Abstract
Simple Summary
Pancreatic cancer has a poor prognosis, which is largely due to resistance to treatment. Tumor heterogeneity is a known cause for treatment failure and has been studied at the molecular level. Morphological heterogeneity is common but has not been investigated, despite the fact that pathology examination is an integral part of clinical diagnostics. This study assessed whether morphological heterogeneity reflects structural and functional diversity in key cancer biological processes. Using archival tissues from resected pancreatic cancer, we selected four common and distinct morphological phenotypes and demonstrated that these differed significantly for a panel of 26 structural and functional features of the cancer-cell and stromal compartments. The strong link between these features and morphological phenotypes allowed prediction of the latter based on the results for the panel of features. The findings of this study indicate that morphological heterogeneity reflects biological diversity and that its assessment may potentially provide clinically relevant information.
Abstract
Inter- and intratumor heterogeneity is an important cause of treatment failure. In human pancreatic cancer (PC), heterogeneity has been investigated almost exclusively at the genomic and transcriptional level. Morphological heterogeneity, though prominent and potentially easily assessable in clinical practice, remains unexplored. This proof-of-concept study aims at demonstrating that morphological heterogeneity reflects structural and functional divergence. From the wide morphological spectrum of conventional PC, four common and distinctive patterns were investigated in 233 foci from 39 surgical specimens. Twenty-six features involved in key biological processes in PC were analyzed (immuno-)histochemically and morphometrically: cancer cell proliferation (Ki67) and migration (collagen fiber alignment, MMP14), cancer stem cells (CD44, CD133, ALDH1), amount, composition and spatial arrangement of extracellular matrix (epithelial proximity, total collagen, collagen I and III, fibronectin, hyaluronan), cancer-associated fibroblasts (density, αSMA), and cancer-stroma interactions (integrins α2, α5, α1; caveolin-1). All features differed significantly between at least two of the patterns. Stromal and cancer-cell-related features co-varied with morphology and allowed prediction of the morphological pattern. In conclusion, morphological heterogeneity in the cancer-cell and stromal compartments of PC correlates with structural and functional diversity. As such, histopathology has the potential to inform on the operationality of key biological processes in individual tumors.
Keywords: human pancreatic ductal adenocarcinoma, morphology, pathology, heterogeneity, stroma, extracellular matrix
1. Introduction
Ductal adenocarcinoma of the pancreas, often referred to as pancreatic cancer (PC), has a dismal prognosis with an overall five-year survival of less than 7% [1,2]. In addition to late diagnosis, limited efficacy of current treatment is the main reason for the poor prognosis. Precision medicine holds the promise of improved outcome through treatment that is tailored to intrinsic properties of the individual tumor. This approach requires a classification system with well-defined tumor subtypes of a cancer entity, as it is established, for example, for breast cancer. Several classification systems have been proposed for PC, which are based on gene expression profiling [3,4,5,6]. However, the latter is not without its practical obstacles, including the need for RNA of sufficient quantity and quality. In addition, there is the unresolved problem of intratumor heterogeneity.
In contrast to active research regarding a molecular taxonomy for PC, morphological variation in PC has received little if any attention, except for a few rare subtypes of PC with unusual morphological features—for example, medullary and colloid carcinomas which are associated with microsatellite instability [7,8]. While these subtypes account for less than 10% of PC, the vast majority of tumors constitute one large group that is denoted as PC not otherwise specified (NOS). In this group, morphological variation is common but considered irrelevant and largely left without further characterization [9]. Only recently, a few attempts have been made to correlate genotype or transcriptional profile with a small number of broad histological categories that encompass a wide range of morphologies of PC [10]. Despite these initial investigations, the relationship between morphological heterogeneity and biological diversity in PC is far from understood. In this study, we aimed at providing proof of concept that differences in morphology are associated with structural and functional divergence in biological processes deemed to be relevant in terms of PC biology and/or clinical outcome. Four common but not previously described morphological patterns of PC were selected from the wide spectrum of morphologies observed in PC NOS [11]. Human PC tissues containing such patterns were investigated with (immuno-)histochemistry and morphometry in order to characterize them for a panel of 26 features that are known to play a role in key aspects of PC biology, including cancer cell proliferation (Ki67) and migration (collagen fiber alignment, matrix metalloproteinase 14), cancer stem cells (CD44, CD133, aldehyde dehydrogenase 1), amount, composition and spatial arrangement of the extracellular matrix (epithelial proximity, total collagen, collagen I and III, fibronectin, hyaluronan), cancer-associated fibroblasts (density, α-smooth muscle actin), and cancer-stroma interactions (integrins α2, α5, β1, caveolin-1). All of the non-structural features analyzed in this study are part of recently compiled comprehensive repertoires of compartment-specific genes in PC [3,12]. The biological roles and clinical relevance of the selected features are summarized in Table 1.
Table 1.
Experimental observations and clinical relevance of the listed features in pancreatic cancer.
| Feature | Experimental Observations | Clinical Relevance |
|---|---|---|
| ALDH1 | ||
| αSMA |
|
|
| CAV1 | ||
| CD44 | ||
| CD133 | ||
| Col I | ||
| Col III |
|
|
| FN |
|
|
| HA | ||
| ITGα2β1 |
|
|
| ITGα5β1 |
|
|
| MMP14 |
ALDH1, aldehyde dehydrogenase 1; αSMA, α-smooth muscle actin; CAV1, caveolin-1; Col, collagen; CSC, cancer stem cell; EMT, epithelial-mesenchymal transition; FN, fibronectin; HA, hyaluronan; ITG, integrin; MMP14, matrix metalloproteinase 14; PCC, pancreatic cancer cell; TGFβ, transforming growth factor β; 5FU, fluorouracil.
2. Results
2.1. Tumor Patterns
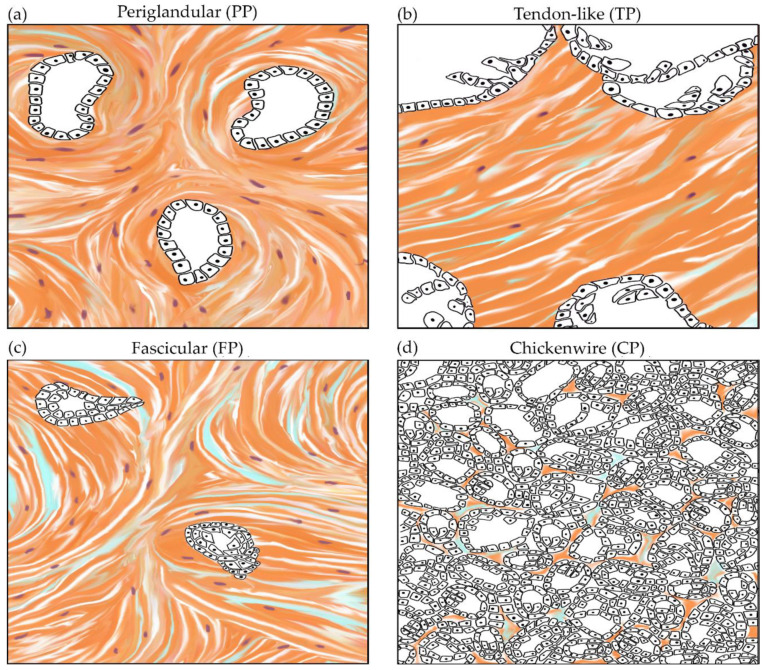
From the wide range of morphologies that can be encountered in PC NOS, four commonly seen patterns were selected and defined by distinctive features that are readily assessable on hematoxylin and eosin (H&E) staining in both the cancer-cell and stromal compartments, as shown in Figure 1 and illustrated in detail in Figure S1. Reflecting the morphology of their stroma, the patterns were coined periglandular (PP), tendon-like (TP), fascicular (FP), and chickenwire (CP). These patterns are not described previously, but the cancer cell-component of TP is reminiscent of the cystic papillary variant mentioned in the World Health Organisation (WHO) classification [7]. The four patterns are characterized by glandular tumor growth and are therefore well differentiated [96].
Figure 1.
Four patterns of pancreatic cancer with distinctive morphological features of the cancer-cell and stromal compartments. (a) The periglandular pattern (PP) is characterized by medium-sized simple glands surrounded by a moderate amount of stroma that encircles the tumor glands. (b) In the tendon-like pattern (TP), the large cystic-papillary tumor glands are embedded in a vast and dense fibrous stroma. (c) The fascicular pattern (FP) typically contains medium-sized tumor glands that are often angulated and lie in a highly cellular, whorled stroma. (d) In the chickenwire pattern (CP), tumor glands are small and lie densely packed with little intervening stroma.
2.2. Study Series
Of the 70 consecutive, treatment-naïve cases that were reviewed, 47 (67%) contained a pattern of interest, either exclusively or in combination with other morphologies that are not investigated in this study. Following exclusion of eight cases because of necrosis or marked inflammation which alter tumor morphology, 39 cases were included in the study series, of which 33 contained one of the four patterns that were analyzed in this study and six cases contained two or three different patterns (Figure 2). In each case of the series, at least five and at most eight regions of interest (ROIs) representative of a particular pattern were analyzed for the panel of features, except in two cases with PP, in which only three ROIs were available for analysis, and in one case of FP with only four ROIs. Overall, investigation of the panel of 26 features in a total number of 233 ROIs (PP: 68; TP: 60; FP: 50; CP: 55) engendered 816 sections with the various stains. Clinicopathological features of the study series are shown in Table S1.
Figure 2.
Case selection process and study series composition. (a) Flow chart depicting the selection process resulting in the study series. (b) Venn-diagram illustrating the number of tumors with a single, two or three different patterns.
2.3. Cancer-Cell and Stromal Features
Significant differences between at least two of the four patterns were observed for all features. The main findings are described below and illustrated in Figure 3. Network visualization is shown in Figure 4. The results of the statistical analysis and all individual data are detailed in Table 2 and Table S2, respectively.
Figure 3.
Divergence of stromal and cancer cell-related features in the four pancreatic cancer patterns. Representative microphotographs of hematoxylin and eosin (H&E) staining and (immuno-)histochemistry for selected features (200x; scale bar = 100 μm) are shown. ALDH1, aldehyde dehydrogenase 1; CAV1, caveolin-1; Col I, collagen I; HA, hyaluronan; H&E, hematoxylin and eosin; FN, fibronectin; ITGα5, integrinα5; MMP14, matrix metalloproteinase 14.
Figure 4.
Complex network visualization of cancer-cell-related and stromal features in the four tumor patterns. For a particular feature, paired Gephi-generated networks visualize the four morphological patterns (left) and the results for that feature (right). Every dot in the network represents a single ROI, and the length of the connecting lines represents the similarity between the connected nodes (n = 233). Absence of connecting lines between dots (e.g., in Gephi for epithelial proximity) indicates the lack of similarity. For each feature, the p-value reflecting the difference between the patterns is stated in Table 2.
Table 2.
