Abstract
Individualizing patient treatment is a core objective of the medical field. Reaching this objective has been elusive owing to the complex set of factors contributing to both disease and health; many factors—from genes to proteins—remain unknown in their role in human physiology. Accurately diagnosing, monitoring, and treating disorders requires advances in biomarker discovery, the subsequent development of accurate signatures that correspond with dynamic disease states, as well as therapeutic interventions that can be continuously optimized and modulated for dose and drug selection. This work highlights key breakthroughs in the development of enabling technologies that further the goal of personalized and precision medicine, and remaining challenges that, when addressed, may forge unprecedented capabilities in realizing truly individualized patient care.
Keywords: Personalized Medicine, Precision Medicine, Therapeutics, Diagnostics, Artificial Intelligence, Clinical Trials
Converging genomic and phenotypic medicine with technology
Personalized and precision medicine seek to build a foundation for actionable health management through a broad spectrum of information. Potential inputs for advancing precision medicine include longitudinal tracking of healthy individuals to better understand the transition from non-diseased to diseased states; more precisely identifying individuals at risk for disease; and tailoring treatments based on diverse and growing data sets from both individual trials and population-based studies [1]. The data flowing into precision medicine will come from genetic databases, medical records, tissue banks, and other clinical sources of “big data.” In parallel with our ability to gather an impressive amount of data from any given patient is the expansion of computing power and, with it, our analytical capabilities and ability to link data sets together to “make sense” of big data. Precision medicine relies on large quantities of population-level data to help determine the appropriate treatment for an individual. Precision medicine, in some respects, deviates from traditional medicine by providing insight into how population-derived genetic, proteomic, or broader biomarker profiles can collectively determine the course of treatment for an individual (Box 1). We should think of human disease as involving “networks” of aberrant activities, such as signaling and cell behavior, as well as modifier genes, “moonlighting” proteins, etc., rather than as a linear progression [2]. In this way, we might uncover new linkages between seemingly disparate diseases that open doors to novel treatment approaches. Precision medicine is already using different sources of patient data to define these new linkages, whether it be through genomics in type 2 diabetes or imaging phenotypes in glioblastoma, for instance [2–4]. There are notable challenges to precision medicine that include data accrual, access to patients, and data analysis, which we will address in subsequent sections.
Text Box 1: Defining Precision and Personalized Medicine.
From an engineering perspective, precision medicine involves the use of technologies to acquire and validate population-wise data, such as -omics based single cell analysis and biomarker discovery, for subsequent application on the individual patient level.
Personalized medicine involves the use of technologies to seriously acquire and assess an individual’s own data for only their own treatment. For example, this may involve the use of artificial intelligence (AI) to both design a drug combination based on a patient’s own biopsy followed by N-of-1 dosing protocols.
Broad deployment of both approaches would rely on their successful integration, with genome-guided drug pairing (driven by population data) followed by AI-guided dynamic dosing (driven by individual data).
Personalized medicine differs from precision medicine. In personalized medicine, which has been considered for some time, the patient is viewed as an individual from diagnosis to therapy, which clearly has some overlaps with precision medicine in execution, but unlike precision medicine does not inherently rely on large data sets or population-based approaches to redefine disease (Box 1). In personalized medicine, engineers have found a stronghold. Engineers have developed enabling technologies ranging from micro/nanofluidics for single circulating tumor cell (CTC, see Glossary) analysis to nanotechnology for isolating extracellular vesicles and exosomes from liquid biopsies to imaging platforms to predict nanomedicine’s effectiveness [5, 6]. These technologies have the opportunity to provide unprecedented diagnostic insight on a personalized level, but are still making their way into routine clinical care. In particular, personalized pharmacokinetics has enabled new intratumoral devices and microfluidic constructs available to track an individual’s response to a particular drug [7, 8]. The recent introduction of phenotypic personalized medicine (PPM) — the harnessing of augmented artificial intelligence (AI) to personalize combination therapy and improve efficacy and safety on the basis of measured end-point phenotypes for specific patients — has enabled continuous, patient-specific optimization of monotherapy and combination therapy as well as the agnostic design of novel, optimized, fixed-dose drug combinations [9, 10]. Pilot clinical trials involving PPM have been launched for tuberculosis, HIV (NCT02632474), liver and kidney transplant immunosuppression, hematologic cancers, and other indications, demonstrating the power of engineering platforms to cut across medicine. To enable patient-specific treatment, a growing pool of technologies is enabling individualized monitoring. These include everything from wearables, like glucose-monitoring, neurostimulation, neuromonitoring, and sweat-monitoring electronic tattoos—representing ‘Small Data’ personalized diagnostics and analytics—to AI-based drug selection and administration; to patient-tailored gene-editing nanoparticles that can both image (diagnostic) and treat (therapy), sometimes called “theranostics” [11–15]. As these data sources grow in scale as well as quality, being richer and more diverse, medicine will be able to more effectively individualize therapy on the basis of population-wide genetic and phenotypic information, with the hope of moving beyond medicine’s ‘one size fits all’ modus operandi.
Bioengineering underpins the implementation of precision and personalized medicine and related enabling technologies in the clinic (Table 1), as knowledge obtained from traditional biomedical fields makes its way into clinical solutions via engineering tools and approaches. Engineers will play an especially crucial role in advancing both personalized and precision medicine, although they may not yet realize how their enabling technologies offer solutions to many unmet clinical needs. In this paper, we explore recent advances in enabling technologies and state-of-the-art bioengineering approaches that will be instrumental in not only continuing to advance personalized and precision medicine into the clinic, but also catalyze its transformation into the standard of care. We also discuss policy considerations that will impact the development, implementation, and adoption of such technologies.
Table 1.
Clinicaltrials.gov identifiers of clinical trials that are assessing emerging technologies for personalized and precision medicine.
| Technology Platform | Status | Key Details of Study | Clinicaltrials.gov Identifier |
|---|---|---|---|
| Artificial Intelligence | Phase II | Harnessing CURATE.AI to optimize N-of-1 combination therapy in multiple myeloma | NCT03759093 |
| Artificial Intelligence | Interventional | Developing N-of-1 training trajectories via CURATE.AI | NCT03832101 |
| Artificial Intelligence | Phase IV | Human Immunodeficiency Virus (HIV): Optimization of dose reduction of Tenofovir (TDF) in antiretroviral therapy (ART). | NCT02632474 |
| Artificial Intelligence | Phase II | Liver and kidney transplant: Dynamic modulation of therapeutic dosing to optimize immunosuppression. | NCT03527238 |
| Artificial Intelligence | Interventional | Radiotherapy: Image-guided adaptive radiotherapy is being studied to modulate intensity to reduce side effects and improve outcomes. | NCT04022018 |
| Genomic Analysis/Companion Diagnostics |
Feasibility | Non-small cell lung cancer (NSCLC): Co-clinical trials with genetic mouse models, NSCLC patient specimens (plasma, serum, tissue), and healthy patient samples to develop liquid biopsies. | NCT02597738 |
| Genome-Guided Therapy | Observational | Pain: Assessment of the cytochrome P450 2D6 (CYP2D6) and μ-opioid receptor gene (OPRM1) genoypes in guiding pain management. | NCT02664350 |
| Genome-Guided Therapy | Observational | Preterm birth: Genomic profiling to predict patient response to 17 hydroxyprogesterone caproate (17OHPC) therapy to prevent preterm birth | NCT02173210 |
| Genome-Guided Therapy | Phase II | Breast cancer: Biomarker analysis and genetic screening to develop individualized treatment regimens. | NCT02624973 |
| Genome-Guided Therapy | Phase II | Multiple tumor types: Matching of drugs to patients based on genomic alterations. | NCT02795156 |
| Genome-Guided Therapy | Phase III | Prostate cancer: Circulating tumor DNA is being used as a biomarker for genome-guided drug selection. | NCT03903835 |
| Microfluidics/Companion Diagnostics | Observational | Multiple Cancer Types/Circulating Tumor Cells: Correlation of titre with treatment response and progression for nasopharyneal carcinoma, breast cancer, colorectal cancer, prostate cancer, and gastric cancer | NCT01022723 |
| Microfluidics/Companon Diagnostics | Observational | Clear Cell Renal Cancer/Circulating Tumor Cells: Comparison of microfluidic technology that can detect CTCs under low epithelial cell adhesion molecule (EpCAM) presence compared to EpCAM-based detection platform for clear cell renal cancer. | NCT02499458 |
| Microfluidics/Companion Diagnostics | Observational | Metastatic Breast Cancer/Circulating Tumor Cells: Application of antibody cocktails to capture CTCs from metastatic breast cancer for microfluidic analysis. | NCT02904135 |
| Genome Editing | Phase I | Mucopolysaccharidosis II (MPS II): Intravenous delivery of SB-913, a zinc finger nuclease, to enable production of Iduronate 2-Sulfatase (IDS) enzyme from the liver. | NCT03041324 |
| Genome Editing | Phase I | Mucopolysaccharidosis I (MPS I): Intravenous delivery of SB-318, a zinc finger nuclease, to insert α-L-iduronidase (IDUA) gene to enable liver-mediated enzyme production. | NCT02702115 |
| Genome Editing | Phase I | Severe Hemophilia: Intravenous delivery of SB-FIX, a zinc finger nuclease, to insert the SB-FIX gene to enable Factor IX clotting factor production. | NCT02695160 |
| Genome Editing/Cell Therapy | Phase I | Human Immunodeficiency Virus (HIV): Administration of zinc finger nuclease CCR5-modified hematopoietic stem/progenitor cells (SB-728mR-HSPC) in patients with undetectable disease/low CD4+ counts. | NCT02500849 |
| Nanomedicine/RNA Therapy | Phase 0 | Glioblastoma: Administration of NU-0129, a spherical nucleic acid (SNA) therapy to target the Bcl2L12 gene to arrest tumor growth. | NCT03020017 |
| Nanomedicine | Phase I | Cervical cancer: Polysiloxane gadolinium chelates are delivered in combination with cisplatin and radiation therapy to address locally advanced disease. | NCT03308604 |
| Wearables | Observational | Perioperative Risk Assessment: Pairing wearable (Garmin VivoSmart HR+) with cardiopulmonary exercise testing (CPET) indicators to develop broadly applicable risk assessment protocol for high-risk elective surgery patient population. | NCT03328039 |
| Wearables | Observational | Chronic obstructive airway disease (COPD): Application of wearable on finger for heart rate variability and oxygen saturation to detect and manage COPD in patients over 60 years in age. | NCT03268538 |
| Wearables | Observational | Cardiology: A shirt-based electrocardiogram (ECG) system is being used to monitor the following patient cohorts: 1) Cardiac rehabilitation following myocardial infarct; 2) Pediatric superventricular tachycardia (SVT); 3) Post-pulmonary vein isolation (PVI); 4) Cardiac resynchronization therapy patients. |
NCT03068169 |
| Digital Health | Observational | Alzheimer’s Disease: Cognitive battery software was used to predict dementia onset. | NCT03676881 |
| Digital Health | Interventional | Health eBrain Study: Application of neurocognitive assessment mobile health software to assess brain health combined with Mindoula intervention to address dementia and depression in Alzheimer’s Disease caregivers. | NCT02903862 |
| Biomaterials | Phase II | Wound healing: A nanodiamond-based biomaterial platform to mediate peri-apical wound healing and prevention of re-infection following root canal therapy. | NCT03376984 |
| Biomaterials | Phase I | 3-D Printing: Personalized 3-D printing of polycaprolactone (PCL) is being explored for breast reconstruction after tumor removal. | NCT03348293 |
| Electronic Health Records/Infrastructure/ Genotyping |
Observational | Pain: CYP2D6 genotyping for pain management/control and integration of pain questionnaire in electronic health record for physician guidance. | NCT02335307 |
Together, personalized and precision medicine, enabling technologies, and smart policy choices will open the doors to optimized targeted therapies, to disease prevention, and to an overall shift in how we think about human disease.
Enabling technologies for precision medicine
The field of precision medicine has already provided important insights into the mechanisms at play at disease onset; into biological targets that can directly inhibit disease progression; and into biomarkers that reflect treatment response. Such understanding has collectively mediated substantial advances towards improving patient treatment outcomes [16–18]. With new data sources and combinations of these data, precision medicine will propel existing drug-selection platforms like pharmacogenomics and patient-derived primary cultures [19–25]. When coupled with big data platforms, these approaches can identify targeted therapies that may predict and/or induce improved response rates over clinical standards. Importantly, these advances are being actively assessed in a clinical setting (NCT02795156, NCT03903835).
