Abstract
Recent months have seen surges of SARS-CoV-2 infection across the globe along with considerable viral evolution. Extensive mutations in the spike protein of variants B.1.1.7, B1.351, and P.1 have raised concerns that the efficacy of current vaccines and therapeutic monoclonal antibodies could be threatened. In vitro studies have shown that one mutation, E484K, plays a crucial role in the loss of neutralizing activity of some monoclonal antibodies as well as most convalescent and vaccinee sera against variant B.1.351. In fact, two vaccine trials have recently reported lower protective efficacy in South Africa, where B.1.351 is dominant. To survey for these novel variants in our patient population in New York City, PCR assays were designed to identify viruses with two signature mutations, E484K and N501Y. We observed a steady increase in the detection rate from late December to mid-February, with an alarming rise to 12.3% in the past two weeks. Whole genome sequencing further demonstrated that most of our E484K isolates (n=49/65) fell within a single lineage: NextStrain clade 20C or Pangolin lineage B.1.526. Patients with this novel variant came from diverse neighborhoods in the metropolitan area, and they were on average older and more frequently hospitalized. Phylogenetic analyses of sequences in the database further reveal that this B.1.526 variant is scattered in the Northeast of US, and its unique set of spike mutations may also pose an antigenic challenge for current interventions.
While evolution of SARS-CoV-2 was deemed to be slow at the beginning of the global pandemic (1), at least three major variants of concern have emerged over the past two months (2–4). These lineages are each characterized by numerous mutations in the spike protein, raising concerns that they may escape from therapeutic monoclonals and vaccine-induced antibodies. The hallmark mutation of B.1.1.7, the first SARS-CoV-2 variant of concern that emerged in the UK, is N501Y located in the receptor-binding domain (RBD) of spike (2). This variant is seemingly more transmissible and possibly more virulent (5–7). The other variants of concern, B.1.351 (first detected in South Africa) (3) and P.1 (first described in Brazilian travelers) (4), share the N501Y mutation with B.1.1.7 but contain an E484K substitution in RBD (3, 4). Epidemiological evidence suggests that P.1 emerged as part of a second surge in Manaus, Brazil despite a high pre-existing seroprevalence to SARS-CoV-2 in the population. Reinfections with P.1, as well as with another related Brazilian variant P.2 that also harbors E484K, have been documented (8, 9).
Our previous study on B.1.351 demonstrated that this variant is refractory to neutralization by a number of monoclonal antibodies directed to the top of RBD, including several that have received emergency use authorization (10). Moreover, this variant was markedly more resistant to neutralization by convalescent plasma and vaccinee sera. Importantly, these effects were largely mediated by the E484K mutation. These finding are worrisome in light of recent reports that two vaccine trials showed a substantial drop in efficacy in South Africa (11, 12). We therefore began an effort to survey our patient population at the Columbia University Irving Medical Center in New York City for B.1.351 and other E484K variants such as P.1 and P.2.
We first developed rapid PCR-based, single-nucleotide-polymorphism assays to search for N501Y and E484K mutations (see schematic in Fig. S1 and Methods in Supplement) in clinical samples known to be positive for SARS-CoV-2 and stored in the Columbia University Biobank, and patient information was extracted from the COVID-Care database (13). Between November 1, 2020 and February 15, 2021, a total of 60,539 nasopharyngeal swabs underwent clinical testing for SARS-CoV-2 at our medical center, with 4,358 positive samples identified. We screened 1,142 samples randomly chosen from this period for the two signature mutations. A total of 927 samples yielded a signal in our genotyping assays. We found that 83 (9.0%) were positive for E484K and 17 (1.8%) were positive for N501Y. Only one sample contained both mutations. The earliest case with E484K was collected in mid-November 2020. Subsequently, there was a substantial increase in E484K-positive cases over time (Fig. 1A), from 1.3% in early November to 5.3% by mid-January, and ultimately to 12.3% between February 8th and 15th. Viruses harboring N501Y also increased over time, from the earliest detection in mid-January to 2.6% of screened isolates by mid-February.
