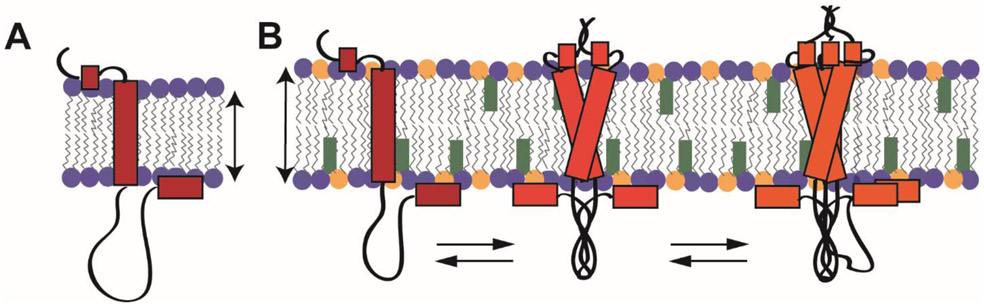
Abstract
How the distinctive lipid composition of mammalian plasma membranes impacts membrane protein structure is largely unexplored, partly because of the dearth of isotropic model membrane systems that contain abundant sphingolipids and cholesterol. This gap is addressed by showing that sphingomyelin and cholesterol-rich (SCOR) lipid mixtures with phosphatidylcholine can be co-solubilized by n-dodecyl-β-melibioside to form bicelles. Small angle X-ray and neutron scattering, as well as cryo-electron microscopy demonstrate that these assemblies are stable over a wide range of conditions and exhibit the bilayered-disc morphology of ideal bicelles even at low lipid-to-detergent mole ratios. SCOR bicelles are shown to be compatible with a wide array of experimental techniques, as applied to the transmembrane human amyloid precursor C99 protein in this medium. These studies reveal an equilibrium between low order oligomer structures that differ significantly from previous experimental structures of C99, providing an example of how ordered membranes alter membrane protein structure.
Graphical Abstract
INTRODUCTION
The plasma membrane (PM) of mammalian cells and related endosome and lysosome membranes is distinguished from other organellar membranes by its high levels of both cholesterol (Chol) and sphingolipids, lipids that are well known to undergo unusually strong lipid-lipid interactions in the membrane that result in local membrane clustering and ordering.1 The observation of local ordering of membrane lipids has led to the lipid raft hypothesis, which posits that phase separation can occur in PMs, resulting in Chol and sphingolipid-rich ordered phase nanodomains (“lipid rafts”) surrounded by disordered phase bilayers.2-4 While the lipid raft hypothesis has been controversial,5 there is no question that PMs are rich in Chol and sphingolipids.6 To test the impact of the unique mammalian PM composition on protein structure and dynamics, there is a need for model membranes that are rich in both Chol and sphingolipids that can be used in biochemical and biophysical studies of isolated membrane proteins and complexes. However, except for bilayered lipid vesicles (liposomes), no such model membrane system has previously been available, mainly due to difficulties in co-solubilizing Chol and sphingolipids to form mixed micelles, bicelles, nanodiscs, or lipodisqs (the latter also known as SMALPs).
Both sphingolipid-rich bicelles and Chol-rich bicelles have been reported,7-10 but not bicelles that are rich in both these lipids. Chol-rich “native” eukaryotic bicelles have been used in studies of peptide structures, but were not morphologically characterized and contained only ~ 1 mole % sphingomyelin (SM).11 Reinforcing the idea of poor SM/Chol co-solubility, recent work by Kot et al. showed that SM and Chol mixtures are recalcitrant to the detergents traditionally used to form bicelles.12 Peptide-based nanodiscs have been used to co-solubilize lipid mixtures containing SM and Chol, but only to 7 and 4 mol % respectively.13 The difficulty of dissolving sphingolipid-Chol mixtures may be related to energetically-favorable complex formation between these two lipids.14-15 In this work we set out to develop a model membrane system that is rich in both SM and Chol and involves the formation of assemblies small enough to be monodisperse and optically clear, such that they can be employed in a wide range of biophysical and biochemical applications requiring these properties. Here, we describe the development and characterization of SM and Chol-rich SCOR bicelles. To demonstrate the utility of this new model membrane system we also report reconstitution and biophysical characterization of the transmembrane C99 domain of the human amyloid precursor protein (APP) into SCOR bicelles. It is shown that this protein exhibits structural properties in this medium that are distinct from what has previously been observed in more conventional model membrane media.
RESULTS
Screening for a Detergent That Can Solubilize Phosphatidylcholine-Sphingolipid-Chol Mixtures
A screen of over 200 mixtures was conducted in search for a detergent that can co-solubilize Chol and SM mixtures in the presence of phosphatidylcholine at a total detergent:lipid of 3:1 molar ratio (q = 0.33), with results summarized in Table S1. The choice of lipid mixtures tested was heavily weighted towards lipid mixtures that either are known to form ordered phase bilayers at near-physiological temperatures or are predicted to do so. Among the cross-section of 35 detergents tested we found a number that could solubilize either Chol or SM. However, only the recently introduced16 n-dodecyl-β-melibioside (DDMB) was successful at solubilizing mixtures of various forms of phosphatidylcholine with both Chol and egg SM (eSM). eSM is predominantly (~86 %) composed of the saturated C16-amidated form (https://avantilipids.com/product/860061) and was used instead of synthetic SM because eSM is much less expensive. It is notable that DDMB, with its melibiose (galactose-α(1→6)-D-glucose) head group, was effective, while n-dodecyl-β-maltoside, with its less flexible glucose-α(1→4)-D-glucose headgroup was not. DDMB was seen to be able to solubilize a range of phospholipid-Chol-eSM mixtures. This study focused on mixtures with a q ratio of 1.0 or below, where the mixtures were optically clear and the assemblies tumbled isotropically. We chose DMPC as the glycerophospholipid for the SCOR bicelles focused on in this work because of its extensive prior use in well-characterized classical bicelle mixtures, including work involving our test protein for this study, C99.17-27 Accordingly, there is a wealth of previous data for classical DMPC-based bicelles that can be compared to our results for the new SCOR bicelles.
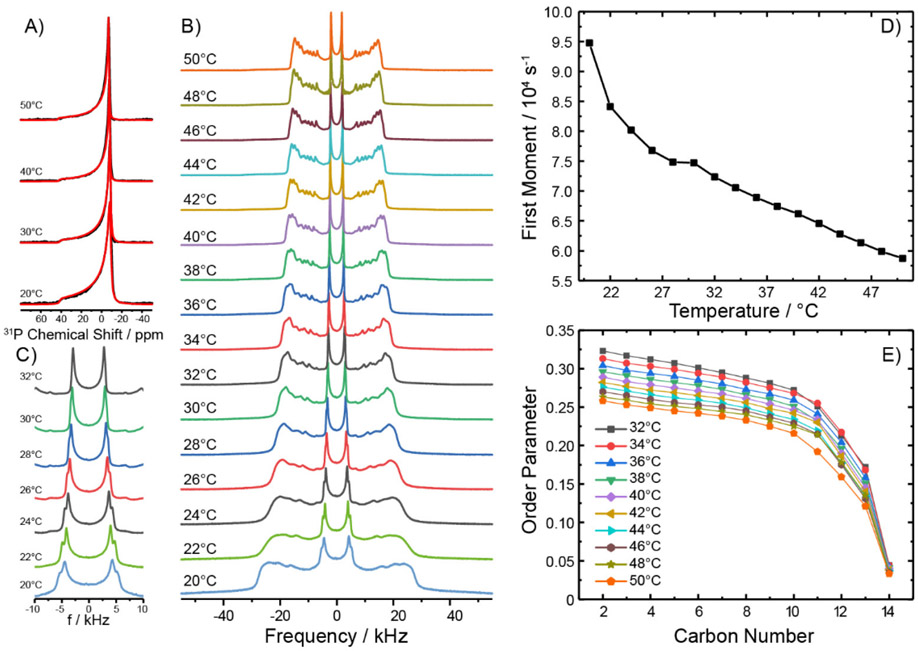
Solid-State NMR Characterization of 4:2:1 DMPC:eSM:Chol Bilayered Vesicles
From the numerous mixtures tested (Table S1) we selected SCOR bicelles composed of DDMB and 4:2:1 DMPC:eSM:Chol to focus on in this study. Because detergent-free 4:2:1 DMPC:eSM:Chol lipid bilayers have not previously been characterized, we examined vesicles of this composition using solid-state NMR spectroscopy. The goal was to illuminate the bilayer phase properties of the parent lipid mixture used for the SCOR bicelles. First, static 31P NMR spectra were acquired as a function of temperature, as shown in Figure 1A. All spectra for the 20 to 50°C temperature range exhibited a single powder pattern characterized by an axially symmetric chemical shift anisotropy tensor spanning ~50 ppm, in which the individual 31P NMR contributions of DMPC and eSM are indistinguishable. Such an NMR powder pattern lineshape is characteristic of lipids undergoing rapid axially symmetric motional averaging, as is typical for lamellar membranes in either the ordered or disordered phase but not the gel phase.28-29 Next, 2H NMR spectra of the mixture containing chain-perdeuterated DMPC-d54 were collected as a function of temperature (Figure 1B). The typical superposition of quadrupolar Pake powder patterns from all deuterons along the myristoyl chains is observed, where each quadrupolar splitting provides information on the degree of molecular order of a particular methylene or methyl segment in the lipid chain. Close inspection of the innermost (and most intense) terminal methyl group splitting reveals the appearance of a second Pake doublet at temperatures below 30°C (Figure 1C). This indicates that DMPC populates two different environments (as would be the case if DMPC is distributed between two different phases).30 No spectral features indicative of a gel (Pβ) or solid-ordered (So) phase were observed even at 20°C, where pure DMPC membranes are well known to form a gel phase. As expected, the widths of the 2H NMR quadrupolar Pake doublets decreased with increasing temperature due to increased motional disorder of the C-D bond vectors along the chain, as also seen in a plot of the first spectral moment as a function of temperature (Figure 1D). The slight discontinuity seen at a temperature of ~30°C coincides with the spectral changes observed for the methyl groups, also consistent with a transition from the ordered Lo phase to the disordered Ld phase above this temperature.29 Above 30°C, the spectral resolution is sufficient for dePakeing31 the 2H NMR spectra, resulting in the order parameter profiles along the carbon chains shown in Figure 1E. These profiles are consistent with a somewhat disordered phase state at temperatures above 30°C. As temperature increases, the order in the acyl chains decrease as well.
Figure 1. Static 31P and 2H NMR spectra of fully hydrated DMPC-d54/eSM/Chol (4/2/1 mol/mol/mol) vesicles at various temperatures.
A) Static 31P NMR spectra exhibiting powder patterns reflecting axially symmetric motional averaging of the lipid in the Lo phase at low and Ld phase at high temperatures. The widths of the 31P NMR spectra exclude that DMPC is in a gel phase. Red lines indicate best fit simulations of the spectral line shape. Samples were observed to be slightly field-oriented. Spectra were simulated using an ellipsoidal distribution function with aspect ratios of 0.65 – 0.75. B) 2H NMR spectra of chain perdeuterated DMPC-d54 in the ternary lipid mixture as a function of temperature. C) Enlarged view of the chain-terminal methyl group quadrupolar splitting of the 2H NMR spectra shown in B, which highlights two sets of quadrupolar splitting at temperatures from 20 to 30°C. D) Plot of the 1st spectral moment of the 2H NMR spectra shown in B as a function of temperature. E) DMPC chain order parameters as a function of chain position in the ternary mixture at various temperatures, as determined after dePakeing the NMR spectra shown in panel B.