Several clinical trials are currently in place to bring precision medicine to the bedside. One study of lung cancer patients is pairing genomic analysis of murine and human specimens, and coupling these data with imaging analysis in a co-clinical trial to identify genomic signatures to improve liquid biopsies (NCT02597738). A precision-guided study for treating cancer pain is being conducted, where patients are screened for the cytochrome p450 2D6 (CYP2D6) and μ-opioid receptor (OPRM1) genotypes to monitor their response to opioid treatment (NCT02664350). Precision medicine is being used to identify genomic and molecular markers to predict patient response to the administration of 17 hydroxyprogesterone caproate (17OHPC) as a potential intervention to prevent preterm birth (NCT02173210). Lastly, one study is combining four predictive markers for breast cancer (HER2; TP53, CHEK2 and RB1) with massive parallel genetic screening to develop datasets that can be leveraged to individualize treatment regimens (NCT02624973).
Precision medicine, however, goes beyond genomic medicine. Instead, it is a “confluence of biological, physical, engineering, computer, and health sciences… toward data-driven, mechanism-based health and health care for each individual” [26]. As such, precision medicine looks at a host of data across a population and defines, on the basis of areas like big data, patient response to a specific disease state and resulting interventional strategies. It is here that engineering plays a powerful role in linking these different “layers”, to redefine disease and search for meaningful mechanistic underpinnings that can inform next-generation therapies.[26] We see engineers continuing to contribute to precision medicine the in the following ways: by enabling biomarker discovery, by creating diagnostic and sensing platforms, and by devising novel drug delivery methods to get precision therapeutics to the patient (Figure 1) [27–37].
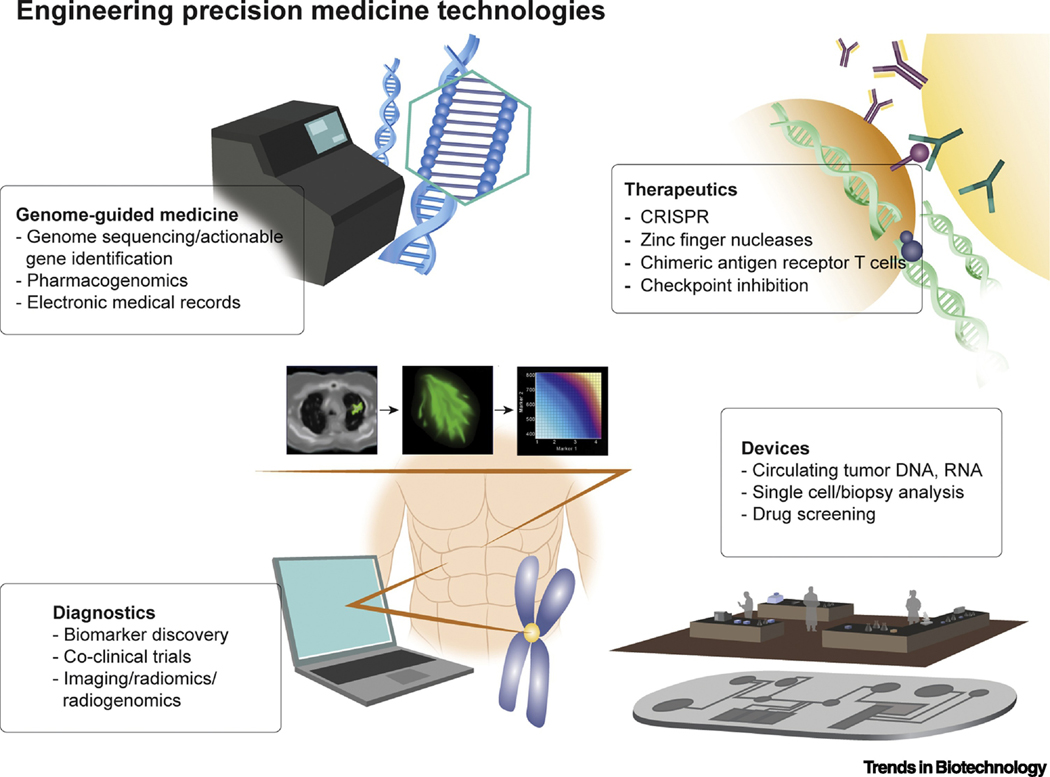
Figure 1. Engineering Precision Medicine Technology Platforms.
From genome-guided medicine to CRISPR, a broad spectrum of technology platforms that bridge engineering with precision medicine are poised to impact clinical outcomes.
Biomarker discovery
Engineering approaches to biomarker discovery and validation have resulted in the development of signatures that may eventually serve as dynamic indicators of patient response to therapeutic intervention [38–43]. The genomics revolution and advent of approaches such as next-generation sequencing (NGS) has played a major role towards broad implementation of precision medicine in the clinic [44, 45]. The fruition of this capability would be vital to enabling individualized treatment regimens in a patient-specific manner. For example, single molecule array (SiMoA) technology, which allows for single molecules to be sequestered by paramagnetic beads for digital readouts in microwells, was able to detect changes in prostate-specific antigen (PSA) levels that spanned orders of magnitude between individual prostate cancer cells [46]. Fiber microarrays have also been produced for multiplexed diagnostics. In these microarrays, a solution of microbeads that contains analogs for different target molecules can be interrogated with an imaging fiber, with etched microwells in the fiber providing positive signals for successfully detected biomarkers [47]. This platform was used to develop a potential signature for cystic fibrosis (CF) using saliva. Via the fiber microarrays, CF patients were observed to have substantially elevated vascular endothelial growth factor (VEGF), interferon-gamma inducible protein (IP-10), interleukin-8 (IL-8), and epidermal growth factor (EGF) levels, while matrix metallopeptidase-9 (MMP-9) levels were lower. The six-biomarker signature, captured by the highly sensitive and multiplexed fiber microarray, was capable of differentiating patient subgroups, and correlated well with the forced expiratory volume (FEV1) readout for disease severity [47]. In addition, long term monitoring of inflammatory cytokines in healthy individuals was conducted, revealing a subset of cytokines that can vary by as much as two orders of magnitude between subjects, while others had substantially lower levels of inter-patient variability [48]. This study demonstrated the importance of taking baseline studies of patient biomarker signatures as way towards highly accurate monitoring of the dynamic states of disease progression and/or treatment response.
Microfluidic technologies have a long history of providing the biomedical community unique, modular platforms for detecting low levels of biomarker in small volumes of fluid. In one instance, a microfluidic device was used to deplete hematopoietic stem cells, thereby allowing circulating tumor cells (CTCs) associated with hepatocellular carcinoma (HCC) to be easily detected. With a pure sample, an RNA-based signature was developed using 10 liver-based transcripts, subsequently resulting in the detection of HCC-derived CTCs in 9 of 16 untreated patients with HCC as well as 1 of 31 patients that had non-malignant liver disease. It is important to note that the digital scoring process also did not correlate to commonly monitored serum alphafetoprotein (AFP) levels, demonstrating the importance of the RNA-based signature in accurately detecting and assessing disease states [49]. This RNA signature would not have been possible without the sorting and purifying capabilities of the microfluidic device.
In addition to the aforementioned approaches, a number of studies have harnessed single cell analysis using approaches such as Raman micro-spectroscopy and multi-plexed imaging using laser particles for biomarker discovery [50–52]. Peptide arrays are now capable of monitoring low-affinity protein-ligand interactions that may not have been captured by conventional methodologies, such as Fourier-transform infrared spectroscopy (FTIR) or surface plasmon resonance spectroscopy, opening doors to novel monitoring and drug discovery strategies [53, 54].
Precision diagnostics and biosensing
A key barrier that confronts precision medicine is the inability, in many cases, to assess quantifiable treatment outcomes due to inadequate biomarkers. This is especially true for solid cancers and certain infectious diseases, such as tuberculosis, and for other indications, such as pain and brain injury. Recent capabilities pertaining to individualized care could especially benefit from frequent biomarker analysis in order to prescribe accurately calibrated treatment responses. Emerging technologies will open the doors to biomarker surveillance with high sensitivity and specificity, as well greater frequency than conventional monitoring approaches such as imaging (Computed tomography (CT), magnetic resonance imaging (MRI)) or blood draws. For example, microfluidics and nanofluidics can process and analyze in parallel nucleic acids, proteins, metabolites, and single cells in various biological fluids, such as urine, blood, and saliva, which increases the frequency of outcome monitoring—a valuable resource for personalized dosing and pharmacokinetics. In fact, guided therapy using microfluidics or nanofluidics for genomic analysis and CTC isolation and characterization for biomarker discovery is starting to make its way into the clinic. CTCs have been isolated using microfluidics from lung adenocarcinoma, undifferentiated Epstein-Barr Virus (EBV) positive nasopharyneal carcinoma, breast cancer, colorectal cancer, prostate cancer, and gastric cancer to develop predictive biomarker panels for the downstream selection of therapeutic regimens (NCT01022723). Circulating endothelial cells and CTCs are also being evaluated as biomarkers for renal cancer (NCT02499458). A study to capture and analyze CTCs from metastatic breast cancer patients using antibody panels and microfluidics is also being conducted (NCT02904135) [5, 55–61].
Surprisingly little is known about the evolution, origin, and metastasis of cancer. Single-cell analysis, an area that is maturing rapidly, promises to deliver new disease insights by parsing out the potential role of cellular heterogeneity in health and disease, including cancer [62, 63] (Figure 1). For example, digital real-time polymerase chain reaction (RT-PCR) for single-cell transcription-factor profiling in hematopoietic stem cells (HSCs) revealed the presence of two HSC subpopulations from a specific type of progenitor cell that were believed to be homogeneous. This finding made it clear that gene expression could be assessed in a copy-number-per-cell context, providing extraordinary insight into transcriptional regulation [57]. Single cell genomics has also used to analyze the clonal structure of tumors in both colon cancer [64] and leukemia [56, 65], and the literature using this approach is growing by leaps and bounds.
Engineers have found ways to peer into biological processes – sometimes in “real-time” – to uncover novel insights into human disease progression. For instance, real-time imaging of colon cancer cell migration in mice has revealed clonal liver metastasis, providing valuable insight into early detection and subsequent treatment of advanced cancers [66]. Imaging dormant breast cancer cells in mice has shown that vascular triggers are responsible for proliferation and growth, revealing new therapeutic targets in the vasculature [67]. Devices can be used to isolate rare cell populations, in some instances to understand individual cell contributions to disease; in others, to make a diagnosis or therapeutic decision. In a recent study using blood samples obtained from metastatic castration-resistant prostate cancer (mCRPC) patients, the presence of androgen-receptor splice variant 7 (AR-V7) on the CTC surface predicted improved response to taxane therapy over aminoacyl-tRNA synthetase (ARS) inhibition. Of note, these systems used antibody cocktails to label and distinguish CTCs. Digital pathology algorithms were then used to track cellular morphology from highly heterogeneous solutions in order to classify CTC subtypes. Pathway activation could also be assessed through the additional detection of surface markers in multiple myeloma [68, 69]. In addition, this integrated approach was capable of isolating both single CTCs and clusters of CTCs positive for AR-V7 to guide treatment selection.[58] In a clever approach to better understanding lung diseases, a microfluidic device was engineered to analyze individual cell deformability as a means of diagnosing noninvasively whether human pleural effusions were malignant or benign [70].
Although these devices can more readily isolate and analyze single cells, we are still left with piecing together their implications in disease. CyTOF (cytometry by time of flight), which can analyze the contribution of a single cell to a given signalling pathway, addresses this point [71]. By using technologies such as CyTOF to assess complex populations of cells, the role of heterogeneity in driver mutations and drug resistance can be comprehensively interrogated. In addition, longitudinal studies need to be designed to decipher clone behavior as well as spatial and temporal genomic signatures, and to resolve evolutionary principles that underpin cancer as well as infectious diseases and other disorders [72]. As a step towards determining the role of heterogeneity in disease progression and treatment, a recent study performed single-cell triple-omic analysis of the genome, DNA methylome, and transcriptome in a population of hepatoceullular carcinoma cells to assess the larger scale contributions of genomic and epigenomic heterogeneity towards transcriptomic heterogeneity. These capabilities in -omics interrogation may provide extraordinary insight into disease heterogeneity, and enable both the prediction of drug responses and individualized therapy [73].
Sample collection poses many limitations for both personalized and precision medicine. Technologies that can analyze small amounts of rare tissues or molecules will allow us to access information previously lost in the bulk. Liquid biopsies, for instance, which can be used to monitor disease, in near real-time, and perhaps in the future be used to predict drug action and continuously modulate combination therapy. Liquid biopsies could extract CTCs, exosomes, cell-free nucleic acids, proteins, and other biological factors for collection and analysis [5, 68, 74]. This approach may enable the identification of effective treatment outcomes and design criteria for drug-regimen selection and predictors of response to the selected therapies.
