To the Editor:
We write in reference to our recent article (1), which describes an early bactericidal activity (EBA) study investigating the optimal dose of isoniazid (INH) for patients with INH-resistant tuberculosis mediated by inhA mutations. Here, we reexamine the measures of sputum bacterial load used with regard to the presence of drug resistance–conferring mutations.
The parent study, AIDS Clinical Trials Group A5312, is an ongoing phase 2A, open-label trial in which individuals with smear-positive pulmonary tuberculosis with INH resistance mediated by inhA mutation were randomized to receive INH at 5, 10, or 15 mg/kg daily for 7 days. Control subjects with no mycobacterial INH resistance received INH at the standard dose of 5 mg/kg daily. Overnight sputum samples were collected daily to assess the fall in log10 colony-forming units (CFUs) and the increase in time to positivity (TTP) for mycobacterial culture. The study’s main outcome, which remains unchallenged by this post hoc analysis, is that INH given at doses of 10–15 and 5 mg/kg to participants with and without inhA-mediated mycobacterial INH resistance, respectively, provide similar 7-day EBA.
Our interest was sparked by the observation that baseline sputum bacillary loads, measured by CFU counts on solid culture medium, were lower in control subjects (median, 5.41 log10CFU) than in participants with mycobacterial inhA mutations (median, 5.65–7.04 log10CFU). However, the baseline TTP for mycobacteria in liquid cultures was shorter overall in control subjects (median, 97 h) than in participants with mycobacterial inhA mutations (median, 124–142 h). This is noteworthy because lower CFU counts would be expected to translate into a higher TTP rather than into the lower TTP observed.
To test whether microbial fitness cost conferred by mycobacterial mutations could explain this incongruence, we reclassified the baseline isolates of participants for the presence of bacterial mutations. Of 59 participants enrolled, 43 participants (73%) had mycobacteria with inhA mutations at least. Thirty-one participants (52%) had mycobacteria with additional mutations conferring rifampicin resistance, three participants (5%) had mycobacteria with mutations conferring fluoroquinolone resistance, and one participant (2%) had mycobacteria with mutations conferring aminoglycoside resistance. Of the 16 control subjects, only 13 subjects had mycobacteria with no detectable resistance, whereas three had mycobacteria with rifampicin monoresistance and were thus reclassified into the mutation group. This resulted in 13 participants (22%) without mycobacterial mutations contributing 168 TTP and CFU results and in 46 participants (88%) with at least one resistance-conferring mycobacterial mutation contributing 643 data points.
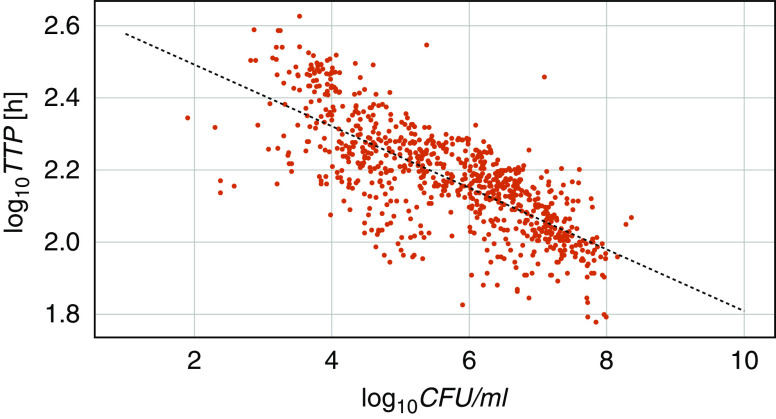
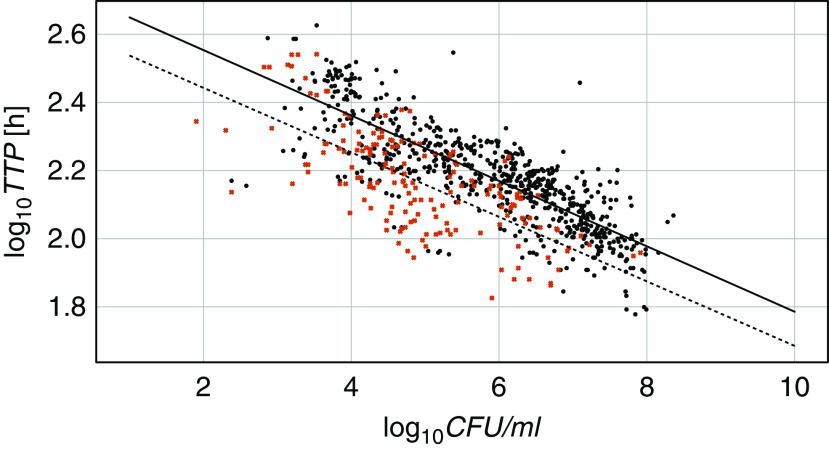
Figure 1 shows all available CFUs and TTPs plotted against each other. A linear ordinary least-squares regression model with log10TTP as a dependent variable, log10CFU as an independent variable, and an intercept term fitted the data well (r2 = 0.555; P < 0.0001 for slope and intercept). The introduction of a mutation variable separated the mutated and nonmutated samples in two almost parallel lines (Figure 2) and improved the correlation (r2 = 0.631). The difference in intercepts was significant (P = 0.002), but there was no significant difference in the steepness of the slopes (P = 0.852). This means that the presence of bacterial mutations significantly prolongs the expected TTP for a given log10CFU by about 50 hours, independent of the CFU count.
