ABSTRACT
Nasal carriage of Staphylococcus aureus by healthcare workers is of great clinical importance as it facilitates the contamination of medical devices and cross-transmission. However, studies regarding the epidemiology and dissemination of S. aureus and Methicillin-resistant S. aureus (MRSA) within the Primary Health Care in Brazil are scarce. The current study aimed to detect and characterize S. aureus and MRSA strains from the nasal cavities of 63 healthcare working in primary health care units in order to determine the prevalence of S. aureus and MRSA, biofilm formation and resistance profile of these isolates. PCR reactions were performed for detecting mecA, icaA and icaD genes. The phenotypic antimicrobial susceptibility was assessed by the disk diffusion method and biofilm formation by the Congo Red Agar (CRA) method. The MRSA isolates were typed for the Staphylococcal Cassette Chromosome mec (SCCmec). The prevalence of nasal carriage of S. aureus was 74.6%, of which 72.3% were MRSA carrying SCCmec type I (24.4%), III (34.1%), IV (36.6%). Two (4.9%) isolates presented a non-typeable cassette by the performed technique. The antimicrobial susceptibility evaluation evidenced penicillin resistance in 66.1% of S. aureus, erythromycin resistance in 49.2%, while 37.3% were resistant to oxacillin, 28.8% to cefoxitin, 5.1% to levofloxacin and 5.1% to clindamycin. All isolates were biofilm producers and 96.6% of the strains contained the ica biofilm-forming genes (icaA and/or icaD). We have demonstrated a high prevalence of S. aureus and MRSA carriage among health care working in Primary Health Care units, the presence of SCCmec types I, III and IV, in addition to their high ability to form biofilm, factors that possibly contribute to the dissemination and persistence of these pathogens within the primary care services. These observations highlight the importance of broadening the perspective of Health Care-Associated Infections prevention, including all health care levels, which are currently little explored. In addition, the dynamics and resistance mechanisms of S. aureus transmission still need to be further clarified to enable the implementation of more effective prevention measures.
Keywords: Healthcare workers, Family Health Strategy, Cross-transmission, Methicillin-resistant, Biofilm, Operon ica, SCCmec
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is recognized as an important cause of infections, with high mortality rates, leading to increased lengths of hospital stay and higher health care costs in recent decades1. S. aureus typically colonizes the skin and mucosae, especially the anterior nares of the nose, living as a commensal within human microbiota of 20-30% of the population2. Nasal carriage of S. aureus is associated with a higher risk of infection, because in most cases, infecting strains match colonizing ones. S. aureus nasal colonization is also associated with the pathogen transmission in health-care settings3. A significant proportion of Health Care-Associated Infections (HAIs) is certainly a result of cross-transmission, that is the transfer of microorganism from one person (or object) to another person, resulting in infection. The health care workers are important vectors, reservoir and victims of health-care-associated MRSA cross-transmission4. The screening of MRSA colonization in health care workers is required for controlling health care facilities and for the appropriate management of these professionals to prevent cross-transmission5.
S. aureus present several virulence factors and toxins, and the acquisition of mechanisms of resistance to antimicrobials can hinder the treatment of infections caused by this bacterium6. MRSA strains produce an altered penicillin-binding protein (PBP2a) that has low affinity for most semisynthetic penicillins. This protein is encoded by the mecA gene, which is carried by a mobile genetic element named staphylococcal cassette chromosome mec (SCCmec). Such genetic element confers a broad-spectrum resistance to the entire class of β-lactam drugs, excepting for ceftaroline and ceftobiprole1. Hence, the acquisition and insertion of these mobile genetic elements into the chromosomes of susceptible strains led to the emergence of MRSA lineages. The earliest cases of MRSA infections were observed among hospitalized patients, being associated with health care settings (HA-MRSA)1.
Biofilm formation represents another virulence factor related to the adhesion and dissemination of S. aureus. It is described as an aggregate of microorganisms surrounded by an extracellular matrix produced by bacteria capable of synthesizing extracellular polymeric substances, thus providing protection for its development, favoring symbiotic relationships and allowing survival in unfavorable environments7. As it is known, there is a direct association between S. aureus antimicrobial resistance and the chances of developing biofilm. According to Bhattacharya et al.8, biofilm-positive S. aureus have a greater ability to cause infections and are less susceptible to antibiotics. In addition, the accumulation of biofilm contributes to the fitness of the most successful MRSA strains worldwide and promotes a continuous source of dissemination for these pathogens8,9.
Investigations regarding the epidemiology and dissemination of MRSA within the dynamics of Primary Health Care units, a key component of the Brazilian health system, are scarce. This is one of the few studies to determine the MRSA prevalence and the characterization of methicillin resistance determinants within the primary health care system in Brazil. Previous studies in these regards were performed to verify the prevalence, the susceptibility profile and the molecular epidemiology of MRSA isolated from wounds and nares of patients within the primary health care system, in Brazil9-11. However, to our knowledge, this is the first study to report and evaluate the prevalence of MRSA nasal carriage by health care workers in Brazilian primary care facilities.