Precision therapeutics
Gene editing tools like CRISPR (clustered regularly interspaced short palindromic repeats) and zinc finger nucleases (ZFN) have the ability to alter specific components of a genome, thereby opening new doors to precision repair of defective genes in indications ranging from cancer to HIV. The democratization of gene editing afforded by CRISPR promises to produce large amounts of data that we expect to shed light on new connections between diseases and on new pathways never before implicated in a particular disease. Gene editing has recently found its way into patients [75]. Recently, the ZFN-based genome editing therapeutic SB-913 was clinically administered to patients diagnosed with Mucopolysaccharidosis II (MPS II), or Hunter Syndrome. One of the objectives of this study was to evaluate the impact of SB-913 on leukocyte and plasma Iduronate 2-Sulfatase (IDS) enzyme activity in an effort to mediate lifelong IDS production (NCT03041324). ZFN genome editing therapies are also being tested for MPS I (SB-318, NCT02702115), and severe hemophilia B (SB-FIX, NCT02695160). Clinical trials evaluating CCR5-modified hematopoietic stem/progenitor cells (via ZFNs) in patients infected with HIV-1 have also been initiated (NCT02500849).
Molecularly targeted and antibody therapies have served as cornerstones in precision medicine, have resulted in a broad spectrum of approved drugs and emerging methods of patient-specific drug prioritization via mutation databases [76–78]. Recent genome- and proteome-guided therapeutic strategies that are in preclinical development have included the use of CRISPR with an enzyme, CRISPR-associated protein-9 nuclease (Cas9), to correct genetic aberrations in diseases such as Duchenne Muscular Dystrophy (DMD). Specifically, CRISPR therapy restored dystrophin production and substantially increased neuronal nitric oxide synthase (nNOS) recruitment in a mouse model compared to the sham cohort, an important advance for targeted gene editing [79]. Tumor profiling to assess the expression of programmed cell death protein 1 (PD-1; which is encoded by the PDCD1 gene), PD-L1, and cytotoxic T lymphocyte–associated protein 4 (CTLA-4) from patient samples was used to predict treatment response for melanoma [80]. Several classes of targeted inhibitors have been paired with genomic profiling due to drug mechanisms of action that can specifically enhance treatment outcomes such as progression-free survival. For example, epidermal growth factor receptor (EGFR) exon 19 deletions and exon 21 point mutations served as indicators for subsequent afatinib administration for non-small cell lung cancer (NSCLC) treatment in both treatment-naïve and refractory NSCLC patients [81–83].
Administering CRISPR with improved efficacy can also be mediated using new delivery mechanisms like nanoparticles. A recent preclinical study using CRISPR/Cas9 and a lipid nanoparticle successfully switched off genes responsible for high cholesterol levels, in cells and mice [84]. Gold nanoparticle-mediated delivery of CRISPR-Cas9 was used to enhance the efficacy of correcting a DNA mutation associated with DMD [85]. In addition to mediating guide RNA-based therapy mediated by CRISPR, novel nanocarrier approaches are also being used for RNA interference. For example, spherical nucleic acids (SNAs) are gold nanoparticles that have been recently synthesized to be densely loaded with RNA duplexes for gene silencing. In multiple in vitro and preclinical studies, SNAs were efficiently taken up in a broad spectrum of cell lines, and capable of mediating efficient RNA interference while remaining well tolerated [86–88]. In a recent murine study, SNAs were utilized to knock down the p53 inhibitor, Bcl2Like12 (Bcl2L12), which is overexpressed in glioblastoma, as a novel form of cancer treatment [89]. The SNA platform (NU-0129) was recently translated into the clinic, where a Phase 0 study is currently underway (NCT03020017).
Bioengineering for personalized medicine
Medicine is moving away from a “one size fits all” mentality. We are realizing that patients with the same disease can respond differently to everything from drugs to biomaterials, and it is now time to better understand this response and treat the patient as an individual. Engineers have the opportunity to make personalized medicine a greater reality in the clinic (Figure 2). Take biomaterials, for instance. Researchers have found that a dextran-dendrimer composite works as an adhesive differently in not only different organs, but also differently in the same organ under different environments (colon cancer versus colitis, for example) [90, 91]. These findings suggest that the application of most biomaterials cannot be generalizable and that the disease indication and organ environment matters in the design of materials that are to reside temporarily within the body. Indeed, biomaterials can have a differential effect on cell fate, survival, and growth, yet there is a dearth of high-throughput approaches to screen for the optimal material composition for a given application. One approach is to use a combination of small and large animal models to determine the best polymer carrier for islet transplantation [92–94]. An approach that avoids animal models altogether looked at how different extracellular matrix (ECM) formulations (different organs, different processing methods) affected stem cell differentiation, cancer cell proliferation, and cell apoptosis [95]. Engineering new platforms for narrowing down the optimal biomaterial formulation could improve personalized biocompatibility and therapeutic outcomes. Organ-on-a-chip platforms are being explored for potential individualized drug screening and toxicology studies. For example, a microfluidics-based model of human intestine was recently developed, where the complex gut microenvironment was recapitulated, allowing for the monitoring of interactions between the gut microbiome, bacteria, and immune cells. The ability to replicate organ-scale complexity using platforms such as this intestine model opens the doors to pharmacokinetics, absorption, and drug metabolism analysis as they relate to drug development and toxicity studies [96, 97]. These capabilities may also serve as the foundation for designing personalized treatments. To accurately personalize treatment, which includes parameters such as drug selection and drug dosing, accurate and timely detection of treatment response is needed. Conventional approaches to acquire these readouts include serum and urine analysis, or imaging modalities such as ultrasound, CT, MRI, and x-rays, among others, which may be limited in terms of testing frequency, especially if treatment response varies on a timescale of hours, or even minutes. Wearables and other classes of emerging technologies may overcome the challenges of infrequent measurements to improve the accuracy of treatment response assessment, thereby improving the design of personalized interventions [98]. As the field of personalized medicine continues to progress, new areas of development have included the use of non-drug based platforms such as digital therapy to address conditions such as mild cognitive impairment, substance abuse, mental health, and attention deficit hyperactivity disorder, among others [99–101].
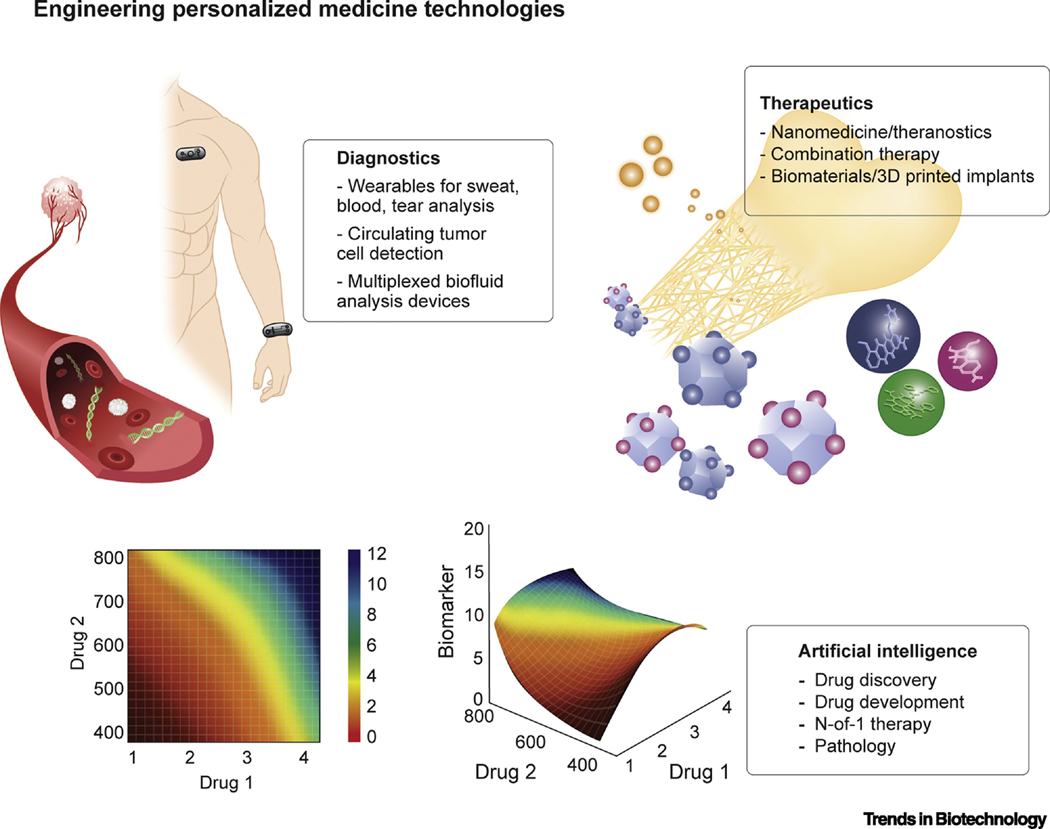
Figure 2. Engineering Personalized Medicine Technology Platforms.
By bridging wearable technologies with artificial intelligence and other engineering platforms, marked enhancements in the development of individualized treatment and monitoring may be realized.
A unifying attribute of these approaches is their potential towards using only a subject’s own data to manage only their own care. This has been apparent with regards to artificial intelligence-driven drug dosing as well as engineered cell therapy. Furthermore, a common attribute at the intersection of engineering and personalized medicine is that intervention and diagnosis can be adjusted in a serial fashion for ongoing treatment optimization.
Artificial intelligence, machine learning, and personalized treatment
A major barrier to the optimization of any targeted therapy has been an inability to determine drug doses that are best suited for an individual patient, especially since dosing requirements can often change during the course of treatment. In the case of combination therapy, this is even more challenging given the virtually infinite dosing parameter space that exists. The same quandary also confronts the concept of pinpointing population-optimized drug-dose ratios. To overcome this challenge, conventional patient dosing and drug development has been guided by first identifying the drugs to be administered, following by dose escalation-based trials to identify the maximum tolerated dose (MTD). To increase the chances of successful treatment outcomes, drug-synergy predictive modelling and other in silico methods have been employed [102]. However, drug synergy and drug antagonism are dose-dependent, and not all genes involved in the uptake, metabolism and activity of any specific drug are known. Therefore, incorrect dosing can substantially decrease or eliminate the efficacy of drug combinations whereas optimized drug–dose ratios can make normally ineffective drugs become potent when co-delivered.
In the drug and dose optimization arena, there remains a perception that correct dosing is important solely for improving efficacy and safety. What is commonly misunderstood is that correct dosing directly impacts the drugs that are selected for treatment in the first place [103, 104]. Furthermore, in lieu of predictive drug discovery, synergy modeling, or monotherapy or combination therapy selection, AI and machine learning (ML)-related platforms are now capable of deterministically identifying, in a continuous fashion, the drug administration parameters [9, 105, 106]. More specifically, the implementation of novel AI-based platforms is precisely demonstrating the key relationship between drug dose and drug selection by reconciling the virtually infinite parameter space created by these factors. AI is also being leveraged to enhance imaging capabilities in the area of diagnostics to further guide patient-specific treatment. In addition, the application of AI is emerging in the area of therapeutics to simultaneously pinpoint the best drugs and doses, even from very large pools of candidate compounds, for optimal combination therapy efficacy and safety [107–118]. In a recent study using the deterministic AI-based quadratic phenotypic optimization platform (QPOP) platform, novel drug combinations were identified from a pool of oncology drug candidates without the need for synergy-based predictions or complex disease mechanism information. In addition, QPOP was used to re-optimize the drug combination dose parameters from the in vitro through preclinical stages of development. These AI-discovered combinations substantially outperformed the clinical standards of care [117]. While QPOP implementation did not require disease mechanism modelling, global optimization via this platform can lead to new insights in the pathways that mediate globally optimized outcomes for comparison with conventional target-based drug combination design. This could in turn lead to the addition of new drug candidates in the combination therapy design pool. Approaches such as QPOP require marked reductions in data requirements as they are based on drug and dose inputs and quantifiable clinical indicators of efficacy and safety (e.g. tumor burden, toxicity panels). This emerging capability of mechanism-agnostic optimization, as well as a continuously increasing body of knowledge in the genome-driven drug selection arenas are expected to address some of the barriers that have been previously encountered with trials such as SHIVA, where molecular alterations serve as a foundation for drug matching. In this previous study, the factors determining drug-patient pairing were driven by an algorithm prioritizing molecular alterations and drug selection, among other factors. Lessons learned from this study, and suggestions provided by the clinical community have shed light on the potential of patient stratification using oligo-mutant tumors in lieu of multi-mutant tumors, while additional suggestions included the use of combination therapy when multiple molecular alterations were identified.
In combination therapy, the ‘sweet spot’ of an optimized drug-dose ratio and the best drugs that comprise the multi-drug treatment is almost always overlooked when dose escalation and MTD are employed [10, 106, 119–124]. As such, the recent clinical application of augmented AI optimization platforms has overcome this challenge (NCT03759093, NCT03832101, NCT02632474, NCT03527238) [105, 106, 119]. In a recent prospective trial, a new technology platform based on PPM successfully personalized post-transplant immunosuppression [9]. The foundation of this capability is based on the PPM-enabled discovery that drug administration (input) can be correlated to phenotypic output (e.g. tumor burden, bacterial/viral load) through a phenotypic response surface (PRS). Without using algorithms or modelling, PPM optimization effectively calibrated optimized dosing regimens for each patient enrolled in the study. Furthermore, patients were continuously calibrated to dynamically adjust their dosing guidelines for the entire duration of care, an unprecedented capability [9, 105]. Intra-patient dose dependent drug synergy and antagonism was also observed, further demonstrating the importance of optimizing personalized care.
