Abstract
Key message
The importance and potential of the multi-purpose crop sorghum in global food security have not yet been fully exploited, and the integration of the state-of-art genomics and high-throughput technologies into breeding practice is required.
Abstract
Sorghum, a historically vital staple food source and currently the fifth most important major cereal, is emerging as a crop with diverse end-uses as food, feed, fuel and forage and a model for functional genetics and genomics of tropical grasses. Rapid development in high-throughput experimental and data processing technologies has significantly speeded up sorghum genomic researches in the past few years. The genomes of three sorghum lines are available, thousands of genetic stocks accessible and various genetic populations, including NAM, MAGIC, and mutagenised populations released. Functional and comparative genomics have elucidated key genetic loci and genes controlling agronomical and adaptive traits. However, the knowledge gained has far away from being translated into real breeding practices. We argue that the way forward is to take a genome-based approach for tailored designing of sorghum as a multi-functional crop combining excellent agricultural traits for various end uses. In this review, we update the new concepts and innovation systems in crop breeding and summarise recent advances in sorghum genomic researches, especially the genome-wide dissection of variations in genes and alleles for agronomically important traits. Future directions and opportunities for sorghum breeding are highlighted to stimulate discussion amongst sorghum academic and industrial communities.
Introduction
A growing global population, coupled with climate change imposing more frequent natural disasters, renders the world’s food security with unprecedented pressure and challenges. The world population is expected to reach 9.7 billion by 2050, and about 70% more food needs to be produced in order to meet the food demand (Tripathi et al. 2019). A recent FAO report indicates that in 2020, an additional 83 to 132 million people will be added to the ranks of the undernourished due to the COVID-19 pandemic, which illustrates the fragility of global food production and supply, and a significant delay on the track to achieve Zero Hunger (The State of Food Security and Nutrition in the World, FAO 2020). Therefore, it is urgent to design proper crop ideotypes to boost more production and avoid global food insecurity.
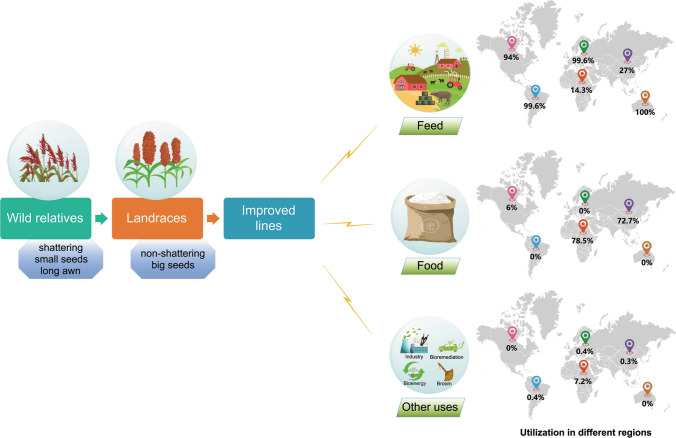
Sorghum (Sorghum bicolor), the fifth most important cereal crop in terms of production and planting area, has attracted widespread attention in recent years as a potential “star” crop to tackle the challenges of global food security. Firstly, cultivated sorghum is remarkably unique in that it has a variety of end uses as food, feed, forage, fuel, beverage and broom (Fig. 1). It also has great potential in the phytoremediation of contaminated soil (Liu et al. 2020). Accordingly, four major groups, including grain sorghum, sweet sorghum, forage sorghum and broom sorghum, are in cultivation worldwide, and the utilisation of different types of sorghum varies considerably in different regions (Batey 2017). Secondly, as a C4 crop with low input requirements and high net return, sorghum is more resilient to adverse environmental conditions and performs well under water and temperature constraints, allowing it to occupy the special niche of marginal land for production without competing with other food crops. It is estimated that there are about 320–702 million hectares of abandoned and degraded cropland are available worldwide (Cai et al. 2011). Planting sorghum on marginal land can not only decrease the pressure in cropland resources, but also improve the soil quality. Thirdly, sorghum is widely cultivated in more than 100 countries around the world. The top 10 sorghum producers, the USA, Sudan, Mexico, Nigeria, India, Niger, Ethiopia, Australia, Brazil and China, contribute about 77% of world sorghum production (Aruna and Cheruku 2018; https://apps.fas.usda.gov). Globally, grain sorghum is a staple food for millions of people in arid and semi-arid regions, particularly in Africa. In developed countries such as the USA, Canada and Australia, it is the main source of livestock feed and industrial uses. In 2018/19, global sorghum production was about 59 million tonnes, of which ~ 64% was used for food seed industrial (FSI), and ~ 36% for feed and residuals consumption (http://apps.fas.usda.gov). In addition, sorghum has received increasing attention as a good source of biologically active compounds which can help prevent chronic diseases and promote human health (Stefoskaneedham et al. 2015). Hence, sorghum is playing a growing pivotal role in global food supply and agro-business.
Fig. 1.
A diagram illustrating the remarkable feature of sorghum as a multiple-purpose crop. Following domestication which changes the traits of domestication syndrome (e.g. seed shattering and gigantism), sorghum is further improved and diversified into various end uses through breeding selection. It can be used not only for forage, food, bioenergy and broom, but also for bioremediation of contaminated cropland. The utilisation of sorghum varies greatly amongst different regions of the word. Generally, it is mainly used for food in developing countries and for feed in developed countries
It should be noted that although great progress has been made in sorghum breeding in the past decades, the average grain yield of sorghum is still very low compared with other cereals. For instance, the global average yield of sorghum in 2018/19 was only ~ 1.49 MT/ha, which is much lower than that of rice (~ 4.58 MT/ha), wheat (~ 3.39 MT/ha) and maize (~ 5.86 MT/ha) (http://www.fao.org/faostat). Moreover, there are serious regional imbalances in global sorghum production. Sorghum yield levels, which are highly influenced by genotype and environment, vary significantly amongst different countries. It is typically high in developed countries, about three times higher than the global average (Upadhyaya et al. 2019). Modern breeding concepts and technologies are urgently needed to accelerate the breeding of sorghum varieties with high genetic yield potential and stress resistance to meet diverse end uses. In this review, we first introduce the concepts of cutting-edge breeding technologies and innovation systems. We then review the recent advances in sorghum genetic and genomic researches. Particular focus is given to essential genes and allelic variations underlying agronomical traits with great potential in sorghum breeding. Additionally, we prospect the future development direction of sorghum molecular breeding using the emerging modern technologies.
State-of-the-art breeding technologies and systems
Crop genetic improvement and breeding aim to increase genetic gain, which is defined as the amount of increase in performance achieved over generations through artificial selection (Xu et al. 2017). Since the invention of DNA technologies, plant breeding has transformed, from conventional breeding which primarily relies on visual phenotypical selection and experience, into the era of molecular breeding (Moose and Mumm 2008). Crop genetic improvement and breeding have been greatly advanced by harnessing and integrating the theories and technologies of DNA markers, marker-assistant selections (MAS) and genetic engineering, as envisaged by more precise foreground and background selection, discovery and utilisation of more diverse genetic stocks and shortening of breeding cycles (Bruce 2012; Collard and Mackill 2008). Yet, agronomically important traits are often so-called complex traits controlled by poly- and/or oligo-genic loci. So far, the success of molecular plant breeding is restricted to limited traits governed by major effect genes due to the intrinsic downsides of low genome coverage of molecular markers, difficulties in exploring a wider range of genetic resources and unavoidable linkage drag associated with selection of large segments of chromosomal regions (Gupta et al. 2010). Hence, breakthroughs in theoretic framework and technologies are required to further speed up breeding practice. Since 2005, the invention of next generation (massively parallel) and third generation (single molecular) sequencing technologies, genome-wide association studies (GWAS), genome editing (GE), molecular modules (MMs), genome selection (GS), as well as non-invasive high throughput phenomics, has revolutionised the scope and toolkits of crop breeding, allowing much more effective exploitation of historically preserved natural and/or artificially generated variation, minor effects of poly- and oligo-genic genes/alleles, and phenotyping and selection of large scale breeding populations in an unprecedented manner (Zhou et al. 2018; Chen et al. 2019; Godwin et al. 2019; Xue et al. 2013; Xu et al. 2020). Here we briefly elaborate the concepts of MMs and GS. Extensive and up-to-date reviews could be found for genetic engineering and GE (Wang and Brummer 2012; Hua et al. 2019).
Molecular modules (MMs)
In molecular genetics, reductionism has been predominantly employed to establish the link between biological trait and the behaviour of molecules, as exemplified by the pioneer work linking the differences in the pea seed colours and fly eyes to the existence and inheritance of variation in genes. The approach has been tremendously successful, leading scientists in the twentieth century to attribute biological phenomena to single molecules. This approach is still and will continue to be powerful. Yet, the facts are that living organisms have a big complex regulatory network with purposes and many components are required to achieve perhaps even a relatively simple biological function. For instance, the perception and signalling of gas phyto-hormone ethylene require multiple genes and gene families, prompting scientists to analyse cellular events through examining interactions.
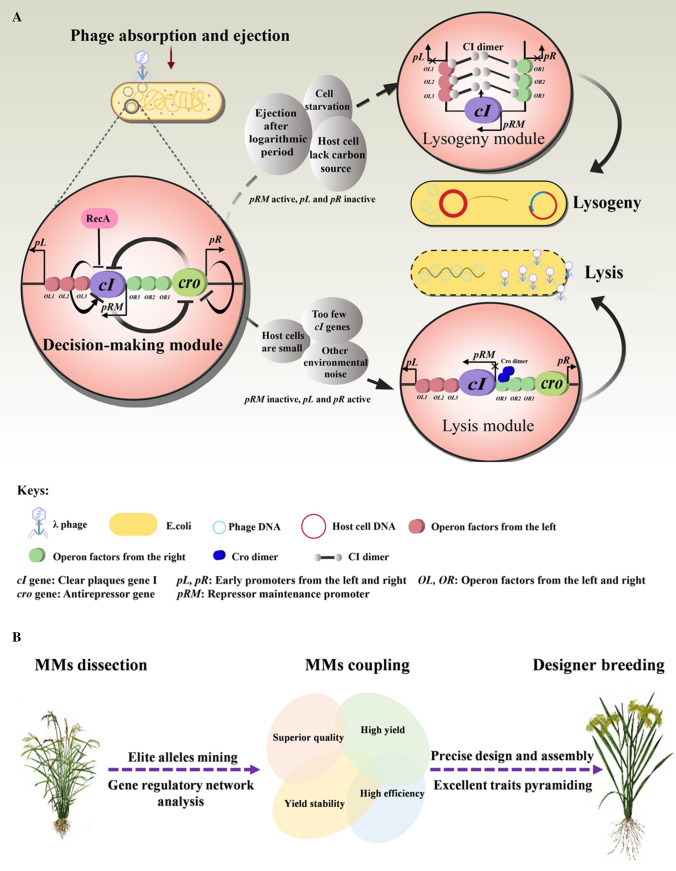
The phage infection process of E. coli is one of the oldest and best-understood MM systems. The first decision-making module in phage was discovered in 1967 (Ptashne et al. 1967). Under different physiological and/or environmental circumstances, the decision-making module determines whether the cell enters a lysogenic or lytic state (Shao et al. 2018) (Fig. 2a). This regulatory process involves two genes (cI, cro) and three promoters (pR, pL and pRM). CI dimers are the key factor of lysogenic state, which can inhibit the transcription of pR by binding to six operators (OL1, OL2, OL3, OR1, OR2, OR3) and form a stable closed loop, causing the phage to enter a stable lysogenic state and to replicate along the bacterial genome (Oppenheim et al. 2005). On the contrary, under conditions where the host cell is nutritious, the phages will be transcribed to a lytic stage through pR to synthesise progeny phage DNA. Cro dimers cut off the closed loop of CI dimers by binding to operator OR3, leading to smooth transcription to the lytic stage (Kobiler et al. 2005).
Fig. 2.
Concepts of the functional module and molecular module. a A diagram deciphering the decision-making functional module in λ phage. This is the first well examined and best understood system of MM. After the phage invades the host cell, the decision-making module determines whether it enters a lysogenic or lytic state in response to physiological environmental signals. Two genes (cI, cro) and three promoters (pR, pL, and pRM) are involved in this regulatory process. CI acts as a regulator of lysogenic state, while Cro protein is the key regulator of lytic state b. A schematic diagram deciphering designer’s breeding by molecular modules (MMs), which consists of three major aspects (modified from Xue et al. (2013)). 1) Mining the functional MMs controlling complex traits, including high yield, superior quality, yield stability and high nutrient efficiency, and analysis of the gene regulatory network and elite allelic variation. 2) Revealing the effects of multi-molecular system coupling and interactions by a multi-module coupling assembly MMs design and breeding innovation system. 3) Achieving the optimal coupling assembly of excellent traits and design breeding of elite varieties.
By analogy, complex agricultural and adaptation traits of crops are products of gene-to-gene and gene-to-environment interactions, rather than being controlled by a single gene or a few genes, which means that complex traits are regulated by a genetic network with "modular" characteristics (Xue et al. 2018a). For instance, the protein–protein interactome network that controls the main biological processes of Arabidopsis is composed of different "modules" (Arabidopsis Interactome Mapping Consortium 2011). In rice, starch synthesis-related genes (SSRGs) could cooperate with each other to form a fine regulating network to control the eating and cooking quality (ECQ), and improved varieties with the desired grain ECQs could be obtained by genetic modification of selected “modules” in this network (Tian et al. 2009). Such MM concepts were further adopted and framed into a so-called “Innovative Designer Breeding System by MMs” (Xue et al. 2013), and successfully exploited in rice breeding (Xue et al. 2018a, b). This breeding system comprehensively utilises molecular biology, genomics and systems biology to study the function of MMs controlling high yield, superior quality, yield stability and high efficiency; then uses computational biology and synthetic biology to couple the MMs and finally achieves the targeted improvement in complex traits and breeding elite varieties (Fig. 2b). Compared with traditional breeding practice, this breeding system has the advantages of less input, higher selection accuracy and effectiveness, and shorter breeding time for pyramiding desirable traits. Through the “Designer Breeding by MMs” strategy, Chinese scientists have successfully designed a series of new elite rice varieties, such as Jiayouzhongke and Zhongke804, which are characterised by high yield, superior quality and resistance to rice blast (Tang and Cheng, 2018).
Genome selection (GS)
GS is a newly developed selection technology, which has been widely used in animal breeding programme (Luan et al. 2009). Unlike traditional marker assistant selection (MAS), GS utilises information from whole genome-wide marker data regardless of whether they are associated with target traits or not (Meuwissen et al. 2001). Thus, GS has at least two advantages over MAS, namely no need to unearth the QTL related to target traits and no need to collect phenotypes of the breeding population, thereby reducing cost (Nakaya and Isobe 2012; Xu et al. 2020). By exploiting the genotype-to-phenotype relationship at the whole-genome level, major and minor gene effects can be captured, reducing cost and time by increasing genetic gain of every cycle (Spindel et al. 2015; Singh and Singh 2015; Guo et al. 2019). GS has been used in crop breeding in recent years, such as wheat, maize and rice (Crossa et al. 2014; Spindel et al. 2015; Schrag et al. 2019). In sorghum, GS mainly focused on model training using a variety of training populations, such as natural population, testcross hybrids and mixed populations (Yu et al. 2016; Dos Santos et al. 2020; Habyarimana et al. 2020).
Over the past two decades, several different statistical models and machine learning methods have been proposed for GS, such as rrBLUP, BayesA/B, random forest and RKHS. Although the optimal models for different traits or purposes may be different, the prediction accuracy values were generally similar and rrBLUP seems to be more popular (Heslot et al. 2012; Arruda et al. 2015; Yu et al. 2016; Dos Santos et al. 2020). However, the effect of the rrBLUP models with peak GWAS markers on prediction accuracy remains controversial (Rice and Lipka 2019; Spindel et al. 2015). A classical cultivar is the combination of many excellent traits; therefore, a multi-trait GS may perform better than single-trait (Budhlakoti et al. 2019; Habyarimana et al. 2020). Marker density is another important factor that affects prediction accuracy. In order to ensure that the maximum number of QTL related to trait is in strong LD with at least one marker, the number of markers should be sufficiently large (Daetwyler et al. 2010). Low-density markers may predict genomic estimated breeding values (GEBVs) with lower accuracy (Singh and Singh 2015). Depending on the trait, the increase in genomic prediction accuracy reaches a plateau as the population size and marker density increases (Arruda et al. 2015).
Sorghum genetic resources and populations
Sorghum germplasm
The two largest sorghum germplasm banks, the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT, http://www.icrisat.org) and the United States Department of Agriculture’s National Plant Germplasm System (USDA-NPGS, http://www.ars-grin.gov), house more than 41,000 accessions, including historic accessions, wild relatives, landraces and improved breeding lines. In addition, about 30 other institutions, such as the Australian Tropical Crops and Forages Genetic Resources Center, which houses the largest collection of Australian wild sorghums, the National Bureau of Plant Genetic Resources in India with about 20,000 collections (www.nbpgr.ernet.in) and the Institute of Crop Germplasm Resources in China, which holds about 16,874 collections (http://www.icgr.caas.net.cn). According to the shape of spikelets/panicles, these sorghum accessions can be classified into five basic races and 10 intermediate types (Dillon et al. 2007). Bicolor is the most primitive of the five races, including primitive forage sorghums with sweet stems; Caudatum is of most recent origin and used for beer brewery; Kafir is insensitive to photoperiod and most male-sterile lines were derived from it; Guinea is the oldest group and mostly photoperiod sensitive; Durra is the most important sorghum type in Ethiopia and is widely used in crop-improvement programs in Asia (Cuevas et al. 2017).
