Abstract
A large number of hormonal biosynthetic or signaling pathways genes controlling shoot branching are widely known for their roles in regulating plant growth and development, operating in synergetic or antagonistic manner. However, their involvement in abiotic stress response mechanism remains unexplored. Initially, we performed an in silico analysis to identify potential transcription binding sites for the basic leucine zipper 62 transcription factor (bZIP62 TF) in the target branching related genes. The results revealed the presence of cis-regulatory elements specific to two bZIP TFs, AtbZIP18 and AtbZIP69, rather than AtbZIP62. Interestingly, these bZIP TFs were previously proposed to be negatively regulated by the AtbZIP62 TF under salinity in Arabidopsis. Therefore, we investigated the transcriptional regulation of more axillary branching (MAX, strigolactone), PIN-FORMED (PINs, auxin carriers), gibberellic acid (GA)-biosynthetic genes as well as isopentenyltransferase (IPT, cytokinin biosynthesis pathway) genes in response to drought stress in Arabidopsis Col-0 wild type. In addition, in the perspective of exploring the transcriptional interplay of the selected genes with the AtbZIP62, we measured their expression by qPCR in the atbzip62 (lacking the AtbZIP62 gene) background under the same conditions. Our findings revealed that the expression of AtMAX2, AtMAX3, and AtMAX4 was differentially regulated by drought stress between the atbzip62 and Col-0 wild type, but not AtMAX1. Similarly, the transcripts accumulation of AtPIN3 and AtPIN7 (known as auxin efflux carriers), and that of the AtAXR1 showed similar regulation patterns in atbzip62. However, AtPIN1 expression was downregulated in Col-0, but no change was observed in atbzip62. Furthermore, AtIPT5 and AtIPT7 exhibited a differential transcripts accumulation pattern in atbzip62 and Col-0 wild type (WT). In the same way, the expression of the GA biosynthetic genes AtGA2ox1 and AtGA20ox2, and that of AtRGA1 were differentially regulated in atbzip62 compared to the Col-0. Meanwhile, AtGA2ox1 showed a similar expression pattern with Col-0. Therefore, all results suggest PIN, MAX, IPT, and GA-biosynthetic genes, which are differentially regulated by AtbZIP62 transcription factor, as emerging candidate genes that could be involved in drought stress response mechanism in Arabidopsis.
Keywords: PIN-FORMED, more axillary branching, isopentenyltransferase, gibberellic acid, AtbZIP62, transcription factor, drought tolerance, Arabidopsis
1. Introduction
Due to their sessile nature, plants are often subjected to various abiotic stresses induced by enviromental stimuli. Abiotic stresses cause major loss to crop yield [1,2,3], and drought stress is considered as one of the major threats to the life and productivity of plants [4]. Drought can impair the growth of the plant in various ways, leading to changes in metabolic functions. One among them is the deteroriation of the photosynthetic pigments resulting in a reduced light harvesting capacity, which ultimately results in the reduction of plants biomass [4]. However, like other abiotic stresses, the key impact of drought stress is the genereation of highly reactive and toxic molecules known as reactive oxygen species (ROS) [5,6], which have the ability to induce oxidative stress. The increased production of ROS during environmental stresses may cause oxidative damage, leading to peroxidation of lipids, oxidation of protiens, inhibition of enzymes, damage to nucleic acids, and induction of programmed cell death (PCD) that culminates in cell death [7,8,9,10].
To survive, plants have developed sophisticated mechanisms, including the activation of antioxidant (enzymatic and non-enzymatic) systems [11,12]. Both systems are assumed crucial to maintain at a controlled level the accumulation of ROS, while tending to maintain a balanced reduction-oxidation state within the cell [13]. Under the same conditions, a transcriptional reprogramming within the cell occurs, which includes the activation or suppression of stress responsive genes, coupled with an active interaction between genes or between proteins and DNA [14,15,16].
Arabidopsis has been recognized as the model plant species for dicots [17,18,19], and has served as an ideal plant species for plant biosciences research in the last two decades. This Brassicaceae offers a wide range of opportunities to study molecular functions of genes, in part due to its relatively small genome size and its short life cycle, coupled with the available T-DNA insertion lines generated by the Arabidopsis Biological Research Center (ABRC).
Transcription factors (TFs) are regulators of the expression of genes in biological systems. Intrinsic to their mode of action is their ability to bind to cis-regulatory elements found in the promoter region of genes [20], operating either alone or in complex with other molecules to activate or repress the recruitment of the basal transcriptional machinery to specific genes [21], thereby determining when and where the target genes are transcribed, how many proteins are synthesized, and what the phenotype looks like. Interactions between proteins and DNA are fundamental to nearly all biological processes of all biological systems [22]. Basic leucine zipper (bZIP) proteins [23] are transcription factors (TFs) involved in diverse developmental processes, including plant growth, flowers development, seeds maturation, and signaling during abiotic and biotic stresses [24].
In Arabidopsis thaliana (Arabidopsis hereafter) genome, about 75 distinct members of the bZIP family have been reported [23], including the AtbZIP62 [24]. AtbZIP62 belongs to the group I of bZIP TFs superfamily having the G-box binding factor 1 (GBF1)-like domain. The GBFs contain an N-terminal proline-rich domain in addition to the bZIP domain. GBFs have been also reported by Ábrahám et al. [25] to be involved in the developmental and physiological processes in response to various stimuli, such as light or hormones.
For several years, phytohormones have been shown to play fundamental and diverse roles in the metabolism of plants, including seed dormancy and germination, plant growth and development, flowering and organogenesis, seed formation and maturation, fruit ripening, and signaling during abiotic or biotic stress occurrence [26]. However, their possible involvement in the adaptive response mechanism under abiotic stress remained unexplored for several decades.
Therefore, this study aimed at investigating the transcriptional regulation of key hormonal biosynthetic or signaling pathways genes, previously known for their roles in the regulation of shoot branching in plants, in response to drought stress. In this perspective, we monitored by qPCR the transcripts accumulation of auxin carriers (PIN-FORMED protein) encoding genes, strigolactone biosynthetic genes (more axillary branching, MAX), gibberellic acid (GA) biosynthetic genes, and cytokinin (CK) biosynthetic genes (isopentenyltransferase, IPT) in Arabidopsis Col-0 wild type under drought stress conditions. In addition, in order to explore the transcriptional interplay of the target genes with the AtbZIP62 TF, their expression levels were analyzed in the atbzip62 knockout plants (lacking the AtbZIP62 gene recently suggested to be involved in the adaptive response towards drought [27] and salinity [28] tolerance) compared to the Col-0 wild type (WT). Moreover, the phenotype of atbzip62 plants was examined under normal growth conditions.