These studies have since been expanded to include the optimization analysis of combination therapy regimens in pediatric acute lymphoblastic leukemia, development of improved combination therapies that maintain undetectable viral loads with substantial dose reduction in patients with HIV, as well as optimization of immunosuppression dosing (NCT02632474, NCT03527238) [106]. Recently, CURATE.AI was harnessed to optimize N-of-1 combination therapy for an advanced prostate cancer patient that resulted in a marked reduction in drug dosage that increased treatment efficacy [125]. In addition, CURATE.AI was recently applied to the emerging field of digital therapeutics, where N-of-1 profiles were constructed for cognitive training (NCT03832101) [99]. In addition to to AI-driven drug treatment, adaptive radiotherapy has also been explored, where images are being used to modulate intensity to reduce treatment toxicity enhance local disease control (NCT04022018). These studies are examples of how AI can expand and optimize an arsenal of interventions against many types of diseases.
As novel platforms for patient-specific testing and treatment continue to evolve, the implementation of AI is expected to expand. For example, organ-on-chip, single cell interrogation platforms, and powerful diagnostic modalities may represent ideal testbeds for N-of-1 or population-scale drug development [126–133]. Specifically, organ-on-a-chip platforms are capable of replicating complex function ranging from the gut, to placental transport and pulmonary function in both healthy and diseased systems [96, 97, 127, 134–136]. Recent advances in organ-on-a-chip development have opened the doors to their applications in drug screening. Since combination therapy is a mainstay for indications ranging from oncology to infectious diseases, among others, these platforms can be integrated with AI-based screening strategies that reduce the number of assays needed to identify optimized drug combinations [110, 120]. In turn, this can accelerate the clinical validation of novel treatment approaches.
Wearable and implantable sensors
Individualizing drug and dose parameters for indications ranging from pulmonary hypertension to cancer therapy require constant and robust readout monitoring. To this end, wearable technology will play a key role in mediating personalized therapy. Capacitive coupling-based flexible electronics that do not require contact between cardiac tissue and metals were recently used for electrophysiological measurements in rabbits.[137] Wearable, battery-free devices for blood oxygenation and heart rate measurements have also been developed [138, 139]. Soft electronics for the analysis of sweat, tears, skin interstitial fluid, and saliva link non-invasive measurements with blood concentrations of these analytes [140–144]. These platforms can be used for glucose sensing and cystic fibrosis monitoring without the barriers associated with fingerstick testing [145, 146]. The clearance of the MiniMed 670G (Medtronic), a closed-loop artificial pancreas, has enabled the algorithm-driven delivery of basal insulin to control blood glucose levels for type 1 diabetes. Recent reporting of real-world data revealed an increased time within the target glucose range with reduced incidence of hyperglycemia and hypoglycemia [147]. Microfluidics-embedded polymer wristbands have also been developed for pulse measurements [148]. Silicone microfiber tubes embedded with electrodes for blood pressure, blood vessel stiffness, and heart rate monitoring have been developed, allowing for non-invasive diagnosis and monitoring of atherosclerosis, veinous ulcers, and potential integration into bandages for diverse applications [149]. Such non-invasive, multiplexed monitoring may ultimately identify personalized biomarker signatures [150].
Wearable devices are being tested in the clinic to potentially predict perioperative risk in patients undergoing high-risk surgery by comparing wearable data (e.g. heart rate and exercise intensity) with cardiopulmonary exercise testing (CPET) standards (NCT03328039). Wearables are also being studied to detect and managed chronic obstructive airway disease (COPD)(NCT03268538). A wearable shirt-based electrocardiogram (ECG) measurement system can detect arrhythmias, and could serve as a complementary real-time readout monitor for dynamically administered cardiovascular therapies (NCT03068169).
In the area of neurocognitive assessment, FDA-cleared mobile application software is also being used to interrogate brain health for the development of interventional solutions in areas like dementia, depression, and Alzheimer’s disease (NCT02903862, NCT03676881) [151]. Studies employing digital medicine in clinical aging research are increasing since several methodological barriers limit conventional in-clinic test administration. First, in-clinic testing is relatively infrequent. In most studies, assessments typically occur on an annual basis in one extended testing session. Second, day to day fluctuations in participant stressors, sleep patterns and time of day effects, and the artificial nature of testing environments can strongly influence cognition, and these sources of variability may be exacerbated in participants with AD pathology [152]. Finally, the cognitive tests most sensitive to the earliest declines in AD also demonstrate high levels of day-to-day variability and can have substantial retest effects [153, 154]. Smartphone assessment approaches afford novel opportunities to overcome these barriers by leveraging the increasing adoption of smartphones by older adults to mitigate the temporal, geographic, space and personnel constraints imposed by in-person testing. Tests are administered in a measurement burst design, in which a “burst” of brief (30–60 seconds per test) cognitive tests are completed at random intervals several times per day over the course of one week on participant’s personal smartphones. One study “visit” thus represents the average performance across 7 days of assessments [155]. Advantages include more frequent measurements at different times of day and a detailed evaluation of variability and practice effects within a day and over a week, which may also serve as sensitive indicators of disease stage [154, 156, 157]. As such, markedly increasing the frequency and scalability of data collection using wearable and mobile health enhances data actionability, reducing the need to use data that has been acquired using conventional approaches, and averaged over several months. When paired with therapeutic strategies that have been properly tailored to this data, wearables and smartphone-based approaches for continuous and robust health monitoring will play a key role in mediating personalized therapy.
Data collection by wearables could allow for patients and their families to take action on their own health and also avoid frequent trips to the doctor to have biomarker analyses performed using conventional approaches. As these data troves grow, too, we can expect a major contribution beyond personalized medicine to precision medicine, opening up new treatment ideas and options for many patients.
Personalized cell therapy and drug delivery
The recent regulatory approval of chimeric antigen receptor-T (CAR-T) cell immunotherapy represents a major advance for personalized cancer treatment. The first of the approved CAR-T therapies, Tisagenlecleucel (Kymriah, Novartis), was developed to treat acute lymphoblastic leukemia in patients 25 years old and younger, and involves removing a patient’s T cells, sending them to a processing facility for reprogramming and expansion, and returning them to the doctor for introduction to the patient [158]. Axicabtagene ciloleucel (Yescarta, KITE Pharma/Gilead Sciences) was recently approved for the treatment of aggressive non-Hodgkin’s lymphomas, including diffuse large B-cell lymphoma (DLBCL) [159, 160]. A global effort to expand the indications that are treated using CAR-T is underway. Furthermore, the regulatory approval of CAR-T therapy represents a paradigm shift for the U.S. FDA, and a gateway towards the continued enhancement of the efficacy and safety of living cell therapies. For example, recent studies have explored the use of non-viral approaches, such as Sleeping Beauty transposition, which can enhance CAR-T scalability for broader deployment [161]. This approach uses simple DNA minicircles to insert CAR genes, potentially reducing the risk of mutagenesis and genotoxicity that may be associated with viral modalities. This strategy may also reduce regulatory hurdles and the cost of CAR-T engineering. Additional engineering approaches have sought to improve CAR-T manufacturing strategies using ‘off-the-shelf’ cell therapy that does not require autologous T cells [162–164]. Through the use of CRISPR/Cas9 to eliminate CD7 and T cell receptor alpha chain, which are also expressed on the malignant T cells, barriers such as fratricide could be avoided and preserved activity against T cell acute lymphoblastic leukemia in vitro and in vivo was demonstrated. In addition, graft-versus-host-disease (GvHD) was not observed. In a recent advance, ZFN editing is being harnessed to modify both autologous and allogeneic cell therapies, further broadening the possibilities of off-the-shelf CAR-T manufacturing, potentially reducing the time to treatment for the patient [165].
Cancer is not the only target of personalized cell therapy. Induced pluripotent stem cells (iPSCs) taken from a patient’s skin or elsewhere can be reprogrammed to a cell of choice, such as one of the kidney’s specialized cells, the beta cell, to treat type 1 diabetes; or into a brain cell, which can be transplanted into the body to – for instance – reduce inflammation in MS, as shown in mice [93, 166, 167]. Additional examples of cell therapy include mitochondrial replacement therapy (MRT), which has also been implemented in the United Kingdom. Mitochondrial disease is caused when mutations in mitochondrial DNA (mtDNA) are maternally transferred to the offspring, which can lead to serious disorders ranging from epilepsy to optic neuropathy and diabetes mellitus and deafness, among others. To implement MRT, the healthy nucleus from the maternal egg with malfunctioning mitochondria is transferred to a healthy egg (and donor mitochondria) without a nucleus. In effect, this approach can result in a fertilized egg that contains nuclear DNA from two parents, and a mitochondrial DNA from a donor to eliminate genetic diseases in offspring [168].
The engineering of cells as biosensors is another particularly relevant area. Synthetic cells have been designed for detecting cancer and for detecting diabetes indicators in urine [169, 170]. Impressively, synthetic biology has advanced from bacteria to mammalian cells, with early biosensors able to not only detect disease markers but also deliver therapeutic payloads to ameliorate symptoms. In this example, acute and chronic psoriasis was held in check by a population of synthetic cells implanted in the mice [171]. This is the ultimate demonstration of personalized medicine, where the cell is an autonomous sensor and therapy, delivering therapeutic levels of “drug” without patient or clinician intervention.
Delivering therapy in a personalized fashion will also serve as an important enabling technology. Knowing that dose modulation, or population optimized fixed dose combination therapy will serve as a cornerstone for improving delivery technologies, methodologies such as personalized 3-D printing or biomaterial-mediated controlled release will play an increasingly important role in personalized medicine (NCT03348293). Tailored release profiles from 3-D printed tablets were recently introduced, where the temporal drug release could be customized [172]. In addition, advances in micro-manufacturing have produced drug-loaded 3-D microstructures with temporal drug release control using an approach termed StampEd Assembly of polymer Layers (SEAL) [173]. Patient-specific responses to combination therapy are often determined by unique dosing profiles. Therefore, these tablets and microstructures may serve as viable drug delivery platforms for personalized medicine.
Combination therapy with nanomedicine has also received substantial attention [174–176]. Augmented AI was also used to design population-optimized combinatorial nanotherapy across multiple breast cancer cell lines (MCF-7, MDA-MB-231, BT-20) and control cell lines (IMR-90, MCF-10A, H9C2). This resulted in the identification of specific drug-dose ratios that mediated optimal treatment efficacy. Specifically, diverse dosing regimens of nanodiamonds (NDs) functionalized with doxorubicin, bleomycin, and mitoxantrone (NDX, ND-BLEO, ND-MTX) and unmodified paclitaxel were applied across the cell lines. Quadratic optimization, an AI-based approach that correlates drug inputs (e.g. drug and dose) and phenotypic outputs (e.g. cancer cell death, control cell viability) using a smooth quadratic surface was conducted to accelerate the identification of the dosages of the four therapies that mediated optimal cancer cell death and control cell survival. Of note, The AI optimization showed that optimized ND-modified combination therapy outperformed ND-modified as well as unmodified single drug therapy, unmodified combination therapy, as well as arbitrarily designed ND-modified combination therapy. While NDs were used as the model delivery system, the augmented AI platform was universally applicable to all classes of nanomaterials [120, 177]. Clinical trials to validate ND-containing biomaterials have since been initiated (NCT03376984) [178]. These studies show that the application of engineering principles to address the barriers to achieving personalized medicine provides a strong foundation for future developments in personalized materials, therapies, and devices.
Policy and infrastructure shifts in personalized and precision medicine
The translation of approaches in personalized and precision medicine into the clinic and marketplace will depend on the evolution of the policy landscape and on the particular disease or indication, with some relying more on genomic/proteomic markers to guide treatment, and others relying more on prognostic variables [179–181]. However, regardless of the indication being addressed, the transition of personalized and precision medicine into widespread use will likely challenge many long-held regulatory policies that are based on single products (rather than platforms) and clinical standards, such as dose escalation, dose expansion, and MTD-based drug evaluation and administration. In addition, the recent approval of vemurafenib was achieved using a basket study (MSK-IMPACT trial), where genomic drivers of the disease indication (Erdheim-Chester Disease, ECD) were used to assign treatment, as opposed to tumor location [182]. The opportunity to regulate -omics profile-specific medicines is likely imminent. Subpopulation-specific diagnostics and therapies are now on our doorstep, suggesting that an emerging regulatory question will be whether (and how) to integrate AI, ML, or other analytical platforms into clinical trial design. In addition, as these platforms can be leveraged to individualize optimal treatment regimens, a move beyond fixed p-value based evaluation may be needed due to the possibility of each patient receiving an individualized regimen. These considerations open the door to potentially broadening the implementation of patient-centered clinical trials and the incorporation of Bayesian decision analysis (BDA) and patient preference into clinical trial design. Trials based on BDA may take into account the balance between benefits of the therapy or device itself on treatment efficacy, risks of the treatment to the patient, and outcomes if the patient is not treated. These, and other factors may influence multiple aspects of the trial design. These include potentially reducing the number of patients recruited to assess trial endpoints, biomarker-based patient selection, and statistical assessment approaches that are implemented, among others [183–185]. As an example, in a recent study of CURATE.AI-based liver transplant immunosuppressant dosing, small cohort statistical analysis was used to confirm that the AI-guided treatment cohort exhibited reduced interpatient variance compared to control cohort patients. Ultimately, applying approaches like AL, ML, and BCA may result in clinical trial designs that identify patient-specific benefits that accelerate the approvals of novel therapies and devices.