The Family Health Strategy (FHS) is considered a model of primary health care focused on the family unit, that works through multidisciplinary teams assigned to specific geographic areas and populations of up to 1000 families12. Studies have also pointed out non-hospital facilities as important reservoirs of MRSA transmission, that are different from the factors identified in hospitals13,14.
Healthcare-associated infections (HAI) is a serious public health problem, and although a third of these infections could be easily prevented through control and hygiene programs, the bacterial resistance to antimicrobials impairs treatment9,15. In addition, many professionals work in more than one health service, moving between basic units and urgency and emergency services, what may facilitate the transmission of S. aureus strains from the hospital environment to FHS Units. The circulation of patients between primary and intensive health systems facilitates S. aureus strains interaction with health professionals, so, as is the case in the health team, patients can also carry and transmit S. aureus.
By performing the present study, we aimed to evaluate the prevalence of MRSA nasal carriage among health care workers of different FHS units, as well as to characterize phenotypically and genotypically the resistance profile of the isolates. Studies carried out in this setting and presenting with this focus can provide important insights for current MRSA research, to help the understanding of its epidemiology and improve the effectiveness of control, prevention and treatment of infections caused by these microorganisms.
MATERIAL AND METHODS
Study design and sample
This prospective, cross-sectional study was approved by the Research Ethics Committee, under the protocol CAAE 50534015.4.0000.5515. The samples were obtained from health workers of 7 FHS units in a city located in the West of Sao Paulo State, Brazil, in June 2017.
All health professionals belonging to the Family Health Strategy program in the city were invited. This municipality has 7 ESF units and each of these units has a minimum team composed of 1 doctor, 1 nurse, 1 nursing technician and 4 to 12 community health workers. From health professionals who agreed to participate in the study, nasal bacterial samples were collected and socio-epidemiological data were recorded. This questionnaire included: demographic information (age, gender); working unit and position; the presence of chronic underlying diseases; use of health services (including hospitalizations and other procedures in the previous 12 months); occurrence of infections in the previous 12 months; antimicrobial use in the previous 12 months; permanence in a closed institution (day care center, nursing home, prison).
Sampling and microbiological analysis
Samples were collected from the FHS staff with the aid of a sterile saline-moistened stuart swab introduced into the nasal cavities with gentle circular movements, three times. Collected samples were immediately sent to the Microbiology Laboratory of the University of Oeste Paulista (UNOESTE) for S. aureus identification.
The bacterial cultures were submitted to phenotypic identification by Gram staining as well as catalase and coagulase tests.
Extraction of DNA
The Illustra Kit (GE healthcare, Chicago, IL, USA) was used for bacterial DNA extraction, following the protocol described by Pereira et al. 16. DNA samples were stored at -20 °C.
Genotypic detection of S. aureus and MRSA by the PCR technique
The genotypic detection of S. aureus by the Polymerase Chain Reaction (PCR) technique was performed for the detection of the Sa442 gene. Amplifications were performed according to the parameters described by Martineau et al. 17. PCR reactions for the detection of the mecA gene were performed according to parameters described by Murakami et al.18. In all reactions, international reference strains were used as positive controls (S. aureus ATCC 33591) and negative controls were also used (S. aureus ATCC 25923). Amplifications for the mecC gene detection were performed according to the parameters of Garcia-Alvarez et al. 19. The amplification products were visualized through electrophoresis in 1% agarose gels stained with ethidium bromide.
Characterization of SCCmec
Determination of the Staphylococcal Cassette Chromosome mec (SCCmec) type was performed in MRSA by multiplex PCR using the primers and protocol described by Oliveira et al.20 and modified by Milheiriço et al.21. Amplification products were visualized through electrophoresis in 2% agarose gels stained with ethidium bromide.
Antimicrobial susceptibility testing by the disk-diffusion technique
The antimicrobial susceptibility testing was performed by the disk-diffusion method employing impregnated disks according to criteria recommended by the Clinical and Laboratory Standards Institute (CLSI)22. The disks used were: Oxacillin (1 µg) and Cefoxitin (30 µg), Penicillin (10 µg), Erythromycin (15 µg), Clindamycin (2 µg), and Levofloxacin (5 µg).
Detection of biofilm formation in Congo Red Agar (CRA)
Biofilm formation was assessed by the Congo Red Agar (CRA) method. Samples were sown on Congo Red Agar and after incubation of the plates at 37 ºC for 24-48 h, biofilm-producing colonies were identified by the black color. Samples that showed red to burgundy colors were considered not to have the ability to produce biofilm23.
Detection of icaA and icaD genes
Amplifications for the detection of icaA and icaD genes involved in biofilm formation were performed according to the parameters described by Arciola et al.24. Amplification products were visualized through electrophoresis in 2% agarose gels stained with ethidium bromide. In all reactions performed, international reference strains were used as positive and negative controls, Staphylococcus epidermidis ATCC 35985 (biofilm producer) and Staphylococcus epidermidis ATCC 12228 (biofilm non-producer).