The region of domestication and diversification of sorghum is in Northeast Africa, a special area extending from Ethiopia to Sudan (Wendorf et al. 1992). Hence, Ethiopia and Sudan possess the oldest wild types and richest diversities. Among the > 41,000 accessions stored in USDA-NPGS, 7,217 accessions were collected from Ethiopia as the largest group, and 2,552 from Sudan (Dahlberg et al. 2004). Although the two regions are the main origins of sorghum, their germplasm resources are distinctively different. A representative Sudan core collection includes five ancestral populations, while the Ethiopia core set has eleven ancestral populations, and the pairwise genetic distance amongst the accessions in Sudan core set is larger than that in Ethiopia, suggesting that Sudan collection has genetically higher diversity (Cuevas et al. 2017; Cuevas and Prom 2020). With the strengthening of international cooperation and germplasm exchange, coupled with the exploitation of regional genomic resources, more and more accessions are being utilised in sorghum breeding and genetic research, including association panels, bi-or multi-parental populations and mutagenised populations.
Diversity panels
According to plant height and flowering time, accessions collected in ICRISAT were clustered into 4 groups, core collections representing 10% of the landraces were selected to assemble World Core Collections (WCC) which consists of 2247 landraces. The size of this collection was too large at the time, which made crop improvement programs costly and inefficient (Grenier et al. 2001a; 2001b). To overcome these difficulties, a mini-core collection (MCC) panel of 242 accessions was further assembled by grouping WCC into 21 clusters based on the phenotypic distance and about 10% of the accessions were selected from every cluster (Upadhyaya et al. 2009). To expedite the use of sorghum in temperate-zone breeding programmes, the USDA established the Sorghum Conversion Programme (SCP) to introduce new genetic variation into modern cultivars (Stephens et al. 1967). BTx406, which carries recessive alleles for photoperiod insensitivity and dwarfism at Ma1 and Dw1-Dw3, was used to convert tropical late-flowering sorghums to photoperiod insensitive, early maturing, and short stature phenotypes. SCP not only bred new grain sorghum varieties, but also created rich germplasm for genetic research.
With the rapid development of GWAS, hundreds of accessions have been collected for association mapping in sorghum. Sorghum Association Panel (SAP) was the first panel assembled for GWAS, consisting of 228 tropical lines of SCP and 149 breeding lines (Casa et al. 2008). To better understand the genetic, geographic, and morphological diversity of sorghum, Morris et al. (2013) combined three different panels, including SAP, mini-core collection, and the sorghum reference set to construct a diverse panel of 971 accessions. 580 conversion lines and their paired exotic progenitor lines were also collected for sorghum adaptation analysis (Thurber et al. 2013). A later study on natural variation in dhurrin synthesis and catabolism genes established another SCP panel, consisting of 700 lines (Hayes et al. 2015). Sweet sorghum is an important bioenergy feedstock. To facilitate the sweet sorghum breeding programme, several panels focusing only on sweet or biomass accessions have been created, such as sweet sorghum panel and bioenergy association panel (Murray et al. 2009; Wang et al. 2009; Brenton et al. 2016). Many regional genomic resources have also been exploited, such as the Nigerian Diversity Population consisting of 516 accessions (Maina et al. 2018), a panel of 421 Senegalese sorghum landraces (Faye et al. 2019), a complex set of 1425 Ethiopian landrace accessions (Nida et al. 2019; Girma et al. 2019), and the Sudan core collection of 318 accessions (Cuevas and Prom 2020).
NAM and MAGIC populations
Nested Association Mapping (NAM) population was first established in maize and became popular soon due to its incredible power for dissecting the genomic architecture of phenotypic variations (Yu et al. 2008). By exploiting the advantages of both linkage analysis and association mapping, it can detect small effect loci but limit the false-positives commonly in GWAS. To date, there is only one true NAM population has been reported in sorghum (Bouchet et al. 2017). An elite line, ‘RTx430′, which has been widely used in public and commercial breeding programs, was used as a common parent. Ten diverse lines representing global sorghum diversity were selected as alternate parents and 2214 lines from 10 RIL families formed the final NAM population.
Multi-parent advanced generation inter-cross (MAGIC) population was firstly reported in Arabidopsis (Kover et al. 2009). It offers great potential both for the dissecting genomic structure and improving breeding populations. Up to now, MAGIC populations have been widely used in crops, such as wheat (Huang et al. 2012), maize (Dell’Acqua et al. 2015), and rice (Bandillo et al. 2013). In sorghum, the first MAGIC population was created using 19 diverse founder lines intercrossed randomly through genetic male sterility system, resulting in 1000 inbred lines used for gene mapping and breeding (Ongom and Ejeta 2018).
Mutagenised populations
Targeting Induced Local Lesions In Genomes (TILLING) was firstly developed in Arabidopsis and has been successfully applied to cereal crops, including rice (Horst et al. 2007), wheat (Slade et al. 2005), and maize (Till et al. 2004). In sorghum, two mutant populations were created with ethyl methane sulphate (EMS) mutagenic sorghum reference ‘BTx623′ (Xin et al. 2008; Addo-Quaye et al. 2018). There are 1600 and 4800 mutant individuals in these two mutant populations, respectively. The latter population used a lower sequence coverage depth (average of 7 ×) from 586 independently mutagenised individuals and detected > 5 million homozygous SNPs. These mutant lines provide important resources for forward and reverse genetic studies. At the same time, more mutants of different genetic background should be created to facilitate breeding and research.
Although USDA-NPGS, ICRISAT and other institutes collected thousands of accessions, most of the accessions lack phenotypic and genotyping data. With the decrease in sequencing cost and the rapid development of phenomics, all collections can be genotyped by resequencing and phenotyped. Accession redundancy is the second major problem in germplasm collections because it requires the maintenance and screening of accessions that do not contribute to collection diversity. The third one is the underutilisation of wild sorghums, which has excellent properties and has great potential in sorghum breeding for various end uses (Abdelhalim et al. 2019).
Genomic research in sorghum
Sorghum reference genomes
Sorghum is a diploid crop with a relatively small and non-duplicated genome. The whole reference genome sequence of BTx623 was first assembled by Sanger sequencing (Paterson et al. 2009) and has been updated thereafter by McCormick et al. (2018). It is characterised by 58.8% retrotransposons and 8.7% DNA transposons and ~ 34,129 annotated genes. Based on the genome information of BTx623, many sequence variations including single nucleotide polymorphism (SNPs) and insertions and deletions (InDels), large genomic copy number variation (CNV) and the presence and absence (PAV) have been identified (Zheng et al. 2011; Mace et al. 2013) and several sorghum SNP databases and microarray datasets are now available (Shakoor et al. 2014; Luo et al. 2016; Hu et al. 2019). With the development of sequencing technologies, other sorghum genome sequences have also been reported. Using an Oxford Nanopore sequencing technology together with Bionano Genomics Direct Label and Stain (DLS) optical maps, Deschamps et al. (2018) generated a chromosome-scale de novo assembly of the genome of a grain sorghum line Tx430. Compared with BTx623, Tx430 is characterised by shorter median length and increased number of predicted genes, reaching 39,510 genes. Subsequently, the chromosome-level sweet sorghum Rio genome was assembled to understand the sugar metabolism. The final genome size of RIO was 729.4 Mb, and a total of 35,476 genes were predicted, 54 of which were unique to Rio. Nevertheless, to further understand the underlying genetic mechanism of phenotypic variation during sorghum domestication and improvement, more reference genomes are needed, which may provide opportunities for sorghum genomic research and molecular breeding.
Genetic diversity and population structure
Population genomics is a popular approach to understand population structure, relatedness, linkage disequilibrium, migration and the genetic basis of adaptation at the genome level. Sorghum is an important cereal crop with a long cultivation history and wide adaptability. Understanding the genetic basis of speciation and agroclimatic adaption of sorghum could facilitate molecular breeding and provide valuable information for other crops as well. The naturally diverse resources available for sorghum, including the genetic (grain, forage, sweet and biofuel types) and geographical resources, provide rich materials for population genomic research.
Genetic diversity is an important basis for genetic dissection of complex traits and sorghum breeding, and population structure is crucial for understanding the historical population dynamic, such as population size, geographical population change and genetic differentiation. To understand the domestication and improvement history of sorghum, the genetic diversity and population structure of 44 diverse sorghum lines were analysed by using resequencing data, including 18 landraces, 17 improved inbreds, 7 wild and weedy sorghums, 2 guinea-margaritiferums and 2 S. propinquum (Mace et al. 2013). There was a strong racial structure in the panel, and guinea-margaritiferums was separated from other sorghums, indicating that at least two distinct domestication events involved in the process of sorghum domestication. High LD value with LD decays to background levels (r2 < 0.1) within ~ 150 kb was observed, and LD of wild sorghums decayed faster than those of landrace and improved inbred, indicating that genetic diversity decreased in both landraces and improved inbred.
To explore the diversity of different sorghum races, Morris et al. (2013) analysed the population structure of 971 diverse accessions from the world germplasm collections. The panel can be divided into five morphological types, including kafir, durra, bicolor, caudatum and guinea. A guinea subgroup, which included guinea-margaritiferums types, formed a distinct separate cluster along with wild genotypes from western Africa. Population structure was found to be associated with sorghum morphology types and geographic regional origins, and both agroclimatic and geographic constraints played essential roles in the sorghum diffusion progress within and across agroclimatic zones of Africa and Asia. A high LD value, which is similar to the previous study (Mace et al. 2013), was also observed. Another study of the relationship of trait fitness and deleterious mutations in a whole genome of 229 diverse accessions showed that sorghum racial groups accumulate considerable amounts of deleterious mutations, which decreased trait fitness (Valluru et al. 2019). Additionally, the pattern of mutation burden varied amongst different races, with caudatum having a significantly higher burden as compared to other groups.
A high level of genetic differentiation has also been found in populations composed of local germplasm. A core subset of 374 USDA-NPGS Ethiopian sorghum germplasm can be divided into 11 populations with pairwise Fst values ranging from 0.11–0.47 (Cuevas et al. 2017). Another complex set of 1425 Ethiopian landrace accessions was also divided into 11 subgroups with rich genetic diversity evolved under the local environmental conditions (Girma et al. 2019). In the NPGS Sudan core set of 318 accessions, there were five ancestral populations and the Fst values amongst these populations ranged from 0.17–0.39. IBS analysis showed that the pairwise genetic distance ranged from 0.70 to 0.95, which is higher than that of NPGS Ethiopian sorghum germplasm, indicating a higher genetic diversity in this panel (Cuevas and Prom 2020). Genetic diversity analysis of a diverse panel of 607 Nigerian accessions and 1785 georeferenced global accessions showed that Nigerian accessions were separated from the global accessions and largely clustered together with West African accessions with a pairwise Fst of 0.007. In the Nigerian germplasm, LD decayed to r2 = 0.1 at 50 kb, while it was 100 kb in the West African germplasm (Olatoye et al. 2018).
Sweet sorghum with rich stem juice and sugar is of polyphyletic origin and there are no criteria to separate sweet sorghum from grain sorghum (Murray et al. 2009; Wang et al. 2009). To characterise the genetic diversity and origin of sweet sorghum, different populations consisting of sweet sorghum and/or grain sorghums have been extensively analysed. Although strong population structure was observed, sweet sorghum cannot be clearly grouped into landraces. And it was found that LD decayed to r2 < 0.2 between 2 and 5 kb and to r2 < 0.1 between 5 and 20 kb (Burks et al. 2015). A recent population genomic study of the Dry gene provided evidence that the stem juiciness trait has multiple origins and that the selection of loss-of function dry mutations was an important step in the origin of sweet sorghum (Zhang et al. 2018).
Sorghum domestication
The site and time of sorghum domestication have been previously reviewed, most of which are based on the archaeological evidence of fossil discoveries (Larson et al. 2014; Winchell et al. 2017). Currently, the widely accepted view is that sorghum was early originated from Sudanic Savannah in Africa during middle Holocene, and its domestication took place at eastern Sudan between 6000 and 4000 B.P. Nevertheless, the inference of sorghum domestication events via fossil data suffers from obvious limitations, including the difficulty of evaluating the fossilised phenotypes, the inaccuracy of estimating the fossil’s age, and the shifts in the domestication syndrome altered by weak selection on the candidate domestication genes (Gaut et al. 2018). In general, the evolution of crops is a complex process involving four stages, namely pre-domestication, purposeful cultivation, geographic expansion of the domesticated species for adapting to new environments, and deliberate breeding (Gaut et al. 2018; Meyer and Purugganan 2013). A candidate method to study the stage of crop domestication from their wild genotypes is population modelling, which follows some predefined evolutionary assumptions (Gaut et al. 2018). The indirect genetic evidence for sustained periods of pre-domestication and purposeful cultivation for few crops have been deduced (Meyer et al. 2016; Wang et al. 2017). Different from rice and maize which underwent a single domestication event and experienced a strong in situ selection in the stage of purposeful cultivation (Purugganan 2019), sorghum had a complex domestication history with at least two distinct domestication events (Lin et al. 2012; Morris et al. 2013; Mace et al. 2013). A recent study via the sequencing of ancient sorghum DNA showed that no obvious bottleneck appeared during the domestication process, instead, an overall increasing trend of deleterious mutation load was found (Smith et al. 2019). This scenario is probably related to the particular multiple end-use breeding episodes and hybridisation events frequently occurring amongst different sorghum subpopulations and even with wild relatives (Ohadi et al. 2017; Pedersen et al. 1998).
The domestication syndrome is commonly defined as a set of traits that marks a crop’s divergence from its wild ancestor (Purugganan 2019). In sorghum, domestication phenotypes are manifested as the reduction in seed dispersal, increase in seed retention (non-shattering) and seed size, loss of seed dormancy, changes in reproductive shoot architecture, and synchronous germination (Meyer and Purugganan 2013; Purugganan 2019). These are very similar to those of other cereals, it is therefore anticipated that common suites of genes are involved (Meyer and Purugganan 2013). However, only a few sorghum domestication genes have been reported so far, including Sh1, SbGBSSI and qDor7, which control seed shattering, starch synthesis and seed dormancy, respectively (Lin et al. 2012; Pedersen et al. 2007; Kawahigashi et al. 2013; Li et al. 2016). Other candidate domestication genes have also been announced via the large-scale variation data generated from the whole genome resequencing (Mace et al. 2013; Morris et al. 2013). Domestication genes can also be discovered through the parallel selection theory (Purugganan 2019; Rendón-Anaya and Herrera-Estrella 2018). Our recent effort shows that the 1:1 orthologues of well-studied domestication genes across multiple crops could be involved in sorghum domestication (unpublished data).
GWAS in sorghum
GWAS, a powerful approach for unlocking the complex traits of crops using genome-wide single nucleotide polymorphism (SNPs) markers, has been successfully applied in the dissection of the genetic architecture of complex agronomical traits. In sorghum, diverse association mapping panels mentioned above have been used for GWAS and important genetic loci and genes controlling the complex agricultural traits have been identified. The most widely used panel was constructed by Morris et al. (2013) and genotyped by genotyping-by-sequencing (GBS), resulting in a total of 265,487 SNPs. This panel has been used to dissect the genetic basis of grain yield components (Boyles et al. 2016; Zhang et al. 2015b), grain quality traits (Boyles et al. 2017; Rhodes et al. 2014; Rhodes et al. 2017a; b; Shakoor et al. 2016), inflorescence morphology and plant height (Zhang et al. 2015a; Zhao et al. 2016), cold and heat stress (Ortiz et al. 2017; Chen et al. 2017; Chopra et al. 2017), anthracnose (Colletotrichum sublineolum) resistance (Cuevas et al. 2018), stalk rot diseases resistance (Adeyanju et al. 2015) and grain mould resistance (Cuevas et al. 2019a).
Environment plays an important role in determining the performance of crop, and improving adaption to the environment is essential for ensuring sustainable food production. Therefore, in addition to the above complex agronomical traits, in-depth studies on genotype-by-environment (G × E) interaction in sorghum have also been performed. Sorghum has been diffused widely across diverse agroecological zones since domesticated, and it is a tractable system to investigate the genotype-by-environment (G × E) interaction. By using 104,627 SNPs, the allelic associations with bioclimatic and soil gradients in a panel of 1943 georeferenced sorghum landraces were analysed and the results showed that environmental variables could explain a substantial portion of SNP variation, and genic SNPs were enriched for environmental (drought and Al toxicity) association (Lasky et al. 2015). They suggested that SNP-level knowledge on adaptation could be used to predict adaption traits under a specific environment. Another genome-wide association mapping analysis of precipitation parameters found a small but significant contribution of clinical adaption shaping nucleotide variation in Nigerian sorghum landraces, and genes underlying variation in morphology and flowering time play essential roles in drought adaptation (Olatoye et al. 2018). Using 213,916 SNPs in a set of 421 Senegalese sorghum landraces, Faye et al. (2019) characterised the population structure of genomic diversity and the genomic regions shaped by climate adaption. Both flora regulatory pathways and stay green contributed the climate adaption in Sahelian and Soudanian zones. Recently, the association analysis of seed mass variation with precipitation gradient in a set of 1901 georeferenced sorghum landraces showed that seed mass variation was adaptive to precipitation gradients (Wang et al. 2020). Comprehensive genomics knowledge of adaptive traits would help predict the performance of sorghum in different environments and genomics-based sorghum climate-resilience breeding.
In fact, agricultural and adaptative traits are complex in nature and controlled by multiple genetic and environmental factors. The data obtained generally demonstrate that most of the genetic loci mined could only explain a small part of phenotypic variation. We should note that GWAS relies mainly on statistical analysis and models, which might generate false positive or negative results. SNPs density, phenotypic accuracy, as well as population size and structure, may influence the GWAS results. Currently, most of the genetic variations in sorghum come from GBS, which is subject to generating large quantities of missing data (Annicchiarico et al. 2015). Therefore, larger population size and a higher density of SNPs are necessary. At the same time, effective statistical methods need to be developed to cope with high marker densities and increased sample sizes. Since traditional GWAS mainly focuses on the common variants with minor allele frequency (MAF) ≥ 0.05, while those rare variants with MAF < 0.05, which might explain the phenotypic variation, is often filtered out before GWAS. Thus, developing the special association study and statistical method of these rare variants is of great importance to explore the missing information.