The aforementioned opportunities to re-engineer the drug and device validation processes may drive the evolution of regulatory science [186]. Next-generation regulatory architecture and greater “regulatory literacy” are needed. Streamlining the regulatory process for the rapidly evolving landscape of interventions and technologies will ultimately play a vital role in enabling personalized care to reach more patients in a timely fashion. Furthermore, issues such as drug pricing, accessibility, repositioning in the context of regulating N-of-1 regimens, and other considerations will need to be addressed [187]. The FDA has acknowledge that there could be a shift in focus from primarily efficacy (because some studies have even been shown to be too efficacious!) to a focus on long-term durability, safety, and product issues related to new technology, such as off-target effects and implications longer-term.
The 21st Century Cures Act, which was signed into law in the United States at the end of 2016, provides funding to the FDA to create new programs that will enhance its ability to expedite approval of certain personalized and precision medicine products, such as cell therapies (Regenerative Medicine Advanced Therapy) and medical devices (Breakthrough Devices). The Act goes beyond the FDA, also allocating money to the NIH for medical research and drug development. AI Singapore was recently unveiled to accelerate drug development and enhance patient-centered care via more efficient hospital triaging processes through the integration of AI and ML with advanced trial design and EMR. A partnership between the European Union (EU) and European pharmaceutical companies led to the recent creation of the Innovative Medicines Initiative (IMI). The IMI is combatting major challenges that will confront EU, including antimicrobial resistance, the need for continued flu vaccine development, and non-alcoholic fatty liver disease (NAFLD), a global issue that affects as much as 30% of the EU population. Furthermore, NAFLD is capable of transitioning to non-alcoholic steatohepatitis, which can lead to hepatocellular carcinoma. In addition, the IMI is also developing novel tools to improve toxicity prediction in drug development as well as establishing a strategy to improve data quality during the preclinical drug development process in an effort to shorten the pathway to approval and improve success rates [188].
Ten requirements for achieving a successful implementation of personalized and precision medicine were recently outlined [189]. Several of these have become increasingly relevant to the role of engineering; in particular, Omics writ large, or the systematic and perpetual monitoring of environmental exposures that may drive disease risk; Computation, which involves the integration of patient electronic medical records as a catalyst for clinical-decision support; and Education, which plays a key role in placing engineering at the foundation of future clinical leadership in personalized and precision medicine. These requirements entail more frequent access to patients and innovative trial design. Trial designs will increasingly need to focus on subpopulations of patients, and pre-specifying these patients for greater signals of efficacy will require a sea change in thinking about designs that are tissue-agnostic and perhaps include a drug-diagnostic combination. It has been opined that even the pathway for approving diagnostics may need to change (from the traditional 510k route) in the face of modern diagnostics. In part, this may come through public awareness of the efforts of precision medicine. For example, the Oncology Precision Network OpeN—started in response to the Cancer Moonshot (rather than the Precision Medicine Initiative, PMI)—aims to reach underserved patients and enrol them in clinical trials, as well as to foster data sharing towards coordinated treatment decision support. Similarly, myriad websites, patient advocacy groups, and nonprofits are geared up for greater patient engagement. Some believe that sharing clinical trial results increases patient recruitment and engagement. Study populations will need to be more diverse if we are to achieve the mission of precision medicine. GWAS are still largely comprised of patients of European descent [190]. In 2009, only 4% of patients in 373 studies were non-European; in 2016, only 19%, which is an improvement but still “persistent bias” [191]. Therefore, re-engineering clinical trial designs, standards, and protocols may have significant benefits for personalized and precision medicine. The Trans-Omics for Precision Medicine (TOPMed) Initiative launched by the U.S. National Heart, Lung, and Blood Institute aims to correct this course, as does its umbrella effort, the National Institutes of Health’s “All of Us” Research Program (as part of the PMI); the Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment (PREDICT) program at Vanderbilt University [192, 193], the UK NHS counterpart, 100,000 Genomes Project; and other programs in China, France, and elsewhere. Data sharing among these initiatives could also bolster the power of these data sets to deliver precision medicine to all, rather than select populations that are over represented in individual PM initiatives. In All of Us, engineers can play an important role in helping to achieve its mission, for example by developing mobile health devices for lifestyle monitoring or designing technologies to discover new biomarkers of health and disease.
With regards to scaling the implementation of personalized and precision medicine in the clinic, the medical community is beginning to establish provider networks that are integrating genomic information with electronic health records in order to enhance point-of-care guidance on diagnostics and/or treatment. One example is the IGNITE (Implementing GeNomics In practice) program [194–197], a network for the analysis of pharmacogenomic data and other family history information for integration into electronic medical records (EMR) to power clinical decision support (CDS) and address a diverse spectrum of indications, such as hypertension renal disease and diabetes. Of note, a recent study pertaining to CYP2C19 genotype-guided antiplatelet therapy in a multisite trial from the IGNITE network was reported [198]. Initiatives such as IGNITE will serve as foundations for the integration of personalized and precision medicine into mainstream healthcare (NCT02335307).
With respect to education policy, integrating engineering and clinical training will cultivate a generation of clinicians who are comfortable with emerging technologies. In turn, they will be well versed in clinical translation, a vital part of education in personalized and precision medicine. In addition, the integration of healthcare economics, regulatory policy, design of user interfaces into programs dedicated to translational medicine will play a key role in driving the next generation of technology transfer in personalized and precision medicine. Institutions that are addressing these issues include the Carle Illinois College of Medicine, which is placing engineering at the forefront of clinical education; EnMed, a collaboration between Texas A&M University and Houston Methodist Hospital; and The University of California, San Francisco (UCSF), which has developed a Masters in Translational Medicine program to bridge engineering and clinic-focused education to bring new technologies to the patient [199, 200]. The integration of cellular and molecular engineering, modelling, simulation and visualization as modalities for medical education, team-based development of engineering solutions to unmet medical needs, as well as technology transfer are examples of components that can be integrated into next generation curriculums to train the next generation of clinicians. These clinicians will ultimately have an enhanced grasp of the requirements for moving emerging technologies and platforms into humans, including regulation, intellectual property, and entrepreneurship, among other skillsets, will ultimately accelerate engineering-based discoveries into patient use [199].
Outlook
Despite the obvious fact that patient physiology varies substantially from one individual to another, drug development and patient care has largely relied on the administration of the same regimen to an almost insurmountably diverse population. While certain scenarios permit bespoke drug administration according to constantly varying patient responses, arbitrary titration also remains the standard of care. These conventional treatment routes pre-dispose patients towards sub-optimal response rates. The era of personalized and precision medicine will likely overcome this challenge. Patients will no longer be confined to target-based drug selection and dose escalation-defined administration protocols that collectively result in the standard one-size fits all treatment approach.
The suite of enabling technologies for personalized and precision medicine has provided unprecedented abilities to identify and surveil disorders. These technologies can accurately diagnose disease, comb large databases of –omics information and EMRs to search for potential therapies, identify biomarkers that may accurately reflect disease states, constantly monitor disease progression or treatment response through wearable technology, and ultimately harness AI and other optimization technologies to dynamically and accurately determine the best drugs (chemotherapy, immunotherapy, nanotherapy, etc.) and corresponding doses that may change as patient physiology evolves during the course of therapy. Achieving this level of treatment personalization would require a seamless integration of biomarker development, and potentially a re-engineering of drug trial design and analysis paradigms. One challenge that arises when this arsenal of enabling of technologies comes to fruition is the seamless integration of their implementation (see Outstanding Questions).
Outstanding Questions.
What are the key technological barriers to the broader deployment of precision and personalized medicine technologies?
What are the potential strategies that can be employed to enhance the degree of integration of precision and personalized medicine technologies?
Will effective clinical validation of precision and personalized medicine technologies require new clinical trial designs to be implemented?
When a patient’s therapeutic regimen is personalized only to their own physiology or disease characteristics during a clinical trial, how will statistical analysis methods be adapted to account for N-of-1, or single patient studies?
Is the regulatory community prepared to utilize novel clinical trial designs, where even a patient’s own regimens will constantly change over time, to bring new technologies towards widespread implementation?
How can innovators, educators, regulators, healthcare systems and other stakeholders better prepare themselves for the potential practice-changing impact of bringing precision and personalized medicine technologies into common clinical practice?
The successful transitioning of validated precision and personalized medicine platforms into clinical practice will require important advances beyond technology-centric innovation. A number of risks and challenges associated with healthcare economics, ethics, and data privacy, which are encountered after the stage of technology validation, need to be addressed [201–205]. In other examples, rising healthcare costs across the globe are simultaneously creating a barrier and opportunity for the clinical implementation of novel technologies. Innovation through the lens of payers, policymakers, will be vital. The minimum threshold to demonstrate patient and societal value in healthcare will require improved treatment outcomes, reduced incidence of acute and long-term treatment complications, and marked reductions in the cost of healthcare delivery and administration. To address these criteria, a specific area of promise in both fundamental and applied innovation is the use of AI-based approaches in drug development. As previously mentioned reconciling data-driven drug combination design into actionable regimens coupled with attaining rapid fundamental insight into the pathways that drive the improved outcomes enabled by these regimens are examples of how engineering platforms for precision and personalized medicine can move the needle in terms of impacting the practice of healthcare. With drug prices reaching record levels, engineering approaches for precision and personalized medicine will ultimately play a vital role in reducing drug development costs, increasing drug accessibility, as well as contributing to cost-effectiveness for patients and healthcare systems, which itself is a topic that may require further evaluation as precision and personalized medicine platforms move towards broad deployability.
Mobilizing personalized and precision medicine for all will require a convergence of the aforementioned suite of enabling technologies and regulatory/public policy with advances in education and coordinated efforts to deploy and fund truly personalized and precision medicine on a global scale. At the core of this effort, biomedical engineering is playing an important role in catalyzing breakthroughs that will ultimately improve the human condition in an individualized fashion. Once we gain a clearer picture of how these new capabilities can be paired and regulated to improve patient outcomes, we can expect even more advances in personalized and precision medicine in the present population, in underrepresented populations, and in future generations to come.
Highlights.
Engineering approaches to precision medicine will harness population-wide data to identify individualized treatment strategies.
Personalized medicine harnesses a subject’s own data to individualize their own care, from diagnosis through treatment selection and monitoring.
Novel clinical trial designs will play a vital role in assessing the efficacy and safety of emerging therapies and diagnostics.
Artificial intelligent platforms will globally optimize combination therapy from the preclinical through clinical stages of validation.
The widespread deployment of precision and personalized medicine technologies will involve the convergence of several factors ranging from evolving education at the interface of engineering and medicine and policies that support new clinical trial designs, to scaling the use of electronic medical records (EMR) to drive clinical decision support.
Acknowledgements
The authors acknowledge the important contributions of Dr. Megan L. Frisk towards this work. We are deeply grateful for her guidance, without which this work would not have been possible. We also thank Dr. Vivian Y. Chang for her valuable expertise pertaining to clinical genomics and the critical review of the manuscript. The authors also acknowledge the innovative discoveries in the personalized and precision medicine community that we were unable to include in this paper due to space limitations. The authors also gratefully acknowledge Peter Wang and Theodore Kee.
Glossary
- Artificial intelligence (AI)
In the context of healthcare, AI uses algorithms to reconcile complex data in an effort to identify actionable strategies for many applications. These range from improving treatment outcomes to accelerating drug discovery, among others.
- Bayesian decision analysis (BDA)
With regards to healthcare, BDA is used to correlate tradeoffs and decision making processes. For example, using BDA towards novel clinical trial designs may involve the correlation of outcome objectives for a patient with the benefits and risks undergoing treatment.
- Chimeric antigen receptor T cell therapy (CAR-T)
This form of immunotherapy modifies a patient’s own T cells, which are derived from their immune system, with chimeric antigen receptors (CAR) on their surfaces. These modified T cells can then selectively target surface markers on the cancer cells using these receptors during treatment.
- Circulating tumor cell (CTC)
This cell is released by a primary tumor into the circulatory system and may serve as a foundation for metastasis.