It has previously been seen that lipids in the Lo phase share a property with those in the Ld phase by undergoing unhindered lateral diffusion and long axis rotational diffusion around the bilayer normal.30, 32 However, the Lo phase state differs from Ld by featuring a dramatic decrease in the number of gauche dihedral angles along the hydrocarbon chains due to strong ordering interactions imposed by proximal Chol.30, 32 This trend is reflected by the wider span of the 2H NMR spectra below 30°C relative to spectra from Ld phase bilayers, as was observed here. The population of gauche conformers can be estimated from the average DMPC chain length determined from each 2H NMR spectrum.33-34 Each gauche excursion reduces the chain length by 1.1 Å.35 At 20°C, the maximum quadrupolar splitting of DMPC in the mixture is 56 kHz, which is close to the maximum of 62.6 kHz, indicating fully extended all trans chains undergoing axially symmetric reorientation along the long axis of DMPC, as expected for an ideal Lo phase state. The average chain order parameter of DMPC-d54 in the mixture at 20°C can roughly be determined to be <S> = 0.36 given the broad nature of the 2H NMR spectrum at this temperature. From the average order parameter of 0.36, the average length of DMPC chains in the mixture was estimated to be 13.1 Å, which can be compared to 15.2 Å for an all trans chain. This suggests that the DMPC chains in the mixture exhibit on average ~2 gauche defects at 20°C. At temperatures above 30°C, the chain length can be determined precisely from the dePaked spectra, yielding chain lengths varying from 11.6 Å at 32°C to 10.7 Å at 50°C. This would correspond to 3.3 to 4.1 gauche defects, which is in agreement with the population of the liquid disordered (Ld) phase state in that temperature range. These results therefore suggest that the 4:2:1 DMPC:eSM:Chol mixture occupies an Lo-like phase state between 20 and 30°C, and the Ld state above 30°C—the transition between these phases is somewhat gradual.
The conclusion that the phase occupied by 4:2:1 DMPC:eSM:Chol at lower temperatures is Lo-like, but not to an ideal extreme, is not surprising. Pure Lo phase formation is most often described only at Chol contents higher than the 14 mol% of the current mixture.30, 36 To confirm that the 4:2:1 mixture truly occupies a Lo-like phase below 30°C we applied 1H MAS NMR to gain additional information. Figure S1 shows a series of 1H MAS NMR spectra of this mixture as a function of temperature. Particularly between 20 - 24°C, the observed NMR spectra closely resembles what has been defined as the pure Lo phase 1H NMR spectrum.36
Taken together, the solid-state NMR characterization of the DMPC/eSM/Chol (4:2:1 mol/mol/mol) mixture indicates the bilayers are in a Lo-like phase state at temperatures below ~30°C and that there is a transition to the Ld state above 30°C. The transition between these states is gradual. The fact that the mixture appears to have properties that are at the borderline between Lo and Ld phases over the 20-40°C temperature range is well suited for mimicking mammalian PMs, which also appear to exhibit properties that also are borderline between these two phases.37
Small Angle Scattering Characterization of SCOR Bicelles
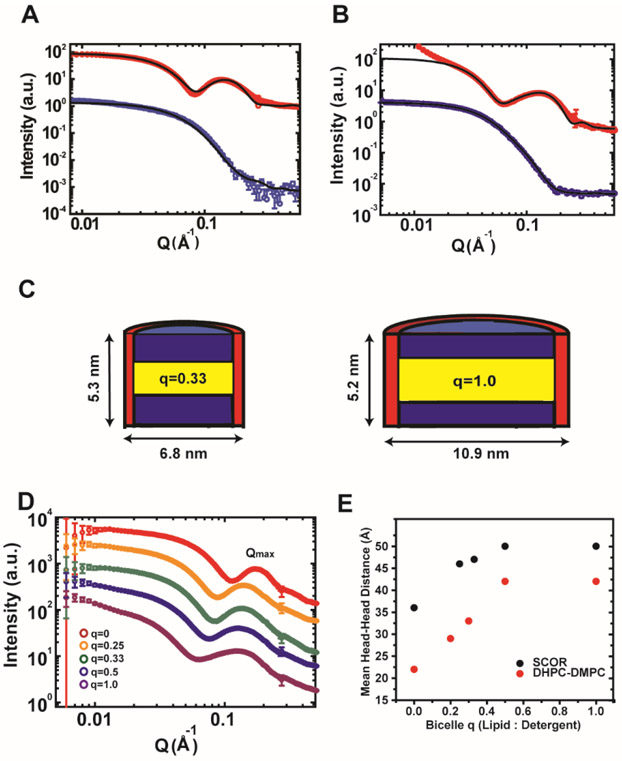
Small angle X-ray and neutron scattering (SAXS & SANS) methods are powerful tools for resolving the ensemble morphology of particles. We therefore used these methods to characterize the shape and structural organization of SCOR bicelles. Measurements were made for mixtures with lipid-to-DDMB q ratios (q is the lipid-to-detergent mole ratio) ranging from 0.25 to 1.0, a range of q in which the bicelles are small and monodisperse. Figure 2 shows that a simultaneous fit of the SAXS and SANS scattering curves with a core shell bicelle model (Figure S2) for both q = 0.33 and 1.0 SCOR bicelles supports the ideal bicelle model in which a discoidal lipid bilayer region is surrounded by a toroid of DDMB detergent. It is noted that the deviation from fit at Q < 0.02 Å−1 in Figure 2B (SAXS data where Q is the scattering vector in SAXS and SANS) was likely due to the presence of air bubbles, but that this did not impact the best fitting parameters, as the simultaneous fit with both SAXS and SANS data greatly reduced the uncertainty. Over q ratios of 0.25 to 1.0, SCOR bicelles were seen to conform to the classical detergent edge-stabilized bilayer disc model with a near constant bilayer thickness of 4.7 nm at q = 0.25 to 5.3 nm at q = 1.0, and with disc diameters ranging from ~7 nm at q = 0.25 to ~11 nm at q = 1.0 (Figures 2C and S2).
Figure 2. Small angle X-ray and neutron scattering of SCOR bicelles.
A) Small-angle scattering measurement for q= 0.33 SCOR bicelles at a total lipid+detergent concentration of 0.5 wt %, a concentration at which bicelles were confirmed to persist (see later section of Results devoted to dynamic light scattering). Fully protonated SCOR bicelles were prepared in H2O (SAXS) or in 100 % D2O (SANS), with scattering results shown in red and blue, respectively. B) Small angle scattering measurements for fully protonated q=1.0 SCOR bicelles at 0.5% w/v prepared in H2O (SAXS) or in 100 % D2O (SANS), shown in red and blue respectively. C) Models of SCOR bicelles at q=0.33 and q=1.0 (left and right, respectively) based on small angle scattering core-shell bicelle model fitting of panels A and B (see Figure S2 and the Materials and Methods for a description of the model). D) SAXS scattering curves for 0.5 wt % DDMB micelles (red) and for SCOR bicelles at lipid:detergent mole (q) ratios of 0.25 (orange), 0.33 (green), 0.5 (blue), and 1.0 (purple) buffered with 10 mM imidazole pH 6.5, 0.1 mM EDTA, and 0.3 mM DDMB. The Qmax peak is around ~ 0.12. E) Mean headgroup-to-headgroup distance determined from scattering results shown in panel D using Qmax ratios for classical and SCOR bicelles at various q ratios. The classical bicelle—DMPC/DHPC—ratios are from Caldwell et al.39
Due to the higher electron density of the phosphate groups, SAXS can provide model-free information about headgroup-to-headgroup distances in bicelles (along the axis orthogonal to the plane of the bicelle) via analysis of the second maxima, referred to as the QMax (Figure 2D).38 As highlighted in Figure 2E, the mean headgroup-to-headgroup distance in the SCOR bicelles increases from 4.6 nm at q = 0.25 to 5.0 nm at q = 1.0 (~ 9 %), while for DMPC-DHPC bicelles the mean headgroup-to-headgroup distance increases by ~44% over a very similar q range.39 Mean headgroup-to-headgroup distances are understood to approximate bilayer thickness and the SCOR bicelles are seen to exhibit only a slight increase in bilayer thickness as a function of q. The SCOR bilayer thickness of ~5.0 nm at q = 1.0 is significantly larger than the ~4.3 nm bilayer thickness of pure DMPC.39 This increase is likely because Chol increases the bilayer thickness, as it is known to do so in DMPC vesicles.40 The thicker SCOR bilayer is also consistent with the higher order of the bilayered domain of the SCOR bicelles, indicating a higher proportion of trans conformations for the acyl chains of DMPC and eSM, as corroborated by our solid state 2H NMR results for vesicles. The decay transition from the low-Q intensity plateau moves toward lower Q values, suggesting that the size of bicelles increases with higher q ratios, as expected. In summary, combined SAXS and SANS results demonstrate that SCOR bicelles have the bilayered disc morphology of ideal bicelles with a ~5 nm thick bilayer core.
Cryo-Electron Microscopy Visualization of SCOR Bicelles
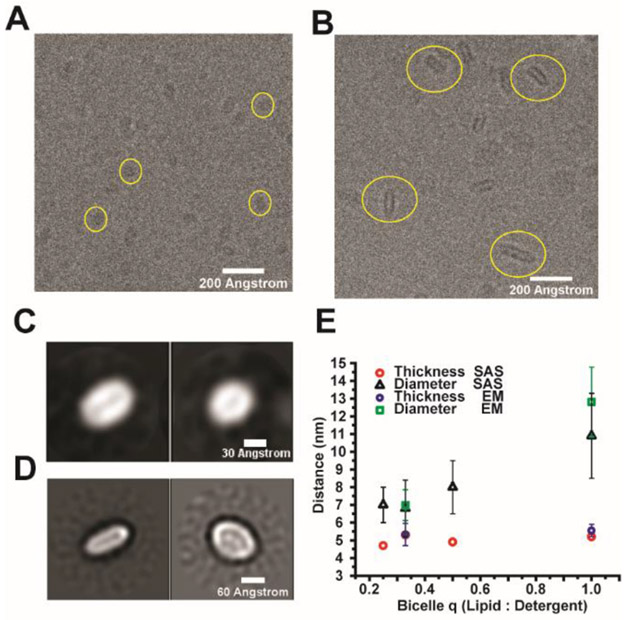
We used transmission cryo-electron microscopy (cryo-EM) on SCOR bicelles frozen in vitreous ice to see if it was possible to obtain direct visualization of bicelle discs. Figures 3A and B show cryo-EM micrographs for q = 0.33 and 1.0 SCOR bicelles.
Figure 3. Cryo-Electron microscopy of SCOR bicelles.
A) Contrast-enhanced cryo-EM image of q= 0.33 SCOR bicelles with yellow circles highlighting individual particles. B) Contrast-enhanced cryo-EM image of q= 1.0 SCOR bicelles with yellow circles highlighting individual particles. Samples in A and B are 0.64 wt % bicelle samples (at both q = 0.33 and q = 1.0) in 75 mM NaCl, 1 mM EDTA, 25 mM sodium acetate, pH 4.5. C) and D) are orthogonal views of class averages for the q = 0.33 and q = 1.0 bicelles, respectively. q=0.33 samples contained the C99 protein at a ~ 2:3 protein-to-bicelle ratio. C99 is not observed owing to its small size. E) Comparison of SCOR bilayer thickness and diameter as determined from cryo-EM (n=50), and small angle scattering fits.
The cryo-EM micrographs did not reveal any self-association of the bicelle assemblies. SCOR bicelles exhibit a hollow discoidal shape reflecting their electron-poor hydrocarbon interior and electron-rich head group surface. Bicelle image populations were sufficiently uniform to be picked and class-averaged, with orthogonal views of the same reconstructions being shown for q = 0.33 and q = 1.0 bicelles in Panels 3C and 3D, respectively. As can be seen, for both compositions the discoidal morphology is confirmed. Moreover the dimensions seen for both the q = 0.33 (thickness of 5.3 ± 0.7 nm, diameter of 6.9 ± 0.8 nm) and q = 1.0 (thickness of 5.5 ± 0.4 nm, diameter of 12.8 ± 1.9 nm) bicelles are in good agreement with the SAXS and SANS results (Figure 3E). It is important to note that the q = 0.33 SCOR bicelle sample, (Figures 3A and C), contained the C99 protein, a small single-span membrane protein (see later section of Results). We could not resolve any protein density, which was as expected given the small (14 kDa) size of our C99 construct and its dynamic oligomeric state (see below). These results confirm the conclusions of the scattering experiments and demonstrated that SCOR bicelles are well-behaved in vitrified conditions, suggesting that they may be suitable for use in cryo-EM studies of larger integral membrane proteins and their complexes, especially those with less dynamic structures. We used the q = 0.33 SCOR bicelles for the C99 studies described below and estimated the aggregate molecular weight of q = 0.33 SCOR bicelles to be around 100 kD with roughly 185 lipid and detergent molecules in each bicelle (see Materials and Methods).