Figure 1.
Daily log10 colony-forming units (log10CFUs) and log10 time to positivity (log10TTP) for mycobacteria over 7 days for all 59 participants. The relationship between TTP and CFUs is determined using an ordinary least-squares regression model: TTP = b0 + b1 × CFUs. The resulting equation is log10TTP = 2.663 – 0.0854 × log10CFU; r2 = 0.555; P < 0.0001 for b0 and b1.
Figure 2.
Daily log10 colony-forming units (log10CFU) and log10 time to positivity (log10TTP) for mycobacteria over 7 days for all 59 participants with mutated (black dots) and nonmutated (red crosses) bacteria. The model includes a mutation variable (0, dashed line, or 1, full line): TTP = b0 + b1 × CFUs + b2 × mutations + b3 × CFUs × mutations. This formula resulted in log10TTP = 2.6324 – 0.0947 × log10CFUs + 0.1133 × mutations – 0.0013 × log10CFUs × mutations; r2 = 0.631; P < 0.0001 for b0, b1; P = 0.002 for b2; P = 0.852 for b3.
Microbial fitness cost due to mutations is a well-known concept (2). Bacterial resistance-conferring mutations are generally costly, but compensatory mutations can alleviate some of the fitness costs of resistance (3). An increased generation time has been described in tuberculosis bacilli with a rpoB mutation under stress (4). Time to culture positivity for bacteria is vulnerable to changes in metabolic activity because its readout is dependent on consumption of nutrients, which in turn depends on both the activity and number of bacteria present. Counting of CFUs, however, only depends on numbers and is thus not as affected. Colonies growing more slowly will be smaller but will still contribute to enumeration.
This finding will not be of much consequence if only differences of the same endpoint marker between time points are measured, such as in EBA studies. The parallel lines in Figure 2 illustrate that a dropping mycobacterial load will change at the same rate if mutated or nonmutated bacteria are measured. A potential pitfall comes in when comparison to an absolute value is made, such as in studies that measure the rate of or time to conversion of culture results from positive to negative.
Consider a clinical study with an all-novel regimen composed of agents with new mechanisms of action that are not susceptible to mutations conferring resistance to conventional agents. This new regimen would be tested in a patient population with disease caused by bacteria resistant to conventional agents. The control group would be on the conventional regimen and would consist only of subjects with mutation-free bacterial infection. The endpoint of having conversion from positive to negative liquid culture results would require the TTP for mycobacteria to increase beyond 42 days for negative results to be declared. According to our findings, if both treatments are equally efficacious, culture conversion is likely to occur sooner in the all-novel treatment group because not only the treatment but also the mutation-associated fitness cost is slowing down the cultures. Mutation-free control cultures would grow faster, and conversion to negative results using the reference treatment would be harder. Faster culture conversion by the all-novel regimen would lead to the conclusion that it is superior to the conventional treatment, even in patients who may be described as “hard to treat.”
Our observations are biologically plausible and based on data collected in a prospective clinical trial, but this post hoc analysis has inherent weaknesses. Mutations were detected by line probes only, thus not allowing quantification of proportions of mutated bacilli and varying degrees of fitness cost or compensatory effects. Larger cross-sectional studies including whole-genome sequencing to detect a broader diversity of strains, mutations, and subpopulations is needed to investigate this further.
Until it is clear how mutation-related fitness cost influences culture-based endpoints, it seems prudent to not use liquid culture results as the only or even the primary endpoint in studies involving participant groups with disease caused by bacteria with different degrees of resistance. Comparisons between groups with disease due to mutated and nonmutated bacilli in clinical trials should be made with caution. Solutions to the problem might come from nucleotide-based alternatives, such as bacterial sputum DNA (5), transrenal DNA (6), or RNA (7, 8); from measurements of bacterial components such as sputum lipoarabinomannan (9); or from genetic epistasis analysis (10). Future clinical trials should test these readouts against each other to better understand their significance.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank the study participants and their families. They thank the INHindsight Study Team (A5312 Study Team) members who contributed to the trial.
