Abstract
Human paraoxonase-1 (PON1) is a high-density lipoprotein-associated enzyme, with antioxidant, anti-inflammatory, and anti-apoptotic roles. The ability of PON1 to hydrolyze specific organophosphate (OP) compounds and prevent accumulation of oxidized lipids in lipoproteins, has prompted a large number of studies investigating PON1’s role in modulating toxicity and disease. Most of these studies, however, have only focused on PON1 single nucleotide polymorphisms (SNPs) analyses and have ignored PON1 activity levels, arguably the most important parameter in determining protection against exposure and disease. We developed a 2-substrate activity assay termed “PON1 status” that reveals both the functional PON1192 genotype and plasma PON1 activity levels. While our previous studies with PON1 status demonstrated that both PON1192 functional genotype and enzymatic activity levels obtained exclusively by determining PON1 status are required for a proper evaluation of PON1’s role in modulating OP exposures and risk of disease, the original PON1 status assay requires the use of highly toxic OP metabolites. As many laboratories are not prepared to handle such toxic compounds and the associated waste generated, determination of PON1 status has been limited to rather few studies. Here, we describe a PON1 status protocol that utilizes non-OP substrates, with a resolution equivalent to that of the original PON1 status approach. We have also included useful suggestions to ensure the assays can easily be carried out in any laboratory. The protocols described here will enable a proper examination of the risk of exposure or susceptibility to disease in PON1 epidemiological studies without the need to handle highly toxic substrates.
Keywords: Paraoxonase-1, PON1 status, PON1 activity assay, non-organophosphate substrates
INTRODUCTION
Human paraoxonase-1 (PON1) is a potent antioxidant, anti-inflammatory, and anti-apoptotic enzyme synthesized in the liver, and is found in circulation tightly bound to high-density lipoproteins (HDLs) (Blatter et al., 1993; Camps et al., 2009a; Furlong et al., 2010, 2016; Hassett et al., 1991). Although there is no evidence of PON1 protein expression in tissues other than liver, the presence of PON1 in multiple human and mouse tissues has been documented (Marsillach et al., 2008, 2011). PON1 is a calcium-dependent lactonase that hydrolyzes a wide range of substrates, from lipo-lactones to certain organophosphorus (OP) metabolites, aromatic carboxylic esters, and quorum sensing factors (Draganov et al., 2005; Khersonsky & Tawfik, 2005; Ozer et al., 2005). While PON1 was first studied for its OP hydrolyzing activities, subsequent reports of PON1’s ability to prevent lipid oxidation of lipoproteins (Aviram et al., 1998; Mackness et al., 1991), as well as to decrease levels of oxidative stress, resulted in multiple studies relating PON1 to prevention of oxidative stress-related diseases, such as cardiovascular disease, diabetes, chronic liver failure, HIV-infection, autism, and Alzheimer’s disease, among others (Besler et al., 2011; Cervellati et al., 2019; Ferré et al., 2013; Gaita et al., 2010; Jornayvaz et al., 2009; Marsillach et al., 2007; Parra et al., 2007).
Polymorphisms in the 5’ and 3’ untranslated regions and in the coding region of human PON1 have been described (Adkins et al., 1993; Brophy et al., 2001a, 2001b; Eckerson et al., 1983; Hassett et al., 1991; Jarvik et al., 2003a; Leviev & James, 2000; Suehiro et al., 2000; Seattle SNPs http://pga.gs.washington.edu/data/pon1/), with the coding region PON1Q192R polymorphism being the most extensively studied due to its substrate-dependent effect on the catalytic efficiency of hydrolysis (Adkins et al., 1993; Costa et al., 2003; Davies et al., 1996; Humbert et al., 1993; Li et al., 2000; Liu et al., 2013). While the PON1Q192R polymorphism does not affect the catalytic efficiency for hydrolysis of the PON1 substrates phenyl acetate and diazoxon, it does affect the catalytic efficiency for the OP metabolites paraoxon and chlorpyrifos oxon (Li et al., 2000). Importantly, animal studies have indicated that low catalytic efficiency for certain OPs is associated with sensitivity to OP exposures and development of neurotoxicity (Cole et al., 2005). Confirmatory findings were reported in our study with a cohort of agricultural pesticide handlers from Washington State (Hofmann et al., 2009). In that study, individuals homozygous for the PON1192Q alloform (lower catalytic efficiency of paraoxon and chlorpyrifos oxon hydrolysis) and with low PON1 activity (expression) had significantly higher levels of plasma butyrylcholinesterase inhibition, a biomarker of OP exposure.
There have several publications examining the relationship of human PON1 polymorphisms and the risk to exposure and susceptibility to disease development, most with conflicting results (Furlong et al., 2010). Even though some polymorphisms have an effect on PON1 activity levels and/or concentration (reviewed in Furlong et al. 2008), there are multiple factors that can affect PON1 activity and concentration, such as age, nutritional and lifestyle habits, and certain drugs (Camps et al., 2009b; Costa et al., 2005, 2011). Thus, in epidemiological studies, it is necessary to take into consideration not only the PON1 gene single nucleotide polymorphisms (SNPs), but also the actual level and functionality of the plasma PON1 protein (Costa et al., 2013; Furlong et al., 2010; Jarvik et al., 2000, 2003b; Mackness et al., 2001).
Our laboratory developed a two-substrate activity assay, termed PON1 status, that determines both the functional PON1192 genotype and the plasma PON1 activity levels simultaneously (Furlong et al., 2006; Richter & Furlong, 1999; Richter et al., 2004, 2008, 2009). PON1 status determination involves two specific PON1 kinetic measurements per sample, and the substrates originally used for these kinetic measurements include the OP compounds diazoxon and paraoxon (Costa et al., 1999; Richter & Furlong, 1999; Richter et al., 2009). The PON1 enzymatic values of all test samples are then plotted in a scatter (x/y) plot, resulting in the generation of three clusters, which represent each of the PON1192 phenotypes (QQ, QR, and RR). Without the need for PON1 genotyping (SNP analysis), PON1 status can reveal both the functional position 192 genotype and the enzymatic activity levels in plasma/serum of each individual analyzed. The PON1 status assay can then resolve all samples into one of the three PON1192 phenotypes or “functional genotypes” (QQ, QR, and RR), and provides the crucially important information about each individual’s PON1 activity values. The assay is said to reveal functional genotypes because it provides both the PON1192 phenotypes and the PON1 activity levels. If only SNP analyses were carried out, there would be no information on PON1 activity levels, which, in most cases, is the most important parameter.
While the method is robust and reproducible, a main drawback of the PON1 status assay is the use of highly toxic substrates (diazoxon and paraoxon), and this might have been a major limiting factor for many laboratories interested in epidemiological studies involving PON1 status for risk of exposure or disease. For this reason, we developed a version of this PON1 status analysis that does not use highly toxic substrates (Richter et al., 2008, 2009). The lack of a detailed protocol to conduct this non-OP version of PON1 status analysis, however, together with the steady increase in publications using only SNP analyses, have hindered the use of both OP and non-OP PON1 status in many epidemiological studies.
In this article, we update our previous detailed PON1 status analyses and genotyping protocol (Richter et al. 2004) to provide a detailed description for determining a person’s PON1 status without the use of highly toxic OP substrates. Basic Protocol 1 describes the enzymatic assays and data analyses required to obtain an individual’s PON1 status without using toxic OP compounds. Support Protocol 1 provides a step-by-step description of how to experimentally determine the pathlength of each sample for when the user’s microplate spectrophotometer software lacks the capability of measuring this automatically. Support Protocol 2 describes a procedure for PON1Q192R genotyping (SNP analyses) for investigators interested in conducting both DNA PON1Q192R SNP analysis and plasma/serum PON1 status.
Given the importance of obtaining functional data for PON1, we hope these protocols will help researchers implement PON1 status instead of performing PON1 SNP analysis alone in epidemiological studies related to PON1 and risk of exposure or susceptibility to disease.
STRATEGIC PLANNING
Prior to blood sample collection for PON1 status assay, it is important to consider which blood specimen collection tube to use (plasma or serum) and use the same collection tubes throughout your study, for all samples. This is particularly important in retrospective studies. PON1 activities from plasma and serum samples from previous studies may not be directly compared.
Type of blood samples to use for PON1 status.
PON1 status assay can be carried out using either plasma, collected in lithium heparin (green-top) tubes, or serum, collected in red-top tubes (no anticoagulant). Blood intended to be used in this assay should not be collected in ethylenediaminetetraacetic acid (EDTA) tubes, as PON1 is a calcium-dependent enzyme and EDTA is a calcium chelator, resulting in irreversible inhibition of PON1 activity (Erdos et al., 1959). Hemolysis, jaundice, and lipemia in plasma/serum samples do not affect the non-OP PON1 status assay described (Richter RJ., personal communication).
Control plasma/serum sample
It is important to control for plate-to-plate variation using a well-characterized plasma/serum sample, which should be included in each 96-well microplate in triplicate and treated as a test sample. Plasma/serum control samples can be purchased commercially or prepared in the laboratory, but they should be available and ready-to-use prior to carrying out the described non-OP PON1 status assay. See the Troubleshooting section for more information.
Type of blood samples to use for PON1 DNA genotyping.
PON1 genotyping requires isolation of DNA, which can be isolated from blood collected in lithium heparin (green-top) or EDTA (purple-top) tubes.
Use of temperature-controlled kinetics.
Temperature can increase or decrease the rates of enzymatic activity. For this reason, even if the described non-OP PON1 status assay is performed at room temperature, prior to proceeding with the assays described below, we recommend setting the microplate spectrophotometer temperature controller to 25°C to minimize the potential effects that changes in the ambient temperature of the laboratory could have on the enzymatic measurements (high-salt arylesterase and CMPAase). We also recommend placing buffers in a 25°C water bath.