2. Results
2.1. AtbZIP62 TF Could Be Involved in the Control of Bud Outgrowth in Plants
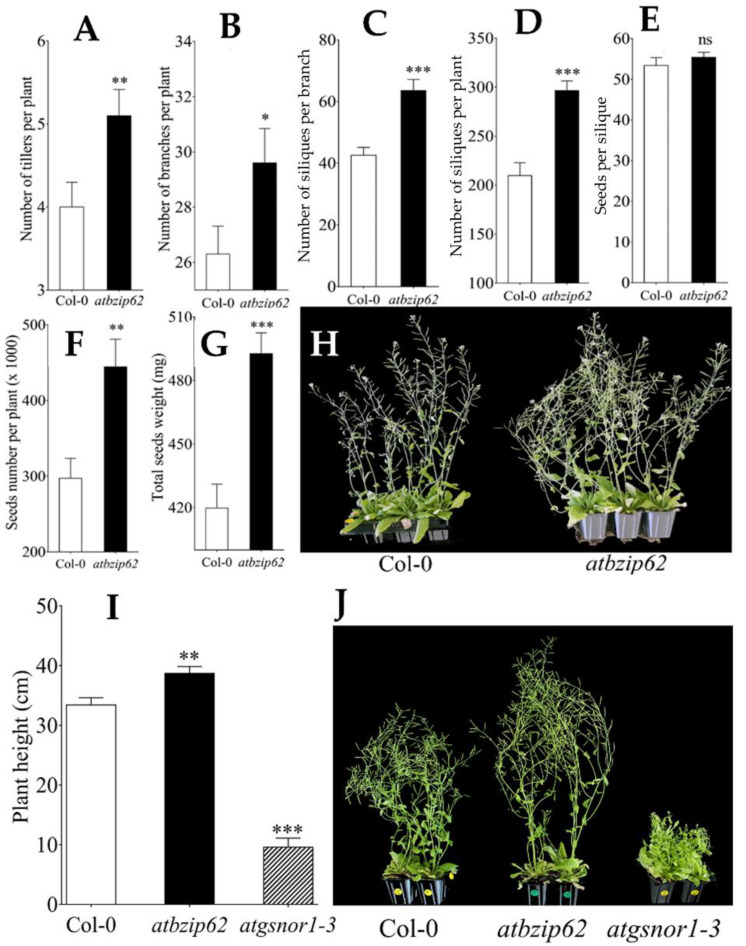
Initially, we were interested to see the phenotype of the atbzip62 knockout plants under normal growth conditions. Therefore, we measured the growth related parameters and the productivity of the atbzip62 compared to that of the Col-0 wild type. Interestingly, our data show that atbzip62 plants had a high branching phenotype under normal growth conditions. In essence, we recorded a significant increase in the number of tillers per plant (27.5%) compared to Col-0 WT (Figure 1A,H) and the number of secondary branches per plant (activated bud outgrowth) (12.6%) (Figure 1B). Consequently, more siliques were formed per branch (49.3%) (Figure 1C) and more siliques were produced per plants (41.4%) (Figure 1D), which profoundly resulted in more seeds per silique (3.7%) (Figure 1E) and more seeds per plant (49.6%) (Figure 1F). In addition, we recorded an increase in seeds weight by 17.4% compared to Col-0 WT (Figure 1G). We also observed that the atbzip62 had taller plants compared to Col-0 WT. For this reason, we were interested to visualise the difference in the plant height between the atbzip62 and the atgsnor1-3 (known to have a stunt and high branching phenotype). As shown in the panels I and J of the Figure 1, atbzip62 and atgsnor1-3 plants exhibited distinctive growth habits (promoted in atbzip62 and inhibited in atgsnor1-3).
Figure 1.
Morphology of atbzip62 plants grown under normal conditions. (A) Number of tillers per plant, (B) number of branches per plant, (C) number of siliques per branch, (D) number of siliques per plant, (E) number of number seeds per silique, (F) number of seeds per plant, (G) seeds weight per plant, (H) phenotype of Arabidopsis atbzip62 showing an increased number of tillers compared to Col-0 wild type, (I) plant height, and (J) phenotype of atbzip62 plants showing an increased in plant height compared to Col-0 and atgsnor1-3. *** p < 0.001, ** p < 0.01, * p < 0.05, ns non-significant.
2.2. In Silico Prediction of Transcription Factor Binding Sites Identified bZIPs Cis-Regulatory Elements
Prior to analyzing the transcripts accumulation of the selected branching related genes, we performed an in silico analysis in order to identify potential binding sites of the bZIP62 TF within the promoter region of the target genes. The results revealed the presence cis-regulatory elements specific to the AtbZIP18 and AtbZIP69, among others (Table 1). These two bZIP TFs were selected because their expression was recently suggested to be negatively regulated by the AtbZIP62 TF in response to salt stress [28]. These results would imply that the AtbZIP62 TF may interact with the AtbZIP18 and/or AtbZIP69 in order to regulate the transcription of each of the target branching genes.
Table 1.
Transcription factor binding sites prediction.
| Gene ID | Gene Name | Target Genes | Position | Strand | p-Value | q-Value | Matched Sequence |
|---|---|---|---|---|---|---|---|
| * | * | AtMAX1 | * | * | * | * | * |
| * | * | AT2G26170 | * | * | * | * | * |
| AtMAX2 | |||||||
| AT2G40620 | AtbZIP18 | AT2G42620 | 1723–1733 | - | 2.14 × 10−5 | 0.046 | CTCGGCTGGCC |
| AT2G40620 | AtbZIP18 | AT2G42620 | 923–933 | + | 7.04 × 10−6 | 0.0303 | TTCAGCTGTCA |
| AT1G06070 | AtbZIP69 | AT2G42620 | 923–933 | + | 1.41 × 10−5 | 0.0583 | TTCAGCTGTCA |
| AtMAX3 | |||||||
| AT2G40620 | At bZIP18 | AT2G44990 | 1704–1714 | - | 6.59 × 10−5 | 0.295 | AATAGCTGTCG |
| AtMAX4 | |||||||
| AT2G40620 | AtbZIP18 | AT4G32810 | 2535–2545 | - | 2.48 × 10−5 | 0.158 | CACGGCTGTCT |
| AT2G40620 | AtbZIP18 | AT4G32810 | 285–295 | + | 8.64 × 10−5 | 0.276 | CAGAGCTGTAA |
| AT1G06070 | AtbZIP69 | AT4G32810 | 2535–2545 | - | 6.02 × 10−5 | 0.369 | CACGGCTGTCT |
| AtPIN1 | |||||||
| AT1G06070 | AtbZIP69 | AT1G73590 | 1935–1945 | - | 2.08 × 10−5 | 0.138 | AAGAGCTGGCA |
| AtPIN3 | |||||||
| AT2G40620 | AtbZIP18 | AT1G70940 | 2256–2266 | - | 1.97 × 10−5 | 0.131 | CCCACCTGTCG |
| AT1G06070 | AtbZIP69 | AT1G70940 | 2256–2266 | - | 5.15 × 10−5 | 0.336 | CCCACCTGTCG |
| AtPIN7 | |||||||
| AT2G40620 | AtbZIP18 | AT1G23080 | 49–59 | + | 9.48 × 10−5 | 0.628 | AAAAGCTGTAA |
| AtAXR1 | |||||||
| AT2G40620 | AtbZIP18 | AT1G05180 | 1160–1170 | + | 4.52 × 10−5 | 0.338 | ACCACCTGTCT |
| AT1G06070 | AtbZIP69 | AT1G05180 | 1035–1045 | + | 4.37 × 10−5 | 0.254 | TGCAGCTGGTG |
| AT1G06070 | AtbZIP69 | AT1G05180 | 1160–1170 | + | 6.95 × 10−5 | 0.254 | ACCACCTGTCT |
| AtIPT5 | |||||||
| AT1G06070 | AtbZIP69 | AT5G19040 | 1294–1304 | - | 8.78 × 10−5 | 0.292 | GGCGGCTGGAA |
| * | * | AtGA2ox1 | * | * | * | * | * |
| * | * | AtGA20ox1 | * | * | * | * | * |
| * | * | AtGA20ox2 | * | * | * | * | * |
| * | * | AtRGA1 | * | * | * | * | * |
(*) indicates that the binding sites for the AtbZIP18 and AtbZIP69 were not detected.