Use of Dynamic Light Scattering to Test the Resilience of SCOR Bicelles to Changes in Concentration, pH, Salt and Temperature.
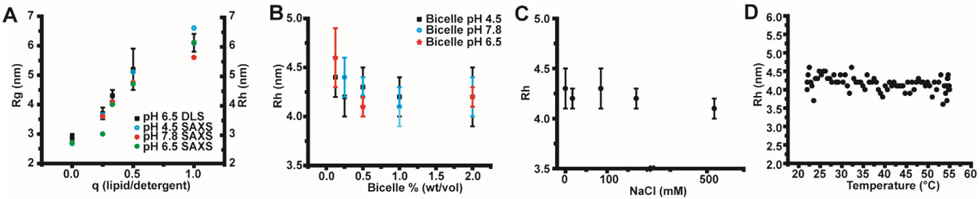
Dynamic light scattering was employed as a convenient way of exploring the range of conditions in which the integrity of SCOR bicelles persists (Figure 4). Figure 4A shows that the radius of hydration (Rh) determined from dynamic light scattering is in general agreement with the radius of gyration (Rg) determined by SAXS from the Guinier region. It was also seen that SCOR bicelle size is not affected by bicelle concentration (total lipid + detergent) above 0.25 wt%, but does exhibit a slight increase in radius of hydration when the total bicelle concentration is reduced to 0.125 wt% (Figure 4B). The persistence of SCOR bicelles at such low concentrations is likely due to the low critical micelle concentration (CMC) of DDMB (0.3 mM16) relative to the higher critical micelle concentration of the most commonly used detergent in conventional bicelles, dihexanoylphosphatidylcholine (DHPC, CMC of 10-15 mM41). It was also seen that SCOR bicelle size is not significantly affected by pH, ionic strength, or temperature. This robustness is well-suited for the possible utility of SCOR bicelles as a membrane mimetic medium (Figure 4B-D).
Figure 4. Dynamic light scattering of SCOR bicelles.
A) DLS-determined apparent hydrodynamic radii (Rh) values are compared to SAXS and SANS-determined apparent radius of gyration (Rg) values for SCOR bicelles at various q (lipid:detergent) ratios. Error bars reflect the standard deviation of Rh values determined from three repetitions of 10 DLS acquisitions for each q ratio. B) DLS-determined Rh for SCOR bicelles at a q of 0.33 for various pH values as a function of bicelle solution wt %. Bicelles at pH 4.5 were buffered with 10 mM sodium acetate, while bicelles at pH 6.5 and 7.8 were buffered with 10 mM imidazole. C) DLS-determined Rh values for SCOR bicelles at a q of 0.33 as a function of ionic strength. D) DLS-determined Rh values for SCOR bicelles at a q of 0.33 as a function of temperature. Temperature ramps to and from 22 to 55 °C were conducted at rate of 1 °C/minute.
Assignment of Backbone NMR Resonances for the Amyloid Precursor C99 Protein in SCOR Bicelles.
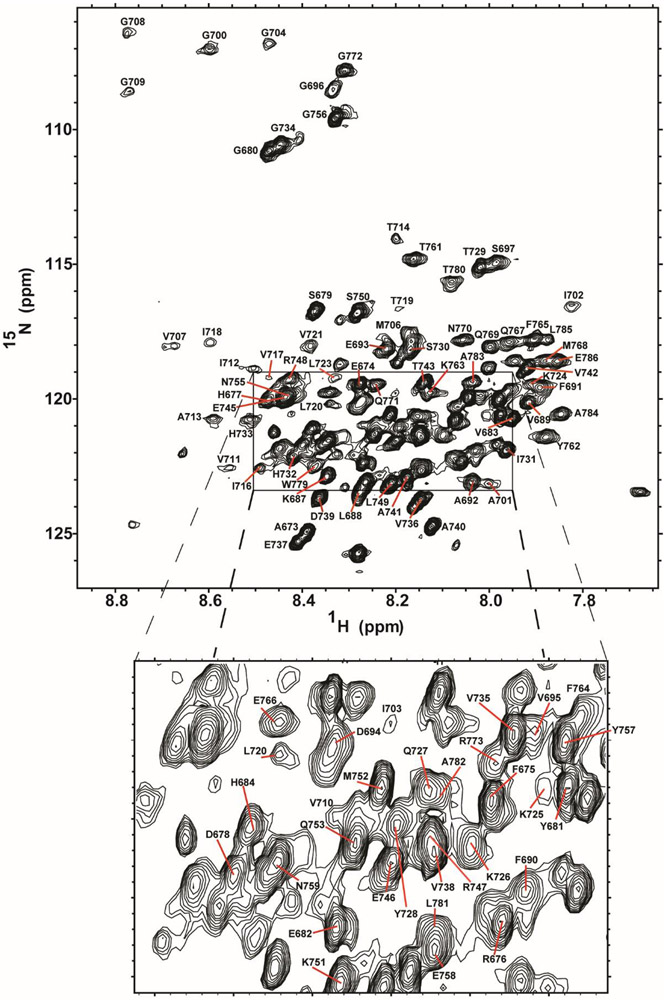
To demonstrate the utility of SCOR bicelles as a medium for biophysical studies of a membrane protein, we selected for study the 99-residue C-terminal domain of the human amyloid precursor protein (APP), the immediate precursor of the amyloid-beta polypeptides that are associated with Alzheimer’s disease. C99 has previously been subjected to structural studies in micelles, bicelles, and lipid vesicles.17-27 When initially screening SCOR bicelle lipid compositions for NMR studies of C99, we found that the 4:2:1 DMPC:eSM:Chol mixture at a q of 0.33 yielded 1H,15N-TROSY NMR spectra of reasonably high spectral quality, reminiscent of previous work with C99 in DHPC/DMPC bicelles.18 Using uniformly 2H,13C,15N-lableled C99 we assigned its backbone 15N and 13C NMR resonances in SCOR bicelles using traditional 3D NMR approaches supplemented with data from amino acid-specific labeling experiments. Assignments were also facilitated by transferring some previously-completed assignments from DDMB micelle conditions (Figures S3-S5). The 1H,15N-TROSY spectrum of C99 in SCOR bicelles is shown in Figure 5. Over 90% of C99 backbone resonances were assigned, with the main hindrance for the remaining resonances being that some transmembrane or juxtamembrane sites yield only very broad peaks.
Figure 5. Assigned 1H,15N-TROSY NMR spectrum of C99 in SCOR bicelles.
The backbone amide 1H-15N peaks are labeled according to amino acid number. This spectrum was collected on a 400 μM C99 sample in 10 wt % SCOR bicelles, q=0.33, in NMR buffer in a shaped tube at 45 °C and 900 MHz.
Structural Studies of C99 in SCOR Bicelles
TALOS-N was used to analyze chemical shifts from C99 to map its secondary structure in SCOR bicelles. In addition to the helical transmembrane domain, there is a short extracellular amphipathic “N-helix” separated from the N-terminus of the transmembrane domain (TMD) by only a few linking residues. There also is an amphipathic “C-helix” at the extreme C-terminus separated from the TMD by a 35 residue “C-loop”. A summary of the secondary structure elements and sequence is shown in Figure 6A. The overall secondary structure of full length C99 in SCOR bicelles is nearly identical to what was previously been seen for full length C99 in micelles and in classical bicelles.18-19
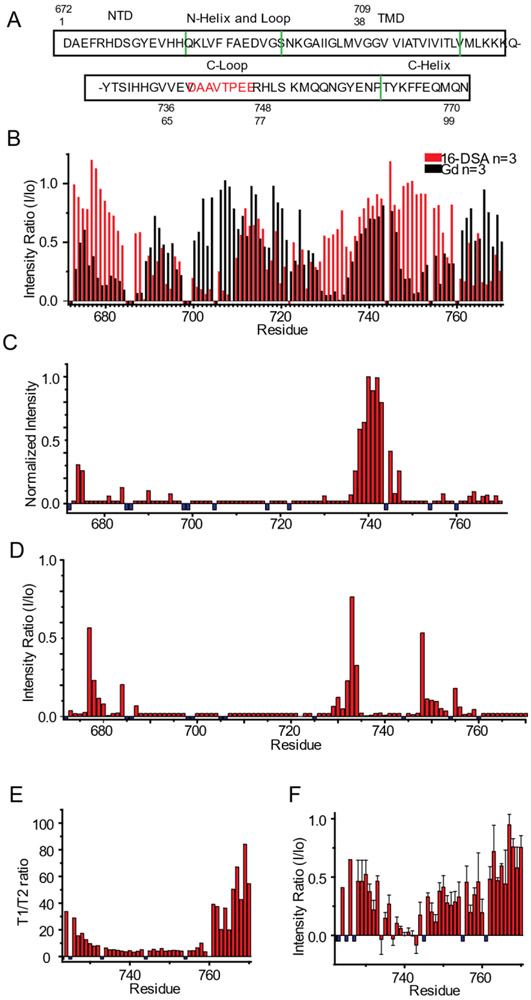
Figure 6. Use of NMR to probe the topology and dynamics of C99 in SCOR bicelles.
A) Sequence of human C99 with numbering based either on C99 only (sites 1-99) or on the parent full-length amyloid precursor protein (672-770). Green lines indicate boundaries of secondary structural elements as determined in this work from analysis of the backbone NMR resonance chemical shifts and paramagnetic data. Amino acids in the C-loop previously identified as being involved in transient amide backbone hydrogen bonding are identified in red.49 B) Topology of C99 in SCOR bicelles as mapped by site-specific TROSY NMR peak linebroadening due to site access by lipophilic (16-DSA) or aqueous (Gd-DTPA) paramagnetic probes. Mean intensity ratios (probe-exposed versus control peak height) are presented for 16-DSA (red) and Gd-DTPA (black). The U-15N-C99 concentrations for all experiments were between ~200 and 400 μM in 5-10 wt% SCOR bicelles, q = 0.33, in NMR buffer. Error bars for these n=3 data are not shown here (to avoid clutter), but are displayed in Figure S6. C) Effect on C99 peak intensities of adding both excess 16-DSA (to 12 mol %), and Gd-DTPA (to 3mM) to a 300μM U-15N-C99 sample in 7 wt % SCOR bicelles and NMR buffer at 45 °C. n=1 D) CLEANEX-PM amide exchange analysis of peak height ratios for a 50 ms mixing time experiment. Ratios are of 50 ms mixing time versus no mixing time. E) NMR 15N T1/T2 relaxation time ratios for the cytosolic domain of C99. T1 and T2 analyses are derived from a 300 μM U-15N-C99 sample in 5 wt % SCOR bicelles q=0.33 in NMR buffer at 45 °C. n=1 F) Intensity ratios for 1H-15N NOE signals from a 300 μM C99 sample in 10 wt % SCOR bicelles, q=0.33, in NMR buffer at 45 °C. n=2 Small negative values in B are residues which were not assigned or were too broad; for C-F, these values are represented by small negative blue bars.