BASIC PROTOCOL 1: Determining PON1 status using non-organophosphate substrates
Here, we describe in detail a protocol for PON1 status analysis that does not use highly toxic OPs. It is described in sufficient detail to allow laboratories to easily adopt it for future research studies. Although the substrates used for the described PON1 status are far less toxic than the highly toxic OP compounds previously used to analyze PON1 status (Costa et al., 1999; Richter & Furlong, 1999; Richter et al., 2009), please follow the personal protective equipment recommendations detailed in the MSDS of each substrate used.
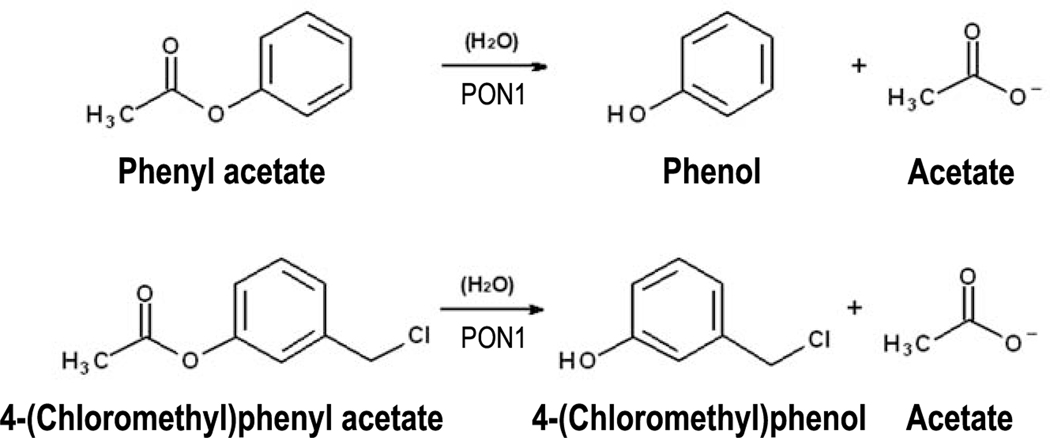
The non-OP determination of PON1 status involves two kinetic measurements of PON1, using the substrates phenyl acetate (arylesterase activity at high-salt conditions) and 4-(chloromethyl)phenyl acetate (CMPAase activity) (Figures 1 and 2). The addition of salt (2 M NaCl) in the assay buffer of PON1 high-salt arylesterase activity assay (see Reagents and Solutions section) greatly improves the resolution of the three PON1192 phenotypes (Richter et al., 2008). Both assays are conducted using an absorbance microplate reader, with controlled temperature (25°C), and with the enzymatic reactions continuously monitored at 270 nm for 4 min. The activity rates from the microplate reader (mOD/min) are then converted to enzymatic activity units (U/mL or μmol/min/mL) using the Beer-Lambert law. The conversion of activity rates (mOD/min) to enzymatic activity units (U/mL) is necessary to enable comparison of the results with other reports using the same enzymatic assay. The obtained enzymatic units are then plotted on a scatter (x/y) plot, with rates of high-salt arylesterase hydrolysis plotted on the y-axis and rates of CPMAase hydrolysis plotted on the x-axis, for each individual. Once plotted, the data will distribute into three clearly resolved groups, representing the three phenotypes of PON1192 (PON1192QQ, PON1192QR and PON1192RR). By following this assay, researchers will obtain plasma/serum PON1 activity values for individuals within each PON1192 functional genotype group.
Figure 1.
PON1-mediated hydrolysis of the substrates used for the non-OP PON1 status assay, phenyl acetate (high-salt arylesterase activity) and 4-(chloromethyl)phenyl acetate (CMPAase activity).
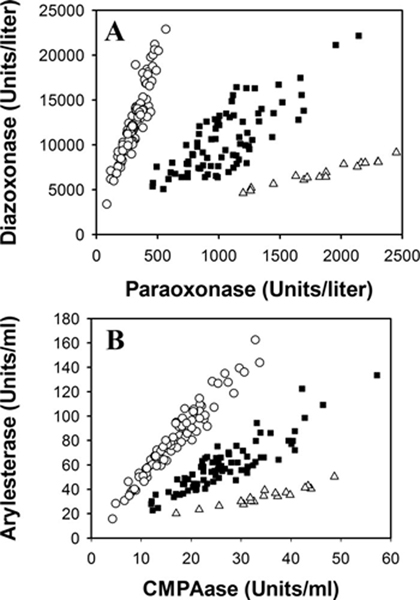
Figure 2. Comparison of the (A) toxic OP and (B) non-OP PON1 status assays using the same population (n = 183).
Samples were run in triplicate, and each data point corresponds to the average value of the pathlength-corrected triplicate. Activity results are displayed in enzymatic units (U/L for panel 2A and U/mL for panel 2B). The PON1 status carried out in panel A corresponds to the original PON1 status that uses toxic OP substrates (paraoxon and diazoxon) (Furlong et al., 2006; Richter & Furlong, 1999; Richter et al., 2004, 2008, 2009). Panel B displays enzymatic activity results using the non-OP PON1 status described here (Richter et al. 2008 and 2009). In both panels, the open circle data points correspond to individuals homozygous for PON1192Q (QQ), the solid square data points correspond to individuals heterozygous for PON1192QR (QR), and the open triangle data points correspond to individuals homozygous for PON1192R (RR). Reproduced with permission from Richter et al., 2008.
Prior to proceeding with the steps below, set the temperature control function of the microplate spectrophotometer to 25°C. All buffers should also be maintained at 25°C.
Materials–
Samples: blood samples (plasma or serum).
Dilution buffer (see Reagents and Solutions)
Phenyl acetate (#108723, Sigma)
4-(Chloromethyl) phenyl acetate, CMPA (#432881, Sigma)
Arylesterase (high salt) assay buffer (see Reagents and Solutions)
CMPAase assay buffer (see Reagents and Solutions)
Equipment–
Refrigerated microcentrifuge (or tabletop microcentrifuge placed in a cold room)
Water bath
UV-transparent 96-well microplates
0.5 mL and 1.5 mL Eppendorf tubes or similar
50 mL screw-cap polypropylene tubes
UV/visible microplate spectrophotometer (e.g. SpectraMax Plus Microplate Spectrophotometer, Molecular Devices), pre-set at 25°C and ideally with plate shaking capabilities
Microplate spectrophotometer acquisition and analysis software (e.g. SoftMax Pro data acquisition and analysis software, version 5.4, if using a Molecular Devices microplate reader).
Protocol Steps
Arylesterase activity (high salt) measurement
-
1
Collect blood using at least 4 mL green-top or red-top blood collection tubes (see Strategic Planning). If blood is collected using red-top tubes (no anticoagulant), allow the blood to clot by leaving it undisturbed at room temperature for about 30 min. This step is not needed if blood was collected with green-top tubes as they contain anticoagulant (lithium heparin).
A total of 5 μL of plasma/serum are needed for PON1 status, but we recommend having at least 10 μL available in case the assay needs to be repeated. From a 4 mL blood tube, users can expect to obtain about 1.5 mL of plasma/serum.
-
2
Centrifuge the blood-containing tubes for 10 min at 2,000 x g, 4°C.After centrifugation, cells (in plasma) or the clot (in serum) will remain at the bottom layer, and plasma/serum (yellow) will be on the top layer.
-
3
Immediately transfer the plasma/serum (top layer) using a Pasteur-type pipette into 0.5 mL polypropylene tubes and keep them frozen at −20°C or −80°C until the researcher is ready to carry out the PON1 status assay. If samples will be used fresh, proceed to Step 5.
In general, it is important to store plasma/serum samples in small aliquots to avoid freeze-thaw cycles because this is detrimental to many serum components. However, freeze-thaw cycles do not seem to affect PON1 activity (Richter RJ., personal communication).
-
4
If plasma/serum samples are frozen, thaw the frozen tubes by placing them at room temperature (18°C – 25°C).
Thawing time can be decreased by gently inverting the tubes a few times or flicking them if the volume is low. This will prevent formation of cryoprecipitates.
-
5
Mix plasma/serum samples by inverting the tubes gently, and then centrifuge plasma/serum samples for 5 min at 10,000 x g, at 4°C.
This step is necessary to avoid pipetting any particulate matter present in the sample.
-
6
Take 2 μL of each plasma/serum sample and mix with 78 μL of dilution buffer (1:40 dilution) in separate tube. Mix well.
-
7
Transfer 20 μL of each diluted plasma/serum sample into a well of a UV-transparent 96-well plate. Do this in triplicate (i.e. do this three times, in three separate wells, per sample).
-
8
In each plate, transfer 20 μL of dilution buffer into an empty well, to be used as blank.
-
9
Prepare the substrate by adding 25 μL of phenyl acetate to 25 mL of arylesterase assay buffer in a 50 mL polypropylene tube (substrate concentration in the final working assay solution will be 6.52 mM). Screw the cap tightly and shake vigorously for 1 min.
Phenyl acetate is an oily solution. For this reason, vigorous shaking is required to ensure that it goes into solution.
Freshly prepared substrate should be used within 1 h, to avoid degradation and evaporation.
-
10
Add 180 μL of the working assay solution to each well in the 96-well plate that contains sample or a blank (final concentration of the substrate in each well will be 5.86 mM).
-
11
Transfer the 96-well plate rapidly into the plate reader (previously set to 25°C).
-
12
Use the mixing program in the plate reader (e.g. SoftMax Pro) to mix the samples for 5 s prior to the start of the readings.
This will ensure an even distribution of the substrate and minimize assay variability.
-
13
Monitor hydrolysis of phenyl acetate at 270 nm. Set the readings for every 15 s and read for at least 4 min. Use only the initial linear rates of hydrolysis for calculating activity values in mOD/min.