Analysis of results
The frequencies of S. aureus and MRSA and the resistance rates of these microorganisms to antimicrobials were described.
A logistic regression model was adjusted to associate the S. aureus carriage outcome (positive or negative) with the following variables: age, second work unit, use of antimicrobial, hospitalization and reporting of other procedures in the last year. Excepting for the variable “age”, all other variables were used assuming a “yes” or “no” answer.
The odds ratio values were presented, as well as their confidence intervals of 95%. The stepwise test was used for the confirmation of results, and the choice of the most appropriate model was based on the values for AIC and BIC criteria, considering that the smaller the value, the better the model adjustment.
RESULTS
A total of 63 healthcare workers gave consent and were included in the study, in the following professions: 22 (34.9%) were community health agents, 10 (15.9%) nursing technicians, 6 (9.5%) receptionists, 5 (7.9%) pharmacy attendants, 5 (7.9%) general services, 3 (4.8%) nurses, 3 (4.8%) dentists, 3 (4.8%) dental assistants, 2 (3.2%) pharmacists, 1 (1.6%) social worker, 1 (1.6%) administrative assistant, 1 (1.6%) doctor, and 1 (1.6%) worker that did not provide information on his profession. (Table 1).
Table 1. Demographic and epidemiological data of the population studied, resistance profile and biofilm genes in S. aureus isolates from the nostrils of health professionals in the seven units of the Family Health Strategy.
| Variables | Carriers | Non-carriers | TOTAL | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MSSA (N = 13) | MRSA (N = 34) | SCCmec type (%a) | |||||||||
| N | % | N | % | I | III | IV | Not typeable | N | % | N | |
| Profession | |||||||||||
| Community health agents | 4 | 18.2 | 12 | 54.5 | 29.4 | 29.4 | 29.4 | 11.8 | 6 | 27.3 | 22 |
| Social worker | 1 | 100 | - | - | - | - | - | - | - | - | 1 |
| Doctor | - | - | - | - | - | - | - | - | 1 | 100 | 1 |
| Nurses | - | - | 3 | 100 | - | 66.7 | 33.3 | - | - | - | 3 |
| Dentists | - | - | 2 | 66.66 | 50.0 | - | 50.0 | - | 1 | 33.3 | 3 |
| Nursing technicians | 2 | 20.0 | 7 | 70.0 | 12.5 | 37.5 | 50.0 | - | 1 | 10.0 | 10 |
| Dental assistants | - | - | 2 | 66.66 | 50.0 | 50.0 | - | - | 1 | 33.3 | 3 |
| Pharmacists | - | - | 1 | 50,0 | - | - | 100 | - | 1 | 50.0 | 2 |
| Pharmacy attendants | 1 | 20.0 | 2 | 40.0 | 33.3 | 33.3 | 33.3 | - | 2 | 40.0 | 5 |
| Administrative assistant | - | - | - | - | - | - | - | - | 1 | 100 | 1 |
| Receptionists | 2 | 33.3 | 3 | 50.0 | 33.3 | 33.3 | 33.3 | - | 1 | 16.7 | 6 |
| General services | 3 | 60.0 | 1 | 20.0 | - | 100 | - | - | 1 | 20.0 | 5 |
| Uninformed | - | - | 1 | 100 | - | - | 100 | - | - | 1 | |
| Total | 13 | 20.7 | 34 | 53.9 | 24.4 | 34.1 | 36.6 | 4.9 | 16 | 25.4 | 63 |
| Works at another health institution | - | - | 8 | 72.7 | - | 62.5 | 37.5 | - | 3 | 27.3 | 11 |
| Family Health Strategy unity | |||||||||||
| FHS-1 | - | - | 2 | 40.0 | - | 50.0 | 50.0 | - | 3 | 60 | 5 |
| FHS-2 | 3 | 30.0 | 5 | 50.0 | 16.7 | 50.0 | 33.3 | - | 2 | 20 | 10 |
| FHS-3 | 2 | 33.3 | 1 | 16.7 | - | 100 | - | - | 3 | 50 | 6 |
| FHS-4 | 2 | 66.7 | 1 | 33.3 | 100 | - | - | - | - | - | 3 |
| FHS-5 | - | - | 4 | 100 | - | 50.0 | 50.0 | - | - | - | 4 |
| FHS-6 | 1 | 11.1 | 8 | 88.9 | 23.1 | 30.8 | 30.8 | 15.4 | - | - | 9 |
| FHS-7 | 5 | 19.2 | 13 | 50.0 | 35.7 | 21.4 | 42.8 | - | 8 | 30.77 | 26 |
| Comorbidity | 3 | 17.6 | 11 | 64.7 | 21.4 | 50.0 | 28.6 | - | 3 | 17.6 | 17 |
| Infection b | |||||||||||
| Upper airways | 4 | 14.3 | 17 | 60.7 | 23.8 | 38.1 | 38.1 | - | 7 | 25.0 | 28 |
| Skin | 3 | 100 | - | - | - | - | - | - | - | - | 3 |
| Urinary | - | - | 6 | 100 | 10.0 | 40.0 | 30.0 | 20.0 | - | - | 6 |
| Antimicrobial b | |||||||||||
| β-lactams | 6 | 22.2 | 15 | 55.6 | 22.2 | 27.8 | 38.9 | 11.1 | 6 | 22.2 | 27 |
| Macrolid | 3 | 25 | 6 | 50.0 | 16.7 | 33.3 | 50.0 | - | 3 | 25.0 | 12 |
| Quinolone | - | - | 2 | 66.7 | - | 33.3 | 66.7 | - | 1 | 33.3 | 3 |
| Hospitalization b | - | - | 3 | 50.0 | 25.0 | 50.0 | 25.0 | - | 3 | - | 6 |
aCalculated over the total number of MRSA isolated strains per profession; bLast 12 months; FHS = Family Health Strategy.