To probe both the membrane topology and possible quaternary structural interfaces for C99 in SCOR bicelles, TROSY NMR spectra were acquired in the presence of paramagnetic probes. The presence of the lipophilic paramagnetic probe, 16-DOXYL-stearic acid (16-DSA), broadens NMR signals from residues in direct contact with the membrane interface or hydrophobic interior. Conversely, the presence of the water-soluble probe gadopentetic acid (Gd-DTPA) broadens the NMR signals from water-exposed sites. Together, these probes permit mapping of which residues are exposed to the membrane and extramembrane regions. As shown in Figures 6B and S6, resonances from both the N- and the C-terminal helices (sites 690-697 and 761-770) were broadened by the presence of 16-DSA and are partially protected from broadening by Gd-DTPA, indicating that these helices are amphipathic and interact with the bicelle surface, similar to what has previously been seen for C99 in micelles,42 vesicles,19 and in DMPC/DHPC bicelles.18 However, a more surprising result for C99 in SCOR bicelles is that a number of sites located in the C-terminal half of the C99 TMD (sites 710-720) were seen to be protected against 16-DSA broadening, while sites in the N-terminal half (sites 700-709) of the TMD were broadened (Figure 6B). This protection of transmembrane sites against access by a lipophilic probe indicates the location of quaternary structural interface. This oligomeric interface does not involve the tandem GXXXG (residues 700-704 and 704-708) motifs seen to be critical for dimerization of C99 in some previous experimental structural studies22, 43-44 all of which were conducted in conventional model membranes bereft of Chol and SM. Instead, the transmembrane oligomerization sites are located in the cytosolic half of the TMD.
Unusual paramagnetic probe NMR results were observed for residues 673-678 at the N-terminus and for residues 735-745 in the C-loop that connects the TMD to the surface-associated C-helix. While analysis of the NMR chemical shifts indicates random coil structure for the 37 residue C-loop (residues 724-760), it was seen that residues 735 to 745 located midway through the loop were protected from both lipophilic and water soluble probes, while being flanked on each end by water-exposed segments (Figure 6B). To verify these results, we collected an NMR spectrum after adding very high concentrations of both Gd-DTPA and 16-DSA probes. As is shown in Figure 6C, this segment of the C-terminal loop is the only part of the protein that was seen to be completely resistant to paramagnetic broadening by the combined influence of excess levels of both probes. Residues 673-678 do not show the same strong protection from excessive paramagnetic probes, indicating that these sites exhibit a weaker or less populated state of the phenomenon occurring for residues 735-745.
To better elucidate the behavior of the C-loop, backbone amide proton exchange with water was probed using the CLEANEX-PM experiment,45 with the results being shown in Figure 6D. As expected, the helical segments of the protein were resistant to amide exchange, while the seemingly random coil segments (much of the N-terminus and C-loop) underwent facile exchange. The notable exception was the 735-745 residue segment of the C-loop. Even though this segment appears to adopt a random coil-like secondary structure, it was seen to be resistant to amide exchange (Figure 6D), results that complement the observation that this segment is inaccessible to both paramagnetic probes (Figures 6B and C). N-terminus residues 673-678 are random coil and also exhibit, albeit to a lesser degree, the same type of probe protection and reduced backbone hydrogen exchange as seen for loop residues 735-745. A similar pattern of backbone amide proton exchange was observed in SCOR bicelles with a 4:1:1 POPC:eSM:Chol lipid mixture (Figure S7), indicating these results pertain even when DMPC is replaced by an unsaturated form of phosphatidylcholine with longer acyl chains.
To gain further insight into the structure and dynamics of the paramagnetic probe-resistant and amide exchange-resistant segment of the C-terminal cytosolic loop, we collected 15N NMR relaxation and 1H-15N NOE data for C99 in SCOR bicelles. The measured T1/T2 ratios and 1H-15N NOE values for the cytosolic domain of C99 are shown in Figures 6E and F, respectively. The relatively high order of the surface-associated C-terminal helix (sites 762-770) is evident from these measurements as is the gradual increase in order as the TMD-anchored N-terminus of the cytosolic domain is approached. Notably, these measurements also reveal that sites 735-745 of the C-loop are extremely mobile. While consistent with the chemical shift/TALOS analysis showing that these sites appear to adopt random coil-like structure, these are also the same sites that were seen to be resistant both to amide exchange and to line broadening by lipophilic and water soluble paramagnetic probes, which in both cases is suggestive of the presence of significant tertiary or quaternary structure. The only previous experimental studies of the C99 cytosolic domain in the context of full-length C9918-19, 42 indicated that the C-loop is largely disordered and revealed no anomalies for the 735-745 segment. Interestingly, previous NMR studies of the isolated cytosolic domain of C99 or of fragments of this domain46-50 in aqueous buffer (no model membranes) led to the conclusion that this segment does exhibit transient structural properties. 51
Oligomeric State of C99 in SCOR Bicelles
C99 has previously been seen to be either a monomer17, 19 or a homodimer,22, 43-44, 52 depending on the model membrane medium used and whether C99 was studied as the full length protein or in a truncated form—most often the residues 1-55 fragment lacking most of its cytosolic domain. Based on the paramagnetic probe-protection results, it seems likely that C99 forms dimers or some other oligomer in SCOR bicelles. Determining the oligomeric state of a small integral membrane protein in a complex model membrane medium is often not straightforward,53 so determination of the C99 oligomeric state offered an ideal opportunity to test the suitability of SCOR bicelles to applications involving a range of biochemical and biophysical techniques. Most measurements were conducted under conditions in which the C99-to-bicelle ratio was carefully controlled to avoid non-specific C99 interactions in crowded bicelles (“forced co-habitation”54). Specifically, sample compositions were usually fixed in the range of 1 C99 molecule for every 1-2 SCOR bicelles. Moreover, all experiments probing oligomerization employed SCOR bicelles with a q of 0.33 so that results using different methods could be compared with each other and with the solution NMR results already presented in this work. Here, we describe experiments conducted using Förster resonance energy transfer (FRET) spectroscopy, electron paramagnetic resonance (EPR) double electron electron resonance spectroscopy (DEER), chemical crosslinking/SDS PAGE, and native ion mobility - mass spectrometry.
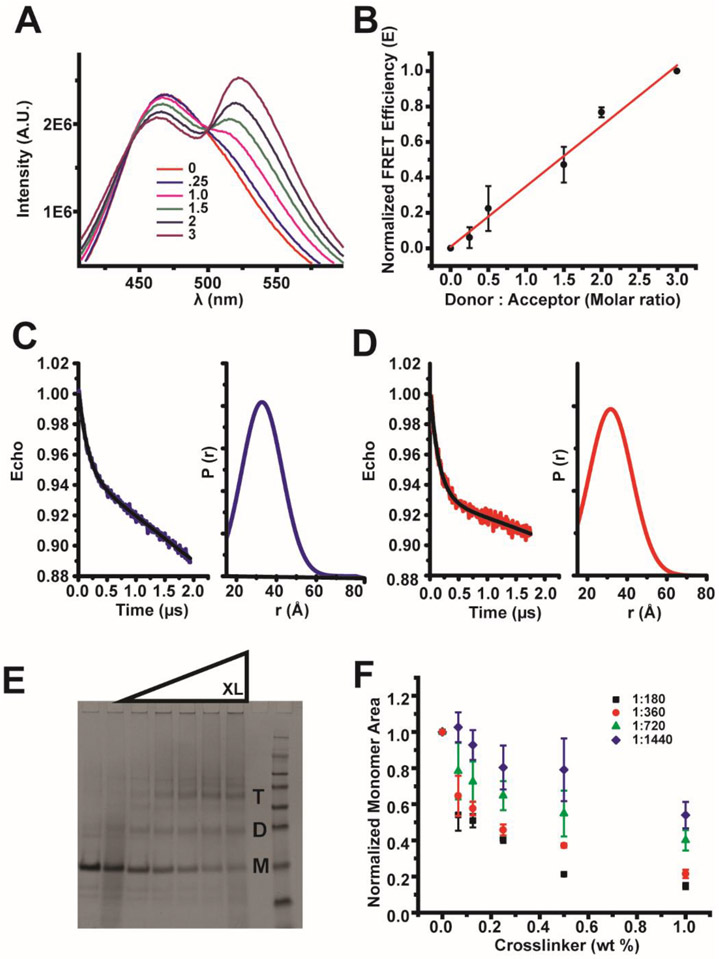
FRET was previously used to detect C99 dimerization in POPC/POPG vesicles.17 The C99 H732C mutation was used to provide a labeling site for IAEDANS (FRET donor) and for IANBD (FRET acceptor). This site is in an unstructured region of the cytosolic loop and outside the C-loop paramagnetic probe-protected region. IADEANS-C99 and IANBD-C99 were mixed in SCOR bicelles at various ratios using conditions in which both the donor-tagged C99 concentration and overall protein:lipid concentration were held constant. A linear relationship between FRET efficiency and the donor-to-acceptor ratio affirms that C99 forms oligomers (Figures 7A and B), but poor labeling efficiency (see Materials and Methods), makes the identification of a specific oligomeric state difficult.55 Nevertheless, the FRET results confirm that C99 does form oligomers in SCOR bicelles. We next turned to EPR DEER measurements.
Figure 7. C99 forms oligomers in SCOR bicelles.
A) FRET spectra of IAEDANS-labeled C99 at a fixed protein to bicelle concentration ratio in the presence of increasing amounts of IANBD-labeled C99. The mole:mole ratios for each scan, in order of decreasing donor intensity, are (donor: acceptor): 1:0, 1:0.25, 1:1, 1:1.5, 1:2, and 1:3. B) Linear fit of normalized FRET efficiency from scans in A. Error bars represent the error in determination of the FRET transfer efficiency. C) DEER spectra of MTSL-labeled 200 μM C99 (L723C mutant) in 5 wt% SCOR bicelles, q=0.33. D) DEER spectra of MTSL-labeled 200 μM C99 (K699C mutant) in 5 wt % SCOR bicelles, q=0.33. E) Example SDS-PAGE (coomassie staining) of purified C99 following glutaraldehyde crosslinking of C99 in SCOR bicelles. Monomer, dimer, and trimer bands are indicated by M, D, and T respectively. F) Quantification of monomeric C99 from SDS-PAGE crosslinking experiments with C99, where the purification tag has been shortened and moved from the C to N terminus. Experiments were conducted with 50 μM C99 and various concentrations of SCOR bicelles in NMR buffer at 25 ° C n=3. The legend indicates the protein to lipid ratio.
C99 with single cysteine sites located at either N- (K699C) or C-terminal (L723C) sides of the transmembrane helix were spin-labeled with MTSL, followed by DEER measurements in SCOR bicelles, as shown in Figures 7C and D. For both labeled forms, the observation of the DEER phenomenon supports oligomerization and indicates a broad distribution of spin label/spin label distances. We did not observe any evidence of aggregation and only one peak is observed in the P(r) distribution, which implies a dimer or trimer oligomeric state.56 The P(r) distribution for each form was centered at ~31-32 Å, which is slightly longer than expected for a tight dimer mediated by a transmembrane interface. While some of the distance distribution almost certainly reflects spin label motions,19 side chain mobility cannot account for the full distance distribution for a tight dimeric state. However, either a flexible dimer or a trimer can account for the slightly larger-than-expected P(r) distances. Another way to determine oligomeric state with DEER is to reference the depth of modulation,57, which we did not do because of incomplete C99 labeling with MTSL. Neither the FRET nor EPR DEER results rule out the possibility that there is also a population of C99 that remains monomeric. Since the FRET and DEER results are consistent with the possibility that C99 may populate either or both dimers and trimers in SCOR bicelles, we next turned to the use of chemical cross-linking in an attempt to covalently trap C99 in dimer and/or trimer forms.
Chemical crosslinking combined with SDS-PAGE is an expeditious way to screen oligomeric states, although results must be validated with an independent technique due to significant risk for false positives/negatives. We cross-linked C99 with glutaraldehyde under near-NMR conditions followed by SDS-PAGE and found that monomer, dimer, and trimer are all observed following cross-linking at high glutaraldehyde concentrations, as shown in Figure 7E. We observed an enrichment in the trimer form as the cross-linker is increased and also confirmed the absence of significant concentrations of higher order oligomers or aggregates. The cross-linking results clarify the FRET and DEER results by suggesting that C99 populates monomer, dimer, and trimer oligomeric states in SCOR bicelles.