Comparative genomics in sorghum
Comparative genomics analysis has been adopted to explore the genomic variation between sweet and grain sorghums. We previously resequenced two sweet and one grain sorghum genomes and identified a large number of SNPs, InDels, PAVs and CNVs. In addition, we detected nearly 1500 genes differentiating sweet and grain-type sorghums, and some of which are involved in starch and sucrose metabolism, lignin and coumarine-synthesis (Zheng et al. 2011). By comparing the genomics of Rio (sweet sorghum) and BTx623 (grain sorghum), Cooper et al. (2019) identified a high rate of nonsynonymous and potential loss of function mutations in sugar metabolism genes. These results provide important clues for understanding the molecular basis of sweet sorghum associated-traits. So far, there are no reports on the comparative genomics of forage and broom types. In-depth study of the genetic mechanisms underlying distinct phenotypic divergence amongst different sorghums will provide important information for future various end-uses sorghum breeding.
Transcriptome analysis in sorghum
The spatio-temporal datasets derived from transcriptome research are essential for understanding the development of sorghum and its response to the environment. There are several sorghum transcriptomic researches focused on the growth and development (McKinley et al. 2016; Kebrom and Mullet 2016; McCormick et al. 2018) and sugar accumulation (Mizuno et al. 2016; Li et al. 2019). The transcriptome profiles of Della stem from floral initiation to 30 days post-grain maturity identified approximately 200 differentially expressed genes involved in the sorghum biomass accumulation and stem composition (McKinley et al. 2016). More comprehensive transcriptome analysis by collecting 47 samples from roots, leaves, stems, seeds and panicles was further performed to annotate the v3.1 reference genome (McCormick et al. 2018), which complemented the transcriptome data reported in previous studies (McKinley et al. 2016; Mizuno et al. 2016). Our comparative transcriptome analysis of development, salt- and dark-induced senescence in sorghum found that a total of 3396 developmental senescence-associated genes (SAGs) were enriched in 13 KEGG pathways, of which 507 common SAGs were found under three conditions (Wu et al. 2016). Gene expression profiles under abiotic and biotic stress have also been reported (Chopra et al. 2015; Marla et al. 2017; Gelli et al. 2017; Yazawa et al. 2013). More recently, Varoquaux et al. (2019) performed a time-series transcriptome analysis of RTx430 (tolerant to pre-flowering drought but sensitive to post-flowering) and BTx642 (a stay-green genotype, tolerant to post-flowering drought). From seedling to post-anthesis, nearly 400 samples of leaf and root were collected and used for RNA-seq and a total of 3977 genes associated with drought stress in a genotype-specific way were found. The two genotypes showed distinct differences in photosynthesis, reactive oxygen species (ROS) scavenging pathways, and symbiotic relationship with AM fungi under drought stress. Another transcriptome analysis of 29 sorghum accessions in the presence of low-P stress revealed 2089 candidate genes, including plant hormone signal transduction related genes and transcriptional factors (Zhang et al. 2019a). These results provide insights into the function and network of genes involved in the growth and development of sorghum. Further integration with the third-generation and single-cell RNA sequencing will significantly improve the accuracy and specificity of transcriptome sequencing.
Genetic analysis of important agronomical and adaptive traits in sorghum
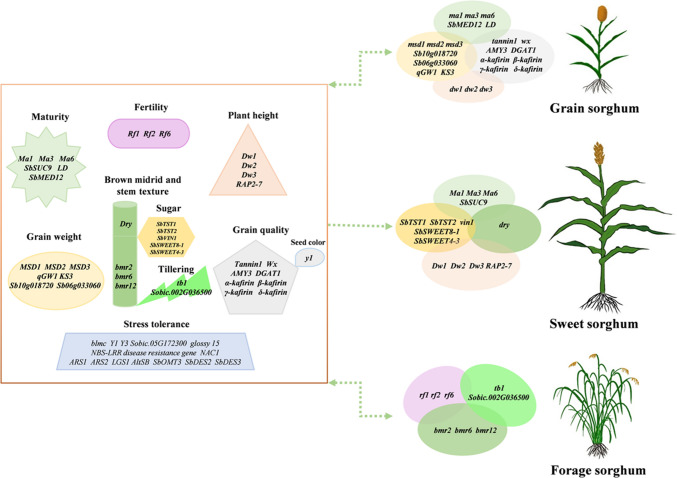
With the rapid development of sequencing and phenotyping technology, a series of important genetic loci and genes controlling sorghum agronomical and adaptive traits have been identified mainly through GWAS, QTL mapping and mutant analysis in the past few decades (Table 1). So far, although the regulation mechanism of these genes remains largely unknown, the knowledge gained and genetic resources generated are fairly good to allow for the design of super sorghum for various end uses (Fig. 3). Below we summarise in detail the progress of the genome-wide dissection of important loci and genes for agronomically important traits.
Table 1.
Major QTL/genes for important agronomical and adaptive traits in sorghum
| Trait | Quantitative trait locus (gene) | Gene ID | Phenotype | Encoding protein | References |
|---|---|---|---|---|---|
| Grain yield | Sh1 | Sobic.001G152901 | Seed shattering | YABBY transcription factor | Lin et al. (2012) |
| SbWRKY | Sobic.001G148000 | Seed shattering | WRKY transcription factor | Tang et al. (2013) | |
| qGW1 | Sobic.001G038900 | Grain weight | Expressed protein | Han et al. (2015) | |
| Unknown | Sb10g018720 | Grain size | Similar to putative fibre protein Fb34 | Zhang et al. (2015b) | |
| Unknown | Sb06g033060 | Grain size | Similar to H0801D08.10 protein | Zhang et al. (2015b) | |
| KS3 | Sb06 g028210 | Seed number | Ent-kaurene synthase | Zhao et al. (2016) | |
| MSD1/SbTCP16 | Sobic.007G135700 | Grain number per panicle | TCP (Teosinte branched/Cycloidea/PCF) transcription factor | Jiao et al. (2018) | |
| MSD2 | Sobic.006G095600 | Grain number per panicle | Lipoxygenase | Gladman et al. (2019) | |
| MSD3 | Sobic.001G407600 | Grain number per panicle | ω-3 Fatty acid desaturase | Dampanaboina et al. (2019) | |
| Grain quality | Unknown | Sobic.005G193000 | Seed protein | α-Kafirin protein | Wu et al. (2013) |
| Unknown | Sobic.009G001600 | Seed protein | β-Kafirin protein | Chamba et al. (2005) | |
| Unknown | Sobic.002G211700 | Seed protein | γ-Kafirin protein | de Freitas et al. (1994) | |
| Unknown | Sb10g013050 | Seed protein | δ-Kafirin protein | Izquierdo and Godwin (2005) | |
| Wx | Sobic.010G022600 | Endosperm texture | Granule-bound ADP-glucose-glucosyl transferase | McIntyre et al. (2008); Sattler et al. (2009) | |
| AMY3 | Sb02g023790 | Fat and protein content | Alpha-amylase debranching enzyme | Rhodes et al. (2017b) | |
| DGAT1 | Sobic.010G170000 | Crude fat | Diacylglyceroal O-acyltransferase 1 | Boyles et al. (2017) | |
| Tannin1/B2 | Sobic.004G280800 | Testa | WD40 protein | Wu et al. (2012); Morris et al. (2013b) | |
| y1 | Sobic.001G398100 | Pericarp color | MYB domain protein | Boddu et al. (2005) | |
| Flowering and height | Ma1/SbPRR37 | Sb06g014570 | Maturity | Pseudoresponse regulator protein 37 | Murphy et al. (2011) |
| Ma3 | Sobic.001G394400 | Maturity | Phytochrome B | Childs et al. (1997) | |
| Ma6 | Sb06g000570 | Maturity | CCT-domain protein | Murphy et al. (2014) | |
| SbSUC9 | Sb06g000520 | Maturity | Sugar transporter | Upadhyaya et al. (2013a) | |
| SbMED12 | Sb01g050280 | Maturity | Flowering regulator | Upadhyaya et al. (2013a) | |
| LD | Sb03g045030 | Maturity | Flowering-time protein | Upadhyaya et al. (2013a) | |
| Dw1/SbHT9.1 | Sobic.009G229800 | Plant height | Membrane protein | Hilley et al. (2016) | |
| Dw2 | Sobic.006G067700 | Plant height | Protein kinase | Hilley et al. (2017) | |
| Dw3 | Sobic.007G163800 | Plant height | Auxin efflux transporter | Multani et al. (2003) | |
| RAP2-7 | Sobic.009G024600 | Plant height | Ethylene responsive transcription factor | Girma et al. (2019) | |
| Brown midrib and stem texture | bmr2 | Sb04g005210 | Brown midrib | 4-Coumarate: coenzyme A ligase | Saballos et al. (2012) |
| bmr6 | Sobic.004G071000 | Brown midrib | Cinnamyl alcohol dehydrogenase | Saballos et al. (2009) | |
| bmr12 | Sobic.007G047300 | Brown midrib | Caffeic O-methyltranferase | Sattler et al. (2012) | |
| Dry | Sobic.006G147400 | Dry stem and midrib | NAC transcription factor | Zhang et al. (2018) | |
| SbTST1 | Sb01g030430 | Sugar accumulation | Tonoplast sugar transporters | Bihmidine et al. (2016) | |
| SbTST2 | Sb04g008150 | Sugar accumulation | Tonoplast sugar transporters | Bihmidine et al. (2016) | |
| SbVIN1 | Sobic.004G004800 | Sugar accumulation | Vacuolar invertase | McKinley et al. (2016) | |
| SbSWEET8-1 | Sobic.008G094000 | Sugar accumulation | Sugar transporters | Mizuno et al. (2016) | |
| SbSWEET4-3 | Sobic.004G136600 | Sugar accumulation | Sugar transporters | Mizuno et al. (2016) | |
| Tillering | tb1 | Sobic.001G121600 | Tillering | Transcription factor | Kebrom et al. (2006; 2010) |
| Cytoplasmic male sterility | Rf1/SbPPR13 | Sb08g019750 | Fertility | Pentatricopeptide repeat protein | Klein et al. (2005) |
| Rf2 | Sobic.002G057050 | Fertility | Pentatricopeptide repeat protein | Madugula et al. (2018) | |
| Rf6 | Sobic.004G004100 | Fertility | Pentatricopeptide repeat protein | Praveen et al. (2015) | |
| Stress tolerance | AltSB | Sobic.003G403000 | Aluminium tolerance and P deficiency | Multidrug and toxic compound extrusion (MATE) transporter | Magalhaes et al. (2007); Leiser et al. (2014) |
| blmc | Sobic.010G001900 | Waxy bloom | Long chain acyl coA oxidase | Burow et al. (2009) | |
| Unknown | Sobic.05G172300 | Anthracnose resistance | F-box domain | Cuevas et al. (2018) | |
| YELLOW SEED1 (Y1) | Sobic.001G398100 | Grain mould resistance | Putative R2R3 MYB transcription factor | Nida et al. (2019) | |
| YELLOW SEED3 (Y3) | Sobic.001G397900 | Grain mould resistance | Putative R2R3 MYB transcription factor | Nida et al. (2019) | |
| glossy 15 | Sb10g025053 | Shoot fly resistance | Similar to Glossy15 | Satish et al. (2009); Aruna et al. (2011) | |
| NBS-LRR disease resistance gene | Sb10g025283 | Shoot fly resistance | NBS-LRR protein | Satish et al. (2009); Aruna et al. (2011) | |
| NAC1 | Sb10g027100 | Shoot fly resistance | NAC domain protein NAC1 | Satish et al. (2009); Aruna et al. (2011) | |
| ARS1 | Sobic.008G036800 | Sorgoleone | Alkylresorcinol synthases | Cook et al. (2010) | |
| ARS2 | Sobic.005G164200 | Sorgoleone | Alkylresorcinol synthases | Cook et al. (2010) | |
| SbOMT3 | Sobic.006G007900 | Sorgoleone | O-Methyltransferase | Baerson et al. (2008) | |
| LGS1 | Sobic.005G213600 | Striga | Sulphotransferase | Gobena et al. (2017) |
Fig. 3.
Tailor design of super sorghum with important genes controlling the complex traits. These genes have been previously reported to be associated with grain yield and quality, fertility, plant height and maturity, juicy and sugar accumulation, tillering, brown midrib, and stress resistance. On the basis of increasing resilience to stress tolerance, super sorghum for various end uses could be accurately designed by pyramiding the super alleles of important genes
Grain yield
The development of non-shattering crop is an essential step to improve sorghum yield. Sh1, encoding a YABBY transcription factor, is the first gene discovered to control seed shattering in sorghum. Non-shattering sorghum harbours three recessive haplotypes of Sh1. Variants in the promoter and intronic region lead to a low expression level of Sh1, a 2.2 kb deletion including exons 2 and 3 results in truncated Sh1, and a GT-to-GG splicing variant in the intron 4 causes the removal of the exon 4 of Sh1 (Lin et al. 2012). SpWRKY is another gene cloned that confers seed shattering in Sorghum propinquum, which was only 300 kb apart from Sh1. SpWRKY is 44 amino acid residues longer than recessive SbWRKY due to the ATG to ATT substitution at the start codon. Besides, there is also a substitution of histidine (H) to glutamine (Q) in amino acid 136, but no amino acids are changed in the WRKY domain. Both Sh1 and SbWRKY might regulate the formation abscission zone in the seed-pedicel junction, but more evidence is necessary to demonstrate the hypothesis (Tang et al. 2013). Besides seed shattering, grain weight, the number of grains per panicle and tiller number are three major factors that influence grain yield. In sorghum, about 340 QTL associated with grain yield have been identified (Mace et al. 2019). qGW1, one important locus for grain weight was fine mapped to a 101 kb region on the short arm of chromosome 1, and the causal gene was Sobic.001G038900, which can be used as a molecular marker for sorghum breeding (Han et al. 2015). Nine hotspots related to seed mass have been identified in a panel of 354 accessions with 265,487 SNPs (Zhang et al. 2015b). Two candidate genes, Sb10g018720 and Sb06g033060, encoding a fibre protein Fb34 and a member of the major facilitator superfamily, respectively, are related to grain size control. Both single SNP allelic variants of these two candidate genes can change the encoding amino acids and lead to an increase in grain size. Boyles et al. (2016) identified 36, 53, 19 significant SNPs, which are associated with grain yield per primary panicle, grain number per primary panicle and 1000-grain weight, respectively. More importantly, there were no overlapping loci for grain number and weight, indicating that these traits can be manipulated independently to increase grain yield. Another GWAS analysis of grain and biomass-related plant architecture traits in sorghum identified 101 SNPs associated with at least one of the nine traits, and KS3, a GA biosynthetic gene, which located in a significant genetic locus on chromosome 6, was associated with seed number (Zhao et al. 2016). Recently, a large-scale GWAS of a panel of 837 sorghum accessions and a BC-NAM population of 1421 individuals dissected 81 QTL related to grain size (Tao et al. 2019). Since the agronomical traits of cereal crops have a similar physiological basis, further comparative studies of different cereal crops can facilitate the research of the genetic basis of sorghum grain yield.
The inflorescence architecture, which affects the number of grains per panicle, is of major importance for sorghum breeding and is more susceptible to environmental influences (Zhang et al. 2015a). Three genes, MSD1, MSD2 and MSD3 have been found to be involved in inflorescence development. They encode a plant-specific transcription factor with a TCP domain, a lipoxygenase, and an ω-3 fatty acid desaturase that catalyses the conversion of linoleic acid (18:2) to linolenic acid (18:3), respectively (Jiao et al. 2018; Dampanaboina et al. 2019; Gladman et al. 2019). MSD1 can bind to the upstream sequences of the transcriptional start site of itself and MSD2 to control the expression. Recessive alleles of MSD1, MSD2 and MSD3 can promote the development of pedicellate spikelets in panicle and make them set grain, resulting in doubling the number of grains per panicle. These three genes are important members in jasmonate (JA) synthesis and can modulate JA pathway during sorghum sex organ development, demonstrating the importance of JA in grain yield. Morris et al. (2013) identified a series of candidate genes homologous to other crops, such as ID1, GDD1 and APO1, which are involved in the transition to flowering in maize (Colasanti et al. 1998), cell elongation (Li et al. 2011), and vegetative and reproductive development (Ikeda et al. 2005) in rice, respectively. In another study, Zhou et al. (2019) identified 35 unique trait-associated SNPs (TASs) associated with inflorescence architecture and nine sorghum homologs of maize and rice candidate genes. The results of Fst and genotype frequencies analysis suggested that chromosomal regions surrounding the TASs might have been targets of breeding selection.