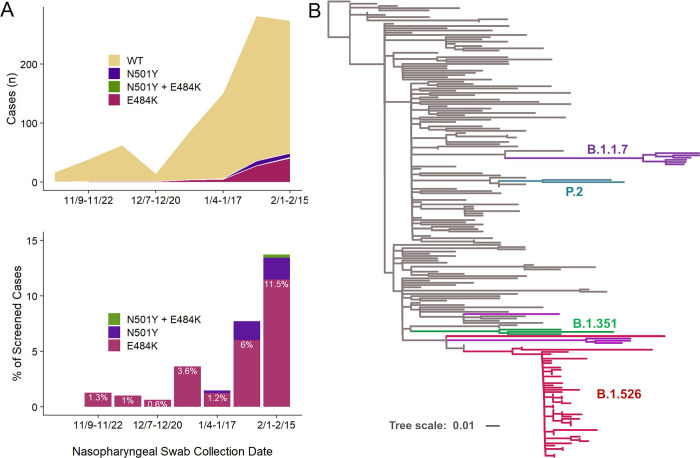
Figure 1. Increasing prevalence of E484K from November 2020 to February 2021 including E484K-harboring isolates within the B.1.526 lineage.
(A) The earliest detected E484K variant from our study period was collected in mid-November 2020, and a total of 83 isolates with this substitution were identified between November 2020 and February 15, 2021, out of 927 total samples that showed a signal in our screening assay (upper panel). The prevalence of E484K (nE484K/(nWT + nE484K)) subsequently increased over time, from 1.3% in early November (11/09/2020–11/22/2020) to 11.5% in early February (2/1/2021–2/15/2021), as shown in the lower panel. (B) To place our E484K isolates within the context of globally circulating SARS-CoV-2 strains, we downloaded 140 genomes from GISAID randomly sampled across 50 distinct lineages. Phylogenetic analysis demonstrated that the majority of our B.1.526 isolates are tightly clustered (bottom; magenta), with the remaining E484K-harboring isolates interspersed within the parent B.1 lineage. The B.1.1.7 (UK; purple), B.1.351 (South Africa; green), and P.2 (Brazil; teal) samples identified fall within these branches, respectively, as expected, as shown. Samples from our collection which harbor E484K or N501Y but do not fall within one of these lineages are shown in magenta and purple, respectively.
We then performed whole genome nanopore sequencing on samples flagged as potential N501Y- or E484K-harboring strains (n=65). We also sequenced samples negative for these signature mutations obtained during the same time period, all with Ct values below 35 (n=65). Sequencing results verified the E484K and N501Y substitutions in all samples identified by our screening PCR assays. Based on phylogenetic analyses including publicly available genomes (Fig. 1B), six cases with N501Y were identified as belonging to the B.1.1.7 lineage, two cases with E484K as P.2, and one sample as B.1.351, which harbored both N501Y and E484K based on our screening assay. However, quite unexpectedly, the large majority (n=49) of the remaining cases with E484K fell within a single lineage, B.1.526 (14). It is this novel variant that is surging, alarmingly, in our patient population over the past few weeks.