2.3. AtbZIP62 Differentially Regulated PIN-FORMED and MAX Encoding Genes in Response to Drought Stress
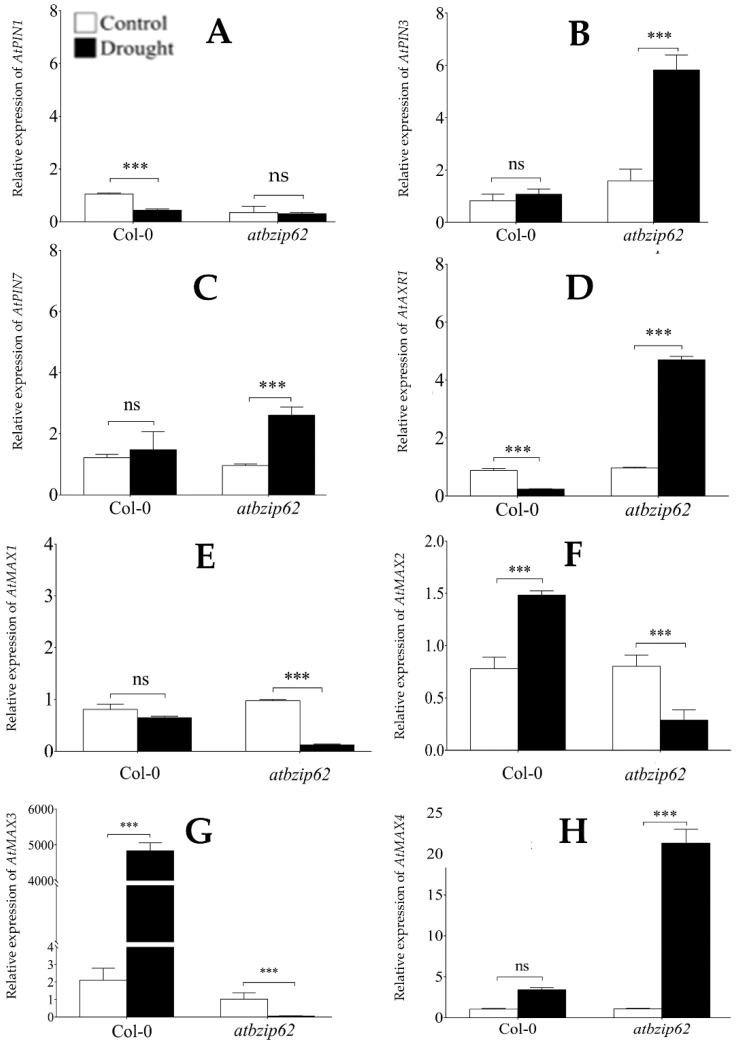
Auxin polar transport in plants involves many efflux carriers (PIN) proteins [29]. Within the cell, PIN proteins are asymmetrically localized and the directional auxin flow is determined by their polarity. In the current research, we studied the expression of the Arabidopsis PIN1, PIN3, and PIN7 in response to drought stress. AtPIN1 is known for playing an active role in the auxin basipetal transport [30], while AtPIN3 was suggested to function in the lateral redistribution of auxin [31,32] and AtPIN7 [33] have been suggested to mediate the lateral re-direction (reflux) of auxin back into the PIN1-dependent auxin transport flow [29,34]. The qPCR results indicate that the expression of AtPIN1 was downregulated (about 0.4-fold change) in Col-0 WT, but a non-significant change was observed in atbzip62 background (Figure 2A). Meanwhile, the expression of AtPIN3 and AtPIN7 was significantly upregulated (3.7 and 2.7-fold change, respectively) in atbzip62 knockout plants, while in Col-0 WT a non-significant change was recorded (Figure 2B,C). Another important gene regulating auxin polar transport is AtAXR1, the Arabidopsis auxin-resistance gene. Here, the expression of AtAXR1 was significantly downregulated (0.3-fold change) by drought stress in Col-0 WT, but an opposite pattern was recorded in atbzip62 (upregulated by 4.9-fold change) (Figure 2D). Thus, AtbZIP62 TF, earlier suggested to positively regulate the adatpive response mechanism towards drought tolerance [27], is believed to negatively control the expression of PIN-FORMED encoding genes in response to drought stress.
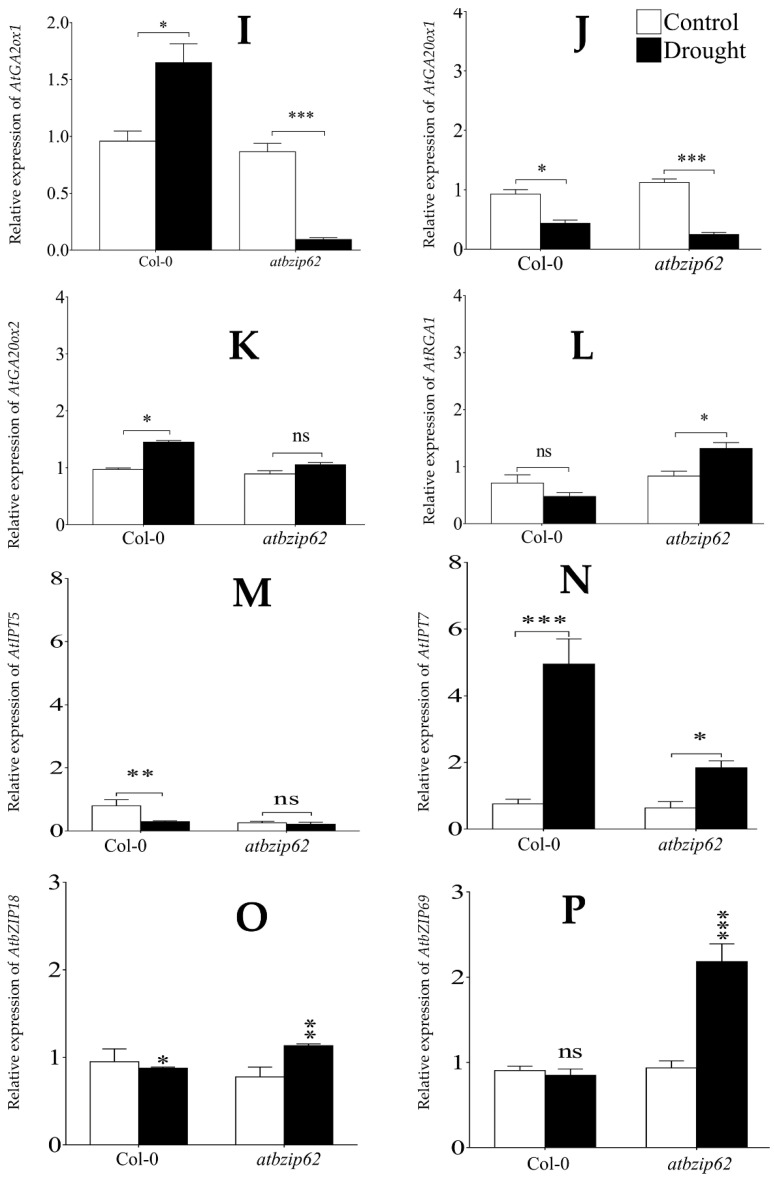
Figure 2.
Transcripts accumulation of hormonal responsive genes under drought stress conditions. (A) AtPIN1. (B) AtPIN3, (C) AtPIN7, (D) AtAXR1, (E) AtMAX1, (F) AtMAX2, (G) AtMAX3, (H) AtMAX4, (I) AtGA2ox1, (J) AtGA20ox1, (K) AtGA20ox2, (L) AtRGA1, (M) AtIPT5, (N) AtIPT7, (O) AtbZIP18, and (P) AtbZIP69 in Arabidopsis Col-0 wild type and atbzip62 knockout plants exposed to drought stress. Bars are mean values ± SE. White bars are controls (routinely watered) plants and black bars are drought-treated plants in triplicate. *** p < 0.001, ** p < 0.01, * p < 0.05, ns non-significant.
