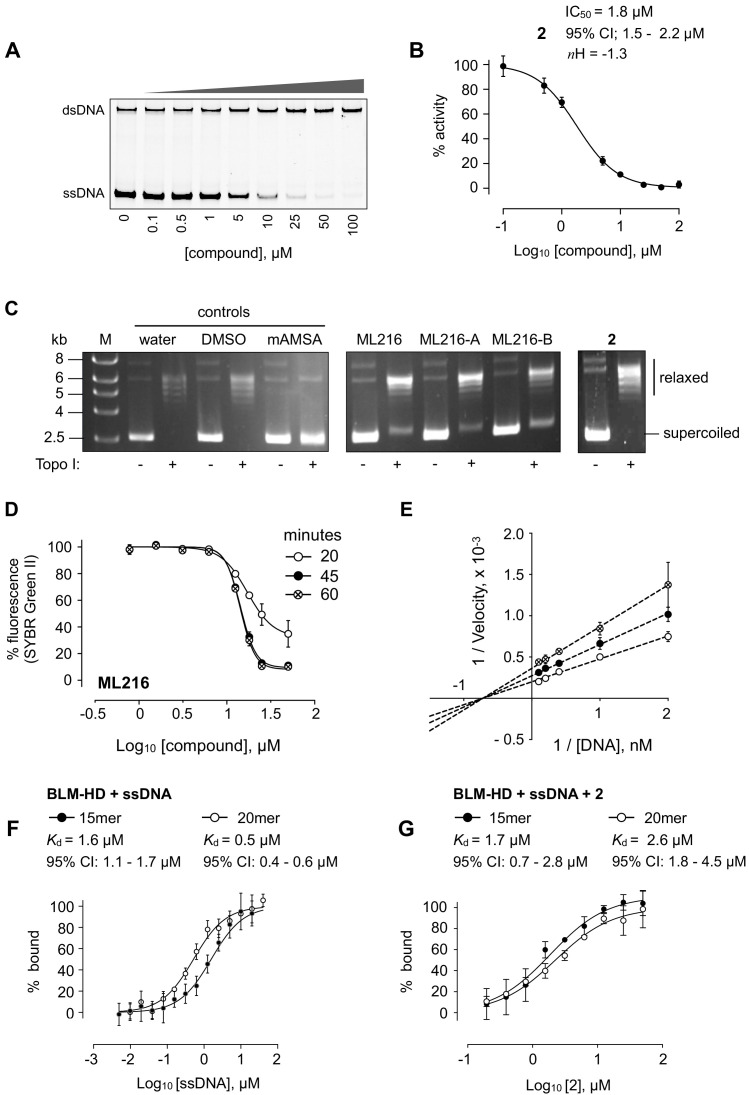
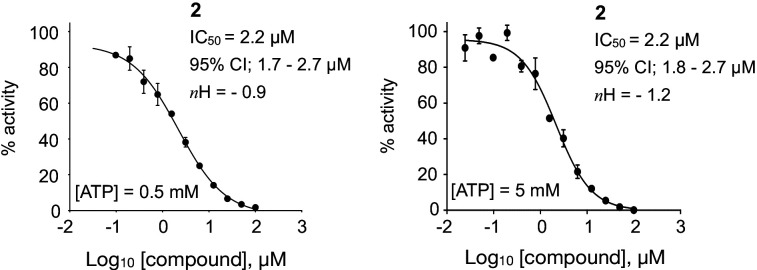
Figure 2. DNA interaction assays.
(A) Titration of BLM-HD with 2 prevents the unwinding of a forked-50mer dsDNA substrate into its component strands, as judged by native gel electrophoresis. (B) Quantification of inhibitory activity by 2 in the gel-based activity assay. Experimental data were fitted with a four parameter, log(inhibitor) vs. response model with variable slope. Calculated values for IC50, nH and 95% CI are given in each case. (C) Representative results from a Topoisomerase I (Topo I) DNA-unwinding assay. M = molecular mass maker; DMSO = buffer supplemented with dimethyl sulfoxide control; mAMSA = mAmsacrine; ML216, ML216-A, ML216-B = refer to the three independent sources of the compound as described in the main text of the manuscript (D) Dose response curves from SYBR-Green II dye displacement assays, using a forked-50mer DNA duplex incubated with ML216 for a period of 20 (open circles), 45 (filled circles), and 60 min (crossed circles). Fitted lines are intended as visual aids only. (E) Lineweaver-Burk plot for data generated at three compound concentrations (0, 5, and 10 µM) in a colourimetric ATP turnover assay. Linear regression produces an intercept of all data on the X-axis indicating that 2 is a non-competitive inhibitor (i.e. same Km, altered Vmax parameter). (F, G) Binding isotherms for binding of BLM-HD to ssDNA-15mer and −20mer, or to compound 2 in the presence of either oligonucleotide, as determined by microscale thermophoresis (MST). Experimental data were fitted with a one-site, specific binding model. Values for Kd and 95% CI are given in each case. For all plots, data represent the mean of three technical replicates with error bars representing 1 SD.