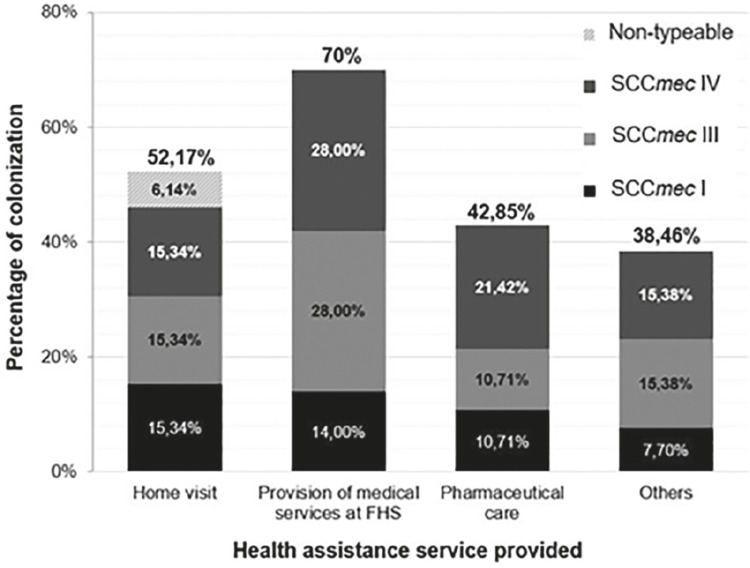
The mean age was 39 years, ranging from 20 to 60 years. Among the group of health workers, genotypic identification through detection of the Sa442 gene confirmed the presence of 59 bacterial samples of S. aureus. The mecA gene detection, the most reliable method for methicillin resistance detection, evidenced 41 (69.5%) MRSA strains among them. The 41 MRSA isolates belonged to 34 participants. Overall, 74.6% (47/63) of the health care workers were S. aureus carriers (MSSA and MRSA). The prevalence of nasal carriers of methicillin-sensitive S. aureus (MSSA) among the participants was 20.7% (13/63) and the overall nasal carriage rate of MRSA was 53.9% (34/63). All 7 units of the FHS included in this study had MRSA and/ or MSSA carriage in health care workers. Figure 1 shows the prevalence and SCCmec typing results of MRSA isolates according to the health care service profession (Table 1).
Figure 1. MRSA colonization and SCCmec type distribution among health workers according to their profession in the FHS. Home Visit (Community health agents and Social worker); Provision of Medical services at FHS (Nurses, Nursing technicians, Dentists, Dental assistants and Doctor); Pharmaceutical care (Pharmacists and Pharmacy attendants); Others (General services, Receptionists, Administrative assistant and Uninformed).
The molecular analysis revealed that 10 (24.4%) of the MRSA isolates carried SCCmec I, 14 (34.1%) had SCCmec III, 15 (36.6%) had SCCmec IV, and 2 (4.9%) isolates presented a non-typeable cassette by the method employed. The distribution of SCCmec types according to the antimicrobial resistance profiles has been shown in Table 2.
Table 2. Antimicrobial resistance profile of MSSA and MRSA according to the type of SCCmec.
| Antimicrobials | MRSA (n=41) | MSSA (n=18) | |||
|---|---|---|---|---|---|
| SCCmec I (n=10) | SCCmec III (n=14) | SCCmec IV (n=15) | Not typed (n=2) | ||
| Penicillin | 40.0% | 71.4% | 80.0% | 50.0% | 66.6% |
| Erythromycin | 30.0% | 50.0% | 86.7% | 0 | 33.3% |
| Clindamycin | 0 | 7.1% | 13.3% | 0 | 0 |
| Levofloxacin | 0 | 14.3% | 6.7% | 0 | 0 |
The antimicrobial susceptibility evaluation found 39 (66.1%) isolates resistant to penicillin, 22 (37.3%) to oxacillin, 17 (28.8%) to cefoxitin and 3 (5.1%) to levofloxacin. Erythromycin and clindamycin resistance was observed in 49.2% (29/59) and 5.1% (3/59) of the isolates, respectively. Concomitant erythromycin and clindamycin resistance was detected in 3 MRSA isolates.