Statistical results for the panel of features compared between the tumor patterns. Low p-values (<0.05) indicate significant differences between the feature data corresponding to the different morphological patterns.
| Feature | Compart-ment | Kruskal-Wallis Test | Mann–Whitney U Test | |||||
|---|---|---|---|---|---|---|---|---|
| PP vs. TP | PP vs. FP | PP vs. CP | TP vs. FP | TP vs. CP | FP vs. CP | |||
| ALDH1 | C | <0.001 | <0.001 | NS | NS | <0.001 | <0.001 | NS |
| αSMA | S | <0.001 | <0.001 | 0.01 | <0.001 | <0.001 | 0.02 | <0.001 |
| CAV1 | C | <0.001 | NS | NS | <0.001 | NS | <0.001 | <0.001 |
| CAV1 | S | <0.001 | <0.001 | 0.006 | <0.001 | NS | <0.001 | <0.001 |
| CD133 | C | <0.001 | <0.001 | NS | <0.001 | <0.001 | NS | <0.001 |
| CD44 | C | 0.004 | 0.003 | 0.007 | 0.001 | NS | NS | NS |
| Col align | S | 0.001 | 0.001 | 0.011 | 0.001 | <0.001 | <0.001 | NS |
| Col I | S | <0.001 | 0.033 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Col I pattern | S | <0.001 | <0.001 | <0.001 | <0.001 | NS | <0.001 | <0.001 |
| Col III | S | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Col III pattern | S | <0.001 | <0.001 | <0.001 | <0.001 | NS | <0.001 | <0.001 |
| Ep prox prox | S | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| FN | S | <0.001 | <0.001 | 0.02 | <0.001 | <0.001 | <0.001 | <0.001 |
| FN pattern | S | <0.001 | <0.001 | <0.001 | 0.009 | <0.001 | <0.001 | <0.001 |
| HA | S | <0.001 | <0.001 | 0.002 | <0.001 | <0.001 | NS | <0.001 |
| ITGα2 | C | <0.001 | <0.001 | 0.001 | <0.001 | NS | NS | NS |
| ITGα2 | S | <0.001 | 0.002 | NS | <0.001 | 0.022 | NS | 0.002 |
| ITGα5 | S | <0.001 | <0.001 | NS | <0.001 | <0.001 | <0.001 | <0.001 |
| ITGα5 pattern | S | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| ITGβ1 | C | <0.001 | <0.001 | <0.001 | <0.001 | NS | <0.001 | <0.001 |
| ITGβ1 | S | <0.001 | <0.001 | NS | <0.001 | <0.001 | NS | <0.001 |
| Ki67 | C | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| MMP14 | C | <0.001 | 0.001 | <0.001 | 0.037 | <0.001 | <0.001 | <0.001 |
| MMP14 | S | <0.001 | <0.001 | <0.001 | NS | <0.001 | <0.001 | <0.001 |
| Stromal cell density | S | <0.001 | <0.001 | 0.008 | <0.001 | <0.001 | <0.001 | <0.001 |
| Total collagen | S | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | NS |
ALDH1, aldehyde dehydrogenase 1; αSMA, α-smooth muscle actin; C, Cancer; CAV1, caveolin-1; Col, collagen; Col align, collagen alignment; CP, chicken-wire pattern; Ep prox, epithelial proximity; FN, fibronectin; FP, fascicular pattern; HA, hyaluronan; ITG, integrin; MMP14, matrix metalloproteinase 14; NS, not significant (p ≥ 0.05); PP, periglandular pattern; S, stroma; TP, tendon-like pattern.
2.3.1. Cancer-Cell Features
Proliferative activity, assessed with the Ki67 index, in the entire series was close to identical with that observed by others (mean: 27%) [97,98]. However, it differed significantly between all four patterns and was found to be highest in CP (42%), followed in descending order by FP (31%), PP (23%), and TP (13%) (p < 0.001).
Three cancer stem cell (CSC) markers were studied. As reported by others, cytoplasmic immunostaining for CD44 was found in the majority of cases (83%) [30,34,35,39,40,51], but expression was significantly less frequent and at a lower level in PP than in the other patterns (p ≤ 0.007). Similarly, expression of CD133, typically limited to the apical cell membrane, was in line with previous reports for the overall series [39,40,48,50,51] but differed between the patterns. It was significantly higher in PP and FP (36% and 46% high, respectively) than in TP and CP (93% and 100% low or negative, respectively) (p < 0.001) Cytoplasmic ALDH1-expression was significantly higher in TP (87% high) than in the three other patterns (0–2% high) (p < 0.001) [13,17,18,51,99] (Figure 3 and Figure 4). There was no co-variation between CD44, CD133 and ALDH1, either overall or for each of the patterns individually.
Expression of integrin (ITG) α2 and β1, which together form the cellular receptor for collagen I, was significantly lower in PP than in the other patterns (p < 0.001). Matrix metalloproteinase 14 (MMP14), which through its ECM-degrading effect promotes cancer cell migration [58], was expressed at significantly different levels by cancer cells in the four patterns: highest in FP (58%), followed by CP (20%) and PP (13%), and TP (2%) (p ≤ 0.037), respectively (Figure 3 and Figure 4). Caveolin-1 (CAV1) can be expressed by both cancer cells and cancer-associated fibroblasts (CAFs) and has multifarious effects, including the promotion of proliferation, invasion and chemoresistance [24,25]. CAV1-expression was significantly higher in cancer cells of CP (82%) than in the other patterns (0%; p < 0.001) (Figure 3 and Figure 4).
2.3.2. Stromal Features
For the purpose of this study, the stroma that lies immediately adjacent to the cancer cells (that is, within a 200x high-power field) was investigated rather than the bulk stroma, as it is the former that plays a pivotal role in the cancer cell biology [19,100,101]. Among stromal features, epithelial proximity, a proxy for the dispersedness of the cancer glands and the amount of intervening stroma, differed significantly between all patterns (p < 0.001), being highest in TP, followed in descending order by FP, PP and CP. The total amount of collagen was highest in TP and moderate in PP (p < 0.001), but similarly low in FP and CP. Collagen I deposition in PP and CP was peritumoral—that is, the fibers formed a sheath that encircled the tumor glands, while it was diffuse in TP and FP (Figure 3 and Figure 4). Results for collagen III were similar, though immunostaining levels were overall lower than for collagen I. Collagen fiber alignment was highest in TP (p ≤ 0.001) and moderate in PP (p ≤ 0.011) but low in FP and CP. Fibronectin content was highest in FP, moderate in PP, low in CP and absent in almost all TP (p < 0.001). Fibronectin deposition was mainly peritumoral in PP (75%) and diffuse in FP (100%), where it was organized in the sweeping bundles that characterize the whorled stromal appearance in this pattern (Figure 3 and Figure 4).
Results for the entire study series regarding hyaluronan were highly similar with those reported by others (high-level expression in 55% [33,63,82,102]). However, levels differed significantly between patterns: they were highest in TP and CP (p < 0.001), and lowest in PP (p ≤ 0.002) (Figure 3 and Figure 4). In all patterns, the vast majority of stromal cells with fibroblast morphology were αSMA-positive, confirming their nature as activated CAFs [103]. Stromal cell density was highest in FP, lowest in TP and CP (p ≤ 0.008).
The ratio between the number of αSMA + CAFs and amount of collagen, the so-called activated stroma ratio has been used to characterize and categorize the stroma in PC [64,104]. Based on the results for both features, each of the four patterns can be assigned to a different stromal subtype. In FP, the stroma is consistent with the fibrolytic subtype (high αSMA/low collagen), while in TP, PP and CP it is of fibrogenic (low αSMA/high collagen), inert (high αSMA/high collagen), and dormant (low αSMA/low collagen) subtype, respectively (Table 3).
Table 3.
Activated stroma index in the four tumor patterns. The index is the ratio between αSMA expression (immunohistochemically assessed) and the total amount of collagen (morphometric evaluation), averaged for each pattern. Assignment to the stromal subtypes is according to [104]. Kruskal-Wallis testing showed significant difference in activated stroma index between the four patterns (p < 0.01). αSMA, α-smooth muscle actin.
| Tumor Pattern |
αSMA (Staining Score) |
Total Collagen (Area Fraction %) | Activated Stroma Index | Stromal Subtype |
|---|---|---|---|---|
| Periglandular | High (2.60) | High (16.95) | 0.27 | Inert |
| Tendon-like | Low (1.50) | High (33.21) | 0.05 | Fibrogenic |
| Fascicular | High (2.90) | Low (7.35) | 1.35 | Fibrolytic |
| Chickenwire | Low (1.20) | Low (3.60) | 0.56 | Dormant |
Stromal MMP14 levels were highest in FP and lowest in TP (p < 0.01) (Figure 3 and Figure 4). CAV1 was absent in nearly all cases of CP but was expressed at similar levels in the other patterns (p < 0.01) (Figure 3 and Figure 4). ITGα5 expression was low or absent in TP and CP but was expressed at high level in PP and FP (p < 0.001). Strong expression limited to the immediate peritumoral stroma was observed in PP, whereas expression was diffuse in the stroma of FP (Figure 3 and Figure 4). ITGα2 and ITGβ1 showed a similar differential expression albeit at lower levels compared to ITGα5.
2.4. Multiple Patterns within a Single Tumor
Five tumors contained two different patterns—that is, a combination of PP and FP (n = 3), PP and TP (n = 1), or FP and CP (n = 1). One tumor contained three patterns (PP, TP, and CP). The different patterns occurred in separate regions within the tumor mass, each pattern showing the features that define the particular morphology of the cancer cells and stroma. Features for the respective patterns did not differ from those found in tumors that contained only a single pattern.
2.5. Prediction of Morphological Pattern
To investigate the robustness of the association between the morphological patterns and the structural and functional features, we tested whether the results for the full set of features would allow prediction of the tumor pattern. Using the k-Nearest Neighbors (k-NN) algorithm, 97% of the ROIs was assigned to the correct morphological pattern when using a separate holdout dataset containing 30% of the samples for testing. When using cancer-cell features only, classification was correct for 84% of the ROIs, and misclassification occurred for ROIs with any of the four patterns. Prediction of the morphological pattern based exclusively on stromal features resulted in correct classification for 96% of the ROIs, with only 10 of 60 ROIs with a PP morphology being misclassified as either TP or FP. Interestingly, when using only the panel of nine features that are related to the ECM, prediction of the morphological pattern was equally good (97%) as when using the full stromal panel (18 features) or the full panel (26 features) (Table S3).
Feature importance computed by the extremely randomized tree (ERT) algorithm showed that features related to collagen I and collagen III as well as fibronectin were the most important ones for the prediction of the morphological pattern (Figure S2). This is in accordance with the finding that the ECM-related features alone were able to predict the morphological pattern with the same accuracy as the full panel of features. Using ERT gave the same classification accuracy for the holdout dataset as obtained with k-NN (97%).
3. Discussion
From the wide spectrum of morphological appearances that exist in PC NOS, four patterns were selected that are common—found in 67% of PC NOS in the current series—and exhibit distinct morphological features of both the cancer-cell and stromal compartments, allowing unequivocal identification on H&E staining. Immuno-/histochemical staining as well as morphometric analysis revealed significant differences between at least two of the four patterns for a panel of 26 features. The latter were investigated in this study given their involvement in structural and functional properties that are deemed important in PC. Furthermore, the features are part of transcriptional signatures that characterize PC subtypes [3,12], with the obvious exception of fiber alignment, epithelial proximity and stromal cell density, as these are topologically defined features.
To the best of our knowledge, this study is the first to report that morphological heterogeneity correlates with structural and functional divergence in tissues of human PC NOS. The findings of this study further demonstrate that the various biological processes, in which the analyzed features are involved, are not uniformly operational in PC and co-vary with the morphological phenotype of the tumor. Indeed, while overall results for the entire study series are in line with observations that are reported in the literature (irrespective of tumor morphology), this study reveals significant divergence between the patterns for each of the analyzed features.
This study is also the first to investigate morphological heterogeneity in the stromal compartment of PC, a phenomenon that has received little attention [9] despite the pivotal role of the stroma in PC [105]. Analysis was limited to the immediate peritumoral stroma rather than the bulk stroma because the former plays a key role in the biology of PC. Moreover, the high prevalence of morphological intratumor heterogeneity also demands that stromal analysis be limited to the juxta-tumoral compartment.
A prominent stroma accounting for up to 80% of the tumor mass is considered a hallmark of PC. However, our study revealed significant differences in stromal content between the four patterns, despite the fact that the study series consisted exclusively of well-differentiated PC with a glandular growth pattern [96,106].
The ECM has a multifarious role by providing structural support as well as biochemical and biophysical cues [107]. While the ECM composition for the overall series was very similar to observations published in the literature [33,63,72,73,82,102,108], our study revealed significant differences between the patterns. In TP, which contained the highest amount of tumor stroma, the ECM was rich in collagens and hyaluronan but lacked fibronectin in nearly all cases. FP and PP, patterns with a moderate amount of stroma, contained comparatively less collagens and hyaluronan but were rich in fibronectin. The very small amount of ECM in CP contained mainly hyaluronan but little collagen and fibronectin. These findings imply that the interstitial pressure, which is mainly determined by the amount of hyaluronan and fibrillary collagen [109,110,111], likely differs between the four patterns. Because hyaluronan is a target of stroma-directed therapy [112,113], the impact of the latter on interstitial pressure and drug delivery may vary considerably between patterns. Recently, fibronectin has been linked to acquired chemoresistance in PC [69]; hence, its high content in FP and PP and absence in TP may be of clinical interest. The stark variation in the amount of collagen between the patterns may explain—at least in part—the conflicting results that have been reported regarding the association between a prominent collagenous stroma and survival [63,64].