In order to understand how the protein-to-lipid ratio affects C99 oligomerization in SCOR bicelles, we also carried out crosslinking experiments at constant C99 but with increasing concentrations of SCOR bicelles present. As shown in Figure 7F, increasing the bicelle concentrations increased the relative population of monomer seen following crosslinking and also led to a reduction in the rate of crosslinking efficiency as the glutaraldehyde concentration was increased. While 1:180 and 1:360 protein-to-(lipid + detergent) ratios show only modest levels of monomer, crosslinking to dimers and trimers appears to be far less efficient at and above 1:720. The monomer/dimer/trimer ratio is responsive to the protein-to-lipid ratio with the ratio shifting to favor monomer when the protein is diluted with additional bicelles. This suggests that monomer, dimer, and trimer forms of C99 are in equilibrium in SCOR bicelles.
We were intrigued that crosslinking revealed that C99 can populate monomer, dimer, and timer oligomeric states in SCOR bicelles over a wide range of both C99-to-bicelle ratios and as detected over a wide range of glutaraldehyde concentrations. These results are fully consistent with both the FRET and DEER EPR results. Given the pitfalls that can accompany the interpretation of chemical cross-linking studies, we turned to one last method, native ion mobility – mass spectrometry (nMS)58-59 to validate the chemical cross-linking results.
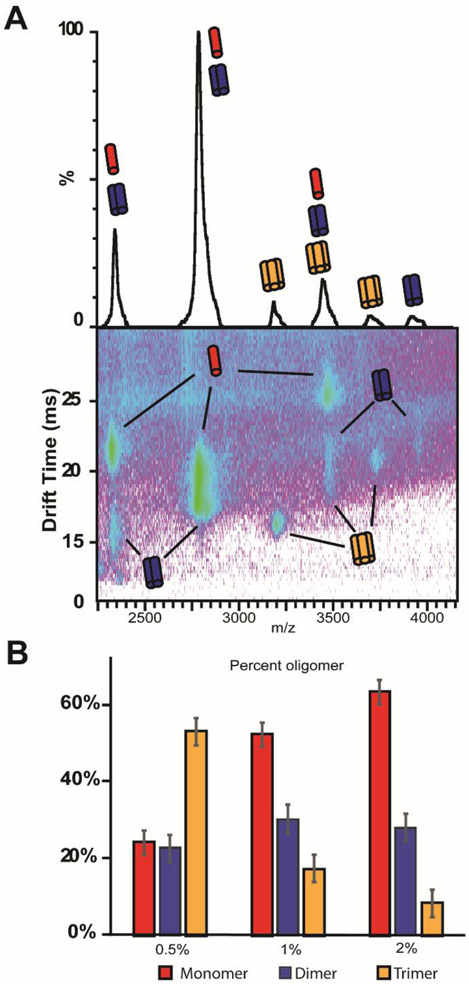
To our knowledge, bicelles of this complexity have not previously been used to study proteins via native mass spectrometry. We were pleased to observe that, with optimization of instrument parameters, intact C99 and C99 complexes could be readily liberated from SCOR bicelles. In agreement with previous experiments there are clear trends in the ion mobility – mass spectrometry data, which can be attributed to monomer, dimer and trimer oligomeric states of C99 liberated from SCOR bicelles (Figure 8A). As shown in Figure 8B, the C99 oligomer populations were found to be impacted by the protein-to-lipid ratio (here defined to include both detergent and lipid), as indicated by a transition from mostly trimer in 0.5 wt % bicelles to mostly monomer in 2.0 wt % bicelles under conditions in which the C99 concentration was held constant. The native MS results are in excellent agreement with the chemical crosslinking data as indicated by the high monomer population seen by native mass spectrometry at a protein to lipid ratio of ~1:1200 (~30 μM C99 in 2 wt % bicelle), and as supported by observation of inefficient crosslinking at ~1:1440 (~50 μM C99 in 4 wt % bicelle).
Figure 8. Determination of the oligomeric composition of C99 in SCOR bicelles with ion mobility – mass spectrometry (native mass spectrometry).
A) Mass spectra of C99 complexes liberated from SCOR bicelles (29 μM C99 in 1% wt/v SCOR bicelle q=0.33) show signals corresponding to the masses of monomer, dimer, and trimer C99 complexes (top). Ion mobility mass spectrometry data sets, which report on the mass, charge, and orientationally averaged size of an analyte, are used to deconvolute signals for which the mass-to-charge ratio alone is ambiguous between C99 complexes and monomer species (bottom). For example, the signal corresponding to 2822 m/z is observed to be comprised of monomeric and dimeric C99 from the ion mobility – mass spectrum shown. This approach also ensures that any lipid noise produced by dissociated bicelles is removed from analysis. B) The relative intensities of each deconvoluted species were used to calculate the percent of each oligomeric species for 29 μM C99 at 0.5%, 1%, and 2% w/v q= 0.33 SCOR bicelles.
DISCUSSION
SCOR Bicelles Represent a New Class of Bicelles
The development and characterization of SCOR bicelles described in this work provides the first isotropic and optically clear model membrane system that is rich in both SM and Chol. It was shown that the parent lipid mixture used for SCOR bicelle formation, 4:2:1 DMPC:eSM:Chol, forms bilayers that are more highly ordered than DMPC-only bilayers and exhibit a Lo to Ld phase transition temperature close to 30°C. It was observed that SCOR bicelles differ from conventional DMPC/DHPC bicelles both by being significantly thicker and in that the bilayer thickness of SCOR bicelles is nearly independent of the q (lipid-to-detergent mole) ratio. This confirms that the increased order of this membrane composition observed in bilayered lipid vesicles composed of 4:2:1 DMPC:eSM:Chol is maintained in SCOR bicelles. The lipid composition of mammalian PMs is, of course, vastly more complex than the lipid composition of SCOR bicelles. However, the inclusion of both Chol and SM in SCOR bicelles leads to increased order in their bilayer domain, representing a significant step closer to native PMs than conventional bicelles formed with glycerophospholipids only. We are also planning to investigate whether there is any segregation of lipid types within the bilayered domains of SCOR bicelles or if the three types of lipid are uniformly mixed.
Besides the DMPC-based 4:2:1 SCOR bicelles that were the focus of this paper, DDMB was found to be capable of solubilizing a range of other Chol and eSM-rich lipid mixtures that also appear to form SCOR-class bicelles, such as 4:1:1 POPC:eSM:Chol and 6:3:1 DPPC:eSM:Chol. This compositional flexibility is advantageous when trying to optimize conditions to suit a particular protein or method. This property will also enable studies to determine if either Chol and/or SM individually, or only as a mixture, modulates membrane protein structure and function.14-15
SCOR Bicelles Have an Ideal Bilayered-Disc Morphology at Low q
It is generally accepted that classical DMPC/DHPC bicelles at q ratios between 2 and 6.5 form a true discoidal morphology with spatial separation between lipid and detergent, with mixtures at even higher q values adopting a “Swiss cheese”-like morphology. However, these “high q” bicelles are not commonly used in solution state NMR studies of integral membrane proteins because of their large size and/or because they fail to tumble isotropically, even sometimes aligning in strong magnetic fields.60-62 Recent work has indicated that at low q ratios suitable for solution NMR—generally less than q = 0.7—DMPC/DHPC bicelles are discoidal but exhibit significant penetration of DHPC into the bilayer core and, as q is reduced, become akin more to classical mixed micelles than ideal DHPC-edge-stabilized bilayered discs.39, 63 Nevertheless, it should be noted that evidence also exists for near-ideal bicelles at low q.41, 64 In this work SAXS, SANS, and cryo-EM are all in accord that the SCOR bicelles contain an ideal discoidal morphology even at a q ratio of 0.33. As expected for ideal bicelles, the diameters of the bilayered discs increase as q is increased from 0.33 to 1.0, while maintaining a relatively stable bilayer thickness of ~5 nm. In this regard, SCOR bicelles resemble nanodiscs65-66 and lipodisqs/SMALPs,67-68 where bilayer discs are edge-stabilized by either amphipathic proteins or amphipathic polymers.
SCOR Bicelles are Robust and Compatible with a Wide Array of Experimental Methods
It was observed in this work that SCOR bicelles are stable over a range of concentrations, pH, ionic strengths, and temperatures. This robustness makes SCOR bicelles well-suited for use as a membrane mimetic because it will permit biophysical and biochemical investigations requiring varying temperature, pH, or ionic strength to approximate the natural conditions of a membrane protein. We found, for example, that SCOR bicelles are morphologically stable down to 0.25 wt % (total lipid + detergent) and only slightly increase in size upon further dilution to 0.125 wt %. The working concentration range of classical DHPC and CHAPSO-based DMPC (and other) bicelles is much more limited because DHPC and CHAPSO have CMC values that are on the order of 40X higher than that of DDMB, such that vesicles begin to form even when the bicelle concentrations are lowered below ~ 2 wt % (see69).
The stability and constancy of SCOR bicelles over a range of conditions enabled successful use of this medium in our studies of the human APP C99 protein using a range of biophysical and biochemical methods, including NMR, DEER, FRET, NMS, and chemical crosslinking. We anticipate that it should be possible to reconstitute many other membrane proteins in SCOR bicelles for analogous studies. Moreover, retention of the bilayered disc morphology even at low concentrations should allow SCOR bicelles to be used in studies that require a high level of dilution, such as cryo-EM. Cryo-EM has recently yielded spectacular progress in determining high resolution structures of large membrane proteins.70 The authors are unaware of any examples of membrane protein structures being determined in bicelles using cryo-EM, possibly because conventional bicelles vesiculate when diluted to concentrations suitable for rapid sample vitrification. The fact that we were able to directly image SCOR bicelles using cryo-EM methods suggests they are suitable model membranes in which to solubilize larger membrane proteins and complexes for cryo-EM analysis.
The Structure of C99 in SCOR Bicelles Exhibits Some Unexpected Features
APP has been shown to mainly reside in bulk membrane domains with a modest population also being observed in lipid raft approximating detergent resistant membranes.71-72,73 The structure of the APP C99 protein or transmembrane domain-containing fragment has been examined by a number of labs in various conventional model membrane systems.17-27, 42, 74-75 This previous body of work made C99 an ideal test protein for study in SCOR bicelles. Full length C99 in SCOR bicelles exhibits a similar secondary structure as seen in previous studies. The helical transmembrane segment of C99 in SCOR bicelles is slightly longer than in DMPC/DHPC bicelles,18 but is similar in length to what was previously seen in DHPC-based POPC/milk sphingomyelin bicelles.18 This result is reasonable in light of the increased thickness of SCOR bicelles relative to DMPC/DHPC bicelles and because DHPC/POPC/milk SM bicelles are also thicker (POPC has longer fatty acids than DMPC and the most common fatty amide chain in milk SM is 23 carbons long, https://avantilipids.com/product/860063).
While the secondary structure of full length C99 in SCOR bicelles is similar to what has previously been observed, there were three aspects of its structure in SCOR bicelles that are surprising. The first is the fact that nearly all previous studies of C99 have, variously, observed that C99 is either a monomer, a dimer, or a mixture of monomer and dimer, depending on sample conditions.27-38, 68-69 In contrast, the results of this work indicate that C99 in SCOR bicelles appears to populate an equilibrium mixture of monomer, dimer, and trimer. This observation is not completely unprecedented, as Higashide et al. previously observed that following extraction of mammalian CHO cells with the non-ionic detergent DDM, C99 was seen to run as a mixture of monomer, dimer, and trimer on blue native PAGE, results that are strikingly similar to those of this paper.76 We do not know why SCOR bicelles promote formation of C99 trimer along with monomer and dimer, but we do note that a common feature of the Higashide study and that of this paper is the presence of Chol and SM in the C99 samples. Recent computational work by Sun et al. suggested that the C99 TM dimer is less stable in a raft-like environment but did not examine possible trimer.77 Of course, the fact that C99 populates three oligomeric states in SCOR bicelles is vexing from a practical standpoint, as it means that most of the C99 samples examined in this work involve a mixture of three different forms of the protein, hindering attempts to interpret the data, especially the NMR data, in terms of a single structural state. At the required protein concentration for NMR, C99 exists in a dynamic mixture of oligomeric states, which—along the relatively large bicelle size—is likely the cause of the additional NMR signal broadening compared to the DDMB micelles environment.