Grain quality
The components of sorghum grain mainly include protein, starch, fat, phenolic compounds and minerals. The proportion of these components directly influences the nutritional value and end use of sorghum grain. Kafirin is the major seed protein in sorghum which can form a tight matrix with starch granules to reduce the digestion of kafirin and starch. α-, β-, γ-, and δ-kafirin are the four subclasses of kafirin. A multigenic family of 20 genes clustered on chromosome 5 are responsible for the synthesis of α-kafirin. A single-point mutation in the signal peptide of α-kafirin renders kafirin the resistance to signal peptide processing and leads to the formation of irregular protein bodies, which significantly promotes protein digestibility and indirectly increase the lysine content (Wu et al. 2013). There are single or low copy number loci for the synthesis of β-, γ-, and δ-kafirin (de Freitas et al. 1994; Chamba et al. 2005; Izquierdo and Godwin 2005). Starch is another important component in sorghum grain. In contrast to normal starch, waxy starch contains little or no amylose and is more digestible. The gene conferring waxy starch, Wx, encodes a granule-bound ADP-glucose-glucosyl transferase. There are three recessive alleles of Wx, of which waxya (wxa) with a large insertion in the third exon, waxyb (wxb) with a missense mutation that results in the conversion of glutamine to histidine at amino acid 268 in the conserved domain, and waxyc(wxc) with a point mutation within the splicing site at the exon–intron boundary (McIntyre et al. 2008; Sattler et al. 2009; Kawahigashi et al. 2013). A GWAS of protein, starch and fat in a panel of 390 SAP accessions identified 4 and 41 significant SNPs associated with protein and fat, respectively. AMY3 (Sb02g023790), encoding a putative homolog of alpha-amylase 3, is associated with fat and protein variation (Rhodes et al. 2017b). Sobic.010G170000, which encodes an important maize DGAT1 homologue gene, is involved in lipid biosynthesis pathway (Boyles et al. 2017). Tannin plays an important role in preventing grain mould and bird predation, but it also has an anti-nutritional effect by reducing the availability of proteins, starch and minerals (de Morais Cardoso et al. 2017). It has been found that orthologue of maize Pr1, homologues of Arabidopsis transparent testa genes, TT4, TT10 and TT16 are related to sorghum polyphenols synthesis (Rhodes et al. 2014; 2017a). Tannin1 (Tan1), which controls the synthesis of anthocyanin and proanthocyanins, encodes a WD40 repeat protein. Haplotype analysis shows that there are two recessive alleles of Tannin1. tan1-a has a 1 bp deletion in an exon and tan1-b has a 10 bp insertion in exon that results in frame shift (Wu et al. 2012). Further research shows that WD40 can form a complex with MYB and bHLH, and promote the expression of GL2, a key negative regulator of fatty acid biosynthesis, thus reducing the level of volatile organic compounds and then resisting the birds attack (Xie et al. 2019). By analysing the natural variation in grain quality in a diversity panel of 196 sorghums, 14, 14, and 492 genetic loci were found to be associated with tannins, starch and amino acids in sorghum, respectively (Kimani et al. 2020). Besides previous known genes associated with grain quality, including Tan1, orthologues of Zm1 and TT16, sucrose phosphate synthases, opaue1 and opaue2, several novel candidate loci were also found, which may be potentially useful for future sorghum breeding.
A Myb gene yellow seed1 (y1), which is homologous to maize p1 gene, determines the pericarp colour in sorghum. In the non-functional alleles of y1 gene (y1-ww), there is a 3218 bp deletion, which corresponds to the 5′ non-coding, putative promoter, exon1, intron1, exon2 and part of the intron 2. Correspondingly, sorghum lines with y1-ww show no detectable accumulation of flavan-4-ols or visible phlobaphenes in the pericarp (Boddu et al. 2005).
Flowering and plant height
Sorghum varies extensively in flowering time and photoperiod sensitivity. Six loci, Ma1 to Ma6, are responsible for flowering time and photoperiod sensitivity and dominant alleles at each locus contribute to late flowering in long days. Ma1, the major repressor of sorghum flowering in long days, encodes the pseudoresponse regulator protein 37 (PRR37). SbPRR37 expression is regulated by the circadian clock and light in a manner consistent with the external coincidence model (Murphy et al. 2011). Ma3 encodes phytochrome B (Childs et al. 1997), and Ma6, as SbGhd7, is a floral repressor regulated by the circadian clock and light signalling (Murphy et al. 2014). SbPRR37 and SbGhd7 act in the additive fashion to downregulate the expression of the floral activator Ehd1 and SbCNs, thereby delaying the floral transition in long days (Murphy et al. 2014). Due to the critical importance of photoperiod sensitivity to crop yield and hybrid seed production, the photoperiod sensitivity loci have been widely used in sorghum breeding. Historically important sorghum accessions, such as SM100 and BTx406, which possess recessive alleles of Sbghd7 and Sbprr37, have been exploited in the SCP to convert the late-flowering photoperiod-sensitive sorghum germplasm to early-flowering photoperiod-insensitive accessions. Genes such as SbSUC9, SbMED12, and LD have also been reported to be related to maturity (Upadhyaya et al. 2013a).
Four loci, Dw1, Dw2, Dw3 and Dw4, which control sorghum height by modifying the internode length have been reported (Quinby and Karper 1954). Dw1 encodes a putative membrane protein of unknown function with highly conserved function in plants (Hilley et al. 2016), and Dw2 encodes a protein kinase that is homologous to the AGCVIII protein kinase KIPK (Hilley et al. 2017). The dw1 allele and dw2 allele have been fully exploited in sorghum breeding, of which dw1 allele could be traced to Dwarf Yellow Milo, the progenitor of grain sorghum, and dw2 allele has been extensively used in U.S. grain sorghum breeding programs and the Sorghum Conversion Program to reduce the length of stems of sorghum genotypes, such as IS3620c and BTx642. Dw3, encoding an ABCB1 auxin efflux transporter, was the first dwarf gene to be cloned in sorghum. Due to the direct duplication in exon 5 of the dw3 allele, dw3 can revert back to Dw3 by unequal crossing-over. However, mutant dw3-sd1 with the disrupted reading frame of DW3, has a stable dwarf phenotype and is expected to be used in sorghum breeding (Multani et al. 2003). For Dw4, although some progress has been made, the corresponding gene has not been cloned yet. In addition, ETHYLENE RESPONSIVE TRANSCRIPTION FACTOR (RAP2-7), which confers flowering time delay, is significantly associated with plant height (Girma et al. 2019).
Brown midrib and stem texture
The brown midrib phenotype has been used to identify mutants that impair lignin content and component, which can improve sorghum biomass for conversion. At least four allelic classes of sorghum bmr mutants, bmr2, bmr6, bmr12, and bmr19, have been identified. bmr2 class leads to a reduction in G-unit and S-unit (Saballos et al. 2008, 2012). bmr6 class results in decreased lignin content, low G-units amount and increased cinnamaldehydes level (Pillonel et al. 1991). bmr12 class causes reduced lignin and positive effect to bioconversion and digestion efficiency (da Silva et al. 2018), and bmr19 class has insignificantly reduced lignin content (Saballos et al. 2008). Among these four allelic classes, bmr6 and bmr12 have been widely used in breeding programs. Genes for Bmr2, Bmr6, and Bmr12 have been reported to encode 4-coumarate coenzyme A ligase, cinnamyl alcohol dehydrogenase, and caffeic O-methyltranferase, respectively (Saballos et al. 2009, 2012; Sattler et al. 2012).
Identifications of genetic loci controlling renewable energy-related traits in sorghum have been developed significantly. The sugar-rich juicy stem is one of the major traits of sweet sorghum. Through an association analysis of sugar yield-related traits, including midrib colour, juice volume, moisture and sugar yield with a total of 42,926 SNPs, Burks et al. (2015) found a significant association at ~ 51.8 Mb on chromosome 6, a region contains a Dry midrib locus. Dry gene, which controls dry versus juicy stems, may affect grain size and drought tolerance in sorghum. It encodes a plant-specific NAC transcription factor, and its loss-of-function mutations in sorghum led to altered stem secondary cell wall composition (Zhang et al. 2018). However, it has not been fully exploited in sorghum breeding due to the lack of information. Dry has 23 different haplotypes in 42 wild sorghums and one S. propinquum, two haplotypes (exactly same with two haplotypes in wild sorghum) in landraces, four haplotypes in improved sorghum. Therefore, it is a good example to study the domestication and improvement in sorghum (Fujimoto et al. 2018; Zhang et al. 2018). The sugars in the juice mainly include sucrose, glucose and fructose, and their content varies significantly amongst different genotypes. Sugar accumulation involves multiple genes, among which SbSWEETs 8–1 and SbSWEETs 4–3 are key sweet genes for sugar transport, respectively, responsible for the efflux of sucrose from leaf and the unloading of sucrose from phloem to stem (Mizuno et al. 2016). tonoplast sugar transporters (SbTST1 and SbTST2) also play essential roles in substantial sugar accumulation (Bihmidine et al. 2016). Down-regulation of vacuolar invertase (SbVIN1) expression has been proved to be inversely associated with sucrose accumulation (McKinley et al. 2016).
To further understand the genetic controls of sorghum bioenergy-related traits, Brenton et al. (2016) performed a GWAS on 390 diverse sorghums composed of sweet and biomass types, and found that a cellulase enzymes and a vacuolar transporter are associated with the non-fibrous carbohydrates accumulation. In the case of forage quality-related traits, Li et al. (2018) analysed the crude protein, neutral detergent fibre, acid detergent fibre, hemicellulose and cellulose contents of 245 sorghum lines, and identified 42 SNPs and 14 candidate genes. More recently, a GWAS of 206 forage sorghum accessions further identified 9 QTLs for lignin content, covering 184 genes. Importantly, 13 of 184 sorghum lignin-related loci were found to have high collinearity with previously reported gene families in other crops (Niu et al. 2020). These studies would facilitate future genetic breeding of sorghum as bioenergy and forage.
Tillering
Tillering is an important agronomical trait which can affect the production and quality of sorghum. The sorghum orthologue of maize domestication gene tb1(Sobic.001G121600), encoding a putative transcription factor, is involved in the regulation of tillering in sorghum and its expression can be affected by phytochrome B (phyB) (Kebrom et al. 2006). Moreover, it might be specifically involved in the shade-mediated repression bud outgrowth (Kebrom et al. 2010). However, its detail function and regulatory mechanism need to be further studied. tin1 has also been reported to control the retention of maize tillering and its function remains conserved among different cereal crops (Zhang et al. 2019b). Thus, its 1:1 orthologue (Sobic.002G036500) might play an important role in in sorghum tillering, which deserves further investigation.
Stress tolerance
Abiotic stress mainly includes drought, extremes of temperature, salinity and heavy metal, and biotic stress mainly refers to disease and pests that may affect the growth and end products of sorghum. Dhurrin, heat shock proteins and antifreeze proteins are associated with drought tolerance (Hayes et al. 2015; Spindel et al. 2018). As a crop of tropical origin, sorghum is sensitive to cold temperatures and increasing the chilling tolerance would help expand sorghum into colder regions. It has been reported that carotenoids, phytohormones, thioredoxin, components of PSI and antioxidants play important roles in chilling tolerance (Ortiz et al. 2017). Another study of cold tolerance identified eight regions associated with final emergence percentage (FEP) and seedling survival (SR) on chromosomes SBI-01, -02, -03, -06, -09 and -10 (Parra-londono et al. 2018b). Colocalisation of chilling tolerance loci with grain tannin and dwarfing genes further indicated that selection for nontannin and dwarfing alleles in early grain sorghum breeding inadvertently resulted in chilling sensitivity (Marla et al. 2019). Similarly, heat stress affects sorghum growth and photosynthesis. Transport proteins, transcription factors, and heat shock proteins are all involved in heat stress (Chen et al. 2017; Chopra et al. 2017).
AltSB is an aluminium-activated citrate transporter identified in sorghum, which can enhance root citrate exudation to form non-toxic complexes with rhizosphere aluminium (Magalhaes et al. 2007). Further research shows that there is a concerted cis–trans interactions mechanism to regulate AltSB expression. Two transcription factors SbWRKY1 and SbZNF1 can bind the MITE repeats of AltSB promoter to regulate its expression. The number of MITE repeats is associated with transactivation activity SbWRKY1 and SbZNF1, and the expression of AltSB. SbWRKY1 and SbZNF1 display different responses to Al3+ toxicity. The expression levels of SbWRKY1 and SbZNF1 in Al-tolerant lines are higher than that in Al-sensitive lines. Fine-tuning the cis–trans interactions will pave the road to enhance plant Al tolerance and crop yield on acidic soils (Melo et al. 2019).
Phosphorus (P) is a vital component of many macromolecules in plant cells. It is an essential nutrient for crop growth and major limiting factors for crop productivity. So far, studies regarding phosphorus assimilation in sorghum are limited. SbMATE, an important gene involved in Al toxicity, is associated with grain yield production under P-limitation, indicating its pleiotropic role in Al toxicity and P deficiency (Leiser et al. 2014). Another study further characterised 19 root-system architecture-related traits of a sorghum panel (n = 194) grown under low and high phosphorus availability and identified a list of genetic loci controlling root development on chromosomes SBI-02, SBI-03, SBI-05 and SBI-09 (Parra-Londono et al. 2018a). Knowledge of the candidate genes might be useful for the breeding of new sorghum cultivars with high phosphorus acquisition.
Anthracnose is one of the major economic constraints restricting sorghum production, especially under warm and humid conditions. Disease resistance-related genes, including two NB-ARC classes of R genes and two hypersensitive response-related genes, have been reported in different panels (Cuevas et al. 2018, 2019b; Cuevas and Prom 2020; Upadhyaya et al. 2013b). In the case of stalk rot diseases, the causal genes included chalcone and stilbene synthase, ROP GTPase, AP2 transcription factor, and pentatricopeptide repeat-containing proteins (Adeyanju et al. 2015). Importantly, the resistant alleles are mainly enriched in durra subpopulations, which is characterised by drought tolerance, indicating that drought tolerance is closely related to stalk rot diseases resistance. As for head smut resistance, Girma et al. (2019) discovered 17 significant SNPs, which can only explain 2% of phenotypic variance. Another GWAS study of 1425 Ethiopian sorghum lines showed that two R2R3 MYB transcription factors, YELLOW SEED1 (Y1) and YELLOW SEED3 (Y3) played essential roles in grain mould resistance (Nida et al. 2019). Shoot fly is one of the most destructive pests in Asia, Africa and the Mediterranean Europe. It can attack sorghum at seedling stage and ultimately reduce the final grain yield (Dhillon et al. 2005). Several candidate genes, such as glossy 15, NBS-LRR disease resistance gene and NAC1 are correlated with sorghum shoot fly resistance (Satish et al. 2009; Aruna et al. 2011).
In Africa and parts of Asia, the parasitic weed Striga is the major biological constraint to sorghum production. The strigolactone is the most potent chemical which can be detected by Striga and results in its germination at the proper time. LGS1 encodes an enzyme annotated as a sulphotransferase and its loss-of-function changed strigolactone chemistry, from 5-deoxystrigol, a highly active Striga germination stimulant, to orobanchol with opposite stereochemistry. In lgs1-1, lgs1-2 and lgs1-3 with low Striga germination stimulant activity, LGS1 gene was completely deleted. In lgs1-4, a 421 bp sequence in the second exon was deleted and in lgs1-5, a 10 bp sequence locating 18 bp upstream of the deleted area in lgs1-4 was deleted. Both lgs1-4 and lgs1-5 caused frame shifts and severely truncated peptides (Gobena et al. 2017). Sorghum breeders can design molecular markers within this gene and introduce into existing lines to effectively control Striga.
Sorghum can produce sorgoleone which confers sorghum the allelopathic properties against many agronomically important weed species. Significant progress has been made in the identification of genes involved in the biosynthetic pathway of sorgoleone. SbDES2 and SbDES3 can catalyse palmitoleic acid (16:1∆9) fatty acyl-CoA into hexadecadienoic acid (16:3∆9, 12, 15) fatty acyl-CoA (Pan et al. 2007). With this long-chain fatty acyl-CoA starter units, ARS1 and ARS2 can produce 5-alkylresorcinols via iterative condensations with malonyl-CoA (Cook et al. 2010). SbOMT3 is capable of catalysing the formation of the 5-pentadecatrienyl resorcinol-3-methyl from 5-pentadecatrienyl resorcinol (Baerson et al. 2008). P450 monooxygenases are likely to catalyse 5-pentadecatrienyl resorcinol-3-methyl into dihydrosorgoleone, which can be converted to the benzoquinone sorgoleone thorough autooxidation, but the gene for P450 monooxygenases have not been cloned (Baerson et al. 2008). Moreover, sorghum can produce epicuticular wax and less permeable cuticle on its culm and leaves, which help itself to tolerate abiotic stress. The BLOOM-CUTICLE (BLMC) locus associated with this phenotype was narrowed down to 153,000 bp region in Chr 10. Sobic.010G001900, encoding the long chain acyl coA oxidase. It is the most significant and interesting candidate gene for profuse wax, but its biological function needs further study (Burow et al. 2009).
Cytoplasmic male sterility
Sorghum is one of the pioneering cereal crops where cytoplasmic genetic male sterility (CMS) is successfully exploited for mass production of F1 hybrid seed. Of the available sorghum cytoplasmic sterility systems, A1 cytoplasm is primarily exploited worldwide, and due to some default of A1 cytoplasm, A2 cytoplasm is the only acceptable alternative to the A1 cytoplasm for commercial hybrid seed production in sorghum. In order to elucidate the molecular mechanisms of fertility restoration, several Rf genes, such as Rf1, Rf2 and Rf6 have been cloned. All the genes encode proteins with a mitochondrial transit peptide and numerous pentatricopeptide repeat (PPR) motifs. Rf1 and Rf2 can restore pollen fertility restoration ability on A1 cytoplasm, and Rf6 controls pollen fertility restoration ability on A1 and A2 cytoplasm in sorghum (Klein et al. 2005; Praveen et al. 2015; Madugula et al. 2018).
In the future, we need to identify more key genes by using modern molecular biology, and examine sorghum orthologues of important genes cloned in other cereals to clarify their roles in sorghum. For cloned genes, exploiting their regulatory mechanism will be helpful to understand their functions. Besides, we should find the elite alleles of cloned genes and arrange them in a more reasonable manner for sorghum breeding.