Not only the composition but also the spatial organization of the ECM is relevant for its biological function. In particular, alignment of collagen and fibronectin fibers promotes directional cancer cell migration in various cancers, including PC [62,114,115]. While marked fiber alignment has been reported in only 12% of PC [65], we found significant variation between the patterns. In the majority of tumors with TP, the stroma showed marked parallel alignment of collagen fibers, resulting in a taut, tendon-like stromal appearance. In contrast, in FP, the whorled fiber arrangement reflected alignment of fibronectin rather than collagen. Interestingly, ITGα5β1, the receptor for fibronectin which is instrumental in the production, assembly and alignment of fibronectin by the CAFs [115], was highly and diffusely expressed in FP. In PP, fibronectin deposition and ITGα5β1 expression were mainly limited to a thin stromal sheath encircling the cancer glands. In contrast, fibronectin and ITGα5β1 were absent or low in TP and CP. These observations are in accordance with in vitro studies revealing the ITGα5β1-dependence of fibronectin deposition and alignment by CAFs [115,116], but at the same time, our findings indicate that this type of ECM organization is not common to all PC. ITGα2, which together with ITGβ1 forms the main receptor for collagen I [117], was present in all patterns, albeit at different levels. Given the multitude of different integrin heterodimer combinations and their biological roles, further studies are needed to understand the relevance of the observed heterogeneity between patterns. Finally, the density of stromal cells, the vast majority of which were αSMA-positive CAFs, differed significantly, being highest in FP and lowest in CP.
Emerging from these findings is a picture of distinct structural and functional diversity of the stroma that co-varies with and is to some extent reflected in the morphology of that compartment (Figure 5).
Figure 5.
Four patterns of pancreatic cancer with distinctive morphological, structural and functional features of the cancer-cell and stromal compartments. (a) In the periglandular pattern, the stromal sheath that encircles the tumor glands contains a moderate amount of CAFs and is rich in fibronectin and MMP14, produced by both CAFs and PCCs. The latter show moderate expression of ALDH1 and Ki67. (b) The tendon-like pattern has a collagen- and hyaluronan-rich stroma that is devoid of fibronectin and shows marked fiber alignment. The majority of cancer cells express ALDH1. Note the low levels of MMP14 and Ki67. (c) The whorled stroma of the fascicular pattern contains a high number of CAFs with high expression of ITGα5 and marked deposition of aligned fibronectin. Note the high-level expression of MMP14 in stroma and PCCs. (d) The small stromal compartment in the chickenwire pattern is relatively rich in hyaluronan but contains little collagen and fibronectin and lacks fiber alignment. CAFs are low in number. Note the high Ki67-expression in the PCCs. ALDH1, aldehyde dehydrogenase 1; CAF, cancer-associated fibroblast; ITGα5, integrin α5; MMP14 matrix metalloproteinase 14; PCC, pancreatic cancer cell.
Interestingly, when applying the recently proposed stromal subclassification of PC that is based on the activated stroma ratio, i.e., the ratio between the number of αSMA + CAFs and amount of collagen, each of the four patterns can be assigned to a different stromal subtype [64,104]. Interestingly, the fibrolytic and fibrogenic subtypes observed in FP and TP, respectively, were reported to associate with shorter and longer progression-free survival, respectively, than the inert (PP) and dormant (CP) subtypes [104]. Further results from this study extend the nature of the stromal subtypes: high MMP14-expression by both CAFs and cancer cells in FP fits with a dynamic stroma, undergoing active remodeling. In contrast, low MMP14-levels in TP support the notion of a stable collagen-rich stroma with little remodeling.
The proliferative activity of cancer cells is clinically relevant, as it relates to the growth capacity of the cancer and its possible response to cytotoxic treatment. While the median value of the Ki67-index in the overall series was near-identical with that reported by others [97,98], the four patterns differed significantly between themselves. It was highest in CP, which would indicate a more aggressive nature of tumors with CP morphology, despite the preserved glandular growth pattern that defines CP as well differentiated [96]. The lack of co-variance between grade of differentiation and proliferative activity in PC was recently highlighted [106]. The high expression level of CAV1 in the cancer cells of CP compared to the other patterns, points also at a more aggressive behavior, as CAV1-expression has been associated with cancer cell proliferation and migration, chemoresistance and worse patient outcome [25,26,27]. The mutually exclusive CAV1-expression in cancer cells and stroma observed in this study has been described previously but remains as yet unexplained [24].
As in other cancers, cancer stem cells in PC drive metastasis and recurrence, the latter by endowing resistance to chemo(radio-)therapy. While expression of the three CSC markers in the overall series was similar to observations by others [13,17,18,30,34,35,39,40,50,51,99], results differed significantly between patterns. The most prominent difference was observed for ALDH1, which was strongly expressed in nearly all TP, but absent or expressed at low level in the other patterns. Furthermore, expression significantly lower than that reported in the literature was observed for CD44 (in PP) and CD133 (in TP and CP). Interestingly, previous reports commented on heterogeneity of CSC marker expression in association with morphological tumor heterogeneity [34,50].
Taken together, the presence of features that are linked to key tumor biological processes varies between the cancer cell compartments of the four patterns. In CP, high proliferative activity and high-level expression of CD44 and CAV1 suggest a more aggressive tumor behavior. While FP shows a lower proliferation than CP, its cancer cells express high levels of two CSC markers (CD44, CD133) as well as MMP14. The latter may have pro-oncogenic effects outside its well-documented role in cell motility and invasion, namely increased metabolism and proliferation [118]. TP is characterized by high-level expression of ALDH1, which is associated with worse patient outcome and resistance to chemo(radio-)therapy. PP shows low proliferation and low-level expression of CSC markers, CAV1 and MMP14.
The analysis shows that the four morphological patterns differ significantly and consistently in most of the features that were analyzed. Conversely, our findings demonstrate that the features form a robust signature that allows correct prediction of the morphological pattern in 97% of ROIs.
In six of the 39 (15%) cases included in the study, two or three of the four patterns were present within the same tumor. The signature of stromal and cancer-cell features of a pattern that co-occurred with other patterns did not differ from that observed in tumors containing only one pattern. Moreover, most cases in this series contained to a varying extent one or several morphological phenotypes other than the four that were investigated. These observations illustrate that morphological intratumor heterogeneity is common, as reported by others [9,10,119]. Collectively, observations in this study concur with the widespread intratumor heterogeneity that has been described recently at the transcriptional and proteomics level [120,121,122]. Moreover, intratumor heterogeneity has also been reported for several of the individual features analyzed in this study: amount of hyaluronan [77,102] and collagen [19], degree of collagen alignment [19,65], expression of CD44 [34], CD133 [50], ALDH1 [99] and MMP14 [123], and proliferative activity [97,106]. While in these studies intratumor heterogeneity for individual features was reported without further characterization of the morphological phenotype of PC, the results of our study indicate that features do not vary randomly between and within tumors but rather co-vary with the tumor morphology.
Phenotypic heterogeneity in cancer is a complex multifactorial phenomenon that results from the integration of genetic, epigenetic and environmental inputs [124,125]. As such, the histomorphology of cancer, including PC, contains rich, highly integrated information that, in contrast to multiomics data, also visualizes the topology of the various tumor features. Deciphering the complex information contained in histomorphology in terms of discrete biological processes at the molecular level has only just started, to some extent prompted by the application of artificial intelligence [126]. Obviously, the association between morphology, tumor behavior and patient outcome is complex and multifactorial, with a possible divergent impact from the cancerous and stromal compartments.
The results of this study have potentially important implications. First, the study reveals significant and possibly clinically relevant differences between morphological phenotypes that currently remain undistinguished as they are lumped together in the large group of PC NOS. While the four patterns analyzed in this study were selected to test the hypothesis that morphology and functional aspects are linked, the development of a morphological classification for PC would require the characterization of a larger number of morphological phenotypes in PC over and above those currently considered by the WHO classification. The prevalence and complexity of morphological heterogeneity in PC is supported by recent studies that reveal marked intratumor heterogeneity at the transcriptional level, implying that current classification systems may offer a too simplistic taxonomy of PC [120,121,127]. By the same token, intratumor heterogeneity hampers the study of the prognostic significance of the various morphological phenotypes.
Second, the findings of this study question the widespread use of a single “representative” tumor block for bulk molecular analysis. The need for higher cancer cell density in order to avoid contamination with nonneoplastic cells may lead to the selection of tumor areas with low stromal volume, such as CP, and result in overrepresentation of biological mechanisms that are active in such phenotypes.
Third, while molecular assessment of intratumor heterogeneity remains challenging, pathology examination allows expedient and low-cost assessment of the extent and nature of morphological heterogeneity within a tumor. Especially the evaluation of heterogeneity in morphological features that are relevant regarding the effect of targeted treatment (e.g., hyaluronan content for hyaluronidase therapy [112,113]) may be of clinical interest.
Fourth, this study reveals that the tumor stroma, which hitherto is ignored in pathology diagnostics for PC, exhibits distinctive morphological features that may inform on relevant underlying processes. Given that accumulating evidence supports a link between the biological properties of the peritumoral stroma and clinical aspects of PC [6,101,105], patient stratification based also on stromal morphology could be clinically more relevant.
Recent studies have demonstrated a mutual shaping of the cancer cell and stromal compartments. While the cancer cell genotype tunes the composition and biophysical properties of the tumor stroma [100], the ECM and CAFs exert epigenetic effects on the cancer cells and shape the tumor architecture [101,107]. Recent transcriptomics-based studies present evidence for a close link between the phenotype of the stroma and the cancer cells [12,128] rather than an independent combination of both [3]. The results of this study show that the morphological phenotype of PC in the four patterns is determined by a particular combination of both stromal and cancer cell-related features rather than either in isolation and independently of each other. Indeed, using exclusively the set of stromal features resulted in a similarly good prediction of the tumor pattern, that is, the morphological phenotype including both cancer cells and stroma.
Our study has a number of limitations. From the wide range of morphological phenotypes, only four were analyzed. As the four selected patterns represent low-grade PC, our observations may not be pertinent to high-grade, that is, poorly differentiated tumors. The analysis was based on a limited set of features that did not represent important processes such as epithelial-mesenchymal transition or immune response. The number of biochemical and biophysical properties that are highlighted by in vitro and in vivo studies as potentially clinically relevant is large and requires extensive analysis beyond this initial proof-of-concept study. Last but not least, investigations at genomic and/or transcriptional level were not included in this study, but 14 of the non-topological features are part of transcription-based signatures that are used for molecular classification of PC [3,12]. Future studies correlating genomics and transcriptomics data with a variety of morphological patterns are needed to understand the molecular basis of morphological heterogeneity.
4. Materials and Methods
4.1. Tissues
Archival H&E-stained histology slides from consecutive cases of treatment-naïve PC resected between December 2017 and April 2019 at Oslo University Hospital, 0424 Oslo, Norway, were reviewed. For each case, tissue blocks containing one of the four patterns were selected. Areas with necrosis or inflammatory cell infiltration were excluded, as this affects the composition of the stromal component [12,104].
4.2. Cancer Cell and Stromal Features
The cancer cells were analyzed for proliferative activity (Ki67) and expression of the CSC markers CD44, CD133, and ALDH1. The stromal compartment was investigated for the total amount of collagen, collagen fiber alignment, amount and deposition pattern of collagen I and III, fibronectin and hyaluronan; density of CAFs, expression of αSMA, and epithelial proximity, the latter as a measure for stromal volume [129]. Cancer-stroma interactions were investigated for expression of ITGα2, α5, and β1. MMP14 and CAV1 were assessed in both compartments.