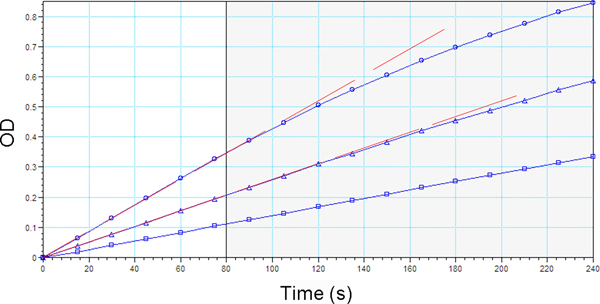
Plotting the signal obtained (y-axis) versus time (x-axis) yields curves that readily identify the initial linear rates of hydrolysis for each assay (Figure 3). Most microplate spectrophotometer software programs have the capability to provide these plots, making it simple to select the initial linear rates. The activity values in mOD/min are calculated by the software. Calculate the standard deviation of the triplicate values obtained for each sample. If the standard deviation is 10 mOD/min or greater, repeat the assay.
It is essential that the enzyme reaction conditions operate within the linear portion of the instrument capacity. Use of non-linear rates can lead to false measurements of hydrolysis rates (Dowd & Riggs, 1965).
-
14
Once the monitoring is complete, use the plate reader to measure the light pathlength (cm) of each well.
Pathlength values are used to normalize absorbance values measured with a microplate reader to correspond with absorbance values measured in a standard precision quartz cuvette. The liquid pathlength in a microplate is not accurate, compared with standard precision quartz cuvettes (1 cm fixed pathlength). Most software programs in microplate spectrophotometers have the capability to correct for the variability of the light pathlength in each well. Refer to Support Protocol 1 if your microplate spectrophotometer lacks this capability.
The pathlength correction feature corrects for volume irregularities and pipetting errors between wells, and normalizes absorbance values for each well.
-
15
Divide the rates of high-salt arylesterase hydrolysis (step 13) by the light pathlength values (step 14) obtained in each well. Then, calculate the average of the pathlength-corrected triplicate values for each sample.
Figure 3. High-salt arylesterase activity rates (mOD/min) over the total duration of the assay (4 min or 240 s).
The activity rates of the top and middle plasma samples are not linear for the entire 4 min. Therefore, a shorter range of time (80 s) that ensures activity rates for all plasma samples are linear should be selected (vertical line). OD: optical density.
Conversion of high-salt arylesterase activity rates (mOD/min) to enzymatic activity (U/mL)
One unit of enzyme activity (U or μmol/min) is defined as the amount of enzyme that hydrolyzes 1 μmol of substrate per min. Enzymatic activity is commonly expressed as U per volume (U/mL), in other words, μmol/min/mL.
-
16Use the Beer-Lambert equation to convert arylesterase activity rates (mOD/min) to activity units (U or μmol/min):
where:A is the rate of change in absorbance/min at 270 nm ( or mOD/min – activity rates), from step 13. Note that the activity rates provided by the Molecular Devices microplate spectrophotometer are in mOD/min;
is the micromolar extinction coefficient of the absorbing product produced during the enzymatic hydrolysis reaction (phenol in the case of phenyl acetate). The micromolar extinction coefficient of phenol at 270 nm is 1.3 × 10−3 μM−1 cm−1 at 25°C (Richter et al., 2008, 2009);
c is the rate of generation of the absorbing product produced during the enzymatic reaction (phenol in the case of phenyl acetate) per 1 min (U/mL), (). This is the unknown value calculated with the Beer-Lambert equation.
l is the pathlength of light through sample in the microplate well (cm), from step 14.
When in the Beer-Lambert equation above are known, can be calculated:Including the units of each parameter, the formula reads:Note that step 15 calculates the normalized average rates of high-salt arylesterase hydrolysis by dividing by 𝑙. We have re-arranged the formula with the parameters to better reflect this:
where is the value from Step 15.
-
17
Take into account the dilution factor of the plasma/serum sample to calculate the high-salt arylesterase activity (U/mL) of the original sample.
The equation above with the units of each parameter specified provides the arylesterase activity in each well. In the protocol, a total of 20 μL of diluted sample is assayed with 180 μL of assay buffer. The total reaction volume is 200 μL, or 0.2 mL. However, each well contains a diluted amount of enzyme (20 μL of a 1:40 plasma/serum dilution). Therefore, the actual amount of plasma/serum used in the assay is 0.5 μL or 0.0005 mL. Thus:where is the value from Step 15, Vt is the total reaction volume (mL), and Vs is the sample volume (dilution-corrected, mL).
-
18
Substitute all known values in the formula above to obtain the high-salt arylesterase enzymatic activity (μmol/min/L or μmol/min/mL).
or:
where and represent the value from Step 15.
CMPAase activity measurement
In addition to measuring arylesterase activity, the samples should also be evaluated for CMPAase activity. Since both enzymatic assays use the same dilution (1:40), we advise diluting the plasma samples just once and using the same sample dilution for both assays.
-
19
Repeat steps 4 to 8.
-
20
Prepare the substrate by adding 4.6 μL of CMPA to 10 mL of CMPAase assay buffer in a 50 mL polypropylene tube (concentration of the substrate in the final assay buffer will be 3 mM). Screw the cap tightly and shake vigorously for 1 min.
CMPA is an oily solution. For this reason, vigorous shaking is required to ensure that it goes into solution.
Freshly prepared substrate should be used within 1 h, to avoid degradation.
-
21
Add 180 μL of the 3 mM CMPA substrate solution (25°C) to each well in the 96-well plate that contains either sample or a blank (final concentration of the substrate in each well is 2.70 mM).
-
22
Transfer the 96-well plate rapidly into the plate reader previously set to 25°C.
-
23
Use the mixing program in the plate reader (e.g. SoftMax Pro) to mix the samples for 5 s prior to the start of readings.
This will ensure an even distribution of the substrate and minimize assay variability.
-
24
Monitor the hydrolysis of CMPA at 280 nm. Set the readings for every 15 s and read for at least 4 min. Use only the initial linear rates of hydrolysis for calculating activity values in mOD/min.
Plotting the signal obtained (y-axis) versus time (x-axis) yields curves that readily identify the initial linear rates of hydrolysis for each assay (Figure 3). Most microplate spectrophotometer software programs have the capability to provide these plots, making it simple to select the initial linear rates. The activity values in mOD/min are calculated by the software. Calculate the standard deviation of the triplicate values obtained for each sample. If the standard deviation is 10 mOD/min or greater, repeat the assay.
It is essential that the enzyme reaction conditions operate within the linear portion of the instrument capacity. Use of non-linear rates can lead to false measurements of hydrolysis rates (Dowd & Riggs, 1965).
-
25
Once the monitoring is complete, use the plate reader to measure the light pathlength (cm) of each well.
Pathlength values are used to normalize absorbance values measured with a microplate reader to correspond with absorbance values measured in a standard precision quartz cuvette. The liquid pathlength in a microplate is not accurate, compared with standard precision quartz cuvettes (1 cm fixed pathlength). Most software programs in microplate spectrophotometers have the capability to correct for the variability of the light pathlength in each well. Refer to Support Protocol 1 if your microplate spectrophotometer lacks this capability.
The pathlength correction feature corrects for volume irregularities and pipetting errors between wells and normalizes absorbance values for each well.
-
26
Divide the rates of CMPAase hydrolysis (step 24) by the light pathlength values (step 25) obtained in each well. Then, calculate the average of the pathlength-corrected triplicate values for each sample.
Conversion of CMPAase activity rates (mOD/min) to enzymatic activity (U/mL, or μmol per min per mL)
-
27
Convert CMPAase activity rates (mOD/min) to enzymatic activity units (U/mL) following the formulas used for high-salt arylesterase (steps 16 to 18), but use the micromolar extinction coefficient of the hydrolysis product 4-(chloromethyl)phenol at 280 nm and 25°C, which is 1.30 × 10−3 μM−1 cm−1 (Richter et al., 2008, 2009). Use the normalized CMPAase hydrolysis rates ( divided by ) calculated in step 26. The dilution factor of the plasma/serum in the CMPAase assay is the same as in the high-salt arylesterase assay (0.005 mL of sample in a final reaction volume of 0.2 mL, step 17). Thus:
resulting in:
or:
where and represent the value from Step 26.
PON1 status analysis using arylesterase (high salt) and CMPAase enzymatic activity values
Now that you have obtained both enzymatic activity values, you can now determine the PON1 status of each sample.
-
28
Plot arylesterase (high salt) enzymatic activity rates (U/mL) (step 18) on the y axis and CMPAase enzymatic activity rates (U/mL) (step 27) on the x axis of a scatter (x/y) plot (see example in Figure 2).
The data in the plot will separate into three distinguishable groups: the top left group corresponds to PON1192QQ individuals, the middle group are PON1192QR individuals, and the bottom right group are PON1192RR subjects (Figure 2). See the Understanding Results section for further details about how to distinguish these three groups.
SUPPORT PROTOCOL 1: Experimental pathlength determination
In standard precision quartz cuvettes, the liquid pathlength is fixed (1 cm), as the absorbance measurement occurs from side to side. However, the liquid pathlength of microplates is not fixed due to the absorbance measurement being taken from the top of the sample to the bottom of the microplate well. Thus, in microplates, this pathlength depends on the sample volume in each well and any variability in the well geometry.
The software programs of modern microplate spectrophotometers typically include a tool to measure the light pathlength in each well. Dividing the activity rates by the pathlength normalizes the absorbance value to a 1-cm pathlength. Pathlength measurement provides a correction of variable microplate well volumes and geometry, providing more accurate results. More importantly, it will also enable calculation of enzymatic activity in international units, using the Beer-Lambert law, as described in Basic Protocol 1, steps 16 and 31. In case the user’s microplate spectrophotometer’s software lacks the capability to measure the light pathlength (steps 14 and 29), this Support Protocol describes how to determine it experimentally.
The described experimental light pathlength determination is based on the fact that water is essentially transparent between 200 and 900 nm, but in the near infrared (NIR), it has a distinctive absorption peak near 977 nm (Parker & Williams, 1974). At 977 nm, most biological molecules have little or no absorbance, so it can be used to measure the light pathlength through an aqueous sample.
Materials–
UV-transparent 96-well microplates used for the high-salt arylesterase and CMPAase assays (Basic Protocol 1) with the diluted plasma/serum samples and substrate added to each well.