Future development of sorghum breeding
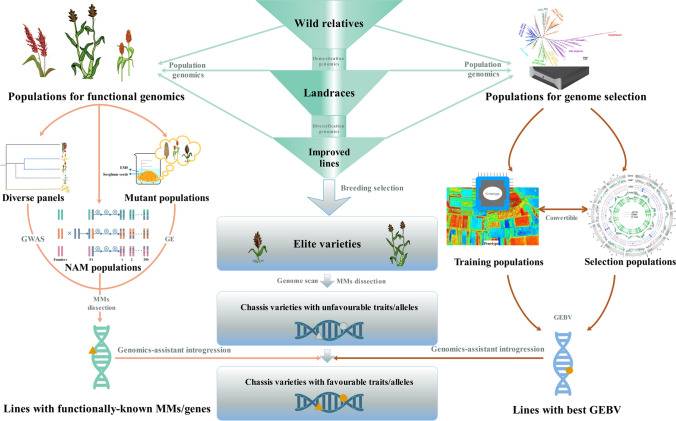
Global demand for food, feed and energy is increasing. It is urgent to explore and develop sustainable food and bioenergy sources. Sorghum is an important versatile crop with a great potential. Compared with other major cereal crops, sorghum breeding has been lagging behind. In the future, genomics should be encouraged or employed as the cornerstone of sorghum breeding programmes. As shown in Fig. 4, genomics could be used to understand the genetic and genomic dynamics of sorghum from wild progenitors to landraces and improved breeding lines by examining the broadness of genetic diversity possessed in the wild species and/or conserved in global germplasm collections, the erosion and decline during domestication and diversification following breeding selection. Such studies of domestication and diversification processes could help identify the “holes” or genomic regions in the designated improved lines and elite varieties contributing negatively to the agronomic performance. Thereafter, populations for functional genomics, either natural diverse panels, structured artificial mapping populations (e.g. the NAM population and its derives) or mutagenised populations, could be compiled to dissect and edit genes and/or functional modules controlling specific traits through GWAS, MM analysis and GE. Simultaneously, populations could be compiled for GS, in which both training and selection populations are constructed and analysed. Both functional genomics and genomic selection will screen for candidate genetic stocks/ breeding lines, either with clearly identified MMs or best Genomic Estimated Breeding Values (GEBVs), which can be used to replace the target genes/alleles/genomic regions in the elite varieties through breeding selection and introgression.
Fig. 4.
Breeding scheme for sorghum improvement using the state-of-the-art genomics-based breeding strategies. The breeding programme consists of four key components. Part I: Diagnosis of changes in genetic diversity during domestication and diversification; Part II: Discovery and characterisation of genetic and genomic variation; Part III: Selection of pre-breeding materials through genome selection; Part IV: Genomics-assistant introgression and improvement in elite varieties. See texts for details
Despite great progress of sorghum genomics and molecular breeding during the past decades, considerably more work is still needed to holistically design sorghum as a multipurpose crop. Firstly, we should pay more attention to the genetic analysis of sorghum complex agronomical traits, especially the identification and validation of important genes and MMs, which are the most important topics in the post-genomic era. For example, plant height and flowering time have been already considered in grain sorghum production at temperate altitude, and major QTL loci and/or genes controlling plant height and flowering have been identified, but the relationship between plant height, photoperiod response and sorghum development is poorly understood. Furthermore, the complex inheritance of juiciness accumulation in sorghum is also far from understanding. Although Dry can control the juicy content in sorghum, its regulation network is unknown yet. Grain sorghums that have juicy stems usually vary in their final grain yields, but it is not clear whether this is related to Dry. There might be a physiological trade-off between juiciness and grain yield. In addition, stay green has been an essential trait in sorghum breeding, and B35, SC56, and E36-1 are generally recognised stay-green sorghums (Reddy Sanjana et al. 2018). Although many stay-green genetic loci including the major four stay-green QTL (Stg1, Stg2, Stg3 and Stg4) have been identified (Xu et al. 2000; Rama Reddy et al. 2014), the causal genes have not been cloned yet. More recently, Kiranmayee et al. (2020) identified seven QTL and several candidate genes controlling stay-green trait in a fine-mapping population, which provides a reliable experimental and theoretical basis for understanding the mechanism of stay green. In the future, in-depth investigations should be performed in terms of unravelling the phenotypic variations and genetic variations, as well as characterising the novel alleles of important genes, MMs and the functional networks conferring important agronomic traits. Further target incorporation of these super alleles of specific genes and functional MMs into superior lines could potentially develop improved varieties.
Secondly, more sorghum genome sequences are needed. Compared with rice and maize, the current exploitation and utilisation of sorghum are far from enough. More genome sequencing of wild, landraces and improved sorghums will provide new genomic variations for the study of sorghum domestication and diversification, as well as pan-genomes of various end-uses. It is suggested that the early domesticated race was bicolor, which then spread to other agroclimatic zones, such as South Asia and Niger Basin (Fuller and Stevens 2018). However, relatively little is known in terms of the origin and evolutionary history of sorghum, which needs more interspecific and intraspecific molecular information. As a unique form of crop species evolution, domestication is a good model for studying the evolutionary processes due to the recent evolution and available archaeological and historical data (Meyer and Purugganan 2013). During this process, dramatic domestication traits are obtained due to the strong selection pressure on the genetic variations. After domestication, crops are subjected to continuously selection and improvement to obtain valuable agronomic traits, such as high yield and quality (Tanksley and McCouch 1997). Both domestications and genetic improvement are evolutionary processes in which the genetic diversity reduction and population expansion occurred, the genetic makeup of wild species was fundamentally altered and complex traits were dramatically improved to meet human needs (Shi and Lai 2015). Thus, genomic variations are of great importance for revealing the genetic diversity, history of domestication, the process of crop speciation, adaptation to local environments, and can facilitate the dissection of key genes and crop breeding. In comparison with other major cereal crops, knowledge of the genomic variation during the domestication and genetic breeding of sorghum is limited. Thus, more wild and cultivated sorghums are necessary for comprehensive analyses of genomic variation, which will provide insights into the basic laws of genetic variation under specific selection conditions, and help us in reconstructing the domestication and improvement events of sorghum and breeding new sorghum varieties.
Understanding the genetic basis of complex agronomic traits is essential for various end-uses sorghum improvement. Nevertheless, a single genome sequence does not adequately represent the entire genomic architecture of a species. Pan-genome analysis, which can collect all the genes at the clad level, provides an opportunity to identify the complete genetic variations within the entire genome repertoire by sequencing multiple individuals of a species. Pan-genome of a species usually includes a core genome and a dispensable genome. The dispensable genome possesses a greater variation than the core genome, and it may be a major contributor to phenotypic variation, plasticity, environmental adaption, organismal interactions and domestication (Hirsch et al. 2014; Li et al. 2014). Progress in pan-genomes of maize, soybean, rice, wheat, Brassica napu and other plants have been recently reviewed (Khan et al. 2019). However, to date, no pan-genome study in sorghum has been reported. Pan-genomic analysis by sequencing and de novo assembling diverse sorghums is of great importance to comprehensively capture the genetic diversity of sorghum gene pool. Further integration of pan-genome analysis and GWAS will significantly improve the characterisation of target genes and genetic variations underlying complex quantitative traits and offer the possibility to develop ideal sorghum varieties for diverse end uses.
Finally, it is urgent to integrate the genome-based technologies and tools into various end-uses sorghum breeding. The genome-based technologies provide the possibility for the development of “tailored super sorghum” with ideal traits. The common main input traits are to improve the resistance to biotic and abiotic stresses, including drought, high and temperature, lodging, diseases and insect pests, while the output traits are to improve grain and biomass yield, grain quality and utilisation efficiency. In fact, sorghum is a climate-smart crop which can be used as food and nonfood uses. Precision molecular breeding of sorghum should be carried out according to different end uses; especially, grain sorghum is characterised by slow digestibility, low cholesterol, antioxidant and other healthy properties and can provide people with rich nutrients. The goal of future breeding is to increase protein (especially lysine) and starch (especially amylopectin) contents and reduce tannins. For energy sorghum, the target traits in breeding programs are to improve its biomass, energy conversion efficiency, sugar content, juicy quality and photosensitivity. At present, low crude protein content, high content of lignin and tannin are the main constraint factors for the end use of sorghum as forage. Improvement in low crude protein, reduction in lignin and tannin content is the main target for forage breeding. And increasing the tillering capacity, quick growth, multicut potential and reducing cyanogenic potential should also be considered in the breeding program.
Although modern approaches can accelerate the breeding cycle of sorghum and assist breeders to develop super-varieties, their application in sorghum breeding is in its infancy and there is still a long way to go. In terms of MM, how to effectively identify and pyramid the super alleles that control the important agronomic traits at the genome-wide level is still a great challenge for sorghum breeding. Some important aspects should be considered according to the end-use of sorghum, such as the number of target genes, population size and choice of parents. GS in sorghum also needs to pay attention to the accuracy of genotyping and phenotyping, the choice and improvement in the model according to different populations and gene-environment interactions. Genome editing plays an essential role in identification of gene function and crop genetic improvement. However, its application heavily depends on the transformation efficiency. Although much progress has been made in Agrobacterium-mediated transformation of sorghum, the transformation efficiency is still much lower than those of other crops. Further optimisation of the transformation system is necessary to promote the application of CRISPR/ Cas9 system in sorghum breeding.
Conclusion
Although great progress has been made in the identification the genetic loci underlying important agronomical and adaptive traits, high-throughput sequencing, phenotyping, pan-genomes, epigenomes and other disciplines should be integrated to identify the genetic diversity, superior alleles and the functional regulation network underlying the complex agronomical traits and the stress resilience of sorghum. Meanwhile, standards for the management, integration and sharing of growing bioscience data should be developed to realise the rational use of data and avoid waste of resources. In addition, efficient breeding strategies are also necessary to effectively combine new breeding approaches to develop new varieties with improved target traits. We believe that these will bring desirable changes to the genetic breeding of sorghum.
Acknowledgements
This study was supported by grants from the National Key R&D Program of China (2019YFD1002701; 2018YFD1000701) and the Chinese Academy of Sciences (151111KYSB20180049; KFJ-FP-201809; KFJ-STS-ZDTP-056).
Author contributions statement
All authors discussed and wrote the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Huaiqing Hao, Email: hqhao@ibcas.ac.cn.
Hai-Chun Jing, Email: hcjing@ibcas.ac.cn.
References
- Abdelhalim TS, Kamal NM, Hassan AB. Nutritional potential of wild sorghum: grain quality of Sudanese wild sorghum genotypes (Sorghum bicolor L. Moench) Food Sci Nutr. 2019;7:1529–1539. doi: 10.1002/fsn3.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C, Tuinstra M, Carraro N, Weil C, Dilkes BP. Whole genome sequence accuracy is improved by replication in a population of mutagenized sorghum. G3 (Bethesda) 2018;28:1079–1094. doi: 10.1534/g3.117.300301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyanju A, Little C, Yu J, Tesso T. Genome-wide association study on resistance to stalk rot diseases in grain sorghum. G3 (Bethesda) 2015;5:1165–1175. doi: 10.1534/g3.114.016394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annicchiarico P, Nazzicari N, Li X, Wei Y, Pecetti L, Brummer EC. Accuracy of genomic selection for alfalfa biomass yield in different reference populations. BMC Genomics. 2015;16:1020–1032. doi: 10.1186/s12864-015-2212-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda MP, Brown PJ, Lipka AE, Krill AM, Thurber C, Kolb FL. Genomic selection for predicting fusarium head blight resistance in a wheat breeding program. Plant Genome. 2015;8:1–12. doi: 10.3835/plantgenome2015.01.0003. [DOI] [PubMed] [Google Scholar]
- Aruna C, Bhagwat VR, Madhusudhana R, Sharma V, Hussain T, Ghorade RB, Khandalkar HG, Audilakshmi S, Seetharama N. Identification and validation of genomic regions that affect shoot fly resistance in sorghum [Sorghum bicolor (L.) Moench] Theor Appl Genet. 2011;122:1617–1630. doi: 10.1007/s00122-011-1559-y. [DOI] [PubMed] [Google Scholar]
- Aruna C, Cheruku D. Genetic improvement of grain sorghum. In: Aruna C, Visarada KBRS, Bhat BV, Tonapi VA, editors. Breeding sorghum for diverse end uses. Cambridge, UK: Woodhead Publishing; 2018. pp. 157–173. [Google Scholar]
- Baerson SR, Dayan FE, Rimando AM, Nanayakkara NPD, Liu CJ, Schroder J, Fishbein M, Pan ZQ, Kagan IA, Pratt LH, Cordonnier-Pratt MM, Duke SO. A functional genomics investigation of allelochemical biosynthesis in Sorghum bicolor root hairs. J Biol Chem. 2008;283:3231–3247. doi: 10.1074/jbc.M706587200. [DOI] [PubMed] [Google Scholar]
- Bandillo N, Raghavan C, Muyco PA, Sevilla MAL, Lobina IT, Dilla-Ermita CJ, Tung CW, McCouch S, Thomson M, Mauleon R, Singh RK, Gregorio G, Redoña E, Leung H. Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice. 2013;6:11–25. doi: 10.1186/1939-8433-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey I. The diversity of uses for cereal grains. In: Wrigley C, Batey I, Miskelly D, editors. Cereal grains. Amsterdam, Netherlands: Elsevier; 2017. pp. 41–53. [Google Scholar]
- Bihmidine S, Julius BT, Dweikat I, Braun DM. Tonoplast sugar transporters (SbTSTs) putatively control sucrose accumulation in sweet sorghum stems. Plant Signal Behav. 2016;11:e1117721. doi: 10.1080/15592324.2015.1117721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddu J, Svabek C, Ibraheem F, Jones DA, Chopra S. Characterization of a deletion allele of a sorghum Myb gene yellow seed1 showing loss of 3-deoxyflavonoids. Plant Sci. 2005;169:542–552. doi: 10.1016/j.plantsci.2005.05.007. [DOI] [Google Scholar]
- Bouchet S, Olatoye MO, Marla SR, Perumal R, Tesso T, Yu J, Tuinstra M, Morris GP. Increased power to dissect adaptive traits in global sorghum diversity using a nested association mapping population. Genetics. 2017;206:573–585. doi: 10.1534/genetics.116.198499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyles RE, Cooper EA, Myers MT, Brenton Z, Rauh BL, Morris GP, Kresovich S. Genome-wide association studies of grain yield components in diverse sorghum germplasm. Plant Genome. 2016;9:1–17. doi: 10.3835/plantgenome2015.09.0091. [DOI] [PubMed] [Google Scholar]
- Boyles RE, Pfeiffer BK, Cooper EA, Rauh BL, Zielinski KJ, Myers MT, Brenton Z, Rooney WL, Kresovich S. Genetic dissection of sorghum grain quality traits using diverse and segregating populations. Theor Appl Genet. 2017;130:697–716. doi: 10.1007/s00122-016-2844-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenton ZW, Cooper EA, Myers MT, Boyles RE, Shakoor N, Zielinski KJ, Rauh BL, Bridges WC, Morris GP, Kresovich S. A genomic resource for the development, improvement, and exploitation of sorghum for bioenergy. Genetics. 2016;204:21–33. doi: 10.1534/genetics.115.183947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce TJA. GM as a route for delivery of sustainable crop protection. J Exp Bot. 2012;63:537–541. doi: 10.1093/jxb/err281. [DOI] [PubMed] [Google Scholar]