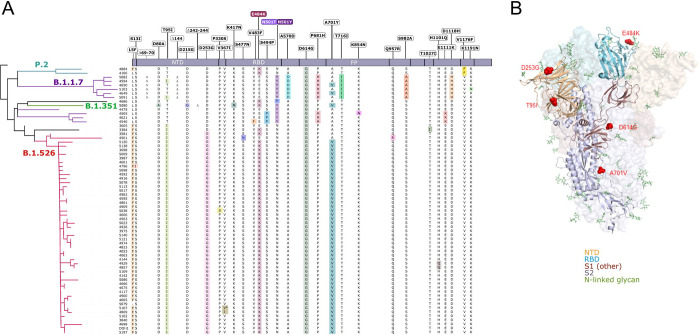
Nearly all of the newly identified B.1.526 variants have a set of common mutations in the spike protein: L5F, T95I, D253G, E484K, D614G, and A701V. Fig. 2A displays all of the spike mutations found in all variant viruses identified in the study, along with their phylogenetic relationship. Fig. 2B shows that D253G resides in the antigenic supersite within the N-terminal domain (15), which is a target for neutralizing antibodies (16), whereas the E484K is situated at the RBD interface with the cellular receptor ACE2. The A701V mutation near the furin cleavage site is also shared with variant B.1.351. The impact of the E484K mutation on antibody neutralization was assessed using 4 monoclonal antibodies with emergency use authorization, 10 convalescent plasma, and 10 vaccinee sera. As shown in Fig. S2, the neutralizing activity of REGN10987 against E484K pseudovirus is unaltered, but the activities of REGN10933, CB6, and LY-CoV555 are either impaired or abolished. Likewise, neutralizing activities of convalescent plasma or vaccinee sera are lower by 7.7-fold or 3.4-fold, respectively, against the E484K variant. These results signify an important antigenic drift in B.1.526 that could have clinical consequences, as have been noted for B.1.351 (10–12).
Figure 2. S-protein amino acid substitutions and structural changes represented in E484K- and N501Y-harboring isolates.
(A) Consensus sequences for 65 genomes were aligned against the Wuhan-Hu-1 reference genome (NC_045512). A phylogenetic reconstruction of these variant genomes is displayed on the left, generated using IQtree with 1,000 bootstrap replicates and maximum-likelihood tree search. Lineage assignments for major clades as determined by Pangolin are shown on the appropriate branches of the phylogenetic tree. On the right, residues at which at least one sample harbored a mutation compared to the reference genome are displayed above the S protein schematic. Amino acid substitutions identified in each sample are highlighted in color and wildtype residues are shown in black; an asterisk (*) indicates a silent nucleotide mutation. (B) Structural changes reflecting substitutions identified in the majority of B.1.526 isolates. Of the amino acid changes largely conserved in the B.1.526 lineage (L5F, T95I, D253G, E484K, D614G, A701V), D253G resides in the antigenic supersite within the N-terminal domain, a target for neutralizing antibodies, E484K at the RBD interface with the cellular receptor ACE2, and A701V near the furin cleavage site. Abbreviations – NTD – N-terminal domain; RBD – receptor binding domain; FP – fusion peptide.
Patients with E484K variant viruses were comparable in gender, race and ethnicity to those with wildtype SARS-CoV-2, but on average were older (58.1 vs 52.4 years, p=0.049) and more likely to present to the ED or be admitted to the hospital (85.9% vs 70.8%, p=0.007, Table S1). There were no differences in the frequency of ICU admissions and length of stay between groups. The majority of patients with E484K variants were geographically concentrated in two distinct neighborhoods in the catchment area of our hospital system, but many others were found scattered throughout the metropolitan area without evidence for a single outbreak (Fig. S3). An early case of our novel B.1.526 variant (NP-3581 in Fig. 2A) was detected in November 2020 in a patient with advanced AIDS who had first presented in August with infection by a SARS-CoV-2 strain (NP-3005 in Fig. 2A) that initially lacked the L5F, D253G, and E484K mutations. This case is reminiscent of reported examples of extensive intra-host evolution due to persistent SARS-CoV-2 infection in immunocompromised individuals (17, 18).
We also investigated SARS-CoV-2 sequences in public databases and found ~140 genomes highly related to the newly identified B.1.526 variant (14) (Fig. 1B). These were predominantly from samples collected in the Northeastern US, suggesting that E484K in the B.1.526 lineage is now widespread in the region, the original epicenter of COVID-19 in the US (19). This supports concerns that novel variants may spread in regions with a relatively high sero-prevalence. Moreover, it appears that the E484K mutation has emerged in at least 59 different lineages of SARS-CoV-2 (20), a real testament to convergent evolution. In conclusion, we identified B.1.526 as a local lineage of concern due to E484K in particular, which could threaten the efficacy of current antibody therapies and vaccines. This discovery also highlights the need for a concerted national surveillance program to track and contain the spread of novel SARS-CoV-2 variants.