Equipment–
UV/visible microplate spectrophotometer (e.g. SpectraMax Plus Microplate Spectrophotometer, Molecular Devices), with the temperature control set to 25°C
Microplate spectrophotometer acquisition and analysis software (e.g.SoftMax Pro data acquisition and analysis software, version 5.4 if using a Molecular Devices microplate reader).
Protocol Steps:
-
As soon as the high-salt arylesterase or CMPAase activity monitoring ends (Basic Protocol 1, steps 13 and 28, respectively), measure absorbance at 900 nm (A900) and at 977 nm (A977) in each well.
Absorbance at 900 nm is used as a baseline absorbance, as it is a wavelength with no water absorption and distant from the water peak.
Calculate means for A900 and A977 for each sample.
-
Determine the pathlength, 𝑙 (cm) of each sample as follows:
The value obtained corresponds to the pathlength of each sample in cm.
The sample is divided by 0.18, because in a 1 cm cuvette, the OD of water at 977 nm – 900 nm is 0.18 OD at room temperature (McGown et al., 1997).
SUPPORT PROTOCOL 2: PON1 DNA genotyping for the 192QR (rs662) polymorphism
We had previously described a detailed protocol for genotyping the PON1Q192R polymorphism (Richter et al., 2004). DNA sequencing technologies have advanced to such point that it is currently much more cost-efficient to send DNA samples to a laboratory with DNA sequencing capabilities.
This Support Protocol describes how to isolate the buffy coat layer using lithium-heparin blood tubes, how to extract DNA from buffy coat, and which primers to use for PON1Q192R genotyping (rs662). Buffy coat is isolated using density gradient centrifugation. DNA from buffy coat is extracted using a commercial kit, following the manufacturer’s recommendations.
Materials–
Samples: 4 mL blood sample collected in lithium heparin or EDTA tubes
Phosphate-buffered saline, pH 7.4, 1X (see Reagents and Solutions)
Ficoll-Paque PLUS (#17–1440-02, GE Healthcare)
Quick-DNA 96 kit (#D3010, Zymo Research)
Molecular Biology grade water (#46–000-CM, Corning)
PON1Q192R forward primer 192: 5′-TAT TGT TGC TGT GGG ACC TGA G-3′
PON1Q192R reverse primer 192: 5′-CAC GCT AAA CCC AAA TAC ATC TC-3′
Equipment–
Centrifuge with the ability to turn the break off, with a swinging rotor that fits 15 mL screw-cap polypropylene conical tubes
Disposable Graduated Transfer Pipettes, 3 mL (#13–711-9AM, ThermoFisher Scientific)
0.5 mL and 1.5 mL Eppendorf tubes
15 mL screw-cap polypropylene conical tubes
Spectrophotometer for nucleic acid quantification (e.g. Nanodrop, ThermoFisher Scientific)
Facility with Sanger sequencing capabilities
Protocol Steps
Buffy coat isolation from blood
-
1
Collect at least 4 mL of blood in heparin lithium (green-top) or EDTA (purple-top) tubes and centrifuge for 10 min at 2,000 x g at 4°C. Separate the top layer containing plasma (yellow) as described in Basic Protocol 1, Steps 1–3. The bottom layer (“cells”) contains white blood cells (buffy coat) and red blood cells. The buffy coat layer (white/pink) typically remains between plasma (yellow) and red blood cells (red).
-
2
Dilute the remaining white and red blood cell pellet with 1X phosphate-buffered saline (PBS) pH 7.4 to twice the original blood volume, using a disposable transfer pipette. Mix very gently by inversion.
For instance, if the original blood volume prior to centrifugation and plasma separation was 4 mL, add a total of 8 mL of 1X PBS.
-
3
Take a 15 mL screw cap polypropylene conical tube and add one original blood volume of Ficoll-Paque, using a disposable transfer pipette.
For instance, if the original blood volume prior to centrifugation and plasma separation was 4 mL, add 4 mL of Ficoll-Paque.
-
4
Carefully and very slowly layer the PBS-diluted cells (8 mL, step 2) on top of the Ficoll-Paque. The cells should layer on top of the transparent Ficoll-Paque. Do not mix.
Ficoll-Paque has a density of 1.007 g/mL. Due to the higher density of red blood cells, they will pass through the Ficoll-Paque layer. Due to the lower density of white blood cells, they will float over the Ficoll-Paque layer.
-
5
Centrifuge the 15 mL tube at 400 x g for 30 min at room temperature with the centrifuge break turned off to avoid lysis of red blood cells.
After centrifugation, the red blood cells will be the bottom layer, the Ficoll-Paque the middle layer, and the white blood cells (buffy coat) the top layer.
-
6
Gently harvest the upper layer of buffy coat using a disposable transfer pipette (roughly between 1–2 mL) and transfer it to a fresh 15 mL conical tube.
-
7
Wash the harvested buffy coat with 3 volumes of 1X PBS, gently suspending the buffy coat in the PBS with a disposable transfer pipette.
If the harvested layer of buffy coat is 2 mL, wash the buffy coat with 6 mL of 1X PBS.
-
8
Centrifuge at 400 x g for 15 min at room temperature, with the brake on. After centrifugation, the buffy coat will be at the bottom of the tube. Discard the top layer of PBS.
-
9
Repeat steps 7 and 8 one more time for a total of 2 washes of the isolated buffy coat.
-
10
Resuspend the washed buffy coat in a small volume (e.g., 1 mL) of 1X PBS and proceed to DNA extraction. Alternatively, store the sample frozen at −80°C until use. When stored frozen, DNA from buffy coat is stable for up to 9 years (Mychaleckyj et al., 2011).
DNA extraction from buffy coat
-
11
Take 100 μL of buffy coat in PBS and follow the manufacturer’s instructions (Zymo Research kit) for extracting the DNA.
-
12
Determine the concentration and purity of the obtained DNA via spectrophotometry (e.g., Nanodrop, ThermoFisher Scientific), following the manufacturer’s instructions.
Using the Zymo Research kit, you will obtain ~30 μL of DNA. The obtained DNA can be used immediately or stored at −80°C until use.
PON1Q192R (rs662) DNA genotyping
-
13
Provide extracted DNA and forward and reverse primers to the sequencing facility, according to the facility’s guidelines, to be used for genotyping PON1Q192R (rs662).
The PCR product obtained by the sequencing facility should have a length of 99 bp, which the sequencing facility will then digest with the restriction enzyme AlwI. If two fragments of 66 and 33 bp are obtained, that corresponds to presence of R allele. The Q allele should remain intact (99 bp) after digestion.
REAGENTS AND SOLUTIONS
Use ultra-pure water or equivalent for the preparation of all buffers, unless otherwise specified.
CMPAase assay buffer (1 L):
Combine 20 mL of UltraPure 1 M Tris-HCl pH 8.0 (20 mM final) and 1 mL of 1 M CaCl2 (1 mM final) and bring volume to 1 L with water. Mix well and store for up to 6 months at room temperature.
Dilution buffer (1 L):
Combine 9 mL of UltraPure 1 M Tris-HCl, pH 8.0 (9 mM final, #T1080, Teknova) with 0.9 mL of 1 M CaCl2 (0.9 mM final, #C0477, Teknova), and add water to a final volume of 1 L. Mix well and store for up to 6 months at room temperature.
Tris-HCl is also known as Tris (hydroxymethyl) aminomethane (THAM) hydrochloride.
You may prefer to purchase Tris-HCl pH 8.0 and CaCl2 as prepared solutions, which are available from vendors including ThermoFisher Scientific (Teknova), MilliporeSigma, and VWR. This is a convenient alternative to preparing these solutions each time they are needed, minimizing inter-assay variability. Both solutions are stable at room temperature and are supplied sterile.
Tris-HCl solutions are generally available at one concentration (1 M) in a variety of pH values, ranging from 6.8 to 10.2.
High-salt arylesterase assay buffer (1 L):
Dissolve 116.8 g NaCl (2 M final; e.g. Mallinckrodt Chemicals, granular) in 800 mL water containing 20 mL of UltraPure 1 M Tris-HCl pH 8.0 (20 mM final) and 1 mL of 1 M CaCl2 (1 mM final). Mix well and bring volume to 1 L with water. Store for up to 6 months at room temperature.
Phosphate-buffered saline (PBS), pH 7.4, 1X (1 L):
Combine 100 mL of PBS pH 7.4 10 X (#46–013-CM, Corning) with 900 mL of Molecular Biology grade water (#46–000-CM, Corning). Mix well and store for up to 6 months at room temperature.
COMMENTARY
Background Information
PON1 was first studied for its ability to hydrolyze aromatic esters and the toxic metabolites of specific OP compounds (Aldridge, 1953a, 1953b). Three decades later, it was suggested that PON1 could also prevent accumulation of oxidized lipids in low-density (Mackness et al., 1991, 1993) and high-density lipoproteins (Aviram et al., 1998). As a consequence, there has been an exponential increase in the number of publications examining the potential role that genetic variability of PON1 plays in the risk of related disease and exposure. Historically, in analyzing PON1 genetic variability, most researchers have ignored the important fact that the functional activity of PON1 is more important than the PON1 SNP status in determining risk of disease or exposure, due to the fact that factors beyond certain PON1 SNPs can influence PON1 activity and levels (Camps et al., 2009b; Costa et al., 2005, 2011). As a result, there have been —and continue to be— conflicting results regarding the involvement of PON1 genetic variability in risk of disease or exposure. Analyzing all of the more than 200 reported SNPs in the PON1 gene will not allow precise prediction of an individual’s level of plasma/serum PON1, arguably the most important parameter for providing risk of exposure or susceptibility to disease. The only way to know the level of an individual’s PON1 plasma/serum activities is to measure them, as described in the PON1 status analyses.