The concordance between cefoxitin and oxacillin disks, and mecA gene detection was found in 34.14% (14/41) of the MRSA strains. Twenty-seven (45.76%) isolates were susceptible to cefoxitin/ oxacillin by disk diffusion, but positive for the mecA gene by PCR. Oxacillin and cefoxitin resistance were simultaneously found in two mecA-negative samples, so these samples were submitted to the mecC gene detection, which did not show any amplification by PCR in any of the isolates.
All 59 S. aureus isolates produced biofilm as shown by the phenotypic detection using the CRA method and 38 (64.4%) of them were positive for the icaA gene, while the icaD gene was found in 54 (91.5%) of the strains by PCR. The concomitant presence of icaA and icaD genes was observed in 35 (59.3%) isolates, 77.1% (27/35) of which were MRSA.
The analysis of S. aureus nasal carriage and the variables of health professionals are shown in Tables 3 and 4.
Table 3. Results of the logistic regression applied to the clinical data of health care professionals with nasal carriage of S. aureus.
| Variable | Odds Ratio [CI 95%] | P-value |
|---|---|---|
| Work at another unit | 0.85 [0.17-4.30] | 0.65 |
| Age | --- | 0.84 |
| Disease | 1.80 [0.35-9.40] | 0.84 |
| Antibiotic | 1.65 [0.43-6.26] | 0.48 |
| Hospitalization | 0.21 [0.03-1.39] | 0.46 |
Table 4. Selection of models by the Stepwise method to explain the colonization of health care professionals with nasal carriage of S. aureus.
| Models | AIC | BIC |
|---|---|---|
| Intercept | 68.96337 | 68.96337 |
| Hospitalization | 69.36967 | 71.44721 |
| Antibiotic + hospitalization | 70.30169 | 72.50241 |
| Disease + hospitalization | 70.30575 | 72.65359 |
| Disease | 70.42487 | 72.91555 |
| Antibiotic | 70.57605 | 72.99117 |
| Age | 70.83801 | 74.45676 |
| Work at another unit | 70.91363 | 74.46082 |
| Age + hospitalization | 70.98617 | 75.14125 |
DISCUSSION
The present study revealed a high rate (74.6%) of S. aureus nasal carriage among health workers included in the dynamic of units of Primary Health Care, while the prevalence of MRSA was 53.9%. The observed prevalence of S. aureus carriage in this study is higher than those reported by most studies carried out with professionals of hospital sectors worldwide. Several studies have reported that the rate of MSSA and MRSA nasal carriage among the health workers ranged from 5.5-34% and 6.1-25.5% respectively25-27. These differences can be related to geographical variations of circulating clones or control practices, as well as to trends of antibiotic prescription.
In this study, 36.6% of the MRSA isolates belonged to the SCCmec IV, which has been a common SCCmec type of community MRSA lineages recovered in Brazil. Recent studies have reported changes in the HA-MRSA population from SCCmec type II to IV, suggesting that some specific SCCmec type IV clones are able to adapt to the hospital sector and persist more easily in these environments28. In Brazil, MRSA isolates carrying SCCmec IV have emerged and have been replacing the previously described widely disseminated Brazilian endemic clone, characterized by the presence of SCCmec III29,30.
Previous analyses have found that the size of the SCCmec element plays an important role in the dissemination of β-lactam resistance among species. SCCmec type IV is one of the smallest SCCmec elements, what makes it the most frequently acquired and selectively favored element by its lower fitness cost with respect to the more complex elements, such as SCCmec III, one of the largest SCCmec elements. Although in earlier years the most frequent SCCmec types were I, II, and III, isolates with SCCmec IV have emerged with potential to become one of the most frequently isolated SCCmec types1. These findings corroborate the frequent recovery of SCCmec type IV and III found in the present investigation.
SCCmec IV is often described as being negative for additional resistance genes besides mecA. In contrast, multiple antibiotic resistance genes have been found in SCCmec type III31,32. However, the known resistance profile of SCCmec IV does not prevent encoded resistance by chromosomal genes or genes carried in plasmids1. In our study, SCCmec type IV had the highest antimicrobial resistance rate among MRSA isolates, followed by type III and type I. The disk-diffusion method revealed a higher resistance rate to clindamycin and erythromycin disks among SCCmec IV isolates compared to the other SCCmec types. The majority of SCCmec type IV isolates (86.6%) were resistant to erythromycin, with two isolates resistant to both, erythromycin and clindamycin, results that suggest a broader resistance phenotype in SCCmec type IV.
The high resistant rates among SCCmec type IV isolates are in accordance with studies which have evidenced that some of these SCCmec strains have acquired resistance to non β-lactam antibiotics to survive in a high antibiotic selective pressure environment, such as the hospital settings. Surprisingly, despite their community origin and have been reported as strains that lack antibiotic resistance genes, many of SCCmec type IV isolates from Brazil and worldwide have already shown multidrug resistance30,33-35.