4.3. Histochemistry and Immunohistochemistry
Histochemical and immunohistochemical staining of formalin-fixed paraffin-embedded, 3.5 µm thick, serially cut, whole-tissue sections was done manually using standard protocols as previously described [130]. Briefly, tissue sections were incubated with primary antibodies at 4 °C overnight, followed by incubation with secondary antibodies as outlined in Table 4. Endogenous peroxidase activity and unspecific antibody binding were blocked with EnVision Flex peroxidase blocking agent (Dako, Glostrup, Denmark, catalogue nr. DM841) and 1% BSA, respectively.
Table 4.
Antibodies and procedures for immunohistochemical and histochemical staining.
| Immunohistochemical Staining | |||||||
|---|---|---|---|---|---|---|---|
| Primary Antibodies | |||||||
| Antigen | Company | Cat # | Antigen Retrieval | Dilution | Incubation | Species | Control Tissue |
| αSMA | Abcam | ab5694 | CB, 100 °C | 1:100 | ON, 4 °C | Rabbit | Blood vessel |
| ALDH1A1 | Abcam | ab52492 | CB, 100 °C | 1:800 | ON, 4 °C | Rabbit | Kidney, lung |
| Caveolin-1 | Abcam | ab32577 | CB, 100 °C | 1:750 | ON, 4 °C | Rabbit | Liver, lung |
| CD133/1 | Miltenyi Biotec | 130–108-062 | CB, 100 °C | 1:100 | ON, 4 °C | Mouse | Gallbladder |
| CD44 | Dako | M7082 | CB, 100 °C | 1:200 | ON, 4 °C | Mouse | Gallbladder |
| Col Iα1 | LifeSpan BioSciences | LS-B5932 | CB, 100 °C | 1:200 | ON, 4 °C | Mouse | Prostate |
| Col III | Abcam | ab7778 | CB, 100 °C | 1:300 | ON, 4 °C | Rabbit | Testis |
| Fibronectin | Abcam | ab6328 | TEB, 100 °C | 1:300 | ON, 4 °C | Mouse | Granulation tissue |
| ITGα2 | Abcam | ab133557 | CB, 100 °C | 1:1000 | ON, 4 °C | Rabbit | Testis |
| ITGα5 | Abcam | ab150361 | CB, 100 °C | 1:200 | ON, 4 °C | Rabbit | Placenta |
| ITGβ1 | ThermoFisher | MA5-17103 | CB, 100 °C | 1:200 | ON, 4 °C | Mouse | Small intestine |
| Ki67 | Dako | M7240 | CB, 100 °C | 1:1000 | ON, 4 °C | Mouse | Small intestine |
| MMP14 | Abcam | ab51074 | TEB, 100 °C | 1:1600 | ON, 4 °C | Rabbit | Placenta |
| Secondary Antibodies | |||||||
| Company | Cat # | Antigen Retrieval | Dilution | Incubation | Detection | Control Tissue | |
| Immpress HRP Anti-Rabbit IgG | Vector Labs | MP-7401 | - | Ready to use | 1 h, RT | DAB Peroxidase substrate kit (SK-4100) | - |
| Immpress HRP Anti-Mouse IgG | Vector Labs | MP-7402 | - | Ready to use | 1 h, RT | DAB Peroxidase substrate kit (SK-4100) | - |
| Histochemical Staining | |||||||
| Hyaluronan | Merck Millipore | 385911 | CB, 60 °C | 1:100 | 1 h, RT | ABC-AP-Kit (Vector Labs, AK-500) and Liquid Permanent Red (Dako, K0640) | Cartilage |
| Picrosirius red | Histolab | HL27150.0500 | - | Ready to use | 10 min, RT | Tendon | |
CB, Citrate buffer (pH 6); TEB, Tris/EDTA buffer (pH 9). Abcam, Cambridge, UK; Histolab, Askim, Sweden; LifeSpan Biosciences, Seattle, WA, USA; Merck Millipore, Burlington, MA, USA; Miltenyi Biotec, Bergisch Gladbach, Germany; Thermo Fisher Scientific, Waltham, MA, USA; Vector Labs, Burlingame, CA, USA; Dako: Agilent Technologies, Glostrup, Denmark; ThermoFisher: Waltham, MA USA.
Histochemistry using hyaluronic acid binding protein was employed for the histochemical detection of hyaluronan, as previously described (see Table 4) [82]. Fibrillar collagen was detected with ready-to-use picrosirius red staining. Hematoxylin was used for counterstaining.
Appropriate positive and negative controls were used to verify specificity of all tested markers. Negative control samples were incubated with PBS instead of the primary antibody.
4.4. Semiquantitative Scoring
Sections were viewed with a light microscope (Nikon eclipse Ni, Amsterdam, Netherlands) using polarized light for visualization of collagen fibers with picrosirius red staining [103,131]. In each tumor, at least five randomly selected ROIs representative of a particular pattern were photographed at 200x magnification (camera: Infinity2–5C Lumenera, Ottawa, ON, Canada). Picture acquisition was done with strict compliance regarding exposure time, light intensity, contrast, aperture, and sensor sensitivity.
From the wealth of different systems and threshold values that have been used previously to score histochemical (hyaluronan) and immunohistochemical staining for any of the markers included in the panel of this study, the following uniform, simple and robust staining score was selected: 0 = no staining, 1 = staining in <25% of the relevant compartment, 2 = staining in 26–50% of the relevant compartment, 3 = staining in >50% of the relevant compartment. Staining scores 0 and 1 were regarded as “low,” score 2 as “medium,” and score 3 as “high,” as previously described [24,82,132,133,134]. Because of major variation in both staining intensity and number of positive cells, immunostaining for CSC markers was scored by multiplying intensity (0 = negative, 1 = low, 2 = moderate, 3 = high) and extent of staining (0 = 0%, 1 = 1–10%, 2 = 11–50%, 3 = 51–80%, 4 ≥ 81%), resulting in an immunoreactive score (IRS) between 0 and 12 [34,36,39,135]. The IRS was categorized as negative (0), low (1–5), and high (6–12). The pattern of deposition of ECM-components was classified as diffuse or peritumoral, the latter denoting deposition immediately surrounding the tumor glands.
4.5. Quantitative Assessment
The total amount of fibrillar collagen and the alignment of collagen fibers were quantified with ImageJ (NIH, Bethesda, MD, USA) and the OrientationJ plugin [136], respectively, using the polarization photographs of picrosirius red-stained sections that were converted to grey-scale 8-bit images. Identical analysis settings and thresholds were used for all cases. Higher values corresponded with higher fibrillar collagen content (reflecting the area fraction of polarized light) and better fiber alignment, respectively. Stromal cells showing features characteristic of fibroblasts, that is, a spindle cell shape and an elongated nucleus without atypical features, were counted on H&E-sections. The average was calculated from cell counts in five random 0.01 mm2 rectangular areas per ROI, excluding foci with inflammatory cell infiltration. Epithelial proximity is a feature that denotes the extent of contiguous areas of stroma that separate cancer cell clusters and has been used previously for the morphological analysis of the stroma in cancer [129]. It was assessed with ImageJ based on the average of shortest distances between any two cancer glands present in an ROI. Immunostaining for Ki67 was counted in the cancer cell population and expressed as the percentage of positive cells.
4.6. Network Visualization
Network visualization of the results for all features was generated with Gephi (v0.9.2) using the Fruchterman–Reingold algorithm [137] and an adjacency matrix (RStudio v 1.453). The former is a force-based approach using small to medium-sized networks (unweighted, undirected graphs) [138,139]. The vertex layout is determined by attractive and repulsive “forces” between the vertices.
4.7. Statistical Analysis
The Kruskal–Wallis multiple comparison test and the Mann–Whitney U test for pair-wise comparison were used to test whether the feature data differed significantly between the four morphological patterns. These are nonparametric methods for analyzing whether two or more datasets come from the same statistical distribution or from different distributions. In this study, p-values < 0.05 were considered significant for detection of a difference. The tests were carried out using the SPSS® software for Windows (v22) (IBM SPSS Statistics, Armonk, NY, USA).
4.8. Prediction of Morphological Pattern
In order to test the strength of the association between morphology and the panel of features, we investigated whether the morphological pattern in each ROI could be predicted based on the features used in this study. The samples were classified with respect to the four different morphological patterns using the k-nearest neighbors algorithm (k-NN) [140], with k equal to 3. According to this algorithm, a sample is classified by a majority vote of its k neighbors, with the object being assigned to the class most common among its k nearest neighbors, and each neighbor is attributed a weight of 1/d, where d is the distance to the neighbor. The classification was done with four different sets of features: (i) all features, (ii) only cancer-cell features, (iii) only stromal features and iv) only ECM-related features. Each classification model was trained using 70% of the samples, while a separate holdout set of 30% of the samples (randomly selected) was used to test the prediction accuracy.
In order to further investigate the importance of the various features for predicting the morphological pattern, the extremely randomized tree (ERT) algorithm for estimation of the feature importance was applied [141]. The ERT algorithm is a tree-based ensemble classification method by which the outcomes from many decision trees are averaged to obtain a final output. Decision trees generate decision rules to divide observations into segments that have the largest difference with respect to the target variable. Each rule selects both the variable and the best breakpoint to separate the resulting subgroups maximally, and one seeks to find the smallest set of rules that is consistent with the training data. The ExtraTreesClassifier implementation in the SCiKit-learn module “ensemble”, where Python v. 3.7.4. was used [142,143].
5. Conclusions
This proof-of-concept study is the first to show that heterogeneity in morphological features reflects structural and functional heterogeneity and that tumors exhibiting the same histological pattern also share functional and structural properties. Furthermore, this study demonstrates that biological processes deemed to be crucial in PC may not be relevant in all PC tumors nor in all parts of a tumor, considering that morphological inter- and intratumor heterogeneity is common. Further systematic investigation of morphological heterogeneity and its link with biological diversity, treatment effect and patient outcome may ultimately allow pathologists to provide new and clinically meaningful information on tumor-intrinsic properties as part of the routine diagnostic work-up.
Abbreviations
| ALDH1 | Aldehyde dehydrogenase 1 |
| αSMA | α-Smooth muscle actin |
| CAF | Cancer-associated fibroblast |
| CAV1 | Caveolin-1 |
| Col | Collagen |
| CP | Chickenwire pattern |
| CSC | Cancer stem cell |
| ECM | Extracellular matrix |
| EMT | Epithelial-mesenchymal transition |
| ERT | Extremely randomized tree |
| FN | Fibronectin |
| FP | Fascicular pattern |
| HA | Hyaluronan |
| H&E | Hematoxylin and eosin |
| IRS | Immunoreactive score |
| ITG | Integrin |
| MMP14 | Matrix metalloproteinase 14 |
| NOS | Not otherwise specified |
| PC | Pancreatic cancer |
| PCC | Pancreatic cancer cell |
| PP | Periglandular pattern |
| ROI | Region of interest |
| SS | Staining score |
| TP | Tendon-like pattern |
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/13/4/895/s1, Figure S1: Histological features of the four morphological patterns, Figure S2: Feature importance estimation based on the ERT algorithm, Table S1: Clinicopathological features of the study series of resected, treatment-naïve human pancreatic cancer (n = 39), Table S2: Results for the panel of features for the series overall and for each tumor pattern individually, Table S3: Correct and incorrect prediction of morphological pattern based on the full set or on subsets of the analyzed features.
Author Contributions
Conceptualization, P.S. and C.V.; methodology, P.S. and C.V.; software, P.S. and K.T.; formal analysis, C.V., P.S. and K.T.; investigation, P.S., D.L., A.F., L.D. and C.V.; writing—original draft preparation, P.S. and C.V.; writing—review and editing, C.V., I.P.G., D.L. and M.A.; visualization, P.S.; supervision, C.V.; project administration, C.V.; funding acquisition, I.P.G. and C.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by a grant from the South-Eastern Norway Regional Health Authority (Helse Sør-Øst, grant number 2016090) and the Norwegian Cancer Society (Kreftforeningen, project number 212734-2019). The funders had no role in the study design, data collection, data analysis and interpretation, or manuscript writing, nor in the decision to submit the manuscript for publication.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Helsinki Declaration of ethical principles involving human subjects and was approved by the Regional Committee for Medical and Health Research Ethics South East (2015/738; date of approval: 16 November 2016).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethical considerations and data regulations.