- Clinical decision support (CDS)
Using a broad spectrum of applicable data, CDS platforms provide actionable guidance to clinicians in areas such as drug selection, dosage modifications, and other courses of treatment.
- Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (CRISPR-Cas9)
This platform is used for genome editing, where genetic material can be added, removed, or modified. This approach can potentially be used to address a multitude of diseases by altering the genetic information that drives the onset of these disorders.
- CURATE.AI
This mechanism-independent artificial intelligence platform is used to dynamically optimize clinical combination therapy dosing during the course of treatment. By using only a patient’s own data to manage their own combination therapy regimen, CURATE.AI can maximize treatment efficacy and safety for a sustained duration on an individualized basis. It is broadly applicable towards oncology, infectious disease, and many other disease indications.
- Cytometry by time of flight (CyTOF)
This is a mass spectrometry methodology that uses heavy metal antibody tags for cell surface and intracellular markers. CyTOF analysis enables multiplexed profiling of single cell responses for applications in drug development and fundamental studies into cellular mechanisms.
- Electronic medical records (EMR)
Electronic medical records can contain a broad spectrum of information pertaining to a patient’s healh history. They can serve as vital platforms for the implementation of treatment and diagnostic paradigms that may integrate emerging technologies such as artificial intelligence, wearables, and other modalities.
- Machine Learning (ML)
Machine learning (ML) platforms use algorithms that are trained with a set of data to subsequently make inferences are identify a course of action without requiring a directed set of instructions. Implementation of ML typically requires minimal human interaction. In the context of healthcare, it can be used for many applications, including the design of drug combinations and the development of biomaterials, among others.
- Maximum Tolerated Dose (MTD)
The maximum tolerated dose is the highest dose of a drug that can be administered to a subject while simultaneously avoiding an unacceptable level of of toxicity. With regards to precision and personalized medicine, emerging studies have shown promise in identifying lower drug doses that result in improved efficacy and safety, potentially avoiding the need to reach the MTD during therapy.
- Mitochontrial replacement therapy (MRT)
This approach is used to address mitochondrial diseases by replacing mitochondria that contain DNA mutations with healthy mitochondria. In the context of reproductive medicine, a mother with mitochondrial disease can have her eggs transferred to a donor egg with healthy mitochondria
- Nanodiamond (ND)
Nanodiamonds are carbon-based nanoparticles that can be used to carry multiple classes of therapeutic and imaging compounds. Their unique surface electrostatic properties have been used to markedly improve magnetic resonance imaging contrast efficiency as well as drug delivery efficacy.
- Phenotypic Personalized Medicine (PPM)
This artificial intelligence-based approach uses quantifiable measures of clinical efficacy and safety, such as tumor burden through imaging or circulating biomarker analysis, as well as toxicity panels to guide drug dosing. This approach can be implemented in a mechanism-independent manner.
- Quadratic Phenotypic Optimization Platform (QPOP)
This AI-based approach simultaneously identifies the right drugs and corresponding doses from large pools of candidate therapies for novel drug combination development. It can be implemented without disease target/mechanism information and does not rely on drug synergy predictions to optimize treatment outcomes.
- StampEd Assembly of polymer Layers (SEAL)
This approach uses 3-D printed microwells that contain multiple drugs, and can be used for the timed release of multiple therapies in a sustained fashion.
- Spherical Nucleic Acids (SNA)
These nanostructures consist of precisely positioned and high density configurations of nucleic acids that have been explored for gene regulation with broad applications across different disease indications. They are currently being evaluated at the clinical level.
- Zinc finger nuclease (ZFN)
This enzyme is comprised of DNA-binding and cleavage domains and is used as a genome editing platform. ZFN-based genome editing therapies are currently being evaluated at the clinical level.
Footnotes
Conflict of Interest Statement
D. Ho, E.K. Chow, X. Ding, C. Ho, and A. Zarrinpar are co-inventors of pending and issued patents pertaining to artificial intelligence-based drug development.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hamburg MA and Collins FS (2010) The Path to Personalized Medicine. New England Journal of Medicine 363, 301–304 [DOI] [PubMed] [Google Scholar]
- 2.Li L, et al. (2015) Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Science Translational Medicine 7, 311ra174–311ra174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Itakura H, et al. (2015) Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Science Translational Medicine 7, 303ra138–303ra138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khoury MJ, et al. (2003) Population Screening in the Age of Genomic Medicine. New England Journal of Medicine 348, 50–58 [DOI] [PubMed] [Google Scholar]
- 5.Sarioglu AF, et al. (2015) A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat Meth 12, 685–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller MA, et al. (2015) Predicting therapeutic nanomedicine efficacy using a companion magnetic resonance imaging nanoparticle. Science Translational Medicine 7, 314ra183–314ra183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonas O, et al. (2015) An implantable microdevice to perform high-throughput in vivo drug sensitivity testing in tumors. Science Translational Medicine 7, 284ra257–284ra257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson BS, et al. (2013) Real-Time, Aptamer-Based Tracking of Circulating Therapeutic Agents in Living Animals. Science Translational Medicine 5, 213ra165–213ra165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zarrinpar A, et al. (2016) Individualizing liver transplant immunosuppression using a phenotypic personalized medicine platform. Science Translational Medicine 8, 333ra349–333ra349 [DOI] [PubMed] [Google Scholar]
- 10.Lim JJ, et al. Maximizing Efficiency of Artificial Intelligence ‐ Driven Drug Combination Optimization through Minimal Resolution Experimental Design. Advanced Therapeutics, 1900122 [Google Scholar]
- 11.Wang Y, et al. (2018) Low-cost, μm-thick, tape-free electronic tattoo sensors with minimized motion and sweat artifacts. npj Flexible Electronics 2, 6 [Google Scholar]
- 12.Koh A, et al. (2016) A soft, wearable microfluidic device for the capture, storage, and colorimetric sensing of sweat. Science translational medicine 8, 366ra165–366ra165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rawson TM, et al. (2019) Microneedle biosensors for real-time, minimally invasive drug monitoring of phenoxymethylpenicillin: a first-in-human evaluation in healthy volunteers. The Lancet Digital Health 1, e335-e343 [DOI] [PubMed] [Google Scholar]
- 14.Pattichis CS and Panayides AS (2019) Connected Health. Frontiers in Digital Health 1, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Meel R, et al. (2019) Smart cancer nanomedicine. Nature Nanotechnology 14, 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neff RT, et al. (2017) BRCA mutation in ovarian cancer: testing, implications and treatment considerations. Therapeutic Advances in Medical Oncology 9, 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng HH, et al. (2017) Prostate Cancer Screening in a New Era of Genetics. Clinical Genitourinary Cancer [DOI] [PubMed] [Google Scholar]
- 18.Bednar EM, et al. (2017) A universal genetic testing initiative for patients with high-grade, non-mucinous epithelial ovarian cancer and the implications for cancer treatment. Gynecologic Oncology 146, 399–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan B, et al. (2017) Single-cell genomic profiling of acute myeloid leukemia for clinical use: A pilot study. Oncology Letters 13, 1625–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pannone L, et al. (2017) Structural, Functional, and Clinical Characterization of a Novel PTPN11 Mutation Cluster Underlying Noonan Syndrome. Human Mutation 38, 451–459 [DOI] [PubMed] [Google Scholar]
- 21.Das K, et al. Genomic predictors of chemotherapy efficacy in advanced or recurrent gastric cancer in the GC0301/TOP002 phase III clinical trial. Cancer Letters 412, 208–215 [DOI] [PubMed] [Google Scholar]
- 22.Mohanty A, et al. (2016) CCND1 mutations increase protein stability and promote ibrutinib resistance in mantle cell lymphoma. Oncotarget 7, 73558–73572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyth E, et al. (2017) Pharmacogenetic analysis of the UK MRC MAGIC trial: association of polymorphisms with toxicity and survival in patients treated with perioperative ECF chemotherapy. Clinical Cancer Research [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinshilboum RM and Wang L (2017) Pharmacogenomics: Precision Medicine and Drug Response. Mayo Clinic Proceedings 92, 1711–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chia S, et al. (2017) Phenotype-driven precision oncology as a guide for clinical decisions one patient at a time. Nature Communications 8, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hawgood S, et al. (2015) Precision medicine: Beyond the inflection point. Science Translational Medicine 7, 300ps317–300ps317 [DOI] [PubMed] [Google Scholar]
- 27.Zhao Z, et al. (2018) Transdermal immunomodulation: Principles, advances and perspectives. Advanced Drug Delivery Reviews [DOI] [PubMed] [Google Scholar]
- 28.Vela Ramirez JE, et al. (2017) Current state and challenges in developing oral vaccines. Advanced Drug Delivery Reviews 114, 116–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeo DC, et al. (2018) Abnormal scar identification with spherical-nucleic-acid technology. Nature Biomedical Engineering 2, 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He L, et al. (2019) Polymer Nanoplatforms at Work in Prostate Cancer Therapy. Advanced Therapeutics 2, 1800122 [Google Scholar]
- 31.Mehta P, et al. (2019) Broad Scale and Structure Fabrication of Healthcare Materials for Drug and Emerging Therapies via Electrohydrodynamic Techniques. Advanced Therapeutics 2, 1800024 [Google Scholar]
- 32.Zhang X, et al. (2019) Supramolecular Nanogel‐Based Universal Drug Carriers Formed by “Soft–Hard” Co‐Assembly: Accurate Cancer Diagnosis and Hypoxia‐Activated Cancer Therapy. Advanced Therapeutics 2, 1800140 [Google Scholar]
- 33.Unbehauen ML, et al. (2019) Tailor‐Made Core‐Multishell Nanocarriers for the Delivery of Cationic Analgesics to Inflamed Tissue. Advanced Therapeutics 2, 1900007 [Google Scholar]
- 34.Wang C, et al. (2019) Cowpea Mosaic Virus Promotes Anti‐Tumor Activity and Immune Memory in a Mouse Ovarian Tumor Model. Advanced Therapeutics 2, 1900003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tieu T, et al. (2019) Advances in porous silicon–based nanomaterials for diagnostic and therapeutic applications. Advanced Therapeutics 2, 1800095 [Google Scholar]
- 36.Lee J, et al. (2019) Self‐Assembled Aptamer Nanoconstruct: A Highly Effective Molecule ‐ Capturing Platform Having Therapeutic Applications. Advanced Therapeutics 2, 1800111 [Google Scholar]
- 37.Yang P, et al. Synthetic, Supramolecular, and Self‐Adjuvanting CD8+ T‐Cell Epitope Vaccine Increases the Therapeutic Antitumor Immunity. Advanced Therapeutics, 1900010 [Google Scholar]
- 38.Leary RJ, et al. (2010) Development of Personalized Tumor Biomarkers Using Massively Parallel Sequencing. Science Translational Medicine 2, 20ra14–20ra14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gootenberg JS, et al. (2017) Nucleic acid detection with CRISPR-Cas13a/C2c2. Science [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng S-B, et al. (2017) EBV-associated primary nodal T/NK-cell lymphoma shows distinct molecular signature and copy number changes. Haematologica [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng IK, et al. (2017) Identifying large indels in targeted next generation sequencing assays for myeloid neoplasms: a cautionary tale of the ZRSR1 pseudogene. Journal of Clinical Pathology [DOI] [PubMed] [Google Scholar]
- 42.Chen X-Q, et al. (2017) Dysregulation of neurotrophin signaling in the pathogenesis of Alzheimer disease and of Alzheimer disease in Down syndrome. Free Radical Biology and Medicine [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yong WP, et al. (2018) Real-Time Tumor Gene Expression Profiling to Direct Gastric Cancer Chemotherapy: Proof-of-Concept “3G” Trial. Clinical Cancer Research 24, 5272–5281 [DOI] [PubMed] [Google Scholar]
- 44.Mohan S, et al. (2019) Profiling of circulating free DNA using targeted and genome wide sequencing in patients with Small Cell Lung Cancer. Journal of Thoracic Oncology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montemayor C, et al. (2019) Banking with precision: transfusion medicine as a potential universal application in clinical genomics. Current opinion in hematology 26, 480–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schubert SM, et al. (2016) Protein Counting in Single Cancer Cells. Analytical Chemistry 88, 2952–2957 [DOI] [PubMed] [Google Scholar]
- 47.Nie S, et al. (2015) Correlations of Salivary Biomarkers with Clinical Assessments in Patients with Cystic Fibrosis. PLOS ONE 10, e0135237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu D, et al. Long-Term Measurements of Human Inflammatory Cytokines Reveal Complex Baseline Variations between Individuals. The American Journal of Pathology [DOI] [PubMed] [Google Scholar]
- 49.Kalinich M, et al. (2017) An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. Proceedings of the National Academy of Sciences 114, 1123–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gawel DR, et al. (2019) A validated single-cell-based strategy to identify diagnostic and therapeutic targets in complex diseases. Genome medicine 11, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu J, et al. (2019) A new approach to find biomarkers in chronic fatigue syndrome/myalgic encephalomyelitis (CFS/ME) by single-cell Raman micro-spectroscopy. Analyst 144, 913–920 [DOI] [PubMed] [Google Scholar]