In the previous paragraphs, atbzip62 is shown to have increased branching phenotype under normal growth conditions. With regard to the observed phenotype, we were interested to see how the strigolactone pathway genes known for being involved in the control of bud outgrowth in Arabidopsis would be regulated in the atbzip62 mutant (showing more branches) in response to drought stress. Here, the expression pattern of AtMAX1 did not change significantly in Col-0, but a significant downregulation (0.13-fold change) was recorded in atbzip62 (Figure 2E). It was interesting to see that the expression of AtMAX2 and that of AtMAX3 genes were significantly upregulated (1.9 and 2,288-fold change, respectively) in Col-0 WT, while being downregulated (0.4 and 0.05-fold change, respectively) in the atbzip62 knockout plants (Figure 2F,G). Additionally, AtMAX4 gene was significantly upregulated (19.5-fold change) in atbzip62, and upregulated by about 3.2-fold change in Col-0 (Figure 2H).
2.4. Drought Stress Differentially Expressed Gibberellic Acid Biosynthetic Genes in Col-0 and atbzip62
The growth-related gibberellins biosynthetic pathway genes, AtGA2ox1 (GA2-oxidase), AtGA20ox1, and AtGA20ox2 control key steps of GAs synthesis in plants. Huang, et al. [35] showed that overexpression of GA20ox did not affect the gibberellic acid (GA4), which is said to be one of the major bioactive GAs in Arabidopsis with GA1, GA9, and GA20 [36]. The authors suggested that other GAs than GA4 would be responsible for the growth phenotype observed in the GA20ox OE plants. Our data in the panel I of the Figure 2 show that AtGA2ox1 was upregulated by about 1.7-fold change in Col-0 WT, while being downregulated (0.11-fold change) in atbzip62.
Because the atbzip62 showed an increased plant height under normal conditions compared to the Col-0 WT, we were interested to see, in addition to the AtGA2ox1, the transcriptional regulation of two other important GA biosynthetic pathway genes, AtGA20ox1 and AtGA20ox2 [37] under drought stress conditions. The activity of AtGA20ox has been earlier reported to be regulated by environmental stimuli [38,39,40]. Our data show that the expression of AtGA20ox1 was downregulated by drought stress in both the Col-0 WT and the atbzip62 (0.5 and 0.2-fold change, respectively) (Figure 2J), contrasting with the recorded expression of AtGA20ox2 in both genotypes (significantly upregulated by 1.5-fold change in WT and a non-significant change was recorded in atbzip62) (Figure 2K). Further investigations revealed that AtRGA1 (a member of the GRAS (GIBBERELLIN-INSENSITIVE (RGA), REPRESSOR of ga1-3 (RGA) and SCARECROW (SCR)) transcription factor familiy protein and the VHIID domain/Deletion of five amino acids (VHIID/DELLA) regulatory family [41], known as a repressor of GA signaling, which inhibits the proliferation and expansion of cell-mediated plant growth [42], was induced (1.6-fold change) by drought stress in atbzip62, but downregulated (0.7-fold change) in Col-0 WT (Figure 2L).
2.5. Drought Stress Differentially Regulated AtIPT5 and AtIPT7 in atbzip62
We expressed three cytokinin biosynthesis pathway genes in Col-0 WT and atbzip62 under drought stress to explore the possibility for the AtbZIP62 TF to mediated the transcriptional regulation of isopenteniltransferase (IPT) genes. It is known that the IPT protein controls the rate-limiting step of cytokinin biosynthesis [43,44]. Our data show, on the one hand, that the expression of AtIPT5 was suppressed at basal level in atbzip62, and remained unchanged under drought stress compared to WT showing higher transcript abundance in well-watered plants and suppressed expression in response to drought stress (Figure 2M). On the other hand, AtIPT7 exhibited an opposite expression pattern under the same conditions. Col-0 plants exposed to drought stress significantly upregulated AtIPT7 (6.6-fold change); however, when expressed in the atbzip62, the transcripts accumulation of AtITP7 dropped significantly (2.9-fold change) under the same conditions (Figure 2N).
2.6. AtbZIP18 and AtbZIP69 Are Differentially Regulated betwen Col-0 and atbzip62
The bZIP transcription factors are said to either operate alone or in complex with other bZIP TFs to regulate the expression of their target genes. In a recent study, the AtbZIP18 and AtbZIP69 were proposed to be negatively regulated by AtbZIP62 in response to salt stress [28]. Similarly, our data show that the expression of AtbZIP18 and AtbZIP69 were upregulated in the atzip62, while showing a downregulation pattern in Col-0 wild type (Figure 2O,P).
3. Discussion
3.1. The AtbZIP62 TF Differentially Regulates the Expression of the Arabidopsis PIN-FORMED Protein, MAX, IPT and GA-Biosynthetic Encoding Genes in Response to Drought Stress
Shoot branching is controlled at different levels by a complex hormonal signaling network, which moves throughout the plant. Auxin efflux carrier PINs play an important role in auxin transport and redistribution to the plant’s organs. PIN1 is the major auxin efflux carrier with a high affinity for auxin polar transport in plants, generating a unidirectional flow of auxin basipetal [34]. Hence, owing to the fact that AtPIN1 expression was significantly suppressed in the atbzip62 knockout plants, at both basal level and under drought stress conditions, our data suggest that auxin polar transport through the plant stem to control axillary bud outgrowth might be restricted in atbzip62 plants. In addition, the recorded transcripts accumulation of AtPIN3 and AtPIN7 suggest a possible co-expression in response to drought stress (Figure 2B,C). In Arabidopsis, AtAXR1 was earlier reported to confer auxin resistance to mutant plants. Moreover, axr1 loss of function mutant has been shown to have a reduced sensitivity to auxin [45]. Therefore, the recorded upregulation of AtAXR1 in atbzip62 knockout plants under drought stress would imply that the AtbZIP62 negatively regulates the expression of the AtAXR1, which would result in auxin sensitivity. The expression pattern of AtAXR1 would also suggest that the increased shoot branching phenotype observed in atbzip62 would be independent of AtAXR1.
From another perspective, a study conducted by Bennett, et al. [46] reported that strigolactone biosynthesis pathway genes, MAX, control shoot branching through the regulation of auxin polar transport-dependent to the PIN1 activity, but independent to the activity of AXR1. Similarly, Ferguson and Beveridge [47] studied the roles of major hormones in the regulation of shoot branching, and supported the assumption that strigolactone (SL) and cytokinin (CK) have an antagonistic effect on shoot branching (SL inhibits axillary bud outgrowth, while CK has an opposite effect).
In a converse approach, a study aiming at characterizing max1 to max4 mutants revealed that all MAX genes act in the same pathway, with no redundancy in their activity [48]. Generally, AtMAX1, AtMAX3, and AtMAX4 are involved in the synthesis of strigolactone (SL), whereas AtMAX2 is more active in the signal perception. AtMAX1 encodes P450, a cytochrome family member, which acts downstream of AtMAX3 and 4 to yield a carotenoids-derived branching inhibiting hormone. The high nitric oxide (NO) producing mutant atgsnor1-3 is also known for its increasing branching phenotype. In our CySNO (S-nitroso L-cysteine) transcriptome [49], AtMAX1 was shown to be about 1.94-fold downregulated by NO among other differentially expressed genes (DEGs). Thus, the recorded downregulation of AtMAX1, AtMAX2, and AtMAX3 expressions in the atbzip62 (Figure 2E–G) would imply that the transcripts accumulation of the three MAX genes could be positively governed by the AtbZIP62 TF. Moreover, the exponential increase in the transcripts of AtMAX3 in Col-0 WT by drought stress proposes AtMAX3 as an emerging candidate gene, which may play a leading role in the adaptive response mechanism towards drought tolerance under the regulatory influence of AtbZIP62 TF. In the same way, the expression pattern of AtMAX4 would indicate that all MAX may not co-express under drought stress.