Supplementary Material
Acknowledgements:
We gratefully acknowledge all the authors, the originating laboratories responsible for obtaining the specimens, and the submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative, on which part of the presented research is based. Biospecimens utilized for this research were obtained from the Columbia University Biobank (CUB) with technical support from Viplan J. Mahadeva, Sebastian Fernando and Sylvia T. Parker-Jones. CUB is supported by the Irving Institute for Clinical and Translational Research, home to Columbia University’s Clinical and Translational Science Award (CTSA) funded through Grant Number UL1TR001873. In particular, we thank Muredach Reilly, Eldad Hod and the CUB COVID-19 Genomics Consortium (CCGC) for facilitating this effort. We are also grateful to Lihong Liu and Sho Iketani for technical support. This work was in part funded by NIH/NIDA grant U01 DA053949 (ACU, MKA) and by support from Andrew & Peggy Cherng, Samuel Yin, Barbara Picower and the JBP Foundation, Brii Biosciences, Roger & David Wu, and the Bill and Melinda Gates Foundation.
References
- 1.Duchene S. et al. , Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol 6, veaa061 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rambaut A. et al. , Preliminary genomic characterisation of an emergent SARSCoV-2 lineage in the UK defined by a novel set of spike mutations. (2020).
- 3.Tegally H. et al. , Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv, 2020.2012.2021.20248640 (2020). [Google Scholar]
- 4.Faria N. R. et al. , Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. (2021).
- 5.Iacobucci G., Covid-19: New UK variant may be linked to increased death rate, early data indicate. BMJ 372, n230 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Volz E. et al. , Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. medRxiv, 2020.2012.2030.20249034 (2021). [Google Scholar]
- 7.Washington N. L. et al. , Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. medRxiv, 2021.2002.2006.21251159 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zucman N., Uhel F., Descamps D., Roux D., Ricard J. D., Severe reinfection with South African SARS-CoV-2 variant 501Y.V2: A case report. Clin Infect Dis, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nonaka C. K. V. et al. , Genomic Evidence of SARS-CoV-2 Reinfection Involving E484K Spike Mutation, Brazil. Emerg Infect Dis 27, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang P. et al. , Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. bioRxiv, 2021.2001.2025.428137 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Wadman M., Cohen J., in Science. (2021).
- 12.Callaway E., Mallapaty S., Novavax offers first evidence that COVID vaccines protect people against variants. Nature 590, 17 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Miller E. H. et al. , Pretest Symptom Duration and Cycle Threshold Values for Severe Acute Respiratory Syndrome Coronavirus 2 Reverse-Transcription Polymerase Chain Reaction Predict Coronavirus Disease 2019 Mortality. Open Forum Infectious Diseases 8, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West A. P., Barnes C. O., Yang Z., Bjorkman P. J., SARS-CoV-2 lineage B.1.526 emerging in the New York region detected by software utility created to query the spike mutational landscape. bioRxiv, 2021.2002.2014.431043 (2021). [Google Scholar]
- 15.Cerutti G. et al. , Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite. bioRxiv, 2021.2001.2010.426120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L. et al. , Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Avanzato V. A. et al. , Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell 183, 1901–1912 e1909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi B. et al. , Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N Engl J Med 383, 2291–2293 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.N. Y. C. D. o. Health. COVID-19: Data; Long Term Trends. Available online: https://www1.nyc.gov/site/doh/covid/covid-19-data-trends.page#antibody (2021)
- 20.Gangavarapu Karthik; Alkuzweny Manar; Cano Marco; Haag Emily; Latif Alaa Abdel; Mullen Julia L.; Rush Benjamin; Tsueng Ginger; Zhou Jerry; Andersen Kristian G.; Wu Chunlei; Su Andrew I.; Hughes Laura D. outbreak.info. Available online: https://outbreak.info/ (2021)
- 21.Smyrlaki I. et al. , Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat Commun 11, 4812 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.