 The amino acid sequence of all the proteins involved in the biosynthesis of the carbohydrates and the alkylnaphthoquinone in the structures 1–3 have more than 84% of similarity with the equivalent proteins in the caniferolides A-D biosynthesis.37
Glucose-1-phosphate is the precursor for L-axenose and D-aminocetose biosynthesis, both share the same firsts four biosynthetic steps, which are performed by a glucose-1-phosphate thymidylyltransferase (cph54), two dehydratases (cph53 and cph71), and a ketoreductase (cph06). The remaining steps for D-aminocetose biosynthesis are performed by a dehydratase (cph50) and an oxidoreductase (cph52 or cph26) and for L-axenose biosynthesis, a methyltransferase (cph24), an epimerase (cph25) and also an oxidoreductase (cph52 or cph26) (Fig. S83).
Alkylnaphthoquinone biosynthesis starts with four proteins encoded by the cph12, cph11, cph09, and cph07 that lead the biosynthesis of 1,4-dihydroxy-6-naphtoic acid (DH6N), an intermediate of menaquinone biosynthesis via futalosine.48 DH6N is methylated in the C3, but there is no methyltransferase gene in the cyphomycin BGC. A gene that encodes a protein with 78.8% of similarity with GonMT, methyltransferase responsible for this methylation in PM100117/8 biosynthesis49, was found in another part of the Streptomyces sp. ISID311 genome. Oxidations of hydroxyl groups are likely performed by some of the four cytochrome P450s, and a PKS coded by cph49 adds a methylmalonyl unit in the carboxylic acid (Fig. S84).
Antifungal and antileishmanial activities of isolated compounds
Compounds 1-3 have a similar macrolactone with sugar units and a methylnaphthoquinone as the natural products axenomycin B38, PM100117 and PM10011840, deplelides A and B41, astolides A and B42, and caniferolides A-D37 (Fig. S34). These compounds have interesting antifungal, antibiotic, antitumor and immunosuppressive properties.37,38,40–42 This explains the antifungal activity of Streptomyces sp. ISID311 and compounds 1-3 against four different Escovopsis sp. strains (Fig. S1–S4 and S85). These metabolites showed inhibition zones similar to that of the positive control miconazole. Streptomyces sp. ISID311 and compounds 1–3 did not inhibit the opportunistic fungi Trichoderma ICBG1100 and Cunninghamella ICBG752 (Fig. S86 and S87), showing selectivity against Escovopsis species.
Macrolides 1-3 showed higher activity against both L. donovani forms than the positive control miltefosine, and are likely the compounds responsible for the high antileishmanial activity observed for the extract. The presence of the epoxide group in C-32 and C-33 in the macrolactone moiety of compounds 2 and 3 increased both the antileishmanial activity and selectivity index (Table 2). This is the first report of the antileishmanial properties of this family of macrolides, and their good selectivity indexes highlights these compounds as new hits for the development of antileishmanial drugs. The most structurally similar compound of 1–3 is amphotericin B, a well-known polyene macrolide therapeutically used as antifungal and antileishmanial agent. However, 36-membered macrocycles 1–3 differ from 38-membered macrocycle amphotericin B in the lower number of double bonds, in the absence of the amino sugar and in the presence of the alkylnaphthoquinone moiety.
Table 2.
Activity of compounds 1–3 on L. donovani intracellular amastigotes, promastigotes and THP-1 cells
| Compounds | IC50 (μM) Intracellular amastigotes | IC50 (μM) Promastigotes | CC50 (μM) THP-1* | Selectivity Index** |
|---|---|---|---|---|
| 1 | 2.32 ± 0.13 | 0.118 ± 0.011 | 11.48 ± 1.11 | 4.94 |
| 2 | 0.091 ± 0.010 | 0.034 ± 0.004 | 6.40 ± 0.46 | 70.32 |
| 3 | 0.073 ± 0.010 | 0.046 ± 0.009 | 6.07 ± 0.82 | 83.15 |
| Doxorubicin | --- | --- | 0.58 ± 0.04 | --- |
| Miltefosine | 5.33 ± 0.74 | 4.98 ± 0.37 | 60.73 ± 2.21 | 11.39 |
Data are shown as mean ± SD (n = 3 biological replicates)
THP-1 human leukemia macrophages (host cells of L. donovani)
Selectivity index = CC50 THP-1/IC50 intracellular amastigotes
Compounds 1-3 were inactive against B. subtilis (NRS 231), E. coli (ATCC 25922), methicillin-sensitive S. aureus (MSSA) (Wichita) and P. aeruginosa (Boston 41501) using an agar-based antibiotic assay.50 So, compounds 1-3 showed higher selectivity against Leishmania and Escovopsis.