- Budhlakoti N, Mishra DC, Rai A, Lal SB, Chaturvedi KK, Kumar RR. A comparative study of single-trait and multi-trait genomic selection. J Comput Biol. 2019;26:1100–1112. doi: 10.1089/cmb.2019.0032. [DOI] [PubMed] [Google Scholar]
- Burks PS, Kaiser CM, Hawkins EM, Brown PJ. Genome-wide association for sugar-yield in sweet sorghum. Crop Sci. 2015;55:2138–2148. doi: 10.2135/cropsci2015.01.0057. [DOI] [Google Scholar]
- Burow GB, Franks CD, Acosta-Martinez V, Xin ZG. Molecular mapping and characterization of BLMC, a locus for profuse wax (bloom) and enhanced cuticular features of Sorghum (Sorghum bicolor (L.) Moench.) Theor Appl Genet. 2009;118:423–431. doi: 10.1007/s00122-008-0908-y. [DOI] [PubMed] [Google Scholar]
- Cai XM, Zhang X, Wang DB. Land availability for biofuel production. Environ Sci Technol. 2011;45:334–339. doi: 10.1021/es103338e. [DOI] [PubMed] [Google Scholar]
- Casa AM, Pressoir G, Brown PJ, Mitchell SE, Rooney WL, Tuinstra MR, Franks CD, Kresovich S. Community resources and strategies for association mapping in sorghum. Crop Sci. 2008;48:30–40. doi: 10.2135/cropsci2007.02.0080. [DOI] [Google Scholar]
- Chamba EB, Halford NG, Forsyth J, Wilkinson M, Shewry PR. Molecular cloning of β-kafirin, a methionine-rich protein of sorghum grain. J Cereal Sci. 2005;41:381–383. doi: 10.1016/j.jcs.2004.09.004. [DOI] [Google Scholar]
- Chen J, Chopra R, Hayes C, Morris G, Marla S, Burke J, Xin Z, Burow G. Genome-wide association study of developing leaves’ heat tolerance during vegetative growth stages in a sorghum association panel. Plant Genome. 2017;10:1–15. doi: 10.3835/plantgenome2016.09.0091. [DOI] [PubMed] [Google Scholar]
- Chen KL, Wang YP, Zhang R, Zhang H, Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- Childs KL, Miller FR, Cordonnier-Pratt MM. The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Phys. 1997;113:611–619. doi: 10.1104/pp.113.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra R, Burow G, Burke JJ, Gladman N, Xin Z. Genome-wide association analysis of seedling traits in diverse sorghum germplasm under thermal stress. BMC Plant Biol. 2017;17:12–26. doi: 10.1186/s12870-016-0966-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra R, Burow G, Hayes C, Emendack Y, Xin Z, Burke J. Transcriptome profiling and validation of gene based single nucleotide polymorphisms (SNPs) in sorghum genotypes with contrasting responses to cold stress. BMC Genom. 2015;16:1040–1050. doi: 10.1186/s12864-015-2268-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell. 1998;93:593–603. doi: 10.1016/S0092-8674(00)81188-5. [DOI] [PubMed] [Google Scholar]
- Collard BC, Mackill DJ. Mark ABERRANT PANICLE ORGANIZATION 1 temporally regulates er-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond B Biol Sci. 2008;363:557–572. doi: 10.1098/rstb.2007.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Rimando AM, Clemente TE, Schroder J, Dayan FE, Dhammika Nanayakkara NP, Pan ZQ, Noonan BP, Fishbein M, Abe I, Duke SO, Baersona SR. Alkylresorcinol synthases expressed in Sorghum bicolor root hairs play an essential role in the biosynthesis of the allelopathic benzoquinone sorgoleone. Plant Cell. 2010;22:867–887. doi: 10.1105/tpc.109.072397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper EA, Brenton ZW, Flinn BS, Jenkins J, Shu SQ, Flowers D, Luo F, Wang YS, Xia P, Barry K, Daum C, Lipzen A, Yoshinaga Y, Schmutz J, Saski C, Vermerris W, Kresovich S. A new reference genome for Sorghum bicolor reveals high levels of sequence similarity between sweet and grain genotypes: implications for the genetics of sugar metabolism. BMC Genom. 2019;20:420–432. doi: 10.1186/s12864-019-5734-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J, Perez P, Hickey J, Burgueno J, Ornella L, Ceron-Rojas J, Zhang X, Dreisigacker S, Babu R, Li Y, Bonnett D, Mathews K. Genomic prediction in CIMMYT maize and wheat breeding programs. Heredity. 2014;112:48–60. doi: 10.1038/hdy.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas HE, Fermin-Perez RA, Prom LK, Cooper EA, Bean S, Rooney WL. Genome-wide association mapping of grain mold resistance in the US sorghum association panel. Plant Genome. 2019;12:1–11. doi: 10.3835/plantgenome2018.09.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas HE, Prom LK, Cooper EA, Knoll JE, Ni X. Genome-wide association mapping of anthracnose (Colletotrichum sublineolum) resistance in the U.S. sorghum association panel. Plant Genome. 2018;11:170099. doi: 10.3835/plantgenome2017.11.0099. [DOI] [PubMed] [Google Scholar]
- Cuevas HE, Prom LK, Cruet-Burgos CM. Genome-wide association mapping of anthracnose (Colletotrichum sublineolum) resistance in NPGS Ethiopian sorghum germplasm. G3 (Bethesda) 2019;9:2879–2885. doi: 10.1534/g3.119.400350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas HE, Prom LK. Evaluation of genetic diversity, agronomic traits, and anthracnose resistance in the NPGS Sudan sorghum core collection. BMC Genom. 2020;21:88–102. doi: 10.1186/s12864-020-6489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas HE, Rosa-Valentin G, Hayes CM, Rooney WL, Hoffmann L. Genomic characterization of a core set of the USDA-NPGS Ethiopian sorghum germplasm collection: implications for germplasm conservation, evaluation, and utilization in crop improvement. BMC Genom. 2017;18:108–124. doi: 10.1186/s12864-016-3475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler HD, Pong-Wong R, Villanueva B, Woolliams JA. The impact of genetic architecture on genome-wide evaluation methods. Genetics. 2010;185:1021–1031. doi: 10.1534/genetics.110.116855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg JA, Burke JJ, Rosenow DT. Development of a sorghum core collection: refinement and evaluation of a subset from Sudan. Econ Bot. 2004;58:556–567. doi: 10.1663/0013-0001(2004)058[0556:DOASCC]2.0.CO;2. [DOI] [Google Scholar]
- Dampanaboina L, Jiao YP, Chen JP, Gladman N, Chopra R, Burow G, Hayes C, Christensen SA, Burke J, Ware D, Xin ZG. Sorghum MSD3 encodes an ω-3 fatty acid desaturase that increases grain number by reducing jasmonic acid levels. Int J Mol Sci. 2019;20:5359. doi: 10.3390/ijms20215359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva MJ, Carneiro PCS, de Souza Carneiro JE, Damasceno CMB, Parrella NNLD, Pastina MM, Simeone MLF, Schaffert RE, da Costa Parrella RA. Evaluation of the potential of lines and hybrids of biomass sorghum. Ind Crops Prod. 2018;125:379–385. doi: 10.1016/j.indcrop.2018.08.022. [DOI] [Google Scholar]
- de Freitas FA, Yunes JA, da Silva MJ, Arruda P, Leite A. Structural characterization and promoter activity analysis of the y-kafirin gene from sorghum. Molec Gen Genet. 1994;245:177–186. doi: 10.1007/BF00283265. [DOI] [PubMed] [Google Scholar]
- Dell’Acqua M, Gatti DM, Pea G, Cattonaro F, Coppens F, Magris G, Hlaing AL, Aung HH, Nelissen H, Baute J, Frascaroli E, Churchill GA, Inzé D, Morgante M, Pè ME. Genetic properties of the MAGIC maize population: a new platform for high definition QTL mapping in Zea mays. Genome Biol. 2015;16:167–189. doi: 10.1186/s13059-015-0716-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Morais CL, Pinheiro SS, Martino HSD, Pinheiro-SantAna HM. Sorghum (Sorghum bicolor L.): Nutrients, bioactive compounds, and potential impact on human health. Crit Rev Food Sci Nutr. 2017;57:372–390. doi: 10.1080/10408398.2014.887057. [DOI] [PubMed] [Google Scholar]
- Deschamps S, Zhang Y, Llaca V, Ye L, Sanyal A, King M, May G, Lin HN. A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping. Nat Commun. 2018;9:4844–4853. doi: 10.1038/s41467-018-07271-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon MK, Sharma HC, Singh R, Naresh JS. Mechanisms of resistance to shoot fly, Atherigona soccata in sorghum. Euphytica. 2005;144:301–312. doi: 10.1007/s10681-005-7400-4. [DOI] [Google Scholar]
- Dillon SL, Shapter FM, Henry RJ, Cordeiro G, Izquierdo L, Lee LS. Domestication to crop improvement: Genetic resources for Sorghum and Saccharum (Andropogoneae) Ann Bot. 2007;100:975–989. doi: 10.1093/aob/mcm192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos JPR, Fernandes SB, McCoy S, Lozano R, Brown PJ, Leakey ADB, Buckler ES, Garcia AAF, Gore MA. Novel bayesian networks for genomic prediction of developmental traits in biomass sorghum. G3 (Bethesda) 2020;10:769–781. doi: 10.1534/g3.119.400759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye JM, Maina F, Hu ZB, Fonceka D, Cisse N, Morris GP. Genomic signatures of adaptation to Sahelian and Soudanian climates in sorghum landraces of Senegal. Ecol Evol. 2019;9:6038–6051. doi: 10.1002/ece3.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Sazuka T, Oda Y, Kawahigashi H, Wu JZ, Takanashi H, Ohnishi T, Yoneda JI, Ishimori M, Kajiya-Kanegae H, Hibara KI, Ishizuna F, Ebine K, Ueda T, Tokunaga T, Iwata H, Matsumoto T, Kasuga S, Yonemaru JI, Tsutsumi N. Transcriptional switch for programmed cell death in pith parenchyma of sorghum stems. Proc Natl Acad Sci USA. 2018;115:E8783–E8792. doi: 10.1073/pnas.1807501115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DQ, Stevens CJ. Sorghum domestication and diversification: A current archaeobotanical perspective. In: Mercuri AM, D'Andrea A, Fornaciari R, Hohn A, editors. Plants and people in the African past. Cham: Springer; 2018. pp. 427–452. [Google Scholar]
- Gaut BS, Seymour DK, Liu QP, Zhou YF. Demography and its effects on genomic variation in crop domestication. Nature Plants. 2018;4:512–520. doi: 10.1038/s41477-018-0210-1. [DOI] [PubMed] [Google Scholar]
- Gelli M, Konda AR, Liu K, Zhang C, Clemente TE, Holding DR, Dweikat IM. Validation of QTL mapping and transcriptome profiling for identification of candidate genes associated with nitrogen stress tolerance in sorghum. BMC Plant Biol. 2017;17:123–140. doi: 10.1186/s12870-017-1064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girma G, Nida H, Seyoum A, Mekonen M, Nega A, Lule D, Dessalegn K, Bekele A, Gebreyohannes A, Adeyanju A, Tirfessa A, Ayana G, Taddese T, Mekbib F, Belete K, Tesso T, Ejeta G, Mengiste T. A large-scale genome-wide association analyses of Ethiopian sorghum landrace collection reveal loci associated with important traits. Front Plant Sci. 2019;10:691–705. doi: 10.3389/fpls.2019.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladman N, Jiao YP, Lee YK, Zhang LF, Chopra R, Regulski M, Burow G, Hayes C, Christensen SA, Dampanaboina L, Chen JP, Burke J, Ware D, Xin ZG. Fertility of pedicellate spikelets in sorghum is controlled by a jasmonic acid regulatory module. Int J Mol Sci. 2019;20:4951–4965. doi: 10.3390/ijms20194951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobena D, Shimels M, Rich PJ, Ruyter-Spira C, Bouwmeester H, Kanuganti S, Mengiste T, Ejeta G. Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. Proc Natl Acad Sci U S A. 2017;114:4471–4476. doi: 10.1073/pnas.1618965114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin ID, Rutkoski J, Varshney RK, Hickey LT. Technological perspectives for plant breeding. Theor Appl Genet. 2019;132:555–557. doi: 10.1007/s00122-019-03321-4. [DOI] [PubMed] [Google Scholar]
- Grenier C, Bramel-Cox PJ, Hamon P. Core collection of sorghum: I. Stratification based on eco-geographical data. Crop Sci. 2001;41:234–240. doi: 10.2135/cropsci2001.411234x. [DOI] [Google Scholar]
- Grenier C, Hamon P, Bramel-Cox PJ. Core collection of sorghum: II. Comparison of three random sampling strategies. Crop Sci. 2001;41:241–246. doi: 10.2135/cropsci2001.411241x. [DOI] [Google Scholar]
- Guo TT, Yu XQ, Li XR, Zhang HZ, Zhu CS, Flint-Garcia S, McMullen MD, Holland JB, Szalma SJ, Wisser RJ, Yu JM. Optimal designs for genomic selection in hybrid crops. Mol Plant. 2019;12:390–401. doi: 10.1016/j.molp.2018.12.022. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Kumar J, Mir RR, Kumar A. Marker-assisted selection as a component of conventional plant breeding. Plant Breed Rev. 2010;33:145–217. [Google Scholar]
- Habyarimana E, Lopez-Cruz M, Baloch FS. Genomic selection for optimum index with dry biomass yield, dry mass fraction of fresh material, and plant height in biomass sorghum. Genes (Basel) 2020;11:61–76. doi: 10.3390/genes11010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han LJ, Chen J, Mace ES, Liu YS, Zhu MJ, Yuyama N, Jordan DR, Cai HW. Fine mapping of qGW1, a major QTL for grain weight in sorghum. Theor Appl Genet. 2015;128:1813–1825. doi: 10.1007/s00122-015-2549-2. [DOI] [PubMed] [Google Scholar]
- Hayes CM, Burow GB, Brown PJ, Thurber C, Xin ZG, Burke JJ. Natural variation in synthesis and catabolism genes influences dhurrin content in sorghum. Plant Genome. 2015;8:1–9. doi: 10.3835/plantgenome2014.09.0048. [DOI] [PubMed] [Google Scholar]
- Heslot N, Yang HP, Sorrells ME, Jannink JL. Genomic selection in plant breeding A comparison of models. Crop Sci. 2012;52:146–161. doi: 10.2135/cropsci2011.06.0297. [DOI] [Google Scholar]
- Hilley J, Truong S, Olson S, Morishige D, Mullet J. Identification of Dw1, a regulator of sorghum stem internode length. PLoS ONE. 2016;11:e0151271. doi: 10.1371/journal.pone.0151271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilley JL, Weers BD, Truong SK, McCormick RF, Mattison AJ, McKinley BA, Morishige DT, Mullet JE. Sorghum Dw2 encodes a protein kinase regulator of stem internode length. Sci Rep. 2017;7:4616–4628. doi: 10.1038/s41598-017-04609-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch CN, Foerster JM, Johnson JM, Sekhon RS, Muttoni G, Vaillancourt B, Peñagaricano F, Lindquist E, Pedraza AM, Barry K, de Leon N, Kaeppler SM, Buell CR. Insights into the maize pan-genome and pantranscriptome. Plant Cell. 2014;26:121–135. doi: 10.1105/tpc.113.119982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst I, Welham T, Kelly S, Kaneko T, Sato S, Tabata S, Parniske M, Wang TL. TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiol. 2007;144:806–820. doi: 10.1104/pp.107.097063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu ZB, Olatoye MO, Marla S, Morris GP. An integrated genotyping-by-sequencing polymorphism map for over 10,000 sorghum genotypes. Plant Genome. 2019;12:180044. doi: 10.3835/plantgenome2018.06.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua K, Zhang J, Botella JR, Ma C, Kong F, Liu B, Zhu JK. Perspectives on the application of genome-editing technologies in crop breeding. Mol Plant. 2019;12:1047–1059. doi: 10.1016/j.molp.2019.06.009. [DOI] [PubMed] [Google Scholar]
- Huang BE, George AW, Forrest KL, Kilian A, Hayden MJ, Morell MK, Cavanagh CR. A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol J. 2012;10:826–839. doi: 10.1111/j.1467-7652.2012.00702.x. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Nagasawa N, Nagato Y. ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev Biol. 2005;282:349–360. doi: 10.1016/j.ydbio.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Izquierdo L, Godwin I. Molecular characterization of a novel methionine-rich δ-kafirin seed storage protein gene in sorghum (Sorghum bicolor L.) Cereal Chem. 2005;82:706–710. doi: 10.1094/CC-82-0706. [DOI] [Google Scholar]
- Jiao YP, Lee YK, Gladman N, Chopra R, Christensen SA, Regulski M, Burow G, Hayes C, Burke J, Ware D, Xin ZG. MSD1 regulates pedicellate spikelet fertility in sorghum through the jasmonic acid pathway. Nat Commun. 2018;9:822–830. doi: 10.1038/s41467-018-03238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahigashi H, Oshima M, Nishikawa T, Okuizumi H, Kasuga S, Yonemaru J. A novel waxy allele in sorghum landraces in East Asia. Plant Breed. 2013;132:305–310. doi: 10.1111/pbr.12054. [DOI] [Google Scholar]
- Kebrom TH, Brutnell TP, Finlayson SA. Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ. 2010;33:48–58. doi: 10.1111/j.1365-3040.2009.02050.x. [DOI] [PubMed] [Google Scholar]