In addition, it has been evidenced that the control of shoot branching is driven by the combinational action of diverse phytohormones, indicating a highly interactive and balanced hormonal signaling cascade [47]. The cytokinin biosynthetic genes, IPTs, were shown to interact with other plant hormones to regulate axillary bud outgrowth. [50]. However, under drought stress, our data shown in panels M and N of Figure 2 suggest a positive regulation of AtIPT5 and AtIPT7 by AtbZIP62 TF, which implies that CK biosynthesis or signaling pathway would be activated under drought stress conditions.
It is well established that plants favor growth and development under normal conditions. Here, we reported an increased plant height phenotype of atbzip62 plants compared to WT, contrasting with that of the atgsnor1-3 under normal growth conditions (Figure 1I). However, in response to an environmental stimulus, plants reallocate their resources by activating defense genes, and hormonal signaling as part of the adaptive response towards stress tolerance [51]. In a recent study, the rice RF2a TF (OsbZIP75) and RF2b (OsbZIP30) were suggested to control GA activity, and consequently plant height [25]. In the same way, Liao, et al. [52] supported that the soybean bZIP TFs GmbZIP44, GmbZIP62, and GmbZIP78 negatively regulated abscisic acid (ABA) signaling in transgenic Arabidopsis under salt stress. In contrast, the recorded downregulation of AGA2ox1, AtGA20ox1, and AtGA20ox2 in atbzip62, which matched the enhanced transcripts abundance of AtRGA1, a member of the GRAS (GAI, RGA, SCR) transcription factor family, encoding a DELLA protein [41], suggests their positive regulation by AtbZIP62 TF under drought stress. In plants, DELLA proteins are growth repressors and modulate all aspects of gibberellic acid responses [53]. This is consistent with the report by Willige, et al. [54], which indicated that a reduction in auxin transport was observed in the inflorescence of Arabidopsis mutants deficient in GA biosynthesis concomitant with a decrease in the PIN transcripts abundance, which facilitates auxin efflux in GA-deficient plants.
The results of the transcription factors binding sites prediction did not detect any specific cis-regulatory element of the AtbZIP62 TF in the promoter regions of the target genes. Rather, we detected cis-regulatory element of two other bZIP TF (AtbZIP18 and AtbZIP69) among others, which have been reported to be negatively regulated by the AtbZIP62 TF in response to salinity stress [28]. Similarly, data shown in the panels O and P of Figure 2 revealed that the expression of AtbZIP18 and AtbZIP69 TFs was significantly upregulated by drought stress in the atbzip62 knockout plants, but downregulated in the Col-0 WT. Therefore, the presence of the potential binding sites for the AtbZIP18 and AtbZIP69 in the promoter of AtPIN1, 3, and 7, AtMAX2, 3, and 4, AtAXR1 and AtIPT5 genes, coupled with the proposed transcriptional interplay between AtbZIP62 and AtbZIP18 and AtbZIP69, associated with the differential expression patterns of the target branching genes between the atbzip62 mutant and the Col-0 WT (Figure 2O,P) suggest that AtbZIP62 TF may require AtbZIP18 and/or AtbZIP69 TFs to regulate the expression of more axillary branching (MAX, strigolactone biosynthetic pathway), PIN-FORMED protein (auxin carriers), gibberellic acid biosynthetic pathway genes (GAs), and isopentenyl transferase (IPT, cytokinin biosynthetic pathway) genes in response to drought stress, which would be mediated by the AtbZIP18 and AtbZIP69 TF.
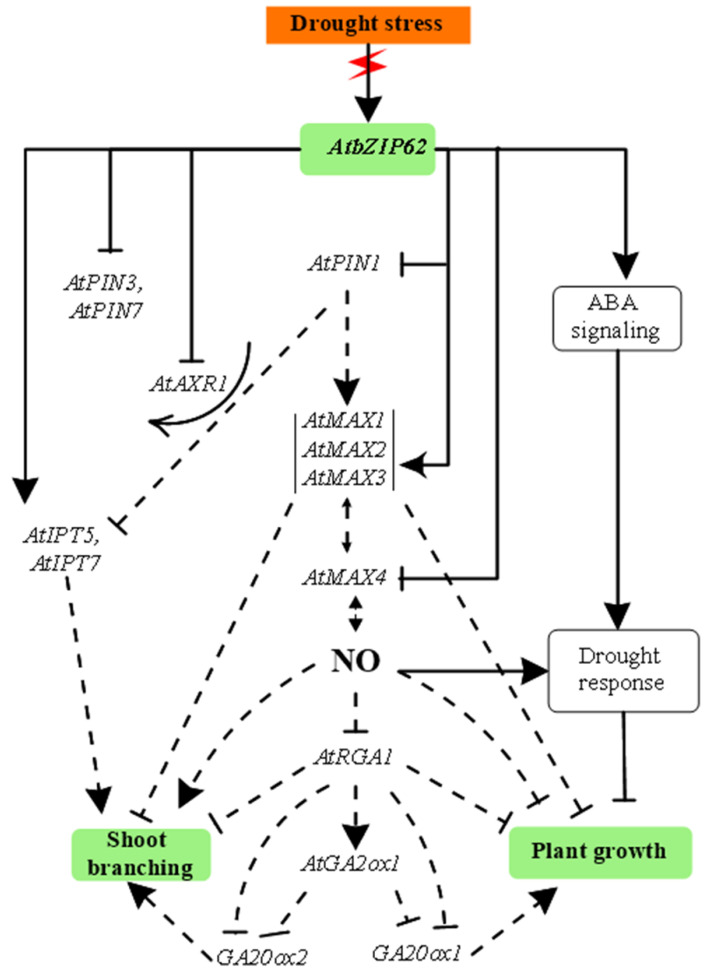
3.2. Proposed Signaling Model of AbZIP62 TF and Homonal Biosynthesic Genes under Drought Stress
Drought stress response mechanisms are complex and involve a very diverse signaling network and metabolic components, which include phytohormones signaling, activation of abiotic stress-responsive transcription factors and genes, stomata regulation, activation of ROS and reactive nitrogen species (RNS) (including nitric oxide, NO) signal pathways. All these metabolic components interact in a very coordinated and sophisticated manner to ensure that the plant uses efficiently the energy produced to maintain its fitness and survival. In Figure 3, this study proposed a signaling model involving the AtbZIP62 TF and other phytohormones biosynthetic pathway genes in response to drought stress of which the transcripts accumulation was measured in the current work. In our CySNO transcriptome [49], an Arabidopsis bZIP transcription factor (AtbZIP53, AT3G62420) encoding gene was shown to be upregulated by 2.61-fold. Similarly, our recent study showed that a significant reduction in SNO (S-nitrosoglutathione) and abscisic acid (ABA) contents was observed in the atbzip62 knockout plants compared to Col-0 WT [27]. Furthermore, NO was reported to interact with auxin to regulate roots growth in rice [55]. In the same way, NO was suggested to mediate the cytokinin functions in cell proliferation and meristem maintenance in Arabidopsis [56], while strigolactone was suggested to interact with NO in regulating root system architecture of Arabidopsis [57]. Moreover, crosstalk between NO and phytohormones during plant development has been widely discussed [58]. Therefore, the signaling model proposed in the present study tends to explain how AtbZIP62 TF would regulate the MAX, PIN-FORMED, GA, and IPT encoding genes under drought stress conditions. This model was designed based on the recorded transcriptional regulation patterns of the genes studied, monitored by qPCR in the atbzip62 knockout plants, coupled with previously reported evidence. Generally, plant hormones are part of the adaptive response mechanisms towards stress tolerance and they interact either in synergy or antagonize each other. Abscisic acid (ABA) has been extensively studied and has been shown to be one of the most responsive phytohormones under abiotic stress in plants. Our findings open new paths towards understanding the role of other hormonal signaling pathways genes in response to stress (abiotic), in addition to their roles in the regulation of plants growth and development.