- 52.Kwok SJ, et al. (2019) Multiplexed laser particles for spatially resolved single-cell analysis. Light: Science & Applications 8, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Kane PT and Mrksich M (2017) An Assay Based on SAMDI Mass Spectrometry for Profiling Protein Interaction Domains. Journal of the American Chemical Society 139, 10320–10327 [DOI] [PubMed] [Google Scholar]
- 54.Szymczak L, et al. (2017) Peptide Arrays: Development and Application. Analytical Chemistry [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Irish JM, et al. (2006) Mapping normal and cancer cell signalling networks: towards single-cell proteomics. Nat Rev Cancer 6, 146–155 [DOI] [PubMed] [Google Scholar]
- 56.Gawad C, et al. (2016) Single-cell genome sequencing: current state of the science. Nat Rev Genet 17, 175–188 [DOI] [PubMed] [Google Scholar]
- 57.Warren L, et al. (2006) Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proceedings of the National Academy of Sciences 103, 17807–17812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scher HI, et al. (2016) ASsociation of ar-v7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu M, et al. (2012) RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature 487, 510–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Warrick J, et al. (2010) A Microfluidic Cell Concentrator. Analytical Chemistry 82, 8320–8326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cima I, et al. (2016) Tumor-derived circulating endothelial cell clusters in colorectal cancer. Science Translational Medicine 8, 345ra389–345ra389 [DOI] [PubMed] [Google Scholar]
- 62.Zanini F, et al. (2018) Single-cell transcriptional dynamics of flavivirus infection. eLife 7, e32942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang W, et al. (2018) High fidelity hypothermic preservation of primary tissues in organ transplant preservative for single cell transcriptome analysis. BMC Genomics 19, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dalerba P, et al. (2011) Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol 29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jan M, et al. (2012) Clonal Evolution of Preleukemic Hematopoietic Stem Cells Precedes Human Acute Myeloid Leukemia. Science Translational Medicine 4, 149ra118–149ra118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ritsma L, et al. (2012) Intravital Microscopy Through an Abdominal Imaging Window Reveals a Pre-Micrometastasis Stage During Liver Metastasis. Science Translational Medicine 4, 158ra145–158ra145 [DOI] [PubMed] [Google Scholar]
- 67.Ghajar CM, et al. (2013) The perivascular niche regulates breast tumour dormancy. Nat Cell Biol 15, 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boffa DJ, et al. (2017) Cellular Expression of PD-L1 in the Peripheral Blood of Lung Cancer Patients is Associated with Worse Survival. Cancer Epidemiology Biomarkers & Prevention 26, 1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang L, et al. (2016) Detection and Characterization of Circulating Tumour Cells in Multiple Myeloma. Journal of Circulating Biomarkers 5, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tse HTK, et al. (2013) Quantitative Diagnosis of Malignant Pleural Effusions by Single-Cell Mechanophenotyping. Science Translational Medicine 5, 212ra163–212ra163 [DOI] [PubMed] [Google Scholar]
- 71.Bendall SC and Nolan GP (2012) From single cells to deep phenotypes in cancer. Nat Biotech 30, 639–647 [DOI] [PubMed] [Google Scholar]
- 72.de Bruin EC, et al. (2014) Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hou Y, et al. (2016) Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res 26, 304–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kenry, et al. (2017) Single-Layer Ternary Chalcogenide Nanosheet as a Fluorescence-Based “Capture-Release” Biomolecular Nanosensor. Small 13, 1601925-n/a [DOI] [PubMed] [Google Scholar]
- 75.Ando D and Meyer K (2017) Gene Editing: Regulatory and Translation to Clinic. Hematology/Oncology Clinics of North America 31, 797–808 [DOI] [PubMed] [Google Scholar]
- 76.Fernández-Navarro P, et al. (2019) The use of PanDrugs to prioritize anticancer drug treatments in a case of T-ALL based on individual genomic data. BMC cancer 19, 1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Russo M, et al. (2019) Adaptive mutability of colorectal cancers in response to targeted therapies. Science [DOI] [PubMed] [Google Scholar]
- 78.Fernandez-Nogueira P, et al. (2019) Tumor Associated Fibroblasts Promote HER2-Targeted Therapy Resistance through FGFR2 Activation. Clinical Cancer Research [DOI] [PubMed] [Google Scholar]
- 79.Nelson CE, et al. (2016) In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351, 403–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Daud AI, et al. Tumor immune profiling predicts response to anti–PD-1 therapy in human melanoma. The Journal of Clinical Investigation 126, 3447–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zenke Y, et al. (2016) Successful treatment with afatinib after grade 3 hepatotoxicity induced by both gefitinib and erlotinib in EGFR mutation-positive non-small cell lung cancer. Lung Cancer 99, 1–3 [DOI] [PubMed] [Google Scholar]
- 82.Venugopalan A, et al. (2016) EGFR-targeted therapy results in dramatic early lung tumor regression accompanied by imaging response and immune infiltration in EGFR mutant transgenic mouse models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hochmair M, et al. (2016) Complete remissions in afatinib-treated non-small-cell lung cancer patients with symptomatic brain metastases. Anti-Cancer Drugs 27, 914–915 [DOI] [PubMed] [Google Scholar]
- 84.Yin H, et al. (2017) Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nature Biotechnology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee K, et al. (2017) Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nature Biomedical Engineering 1, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng D, et al. (2012) Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proceedings of the National Academy of Sciences 109, 11975–11980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Randeria PS, et al. (2015) siRNA-based spherical nucleic acids reverse impaired wound healing in diabetic mice by ganglioside GM3 synthase knockdown. Proceedings of the National Academy of Sciences 112, 5573–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Radovic-Moreno AF, et al. (2015) Immunomodulatory spherical nucleic acids. Proceedings of the National Academy of Sciences 112, 3892–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jensen SA, et al. (2013) Spherical Nucleic Acid Nanoparticle Conjugates as an RNAi-Based Therapy for Glioblastoma. Science Translational Medicine 5, 209ra152–209ra152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Artzi N, et al. (2009) Aldehyde-Amine Chemistry Enables Modulated Biosealants with Tissue-Specific Adhesion. Advanced Materials 21, 3399–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oliva N, et al. (2015) Regulation of dendrimer/dextran material performance by altered tissue microenvironment in inflammation and neoplasia. Science Translational Medicine 7, 272ra211–272ra211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vegas AJ, et al. (2016) Combinatorial hydrogel library enables identification of materials that mitigate the foreign body response in primates. Nat Biotech 34, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vegas AJ, et al. (2016) Long-term glycemic control using polymer-encapsulated human stem cell-derived beta cells in immune-competent mice. Nat Med 22, 306–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lind JU, et al. (2016) Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nature Materials 16, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Beachley VZ, et al. (2015) Tissue matrix arrays for high-throughput screening and systems analysis of cell function. Nat Meth 12, 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bein A, et al. (2018) Microfluidic Organ-on-a-Chip Models of Human Intestine. Cellular and Molecular Gastroenterology and Hepatology 5, 659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Prantil-Baun R, et al. (2018) Physiologically Based Pharmacokinetic and Pharmacodynamic Analysis Enabled by Microfluidically Linked Organs-on-Chips. Annual Review of Pharmacology and Toxicology 58, 37–64 [DOI] [PubMed] [Google Scholar]
- 98.Blicharz TM, et al. (2018) Microneedle-based device for the one-step painless collection of capillary blood samples. Nature Biomedical Engineering 2, 151–157 [DOI] [PubMed] [Google Scholar]
- 99.Kee T, et al. Harnessing CURATE. AI as a Digital Therapeutics Platform by Identifying N‐of‐1 Learning Trajectory Profiles. Advanced Therapeutics, 1900023 [Google Scholar]
- 100.Davis NO, et al. (2018) Proof-of-concept study of an at-home, engaging, digital intervention for pediatric ADHD. PloS one 13, e0189749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cho C-H and Lee H-J (2019) Could digital therapeutics be a game changer in psychiatry? Psychiatry investigation 16, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zimmer A, et al. (2017) Prediction of drug cocktail effects when the number of measurements is limited. PLOS Biology 15, e2002518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang J, et al. (2017) Integrating evolutionary dynamics into treatment of metastatic castrate-resistant prostate cancer. Nature communications 8, 1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gatenby RA, et al. (2009) Adaptive therapy. Cancer research 69, 4894–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chakradhar S (2017) Predictable response: Finding optimal drugs and doses using artificial intelligence. Nature Medicine 23, 1244. [DOI] [PubMed] [Google Scholar]
- 106.Lee D-K, et al. Optimizing Combination Therapy for Acute Lymphoblastic Leukemia Using a Phenotypic Personalized Medicine Digital Health Platform. Journal of Laboratory Automation 0, 2211068216681979 [DOI] [PubMed] [Google Scholar]
- 107.Wong PK, et al. (2008) Closed-loop control of cellular functions using combinatory drugs guided by a stochastic search algorithm. Proceedings of the National Academy of Sciences 105, 5105–5110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Honda Y, et al. (2013) Guiding the osteogenic fate of mouse and human mesenchymal stem cells through feedback system control. 3, 3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xianting D, et al. (2014) Discovery of a low order drug-cell response surface for applications in personalized medicine. Physical Biology 11, 065003 [DOI] [PubMed] [Google Scholar]
- 110.Mohd Abdul Rashid MB, et al. (2015) Identification and Optimization of Combinatorial Glucose Metabolism Inhibitors in Hepatocellular Carcinomas. Journal of Laboratory Automation 20, 423–437 [DOI] [PubMed] [Google Scholar]
- 111.Weiss A, et al. (2015) Rapid optimization of drug combinations for the optimal angiostatic treatment of cancer. Angiogenesis 18, 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu Q, et al. (2015) Preclinical optimization of a broad-spectrum anti-bladder cancer tri-drug regimen via the Feedback System Control (FSC) platform. Scientific Reports 5, 11464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Silva A, et al. (2016) Output-driven feedback system control platform optimizes combinatorial therapy of tuberculosis using a macrophage cell culture model. Proceedings of the National Academy of Sciences 113, E2172-E2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee B-Y, et al. (2017) Drug regimens identified and optimized by output-driven platform markedly reduce tuberculosis treatment time. Nature Communications 8, 14183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jha S and Topol EJ (2018) Information and Artificial Intelligence. Journal of the American College of Radiology 15, 509–511 [DOI] [PubMed] [Google Scholar]
- 116.Al-Shyoukh I, et al. (2011) Systematic quantitative characterization of cellular responses induced by multiple signals. BMC Systems Biology 5, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rashid MBMA, et al. (2018) Optimizing drug combinations against multiple myeloma using a quadratic phenotypic optimization platform (QPOP). Science translational medicine 10, eaan0941 [DOI] [PubMed] [Google Scholar]
- 118.Ho D, et al. (2019) Artificial intelligence in nanomedicine. Nanoscale Horizons 4, 365–377 [DOI] [PubMed] [Google Scholar]
- 119.Ho D, et al. (2016) Diamonds, Digital Health, and Drug Development: Optimizing Combinatorial Nanomedicine. ACS Nano [DOI] [PubMed] [Google Scholar]
- 120.Wang H, et al. (2015) Mechanism-Independent Optimization of Combinatorial Nanodiamond and Unmodified Drug Delivery Using a Phenotypically Driven Platform Technology. ACS Nano 9, 3332–3344 [DOI] [PubMed] [Google Scholar]
- 121.Ho D, et al. (2015) Nanodiamonds: The intersection of nanotechnology, drug development, and personalized medicine. Science Advances 1, e1500439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shen Y, et al. Harnessing Artificial Intelligence to Optimize Long ‐ Term Maintenance Dosing for Antiretroviral‐Naive Adults with HIV‐1 Infection. Advanced Therapeutics [Google Scholar]
- 123.Ding X, et al. (2019) Harnessing an Artificial Intelligence Platform to Dynamically Individualize Combination Therapy for Treating Colorectal Carcinoma in a Rat Model. Advanced Therapeutics [Google Scholar]
- 124.Ho D and Zarrinpar A (2017) Making N-of-1 medicine a reality. SAGE Publications Sage CA: Los Angeles, CA: [DOI] [PubMed] [Google Scholar]
- 125.Pantuck AJ, et al. (2018) Modulating BET Bromodomain Inhibitor ZEN‐3694 and Enzalutamide Combination Dosing in a Metastatic Prostate Cancer Patient Using CURATE. AI, an Artificial Intelligence Platform. Advanced Therapeutics 1, 1800104 [Google Scholar]
- 126.Guan A, et al. (2017) Medical devices on chips. Nature Biomedical Engineering 1, 0045 [Google Scholar]