4. Conclusion
The absolute configuration of 1 was elucidated by combination of NMR with bioinformatics analysis of the biosynthetic gene cluster. It has the same configuration of all common chiral centers as caniferolides A-D37.
Modifying the alkene group in C-32 and C-33 of 1 by an epoxide group, as in 2 and 3, improved considerable the antileishmanial activity and selectivity index. The three compounds were more active than miltefosine, the last drug approved for treatment of different types of Leishmaniasis. Actinobacteria in the genus Streptomyces have already proved their value as sources of antileishmanial drugs amphotericin and paromomycin. Our research group also had previously reported the identification of antileishmanial natural products from other Streptomyces strains associated with fungus-growing ants.51 The isolation of compounds 1–3 from a fungus-growing ant associated Streptomyces supports the concept of insect microbiomes as sources of antimicrobial natural products.
Supplementary Material
Acknowledgements
We acknowledge the financial support from São Paulo Research Foundation (FAPESP) grants #2013/50954-0 (MTP), #2014/14095-6 (HEO), #2016/20154-0 (HEO), # 2019/16559-3 (VBL), #2015/01001-6 (WGPM), #2013/07600-3 (ADA, CIBFar), #2013/25658-9 (LLGF), and #2016/17614-0 (WGPM); and Fogarty International Center, National Institutes of Health (FIC-NIH) grant U19TW009872; Conselho Nacional de Desenvolvimento Científico e Tecnológico – Brasil (CNPq), grants #409514/2016-0 and #303792/2018-2 (MTP); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) – Finance Code 001; Additional support for MGC provided by grant 2020-67012-31772 (accession 1022881) from the USDA National Institute of Food and Agriculture.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary material
NMR and HRMS spectra of compounds 1-3. This material is available free of charge via the Internet at https://doi.org/10.1016/j.bmc.2021.116016.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chappuis F, Sundar S, Hailu A, et al. Visceral leishmaniasis: what are the needs for diagnosis, treatment and control? Nat Rev Microbiol. 2007;5:873–882. doi: 10.1038/nrmicro1748 [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. https://www.who.int/news-room/fact-sheets/detail/leishmaniasis; 2020. Accessed 12 December 2020.
- 3.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055 [DOI] [PubMed] [Google Scholar]
- 4.Sunyoto T, Potet J, Boelaert M. Why miltefosine—a life-saving drug for leishmaniasis—is unavailable to people who need it the most. BMJ Glob Heal. 2018;3:e000709. doi: 10.1136/bmjgh-2018-000709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod. 2020;83:770–803. doi: 10.1021/acs.jnatprod.9b01285 [DOI] [PubMed] [Google Scholar]
- 6.Barka EA, Vatsa P, Sanchez L, et al. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol Rev. 2016;80:1–43. doi: 10.1128/MMBR.00019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiwari N, Gedda MR, Tiwari VK, Singh SP, Singh RK. Limitations of current therapeutic options, possible drug targets and scope of natural products in control of leishmaniasis. Mini-Reviews Med Chem. 2018;18:26–41. doi: 10.2174/1389557517666170425105129 [DOI] [PubMed] [Google Scholar]
- 8.Rodríguez-Hernández D, Melo WGP, Menegatti C, Lourenzon VB, Do Nascimento FS, Pupo MT. Actinobacteria associated with stingless bees biosynthesize bioactive polyketides against bacterial pathogens. New J Chem. 2019;43:10109–10117. doi: 10.1039/c9nj01619h [DOI] [Google Scholar]
- 9.Menegatti C, Lourenzon VB, Rodríguez-Hernández D, et al. Meliponamycins: antimicrobials from stingless bee-associated Streptomyces sp. J Nat Prod. 2020;83:610–616. doi: 10.1021/acs.jnatprod.9b01011 [DOI] [PubMed] [Google Scholar]
- 10.Beemelmanns C, Ramadhar TR, Kim KH, et al. Macrotermycins A-D, Glycosylated macrolactams from a termite-associated Amycolatopsis sp. M39. Org Lett. 2017;19:1000–1003. doi: 10.1021/acs.orglett.6b03831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim KH, Ramadhar TR, Beemelmanns C, et al. Natalamycin A, an ansamycin from a termite-associated Streptomyces sp. Chem Sci. 2014;5:4333–4338. doi: 10.1039/c4sc01136h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroiss J, Kaltenpoth M, Schneider B, et al. Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat Chem Biol. 2010;6:261–263. doi: 10.1038/nchembio.331 [DOI] [PubMed] [Google Scholar]