Moreover, erythromycin/clindamycin resistance was higher in MRSA (56.09%/7.31%) than in MSSA isolates (33.33%/0%), and one isolate belonging to SCCmec type III was multi-resistant to oxacillin, cefoxitin, penicillin, erythromycin and clindamycin. It is reasonable to suppose that the widespread antibiotic use has led to a selection pressure of antimicrobials that facilitates the acquisition of cross-resistance to macrolides, lincosamides and streptogramins (MLS) by staphylococci36.
The MRSA isolates obtained in the present study showed an increased susceptibility to cefoxitin/oxacillin disks. The reason for this phenotype remains to be elucidated, but it seems to be associated with mutations in regions of nucleotide repeats within the mecA gene sequence, which can produce “stealth” MRSA, strains, that are able to restore the gene function and develop a high resistance under an antibiotic selection pressure37. This phenomenon represents a challenge for diagnostic laboratories, since it contributes to MRSA misidentification by conventional susceptibility tests and may lead to potential therapeutic failure.
The absence of the mecA gene was found in two phenotypically resistant samples to oxacillin and cefoxitin. Therefore, the mecC gene was considered as an alternative genetic possibility of resistance. Such isolates, however, were not positive for this gene, suggesting the presence of other intrinsic factors that may compete with mecA and mecC genes to induce resistance.
Although all S. aureus isolates were biofilm producers, the genes associated with polysaccharide intercellular adhesin (PIA) (icaA and icaD) were both or individually detected in 57 (96.6%) S. aureus isolates. The majority of MRSA isolates (65.8%) were positive for both, the icaA and icaD genes. These results indicate a successful strategy behind bacterial tolerance and persistence, raising concerns about the continuous source of spread of these pathogens, with potential multiresistance in health care facilities and the community assisted by the health team investigated.
There is a positive relationship between antibiotic resistance and biofilm production in S. aureus, and this can be explained by the reduced metabolic and growth rates of bacteria embedded in the biofilm matrix38. Moreover, the high population densities and close proximity of cells within biofilm communities allow bacteria to efficiently acquire antibiotic resistance genes in a process known as the horizontal gene transfer39. However, in the current study all strains, from both, MSSA and MRSA groups, were biofilm producers with the presence of icaA and/ or icaD genes being reported in the vast majority. Therefore, the results presented here suggest that all biofilm-producing strains of MRSA may be more resistant to the action of antimicrobials if they are producing biofilm during an infectious process.
The results analysis demonstrated that the antimicrobial use in the last year and the presence of an underlying disease were the only variables with odds ratio higher than 1, what might indicate a risk factor for S. aureus nasal carriage. Nevertheless, their confidence intervals of 95% contain the value 1, what invalidates such conclusion. Moreover, the p-values were all higher than 5%, so we conclude that none of the variables studied were significant for S. aureus nasal carriage among the ESF health workers. Further studies investigating different variables are necessary.
Regarding the MRSA carriage by health workers according to the job performed in the FHS, and the supposed risk factors considered in the questionnaire, the results were inconclusive and did not establish a solid relationship between the variables, what might be due to the small sample size. However, in the present study, the highest prevalence of MRSA carriage was identified in health workers performing tasks that require a higher level of contact with the assisted community, such as community health agents, nurses, nursing technicians, dentists and dental assistants. A study by Franchi et al. 11 included 171 patients of basic health units that perform primary care in Brazil, determining prevalence rates of S. aureus and MRSA of 51.5% and 8.7%, respectively. The authors called attention to the circulation and potential reservoir of resistant strains in patients without the usual risk factors or with exposures limited to the hospital settings.
It is worth noting the lack of studies about the transmission and antimicrobial resistance of these strains in the primary health care environment. Actually, the FHS have a different dynamic when compared to hospitals, with health promotion activities taking place at health facilities, in the patients’ homes, and in the community. These care units provide primary health care services to specific populations, which generates a deeper connection between patients and healthcare workers, creating a different pattern of interpersonal contact that does not apply to the hospital setting, possibly contributing to the extensively community level transmission of S. aureus.
The use of personal protective equipment, hygiene techniques and continuing education, as well as the rational use of antibacterials to reduce the selective pressure of antibiotics are strategies that should be adopted, focusing on the health of the professional and, consequently, of the patients and the community.
CONCLUSION
In summary, we have demonstrated a high prevalence of S. aureus and MRSA carriage among health care workers of Primary Health Care units, the presence of SCCmec types I, III and IV, and their high ability to form biofilm, possibly contributing to the dissemination and persistence of these pathogens within the primary care services. These observations highlight the importance of broadening the perspective of HAI prevention, including all health care levels, which are currently little explored. In addition, the transmission dynamics and resistance mechanisms of S. aureus still need to be further clarified to enable implementation of more effective prevention measures.