Figure 3.
Signaling model involving AtbZIP62 transcription factor (TF) and more axillary branching (MAX), PIN-FORMED, gibberellic acid (GA), and isopentenyl transferase (IPT) encoded genes under drought stress. Upon drought stress induction, a variety of signals transduction and hormonal pathways are activated, followed by the induction of an array of drought-responsive genes, including transcription factors, such as AtbZIP62 which interact with other proteins or DNA to regulate the expression of stress inducible genes. As part of the drought response mechanism, plants activate ABA signaling components, which interact with other signaling pathways and specific genes in order to provide an appropriate response towards stress tolerance. Consequently, the regular plant growth-related metabolism is reduced. In the proposed signaling model, AtbZIP62 TF is shown to differentially regulate the transcripts accumulation of AtMAX1, AtMAX2, AtMAX3, AtMAX4 (SL), AtPIN1, AtPIN3, AtPIN7 (auxin carriers), AtGA2ox1, AtGA20ox1, AtGA20ox1, and AtRGA1, AtIPT5, and AtIPT7 (Cytokinin) genes. Arrows with continuous lines indicate positive regulation (of gene expression or induction of plant growth/shoot branching). Continuous lines with a perpendicular bar or an arrow suggest a negative regulation/inhibition or positive regulation/induction (of gene expression or plant growth/shoot branching) by our studies (current and previous). Dotted lines with a perpendicular bar or an arrow suggest a negative regulation/inhibition or positive regulation/induction (of gene expression or plant growth/shoot branching) by previous evidence by other research groups. NO, nitric oxide.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
The seeds of the wild type (WT) Arabidopsis Col-0 and the atbzip62 (AT1G19490: SALK_053908C) knockout line derived from it were obtained from the Arabidopsis Biological Resource Center (ABRC) (https://abrc.osu.edu/ (accessed on 31 January 2021)). In addition, the atgsnor1-3 mutant lacking the S-nitrosoglutathione (GSNO) reductase 1 (GSNOR1), known to regulate the cellular S-nitrosothiols (SNO) levels, was included for its stunt, and high branching phenotype was identified from the GABI-Kats T-DNA insertion collection [59,60]. Plants were grown on a peat moss soil mixture at 22°C with 16 h light and 8 h dark cycles. The atbzip62 plants were previously genotyped to identify homozygous transfer DNA (T-DNA) insertion lines by polymerase chain reaction (PCR) for further experiments. The T-DNA insertion lines were confirmed as described earlier [27]. The list of primers and their corresponding forward and reverse sequences is given in Table 2.
Table 2.
List of primers for expression of target genes used in the study.
| Gene Name/Genotype | Locus/ SALK |
Forward Primer (5′->3′) | Reverse Primer (5′->3′) | Gene Name |
|---|---|---|---|---|
| atbzip62 | SALK_053908C | TGGCACTTTTAACTTTGTGCC | TACGTTTCCATCGAGTGAACC | Arabidopsis bzip62 loss of function mutant |
| Drought responsive gene in Arabidopsis | ||||
| AtbZIP62 | AT1G19490 | CATCGAGTTGTTGCTCGTCG | AAATCCGCCAATGCTTCTGC | Basic-leucine zipper transcription factor encoding gene 62 |
| Genes involved in strigolactone biosynthesis pathway and controlling shoot branching (bud outgrowth) | ||||
| AtMAX1 | AT2G26170 | TGGTCACTTGCCCTTGATGG | GGTTGCCTCCCCATCTGAAA | More axillary branching 1 gene |
| AtMAX2 | AT2G42620 | CCGAGCCAGAGTTTGGGTTA | GTGCGAAACCGATTGTGTCC | More axillary branching 2 gene |
| AtMAX3 | AT2G44990 | CGTTGGTGAGCCCATGTTTG | TCCACCGAAACCGCATACTC | More axillary branching 3 gene |
| AtMAX4 | AT4G32810 | TATCGGGTCGTGAGGATGGA | GCAAACGAATGGACCCAACC | More axillary branching 4 gene |
| Genes involved in Auxin polar transport (efflux carrier) and controlling shoot branching | ||||
| AtPIN1 | AT1G73590 | ACGACAACCAGTACGTGGAG | TATGTTGTTCCCACCGTCCG | PIN-FORMED (PIN) protein encoding gene1 |
| AtPIN3 | AT1G70940 | TGGCCATGATCCTCGCTTAC | CGAAGATGGCGACAAAACGG | PIN-FORMED (PIN) protein encoding gene 3 |
| AtPIN7 | AT1G23080 | AGCCATGATCCTCGCTTACG | AGAGGGACGGCGAAAATAGC | PIN-FORMED (PIN) protein encoding gene 7 |
| Cytokinin biosynthetic pathway genes | ||||
| AtIPT5 | AT5G19040 | CGACGGAGGTTTTTCTCCGA | GAACTTTTCGACGGCGAGTG | Isopentenyl transferase encoding gene 5 |
| AtIPT7 | AT3G23630 | GACGCCACTGAGGTGTTCTT | CGACGATTCTCTCGCTTGGT | Isopentenyl transferase encoding gene 7 |
| Genes involved in Gibberellic acid biosynthesis pathway | ||||
| AtGA2ox1 | AT1G78440 | CTCGTTGCCCAAGTCAGAGA | TACTCAACCCAACCCACGTC | Gibberellic acid 2 oxidase 1 |
| AtGA20ox1 | AT4G25420 | GTGAGAGTGTTGGCTACGCA | CTCATGTCGTCGCAAAACCG | Gibberellic acid 20 oxidase 1 |
| AtGA20ox2 | AT5G51810 | TGGCCAGACGAAGAGAAACC | TTGACGACGAGGAAGAAGCC | Gibberellic acid 20 oxidase 2 |
| AtAXR1 | AT1G05180 | CGGACAGATTTGCTGCCAAC | ATCTGGGAGTACTGAGCCGT | Arabidopsis Auxin repressor 1 |
| AtRGA1 | AT2G01570 | TTGTCCAACCACGGGACTTC | AGCTCGTCGTCCATGTTACC | Arabidopsis Repressor of GA 1 |
| Arabidopsis housekeeping gene | ||||
| AtACT2 | AT3G18780 | AGGTTCTGTTCCAGCCATC | TTAGAAGCATTTCCTGTGAAC | Arabidopsis Actin coding gene 2 |
4.2. In Silico Transcription Factor Binding Site Prediction
With regards to the transcripts accumulation levels of the target branching related genes, we performed an in silico analysis in order to explore the possibility for the AtbZIP62 TF to be involved in their transcriptional initiation or expression, using bioinformatics approach. To achieve that, we predicted the transcription factor binding sites in the DNA sequence of each of the selected hormonal signaling pathway genes included in the study using the Binding site prediction tool available at http://plantregmap.gao-lab.org (accessed on 31 January 2021).
4.3. Drought Stress Induction by Water Withholding
Plants were subjected to drought stress at rossette stage (4-week-old plants) following the water withholding method as described earlier [61], with slight modifications. Briefly, the moisture content of the soil was routinely monitored by measuring the weight of each of the 50-well trays in triplicate to evaluate the water loss. The soil moisture percentage (about 30%) was calculated as the percentage of the actual weight loss relative to the initial weight of the saturated soil considered as having 100% moisture content. Leaf samples for gene expression analysis were collected as soon as the loss of turgidity and wilting of leaves were apparent (9 days after water withholding).