- 127.Zhang YS, et al. (2017) Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proceedings of the National Academy of Sciences 114, E2293-E2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shalek AK and Benson M (2017) Single-cell analyses to tailor treatments. Science Translational Medicine 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ting DS, et al. (2018) AI for medical imaging goes deep. Nature medicine 24, 539. [DOI] [PubMed] [Google Scholar]
- 130.Ardila D, et al. (2019) End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nature medicine 25, 954. [DOI] [PubMed] [Google Scholar]
- 131.Ngiam KY and Khor W (2019) Big data and machine learning algorithms for health-care delivery. The Lancet Oncology 20, e262-e273 [DOI] [PubMed] [Google Scholar]
- 132.Coudray N, et al. (2018) Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nature medicine 24, 1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Titano JJ, et al. (2018) Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat Med 24, 1337–1341 [DOI] [PubMed] [Google Scholar]
- 134.Huh D, et al. (2010) Reconstituting Organ-Level Lung Functions on a Chip. Science 328, 1662–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cassidy B, et al. (2018) Placental Drug Transport ‐ on ‐ a ‐ Chip: A Microengineered In Vitro Model of Transporter‐Mediated Drug Efflux in the Human Placental Barrier. Advanced Healthcare Materials 7, 1700786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.K.M.A., et al. Microfluidics‐Enabled Multimaterial Maskless Stereolithographic Bioprinting. Advanced Materials 0, 1800242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fang H, et al. (2017) Capacitively Coupled Arrays of Multiplexed Flexible Silicon Transistors for Long-Term Cardiac Electrophysiology. Nature biomedical engineering 1, 0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kim J, et al. (2017) Miniaturized Battery-Free Wireless Systems for Wearable Pulse Oximetry. Advanced Functional Materials 27, 1604373-n/a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rogers JA (2017) Nanomesh on-skin electronics. Nature Nanotechnology 12, 839. [DOI] [PubMed] [Google Scholar]
- 140.Tian X, et al. (2019) Wireless body sensor networks based on metamaterial textiles. Nature Electronics, 1 [Google Scholar]
- 141.Cao Y, et al. (2019) Self-healing electronic skins for aquatic environments. Nature Electronics 2, 75 [Google Scholar]
- 142.Zhao Y, et al. (2019) Design and applications of stretchable and self-healable conductors for soft electronics. Nano convergence 6, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao Y, et al. (2019) Highly conductive 3D metal-rubber composites for stretchable electronic applications. APL Materials 7, 031508 [Google Scholar]
- 144.Djenizian T, et al. (2019) Advances in flexible and soft electronics. AIP Publishing [Google Scholar]
- 145.Gao W, et al. (2017) Wearable Physiological Systems and Technologies for Metabolic Monitoring. Journal of Applied Physiology [DOI] [PubMed] [Google Scholar]
- 146.Emaminejad S, et al. (2017) Autonomous sweat extraction and analysis applied to cystic fibrosis and glucose monitoring using a fully integrated wearable platform. Proceedings of the National Academy of Sciences 114, 4625–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Agrawal P, et al. (2018) Real-World Data from the MiniMed™ 670G System Commercial Launch. Diabetes 67(Supplement 1). 10.2337/db18-960-P [DOI] [Google Scholar]
- 148.Gao Y, et al. (2017) Wearable Microfluidic Diaphragm Pressure Sensor for Health and Tactile Touch Monitoring. Advanced Materials 29, 1701985-n/a [DOI] [PubMed] [Google Scholar]
- 149.Xi W, et al. (2017) Soft tubular microfluidics for 2D and 3D applications. Proceedings of the National Academy of Sciences 114, 10590–10595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gao W, et al. (2016) Fully integrated wearable sensor arrays for multiplexed in situ perspiration analysis. Nature 529, 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wallace A, et al. (2015) Psychometric properties of a mobile neurocognitive assessment tool. Alzheimer’s & Dementia: The Journal of the Alzheimer’s Association 11, P703 [Google Scholar]
- 152.Bangert AS and Balota DA (2012) Keep up the pace: Declines in simple repetitive timing differentiate healthy aging from the earliest stages of Alzheimer’s disease. Journal of the International Neuropsychological Society : JINS 18, 1052–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Calamia M, et al. (2013) The Robust Reliability of Neuropsychological Measures: Meta-Analyses of Test–Retest Correlations. The Clinical Neuropsychologist 27, 1077–1105 [DOI] [PubMed] [Google Scholar]
- 154.Hassenstab J, et al. (2015) Absence of Practice Effects in Preclinical Alzheimer’s Disease. Neuropsychology 29, 940–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rast P, et al. (2012) Intensive Measurement Designs for Research on Aging. GeroPsych 25, 45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.J.S.M. (2008) Measurement‐Burst Designs for Social Health Research. Social and Personality Psychology Compass 2, 245–261 [Google Scholar]
- 157.Duchek JM, et al. (2013) Relationship Between Stroop Performance and Resting State Functional Connectivity in Cognitively Normal Older Adults. Neuropsychology 27, 10.1037/a0033402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Prasad V (2017) Tisagenlecleucel — the first approved CAR-T-cell therapy: implications for payers and policy makers. Nature Reviews Clinical Oncology [DOI] [PubMed] [Google Scholar]
- 159.Roberts ZJ, et al. (2017) Axicabtagene ciloleucel, a first-in-class CAR T cell therapy for aggressive NHL. Leukemia & Lymphoma, 1–12 [DOI] [PubMed] [Google Scholar]
- 160.(2017) FDA Approves Second CAR T-cell Therapy. Cancer Discovery [DOI] [PubMed] [Google Scholar]
- 161.Monjezi R, et al. (2016) Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors. Leukemia 31, 186. [DOI] [PubMed] [Google Scholar]
- 162.Sadelain M, et al. (2017) Therapeutic T cell engineering. Nature 545, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cooper ML, et al. (2018) An “off-the-shelf” fratricide-resistant CAR-T for the treatment of T cell hematologic malignancies. Leukemia [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ruella M and Kenderian SS (2017) Next-Generation Chimeric Antigen Receptor T-Cell Therapy: Going off the Shelf. BioDrugs 31, 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.(2018) Kite, Sangamo Partner on Gene-Edited Cell Therapies. Cancer Discovery 8, 379–380 [DOI] [PubMed] [Google Scholar]
- 166.Peruzzotti-Jametti L, et al. (2018) Macrophage-Derived Extracellular Succinate Licenses Neural Stem Cells to Suppress Chronic Neuroinflammation. Cell Stem Cell 22, 355–368.e313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ma H, et al. (2018) Establishment of human pluripotent stem cell-derived pancreatic β-like cells in the mouse pancreas. Proceedings of the National Academy of Sciences 115, 3924–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Saxena N, et al. (2018) Mitochondrial Donation: A Boon or Curse for the Treatment of Incurable Mitochondrial Diseases. Journal of Human Reproductive Sciences 11, 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Danino T, et al. (2015) Programmable probiotics for detection of cancer in urine. Science Translational Medicine 7, 289ra284–289ra284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Courbet A, et al. (2015) Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Science Translational Medicine 7, 289ra283–289ra283 [DOI] [PubMed] [Google Scholar]
- 171.Schukur L, et al. (2015) Implantable synthetic cytokine converter cells with AND-gate logic treat experimental psoriasis. Science Translational Medicine 7, 318ra201–318ra201 [DOI] [PubMed] [Google Scholar]
- 172.Sun Y and Soh S (2015) Printing Tablets with Fully Customizable Release Profiles for Personalized Medicine. Advanced Materials 27, 7847–7853 [DOI] [PubMed] [Google Scholar]
- 173.McHugh KJ, et al. (2017) Fabrication of fillable microparticles and other complex 3D microstructures. Science 357, 1138–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tai W, et al. (2014) Folding graft copolymer with pendant drug segments for co-delivery of anticancer drugs. Biomaterials 35, 7194–7203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.von Maltzahn G, et al. (2011) Nanoparticles that communicate in vivo to amplify tumour targeting. Nat Mater 10, 545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Deng ZJ, et al. (2013) Layer-by-Layer Nanoparticles for Systemic Codelivery of an Anticancer Drug and siRNA for Potential Triple-Negative Breast Cancer Treatment. ACS Nano 7, 9571–9584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ding X, et al. (2017) Effective drug combination for Caenorhabditis elegans nematodes discovered by output-driven feedback system control technique. Science Advances 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lee D-K, et al. (2017) Clinical validation of a nanodiamond-embedded thermoplastic biomaterial. Proceedings of the National Academy of Sciences 114, E9445-E9454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Blaus A, et al. (2015) Personalized Cardiovascular Medicine Today. A Food and Drug Administration/Center for Drug Evaluation and Research Perspective 132, 1425–1432 [DOI] [PubMed] [Google Scholar]
- 180.Vicini P, et al. (2016) Precision medicine in the age of big data: The present and future role of large-scale unbiased sequencing in drug discovery and development. Clinical Pharmacology & Therapeutics 99, 198–207 [DOI] [PubMed] [Google Scholar]
- 181.Fang H, et al. FDA drug labeling: rich resources to facilitate precision medicine, drug safety, and regulatory science. Drug Discovery Today [DOI] [PubMed] [Google Scholar]
- 182.Hyman DM, et al. (2015) Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. New England Journal of Medicine 373, 726–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chaudhuri SE, et al. (2018) Patient-centered clinical trials. Drug Discovery Today 23, 395–401 [DOI] [PubMed] [Google Scholar]
- 184.Das S and Lo AW (2017) Re-inventing drug development: A case study of the I-SPY 2 breast cancer clinical trials program. Contemporary Clinical Trials 62, 168–174 [DOI] [PubMed] [Google Scholar]
- 185.Wong CH, et al. (2018) Estimation of clinical trial success rates and related parameters. Biostatistics, kxx069-kxx069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Altman RB, et al. (2016) A research roadmap for next-generation sequencing informatics. Science Translational Medicine 8, 335ps310–335ps310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.van der Gronde T, et al. (2017) Addressing the challenge of high-priced prescription drugs in the era of precision medicine: A systematic review of drug life cycles, therapeutic drug markets and regulatory frameworks. PLoS ONE 12, e0182613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.M G (2012) The Innovative Medicines Initiative: A European Response to the Innovation Challenge. Clinical Pharmacology & Therapeutics 91, 418–425 [DOI] [PubMed] [Google Scholar]
- 189.Kohane IS (2015) Ten things we have to do to achieve precision medicine. Science 349, 37–38 [DOI] [PubMed] [Google Scholar]
- 190.Rosenberg NA, et al. (2010) Genome-wide association studies in diverse populations. Nature reviews. Genetics 11, 356–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Popejoy AB and Fullerton SM (2016) Genomics is failing on diversity. Nature 538, 161–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.CARLSON B (2012) Vanderbilt pioneers bedside genetics. Biotechnology healthcare 9, 31. [PMC free article] [PubMed] [Google Scholar]
- 193.Wells QS, et al. (2019) Accelerating Biomarker Discovery Through Electronic Health Records, Automated Biobanking, and Proteomics. Journal of the American College of Cardiology 73, 2195–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Weitzel KW, et al. (2016) The IGNITE network: a model for genomic medicine implementation and research. BMC Medical Genomics 9, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Orlando LA, et al. (2017) Developing a common framework for evaluating the implementation of genomic medicine interventions in clinical care: the IGNITE Network’s Common Measures Working Group. Genetics In Medicine [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sperber NR, et al. (2017) Challenges and strategies for implementing genomic services in diverse settings: experiences from the Implementing GeNomics In pracTicE (IGNITE) network. BMC Medical Genomics 10, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cavallari LH, et al. (2017) The IGNITE Pharmacogenetics Working Group: An Opportunity for Building Evidence with Pharmacogenetic Implementation in a Real‐World Setting. Clinical and Translational Science 10, 143–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Cavallari LH, et al. (2017) Multisite Investigation of Outcomes With Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy After Percutaneous Coronary Intervention. JACC: Cardiovascular Interventions [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chien S, et al. (2015) Engineering as a new frontier for translational medicine. Science Translational Medicine 7, 281fs213–281fs213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kurpinski K, et al. (2014) Mastering Translational Medicine: Interdisciplinary Education for a New Generation. Science Translational Medicine 6, 218fs212–218fs212 [DOI] [PubMed] [Google Scholar]
- 201.Reddy S, et al. (2019) A governance model for the application of AI in health care. Journal of the American Medical Informatics Association [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cohen J (2019) Did CRISPR help—or harm—the first-ever gene-edited babies?, Science [Google Scholar]
- 203.Dinh-Le C, et al. (2019) Wearable Health Technology and Electronic Health Record Integration: Scoping Review and Future Directions. JMIR mHealth and uHealth 7, e12861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tafuri WL, et al. (1988) Dogs as experimental models for the study of the natural course of Chagas disease. Revista da Sociedade Brasileira de Medicina Tropical 21, 77. [DOI] [PubMed] [Google Scholar]
- 205.Lee SS-J, et al. (2019) Ethics of inclusion: Cultivate trust in precision medicine. Science 364, 941–942 [DOI] [PMC free article] [PubMed] [Google Scholar]