A second important structural observation for C99 in SCOR bicelles concerns the location(s) of its oligomeric interface(s). There has previously been much interest in the dimerization of C99 due the possibility that dimerization is linked to its cleavage (either promoting or hindering amyloid β (Aβ) production).52, 78-83 Previous biophysical studies in which dimerization of C99 or of derived TMD-containing fragments have usually pointed to either or both GXXXG motifs located in the tandem G700XXXG704XXXG708 segment,22, 44, 75, 84-85 to G709XXXA713,27, 86 or to both GXXXG and GXXXA motifs43, 87-88 as being the location of the dimerization interface. Studies highlighting the G709XXXA713 oligomerization motif often additionally implicate the following TV715 residues.27, 43, 86-88 These previous experimental studies were often conducted in conventional model membranes bereft of Chol and SM and often did not use full length C99. However, in the present work with SCOR bicelles, paramagnetic probe accessibility studies indicated that the oligomer interface is located in the cytosolically disposed C-terminal half of the TMD, roughly spanning residues 710-716. The fact that C99 populates both dimer and trimer states in SCOR bicelles makes exact identification of the specific residues involved difficult, but our results are very clear that the upper end of the TMD containing the two GXXXG motifs is not involved. These conclusions are strongly supported by a previous biochemical (Tango assays) and biophysical (BRET) study of C99 oligomer formation (assumed to be dimer) in mammalian cells.89 In that study, Yan et al. showed that C99 oligomerization is mediated by residues 714-717, in agreement with the results in SCOR bicelles. A computational examination of the C99 TM dimer in a raft-like environment also found the dimer is mediated by residues V711 and V715,77 also in close agreement with our SCOR bicelle data. The fact that the oligomerization interface for C99 in SCOR bicelles appears to be similarly located to the interface previously observed for this protein in mammalian PMs supports the notion that SCOR bicelles may be a better mimic for mammalian plasma membranes than conventional model membranes.
A third surprising structural observation made in this paper is that there is a segment within the long cytosolic loop of C99 (resides 735-745, based on full length APP numbering) that seems to populate a highly dynamic random coil ensemble of conformations and yet is both inaccessible to paramagnetic probes and resistant to backbone amide-water hydrogen exchange. N-terminal residues 673-678 indicate a similar phenomenon, albeit to a lesser degree and this region is likely sampling beta sheet secondary structure as seen in previous simulations20 and known amyloid beta structure.90 We hypothesize that these results reflect participation of these segments in a dimer and/or trimer interface that is predominantly random coil but can sample alpha helical and beta sheet states.20 It should be added that previous solution NMR studies of a soluble APP intracellular domain (AICD) fragment found that residues V742TPEER747 of AICD forms a transient N-terminal helix capping box49-50, hinting at a predilection of this segment for complex dynamics. Moreover, the non-conventional nature of the dynamic structure formed by this segment is likely reflected by the fact that the PEER segment yielded only broad and unassignable resonances in our previous NMR studies of monomeric C99 in detergent micelles.18-19, 42
While unusual, the formation of non-canonical tertiary or quaternary structure seen for the C-loop 735-745 segment is not unprecedented. Borgia et al recently reported a high-affinity heterodimeric structural ensemble where two oppositely charged IDPs, histone H1 and its nuclear chaperone, the prothymosin α protein, interact while retaining structural disorder.51 They report a lack of peak dispersion in the HSQC spectrum upon complex formation and no obvious indication of secondary structure in the Cα chemical shifts, both classic indications of random coil. We find that the C99 C-loop also possesses these classic random coil-like traits and yet, paramagnetic and hydrogen exchange experiments indicate the presence of structure for residues 735-745. The C99 C-loop does not contain the high charge densities found in Borgia et al; however, it is likely that the transmembrane oligomerization facilitates co-localization of the C-loop interfaces. Our homo-oligomeric C99 C-loop interface and the work of Borgia et al on a hetero-dimeric system appear to be similar in nature as both represent oligomeric interfaces that lack classical structure.
Many proteins are known to interact with the intracellular portion of APP/C99 and we posit that this paradoxical C-loop interaction may play a role in a regulatory mechanism that modulates interactions of the 735-745 segment with cytoplasmic binding partners such as caspases and trafficking adaptor proteins.91 Phosphorylation in the C-loop oligomeric interface at T743 has been shown to impact Aβ production46-47, 92 and it will be interesting to explore how the unstructured and dynamic oligomeric interfaces impacts its interaction with other proteins and how this impacts Aβ production. A summary of the oligomeric interfaces and dynamic oligomeric states found for C99 in SCOR bicelles is highlighted in Figure 9.
Figure 9. Schematic model for C99 in SCOR Bicelles.
A) Model for monomeric C99 based on previous studies of C99 in conventional DMPC/DHPC bicelles.19 B) C99 in SCOR bicelles populates a dynamic mixture of monomer/dimer/trimer. Two oligomeric interfaces were identified, one in the cytosolic half of the TM and another in the disordered C-loop. The double headed arrow highlights that the bilayer thickness of the SCOR lipid bilayer shown in B is thicker than the DMPC-only bilayer shown in A (not to scale). The blue and orange lipids symbolize DMPC and eSM respectively. Green blocks represent Chol.
CONCLUSIONS
Here, we introduced a new bicelle platform that is both eSM and Chol-rich (SCOR). The bicelles are ~5 nm thick and are edge-stabilized by a DDMB annulus. We found the SCOR bicelles to be well-behaved, even at concentrations as low as 0.25 % w/v, making them well-suited for cryo-EM studies. SCOR bicelles are stable over a wide range of pH, ionic strength, concentration, and temperature, such that they are well-suited for implementation of a wide range of biochemical and biophysical techniques, as demonstrated herein for the well-characterized protein C99. Our studies of the C99 protein in SCOR bicelles indicate notable differences in the structure and dynamics of this important protein relative to what had previously been documented from studies of C99 in micelles, conventional bicelles or glycerophospholipid vesicles. Moreover, the C99 oligomeric interfaces present in SCOR bicelles seem to closely approximate the oligomeric interface documented in recent studies of the protein in mammalian plasma membrane. This supports both the possibilities that the presence of abundant Chol and sphingolipids in membranes may sometime significantly alter the structure and dynamics of membrane proteins and that SCOR bicelles may be a closer mimetic for mammalian plasma membranes than commonly-used model membrane systems.
Supplementary Material
Grant Information and Acknowledgements
This work was supported by NIH grants RF1 AG056147 (CRS), R01 GM106672 (CRS), R01 NS095989 (CRS), and R01 GM105942 (BTR). JMH was supported by NIH T32 CA00958229 and by F31 AG061984. KFM was supported by NIH T32 GM08320 and NSF Predoctoral Research Fellowship DGE090667. Special thanks to Dr. Ricardo Capone for useful discussion and Dr. Geoffrey Li for proof-reading. NMR instrumentation was supported by NSF (0922862), NIH (S10 RR025677) and Vanderbilt University matching funds. A portion of this research used resources at the High Flux Isotope Reactor, a DOE Office of Science User Facility operated by the Oak Ridge National Laboratory under contract no. DE-AC05 00OR2275. The Bio-SANS instrument is supported by the Office of Biological and Environmental Research of the US DOE. This work employed SASView 4.2.0 software, which was originally developed through the support of NSF Award DMR-0520547. SasView also contains code developed with funding from the EU Horizon 2020 program under the SINE2020 project Grant No 654000. The LiX beamline is part of the Life Science Biomedical Technology Research resource, primarily supported by US National Institute of General Medical Sciences grant P41 GM111244, and by the DOE Office of Biological and Environmental Research grant KP1605010, with additional support from the NIH ( S10 OD012331). NSLS-II is a User Facility operated for the US Department of Energy, Office of Science, by Brookhaven National Laboratory (contract DE-SC0012704). DH acknowledges a grant by the Deutsche Forschungsgemeinsachft, DFG (HU 720 15-2).
Footnotes
The authors declare no competing financial interest.
Contributor Information
James M. Hutchison, Chemical and Physical Biology Graduate Program and Center for Structural Biology, Vanderbilt University, Nashville, TN 37240, United States
Kuo-Chih Shih, Polymer Program, Department of Chemical & Biomolecular Engineering, and Department of Biomedical Engineering, University of Connecticut, Storrs, CT 06269, United States.
Holger A. Scheidt, Institute for Medical Physics and Biophysics, Leipzig University, Leipzig 16-18, 04107, Germany
Sarah M. Fantin, Department of Chemistry, University of Michigan, Ann Arbor 48109, MI United States
Kristine F. Parson, Department of Chemistry, University of Michigan, Ann Arbor 48109, MI United States
George A. Pantelopulos, Department of Chemistry, Boston University, Boston, MA 02215, United States
Haley R. Harrington, Center for Structural Biology and Department of Biochemistry, Vanderbilt University School of Medicine Basic Sciences, Nashville, TN 37240, United States
Kathleen F. Mittendorf, Center for Structural Biology and Department of Biochemistry, Vanderbilt University School of Medicine Basic Sciences, Nashville, TN 37240, United States. Department of Translational and Applied Genomics, Center for Health Research, Kaiser Permanente Northwest, Portland, OR, 97227, United States
Shuo Qian, Neutron Scattering Division, Oak Ridge National Laboratory, Oak Ridge, TN 37831, United States.
Richard A. Stein, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, TN 37240, United States
Scott E. Collier, Center for Structural Biology, Vanderbilt University, Nashville, TN 37240, United States
Melissa G. Chambers, Center for Structural Biology, Vanderbilt University, Nashville, TN 37240, United States
John Katsaras, Neutron Scattering Division and Shull Wollan Center, Oak Ridge National Laboratory, Oak Ridge, TN 37831, United States.
Markus W. Voehler, Center for Structural Biology and Department of Chemistry, Vanderbilt University, Nashville, TN 37240, United States
Brandon T. Ruotolo, Department of Chemistry, University of Michigan, Ann Arbor MI 48109, United States
Daniel Huster, Institute for Medical Physics and Biophysics, Leipzig University, Leipzig 16-18, 04107, Germany.
Robert L. McFeeters, Department of Chemistry, University of Alabama, Huntsville AL 35899, United States
John E. Straub, Department of Chemistry, Boston University, Boston, MA 02215, United States
Mu-Ping Nieh, Polymer Program, Department of Chemical & Biomolecular Engineering, and Department of Biomedical Engineering, University of Connecticut, Storrs, CT 06269, United States.