4.4. Total RNA Isolation and Gene Expression Analysis by qPCR
Total RNA was isolated from leaf samples collected a rosette stage, using the TRI-SolutionTM Reagent (Cat. No: TS200-001, Virginia Tech Biotechnology, Lot: 337871401001) following the manufacturer’s instructions. Thereafter, the complementary DNA (cDNA) was synthesized as previously described [62]. The cDNA was then used as a template in qPCR to study the transcripts accumulation of selected genes (Table 2) using SYBR green (BioFact, Daejeon, Korea) in a real-time PCR machine (Eco™ Illumina, San Diego, CA, USA). The relative expression of each gene was normalized to that of the Arabidopsis actin.
5. Conclusions
For the last two decades, the focus of plant biosciences-related research has been marked by an increasing interest in the use of a genome-wide approach while studying the function of genes in response to adverse environmental conditions, rather than a single gene–gene approach. The output from these years of research helped elucidate factors involved in the regulation of gene expression, including the cross-talk mechanism.
This study investigated the transcriptional regulation of key branching related genes in response to drought stress, including AtPIN1, 3, and 7, AtMAX1–4, AtXR1, AtIPT5 and 7, AtGA2ox1, AtGA20ox1 and 2, and AtRGA1. The results showed that all the branching-related genes were differentially regulated between Col-0 WT and the atbzip62 mutant in response to drought stress. In addition, with regard to the transcripts accumulation patterns of the two bZIP TFs, AtbZIP18 and AbZIPT69, in the atbzip62 background, and their proposed transcriptional interplay, coupled with the detected potential binding sites specific to the AtbZIP18 and AtbZIP69 in the target genes, all the results suggest that the AtbZIP62 TF may require AtbZIP18 and/or AbZIP69 to regulate the expression of the analyzed branching related genes under drought stress conditions. Future studies may include the use of loss of function mutant lines of selected hormonal signaling pathways genes, other than abscisic acid (atmax, atpin, or atipt) reported in the current study to elucidate their roles in drought stress response.
Author Contributions
Conceptualization, methodology, and validation, B.-W.Y.; formal analysis and investigation and data curation, N.K.R. and B.-G.M.; writing—original draft preparation, N.K.R.; writing—review and editing, N.K.R. and B.-G.M.; visualization, supervision, project administration, and funding acquisition, B.-W.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the grants from the Next-Generation BioGreen 21 Program (SSAC, Grant No., PJ01342501), Rural Development Administration and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (Grant number 2020R1I1A3073247), Republic of Korea.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Atkinson N.J., Urwin P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012;63:3523–3543. doi: 10.1093/jxb/ers100. [DOI] [PubMed] [Google Scholar]
- 2.Lobell D.B., Schlenker W., Costa-Roberts J. Climate trends and global crop production since 1980. Science. 2011;333:616–620. doi: 10.1126/science.1204531. [DOI] [PubMed] [Google Scholar]
- 3.Mafakheri A., Siosemardeh A., Bahramnejad B., Struik P., Sohrabi Y. Effect of drought stress on yield, proline and chlorophyll contents in three chickpea cultivars. Aust. J. Crop Sci. 2010;4:580–585. [Google Scholar]
- 4.Jaleel C.A., Manivannan P., Wahid A., Farooq M., Al-Juburi H.J., Somasundaram R., Panneerselvam R. Drought stress in plants: A review on morphological characteristics and pigments composition. Int. J. Agric. Biol. 2009;11:100–105. [Google Scholar]
- 5.Gill S.S., Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 6.Borsani O., Valpuesta V., Botella M.A. Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiol. 2001;126:1024–1030. doi: 10.1104/pp.126.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma P., Jha A.B., Dubey R.S., Pessarakli M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012;2012 doi: 10.1155/2012/217037. [DOI] [Google Scholar]
- 8.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7:405–410. doi: 10.1016/S1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 9.Verma S., Dubey R. Lead toxicity induces lipid peroxidation and alters the activities of antioxidant enzymes in growing rice plants. Plant Sci. 2003;164:645–655. doi: 10.1016/S0168-9452(03)00022-0. [DOI] [Google Scholar]
- 10.Meriga B., Reddy B.K., Rao K.R., Reddy L.A., Kishor P.K. Aluminium-induced production of oxygen radicals, lipid peroxidation and DNA damage in seedlings of rice (Oryza sativa) J. Plant Physiol. 2004;161:63–68. doi: 10.1078/0176-1617-01156. [DOI] [PubMed] [Google Scholar]
- 11.Jaleel C.A., Riadh K., Gopi R., Manivannan P., Ines J., Al-Juburi H.J., Chang-Xing Z., Hong-Bo S., Panneerselvam R. Antioxidant defense responses: Physiological plasticity in higher plants under abiotic constraints. Acta Physiol. Plant. 2009;31:427–436. doi: 10.1007/s11738-009-0275-6. [DOI] [Google Scholar]
- 12.Nimse S.B., Pal D. Free radicals, natural antioxidants, and their reaction mechanisms. RSC Adv. 2015;5:27986–28006. doi: 10.1039/C4RA13315C. [DOI] [Google Scholar]
- 13.Corpas F.J. What is the role of hydrogen peroxide in plant peroxisomes? Plant Biol. 2015;17:1099–1103. doi: 10.1111/plb.12376. [DOI] [PubMed] [Google Scholar]
- 14.Hoang X.L.T., Du Nhi N.H., Binh Anh Thu N., Phuong Thao N., Phan Tran L.-S.J.C.g. Transcription factors and their roles in signal transduction in plants under abiotic stresses. Curr. Genom. 2017;18:483–497. doi: 10.2174/1389202918666170227150057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan S.-A., Li M.-Z., Wang S.-M., Yin H.J. Revisiting the role of plant transcription factors in the battle against abiotic stress. Int. J. Mol. Sci. 2018;19:1634. doi: 10.3390/ijms19061634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford T., Karamat F., Lehotai N., Rentoft M., Blomberg J., Strand Å., Björklund S. Specific functions for Mediator complex subunits from different modules in the transcriptional response of Arabidopsis thaliana to abiotic stress. Sci. Rep. 2020;10:1–18. doi: 10.1038/s41598-020-61758-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gale M., Devos K. Plant comparative genetics after 10 years. Science. 1998;282:656–659. doi: 10.1126/science.282.5389.656. [DOI] [PubMed] [Google Scholar]
- 18.Dodeweerd A.-M.v., Hall C.R., Bent E.G., Johnson S.J., Bevan M.W., Bancroft I. Identification and analysis of homoeologous segments of the genomes of rice and Arabidopsis thaliana. Genome. 1999;42:887–892. doi: 10.1139/g99-033. [DOI] [PubMed] [Google Scholar]
- 19.Liu H., Sachidanandam R., Stein L. Comparative genomics between rice and Arabidopsis shows scant collinearity in gene order. Genome Res. 2001;11:2020–2026. doi: 10.1101/gr.194501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson D.K., Kitching R., Vary C., Kola I., Seth A. Isolation of target gene promoter/enhancer sequences by whole genome PCR method. Methods Mol. Biol. (Clifton, N.J.) 2000;130:1–11. doi: 10.1385/1-59259-686-x:1. [DOI] [PubMed] [Google Scholar]
- 21.Perry S.E. Plant Transcription Factors: Methods and Protocols. Springer; Berlin, Germany: 2011. [Google Scholar]
- 22.Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. Molecular Biology of the Cell. 4th ed. Volume 91. Garland Science; New York, NY, USA: 2002. p. 401. [Google Scholar]
- 23.Jakoby M., Weisshaar B., Droge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/S1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 24.Ali Z., Sarwat S.S., Karim I., Faridi R., Jaskani M.J., Khan A.A. Functions of plant’s bZIP transcription factors. Pak. J. Agric. Sci. 2016;53 doi: 10.21162/pakjas/16.2043. [DOI] [Google Scholar]