Conflicts of Interest
C.V. has received honoraria for consulting for FibroGen Inc. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kamisawa T., Wood L.D., Itoi T., Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 2.Kleeff J., Korc M., Apte M., La Vecchia C., Johnson C.D., Biankin A.V., Neale R.E., Tempero M., Tuveson D.A., Hruban R.H., et al. Pancreatic cancer. Nat. Rev. Dis. Primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 3.Moffitt R.A., Marayati R., Flate E.L., Volmar K.E., Loeza S.G., Hoadley K.A., Rashid N.U., Williams L.A., Eaton S.C., Chung A.H., et al. Virtual microdissection identifies distinct tumor-and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C., Miller D.K., Christ A.N., Bruxner T.J., Quinn M.C., et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 5.Collisson E.A., Sadanandam A., Olson P., Gibb W.J., Truitt M., Gu S., Cooc J., Weinkle J., Kim G.E., Jakkula L., et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puleo F., Nicolle R., Blum Y., Cros J., Marisa L., Demetter P., Quertinmont E., Svrcek M., Elarouci N., Iovanna J., et al. Stratification of pancreatic ductal adenocarcinomas based on tumor and microenvironment features. Gastroenterology. 2018;155:1999–2013.e3. doi: 10.1053/j.gastro.2018.08.033. [DOI] [PubMed] [Google Scholar]
- 7.Hruban R.H., Adsay N.V., Esposito I., Fukushima N., Klöppel G., Maitra A., Notohara K., Offerhaus G.J.A., Ohike N., Pitman M.B., et al. Digestive System Cancers. 5th ed. Volume 1. International Agency for Research on Cancer; Lyon, France: 2019. Tumours of the pancreas; pp. 322–332. WHO Classification of Tumours. [Google Scholar]
- 8.Luchini C., Brosens L.A.A., Wood L.D., Chatterjee D., Shin J.I., Sciammarella C., Fiadone G., Malleo G., Salvia R., Kryklyva V., et al. Comprehensive characterisation of pancreatic ductal adenocarcinoma with microsatellite instability: Histology, molecular pathology and clinical implications. Gut. 2020;70:148–156. doi: 10.1136/gutjnl-2020-320726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verbeke C. Morphological heterogeneity in ductal adenocarcinoma of the Pancreas-Does it matter? Pancreatology. 2016;16:295–301. doi: 10.1016/j.pan.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Kalimuthu S.N., Wilson G.W., Grant R.C., Seto M., O’Kane G., Vajpeyi R., Notta F., Gallinger S., Chetty R. Morphological classification of pancreatic ductal adenocarcinoma that predicts molecular subtypes and correlates with clinical outcome. Gut. 2020;69:317–328. doi: 10.1136/gutjnl-2019-318217. [DOI] [PubMed] [Google Scholar]
- 11.Santha P., Dorg L., Finstadsveen A., Lenggenhager D., Amrutkar M., Gladhaug I.P., Verbeke C.S. Morphological heterogeneity in pancreatic ductal adenocarcinoma is associated with structural and functional divergence in the cancer cell population and stroma. Pancreatology. 2019;19:S85–S86. doi: 10.1016/j.pan.2019.05.225. [DOI] [Google Scholar]
- 12.Maurer C., Holmstrom S.R., He J., Laise P., Su T., Ahmed A., Hibshoosh H., Chabot J.A., Oberstein P.E., Sepulveda A.R., et al. Experimental microdissection enables functional harmonisation of pancreatic cancer subtypes. Gut. 2019;68:1034–1043. doi: 10.1136/gutjnl-2018-317706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S.K., Kim H., Lee D.H., Kim T.S., Kim T., Chung C., Koh G.Y., Kim H., Lim D.S. Reversing the intractable nature of pancreatic cancer by selectively targeting ALDH-High, therapy-resistant cancer cells. PLoS ONE. 2013;8:e78130. doi: 10.1371/journal.pone.0078130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duong H.-Q., Hwang J.S., Kim H.J., Kang H.J., Seong Y.-S., Bae I. Aldehyde dehydrogenase 1A1 confers intrinsic and acquired resistance to gemcitabine in human pancreatic adenocarcinoma MIA PaCa-2 cells. Int. J. Oncol. 2012;41:855–861. doi: 10.3892/ijo.2012.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oria V., Bronsert P., Thomsen A., Föll M., Zamboglou C., Hannibal L., Behringer S., Biniossek M., Schreiber C., Grosu A., et al. Proteome Profiling of Primary Pancreatic Ductal Adenocarcinomas Undergoing Additive Chemoradiation Link ALDH1A1 to Early Local Recurrence and Chemoradiation Resistance. Transl. Oncol. 2018;11:1307–1322. doi: 10.1016/j.tranon.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jimeno A., Feldmann G., Suarez-Gauthier A., Rasheed Z., Solomon A., Zou G.M., Rubio-Viqueira B., Garcia-Garcia E., Lopez-Rios F., Matsui W., et al. A direct pancreatic cancer xenograft model as a platform for cancer stem cell therapeutic development. Mol. Cancer Ther. 2009;8:310–314. doi: 10.1158/1535-7163.MCT-08-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasheed Z.A., Yang J., Wang Q., Kowalski J., Freed I., Murter C., Hong S.-M., Koorstra J.-B., RajeshKumar N.V., He X., et al. Prognostic Significance of Tumorigenic Cells With Mesenchymal Features in Pancreatic Adenocarcinoma. J. Natl. Cancer Inst. 2010;102:340–351. doi: 10.1093/jnci/djp535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahlert C., Bergmann F., Beck J., Welsch T., Mogler C., Herpel E., Dutta S., Niemietz T., Koch M., Weitz J. Low expression of aldehyde dehydrogenase 1A1 (ALDH1A1) is a prognostic marker for poor survival in pancreatic cancer. BMC Cancer. 2011;11:275. doi: 10.1186/1471-2407-11-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drifka C.R., Tod J., Loeffler A.G., Liu Y., Thomas G.J., Eliceiri K.W., Kao W.J. Periductal stromal collagen topology of pancreatic ductal adenocarcinoma differs from that of normal and chronic pancreatitis. Mod. Pathol. 2015;28:1470–1480. doi: 10.1038/modpathol.2015.97. [DOI] [PubMed] [Google Scholar]
- 20.Sinn M., Denkert C., Striefler J.K., Pelzer U., Stieler J.M., Bahra M., Lohneis P., Dorken B., Oettle H., Riess H., et al. Alpha-Smooth muscle actin expression and desmoplastic stromal reaction in pancreatic cancer: Results from the CONKO-001 study. Br. J. Cancer. 2014;111:1917–1923. doi: 10.1038/bjc.2014.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuninty P.R., Bansal R., De Geus S.W.L., Mardhian D.F., Schnittert J., Van Baarlen J., Storm G., Bijlsma M.F., Van Laarhoven H.W., Metselaar J.M., et al. ITGA5 inhibition in pancreatic stellate cells attenuates desmoplasia and potentiates efficacy of chemotherapy in pancreatic cancer. Sci. Adv. 2019;5:eaax2770. doi: 10.1126/sciadv.aax2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno-Vicente R., Pavón D.M., Martín-Padura I., Català-Montoro M., Díez-Sánchez A., Quílez-Álvarez A., López J.A., Sánchez-Álvarez M., Vázquez J., Strippoli R., et al. Caveolin-1 Modulates Mechanotransduction Responses to Substrate Stiffness through Actin-Dependent Control of YAP. Cell Rep. 2018;25:1622–1635.e6. doi: 10.1016/j.celrep.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goetz J.G., Minguet S., Navarro-Lerida I., Lazcano J.J., Samaniego R., Calvo E., Tello M., Osteso-Ibanez T., Pellinen T., Echarri A., et al. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell. 2011;146:148–163. doi: 10.1016/j.cell.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamposioras K., Tsimplouli C., Verbeke C., Anthoney A., Daoukopoulou A., Papandreou C.N., Sakellaridis N., Vassilopoulos G., Potamianos S.P., Liakouli V., et al. Silencing of caveolin-1 in fibroblasts as opposed to epithelial tumor cells results in increased tumor growth rate and chemoresistance in a human pancreatic cancer model. Int. J. Oncol. 2019;54:537–549. doi: 10.3892/ijo.2018.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatterjee M., Ben-Josef E., Thomas D.G., Morgan M.A., Zalupski M.M., Khan G., Robinson C.A., Griffith K.A., Chen C.-S., Ludwig T., et al. Caveolin-1 is Associated with Tumor Progression and Confers a Multi-Modality Resistance Phenotype in Pancreatic Cancer. Sci. Rep. 2015;5:10867. doi: 10.1038/srep10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuoki M., Miyamoto M., Kato K., Hiraoka K., Oshikiri T., Nakakubo Y., Fukunaga A., Shichinohe T., Shinohara T., Itoh T., et al. Impact of caveolin-1 expression on prognosis of pancreatic ductal adenocarcinoma. Br. J. Cancer. 2002;87:1140–1144. doi: 10.1038/sj.bjc.6600619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shan T., Lu H., Ji H., Li Y., Guo J., Chen X., Wu T. Loss of stromal caveolin-1 expression: A novel tumor microenvironment biomarker that can predict poor clinical outcomes for pancreatic cancer. PLoS ONE. 2014;9:e97239. doi: 10.1371/journal.pone.0097239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang W., Zhang Y., Kane K.T., Collins M.A., Simeone D.M., Di Magliano M.P., Nguyen K.T. CD44 Regulates Pancreatic Cancer Invasion through MT1-MMP. Mol. Cancer Res. 2015;13:9–15. doi: 10.1158/1541-7786.MCR-14-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toole B.P., Slomiany M.G. Hyaluronan: A constitutive regulator of chemoresistance and malignancy in cancer cells. Semin. Cancer Biol. 2008;18:244–250. doi: 10.1016/j.semcancer.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong S.P., Wen J., Bang S., Park S., Song S.Y. CD44-Positive cells are responsible for gemcitabine resistance in pancreatic cancer cells. Int. J. Cancer. 2009;125:2323–2331. doi: 10.1002/ijc.24573. [DOI] [PubMed] [Google Scholar]
- 31.Günthert U., Hofmann M., Rudy W., Reber S., Zöller M., Hauβmann I., Matzku S., Wenzel A., Ponta H., Herrlich P. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell. 1991;65:13–24. doi: 10.1016/0092-8674(91)90403-L. [DOI] [PubMed] [Google Scholar]
- 32.Zhao S., Chen C., Chang K., Karnad A., Jagirdar J., Kumar A.P., Freeman J.W. CD44 Expression Level and Isoform Contributes to Pancreatic Cancer Cell Plasticity, Invasiveness, and Response to Therapy. Clin. Cancer Res. 2016;22:5592–5604. doi: 10.1158/1078-0432.CCR-15-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franklin O., Billing O., Öhlund D., Berglund A., Herdenberg C., Wang W., Hellman U., Sund M. Novel prognostic markers within the CD44-stromal ligand network in pancreatic cancer. J. Pathol. Clin. Res. 2019;5:130–141. doi: 10.1002/cjp2.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Immervoll H., Hoem D., Steffensen O.J., Miletic H., Molven A. Visualization of CD44 and CD133 in normal pancreas and pancreatic ductal adenocarcinomas: Non-Overlapping membrane expression in cell populations positive for both markers. J. Histochem. Cytochem. 2011;59:441–455. doi: 10.1369/0022155411398275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X.-P., Zhang X.-W., Zheng L.-Z., Guo W.-J. Expression of CD44 in pancreatic cancer and its significance. Int. J. Clin. Exp. Pathol. 2015;8:6724–6731. [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu C.-P., Lee L.-Y., Hsu J.-T., Hsu Y.-P., Wu Y.-T., Wang S.-Y., Yeh C.-N., Chen T.-C., Hwang T.-L. CD44 Predicts Early Recurrence in Pancreatic Cancer Patients Undergoing Radical Surgery. In Vivo. 2018;32:1533–1540. doi: 10.21873/invivo.11411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumazoe M., Takai M., Bae J., Hiroi S., Huang Y., Takamatsu K., Won Y., Yamashita M., Hidaka S., Yamada S., et al. FOXO3 is essential for CD44 expression in pancreatic cancer cells. Oncogene. 2016;36:2643–2654. doi: 10.1038/onc.2016.426. [DOI] [PubMed] [Google Scholar]