- Kebrom TH, Burson BL, Finlayson SA. Phytochrome B represses teosinte branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 2006;140:1109–1117. doi: 10.1104/pp.105.074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom TH, Mullet JE. Transcriptome profiling of tiller buds provides new insights into PhyB regulation of tillering and indeterminate growth in sorghum. Plant Physiol. 2016;170:2232–2250. doi: 10.1104/pp.16.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AW, Garg V, Roorkiwal M, Golicz AA, Edwards D, Varshney RK. Super-pangenome by integrating the wild side of a species for accelerated crop improvement. Trends Plant Sci. 2019;25:148–158. doi: 10.1016/j.tplants.2019.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimani W, Zhang LM, Wu XY, Hao HQ, Jing HC. Genome-wide association study reveals that different pathways contribute to grain quality variation in sorghum (Sorghum bicolor) BMC Genom. 2020;21:112–130. doi: 10.1186/s12864-020-6538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiranmayee KNSU, Hash CT, Sivasubramani S, Ramu P, Amindala BP, Rathore A, Kishor PBK, Gupta R, Deshpande SP. Fine-mapping of sorghum stay-green QTL on chromosome10 revealed genes associated with delayed senescence. Genes (Basel) 2020;11:1026. doi: 10.3390/genes11091026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RR, Klein PE, Mullet JE, Minx P, Rooney WL, Schertz KF. Fertility restorer locus Rf1 of sorghum (Sorghum bicolor L.) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12. Theor Appl Genet. 2005;111:994–1012. doi: 10.1007/s00122-005-2011-y. [DOI] [PubMed] [Google Scholar]
- Kobiler O, Rokney A, Friedman N, Court DL, Stavans J, Oppenheim AB. Quantitative kinetic analysis of the bacteriophage λ genetic network. Proc Natl Acad Sci USA. 2005;102:4470–4475. doi: 10.1073/pnas.0500670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, Durrant C, Mott R. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000551. doi: 10.1371/journal.pgen.1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Piperno DR, Allaby RG, Purugganan MD, Andersson L, Arroyo-Kalin M, Barton L, Climer Vigueira C, Denham T, Dobney K, Doust AN, Gepts P, Gilbert MT, Gremillion KJ, Lucas L, Lukens L, Marshall FB, Olsen KM, Pires JC, Richerson PJ, Rubio de Casas R, Sanjur OI, Thomas MG, Fuller DQ. Current perspectives and the future of domestication studies. Proc Natl Acad Sci U S A. 2014;111:6139–6146. doi: 10.1073/pnas.1323964111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky JR, Upadhyaya HD, Ramu P, Deshpande S, Hash CT, Bonnette J, Juenger TE, Hyma K, Acharya C, Mitchell SE, Buckler ES, Brenton Z, Kresovich S, Morris GP. Genome-environment associations in sorghum landraces predict adaptive traits. Sci Adv. 2015;1:e1400218. doi: 10.1126/sciadv.1400218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiser WL, Rattunde HFW, Weltzien E, Cisse N, Abdou M, Diallo A, Toure AO, Magalhaes JV, Haussmann BI. Two in one sweep: aluminum tolerance and grain yield in P-limited soils are associated to the same genomic region in West African sorghum. BMC Plant Biol. 2014;14:206–218. doi: 10.1186/s12870-014-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jiang JF, Qian Q, Xu YY, Zhang C, Xiao J, Du C, Luo W, Zou GX, Chen ML, Huang YQ, Feng YQ, Cheng ZK, Yuan M, Chong K. Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell. 2011;23:628–640. doi: 10.1105/tpc.110.081901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JQ, Tang WJ, Zhang YW, Chen KN, Wang CC, Liu YL, Zhan QW, Wang CM, Wang SB, Xie SQ, Wang LH. Genome-wide association studies for five forage quality-related traits in sorghum (Sorghum bicolor L.) Front Plant Sci. 2018;9:1146–1153. doi: 10.3389/fpls.2018.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Mace ES, Guo Y, Han LJ, Wang MM, He YF, Chen J, Yuyama N, Jordan DR, Cai HW. Fine mapping of qDor7, a major QTL affecting seed dormancy in sorghum (Sorghum bicolor (L.) Moench) Tropical Plant Biol. 2016;9:109–116. doi: 10.1007/s12042-016-9169-0. [DOI] [Google Scholar]
- Li Y, Wang WQ, Feng YP, Tu M, Wittich PE, Bate NJ, Messing J. Transcriptome and metabolome reveal distinct carbon allocation patterns during internode sugar accumulation in different sorghum genotypes. Plant Biotechnol J. 2019;17:472–487. doi: 10.1111/pbi.12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Zhou GY, Ma JX, Jiang LG, Jin LG, Zhang ZH, Guo Y, Zhang JB, Sui Y, Zheng LT, Zhang SS, Zuo QY, Shi XH, Li YF, Zhang WK, Hu YY, Kong GY, Hong HL, Tan B, Song J, Liu ZX, Wang YS, Ruan H, Yeung CKL, Liu J, Wang HL, Zhang LJ, Guan RX, Wang KJ, Li WB, Chen SY, Chang RZ, Jiang Z, Jackson SA, Li RQ, Qiu LJ. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat Biotechnol. 2014;32:1045–1052. doi: 10.1038/nbt.2979. [DOI] [PubMed] [Google Scholar]
- Lin ZW, Li XE, Shannon LM, Yeh CT, Wang ML, Bai GH, Peng Z, Li JR, Trick HN, Clemente TE, Doebley J, Schnable PS, Tuinstra MR, Tesso TT, White F, Yu JM. Parallel domestication of the Shattering1 genes in cereals. Nat Genet. 2012;44:720–724. doi: 10.1038/ng.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZQ, Li HL, Zeng XJ, Lu C, Fu JY, Guo LJ, Kimani WM, Yan HL, He ZY, Hao HQ, Jing HC. Coupling phytoremediation of cadmium-contaminated soil with safe crop production based on a sorghum farming system. J Clean Prod. 2020;275:123002. doi: 10.1016/j.jclepro.2020.123002. [DOI] [Google Scholar]
- Luan T, Woolliams JA, Lien S, Kent M, Svendsen M, Meuwissen THE. The accuracy of genomic selection in Norwegian red cattle assessed by cross-validation. Genetics. 2009;183:1119–1126. doi: 10.1534/genetics.109.107391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Zhao WM, Wang YQ, Xia Y, Wu XY, Zhang LM, Tang BX, Zhu JW, Fang L, Du ZL, Bekele WA, Tai SS, Jordan DR, Godwin ID, Snowdon RJ, Mace ES, Luo JC, Jing HC. SorGSD: a sorghum genome SNP database. Biotechnol Biofuels. 2016;9:6–14. doi: 10.1186/s13068-015-0415-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace E, Innes D, Hunt C, Wang XM, Tao YF, Baxter J, Hassall M, Hathorn A, Jordan D. The sorghum QTL Atlas: a powerful tool for trait dissection, comparative genomics and crop improvement. Theor Appl Genet. 2019;132:751–766. doi: 10.1007/s00122-018-3212-5. [DOI] [PubMed] [Google Scholar]
- Mace ES, Tai SS, Gilding EK, Li YH, Prentis PJ, Bian LL, Campbell BC, Hu WS, Innes DJ, Han XL, Cruickshank A, Dai CM, Frère C, Zhang HK, Hunt CH, Wang XY, Shatte T, Wang MM, Su Z, Li J, Lin XZ, Godwin ID, Jordan DR, Wang J. Whole-genome sequencing reveals untapped genetic potential in Africa's indigenous cereal crop sorghum. Nat Commun. 2013;4:2320–2328. doi: 10.1038/ncomms3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madugula P, Uttam AG, Tonapi VA, Ragimasalawada M. Fine mapping of Rf2, a major locus controlling pollen fertility restoration in sorghum A1 cytoplasm, encodes a PPR gene and its validation through expression analysis. Plant Breed. 2018;137:148–161. doi: 10.1111/pbr.12569. [DOI] [Google Scholar]
- Magalhaes JV, Liu JP, Guimaraes CT, Lana UGP, Alves VMC, Wang YH, Schaffert RE, Hoekenga OA, Piñeros MA, Shaff JE, Klein PE, Carneiro NP, Coelho CM, Trick HN, Kochian LV. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet. 2007;39:1156–1161. doi: 10.1038/ng2074. [DOI] [PubMed] [Google Scholar]
- Maina F, Bouchet S, Marla SR, Hu ZB, Wang JN, Mamadou A, Abdou M, Saidou AA, Morris GP. Population genomics of sorghum (Sorghum bicolor) across diverse agroclimatic zones of Niger. Genome. 2018;61:223–232. doi: 10.1139/gen-2017-0131. [DOI] [PubMed] [Google Scholar]
- Marla SR, Shiva S, Welti R, Liu SZ, Burke JJ, Morris GP. Comparative transcriptome and lipidome analyses reveal molecular chilling responses in chilling-tolerant sorghums. Plant Genome. 2017;10:1–16. doi: 10.3835/plantgenome2017.03.0025. [DOI] [PubMed] [Google Scholar]
- Marla SR, Burow G, Chopra R, Hayes C, Olatoye MO, Felderhoff T, Hu ZB, Raymundo R, Perumal R, Morris GP. Genetic architecture of chilling tolerance in sorghum dissected with a nested association mapping population. G3 (Bethesda) 2019;9:4045–4057. doi: 10.1534/g3.119.400353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick RF, Truong SK, Sreedasyam A, Jenkins J, Shu SQ, Sims D, Kennedy M, Amirebrahimi M, Weers BD, McKinley B, Mattison A, Morishige DT, Grimwood J, Schmutz J, Mullet JE. The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 2018;93:338–354. doi: 10.1111/tpj.13781. [DOI] [PubMed] [Google Scholar]
- McIntyre CL, Drenth J, Gonzalez N, Henzell RG, Jordan DR. Molecular characterization of the waxy locus in sorghum. Genome. 2008;51:524–533. doi: 10.1139/G08-035. [DOI] [PubMed] [Google Scholar]
- McKinley B, Rooney W, Wilkerson C, Mullet J. Dynamics of biomass partitioning, stem gene expression, cell wall biosynthesis, and sucrose accumulation during development of Sorghum bicolor. Plant J. 2016;88:662–680. doi: 10.1111/tpj.13269. [DOI] [PubMed] [Google Scholar]
- Melo JO, Martins LGC, Barros BA, Pimenta MR, Lana UGP, Duarte CEM, Pastina MM, Guimaraes CT, Schaffert RE, Kochian LV, Fontes EPB, Magalhaes JV. Repeat variants for the SbMATE transporter protect sorghum roots from aluminum toxicity by transcriptional interplay in cis and trans. Proc Natl Acad Sci USA. 2019;116:313–318. doi: 10.1073/pnas.1808400115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen TH, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RS, Choi JY, Sanches M, Plessis A, Flowers JM, Amas J, Dorph K, Barretto A, Gross B, Fuller DQ, Bimpong IK, Ndjiondjop MN, Hazzouri KM, Gregorio GB, Purugganan MD. Domestication history and geographical adaptation inferred from a SNP map of African rice. Nat Genet. 2016;48:1083–1088. doi: 10.1038/ng.3633. [DOI] [PubMed] [Google Scholar]
- Meyer RS, Purugganan MD. Evolution of crop species: genetics of domestication and diversification. Nat Rev Genet. 2013;14:840–852. doi: 10.1038/nrg3605. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Kasuga S, Kawahigashi H. The sorghum SWEET gene family: stem sucrose accumulation as revealed through transcriptome profiling. Biotechnol Biofuels. 2016;9:127–138. doi: 10.1186/s13068-016-0546-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose SP, Mumm RH. Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol. 2008;147:969–977. doi: 10.1104/pp.108.118232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, Riera-Lizarazu O, Brown PJ, Acharya CB, Mitchell SE, Harriman J, Glaubitz JC, Buckler ES, Kresovich S. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci USA. 2013;110:453–458. doi: 10.1073/pnas.1215985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science. 2003;302:81–84. doi: 10.1126/science.1086072. [DOI] [PubMed] [Google Scholar]
- Murphy RL, Klein RR, Morishige DT, Brady JA, Rooney WL, Miller FR, Dugas DV, Klein PE, Mullet JE. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci USA. 2011;108:16469–16474. doi: 10.1073/pnas.1106212108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RL, Morishige DT, Brady JA, Rooney WL, Yang S, Klein PE, Muller E. Ghd7 (Ma6) represses sorghum flowering in long days: Ghd7 alleles enhance biomass accumulation and grain production. Plant Genome. 2014;7:1–10. doi: 10.3835/plantgenome2013.11.0040. [DOI] [Google Scholar]
- Murray SC, Rooney WL, Hamblin MT, Mitchell SE, Kresovich S. Sweet sorghum genetic diversity and association mapping for brix and height. Plant Genome. 2009;2:48–62. doi: 10.3835/plantgenome2008.10.0011. [DOI] [Google Scholar]
- Nakaya A, Isobe SN. Will genomic selection be a practical method for plant breeding? Ann Bot. 2012;110:1303–1316. doi: 10.1093/aob/mcs109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nida H, Girma G, Mekonen M, Lee S, Seyoum A, Dessalegn K, Tadesse T, Ayana G, Senbetay T, Tesso T, Ejeta G, Mengiste T. Identification of sorghum grain mold resistance loci through genome wide association mapping. J Cereal Sci. 2019;85:295–304. doi: 10.1016/j.jcs.2018.12.016. [DOI] [Google Scholar]
- Niu H, Ping JN, Wang YB, Lv X, Li HM, Zhang FY, Chu JQ, Han YH. Population genomic and genome-wide association analysis of lignin content in a global collection of 206 forage sorghum accessions. Mol Breed. 2020;40:73. doi: 10.1007/s11032-020-01151-7. [DOI] [Google Scholar]
- Ohadi S, Hodnett G, Rooney W, Bagavathiannan M. Gene flow and its consequences in Sorghum spp. Crit Rev Plant Sci. 2017;36:367–385. doi: 10.1080/07352689.2018.1446813. [DOI] [Google Scholar]
- Olatoye MO, Hu ZB, Maina F, Morris GP. Genomic signatures of adaptation to a precipitation gradient in Nigerian sorghum. G3 (Bethesda) 2018;8:3269–3281. doi: 10.1534/g3.118.200551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongom PO, Ejeta G. Mating design and genetic structure of a multi-parent advanced generation intercross (MAGIC) population of sorghum (Sorghum bicolor(L.) Moench) G3 (Bethesda) 2018;8:331–341. doi: 10.1534/g3.117.300248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim AB, Kobiler O, Stavans J, Court DL, Adhya S. Switches in bacteriophage λ development. Annu Rev Genet. 2005;39:409–429. doi: 10.1146/annurev.genet.39.073003.113656. [DOI] [PubMed] [Google Scholar]
- Ortiz D, Hu JY, Salas Fernandez MG. Genetic architecture of photosynthesis in Sorghum bicolor under non-stress and cold stress conditions. J Exp Bot. 2017;68:4545–4557. doi: 10.1093/jxb/erx276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan ZQ, Rimando AM, Baerson SR, Fishbein M, Duke SO. Functional characterization of desaturases involved in the formation of the terminal double bond of an unusual 16: 3Delta 9,12,150 fatty acid isolated from Sorghum bicolor root hairs. J Biol Chem. 2007;282:4326–4335. doi: 10.1074/jbc.M606343200. [DOI] [PubMed] [Google Scholar]
- Parra-Londono S, Fiedler K, Kavka M, Samans B, Wieckhorst S, Zacharias A, Uptmoor R. Genetic dissection of early-season cold tolerance in sorghum: genome-wide association studies for seedling emergence and survival under field and controlled environment conditions. Theor Appl Genet. 2018;13:581–595. doi: 10.1007/s00122-017-3021-2. [DOI] [PubMed] [Google Scholar]
- Parra-Londono S, Kavka M, Samans B, Snowdon R, Wieckhorst S, Uptmoor R. Sorghum root-system classification in contrasting P environments reveals three main rooting types and root-architecture-related marker-trait associations. Ann Bot. 2018;121:267–280. doi: 10.1093/aob/mcx157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Pedersen JF, Graybosch RA, Funnell DL. Occurrence of the waxy alleles wxa and wxb in waxy sorghum plant introductions and their effect on starch thermal properties. Crop Sci. 2007;47:1927–1933. doi: 10.2135/cropsci2006.10.0652. [DOI] [Google Scholar]
- Pedersen JF, Toy JJ, Johnson B. Natural outcrossing of sorghum and sudangrass in the central great plains. Crop Sci. 1998;38:937–939. doi: 10.2135/cropsci1998.0011183X003800040009x. [DOI] [Google Scholar]
- Pillonel C, Mulder MM, Boon JJ, Forster B, Binder A. Involvement of cinnamyl-alcohol dehydrogenase in the control of lignin formation in Sorghum bicolor L. Moench Planta. 1991;185:538–544. doi: 10.1007/BF00202964. [DOI] [PubMed] [Google Scholar]
- Praveen M, Anurag Uttam G, Suneetha N, Umakanth A, Patil JV, Madhusudhana R. Inheritance and molecular mapping of Rf6 locus with pollen fertility restoration ability on A1 and A2 cytoplasms in sorghum. Plant Sci. 2015;238:73–80. doi: 10.1016/j.plantsci.2015.05.020. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Specific binding of the λ phage repressor to λ DNA. Nature. 1967;214:232–234. doi: 10.1038/214232a0. [DOI] [PubMed] [Google Scholar]
- Purugganan MD. Evolutionary insights into the nature of plant domestication. Cur Biol. 2019;29:R705–R714. doi: 10.1016/j.cub.2019.05.053. [DOI] [PubMed] [Google Scholar]
- Quinby JR, Karper RE. Inheritance of height in sorghum. Agron J. 1954;46:211–216. doi: 10.2134/agronj1954.00021962004600050007x. [DOI] [Google Scholar]
- Rama Reddy NR, Ragimasalawada M, Sabbavarapu MM, Nadoor S, Patil JV. Detection and validation of stay-green QTL in post-rainy sorghum involving widely adapted cultivar, M35–1 and a popular stay-green genotype B35. BMC Genom. 2014;15:909–930. doi: 10.1186/1471-2164-15-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy Sanjana P. Breeding for abiotic stress resistance in sorghum. In: Aruna C, Visarada KBRS, Bhat BV, Tonapi VA, editors. Breeding sorghum for diverse end uses. Cambridge, UK: Woodhead Publishing; 2018. pp. 325–340. [Google Scholar]
- Rendón-Anaya M, Herrera-Estrella A. The advantage of parallel selection of domestication genes to accelerate crop improvement. Genome Biol. 2018;19:147–149. doi: 10.1186/s13059-018-1537-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D, Gadgil P, Perumal R, Tesso T, Herald TJ. Natural variation and genome-wide association study of antioxidants in a diverse sorghum collection. Cereal Chem. 2017;94:190–198. doi: 10.1094/CCHEM-03-16-0075-R. [DOI] [Google Scholar]
- Rhodes DH, Hoffmann L, Rooney WL, Ramu P, Morris GP, Kresovich S. Genome-wide association study of grain polyphenol concentrations in global sorghum [Sorghum bicolor (L.) Moench] germplasm. J Agric Food Chem. 2014;62:10916–10927. doi: 10.1021/jf503651t. [DOI] [PubMed] [Google Scholar]