There is only one common polymorphism that affects the catalytic efficiency of PON1 for hydrolyzing specific substrates, the PON1Q192R polymorphism (Humbert et al., 1993; Li et al., 2000). These effects are substrate-dependent, as the following examples illustrate. While both PON1192 alloforms hydrolyze phenyl acetate with equal rates (Furlong et al., 2006), there are dramatic differences between the PON1192 alloforms over the catalytic efficiency of hydrolysis of other substrates, such as paraoxon, the compound for which PON1 received its name. PON1R192 hydrolyzes paraoxon (the oxon form of the OP insecticide parathion) with approximately 7-times the rate of the PON1Q192 alloform. This was the property that first elucidated the polymorphic distribution of rates of paraoxon hydrolysis in human populations (Geldmacher-von Mallinckrodt and Diepgen, 1988). The catalytic efficiency of the PON1R192 alloform for hydrolyzing chlorpyrifos oxon (the oxon form of the OP insecticide chlorpyrifos) is higher than that of the PON1Q192 alloform, while the catalytic efficiencies of the two alloforms for hydrolyzing diazoxon (the oxon form of the OP insecticide diazinon) are the same (Li et al., 2000). These findings were validated by examining the protection against OP toxicity afforded by injecting purified human PON1192 alloforms into Pon1 knockout mice (Li et al., 2000; Cole et al., 2005).
Our laboratory has developed SNP assays for PON155 and PON1192 (Humbert et al., 1993), has characterized the effects of PON1 promoter region SNPs (Brophy et al., 2001a, 2001b), and has developed PON1 status analyses. Hence, we could easily use and recommend any of these approaches for epidemiological studies, and if we found evidence that, for instance, the SNP assays provided sufficient and adequate information for epidemiological studies, we would certainly be using and recommending SNP analyses for them. However, we and others have found that determining the PON1 status with the protocol herein described is the only approach among these that provides the necessary data for a proper epidemiological study. The PON1 status studies already published have demonstrated that PON1 activity levels can vary significantly among individuals by at least 13-fold, or by 60-fold if newborns are included in the analyses (Furlong et al., 2006, 2010). This large, stable variability of PON1 activity levels among individuals with the same PON1192 functional genotype can also be seen in the example shown in Figure 2, and should be an incentive for investigators to make use of PON1 status analysis as opposed to simple SNP analysis for any epidemiological study examining the genetic variability of PON1 activity related to risk of exposure or disease. It is, after all, the level of PON1 activity that determines risk of disease or exposure, and not specific SNPs. Individuals with lower PON1 activity levels have been reported to be more sensitive to OP exposures (Hofmann et al., 2009; Furlong et al., 2008) and at higher risk for disease development (Besler et al., 2011; Cervellati et al., 2019; Ferré et al., 2013; Furlong et al., 2010; Gaita et al., 2010; Jarvik et al., 2000, 2003b; Jornayvaz et al., 2009; Marsillach et al., 2007; Parra et al., 2007). The SNPs may contribute to differences in levels, but it is necessary to directly measure PON1 activity levels via, for instance, PON1 status assays, to determine activity levels.
The PON1 status assays were developed to provide the research community with a high-throughput method for determining both the functional PON1192 genotype and the plasma PON1 activity levels (Furlong et al., 2002; Richter & Furlong, 1999; Richter et al., 2004, 2008, 2009). The original PON1 status analyses make use of the toxic OP metabolites paraoxon and diazoxon (Furlong et al., 2002; Richter & Furlong, 1999; Richter et al., 2004) and not all laboratories are equipped to handle these highly toxic OP substrates. The use of OP compounds requires training of laboratory personnel in safety practices and management of associated waste, as well as preparing a laboratory safety plan to follow in the case of accidental exposures and having specific antidotes available.
Just as its OP-based counterpart, the non-OP version of the PON1 status (Richter et al., 2008, 2009) provides excellent resolution of the three PON1192 phenotypes, while additionally allowing any laboratory to safely handle the required substrates. Here, we have described this non-toxic PON1 status analysis in detail to facilitate the use of this analysis in laboratories interested in carrying out meaningful epidemiological studies. As seen in Figure 2, the plasma activity levels of PON1 can vary significantly among individuals within the same PON1192 genotype, which means that individuals within the same polymorphic group may be significantly more or less protected against risk of exposure or disease not based on their PON1192 genotype, but on their plasma/serum PON1 activities.
Although we highly favor the use of PON1 status in epidemiological studies, we do not discourage analyzing both PON1 status and PON1192 SNPs. Users interested in carrying out an epidemiological study aimed at exploring the effects of PON1 variability on resistance to disease or exposure, should not base the study on SNP analysis alone. PON1 status is the most important parameter to measure, as it provides the functional PON1192 genotype as well as the plasma PON1 activity levels. However, if users are looking for disabling mutations in the PON1 gene, it will be useful to carry out both PON1 status analyses and SNP genotyping for position 192, to flag the possibility of a discrepancy between PON1192 functional genotypes and SNP analyses, but again, users should not examine SNP analyses alone due to the large variability of PON1 activity within the three PON1192 phenotypes. When run in parallel, PON1 status functional phenotyping results agree, in most cases, with the results obtained by DNA SNP analysis (Haley et al., 1999; Jarvik et al., 2000, 2003a, 2003b; Carlson et al., 2006). In the few analyses in which we have observed discordant results, sequencing of the entire PON1 gene detected genetic mutations or deletions which explained them (Jarvik et al., 2003a). For example, in Jarvik and colleagues (2003a), 4 subjects out of 704 subjects analyzed had discordant PON1 status and PON1Q192R genotyping results or very low PON1 activity. One individual genotyped as PON1192QR but phenotyped functionally as PON1192QQ. Full sequencing of the subject’s PON1 genes revealed a rare PON1Trp194stop mutation, resulting in no active PON1R192 protein product. This mutation failed to be detected by PON1 SNP analysis. Another subject genotyped as PON1192QR and phenotyped as PON1192RR. This individual appeared to have a partial deletion of their PONQ192 allele. Sequencing the PON1 genes of one individual who genotyped as a PON1Q192 homozygote but had very low PON1 associated activities revealed a PON1P90L mutation in one PON1Q192 allele, accounting for the low activity. Sequencing of the PON1 genes of one other individual who genotyped and phenotyped as a PON1R192 individual but had low activity revealed a PON1asp124 missplice mutation accounting for the low activity (Jarvik et al., 2003a).
CRITICAL PARAMETERS
PON1 is a calcium-dependent enzyme. Calcium chloride must be included in all buffers and is a critical component in maintaining the activity of plasma PON1. In this regard, PON1 status cannot be carried out using plasma collected using ethylenediaminetetraacetic acid tubes (EDTA, purple or pink-top tubes), as EDTA is a calcium chelator and it results in irreversible inhibition of human plasma PON1 (Erdos et al., 1959). Sodium heparin or lithium heparin plasma (green-top tube, plasma) or serum are preferred specimens (Ferré et al., 2005). Sodium citrate plasma (blue-top tube) can also be used (Richter et al., 2004).
The substrates used in this non-OP PON1 status analysis, phenyl acetate and CMPA, are oily liquids. For this reason, when added to the assay buffer, they do not readily dissolve. When pipetting the substrates, always wash the pipet tips 3–4 times in the assay buffer.. Then, mix the tube containing the substrate solution (substrate plus assay buffer) vigorously for at least 30 s. To prevent unnecessary splashes due to a faulty tube or not-well-capped tube, we suggest adding parafilm outside the cap and performing the vigorous shaking with the tube facing the laboratory sink. Since the substrates degrade or volatilize over time once in an aqueous solution, discard the substrate solution after 1 h.
Plasma/serum PON1 activity is very stable over time and is not highly affected by multiple freeze-thaw cycles when stored at −80°C for 2 years (Huen et al., 2009). Previous studies have shown minimal changes in PON1 activity assays in samples stored at −80°C for up to 2 years (Beekhof et al., 2012; Huen et al., 2009). We have found PON1 activity to be stable for at least 9 years when plasma samples remain frozen at −80°C (Richter RJ., personal communication). This is particularly relevant for longitudinal studies.
The viscosity of plasma/serum is much different than that of water and other aqueous solutions, thus, careful mixing of plasma/serum samples is important. If users do not take this into account when pipetting plasma/serum samples, it can result in inaccurate sample transfers. Ensure use of standard or wide orifice tips, slow down the dispense speed, and only immerse the pipette tip 1–2 mm below plasma/serum surface to decrease presence of excess droplets on the tip that can lead to loading more plasma/serum sample than indicated in the pipette setting.
As soon as samples are diluted, they should be analyzed, as diluted plasma PON1 degrades fast over time. Leftover diluted samples should not be stored or re-used in the future. If PON1 status needs to be repeated for any reason, fresh plasma/serum dilutions should be prepared. The use of multichannel pipettes is highly recommended to speed up the process of pipetting both diluted plasma/serum samples and the enzymatic substrate to the 96-well microplate, to minimize the time from the samples mixing with substrates to the time of the determination of rates of hydrolysis in the microplate reader. Our laboratory uses the E1-ClipTip Equalizer pipettes (ThermoFisher Scientific), which are adjustable tip spacing electronic pipettes, allowing volume transfers between any type of tube, rack, or microplate.
It is critical to ensure that the activity rates (mOD/min) obtained from the spectrophotometer that will be used in calculations are derived from only the linear portion of the rate vs. time plots generated during the kinetic analyses (Dowd & Riggs, 1965). If rates become nonlinear during the 4 min enzymatic assay, it is necessary to select a shorter range of time during which the rate remains linear and adjust for the time change in your calculations. An example is provided in Figure 3.
It is recommended to run both enzymatic assays in triplicate and to use the same brand of 96-well microplates throughout the study. This will help to minimize any plate-to-plate variability.
TROUBLESHOOTING
Ensure that the blank sample activity rate (mOD/min) is close to zero before using it to subtract from the activity rates of the plasma/serum samples. The blank absorbance rates should be close to zero in all cases. If the blank sample shows a significant absorbance, we recommend preparing new dilution buffer and substrate buffer and repeating the assay. In addition, check the expiration date of the substrates used. Contamination of either the dilution buffer or the microplate well containing the blank sample with plasma/serum would cause unexpected increase in an activity rate. In the case of the substrate buffer, if the substrate has been in an aqueous solution for a prolonged time, it could be degraded and result in increased absorbance. This would show up as an increased starting absorbance. For this reason, it is always best to prepare the substrate buffer shortly before it is needed and to not use it after 1 h of preparation.