- 13.Poulsen M, Oh DC, Clardy J, Currie CR. Chemical analyses of wasp-associated Streptomyces bacteria reveal a prolific potential for natural products discovery. PLoS One. 2011;6:e16763. doi: 10.1371/journal.pone.0016763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haeder S, Wirth R, Herz H, Spiteller D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc Natl Acad Sci. 2009;106:4742–4746. doi: 10.1073/pnas.0812082106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chevrette MG, Carlson CM, Ortega HE, et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat Commun. 2019;10:516. doi: 10.1038/s41467-019-08438-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chevrette MG, Currie CR. Emerging evolutionary paradigms in antibiotic discovery. J Ind Microbiol Biotechnol. 2019;46:257–271. doi: 10.1007/s10295-018-2085-6 [DOI] [PubMed] [Google Scholar]
- 17.Schultz TR, Brady SG. Major evolutionary transitions in ant agriculture. Proc Natl Acad Sci. 2008; 2008;105:5435–5440. doi: 10.1073/pnas.0711024105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh DC, Poulsen M, Currie CR, Clardy J. Dentigerumycin: A bacterial mediator of an ant-fungus symbiosis. Nat Chem Biol. 2009;5:391–393. doi: 10.1038/nchembio.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Currie CR, Scott JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic producing bacteria tocontrol garden parasites. Nature. 1999;398:701–704. doi: 10.1038/19519 [DOI] [Google Scholar]
- 20.Sit CS, Ruzzini AC, Van Arnam EB, Ramadhar TR, Currie CR, Clardy J. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc Natl Acad Sci. 2015;112:13150–13154. doi: 10.1073/pnas.1515348112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Arnam EB, Ruzzini AC, Sit CS, Currie CR, Clardy J. A rebeccamycin analog provides plasmid-encoded niche defense. J Am Chem Soc. 2015;137:14272–14274. doi: 10.1021/jacs.5b09794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chevrette MG, Gutiérrez-García K, Selem-Mojica N, et al. Evolutionary dynamics of natural product biosynthesis in bacteria. Nat Prod Rep. 2020;37:566–599. doi: 10.1039/c9np00048h [DOI] [PubMed] [Google Scholar]
- 23.Caldera EJ, Chevrette MG, McDonald BR, Currie CR. Local adaptation of bacterial symbionts within a geographic mosaic of antibiotic coevolution. Appl Environ Microbiol. 2019;85:e01580–19. doi: 10.1128/AEM.01580-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pupo MT, Currie CR, Clardy J. Microbial symbionts of insects are the focus of the first International Cooperative Biodiversity Group (ICBG) in Brazil. J Braz Chem Soc. 2017;28:393–401. doi: 10.21577/0103-5053.20160284 [DOI] [Google Scholar]
- 25.Menegatti C, Fukuda TTH, Pupo MT. Chemical Ecology in insect-microbe interactions in the Neotropics. Planta Med. 2020. doi: 10.1055/a-1229-9435 [DOI] [PubMed] [Google Scholar]
- 26.Poulsen M, Currie CR. Symbiont interactions in a tripartite mutualism: exploring the presence and impact of antagonism between two fungus-growing ant mutualists. PLoS One. 2010;5:e8748. doi: 10.1371/journal.pone.0008748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar V, Bharti A, Gusain O, Bisht GS. An improved method for isolation of genomic DNA from filamentous actinomycetes. J Eng Technol Manag. 2010;2:10–13. [Google Scholar]
- 28.Blin K, Wolf T, Chevrette MG, et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36–W41. doi: 10.1093/nar/gkx319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta S Visceral leishmaniasis: experimental models for drug discovery. Indian J Med Res. 2011;133:27–39. doi:IndianJMedRes_2011_133_1_27_76700 [pii] [PMC free article] [PubMed] [Google Scholar]
- 32.Ioset JR, Brun R, Wenzler T, Kaiser M, Yardley V. Drug screening for Kinetoplastids diseases. A training manual for screening in neglected diseases. DNDi and Pan-Asian Screening Network. 2009. [Google Scholar]
- 33.Shimony O, Jaffe CL. Rapid fluorescent assay for screening drugs on Leishmania amastigotes. J Microbiol Methods. 2008;75:196–200. doi: 10.1016/j.mimet.2008.05.026 [DOI] [PubMed] [Google Scholar]