Biosafety measures, together with the monitoring of health professionals carrying multi-resistant S. aureus, are some important actions to be adopted in primary health units. Nasal decolonization of MRSA may also be a measure to be implemented, although this is a controversial topic among health professionals. There is a need for more research to be conducted in the Family Health Strategies to better understand the prevalence and dynamics of the dissemination of drug-resistant microorganisms, and their role in health-related infections.
ACKNOWLEDGMENTS
To Sao Paulo State Research Foundation for Scientific and Technological Development (FAPESP), grant Nº 2017/01104-5.
REFERENCES
- 1.Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus : molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31:e00020–e00018. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 3.Albrich WC, Harbarth S. Health-care workers: source, vector, or victim of MRSA? Lancet Infect Dis. 2008;8:289–301. doi: 10.1016/S1473-3099(08)70097-5. [DOI] [PubMed] [Google Scholar]
- 4.Bhatta DR, Hamal D, Shrestha R, Hosuru Subramanya S, Baral N, Singh RK, et al. Bacterial contamination of frequently touched objects in a tertiary care hospital of Pokhara, Nepal: how safe are our hands? 97Antimicrob Resist Infect Control. 2018;7 doi: 10.1186/s13756-018-0385-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rongpharpi SR, Hazarika NK, Kalita H. The prevalence of nasal carriage of Staphylococcus aureus among healthcare workers at a tertiary care hospital in Assam with special reference to MRSA. J Clin Diagn Res. 2013;7:257–260. doi: 10.7860/JCDR/2013/4320.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eshetie S, Tarekegn F, Moges F, Amsalu A, Birhan W, Huruy K. Methicillin resistant Staphylococcus aureus in Ethiopia: a meta-analysis. 689BMC Infect Dis. 2016;16 doi: 10.1186/s12879-016-2014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliveira A, Pereira VC, Pinheiro L, Riboli DM, Martins KB, Cunha ML. Antimicrobial resistance profile of planktonic and biofilm cells of Staphylococcus aureus and coagulase-negative Staphylococci. Int J Mol Sci. 2016;17:1423. doi: 10.3390/ijms17091423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharya M, Wozniak DJ, Stoodley P, Hall-Stoodley L. Prevention and treatment of Staphylococcus aureus biofilms. Expert Rev Anti Infect Ther. 2015;13:1499–1516. doi: 10.1586/14787210.2015.1100533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pereira-Franchi EP, Barreira MR, Costa NS, Riboli DF, Abraão LM, Martins KB, et al. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in the Brazilian primary health care system. Trop Med Int Health. 2019;24:339–347. doi: 10.1111/tmi.13192. [DOI] [PubMed] [Google Scholar]
- 10.Martins MA, Santos LV, Leão LS, Araújo NP, Bachion MM. Prevalence of resistance phenotypes in Staphylococcus aureus and coagulase-negative isolates of venous ulcers of primary healthcare patients. Rev Soc Bras Med Trop. 2012;45:717–722. doi: 10.1590/s0037-86822012000600012. [DOI] [PubMed] [Google Scholar]
- 11.Pereira-Franchi EP, Barreira MR, Costa NS, Fortaleza CM, Cunha ML. Prevalence of and risk factors associated with the presence of Staphylococcus aureus in the chronic wounds of patients treated in primary health care settings in Brazil. Rev Soc Bras Med Trop. 2017;50:833–838. doi: 10.1590/0037-8682-0205-2017. [DOI] [PubMed] [Google Scholar]
- 12.Paim J, Travassos C, Almeida C, Bahia L, Macinko J. The Brazilian health system: history, advances, and challenges. Lancet. 2011;377:1778–1797. doi: 10.1016/S0140-6736(11)60054-8. [DOI] [PubMed] [Google Scholar]
- 13.Schubert M, Kämpf D, Jatzwauk L, Kynast F, Stein A, Strasser R, et al. Prevalence and predictors of MRSA carriage among employees in a non-outbreak setting: a cross-sectional study in an acute care hospital. 7J Occup Med Toxicol. 2019;14 doi: 10.1186/s12995-019-0226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwok KO, Read JM, Tang A, Chen H, Riley S, Kam KM. A systematic review of transmission dynamic studies of methicillin-resistant Staphylococcus aureus in non-hospital residential facilities. 188BMC Infect Dis. 2018;18 doi: 10.1186/s12879-018-3060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alonso CM, Béguin PD, Duarte FJ. Work of community health agents in the Family Health Strategy: meta-synthesis. Rev Saude Publica. 2018;52:14. doi: 10.11606/S1518-8787.2018052000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira VC, Martins A, Rugolo LM, Cunha ML. Detection of Oxacillin resistance in Staphylococcus aureus isolated from the neonatal and pediatric units of a Brazilian teaching hospital. Clin Med Pediatr. 2009;3:23–31. doi: 10.4137/cmped.s2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martineau F, Picard FJ, Roy PH, Ouellette M, Bergeron MG. Species-specific and ubiquitous-DNA-based assays for rapid identification of Staphylococcus aureus. J Clin Microbiol. 