- 38.Xiaoping L., Xiaowei Z., Leizhen Z., Weijian G. Expression and significance of CD44 and p-AKT in pancreatic head cancer. World J. Surg. Oncol. 2015;13:1–7. doi: 10.1186/s12957-015-0746-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durko L., Wlodarski W., Stasikowska-Kanicka O., Wagrowska-Danilewicz M., Danilewicz M., Hogendorf P., Strzelczyk J., Malecka-Panas E. Expression and clinical significance of cancer stem cell markers CD24, CD44, and CD133 in pancreatic ductal adenocarcinoma and chronic pancreatitis. Dis. Markers. 2017;2017:3276806. doi: 10.1155/2017/3276806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou Y., Chao Y., Tung H., Wang H., Shan Y. Coexpression of CD44-positive/CD133-positive cancer stem cells and CD204-positive tumor-associated macrophages is a predictor of survival in pancreatic ductal adenocarcinoma. Cancer. 2014;120:2766–2777. doi: 10.1002/cncr.28774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molejon M.I., Tellechea J.I., Moutardier V., Gasmi M., Ouaissi M., Turrini O., Delpero J.R., Dusetti N., Iovanna J. Targeting CD44 as a novel therapeutic approach for treating pancreatic cancer recurrence. Oncoscience. 2015;2:572–575. doi: 10.18632/oncoscience.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quint K., Tonigold M., di Fazio P., Montalbano R., Lingelbach S., Ruckert F., Alinger B., Ocker M., Neureiter D. Pancreatic cancer cells surviving gemcitabine treatment express markers of stem cell differentiation and epithelial-mesenchymal transition. Int. J. Oncol. 2012;41:2093–2102. doi: 10.3892/ijo.2012.1648. [DOI] [PubMed] [Google Scholar]
- 43.Hermann P.C., Huber S.L., Herrler T., Aicher A., Ellwart J.W., Guba M., Bruns C.J., Heeschen C. Distinct Populations of Cancer Stem Cells Determine Tumor Growth and Metastatic Activity in Human Pancreatic Cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 44.Weng C.C., Kuo K.K., Su H.T., Hsiao P.J., Chen Y.W., Wu D.C., Hung W.C., Cheng K.H. Pancreatic tumor progression associated with CD133 overexpression: Involvement of increased TERT expression and epidermal growth factor receptor-dependent Akt activation. Pancreas. 2016;45:443–457. doi: 10.1097/MPA.0000000000000460. [DOI] [PubMed] [Google Scholar]
- 45.Ding Q., Miyazaki Y., Tsukasa K., Matsubara S., Yoshimitsu M., Takao S. CD133 facilitates epithelial-mesenchymal transition through interaction with the ERK pathway in pancreatic cancer metastasis. Mol. Cancer. 2014;13:15. doi: 10.1186/1476-4598-13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding Q., Yoshimitsu M., Kuwahata T., Maeda K., Hayashi T., Obara T., Miyazaki Y., Matsubara S., Natsugoe S., Takao S. Establishment of a highly migratory subclone reveals that CD133 contributes to migration and invasion through epithelial-mesenchymal transition in pancreatic cancer. Hum. Cell. 2012;25:1–8. doi: 10.1007/s13577-011-0037-9. [DOI] [PubMed] [Google Scholar]
- 47.Nomura A., Dauer P., Gupta V., McGinn O., Arora N., Majumdar K., Uhlrich C., 3rd, Dalluge J., Dudeja V., Saluja A. Microenvironment mediated alterations to metabolic pathways confer increased chemo-resistance in CD133+ tumor initiating cells. Oncotarget. 2016;7:56324–56337. doi: 10.18632/oncotarget.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen K., Li Z., Jiang P., Zhang X., Zhang Y., Jiang Y., He Y., Li X. Co-expression of CD133, CD44v6 and human tissue factor is associated with metastasis and poor prognosis in pancreatic carcinoma. Oncol. Rep. 2014;32:755–763. doi: 10.3892/or.2014.3245. [DOI] [PubMed] [Google Scholar]
- 49.Maeda S., Shinchi H., Kurahara H., Mataki Y., Maemura K., Sato M., Natsugoe S., Aikou T., Takao S. CD133 expression is correlated with lymph node metastasis and vascular endothelial growth Factor-C expression in pancreatic cancer. Br. J. Cancer. 2008;98:1389–1397. doi: 10.1038/sj.bjc.6604307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Immervoll H., Hoem D., Sakariassen P.Ø., Steffensen O.J., Molven A. Expression of the “stem cell marker” CD133 in pancreas and pancreatic ductal adenocarcinomas. BMC Cancer. 2008;8:1–14. doi: 10.1186/1471-2407-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mizukami T., Kamachi H., Mitsuhashi T., Tsuruga Y., Hatanaka Y., Kamiyama T., Matsuno Y., Taketomi A. Immunohistochemical analysis of cancer stem cell markers in pancreatic adenocarcinoma patients after neoadjuvant chemoradiotherapy. BMC Cancer. 2014;14:687. doi: 10.1186/1471-2407-14-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koenig A., Mueller C., Hasel C., Adler G., Menke A. Collagen Type I Induces Disruption of E-Cadherin–Mediated Cell-Cell Contacts and Promotes Proliferation of Pancreatic Carcinoma Cells. Cancer Res. 2006;66:4662–4671. doi: 10.1158/0008-5472.CAN-05-2804. [DOI] [PubMed] [Google Scholar]
- 53.Shintani Y., Hollingsworth M.A., Wheelock M.J., Johnson K.R. Collagen I Promotes Metastasis in Pancreatic Cancer by Activating c-Jun NH2-Terminal Kinase 1 and Up-regulating N-Cadherin Expression. Cancer Res. 2006;66:11745–11753. doi: 10.1158/0008-5472.CAN-06-2322. [DOI] [PubMed] [Google Scholar]
- 54.Armstrong T., Packham G., Murphy L.B., Bateman A.C., Conti J.A., Fine D.R., Johnson C.D., Benyon R.C., Iredale J.P. Type I Collagen Promotes the Malignant Phenotype of Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2004;10:7427–7437. doi: 10.1158/1078-0432.CCR-03-0825. [DOI] [PubMed] [Google Scholar]
- 55.Ryschich E., Khamidjanov A., Kerkadze V., Büchler M.W., Zöller M., Schmidt J. Promotion of Tumor Cell Migration by Extracellular Matrix Proteins in Human Pancreatic Cancer. Pancreas. 2009;38:804–810. doi: 10.1097/MPA.0b013e3181b9dfda. [DOI] [PubMed] [Google Scholar]
- 56.Shields M.A., Dangi-Garimella S., Redig A.J., Munshi H.G. Biochemical role of the collagen-rich tumour microenvironment in pancreatic cancer progression. Biochem. J. 2012;441:541–552. doi: 10.1042/BJ20111240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu J., Zhou S., Siech M., Habisch H., Seufferlein T., Bachem M.G. Pancreatic stellate cells promote hapto-migration of cancer cells through collagen I-mediated signalling pathway. Br. J. Cancer. 2013;110:409–420. doi: 10.1038/bjc.2013.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyamoto H., Murakami T., Tsuchida K., Sugino H., Miyake H., Tashiro S. Tumor-Stroma interaction of human pancreatic cancer: Acquired resistance to anticancer drugs and proliferation regulation is dependent on extracellular matrix proteins. Pancreas. 2004;28:38–44. doi: 10.1097/00006676-200401000-00006. [DOI] [PubMed] [Google Scholar]
- 59.Grzesiak J.J., Bouvet M. The alpha2beta1 integrin mediates the malignant phenotype on type I collagen in pancreatic cancer cell lines. Br. J. Cancer. 2006;94:1311–1319. doi: 10.1038/sj.bjc.6603088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DuFort C.C., DelGiorno K.E., Hingorani S.R. Mounting Pressure in the Microenvironment: Fluids, Solids, and Cells in Pancreatic Ductal Adenocarcinoma. Gastroenterology. 2016;150:1545–1557.e2. doi: 10.1053/j.gastro.2016.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gritsenko P.G., Ilina O., Friedl P. Interstitial guidance of cancer invasion. J. Pathol. 2011;226:185–199. doi: 10.1002/path.3031. [DOI] [PubMed] [Google Scholar]
- 62.Lee H.O., Mullins S.R., Franco-Barraza J., Valianou M., Cukierman E., Cheng J.D. FAP-Overexpressing fibroblasts produce an extracellular matrix that enhances invasive velocity and directionality of pancreatic cancer cells. BMC Cancer. 2011;11:245. doi: 10.1186/1471-2407-11-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whatcott C.J., Diep C.H., Jiang P., Watanabe A., LoBello J., Sima C., Hostetter G., Shepard H.M., Von Hoff D.D., Han H. Desmoplasia in Primary Tumors and Metastatic Lesions of Pancreatic Cancer. Clin. Cancer Res. 2015;21:3561–3568. doi: 10.1158/1078-0432.CCR-14-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Erkan M., Michalski C.W., Rieder S., Reiser–Erkan C., Abiatari I., Kolb A., Giese N.A., Esposito I., Friess H., Kleeff J. The Activated Stroma Index Is a Novel and Independent Prognostic Marker in Pancreatic Ductal Adenocarcinoma. Clin. Gastroenterol. Hepatol. 2008;6:1155–1161. doi: 10.1016/j.cgh.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 65.Drifka C.R., Loeffler A.G., Mathewson K., Keikhosravi A., Eickhoff J.C., Liu Y., Weber S.M., Kao W.J., Eliceiri K.W. Highly aligned stromal collagen is a negative prognostic factor following pancreatic ductal adenocarcinoma resection. Oncotarget. 2016;7:76197–76213. doi: 10.18632/oncotarget.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Menke A., Philippi C., Vogelmann R., Seidel B., Lutz M.P., Adler G., Wedlich D. Down-regulation of E-cadherin gene expression by collagen type I and type III in pancreatic cancer cell lines. Cancer Res. 2001;61:3508–3517. [PubMed] [Google Scholar]
- 67.Chen I.M., Willumsen N., Dehlendorff C., Johansen A.Z., Jensen B.V., Hansen C.P., Hasselby J.P., Bojesen S.E., Pfeiffer P., Nielsen S.E., et al. Clinical value of serum hyaluronan and propeptide of type III collagen in patients with pancreatic cancer. Int. J. Cancer. 2020;146:2913–2922. doi: 10.1002/ijc.32751. [DOI] [PubMed] [Google Scholar]
- 68.Willumsen N., Ali S.M., Leitzel K., Drabick J.J., Yee N., Polimera H.V., Nagabhairu V., Krecko L., Ali A., Maddukuri A., et al. Collagen fragments quantified in serum as measures of desmoplasia associate with survival outcome in patients with advanced pancreatic cancer. Sci. Rep. 2019;9:19761. doi: 10.1038/s41598-019-56268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amrutkar M., Aasrum M., Verbeke C.S., Gladhaug I.P. Secretion of fibronectin by human pancreatic stellate cells promotes chemoresistance to gemcitabine in pancreatic cancer cells. BMC Cancer. 2019;19:596. doi: 10.1186/s12885-019-5803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Javle M.M., Gibbs J.F., Iwata K.K., Pak Y., Rutledge P., Yu J., Black J.D., Tan D., Khoury T. Epithelial-Mesenchymal Transition (EMT) and Activated Extracellular Signal-regulated Kinase (p-Erk) in Surgically Resected Pancreatic Cancer. Ann. Surg. Oncol. 2007;14:3527–3533. doi: 10.1245/s10434-007-9540-3. [DOI] [PubMed] [Google Scholar]