- 25.Ábrahám E., Hourton-Cabassa C., Erdei L., Szabados L. Methods in Molecular Biology. Volume 639. Springer International Publishing; Berlin, Germany: 2010. Methods for determination of proline in plants; pp. 317–331. [DOI] [PubMed] [Google Scholar]
- 26.Peleg Z., Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011;14:290–295. doi: 10.1016/j.pbi.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Rolly N.K., Imran Q.M., Shahid M., Imran M., Khan M., Lee S.-U., Hussain A., Lee I.-J., Yun B.-W. Drought-induced AtbZIP62 transcription factor regulates drought stress response in Arabidopsis. Plant Physiol. Biochem. 2020;156:384–395. doi: 10.1016/j.plaphy.2020.09.013. [DOI] [PubMed] [Google Scholar]
- 28.Rolly N.K., Imran Q.M., Lee I.-J., Yun B.-W. Salinity Stress-Mediated Suppression of Expression of Salt Overly Sensitive Signaling Pathway Genes Suggests Negative Regulation by AtbZIP62 Transcription Factor in Arabidopsis thaliana. Int. J. Mol. Sci. 2020;21:1726. doi: 10.3390/ijms21051726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nat. Cell Biol. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 30.Křeček P., Skůpa P., Libus J., Naramoto S., Tejos R., Friml J., Zažímalová E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009;10:1–11. doi: 10.1186/gb-2009-10-12-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keuskamp D.H., Pollmann S., Voesenek L.A., Peeters A.J., Pierik R. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc. Nat. Acad. Sci. USA. 2010;107:22740–22744. doi: 10.1073/pnas.1013457108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nat. Cell Biol. 2002;415:806. doi: 10.1038/415806a. [DOI] [PubMed] [Google Scholar]
- 33.Blakeslee J.J., Peer W.A., Murphy A.S. Auxin transport. Curr. Opin. Plant Biol. 2005;8:494–500. doi: 10.1016/j.pbi.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 34.Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nat. Cell Biol. 2003;426:147. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 35.Huang S., Raman A.S., Ream J.E., Fujiwara H., Cerny R.E., Brown S.M. Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol. 1998;118:773–781. doi: 10.1104/pp.118.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharya A. Effect of High Temperature on Crop Productivity and Metabolism of Macro Molecules. Elsevier BV; Amsterdam, The Netherlands: 2019. [Google Scholar]
- 37.Phillips A.L., Ward D.A., Uknes S., Appleford N.E., Lange T., Huttly A.K., Gaskin P., Graebe J.E., Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilmour S.J., Zeevaart J.A., Schwenen L., Graebe J.E. Gibberellin metabolism in cell-free extracts from spinach leaves in relation to photoperiod. Plant Physiol. 1986;82:190–195. doi: 10.1104/pp.82.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hedden P., Croker S.J. Red. Clover Science. Springer International Publishing; Berlin, Germany: 1992. Regulation of gibberellin biosynthesis in maize seedlings; pp. 534–544. [Google Scholar]
- 40.Xu Y.-L., Li L., Wu K., Peeters A., Gage D.A., Zeevaart J. The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: Molecular cloning and functional expression. Proc. Nat. Acad. Sci. USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pysh L.D., Wysocka-Diller J.W., Camilleri C., Bouchez D., Benfey P.N. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313X.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- 42.Silverstone A.L., Ciampaglio C.N., Sun T.-p. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kakimoto T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferases. Plant Cell Physiol. 2001;42:677–685. doi: 10.1093/pcp/pce112. [DOI] [PubMed] [Google Scholar]
- 44.Takei K., Sakakibara H., Sugiyama T. Genes: Structure and Regulation-Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 2001;276:26405–26410. doi: 10.1074/jbc.M102130200. [DOI] [PubMed] [Google Scholar]
- 45.Leyser H.O., Lincoln C.A., Timpte C., Lammer D., Turner J., Estelle M. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- 46.Bennett T., Sieberer T., Willett B., Booker J., Luschnig C., Leyser O. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 2006;16:553–563. doi: 10.1016/j.cub.2006.01.058. [DOI] [PubMed] [Google Scholar]
- 47.Ferguson B.J., Beveridge C.A. Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiol. 2009;149:1929–1944. doi: 10.1104/pp.109.135475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Booker J., Sieberer T., Wright W., Williamson L., Willett B., Stirnberg P., Turnbull C., Srinivasan M., Goddard P., Leyser O. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell. 2005;8:443–449. doi: 10.1016/j.devcel.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 49.Hussain A., Mun B.-G., Imran Q.M., Lee S.-U., Adamu T.A., Shahid M., Kim K.-M., Yun B.-W. Nitric oxide mediated transcriptome profiling reveals activation of multiple regulatory pathways in Arabidopsis thaliana. Front. Plant Sci. 2016;7:975. doi: 10.3389/fpls.2016.00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller D., Leyser O. Auxin, cytokinin and the control of shoot branching. Ann. Bot. 2011;107:1203–1212. doi: 10.1093/aob/mcr069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hauser F., Waadt R., Schroeder J.I. Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 2011;21:R346–R355. doi: 10.1016/j.cub.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao Y., Zou H.-F., Wei W., Hao Y.-J., Tian A.-G., Huang J., Liu Y.-F., Zhang J.-S., Chen S.-Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta. 2008;228:225–240. doi: 10.1007/s00425-008-0731-3. [DOI] [PubMed] [Google Scholar]
- 53.Penfield S., Gilday A.D., Halliday K.J., Graham I.A. DELLA-mediated cotyledon expansion breaks coat-imposed seed dormancy. Curr. Biol. 2006;16:2366–2370. doi: 10.1016/j.cub.2006.10.057. [DOI] [PubMed] [Google Scholar]
- 54.Willige B.C., Isono E., Richter R., Zourelidou M., Schwechheimer C. Gibberellin regulates PIN-FORMED abundance and is required for auxin transport–dependent growth and development in Arabidopsis thaliana. Plant Cell. 2011;23:2184–2195. doi: 10.1105/tpc.111.086355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun H., Feng F., Liu J., Zhao Q. The interaction between auxin and nitric oxide regulates root growth in response to iron deficiency in rice. Front. Plant Sci. 2017;8:2169. doi: 10.3389/fpls.2017.02169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen Q., Wang Y.-T., Tian H., Guo F.-Q. Nitric oxide mediates cytokinin functions in cell proliferation and meristem maintenance in Arabidopsis. Mol. Plant. 2013;6:1214–1225. doi: 10.1093/mp/sss148. [DOI] [PubMed] [Google Scholar]
- 57.Oláh D., Feigl G., Molnár Á., Ördög A., Kolbert Z. Strigolactones interact with nitric oxide in regulating root system architecture of Arabidopsis thaliana. Front. Plant Sci. 2020;11:1019. doi: 10.3389/fpls.2020.01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanz L., Albertos P., Mateos I., Sánchez-Vicente I., Lechón T., Fernández-Marcos M., Lorenzo O. Nitric oxide (NO) and phytohormones crosstalk during early plant development. J. Exp. Bot. 2015;66:2857–2868. doi: 10.1093/jxb/erv213. [DOI] [PubMed] [Google Scholar]
- 59.Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- 60.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Tech. Adv. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 61.Harb A., Pereira A. Screening Arabidopsis genotypes for drought stress resistance. Methods Mol. Biol. 2010;678:191–198. doi: 10.1007/978-1-60761-682-5_14. [DOI] [PubMed] [Google Scholar]
- 62.Mun B.-G., Lee S.-U., Hussain A., Kim H.-H., Rolly N.K., Jung K.-H., Yun B.-W. S-nitrosocysteine-responsive genes modulate diverse regulatory pathways in Oryza sativa: A transcriptome profiling study. Funct. Plant Biol. 2018;45:630–644. doi: 10.1071/FP17249. [DOI] [PubMed] [Google Scholar]