- 34.Mikus J, Steverding D. A simple colorimetric method to screen drug cytotoxicity against Leishmania using the dye Alamar Blue®. Parasitol Int. 2000;48:265–269. doi: 10.1016/S1383-5769(99)00020-3 [DOI] [PubMed] [Google Scholar]
- 35.Zulfiqar B, Shelper TB, Avery VM. Leishmaniasis drug discovery: recent progress and challenges in assay development. Drug Discov Today. 2017;22:1516–1531. doi: 10.1016/j.drudis.2017.06.004 [DOI] [PubMed] [Google Scholar]
- 36.Karwaciak I, Gorzkiewicz M, Bartosz G, Pulaski L. TLR2 activation induces antioxidant defence in human monocyte-macrophage cell line models. Oncotarget. 2017;8:54243–54264. doi: 10.18632/oncotarget.17342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pérez-Victoria I, Oves-Costales D, Lacret R, et al. Structure elucidation and biosynthetic gene cluster analysis of caniferolides A-D, new bioactive 36-membered macrolides from the marine-derived: Streptomyces caniferus CA-271066. Org Biomol Chem. 2019;17:2954–2971. doi: 10.1039/c8ob03115k [DOI] [PubMed] [Google Scholar]
- 38.Arcamone F, Barbieri W, Franceschi G, Penco S, Vigevani A. Axenomycins. I. The structure of chromophore and sugar moieties. J Am Chem Soc. 1973;95:2008–2009. doi: 10.1021/ja00787a048 [DOI] [PubMed] [Google Scholar]
- 39.Takahashi I, Seike Y, Uosaki Y, Ochiai K. Fungicidal GT35 manufacture with Streptomyces. JP 09100290. 1997. [Google Scholar]
- 40.Pérez M, Schleissner C, Fernández R, et al. PM100117 and PM100118, new antitumor macrolides produced by a marine Streptomyces caniferus GUA-06–05-006A. J Antibiot (Tokyo). 2016;69:388–394. doi: 10.1038/ja.2015.121 [DOI] [PubMed] [Google Scholar]
- 41.Takeuchi T, Hatano M, Umekita M, et al. ATP Depletion assay led to the isolation of new 36-membered polyol macrolides deplelides A and B from Streptomyces sp. MM581-NF15. Org Lett. 2017;19(16):4207–4210. doi: 10.1021/acs.orglett.7b01807 [DOI] [PubMed] [Google Scholar]
- 42.Alferova VA, Novikov RA, Bychkova OP, et al. Astolides A and B, antifungal and cytotoxic naphthoquinone-derived polyol macrolactones from Streptomyces hygroscopicus. Tetrahedron. 2018;74:7442–7449. doi: 10.1016/j.tet.2018.11.015 [DOI] [Google Scholar]
- 43.Yoshihisa K, Jinhwa L, Kenichi T, Kishi Y. Toward creation of a universal NMR database for the stereochemical assignment of acyclic compounds: the case of two contiguous propionate units. 1999;1:2177–2180. doi: 10.1021/OL9903786 [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi Y, Tan CH, Kishi Y. Toward creation of a universal NMR database for stereochemical assignment: the case of 1,3,5-trisubstituted acyclic systems. Helv Chim Acta. 2000;83:2562–2571. doi: [DOI] [Google Scholar]
- 45.Yadav G, Gokhale RS, Mohanty D. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J Mol Biol. 2003;328:335–363. doi: 10.1016/S0022-2836(03)00232-8 [DOI] [PubMed] [Google Scholar]
- 46.Bachmann BO, Ravel J. Chapter 8 Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 2009;458:181–217. doi: 10.1016/S0076-6879(09)04808-3 [DOI] [PubMed] [Google Scholar]
- 47.Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009 [DOI] [PubMed] [Google Scholar]
- 48.Keatinge-Clay A Crystal structure of the erythromycin polyketide synthase dehydratase. J Mol Biol. 2008;384:941–953. doi: 10.1016/j.jmb.2008.09.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salcedo RG, Olano C, Fernández R, et al. Elucidation of the glycosylation steps during biosynthesis of antitumor macrolides PM100117 and PM100118 and engineering for novel derivatives. Microb Cell Fact. 2016;15:187. doi: 10.1186/s12934-016-0591-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adnani N, Vazquez-Rivera E, Adibhatla SN, et al. Investigation of interspecies interactions within marine Micromonosporaceae using an improved co-culture approach. Mar Drugs. 2015;13:6082–6098. doi: 10.3390/md13106082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ortega HE, Ferreira LLG, Melo WGP, et al. Antifungal compounds from Streptomyces associated with attine ants also inhibit Leishmania donovani. Fischer K, ed. PLoS Negl Trop Dis. 2019;13:e0007643. doi: 10.1371/journal.pntd.0007643 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.