- Rhodes DH, Hoffmann L, Rooney WL, Herald TJ, Bean S, Boyles R, Brenton ZW, Kresovich S. Genetic architecture of kernel composition in global sorghum germplasm. BMC Genom. 2017;18:15–22. doi: 10.1186/s12864-016-3403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice B, Lipka AE. Evaluation of RR-BLUP genomic selection models that incorporate peak genome-wide association study signals in maize and sorghum. Plant Genome. 2019;12:180052. doi: 10.3835/plantgenome2018.07.0052. [DOI] [PubMed] [Google Scholar]
- Saballos A, Ejeta G, Sanchez E, Kang C, Vermerris W. A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the Brown midrib6 gene. Genetics. 2009;181:783–795. doi: 10.1534/genetics.108.098996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saballos A, Sattler SE, Sanchez E, Foster TP, Xin ZG, Kang C, Pedersen JF, Vermerris W. Brown midrib2 (Bmr2) encodes the major 4-coumarate: coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench) Plant J. 2012;70:818–830. doi: 10.1111/j.1365-313X.2012.04933.x. [DOI] [PubMed] [Google Scholar]
- Saballos A, Vermerris W, Rivera L, Ejeta G. Allelic association, chemical characterization and saccharification properties of brown midrib mutants of sorghum (Sorghum bicolor (L.) Moench) Bioenergy Res. 2008;1:193–204. doi: 10.1007/s12155-008-9025-7. [DOI] [Google Scholar]
- Satish K, Srinivas G, Madhusudhana R, Padmaja PG, Nagaraja Reddy R, Murali Mohan S, Seetharama N. Identification of quantitative trait loci for resistance to shoot fly in sorghum [Sorghum bicolor (L.) Moench] Theo Appl Genet. 2009;119:1425–1439. doi: 10.1007/s00122-009-1145-8. [DOI] [PubMed] [Google Scholar]
- Sattler SE, Palmer NA, Saballos A, Greene AM, Xin ZG, Sarath G, Vermerris W, Pedersen JF. Identification and characterization of four missense mutations in brown midrib 12 (bmr12), the Caffeic O-Methyltranferase (COMT) of sorghum. Bioenerg Res. 2012;5:855–865. doi: 10.1007/s12155-012-9197-z. [DOI] [Google Scholar]
- Sattler SE, Singh J, Haas EJ, Guo LN, Sarath G, Pedersen JF. Two distinct waxy alleles impact the granule-bound starch synthase in sorghum. Mol Breed. 2009;24:349–359. doi: 10.1007/s11032-009-9296-5. [DOI] [Google Scholar]
- Schrag TA, Schipprack W, Melchinger AE. Across-years prediction of hybrid performance in maize using genomics. Theor Appl Genet. 2019;132:933–946. doi: 10.1007/s00122-018-3249-5. [DOI] [PubMed] [Google Scholar]
- Shakoor N, Nair R, Crasta O, Morris G, Feltus A, Kresovich S. A Sorghum bicolor expression atlas reveals dynamic genotype-specific expression profiles for vegetative tissues of grain, sweet and bioenergy sorghums. BMC Plant Biol. 2014;14:35–48. doi: 10.1186/1471-2229-14-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakoor N, Ziegler G, Dilkes BP, Brenton Z, Boyles R, Connolly EL, Kresovich S, Baxter I. Integration of experiments across diverse environments identifies the genetic determinants of variation in Sorghum bicolor seed element composition. Plant Physiol. 2016;170:1989–1998. doi: 10.1104/pp.15.01971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao QY, Trinh JT, Zeng LY. High-resolution studies of lysis-lysogeny decision making in bacteriophage λ. J Biol Chem. 2018;294:3343–3349. doi: 10.1074/jbc.TM118.003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JP, Lai JS. Patterns of genomic changes with crop domestication and breeding. Curr Opin Plant Biol. 2015;24:47–53. doi: 10.1016/j.pbi.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Singh BD, Singh AK. Genomic selection Marker-assisted plant breeding: principles and practices. New Delhi: Springer; 2015. pp. 295–312. [Google Scholar]
- Slade AJ, Fuerstenberg SI, Loeffler D, Steine MN, Facciotti D. A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat Biotechnol. 2005;23:75–81. doi: 10.1038/nbt1043. [DOI] [PubMed] [Google Scholar]
- Smith O, Nicholson WV, Kistler L, Mace E, Clapham A, Rose P, Stevens C, Ware R, Samavedam S, Barker G, Jordan D, Fuller DQ, Allaby RG. A domestication history of dynamic adaptation and genomic deterioration in sorghum. Nat Plants. 2019;5:369–379. doi: 10.1038/s41477-019-0397-9. [DOI] [PubMed] [Google Scholar]
- Spindel J, Begum H, Akdemir D, Virk P, Collard B, Redona E, Atlin G, Jannink J, McCouch SR. Genomic selection and association mapping in rice (Oryza sativa): effect of trait genetic architecture, training population composition, marker number and statistical model on accuracy of rice genomic selection in elite, tropical rice breeding lines. PLoS Genet. 2015;11:e1004982. doi: 10.1371/journal.pgen.1004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindel JE, Dahlberg J, Colgan M, Hollingsworth J, Sievert J, Staggenborg SH, Hutmacher R, Jansson C, Vogel JP. Association mapping by aerial drone reveals 213 genetic associations for Sorghum bicolor biomass traits under drought. BMC Genom. 2018;19:679–696. doi: 10.1186/s12864-018-5055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefoskaneedham A, Beck EJ, Johnson SK, Tapsell LC. Sorghum: an underutilized cereal whole grain with the potential to assist in the prevention of chronic disease. Food Rev Int. 2015;31:401–437. doi: 10.1080/87559129.2015.1022832. [DOI] [Google Scholar]
- Stephens JC, Miller FR, Rosenow DT. Conversion of alien sorghums to early combine genotypes. Crop Sci. 1967;7:396. doi: 10.2135/cropsci1967.0011183X000700040036x. [DOI] [Google Scholar]
- Tang D, Cheng ZK. From basic research to molecular breeding - Chinese scientists play a central role in boosting world rice production. Genom Proteom Bioinf. 2018;16:389–392. doi: 10.1016/j.gpb.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Cuevas HE, Das S, Sezen UU, Zhou C, Guo H, Goff VH, Ge ZX, Clemente TE, Paterson AH. Seed shattering in a wild sorghum is conferred by a locus unrelated to domestication. Proc Natl Acad Sci USA. 2013;110:15824–15829. doi: 10.1073/pnas.1305213110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, McCouch SR. Seed banks and molecular maps: unlocking genetic potential from the wild. Science. 1997;277:1063–1066. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- Tao YF, Zhao XR, Wang XM, Hathorn A, Hunt CH, Cruickshank AW, van Oosterom EJ, Godwin ID, Mace ES, Jordan DR. Large-scale GWAS in sorghum reveals common genetic control of grain size among cereals. Plant Biotechnol J. 2019;18:1093–1105. doi: 10.1111/pbi.13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber CS, Ma JM, Higgins RH, Brown PJ. Retrospective genomic analysis of sorghum adaptation to temperate-zone grain production. Genome Biol. 2013;14:R68. doi: 10.1186/gb-2013-14-6-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian ZX, Qian Q, Liu QQ, Yan MX, Liu XF, Yan CJ, Liu GF, Gao ZY, Tang SZ, Zeng DL, Wang YH, Yu JM, Gu MH, Li JY. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc Natl Acad Sci U S A. 2009;106:21760–21765. doi: 10.1073/pnas.0912396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Weil C, Springer N, Burtner C, Young K, Bowers E, Codomo CA, Enns LC, Odden AR, Greene EA, Comai L, Henikoff S. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 2004;4:12–19. doi: 10.1186/1471-2229-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi AD, Mishra R, Maurya KK, Singh RB, Wilson DW. Estimates for world population and global food availability for global health. In: Singh RB, Watson RR, Takahashi T, editors. The role of functional food security in global health. Cambridge: Academic Press; 2019. pp. 3–24. [Google Scholar]
- Upadhyaya HD, Pundir RPS, Dwivedi SL, Gowda LL, Reddy VG, Singh S. Developing a mini core collection of sorghum for diversified utilization of germplasm. Crop Sci. 2009;49:1769–1780. doi: 10.2135/cropsci2009.01.0014. [DOI] [Google Scholar]
- Upadhyaya HD, Vetriventhan M, Asiri AM, Azevedo VCR, Sharma HC, Sharma R, Suraj Prasad Sharma SP, Wang YH. Multi-trait diverse germplasm sources from mini core collection for sorghum improvement. Agriculture. 2019;9:121–137. doi: 10.3390/agriculture9060121. [DOI] [Google Scholar]
- Upadhyaya UD, Wang YH, Gowda CLL, Sharma S. Association mapping of maturity and plant height using SNP markers with the sorghum mini core collection. Theor Appl Genet. 2013;126:2003–2015. doi: 10.1007/s00122-013-2113-x. [DOI] [PubMed] [Google Scholar]
- Upadhyaya HD, Wang YH, Sharma R, Sharma S. Identification of genetic markers linked to anthracnose resistance in sorghum using association analysis. Theor Appl Genet. 2013;126:1649–1657. doi: 10.1007/s00122-013-2081-1. [DOI] [PubMed] [Google Scholar]
- Valluru R, Gazave EE, Fernandes SB, Ferguson JN, Lozano R, Hirannaiah P, Zuo T, Brown PJ, Leakey ADB, Gore MA, Buckler ES, Bandillo N. Deleterious mutation burden and its association with complex traits in sorghum (Sorghum bicolor) Genetics. 2019;211:1075–1087. doi: 10.1534/genetics.118.301742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux N, Cole B, Gao C, Pierroz G, Baker CR, Patel D, Madera M, Jeffers T, Hollingsworth J, Sievert J, Yoshinaga Y, Owiti JA, Singan VR, DeGraaf S, Xu L, Blow MJ, Harrison MJ, Visel A, Jansson C, Niyogi KK, Hutmacher R, Coleman-Derr D, O'Malley RC, Taylor JW, Dahlberg J, Vogel JP, Lemaux PG, Purdom E. Transcriptomic analysis of field-droughted sorghum from seedling to maturity reveals biotic and metabolic responses. Proc Natl Acad Sci USA. 2019;116:27124–27132. doi: 10.1073/pnas.1907500116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JN, Hu ZB, Upadhyaya HD, Morris GP. Genomic signatures of seed mass adaptation to global precipitation gradients in sorghum. Heredity. 2020;124:108–121. doi: 10.1038/s41437-019-0249-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Beissinger TM, Lorant A, Ross-Ibarra C, Ross-Ibarra J, Hufford MB. The interplay of demography and selection during maize domestication and expansion. Genome Biol. 2017;18:215. doi: 10.1186/s13059-017-1346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ML, Zhu CS, Barkley NA, Chen ZB, Erpelding JE, Murray SC, Tuinstra MR, Tesso T, Pederson GA, Yu JM. Genetic diversity and population structure analysis of accessions in the US historic sweet sorghum collection. Theor Appl Genet. 2009;120:13–23. doi: 10.1007/s00122-009-1155-6. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Brummer EC. Is genetic engineering ever going to take off in forage, turf and bioenergy crop breeding? Ann Bot. 2012;110:1317–1325. doi: 10.1093/aob/mcs027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendorf F, Close AE, Schild R, Wasylikowa K, Housley RA, Harlan JR, Królik H. Saharan exploitation of plants 8,000 years BP. Nature. 1992;359:721–724. doi: 10.1038/359721a0. [DOI] [Google Scholar]
- Winchell F, Stevens CJ, Murphy C, Champion L, Fuller DQ. Evidence for sorghum domestication in fourth millennium BC Eastern Sudan: spikelet morphology from ceramic impressions of the Butana group. Cur Anthr. 2017;58:673–683. doi: 10.1086/693898. [DOI] [Google Scholar]
- Wu XY, Hu WJ, Luo H, Xia Y, Zhao Y, Wang LD, Zhang LM, Luo JC, Jing HC. Transcriptome profiling of developmental leaf senescence in sorghum (Sorghum bicolor) Plant Mol Biol. 2016;92:555–580. doi: 10.1007/s11103-016-0532-1. [DOI] [PubMed] [Google Scholar]
- Wu YY, Li XR, Xiang WW, Zhu CS, Lin ZW, Wu Y, Li JR, Pandravada S, Ridder DD, Bai GH, Wang ML, Trick HN, Bean SR, Tuinstra MR, Tesso TT, Yu JM. Presence of tannins in sorghum grains is conditioned by different natural alleles of Tannin1. Proc Natl Acad Sci USA. 2012;109:10281–10286. doi: 10.1073/pnas.1201700109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YR, Yuan LL, Guo XM, Holding DR, Messing J. Mutation in the seed storage protein kafirin creates a high-value food trait in sorghum. Nat Commun. 2013;4:2217–2223. doi: 10.1038/ncomms3217. [DOI] [PubMed] [Google Scholar]
- Xie P, Shi JY, Tang SY, Chen CX, Khan A, Zhang FX, Xiong Y, Li C, He W, Wang GD, Lei FM, Wu YR, Xie Q. Control of bird feeding behavior by Tannin1 through modulating the biosynthesis of polyphenols and fatty acid-derived volatiles in sorghum. Mol Plant. 2019;12:1315–1324. doi: 10.1016/j.molp.2019.08.004. [DOI] [PubMed] [Google Scholar]
- Xin ZG, Wang ML, Barkley NA, Burow G, Frank C, Pederson G, Burke J. Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol. 2008;8:103–116. doi: 10.1186/1471-2229-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Subudhi PK, Crasta OR, Rosenow DT, Mullet JE, Nguyen HT. Molecular mapping of QTLs conferring stay-green in grain sorghum (Sorghum bicolor L. Moench) Genome. 2000;43:461–469. doi: 10.1139/g00-003. [DOI] [PubMed] [Google Scholar]
- Xu YB, Li P, Zou C, Lu YL, Xie CX, Zhang XC, Prasanna BM, Olsen MS. Enhancing genetic gain in the era of molecular breeding. J Exp Bot. 2017;83:2641–2666. doi: 10.1093/jxb/erx135. [DOI] [PubMed] [Google Scholar]
- Xu YB, Liu XG, Fu JJ, Wang HW, Wang JK, Huang CL, Prasanna BM, Olsen MS, Wang GY, Zhang AM. Enhancing genetic gain through genomic selection: from livestock to plants. Plant Commu. 2020;1:100005. doi: 10.1016/j.xplc.2019.100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue YB, Chong K, Han B, Gui JF, Jing HC. Innovation and achievements of designer breeding by molecular modules in China. Bull Chin Acad Sci. 2018;33:893–899. [Google Scholar]
- Xue YB, Duan ZZ, Chong K, Yao Y. Next-generation biotechnological breeding technologies for the future — Designer breeding by molecular modules. Bull Chin Acad Sci. 2013;28:308–314. [Google Scholar]
- Xue YB, Han B, Chong K, Wang T, He ZH, Fu XD, Chu CC, Cheng ZK, Xu YY, Li M. Achievements and prospect of designer breeding by molecular modules in rice. Bull Chin Acad Sci. 2018;33:890–898. [Google Scholar]
- Yazawa T, Kawahigashi H, Matsumoto T, Mizuno H. Simultaneous transcriptome analysis of sorghum and Bipolaris sorghicola by using RNA-seq in combination with de novo transcriptome assembly. PLoS ONE. 2013;8:e62460. doi: 10.1371/journal.pone.0062460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JM, Holland JB, McMullen MD, Buckler ES. Genetic design and statistical power of nested association mapping in maize. Genetics. 2008;178:539–551. doi: 10.1534/genetics.107.074245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XQ, Li XR, Guo TT, Zhu CS, Wu YY, Mitchell SE, Roozeboom KL, Wang DH, Wang ML, Pederson GA, Tesso TT, Schnable PS, Bernardo R, Yu JM. Genomic prediction contributing to a promising global strategy to turbocharge gene banks. Nat Plants. 2016;2:16150. doi: 10.1038/nplants.2016.150. [DOI] [PubMed] [Google Scholar]
- Zhang D, Kong WQ, Robertson J, Goff VH, Epps E, Kerr A, Mills G, Cromwell J, Lugin Y, Phillips C, Paterson AH. Genetic analysis of inflorescence and plant height components in sorghum (Panicoidae) and comparative genetics with rice (Oryzoidae) BMC Plant Biol. 2015;15:107–121. doi: 10.1186/s12870-015-0477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Li J, Compton RO, Robertson J, Goff VH, Epps E, Kong W, Kim C, Paterson AH. Comparative genetics of seed size traits in divergent cereal lineages represented by sorghum (Panicoidae) and rice (Oryzoidae) G3 (Bethesda) 2015;5:1117–1128. doi: 10.1534/g3.115.017590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jiang F, Shen Y, Zhan Q, Bai B, Chen W, Chi Y. Transcriptome analysis reveals candidate genes related to phosphorus starvation tolerance in sorghum. BMC Plant Biol. 2019;19:306–323. doi: 10.1186/s12870-019-1914-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LM, Leng CY, Luo H, Wu XY, Liu ZQ, Zhang YM, Zhang H, Xia Y, Shang L, Liu CM, Hao DY, Zhou YH, Chu CC, Cai HW, Jing HC. Sweet sorghum originated through selection of Dry, a plant-specific NAC transcription factor gene. Plant Cell. 2018;30:2286–2307. doi: 10.1105/tpc.18.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lin ZL, Wang J, Liu HQ, Zhou LN, Zhong SY, Yan L, Zhu C, Liu JC, Lin ZW. The tin1 gene retains the function of promoting tillering in maize. Nat Commun. 2019;10:5608. doi: 10.1038/s41467-019-13425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Mantilla Perez MB, Hu J, Salas Fernandez MG. Genome-wide association study for nine plant architecture traits in sorghum. Plant Genome. 2016;9:1–14. doi: 10.3835/plantgenome2015.06.0044. [DOI] [PubMed] [Google Scholar]
- Zheng LY, Guo XS, He B, Sun LJ, Peng Y, Dong SS, Liu TF, Jiang S, Ramachandran S, Liu CM, Jing HC. Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolor) Genome Biol. 2011;12:R114. doi: 10.1186/gb-2011-12-11-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XC, Bai XF, Xing YZ. A rice genetic improvement boom by next generation sequencing. Curr Issues Mol Biol. 2018;27:109–126. doi: 10.21775/cimb.027.109. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Srinivasan S, Mirnezami SV, Kusmec A, Fu Q, Attigala L, Salas Fernandez MG, Ganapathysubramanian B, Schnable PS. Semiautomated feature extraction from RGB images for sorghum panicle architecture GWAS. Plant Physiol. 2019;179:24–37. doi: 10.1104/pp.18.00974. [DOI] [PMC free article] [PubMed] [Google Scholar]