Good techniques in order to avoid non-acceptable/questionable values are: 1) using only the initial linear part of the assay rate curves (Basic Protocol 1, steps 13 and 28; Figure 3); 2) assaying samples in triplicate; 3) paying attention to the standard deviation of the obtained hydrolysis rates between triplicates (as described in Basic Protocol 1, steps 13 and 28); 4) making sure the plasma/serum sample does not include any precipitates or unusual color; and 5) not using samples collected in EDTA-containing blood tubes.
To control for plate-to-plate variation, it is useful to include a well-characterized control plasma/serum sample in triplicate in each 96-well microplate. There are excellent commercially- available control plasma and serum samples, such as Golden West Diagnostics (e.g. catalog #UN1000 or #SP1000) or MilliporeSigma (e.g. catalog #S1–100ML or P9523–5ML), or you can create one from your own collection of plasma/serum samples. We advise aliquoting the control samples in small aliquots to avoid multiple freeze-thaw cycles, and to treat this control sample as if it is a study sample. The resulting mOD/min should be similar between microplates assayed.
The inter-assay coefficient of variation between triplicate values should be less than 15%. If it is higher, ensure that the enzymatic assays are carried out as soon as plasma/serum samples are diluted and with the substrate assay buffer as freshly prepared as possible. In addition, make sure the pipettes used are calibrated.
The temperature of the laboratory where analyses are conducted should stay at ≤ 25°C when possible, and the plate reader temperature controller should be set at 25°C. Higher temperatures will result in increased activities. In the case that the ambient temperature of the laboratory is higher than 25°C, the buffers should be placed in 25°C water baths.
If the activity assays do not show any rates of hydrolysis, ensure that the 96-well plates used are UV-rated. The absorbance of light in regular 96-well plates (made usually of polystyrene or polypropylene) in the visible range of the electromagnetic spectrum is very low. However, these materials absorb light in the UV range. UV transparent 96-well plates do not absorb light in the UV spectrum, and provide low background and consistent performance for absorbances between 260 and 280 nm.
UNDERSTANDING RESULTS
The non-OP PON1 status assays described in this protocol provide easy-to-follow steps for obtaining high-salt arylesterase and CMPAase activity rates (mOD/min) that are then normalized with the pathlength and converted into enzymatic rate activity units (U/mL) prior to visualizing the results in 2-D plots (Figure 2).
When the high-salt arylesterase rates of hydrolysis (y axis) are plotted against the CMPAase rates of hydrolysis (x axis), the resulting x/y plot should generate a clear resolution of the samples into the three PON1192 phenotypes, similar to what is shown in Figure 2. As mentioned in Basic Protocol 1, step 32, the group of dots/activity values represented at the top left of the x/y plot corresponds to PON1192QQ individuals, the middle group are PON1192QR, and the bottom right group values correspond to PON1192RR. If the researcher carrying out the assays is not experienced in PON1 status analyses, distinguishing the three groups may not seem very obvious. In those cases, we recommend carrying out both non-OP PON1 status (Basic Protocol 1) and PON1Q192R genotyping (Support Protocol 2). The results from both assays will be helpful in identifying which individuals belong to each of the three groups (QQ, QR, and RR), when in doubt. However, in designing this non-OP PON1 status assay, we found that the use of the two substrates (phenyl acetate and CMPA) with the assay conditions described above, results in plots that typically provide excellent resolution of the three PON1192 phenotypes (Richter et al., 2008). The plasma samples analyzed in both plots (Figure 2A and 2B) are the same, and the performance between the original PON1 status (Figure 2A) and the non-OP PON1 status (Figure 2B) is comparable. In addition, all PON1192 functional genotypes obtained with the two PON1 status analyses (original and non-OP) were consistent with the PON1Q192R polymorphism genotyping data DNA obtained from the same set of samples (Richter et al., 2008).
PON1 status does not require the use of positive plasma/serum controls for each PON1Q192R genotype. When the 2-D plots are obtained, samples always fall within one of the three visualized PON1192 phenotypes (QQ, QR or RR) (Figure 2). We do recommend, however, including a plasma/serum control sample in each analyzed 96-well plate, treated as if it is a study sample, to account for plate-to-plate variability, as described in the Troubleshooting section. Regarding acceptable and non-acceptable PON1 status values, an acceptable value is one that when plotted, falls within either of the three PON1192 phenotyping groups (QQ, QR, RR), as seen in Figure 2. A non-acceptable/questionable value would be one that does not clearly fall within either of the three PON1192 phenotyping groups, or a value with really low activity values (at 15 U/mL or less high-salt arylesterase activity). Samples with non-acceptable values should be re-assayed. As already noted in the Strategic Planning section, hemolysis, jaundice, and lipemia in plasma/serum samples should not affect the described non-OP PON1 status assay due to the sample dilution used in the activity assays. If the assay is repeated and a non-acceptable value is once again obtained, we recommend carrying out PON1Q192R (rs662) DNA genotyping (Support Protocol 2) or contacting a DNA Sequencing Service for sequencing the entire PON1 gene. It is likely that there is a nonsense or missense mutation or a partial gene deletion in a PON1 gene of that individual (Jarvik et al., 2003a).
In epidemiological studies, once PON1 status assays are completed, PON1 phenotypic distributions between controls and cases should be stratified by PON1192 genotype, as obtained when plotting the PON1 status results on a scatter (x/y) plot (Jarvik et al., 2000). Gene frequencies will vary between groups of different ethnic origin; for example, individuals of Northern European origin will have gene frequencies of ~0.72 for PON1192Q and ~0.28 for PON1192R, whereas individuals from Asian and African populations will have frequencies closer to 0.5 for each PON1192 allele (Diepgen et al., 1986; Furlong, 2007). For this reason, a minimum number of samples to be assayed should be considered, to ensure that there are individuals represented in each of the three PON1192 groups in the plot. For instance, in cohorts with individuals of primarily White ethnicity, a low representation of PON1192RR individuals should be expected due to their reported low R allele frequency (Davies et al., 2009; Diepgen et al., 1986; Furlong, 2007; Furlong et al., 2016). If there are not enough samples analyzed, the resulting PON1 status x/y scatter plot may only show two groups instead of three, and that can be confusing, especially for a researcher less experienced in PON1 status analyses. From our experience, we recommend analyzing a minimum of 50–100 samples. This minimum number of samples should provide dots/values in the plot corresponding to each of the PON1Q192R groups. Additional information in epidemiological studies may be obtained by also stratifying the data by sex in addition to by functional PON1192 genotype. We also recommend using different symbols for each genotype, for example, open circles for PON1QQ individuals, solid squares for PON1QR individuals, and open triangles for PON1RR individuals, or different colors for each PON1192 phenotype (Figure 2).
To facilitate comparison of non-OP PON1 status results with data acquired using the original toxic PON1 status, rates of high-salt phenyl acetate and CMPA hydrolysis can be converted to rates of diazoxon or paraoxon hydrolysis (the toxic OP substrates used in the original PON1 status analysis) from tables presented in Richter et al. (2008 & 2009). This would be relevant for researchers that have older data acquired with the original PON1 status and would like to use the non-OP PON1 status in future cohorts but retain the capability of comparing the PON1 status results from earlier studies.
TIME CONSIDERATIONS
A total of 30 samples, plus a control sample and a blank sample, can be run in triplicate in a single 96-well microplate. Taking into account that the dilution of the sample for both assays is the same, and the use of multichannel pipettes, the determination of the non-OP PON1 status for 30 individuals can easily be completed in a half-day. If the person carrying out the non-OP PON1 status analysis is experienced with this method, 90 samples (three 96-well microplates) can be assayed in a single day.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (grants R01 ES009883, P42 ES004696, and P30 ES007033).
Footnotes
Conflicts of Interest
The authors declare no conflict of interest.