1998;36:618–623. doi: 10.1128/jcm.36.3.618-623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García-Álvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveira DC, Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milheiriço C, Oliveira DC, Lencastre H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex. J Antimicrob Chemother. 2007;60:42–48. doi: 10.1093/jac/dkm112. [DOI] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility testing: M100-S23. Wayne: CLSI; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol. 1989;42:872–874. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arciola CR, Baldassarri L, Montanaro L. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J Clin Microbiol. 2001;39:2151–2156. doi: 10.1128/JCM.39.6.2151-2156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agarwal L, Singh AK, Sengupta C, Agarwal A. Nasal carriage of Methicillin- and Mupirocin-resistant S. aureus among health care workers in a tertiary care hospital. J Res Pharm Pract. 2015;4:182–186. doi: 10.4103/2279-042X.167046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El Aila NA, Al Laham NA, Ayesh BM. Nasal carriage of methicillin resistant Staphylococcus aureus among health care workers at Al Shifa hospital in Gaza Strip. 28BMC Infect Dis. 2017;17 doi: 10.1186/s12879-016-2139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu TH, Lee CY, Yang HJ, Fang YP, Chang YF, Tzeng SL, et al. Prevalence and molecular characteristics of methicillin-resistant Staphylococcus aureus among nasal carriage strains isolated from emergency department patients and healthcare workers in central Taiwan. J Microbiol Immunol Infect. 2019;52:248–254. doi: 10.1016/j.jmii.2018.08.015. [DOI] [PubMed] [Google Scholar]
- 28.Harada D, Nakaminami H, Miyajima E, Sugiyama T, Sasai N, Kitamura Y, et al. Change in genotype of methicillin-resistant Staphylococcus aureus (MRSA) affects the antibiogram of hospital-acquired MRSA. J Infect Chemother. 2018;24:563–569. doi: 10.1016/j.jiac.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Chamon RC, Ribeiro SD, Costa TM, Nouér SA, Santos KR. Complete substitution of the Brazilian endemic clone by other methicillin-resistant Staphylococcus aureus lineages in two public hospitals in Rio de Janeiro, Brazil. Braz J Infect Dis. 2017;21:185–189. doi: 10.1016/j.bjid.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caboclo RM, Cavalcante FS, Iorio NL, Schuenck RP, Olendzki AN, Felix MJ, et al. Methicillin-resistant Staphylococcus aureus in Rio de Janeiro hospitals: Dissemination of the USA400/ST1 and USA800/ST5 SCCmec type IV and USA100/ST5 SCCmec type II lineages in a public institution and polyclonal presence in a private one. Am J Infect Control. 2013;41:e21–e26. doi: 10.1016/j.ajic.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Robinson DA, Enright MC. Evolutionary models of the emergence of Methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47:3926–3934. doi: 10.1128/AAC.47.12.3926-3934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, et al. Structural comparison of three types of Staphylococcal cassette chromosome mec integrated in the chromosome in Methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva-Carvalho MC, Bonelli RR, Souza RR, Moreira S, Santos LC, Souza Conceição M, et al. Emergence of multiresistant variants of the community-acquired methicillin-resistant Staphylococcus aureus lineage ST1-SCCmecIV in 2 hospitals in Rio de Janeiro, Brazil. Diagn Microbiol Infect Dis. 2009;65:300–305. doi: 10.1016/j.diagmicrobio.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 34.Matos PD, Oliveira TL, Cavalcante FS, Ferreira DC, Iorio NL, Pereira EM, et al. Molecular markers of antimicrobial resistance in Methicillin-resistant Staphylococcus aureus SCC mec IV presenting different genetic backgrounds. Microb Drug Resist. 2016;22:700–706. doi: 10.1089/mdr.2015.0255. [DOI] [PubMed] [Google Scholar]
- 35.Lim KT, Hanifah YA, Mohd Yusof MY, Ito T, Thong KL. Comparison of methicillin-resistant Staphylococcus aureus strains isolated in 2003 and 2008 with an emergence of multidrug resistant ST22: SCCmec IV clone in a tertiary hospital, Malaysia. J Microbiol Immunol Infect. 2013;46:224–233. doi: 10.1016/j.jmii.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Mišić M, Čukić J, Vidanović D, Šekler M, Matić S, Vukašinović M, et al. Prevalence of genotypes that determine resistance of Staphylococci to macrolides and lincosamides in Serbia. 200Front Public Health. 2017;5 doi: 10.3389/fpubh.2017.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goering RV, Swartzendruber EA, Obradovich AE, Tickler IA, Tenover FC. Emergence of oxacillin resistance in stealth Methicillin-resistant Staphylococcus aureus due to mecA sequence instability. Antimicrob Agents Chemother. 2019;63:e00558-19. doi: 10.1128/AAC.00558-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;(2):114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 39.Aminov RI. Horizontal gene exchange in environmental microbiota. 158Front Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]