A5312 Study Team members: Sachiko Miyahara, Florian von Groote-Bidlingmaier, Xin Sun, Richard Hafner, Susan L. Rosenkranz, Elisa H. Ignatius, Eric L. Nuermberger, Laura Moran, Kathleen Donahue, Susan Swindells, Rachel Issa, Christopher Lane, Mark Lojacono, Rachel Mahachi, William Murtaugh, Lynette Purdue, Akbar Shahkolahi, and Ronald Ssenyonga.
Footnotes
Supported by the Division of AIDS, NIH (TASK Clinical Research Site grant UM1 AI069521). Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the NIH under award numbers UM1 AI068634, UM1 AI068636, and UM1 AI106701. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author Contributions: All authors designed the study. S.P.l.R. conducted the analysis and drafted the manuscript. All authors provided input and reviewed and approved the final manuscript.
Originally Published in Press as DOI: 10.1164/rccm.202007-2740LE on October 20, 2020
Author disclosures are available with the text of this letter at www.atsjournals.org.
Contributor Information
Collaborators: for the A5312 Study Team, Sachiko Miyahara, Florian von Groote-Bidlingmaier, Xin Sun, Richard Hafner, Susan L. Rosenkranz, Elisa H. Ignatius, Eric L. Nuermberger, Laura Moran, Kathleen Donahue, Susan Swindells, Rachel Issa, Christopher Lane, Mark Lojacono, Rachel Mahachi, William Murtaugh, Lynette Purdue, Akbar Shahkolahi, and Ronald Ssenyonga
References
- 1.Dooley KE, Miyahara S, von Groote-Bidlingmaier F, Sun X, Hafner R, Rosenkranz SL, et al. A5312 Study Team. Early bactericidal activity of different isoniazid doses for drug-resistant TB (INHindsight): a randomized open-label clinical trial. Am J Respir Crit Care Med. 2020;201:1416–1424. doi: 10.1164/rccm.201910-1960OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Sullivan DM, McHugh TD, Gillespie SH. Mapping the fitness of Mycobacterium tuberculosis strains: a complex picture. J Med Microbiol. 2010;59:1533–1535. doi: 10.1099/jmm.0.019091-0. [DOI] [PubMed] [Google Scholar]
- 3.Melnyk AH, Wong A, Kassen R. The fitness costs of antibiotic resistance mutations. Evol Appl. 2015;8:273–283. doi: 10.1111/eva.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rifat D, Campodónico VL, Tao J, Miller JA, Alp A, Yao Y, et al. In vitro and in vivo fitness costs associated with Mycobacterium tuberculosis RpoB mutation H526D. Future Microbiol. 2017;12:753–765. doi: 10.2217/fmb-2017-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedrich SO, Rachow A, Saathoff E, Singh K, Mangu CD, Dawson R, et al. Pan African Consortium for the Evaluation of Anti-tuberculosis Antibiotics (PanACEA) Assessment of the sensitivity and specificity of Xpert MTB/RIF assay as an early sputum biomarker of response to tuberculosis treatment. Lancet Respir Med. 2013;1:462–470. doi: 10.1016/S2213-2600(13)70119-X. [DOI] [PubMed] [Google Scholar]
- 6.Patel K, Nagel M, Wesolowski M, Dees S, Rivera-Milla E, Geldmacher C, et al. Evaluation of a urine-based rapid molecular diagnostic test with potential to be used at point-of-care for pulmonary tuberculosis: Cape Town cohort. J Mol Diagn. 2018;20:215–224. doi: 10.1016/j.jmoldx.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Svensson RJ, Sabiiti W, Kibiki GS, Ntinginya NE, Bhatt N, Davies G, et al. Model-based relationship between the molecular bacterial load assay and time to positivity in liquid culture. Antimicrob Agents Chemother. 2019;63:e00652-19. doi: 10.1128/AAC.00652-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter ND, Moore CM, Kayigire XA, Dide-Agossou C, Worodria W, Huang L, et al. Does discovery of differentially culturable M tuberculosis really demand a new treatment paradigm? Longitudinal analysis of DNA clearance from sputum. BMC Infect Dis. 2018;18:293. doi: 10.1186/s12879-018-3213-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawasaki M, Echiverri C, Raymond L, Cadena E, Reside E, Gler MT, et al. Lipoarabinomannan in sputum to detect bacterial load and treatment response in patients with pulmonary tuberculosis: analytic validation and evaluation in two cohorts. PLoS Med. 2019;16:e1002780. doi: 10.1371/journal.pmed.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kavvas ES, Catoiu E, Mih N, Yurkovich JT, Seif Y, Dillon N, et al. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat Commun. 2018;9:4306. doi: 10.1038/s41467-018-06634-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.