LITERATURE CITED
- Adkins S, Gan KN, Mody M, & La Du BN (1993). Molecular basis for the polymorphic forms of human serum paraoxonase/arylesterase: Glutamine or arginine at position 191, for the respective A or B allozymes. American Journal of Human Genetics, 52(3), 598–608. [PMC free article] [PubMed] [Google Scholar]
- Aldridge WN (1953a). Serum esterases. I. Two types of esterase (A and B) hydrolysing p-nitrophenyl acetate, propionate and butyrate, and a method for their determination. The Biochemical Journal, 53(1), 110–117. 10.1042/bj0530110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge WN (1953b). Serum esterases. II. An enzyme hydrolysing diethyl p-nitrophenyl phosphate (E600) and its identity with the A-esterase of mammalian sera. The Biochemical Journal, 53(1), 117–124. 10.1042/bj0530117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviram M, Rosenblat M, Bisgaier CL, Newton RS, Primo-Parmo SL, & La Du BN (1998). Paraoxonase inhibits high-density lipoprotein oxidation and preserves its functions. A possible peroxidative role for paraoxonase. The Journal of Clinical Investigation, 101(8), 1581–1590. 10.1172/JCI1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekhof PK, Gorshunska M, & Jansen EH (2012). Long term stability of paraoxonase-1 and high-density lipoprotein in human serum. Lipids in Health and Disease, 11, 53. 10.1186/1476-511X-11-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besler C, Heinrich K, Rohrer L, Doerries C, Riwanto M, Shih DM, Chroni A, Yonekawa K, Stein S, Schaefer N, Mueller M, Akhmedov A, Daniil G, Manes C, Templin C, Wyss C, Maier W, Tanner FC, Matter CM, …Landmesser U. (2011). Mechanisms underlying adverse effects of HDL on eNOS-activating pathways in patients with coronary artery disease. The Journal of Clinical Investigation, 121(7), 2693–2708. 10.1172/JCI42946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatter MC, James RW, Messmer S, Barja F, & Pometta D. (1993). Identification of a distinct human high-density lipoprotein subspecies defined by a lipoprotein-associated protein, K-45. Identity of K-45 with paraoxonase. European Journal of Biochemistry, 211(3), 871–879. 10.1111/j.1432-1033.1993.tb17620.x [DOI] [PubMed] [Google Scholar]
- Brophy VH, Hastings MD, Clendenning JB, Richter RJ, Jarvik GP, & Furlong CE (2001a). Polymorphisms in the human paraoxonase (PON1) promoter. Pharmacogenetics, 11(1), 77–84. 10.1097/00008571-200102000-00009 [DOI] [PubMed] [Google Scholar]
- Brophy VH, Jampsa RL, Clendenning JB, McKinstry LA, Jarvik GP, & Furlong CE (2001. b). Effects of 5’ regulatory-region polymorphisms on paraoxonase-gene (PON1) expression. American Journal of Human Genetics, 68(6), 1428–1436. 10.1086/320600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps J, Marsillach J, & Joven J. (2009. a). The paraoxonases: Role in human diseases and methodological difficulties in measurement. Critical Reviews in Clinical Laboratory Sciences, 46(2), 83–106. 10.1080/10408360802610878 [DOI] [PubMed] [Google Scholar]
- Camps J, Marsillach J, & Joven J. (2009. b). Pharmacological and lifestyle factors modulating serum paraoxonase-1 activity. Mini Reviews in Medicinal Chemistry, 9(8), 911–920. 10.2174/138955709788681591 [DOI] [PubMed] [Google Scholar]
- Carlson CS, Heagerty PJ, Hatsukami TS, Richter RJ, Ranchalis J, Lewis J, Bacus TJ, McKinstry LA, Schellenberg GD, Rieder M, Nickerson D, Furlong CE, Chait A, & Jarvik GP (2006). TagSNP analyses of the PON gene cluster: Effects on PON1 activity, LDL oxidative susceptibility, and vascular disease. Journal of Lipid Research, 47(5), 1014–1024. 10.1194/jlr.M500517-JLR200 [DOI] [PubMed] [Google Scholar]
- Cervellati C, Valacchi G, Tisato V, Zuliani G, & Marsillach J. (2019). Evaluating the link between Paraoxonase-1 levels and Alzheimer’s disease development. Minerva Medica, 110(3), 238–250. 10.23736/S0026-4806.18.05875-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TB, Walter BJ, Shih DM, Tward AD, Lusis AJ, Timchalk C, Richter RJ, Costa LG, & Furlong CE (2005). Toxicity of chlorpyrifos and chlorpyrifos oxon in a transgenic mouse model of the human paraoxonase (PON1) Q192R polymorphism. Pharmacogenetics and Genomics, 15(8), 589–598. 10.1097/01.fpc.0000167327.08034.d2 [DOI] [PubMed] [Google Scholar]
- Costa LG, Li WF, Richter RJ, Shih DM, Lusis A, & Furlong CE (1999). The role of paraoxonase (PON1) in the detoxication of organophosphates and its human polymorphism. Chemico-Biological Interactions, 119–120, 429–438. 10.1016/s0009-2797(99)00055-1 [DOI] [PubMed] [Google Scholar]
- Costa LG, Cole TB, Jarvik GP, & Furlong CE (2003). Functional genomic of the paraoxonase (PON1) polymorphisms: Effects on pesticide sensitivity, cardiovascular disease, and drug metabolism. Annual Review of Medicine, 54, 371–392. 10.1146/annurev.med.54.101601.152421 [DOI] [PubMed] [Google Scholar]
- Costa LG, Vitalone A, Cole TB, & Furlong CE (2005). Modulation of paraoxonase (PON1) activity. Biochemical Pharmacology, 69(4), 541–550. 10.1016/j.bcp.2004.08.027 [DOI] [PubMed] [Google Scholar]
- Costa LG, Giordano G, & Furlong CE (2011). Pharmacological and dietary modulators of paraoxonase 1 (PON1) activity and expression: The hunt goes on. Biochemical Pharmacology, 81(3), 337–344. 10.1016/j.bcp.2010.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa LG, Giordano G, Cole TB, Marsillach J, & Furlong CE (2013). Paraoxonase 1 (PON1) as a genetic determinant of susceptibility to organophosphate toxicity. Toxicology, 307, 115–122. 10.1016/j.tox.2012.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies HG, Richter RJ, Keifer M, Broomfield CA, Sowalla J, & Furlong CE (1996). The effect of the human serum paraoxonase polymorphism is reversed with diazoxon, soman and sarin. Nature Genetics, 14(3), 334–336. 10.1038/ng1196-334 [DOI] [PubMed] [Google Scholar]
- Davis KA, Crow JA, Chambers HW, Meek EC, & Chambers JE (2009). Racial differences in paraoxonase-1 (PON1): a factor in the health of southerners? Environmental Health Perspectives, 117(8), 1226–1231. doi: 10.1289/ehp.0900569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepgen TL, & Geldmacher-von Mallinckrodt M. (1986). Interethnic differences in the detoxification of organophosphates: the human serum paraoxonase polymorphism. Archives of Toxicology. Supplement, 9, 154–158. 10.1007/978-3-642-71248-7_18 [DOI] [PubMed] [Google Scholar]
- Draganov DI, Teiber JF, Speelman A, Osawa Y, Sunahara R, & La Du BN (2005). Human paraoxonases (PON1, PON2, and PON3) are lactonases with overlapping and distinct substrate specificities. Journal of Lipid Research, 46(6), 1239–1247. 10.1194/jlr.M400511-JLR200 [DOI] [PubMed] [Google Scholar]
- Dowd JE, & Riggs DS (1965). A comparison of estimates of Michaelis-Menten kinetic constants from various linear transformations. Journal of Biological Chemistry, 240(2), 863–9. [PubMed] [Google Scholar]
- Eckerson HW, Wyte CM, & La Du BN (1983). The human serum paraoxonase/arylesterase polymorphism. American Journal of Human Genetics, 35(6), 1126–1138. [PMC free article] [PubMed] [Google Scholar]
- Erdos EG, Debay CR, & Westerman MP (1959). Activation and inhibition of the arylesterase of human serum. Nature, 8(184), 430–431. 10.1038/184430a0. [DOI] [PubMed] [Google Scholar]
- Ferré N, Camps J, Marsillach J, Mackness B, Mackness M, Coll B, Tous M, & Joven J. (2005). Comparison of paraoxonase 1 measurements in serum and in lithium-heparin-anticoagulated plasma samples. Clinical Chemistry, 51(5), 922–923. 10.1373/clinchem.2005.048231 [DOI] [PubMed] [Google Scholar]
- Ferré N, Feliu A, García-Heredia A, Marsillach J, París N, Zaragoza-Jordana M, Mackness B, Mackness M, Escribano J, Closa-Monasterolo R, Joven J, & Camps J. (2013). Impaired paraoxonase-1 status in obese children. Relationships with insulin resistance and metabolic syndrome. Clinical Biochemistry, 46(18), 1830–1836. 10.1016/j.clinbiochem.2013.08.020 [DOI] [PubMed] [Google Scholar]
- Furlong CE, Cole TB, Jarvik GP, & Costa LG (2002). Pharmacogenomic considerations of the paraoxonase polymorphisms. Pharmacogenomics, 3(3), 341–348. 10.1517/14622416.3.3.341 [DOI] [PubMed] [Google Scholar]
- Furlong CE, Holland N, Richter RJ, Bradman A, Ho A, & Eskenazi B. (2006). PON1 status of farmworker mothers and children as a predictor of organophosphate sensitivity. Pharmacogenetics and Genomics, 16(3), 183–190. 10.1097/01.fpc.0000189796.21770.d3 [DOI] [PubMed] [Google Scholar]
- Furlong CE (2007). Genetic variability in the cytochrome P450-paraoxonase 1 (PON1) pathway for detoxication of organophosphorus compounds. Journal of Biochemical and Molecular Toxicology, 21(4), 197–205. 10.1002/jbt.20181 [DOI] [PubMed] [Google Scholar]
- Furlong CE, Richter RJ, Li W-F, Brophy VH, Carlson C, Rieder M, Nickerson D, Costa LG, Ranchalis J, Lusis AJ, Shih DM, Tward A, & Jarvik GP (2008). The Functional Consequences of Polymorphisms in the Human PON1 Gene. In Mackness B, Mackness M, Aviram M, & Paragh G (Eds.), The Paraoxonases: Their Role in Disease Development and Xenobiotic Metabolism (pp. 267–281). Springer Netherlands. 10.1007/978-1-4020-6561-3_18 [DOI] [Google Scholar]