Charles R. Sanders, Center for Structural Biology, Department of Biochemistry, and Department of Medicine, Vanderbilt University School of Medicine, Nashville, TN 37240, United States
REFERENCES
- 1.van Meer G; de Kroon AI, Lipid Map of the Mammalian Cell. J. Cell. Sci 2011, 124 (Pt 1), 5–8. [DOI] [PubMed] [Google Scholar]
- 2.Simons K; Ikonen E, Functional Rafts in Cell Membranes. Nature 1997, 387 (6633), 569–72. [DOI] [PubMed] [Google Scholar]
- 3.Brown DA; London E, Functions of Lipid Rafts in Biological Membranes. Annu. Rev. Cell Dev. Biol 1998, 14, 111–36. [DOI] [PubMed] [Google Scholar]
- 4.Sezgin E; Levental I; Mayor S; Eggeling C, The Mystery of Membrane Organization: Composition, Regulation and Roles of Lipid Rafts. Nat. Rev. Mol. Cell. Biol 2017, 18 (6), 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraft ML, Plasma Membrane Organization and Function: Moving Past Lipid Rafts. Mol. Biol. Cell 2013, 24 (18), 2765–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Meer G; Voelker DR; Feigenson GW, Membrane Lipids: Where They are and how They Behave. Nat. Rev. Mol. Cell. Biol 2008, 9 (2), 112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt ML; Davis JH, Liquid Disordered-liquid Ordered Phase Coexistence in Bicelles Containing Unsaturated Lipids and Cholesterol. Biochim. Biophys. Acta 2016, 1858 (4), 619–26. [DOI] [PubMed] [Google Scholar]
- 8.Cho HS; Dominick JL; Spence MM, Lipid Domains in Bicelles Containing Unsaturated Lipids and Cholesterol. J. Phys. Chem. B 2010, 114 (28), 9238–45. [DOI] [PubMed] [Google Scholar]
- 9.Minto RE; Adhikari PR; Lorigan GA, A 2H Solid-state NMR Spectroscopic Investigation of Biomimetic Bicelles Containing Cholesterol and Polyunsaturated Phosphatidylcholine. Chem. Phys. Lipids 2004, 132 (1), 55–64. [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi T; Suzuki T; Yasuda T; Oishi T; Matsumori N; Murata M, NMR-based Conformational Analysis of Sphingomyelin in Bicelles. Bioorg. Med. Chem 2012, 20 (1), 270–8. [DOI] [PubMed] [Google Scholar]
- 11.Smrt ST; Draney AW; Singaram I; Lorieau JL, Structure and Dynamics of Membrane Proteins and Membrane Associated Proteins with Native Bicelles from Eukaryotic Tissues. Biochemistry 2017, 56 (40), 5318–5327. [DOI] [PubMed] [Google Scholar]
- 12.Kot EF; Arseniev AS; Mineev KS, Behavior of Most Widely Spread Lipids in Isotropic Bicelles. Langmuir 2018, 34 (28), 8302–8313. [DOI] [PubMed] [Google Scholar]
- 13.Barnaba C; Sahoo BR; Ravula T; Medina-Meza IG; Im SC; Anantharamaiah GM; Waskell L; Ramamoorthy A, Cytochrome-P450-Induced Ordering of Microsomal Membranes Modulates Affinity for Drugs. Angew. Chem. Int. Ed. Engl 2018, 57 (13), 3391–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsamaloukas A; Szadkowska H; Heerklotz H, Thermodynamic Comparison of the Interactions of Cholesterol with Unsaturated Phospholipid and Sphingomyelins. Biophys. J 2006, 90 (12), 4479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Endapally S; Frias D; Grzemska M; Gay A; Tomchick DR; Radhakrishnan A, Molecular Discrimination between Two Conformations of Sphingomyelin in Plasma Membranes. Cell 2019, 176 (5), 1040–1053.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchison JM; Lu Z; Li GC; Travis B; Mittal R; Deatherage CL; Sanders CR, Dodecyl-beta-melibioside Detergent Micelles as a Medium for Membrane Proteins. Biochemistry 2017, 56 (41), 5481–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song Y; Hustedt EJ; Brandon S; Sanders CR, Competition Between Homodimerization and Cholesterol Binding to the C99 Domain of the Amyloid Precursor Protein. Biochemistry 2013, 52 (30), 5051–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Y; Mittendorf KF; Lu Z; Sanders CR, Impact of Bilayer Lipid Composition on the Structure and Topology of the Transmembrane Amyloid Precursor C99 Protein. J. Am. Chem. Soc 2014, 136 (11), 4093–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett PJ; Song Y; Van Horn WD; Hustedt EJ; Schafer JM; Hadziselimovic A; Beel AJ; Sanders CR, The Amyloid Precursor Protein has a Flexible Transmembrane Domain and Binds Cholesterol. Science 2012, 336 (6085), 1168–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantelopulos GA; Straub JE; Thirumalai D; Sugita Y, Structure of APP-C991-99 and Implications for Role of Extra-membrane Domains in Function and Oligomerization. Biochim. Biophys. Acta Biomembr 2018, 1860 (9), 1698–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemmin T; Dimitrov M; Fraering PC; Dal Peraro M, Perturbations of the Straight Transmembrane Alpha-helical Structure of the Amyloid Precursor Protein Affect its Processing by Gamma-secretase. J. Biol. Chem 2014, 289 (10), 6763–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato T; Tang TC; Reubins G; Fei JZ; Fujimoto T; Kienlen-Campard P; Constantinescu SN; Octave JN; Aimoto S; Smith SO, A Helix-to-coil Transition at the Epsilon-cut Site in the Transmembrane Dimer of the Amyloid Precursor Protein is Required for Proteolysis. Proc. Natl. Acad. Sci. U. S. A 2009, 106 (5), 1421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nierzwicki L; Czub J, Specific Binding of Cholesterol to the Amyloid Precursor Protein: Structure of the Complex and Driving Forces Characterized in Molecular Detail. J. Phys. Chem. Lett 2015, 6 (5), 784–90. [DOI] [PubMed] [Google Scholar]
- 24.Scharnagl C; Pester O; Hornburg P; Hornburg D; Gotz A; Langosch D, Side-chain to Main-chain Hydrogen Bonding Controls the Intrinsic Backbone Dynamics of the Amyloid Precursor Protein Transmembrane Helix. Biophys. J 2014, 106 (6), 1318–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu JX; Yau WM; Tycko R, Evidence from Solid-state NMR for Nonhelical Conformations in the Transmembrane Domain of the Amyloid Precursor Protein. Biophys. J 2011, 100 (3), 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Audagnotto M; Lemmin T; Barducci A; Dal Peraro M, Effect of the Synaptic Plasma Membrane on the Stability of the Amyloid Precursor Protein Homodimer. J. Phys. Chem. Lett 2016, 7 (18), 3572–8. [DOI] [PubMed] [Google Scholar]
- 27.Nadezhdin KD; Bocharova OV; Bocharov EV; Arseniev AS, Dimeric Structure of Transmembrane Domain of Amyloid Precursor Protein in Micellar Environment. FEBS Lett. 2012, 586 (12), 1687–92. [DOI] [PubMed] [Google Scholar]
- 28.Seelig J, 31P Nuclear Magnetic Resonance and the Head Group Structure of Phospholipids in Membranes. Biochim. Biophys. Acta 1978, 515 (2), 105–40. [DOI] [PubMed] [Google Scholar]
- 29.Bunge A; Müller P; Stöckl M; Herrmann A; Huster D, Characterization of the Ternary Mixture of Sphingomyelin, POPC, and Cholesterol: Support for an Inhomogeneous Lipid Distribution at High Temperatures. Biophys. J 2008, 94 (7), 2680–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veatch SL; Soubias O; Keller SL; Gawrisch K, Critical Fluctuations in Domain-forming Lipid Mixtures. Proc. Natl. Acad. Sci. U. S. A 2007, 104 (45), 17650–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sani MA; Weber DK; Delaglio F; Separovic F; Gehman JD, A Practical Implementation of de-Pake-ing via Weighted Fourier Transformation. PeerJ 2013, 1, e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clarke JA; Heron AJ; Seddon JM; Law RV, The Diversity of the Liquid Ordered (Lo) Phase of Phosphatidylcholine/Cholesterol Membranes: A Variable Temperature Multinuclear Solid-state NMR and X-ray Diffraction Study. Biophys. J 2006, 90 (7), 2383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogel A; Reuther G; Weise K; Triola G; Nikolaus J; Tan KT; Nowak C; Herrmann A; Waldmann H; Winter R; Huster D, The Lipid Modifications of Ras that Sense Membrane Environments and Induce Local Enrichment. Angew. Chem. Int. Ed. Engl 2009, 48 (46), 8784–7. [DOI] [PubMed] [Google Scholar]
- 34.Huster D; Paasche G; Dietrich U; Zschornig O; Gutberlet T; Gawrisch K; Arnold K, Investigation of Phospholipid Area Compression Induced by Calcium-Mediated Dextran Sulfate Interaction. Biophys. J 1999, 77 (2), 879–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feller SE; Gawrisch K; MacKerell AD Jr., Polyunsaturated Fatty Acids in Lipid Bilayers: Intrinsic and Environmental Contributions to their Unique Physical Properties. J. Am. Chem. Soc 2002, 124 (2), 318–26. [DOI] [PubMed] [Google Scholar]
- 36.Polozov IV; Gawrisch K, Characterization of the Liquid-ordered State by Proton MAS NMR. Biophys. J 2006, 90 (6), 2051–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honerkamp-Smith AR; Veatch SL; Keller SL, An Introduction to Critical Points for Biophysicists; Observations of Compositional Heterogeneity in Lipid Membranes. Biochim. Biophys. Acta 2009, 1788 (1), 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipfert J; Columbus L; Chu VB; Lesley SA; Doniach S, Size and Shape of Detergent Micelles Determined by Small-angle X-ray Scattering. J. Phys. Chem. B 2007, 111 (43), 12427–38. [DOI] [PubMed] [Google Scholar]
- 39.Caldwell TA; Baoukina S; Brock AT; Oliver RC; Root KT; Krueger JK; Glover KJ; Tieleman DP; Columbus L, Low- q Bicelles are Mixed Micelles. J. Phys. Chem. Lett 2018, 9 (15), 4469–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pencer J; Nieh MP; Harroun TA; Krueger S; Adams C; Katsaras J, Bilayer Thickness and Thermal Response of Dimyristoylphosphatidylcholine Unilamellar Vesicles Containing Cholesterol, Ergosterol and Lanosterol: A Small-angle Neutron Scattering Study. Biochim. Biophys. Acta 2005, 1720 (1-2), 84–91. [DOI] [PubMed] [Google Scholar]
- 41.Mineev KS; Nadezhdin KD; Goncharuk SA; Arseniev AS, Characterization of Small Isotropic Bicelles with Various Compositions. Langmuir 2016, 32 (26), 6624–37. [DOI] [PubMed] [Google Scholar]
- 42.Beel AJ; Mobley CK; Kim HJ; Tian F; Hadziselimovic A; Jap B; Prestegard JH; Sanders CR, Structural Studies of the Transmembrane C-terminal Domain of the Amyloid Precursor Protein (APP): Does APP Function as a Cholesterol Sensor? Biochemistry 2008, 47 (36), 9428–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H; Barreyro L; Provasi D; Djemil I; Torres-Arancivia C; Filizola M; Ubarretxena-Belandia I, Molecular Determinants and Thermodynamics of the Amyloid Precursor Protein Transmembrane Domain Implicated in Alzheimer's Disease. J. Mol. Biol 2011, 408 (5), 879–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munter LM; Voigt P; Harmeier A; Kaden D; Gottschalk KE; Weise C; Pipkorn R; Schaefer M; Langosch D; Multhaup G, GxxxG Motifs Within the Amyloid Precursor Protein Transmembrane Sequence are Critical for the Etiology of Abeta42. Embo. J 2007, 26 (6), 1702–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hwang TL; van Zijl PC; Mori S, Accurate Quantitation of Water-Amide Proton Exchange Rates using the Phase-Modulated CLEAN Chemical EXchange (CLEANEX-PM) Approach with a Fast-HSQC (FHSQC) Detection Scheme. J. Biomol. NMR 1998, 11 (2), 221–6. [DOI] [PubMed] [Google Scholar]
- 46.De S; Greenwood AI; Rogals MJ; Kovrigin EL; Lu KP; Nicholson LK, Complete Thermodynamic and Kinetic Characterization of the Isomer-specific Interaction Between Pin1-WW Domain and the Amyloid Precursor Protein Cytoplasmic Tail Phosphorylated at Thr668. Biochemistry 2012, 51 (43), 8583–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pastorino L; Sun A; Lu PJ; Zhou XZ; Balastik M; Finn G; Wulf G; Lim J; Li SH; Li X; Xia W; Nicholson LK; Lu KP, The Prolyl Isomerase Pin1 Regulates Amyloid Precursor Protein Processing and Amyloid-beta Production. Nature 2006, 440 (7083), 528–34. [DOI] [PubMed] [Google Scholar]