- 71.Hiroshima Y., Kasajima R., Kimura Y., Komura D., Ishikawa S., Ichikawa Y., Bouvet M., Yamamoto N., Oshima T., Morinaga S., et al. Novel targets identified by integrated cancer-stromal interactome analysis of pancreatic adenocarcinoma. Cancer Lett. 2020;469:217–227. doi: 10.1016/j.canlet.2019.10.031. [DOI] [PubMed] [Google Scholar]
- 72.Leppänen J., Lindholm V., Isohookana J., Haapasaari K.-M., Karihtala P., Lehenkari P.P., Saarnio J., Kauppila J.H., Karttunen T.J., Helminen O., et al. Tenascin C, Fibronectin, and Tumor-Stroma Ratio in Pancreatic Ductal Adenocarcinoma. Pancreas. 2019;48:43–48. doi: 10.1097/MPA.0000000000001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu D., Ansari D., Zhou Q., Sasor A., Hilmersson K.S., Andersson R. Stromal fibronectin expression in patients with resected pancreatic ductal adenocarcinoma. World J. Surg. Oncol. 2019;17:1–8. doi: 10.1186/s12957-019-1574-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Provenzano P.P., Cuevas C., Chang A.E., Goel V.K., Von Hoff D.D., Hingorani S.R. Enzymatic Targeting of the Stroma Ablates Physical Barriers to Treatment of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2012;21:418–429. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chauhan V.P., Boucher Y., Ferrone C.R., Roberge S., Martin J.D., Stylianopoulos T., Bardeesy N., Depinho R.A., Padera T.P., Munn L.L., et al. Compression of Pancreatic Tumor Blood Vessels by Hyaluronan Is Caused by Solid Stress and Not Interstitial Fluid Pressure. Cancer Cell. 2014;26:14–15. doi: 10.1016/j.ccr.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Provenzano P.P., Hingorani S.R. Hyaluronan, fluid pressure, and stromal resistance in pancreas cancer. Br. J. Cancer. 2013;108:1–8. doi: 10.1038/bjc.2012.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang H., Mislati R., Ahmed R., Vincent P., Nwabunwanne S.F., Gunn J.R., Pogue B.W., Doyley M.M. Elastography can map the local inverse relationship between shear modulus and drug delivery within the pancreatic ductal adenocarcinoma microenvironment. Clin. Cancer Res. 2019;25:2136–2143. doi: 10.1158/1078-0432.CCR-18-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng X.-B., Kohi S., Koga A., Hirata K., Sato N. Hyaluronan stimulates pancreatic cancer cell motility. Oncotarget. 2015;7:4829–4840. doi: 10.18632/oncotarget.6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheng X., Sato N., Kohi S., Koga A., Hirata K. 4-Methylumbelliferone inhibits enhanced hyaluronan synthesis and cell migration in pancreatic cancer cells in response to tumor-stromal interactions. Oncol. Lett. 2018;15:6297–6301. doi: 10.3892/ol.2018.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scaife C.L., Shea J.E., Dai Q., Firpo M.A., Prestwich G.D., Mulvihill S.J. Synthetic Extracellular Matrix Enhances Tumor Growth and Metastasis in an Orthotopic Mouse Model of Pancreatic Adenocarcinoma. J. Gastrointest. Surg. 2007;12:1074–1080. doi: 10.1007/s11605-007-0425-3. [DOI] [PubMed] [Google Scholar]
- 81.Junliang L., Lili W., Xiaolong L., Xuguang L., Huanwen W., Zhiyong L. High-molecular-weight hyaluronan produced by activated pancreatic stellate cells promotes pancreatic cancer cell migration via paracrine signaling. Biochem. Biophys. Res. Commun. 2019;515:493–498. doi: 10.1016/j.bbrc.2019.05.167. [DOI] [PubMed] [Google Scholar]
- 82.Gebauer F., Kemper M., Sauter G., Prehm P., Schumacher U. Is hyaluronan deposition in the stroma of pancreatic ductal adenocarcinoma of prognostic significance? PLoS ONE. 2017;12:e0178703. doi: 10.1371/journal.pone.0178703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cheng X.-B., Sato N., Kohi S., Yamaguchi K. Prognostic Impact of Hyaluronan and Its Regulators in Pancreatic Ductal Adenocarcinoma. PLoS ONE. 2013;8:e80765. doi: 10.1371/journal.pone.0080765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hingorani S.R., Harris W.P., Beck J.T., Berdov B.A., Wagner S.A., Pshevlotsky E.M., Tjulandin S.A., Gladkov O.A., Holcombe R.F., Korn R., et al. Phase Ib Study of PEGylated Recombinant Human Hyaluronidase and Gemcitabine in Patients with Advanced Pancreatic Cancer. Clin. Cancer Res. 2016;22:2848–2854. doi: 10.1158/1078-0432.CCR-15-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grzesiak J.J., Bouvet M. Determination of the ligand-binding specificities of the alpha2beta1 and alpha1beta1 integrins in a novel 3-dimensional in vitro model of pancreatic cancer. Pancreas. 2007;34:220–228. doi: 10.1097/01.mpa.0000250129.64650.f6. [DOI] [PubMed] [Google Scholar]
- 86.Zhuang H., Zhou Z., Ma Z., Li Z., Liu C., Huang S., Zhang C., Hou B. Characterization of the prognostic and oncologic values of ITGB superfamily members in pancreatic cancer. J. Cell. Mol. Med. 2020;24:13481–13493. doi: 10.1111/jcmm.15990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grzesiak J.J., Ho J.C., Moossa A.R., Bouvet M. The Integrin-Extracellular Matrix Axis in Pancreatic Cancer. Pancreas. 2007;35:293–301. doi: 10.1097/mpa.0b013e31811f4526. [DOI] [PubMed] [Google Scholar]
- 88.Miyazaki K., Oyanagi J., Hoshino D., Togo S., Kumagai H., Miyagi Y. Cancer cell migration on elongate protrusions of fibroblasts in collagen matrix. Sci. Rep. 2019;9:1–15. doi: 10.1038/s41598-018-36646-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Conlon G., Murray G.I. Recent advances in understanding the roles of matrix metalloproteinases in tumour invasion and metastasis. J. Pathol. 2019;247:629–640. doi: 10.1002/path.5225. [DOI] [PubMed] [Google Scholar]
- 90.Krantz S.B., Shields M.A., Dangi-Garimella S., Cheon E.C., Barron M.R., Hwang R.F., Rao M.S., Grippo P.J., Bentrem D.J., Munshi H.G. MT1-MMP cooperates with Kras(G12D) to promote pancreatic fibrosis through increased TGF-beta signaling. Mol. Cancer Res. 2011;9:1294–1304. doi: 10.1158/1541-7786.MCR-11-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kajita M., Itoh Y., Chiba T., Mori H., Okada A., Kinoh H., Seiki M. Membrane-Type 1 Matrix Metalloproteinase Cleaves Cd44 and Promotes Cell Migration. J. Cell Biol. 2001;153:893–904. doi: 10.1083/jcb.153.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dangi-Garimella S., Krantz S.B., Barron M.R., Shields M.A., Heiferman M.J., Grippo P.J., Bentrem D.J., Munshi H.G. Three-Dimensional Collagen I Promotes Gemcitabine Resistance in Pancreatic Cancer through MT1-MMP–Mediated Expression of HMGA2. Cancer Res. 2011;71:1019–1028. doi: 10.1158/0008-5472.CAN-10-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shields M.A., Krantz S.B., Bentrem D.J., Dangi-Garimella S., Munshi H.G. Interplay between beta1-integrin and Rho signaling regulates differential scattering and motility of pancreatic cancer cells by snail and slug proteins. J. Biol. Chem. 2012;287:6218–6229. doi: 10.1074/jbc.M111.308940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bramhall S.R., Neoptolemos J.P., Stamp G.W.H., Lemoine N.R. Imbalance of expression of matrix metalloproteinases (MMPs) and tissue inhibitors of the matrix metalloproteinases (TIMPs) in human pancreatic carcinoma. J. Pathol. 1997;182:347–355. doi: 10.1002/(SICI)1096-9896(199707)182:3<347::AID-PATH848>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 95.Mamura T., Ohshio G., Mise M., Harada T., Suwa H., Okada N., Wang Z.-H., Yoshitomi S., Tanaka T., Sato H., et al. Expression of membrane-type matrix metalloproteinase-1 in human pancreatic adenocarcinomas. J. Cancer Res. Clin. Oncol. 1998;124:65–72. doi: 10.1007/s004320050137. [DOI] [PubMed] [Google Scholar]
- 96.Kakar S., Pawlik T.M., Allen P.J., Vauthey J.-N. AJCC Cancer Staging Manual. Springer International Publishing; New York, NY, USA: 2016. Exocrine Pancreas; pp. 337–347. [Google Scholar]
- 97.Stanton K.J., Sidner R.A., Miller G.A., Cummings O.W., Schmidt C., Howard T.J., Wiebke E.A. Analysis of Ki-67 antigen expression, DNA proliferative fraction, and survival in resected cancer of the pancreas. Am. J. Surg. 2003;186:486–492. doi: 10.1016/j.amjsurg.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 98.Pergolini I., Crippa S., Pagnanelli M., Belfiori G., Pucci A., Partelli S., Rubini C., Castelli P., Zamboni G., Falconi M. Prognostic impact of Ki-67 proliferative index in resectable pancreatic ductal adenocarcinoma. BJS Open. 2019;3:646–655. doi: 10.1002/bjs5.50175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim M.P., Fleming J.B., Wang H., Abbruzzese J.L., Choi W., Kopetz S., McConkey D.J., Evans D.B., Gallick G.E. ALDH Activity Selectively Defines an Enhanced Tumor-Initiating Cell Population Relative to CD133 Expression in Human Pancreatic Adenocarcinoma. PLoS ONE. 2011;6:e20636. doi: 10.1371/journal.pone.0020636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Laklai H., Miroshnikova Y.A., Pickup M.W., Collisson E.A., Kim G.E., Barrett A.S., Hill R.C., Lakins J.N., Schlaepfer D.D., Mouw J.K. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat. Med. 2016;22:497–505. doi: 10.1038/nm.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ligorio M., Sil S., Malagon-Lopez J., Nieman L.T., Misale S., Di Pilato M., Karabacak M., Kulkarni A.S., Jordan N.V., Franses J., et al. Stromal Microenvironment Shapes the Intratumoral Architecture of Pancreatic Cancer. SSRN Electron. J. 2018;178:160. doi: 10.2139/ssrn.3249464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Larson B.K., Guan M., Placencio V., Tuli R., Hendifar A.E. Stromal hyaluronan accumulation is associated with low tumor grade and nodal metastases in pancreatic ductal adenocarcinoma. Hum. Pathol. 2019;90:37–44. doi: 10.1016/j.humpath.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 103.Apte M.V., Park S., Phillips P.A., Santucci N., Goldstein D., Kumar R.K., Ramm G.A., Buchler M., Friess H., McCarroll J.A. Desmoplastic reaction in pancreatic cancer: Role of pancreatic stellate cells. Pancreas. 2004;29:179–187. doi: 10.1097/00006676-200410000-00002. [DOI] [PubMed] [Google Scholar]
- 104.Mahajan U.M., Langhoff E., Goni E., Costello E., Greenhalf W., Halloran C., Ormanns S., Kruger S., Boeck S., Ribback S., et al. Immune Cell and Stromal Signature Associated With Progression-Free Survival of Patients With Resected Pancreatic Ductal Adenocarcinoma. Gastroenterology. 2018;155:1625–1639.e2. doi: 10.1053/j.gastro.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 105.Neesse A., Bauer C.A., Öhlund D., Lauth M., Buchholz M., Michl P., Tuveson D.A., Gress T.M. Stromal biology and therapy in pancreatic cancer: Ready for clinical translation? Gut. 2019;68:159–171. doi: 10.1136/gutjnl-2018-316451. [DOI] [PubMed] [Google Scholar]