- Furlong CE, Suzuki SM, Stevens RC, Marsillach J, Richter RJ, Jarvik GP, Checkoway H, Samii A, Costa LG, Griffith A, Roberts JW, Yearout D, & Zabetian CP (2010). Human PON1, a biomarker of risk of disease and exposure. Chemico-Biological Interactions, 187(1–3), 355–361. 10.1016/j.cbi.2010.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong CE, Marsillach J, Jarvik GP, & Costa LG (2016). Paraoxonases-1, −2 and −3: What are their functions? Chemico-Biological Interactions, 259(Pt B), 51–62. doi: 10.1016/j.cbi.2016.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaita L, Manzi B, Sacco R, Lintas C, Altieri L, Lombardi F, Pawlowski TL, Redman M, Craig DW, Huentelman MJ, Ober-Reynolds S, Brautigam S, Melmed R, Smith CJ, Marsillach J, Camps J, Curatolo P, & Persico AM (2010). Decreased serum arylesterase activity in autism spectrum disorders. Psychiatry Research, 180(2–3), 105–113. 10.1016/j.psychres.2010.04.010 [DOI] [PubMed] [Google Scholar]
- Geldmacher-von Mallinckrodt M, & Diepgen TL (1988). The human serum paraoxonase – Polymorphism and specificity. Toxicological and Environmental Chemistry, 18(2–3), 79–196. 10.1080/02772248809357310 [DOI] [Google Scholar]
- Haley RW, Billecke S, & La Du BN (1999). Association of low PON1 type Q (type A) arylesterase activity with neurologic symptom complexes in Gulf War veterans. Toxicology and Applied Pharmacology, 157(3), 227–233. 10.1006/taap.1999.8703 [DOI] [PubMed] [Google Scholar]
- Hassett C, Richter RJ, Humbert R, Chapline C, Crabb JW, Omiecinski CJ, & Furlong CE (1991). Characterization of cDNA clones encoding rabbit and human serum paraoxonase: The mature protein retains its signal sequence. Biochemistry, 30(42), 10141–10149. 10.1021/bi00106a010 [DOI] [PubMed] [Google Scholar]
- Hofmann JN, Keifer MC, Furlong CE, De Roos AJ, Farin FM, Fenske RA, van Belle G, & Checkoway H. (2009). Serum cholinesterase inhibition in relation to paraoxonase-1 (PON1) status among organophosphate-exposed agricultural pesticide handlers. Environmental Health Perspectives, 117(9), 1402–1408. 10.1289/ehp.0900682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen K, Richter R, Furlong C, Eskenazi B, & Holland N. (2009). Validation of PON1 enzyme activity assays for longitudinal studies. Clinica Chimica Acta; International Journal of Clinical Chemistry, 402(1–2), 67–74. 10.1016/j.cca.2008.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert R, Adler DA, Disteche CM, Hassett C, Omiecinski CJ, & Furlong CE (1993). The molecular basis of the human serum paraoxonase activity polymorphism. Nature Genetics, 3(1), 73–76. 10.1038/ng0193-73 [DOI] [PubMed] [Google Scholar]
- Jarvik GP, Rozek LS, Brophy VH, Hatsukami TS, Richter RJ, Schellenberg GD, & Furlong CE (2000). Paraoxonase (PON1) phenotype is a better predictor of vascular disease than is PON1(192) or PON1(55) genotype. Arteriosclerosis, Thrombosis, and Vascular Biology, 20(11), 2441–2447. 10.1161/01.atv.20.11.2441 [DOI] [PubMed] [Google Scholar]
- Jarvik GP, Jampsa R, Richter RJ, Carlson CS, Rieder MJ, Nickerson DA, & Furlong CE (2003a). Novel paraoxonase (PON1) nonsense and missense mutations predicted by functional genomic assay of PON1 status. Pharmacogenetics, 13(5), 291–295. 10.1097/00008571-200305000-00009 [DOI] [PubMed] [Google Scholar]
- Jarvik GP, Hatsukami TS, Carlson C, Richter RJ, Jampsa R, Brophy VH, Margolin S, Rieder M, Nickerson D, Schellenberg GD, Heagerty PJ, & Furlong CE (2003b). Paraoxonase activity, but not haplotype utilizing the linkage disequilibrium structure, predicts vascular disease. Arteriosclerosis, Thrombosis, and Vascular Biology, 23(8), 1465–1471. 10.1161/01.ATV.0000081635.96290.D3 [DOI] [PubMed] [Google Scholar]
- Jornayvaz FR, Brulhart-Meynet M-C, & James RW (2009). Myeloperoxidase and paraoxonase-1 in type 2 diabetic patients. Nutrition, Metabolism, and Cardiovascular Diseases: NMCD, 19(9), 613–619. 10.1016/j.numecd.2008.12.005 [DOI] [PubMed] [Google Scholar]
- Khersonsky O, & Tawfik DS (2005). Structure-reactivity studies of serum paraoxonase PON1 suggest that its native activity is lactonase. Biochemistry, 44(16), 6371–6382. 10.1021/bi047440d [DOI] [PubMed] [Google Scholar]
- Leviev I, & James RW (2000). Promoter polymorphisms of human paraoxonase PON1 gene and serum paraoxonase activities and concentrations. Arteriosclerosis, Thrombosis, and Vascular Biology, 20(2), 516–521. 10.1161/01.atv.20.2.516 [DOI] [PubMed] [Google Scholar]
- Li WF, Costa LG, Richter RJ, Hagen T, Shih DM, Tward A, Lusis AJ, & Furlong CE (2000). Catalytic efficiency determines the in-vivo efficacy of PON1 for detoxifying organophosphorus compounds. Pharmacogenetics, 10(9), 767–779. 10.1097/00008571-200012000-00002 [DOI] [PubMed] [Google Scholar]
- Liu M-E, Liao Y-C, Lin R-T, Wang Y-S, Hsi E, Lin H-F, Chen K-C, & Juo S-HH (2013). A functional polymorphism of PON1 interferes with microRNA binding to increase the risk of ischemic stroke and carotid atherosclerosis. Atherosclerosis, 228(1), 161–167. 10.1016/j.atherosclerosis.2013.01.036 [DOI] [PubMed] [Google Scholar]
- Mackness MI, Arrol S, & Durrington PN (1991). Paraoxonase prevents accumulation of lipoperoxides in low-density lipoprotein. FEBS Letters, 286(1–2), 152–154. 10.1016/0014-5793(91)80962-3 [DOI] [PubMed] [Google Scholar]
- Mackness MI, Arrol S, Abbott C, & Durrington PN (1993). Protection of low-density lipoprotein against oxidative modification by high-density lipoprotein associated paraoxonase. Atherosclerosis, 104(1–2), 129–135. 10.1016/0021-9150(93)90183-u [DOI] [PubMed] [Google Scholar]
- Mackness B, Davies GK, Turkie W, Lee E, Roberts DH, Hill E, Roberts C, Durrington PN, & Mackness MI (2001). Paraoxonase Status in Coronary Heart Disease: Are activity and concentration more important than genotype? Arteriosclerosis, Thrombosis, and Vascular Biology, 21(9), 1451–1457. 10.1161/hq0901.094247 [DOI] [PubMed] [Google Scholar]
- Marsillach J, Ferré N, Vila MC, Lligoña A, Mackness B, Mackness M, Deulofeu R, Solá R, Parés A, Pedro-Botet J, Joven J, Caballeria J, & Camps J. (2007). Serum paraoxonase-1 in chronic alcoholics: Relationship with liver disease. Clinical Biochemistry, 40(9–10), 645–650. 10.1016/j.clinbiochem.2007.01.020 [DOI] [PubMed] [Google Scholar]
- Marsillach J, Mackness B, Mackness M, Riu F, Beltrán R, Joven J, & Camps J. (2008). Immunohistochemical analysis of paraoxonases-1, 2, and 3 expression in normal mouse tissues. Free Radical Biology & Medicine, 45(2), 146–157. 10.1016/j.freeradbiomed.2008.03.023 [DOI] [PubMed] [Google Scholar]
- Marsillach J, Camps J, Beltran-Debón R, Rull A, Aragones G, Maestre-Martínez C, Sabench F, Hernández M, Castillo DD, Joven J, Mackness M, & Mackness B. (2011). Immunohistochemical analysis of paraoxonases-1 and 3 in human atheromatous plaques. European Journal of Clinical Investigation, 41(3), 308–314. 10.1111/j.1365-2362.2010.02411.x [DOI] [PubMed] [Google Scholar]
- McGown EL, Nelson JW, Williams J, & Fraser C. (1997). UV-VIS Spectrophotometry: Automated Determination of Pathlength in Microplate Samples. American Laboratory, 29(10), 21–24. [Google Scholar]
- Mychaleckyj JC, Farber EA, Chmielewski J, Artale J, Light LS, Bowden DW, Hou X, & Marcovina SM (2011). Buffy coat specimens remain viable as a DNA source for highly multiplexed genome-wide genetic tests after long term storage. Journal of Translational Medicine, 9, 91. 10.1186/1479-5876-9-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer EA, Pezzulo A, Shih DM, Chun C, Furlong C, Lusis AJ, Greenberg EP, & Zabner J. (2005). Human and murine paraoxonase 1 are host modulators of Pseudomonas aeruginosa quorum-sensing. FEMS Microbiology Letters, 253(1), 29–37. 10.1016/j.femsle.2005.09.023 [DOI] [PubMed] [Google Scholar]
- Palmer KF, & Williams D. (1974). Optical properties of water in the near infrared. Journal of the Optical Society of America, 64(8), 1107–1110. 10.1364/JOSA.64.001107 [DOI] [Google Scholar]
- Parra S, Alonso-Villaverde C, Coll B, Ferré N, Marsillach J, Aragonès G, Mackness M, Mackness B, Masana L, Joven J, & Camps J. (2007). Serum paraoxonase-1 activity and concentration are influenced by human immunodeficiency virus infection. Atherosclerosis, 194(1), 175–181. 10.1016/j.atherosclerosis.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Richter RJ, & Furlong CE (1999). Determination of paraoxonase (PON1) status requires more than genotyping. Pharmacogenetics, 9(6), 745–753. [PubMed] [Google Scholar]
- Richter RJ, Jampsa RL, Jarvik GP, Costa LG, & Furlong CE (2004). Determination of paraoxonase 1 status and genotypes at specific polymorphic sites. Current Protocols in Toxicology, Chapter 4, Unit4.12. 10.1002/0471140856.tx0412s19 [DOI] [PubMed] [Google Scholar]
- Richter RJ, Jarvik GP, & Furlong CE (2008). Determination of paraoxonase 1 status without the use of toxic organophosphate substrates. Circulation. Cardiovascular Genetics, 1(2), 147–152. 10.1161/CIRCGENETICS.108.811638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter RJ, Jarvik GP, & Furlong CE (2009). Paraoxonase 1 (PON1) status and substrate hydrolysis. Toxicology and Applied Pharmacology, 235(1), 1–9. 10.1016/j.taap.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suehiro T, Nakamura T, Inoue M, Shiinoki T, Ikeda Y, Kumon Y, Shindo M, Tanaka H, & Hashimoto K. (2000). A polymorphism upstream from the human paraoxonase (PON1) gene and its association with PON1 expression. Atherosclerosis, 150(2), 295–298. 10.1016/s0021-9150(99)00379-2 [DOI] [PubMed] [Google Scholar]