- 48.Ramelot TA; Nicholson LK, Phosphorylation-induced Structural Changes in the Amyloid Precursor Protein Cytoplasmic Tail Detected by NMR. J. Mol. Biol 2001, 307 (3), 871–84. [DOI] [PubMed] [Google Scholar]
- 49.Ramelot TA; Gentile LN; Nicholson LK, Transient Structure of the Amyloid Precursor Protein Cytoplasmic Tail Indicates Preordering of Structure for Binding to Cytosolic Factors. Biochemistry 2000, 39 (10), 2714–25. [DOI] [PubMed] [Google Scholar]
- 50.Kroenke CD; Ziemnicka-Kotula D; Xu J; Kotula L; Palmer AG 3rd, Solution Conformations of a Peptide Containing the Cytoplasmic Domain Sequence of the Beta Amyloid Precursor Protein. Biochemistry 1997, 36 (26), 8145–52. [DOI] [PubMed] [Google Scholar]
- 51.Borgia A; Borgia MB; Bugge K; Kissling VM; Heidarsson PO; Fernandes CB; Sottini A; Soranno A; Buholzer KJ; Nettels D; Kragelund BB; Best RB; Schuler B, Extreme Disorder in an Ultrahigh-affinity Protein Complex. Nature 2018, 555 (7694), 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Decock M; Stanga S; Octave JN; Dewachter I; Smith SO; Constantinescu SN; Kienlen-Campard P, Glycines from the APP GXXXG/GXXXA Transmembrane Motifs Promote Formation of Pathogenic Abeta Oligomers in Cells. Front. Aging. Neurosci 2016, 8, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu Q; Piai A; Chen W; Xia K; Chou JJ, Structure Determination Protocol for Transmembrane Domain Oligomers. Nat. Protoc 2019, 14 (8), 2483–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhuang T; Jap BK; Sanders CR, Solution NMR Approaches for Establishing Specificity of Weak Heterodimerization of Membrane Proteins. J. Am. Chem. Soc 2011, 133 (50), 20571–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li M; Reddy LG; Bennett R; Silva ND Jr.; Jones LR; Thomas DD, A Fluorescence Energy Transfer Method for Analyzing Protein Oligomeric Structure: Application to Phospholamban. Biophys. J 1999, 76 (5), 2587–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giannoulis A; Ward R; Branigan E; Naismith JH; Bode BE, PELDOR in Rotationally Symmetric Homo-oligomers. Mol. Phys 2013, 111 (18-19), 2845–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt T; Ghirlando R; Baber J; Clore GM, Quantitative Resolution of Monomer-Dimer Populations by Inversion Modulated DEER EPR Spectroscopy. Chemphyschem 2016, 17 (19), 2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marty MT; Hoi KK; Robinson CV, Interfacing Membrane Mimetics with Mass Spectrometry. Acc. Chem. Res 2016, 49 (11), 2459–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Osterlund N; Moons R; Ilag LL; Sobott F; Graslund A, Native Ion Mobility-Mass Spectrometry Reveals the Formation of beta-Barrel Shaped Amyloid-beta Hexamers in a Membrane-Mimicking Environment. J. Am. Chem. Soc 2019, 141 (26), 10440–10450. [DOI] [PubMed] [Google Scholar]
- 60.Dürr UH; Gildenberg M; Ramamoorthy A, The Magic of Bicelles Lights up Membrane Protein sStructure. Chem. Rev 2012, 112 (11), 6054–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanders CR; Prosser RS, Bicelles: a Model Membrane System for All Seasons? Structure 1998, 6 (10), 1227–34. [DOI] [PubMed] [Google Scholar]
- 62.Katsaras J; Harroun TA; Pencer J; Nieh MP, "Bicellar" Lipid Mixtures as used in Biochemical and Biophysical Studies. Naturwissenschaften 2005, 92 (8), 355–66. [DOI] [PubMed] [Google Scholar]
- 63.Piai A; Fu Q; Dev J; Chou JJ, Optimal Bicelle Size q for Solution NMR Studies of the Protein Transmembrane Partition. Chemistry 2017, 23 (6), 1361–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt T; Situ AJ; Ulmer TS, Direct Evaluation of Protein-Lipid Contacts Reveals Protein Membrane Immersion and Isotropic Bicelle Structure. J. Phys. Chem. Lett 2016, 7 (21), 4420–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Denisov IG; Sligar SG, Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev 2017, 117 (6), 4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rouck JE; Krapf JE; Roy J; Huff HC; Das A, Recent Advances in Nanodisc Technology for Membrane Protein Studies (2012-2017). FEBS Lett. 2017, 591 (14), 2057–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simon KS; Pollock NL; Lee SC, Membrane Protein Nanoparticles: the Shape of Things to Come. Biochem. Soc. Trans 2018, 46 (6), 1495–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Postis V; Rawson S; Mitchell JK; Lee SC; Parslow RA; Dafforn TR; Baldwin SA; Muench SP, The use of SMALPs as a Novel Membrane Protein Scaffold for Structure Study by Negative Stain Electron Microscopy. Biochim. Biophys. Acta 2015, 1848 (2), 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu Z; Van Horn WD; Chen J; Mathew S; Zent R; Sanders CR, Bicelles at Low Concentrations. Mol. Pharm 2012, 9 (4), 752–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheng Y, Membrane Protein Structural Biology in the Era of Single Particle cryo-EM. Curr. Opin. Struct. Biol 2018, 52, 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simons M; Keller P; De Strooper B; Beyreuther K; Dotti CG; Simons K, Cholesterol depletion inhibits the generation of beta-amyloid in hippocampal neurons. Proc. Natl. Acad. Sci. U. S. A 1998, 95 (11), 6460–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bouillot C; Prochiantz A; Rougon G; Allinquant B, Axonal amyloid precursor protein expressed by neurons in vitro is present in a membrane fraction with caveolae-like properties. J. Biol. Chem 1996, 271 (13), 7640–4. [DOI] [PubMed] [Google Scholar]
- 73.Hare J, Trafficking of amyloid β-precursor protein products C83 and C99 on the endocytic pathway. Biochem. Biophys. Res. Commun 2010, 401 (2), 219–24. [DOI] [PubMed] [Google Scholar]
- 74.Chen W; Gamache E; Rosenman DJ; Xie J; Lopez MM; Li YM; Wang C, Familial Alzheimer's Mutations within APPTM Increase Aβ42 Production by Enhancing Accessibility of ε-Cleavage Site. Nat. Commun 2014, 5, 3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Itkin A; Salnikov ES; Aisenbrey C; Raya J; Glattard E; Raussens V; Ruysschaert JM; Bechinger B, Structural Characterization of the Amyloid Precursor Protein Transmembrane Domain and Its γ-Cleavage Site. ACS Omega 2017, 2 (10), 6525–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Higashide H; Ishihara S; Nobuhara M; Ihara Y; Funamoto S, Alanine Substitutions in the GXXXG Motif alter C99 Cleavage by Gamma-secretase but not its Dimerization. J. Neurochem 2017, 140 (6), 955–962. [DOI] [PubMed] [Google Scholar]
- 77.Sun F; Chen L; Wei P; Chai M; Ding X; Xu L; Luo SZ, Dimerization and Structural Stability of Amyloid Precursor Proteins Affected by the Membrane Microenvironments. J. Chem. Inf. Model 2017, 57 (6), 1375–1387. [DOI] [PubMed] [Google Scholar]
- 78.Khalifa NB; Van Hees J; Tasiaux B; Huysseune S; Smith SO; Constantinescu SN; Octave JN; Kienlen-Campard P, What is the Role of Amyloid Precursor Protein Dimerization? Cell. Adh. Migr 2010, 4 (2), 268–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Scheuermann S; Hambsch B; Hesse L; Stumm J; Schmidt C; Beher D; Bayer TA; Beyreuther K; Multhaup G, Homodimerization of Amyloid Precursor Protein and its Implication in the Amyloidogenic Pathway of Alzheimer's Disease. J. Biol. Chem 2001, 276 (36), 33923–9. [DOI] [PubMed] [Google Scholar]
- 80.Winkler E; Julius A; Steiner H; Langosch D, Homodimerization Protects the Amyloid Precursor Protein C99 Fragment from Cleavage by gamma-Secretase. Biochemistry 2015, 54 (40), 6149–52. [DOI] [PubMed] [Google Scholar]
- 81.Jung JI; Premraj S; Cruz PE; Ladd TB; Kwak Y; Koo EH; Felsenstein KM; Golde TE; Ran Y, Independent Relationship Between Amyloid Precursor Protein (APP) Dimerization and γ-Secretase Processivity. PLoS One 2014, 9 (10), e111553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Decock M; El Haylani L; Stanga S; Dewachter I; Octave JN; Smith SO; Constantinescu SN; Kienlen-Campard P, Analysis by a Highly Sensitive Split Luciferase Assay of the Regions Involved in APP Dimerization and its Impact on Processing. FEBS Open Bio 2015, 5, 763–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Munter LM; Botev A; Richter L; Hildebrand PW; Althoff V; Weise C; Kaden D; Multhaup G, Aberrant amyloid precursor protein (APP) processing in hereditary forms of Alzheimer disease caused by APP familial Alzheimer disease mutations can be rescued by mutations in the APP GxxxG motif. J Biol Chem 2010, 285 (28), 21636–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pester O; Barrett PJ; Hornburg D; Hornburg P; Pröbstle R; Widmaier S; Kutzner C; Dürrbaum M; Kapurniotu A; Sanders CR; Scharnagl C; Langosch D, The Backbone Dynamics of the Amyloid Precursor Protein Transmembrane Helix Provides a Rationale for the Sequential Cleavage Mechanism of γ-Secretase. J. Am. Chem. Soc. 2013, 135 (4), 1317–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kienlen-Campard P; Tasiaux B; Van Hees J; Li M; Huysseune S; Sato T; Fei JZ; Aimoto S; Courtoy PJ; Smith SO; Constantinescu SN; Octave JN, Amyloidogenic Processing but not Amyloid Precursor Protein (APP) Intracellular C-terminal Domain Production Requires a Precisely Oriented APP Dimer Assembled by Transmembrane GXXXG Motifs. J. Biol. Chem 2008, 283 (12), 7733–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gorman PM; Kim S; Guo M; Melnyk RA; McLaurin J; Fraser PE; Bowie JU; Chakrabartty A, Dimerization of the Transmembrane Domain of Amyloid Precursor Proteins and Familial Alzheimer's Disease Mutants. BMC Neurosci. 2008, 9, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dominguez L; Foster L; Straub JE; Thirumalai D, Impact of Membrane Lipid Composition on the Structure and Stability of the Transmembrane Domain of Amyloid Precursor Protein. Proc. Natl. Acad. Sci. U. S. A 2016, 113 (36), E5281–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dominguez L; Foster L; Meredith SC; Straub JE; Thirumalai D, Structural Heterogeneity in Transmembrane Amyloid Precursor Protein Homodimer is a Consequence of Environmental Selection. J. Am. Chem. Soc 2014, 136 (27), 9619–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan Y; Xu TH; Harikumar KG; Miller LJ; Melcher K; Xu HE, Dimerization of the Transmembrane Domain of Amyloid Precursor Protein is Determined by Residues Around the Gamma-secretase Cleavage Sites. J. Biol. Chem 2017, 292 (38), 15826–15837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kollmer M; Close W; Funk L; Rasmussen J; Bsoul A; Schierhorn A; Schmidt M; Sigurdson CJ; Jucker M; Fändrich M, Cryo-EM Structure and Polymorphism of Aβ Amyloid Fibrils Purified from Alzheimer's Brain Tissue. Nat. Commun 2019, 10 (1), 4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bukhari H; Glotzbach A; Kolbe K; Leonhardt G; Loosse C; Muller T, Small Things Matter: Implications of APP Intracellular Domain AICD Nuclear Signaling in the Progression and Pathogenesis of Alzheimer's Disease. Prog. Neurobiol 2017, 156, 189–213. [DOI] [PubMed] [Google Scholar]
- 92.Akiyama H; Shin RW; Uchida C; Kitamoto T; Uchida T, Pin1 Promotes Production of Alzheimer's Amyloid Beta from Beta-cleaved Amyloid Precursor Protein. Biochem. Biophys. Res. Commun 2005, 336 (2), 521–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.