- 106.Yoon S., Li H., Quintanar L., Armstrong B., Rossi J.J. Uncovering Differently Expressed Markers and Heterogeneity on Human Pancreatic Cancer. Transl. Oncol. 2020;13:100749. doi: 10.1016/j.tranon.2020.100749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weniger M., Honselmann K.C., Liss A.S. The Extracellular Matrix and Pancreatic Cancer: A Complex Relationship. Cancers. 2018;10:316. doi: 10.3390/cancers10090316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Linder S., Castanos-Velez E., von Rosen A., Biberfeld P. Immunohistochemical expression of extracellular matrix proteins and adhesion molecules in pancreatic carcinoma. Hepatogastroenterology. 2001;48:1321–1327. [PubMed] [Google Scholar]
- 109.Chauhan V.P., Martin J.D., Liu H., Lacorre D.A., Jain S.R., Kozin S.V., Stylianopoulos T., Mousa A.S., Han X., Adstamongkonkul P., et al. Angiotensin inhibition enhances drug delivery and potentiates chemotherapy by decompressing tumour blood vessels. Nat. Commun. 2013;4:2516. doi: 10.1038/ncomms3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nieskoski M.D., Marra K., Gunn J.R., Kanick S.C., Doyley M.M., Hasan T., Pereira S.P., Trembly B.S., Pogue B.W. Separation of Solid Stress From Interstitial Fluid Pressure in Pancreas Cancer Correlates With Collagen Area Fraction. J. Biomech. Eng. 2017;139:061002–0610028. doi: 10.1115/1.4036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nieskoski M.D., Marra K., Gunn J.R., Hoopes P.J., Doyley M.M., Hasan T., Trembly B.S., Pogue B.W. Collagen complexity spatially defines microregions of total tissue pressure in pancreatic cancer. Sci. Rep. 2017;7:10093. doi: 10.1038/s41598-017-10671-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hingorani S.R., Zheng L., Bullock A.J., Seery T.E., Harris W.P., Sigal D.S., Braiteh F., Ritch P.S., Zalupski M.M., Bahary N., et al. HALO 202: Randomized Phase II Study of PEGPH20 Plus Nab-Paclitaxel/Gemcitabine Versus Nab-Paclitaxel/Gemcitabine in Patients With Untreated, Metastatic Pancreatic Ductal Adenocarcinoma. J. Clin. Oncol. 2018;36:359–366. doi: 10.1200/JCO.2017.74.9564. [DOI] [PubMed] [Google Scholar]
- 113.Van Cutsem E., Tempero M.A., Sigal D., Oh D.Y., Fazio N., Macarulla T., Hitre E., Hammel P., Hendifar A.E., Bates S.E., et al. Randomized phase III trial of pegvorhyaluronidase alfa with nab-paclitaxel plus gemcitabine for patients with hyaluronan-high metastatic pancreatic adenocarcinoma. J. Clin. Oncol. 2020;38:3185–3194. doi: 10.1200/JCO.20.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Drifka C.R., Loeffler A.G., Esquibel C.R., Weber S.M., Eliceiri K.W., Kao W.J. Human pancreatic stellate cells modulate 3D collagen alignment to promote the migration of pancreatic ductal adenocarcinoma cells. Biomed. Microdevices. 2016;18:105. doi: 10.1007/s10544-016-0128-1. [DOI] [PubMed] [Google Scholar]
- 115.Erdogan B., Ao M., White L.M., Means A.L., Brewer B.M., Yang L., Washington M.K., Shi C., Franco O.E., Weaver A.M., et al. Cancer-associated fibroblasts promote directional cancer cell migration by aligning fibronectin. J. Cell Biol. 2017;216:3799–3816. doi: 10.1083/jcb.201704053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jang I., Beningo K. Integrins, CAFs and Mechanical Forces in the Progression of Cancer. Cancers. 2019;11:721. doi: 10.3390/cancers11050721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jokinen J., Dadu E., Nykvist P., Käpylä J., White D.J., Ivaska J., Vehviläinen P., Reunanen H., Larjava H., Häkkinen L., et al. Integrin-mediated Cell Adhesion to Type I Collagen Fibrils. J. Biol. Chem. 2004;279:31956–31963. doi: 10.1074/jbc.M401409200. [DOI] [PubMed] [Google Scholar]
- 118.Sakamoto T., Seiki M. Integrated functions of membrane-type 1 matrix metalloproteinase in regulating cancer malignancy: Beyond a proteinase. Cancer Sci. 2017;108:1095–1100. doi: 10.1111/cas.13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schlitter A.M., Segler A., Steiger K., Michalski C.W., Jager C., Konukiewitz B., Pfarr N., Endris V., Bettstetter M., Kong B., et al. Molecular, morphological and survival analysis of 177 resected pancreatic ductal adenocarcinomas (PDACs): Identification of prognostic subtypes. Sci. Rep. 2017;7:41064. doi: 10.1038/srep41064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chan-Seng-Yue M., Kim J.C., Wilson G.W., Ng K., Figueroa E.F., O’Kane G.M., Connor A.A., Denroche R.E., Grant R.C., McLeod J., et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat. Genet. 2020;52:231–240. doi: 10.1038/s41588-019-0566-9. [DOI] [PubMed] [Google Scholar]
- 121.Juiz N., Elkaoutari A., Bigonnet M., Gayet O., Roques J., Nicolle R., Iovanna J., Dusetti N. Basal-like and classical cells coexist in pancreatic cancer revealed by single-cell analysis on biopsy-derived pancreatic cancer organoids from the classical subtype. FASEB J. 2020;34:12214–12228. doi: 10.1096/fj.202000363RR. [DOI] [PubMed] [Google Scholar]
- 122.Le Large T.Y., Mantini G., Meijer L.L., Pham T.V., Funel N., van Grieken N.C., Kok B., Knol J., van Laarhoven H.W., Piersma S.R., et al. Microdissected pancreatic cancer proteomes reveal tumor heterogeneity and therapeutic targets. JCI Insight. 2020;5:5. doi: 10.1172/jci.insight.138290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ottaviano A.J., Sun L., Ananthanarayanan V., Munshi H.G. Extracellular matrix-mediated membrane-type 1 matrix metalloproteinase expression in pancreatic ductal cells is regulated by transforming growth factor-beta1. Cancer Res. 2006;66:7032–7040. doi: 10.1158/0008-5472.CAN-05-4421. [DOI] [PubMed] [Google Scholar]
- 124.Marusyk A., Janiszewska M., Polyak K. Intratumor Heterogeneity: The Rosetta Stone of Therapy Resistance. Cancer Cell. 2020;37:471–484. doi: 10.1016/j.ccell.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Juiz N.A., Iovanna J., Dusetti N. Pancreatic Cancer Heterogeneity Can Be Explained Beyond the Genome. Front. Oncol. 2019;9:246. doi: 10.3389/fonc.2019.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schmauch B., Romagnoni A., Pronier E., Saillard C., Maillé P., Calderaro J., Kamoun A., Sefta M., Toldo S., Zaslavskiy M., et al. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat. Commun. 2020;11:1–15. doi: 10.1038/s41467-020-17678-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nicolle R., Blum Y., Duconseil P., Vanbrugghe C., Brandone N., Poizat F., Roques J., Bigonnet M., Gayet O., Rubis M., et al. Establishment of a pancreatic adenocarcinoma molecular gradient (PAMG) that predicts the clinical outcome of pancreatic cancer. EBioMedicine. 2020;57:102858. doi: 10.1016/j.ebiom.2020.102858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nicolle R., Blum Y., Marisa L., Loncle C., Gayet O., Moutardier V., Turrini O., Giovannini M., Bian B., Bigonnet M., et al. Pancreatic Adenocarcinoma Therapeutic Targets Revealed by Tumor-Stroma Cross-Talk Analyses in Patient-Derived Xenografts. Cell Rep. 2017;21:2458–2470. doi: 10.1016/j.celrep.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Beck A.H., Sangoi A.R., Leung S., Marinelli R.J., Nielsen T.O., Van De Vijver M.J., West R.B., Van De Rijn M., Koller D. Systematic Analysis of Breast Cancer Morphology Uncovers Stromal Features Associated with Survival. Sci. Transl. Med. 2011;3:108ra113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 130.Amrutkar M., Vethe N.T., Verbeke C.S., Aasrum M., Finstadsveen A.V., Sántha P., Gladhaug I.P. Differential Gemcitabine Sensitivity in Primary Human Pancreatic Cancer Cells and Paired Stellate Cells Is Driven by Heterogenous Drug Uptake and Processing. Cancers. 2020;12:3628. doi: 10.3390/cancers12123628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lattouf R., Younes R., Lutomski D., Naaman N., Godeau G., Senni K., Changotade S. Picrosirius red staining: A useful tool to appraise collagen networks in normal and pathological tissues. J. Histochem. Cytochem. 2014;62:751–758. doi: 10.1369/0022155414545787. [DOI] [PubMed] [Google Scholar]
- 132.Jiang P., Li X., Thompson C.B., Huang Z., Araiza F., Osgood R., Wei G., Feldmann M., Frost G.I., Shepard H.M. Effective targeting of the tumor microenvironment for cancer therapy. Anticancer Res. 2012;32:1203–1212. [PubMed] [Google Scholar]
- 133.Haeberle L., Steiger K., Schlitter A.M., Safi S.A., Knoefel W.T., Erkan M., Esposito I. Stromal heterogeneity in pancreatic cancer and chronic pancreatitis. Pancreatology. 2018;18:536–549. doi: 10.1016/j.pan.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 134.Wegner C.S., Gaustad J.-V., Andersen L.M.K., Simonsen T.G., Rofstad E.K. Diffusion-weighted and dynamic contrast-enhanced MRI of pancreatic adenocarcinoma xenografts: Associations with tumor differentiation and collagen content. J. Transl. Med. 2016;14:1–11. doi: 10.1186/s12967-016-0920-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Andrlova H., Mastroianni J., Madl J., Kern J.S., Melchinger W., Dierbach H., Wernet F., Follo M., Technau-Hafsi K., Has C., et al. Biglycan expression in the melanoma microenvironment promotes invasiveness via increased tissue stiffness inducing integrin-beta1 expression. Oncotarget. 2017;8:42901–42916. doi: 10.18632/oncotarget.17160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fonck E., Feigl G.G., Fasel J., Sage D., Unser M., Stergiopulos N., Rufenacht D.A. Effect of Aging on Elastin Functionality in Human Cerebral Arteries. Stroke. 2009;40:2552–2556. doi: 10.1161/STROKEAHA.108.528091. [DOI] [PubMed] [Google Scholar]
- 137.Fruchterman T.M.J., Reingold E.M. Graph drawing by force-directed placement. Softw. Pr. Exp. 1991;21:1129–1164. doi: 10.1002/spe.4380211102. [DOI] [Google Scholar]
- 138.Cherven K. Mastering Gephi Network Visualization. Packt Publishing; Birmingham, UK: 2015. [Google Scholar]
- 139.Bastian M., Heymann S., Jacomy M. Gephi: An open source software for exploring and manipulating networks; Proceedings of the International AAAI Conference on Web and Social Media; San Jose, CA, USA. 17–20 May 2009; No. 1. [Google Scholar]
- 140.Altman N.S. An introduction to kernel and nearest-neighbor nonparametric regression. Am. Stat. 1992;46:175–185. [Google Scholar]
- 141.Geurts P., Ernst D., Wehenkel L. Extremely randomized trees. Mach. Learn. 2006;63:3–42. doi: 10.1007/s10994-006-6226-1. [DOI] [Google Scholar]
- 142.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Sciait-learn: Machine Learning in Python. JMLR. 2011;12:2825–2830. [Google Scholar]
- 143.Brierley J.D., Gospodarowicz M.K., Wittekind C., editors. TNM Classification of Malignant Tumours. 8th ed. Wiley Blackwell; Oxford, UK: 2017. pp. 93–95. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethical considerations and data regulations.