Abstract
Alterations in global epigenetic signatures on chromatin are well established to contribute to tumor initiation and progression. Chromatin methylation status modulates several key cellular processes that maintain the integrity of the genome. KDM4A, a demethylase that belongs to the Fe-II dependent dioxygenase family that uses α-ketoglutarate and molecular oxygen as cofactors, is overexpressed in several cancers and is associated with an overall poor prognosis. KDM4A demethylates lysine 9 (H3K9me2/3) and lysine 36 (H3K36me3) methyl marks on histone H3. Given the complexity that exists with these marks on chromatin and their effects on transcription and proliferation, it naturally follows that demethylation serves an equally important role in these cellular processes. In this review, we highlight the role of KDM4A in transcriptional modulation, either dependent or independent of its enzymatic activity, arising from the amplification of this demethylase in cancer. KDM4A modulates re-replication of distinct genomic loci, activates cell cycle inducers, and represses proteins involved in checkpoint control giving rise to proliferative damage, mitotic disturbances and chromosomal breaks, ultimately resulting in genomic instability. In parallel, emerging evidence of non-nuclear substrates of epigenetic modulators emphasize the need to investigate the role of KDM4A in regulating non-nuclear substrates and evaluate their contribution to genomic instability in this context. The existence of promising KDM-specific inhibitors makes these demethylases an attractive target for therapeutic intervention in cancers.
Keywords: cancer, chromatin, genome integrity, KDM4A
Introduction
The three-dimensional structure of chromatin is modulated by processes including nucleosome positioning, histone composition, and histone modifications [1]. Modifications on positively charged histone proteins directly impact processes such as replication, gene transcription, and DNA damage repair. Post-translational modifications (PTMs) on histone tails play a crucial role in compaction of DNA around the nucleosome, i.e. formation of heterochromatin or euchromatin, and thereby impact the accessibility of DNA [2]. For example, histone acetylation (at lysine residues) neutralizes the positive charge of the histone core, reducing binding of histones to negatively charged DNA, thereby modestly releasing DNA into an open conformation rendering it more accessible to various DNA-binding proteins [3,4]. In contrast, histone methylation, which occurs in a complex pattern as mono-, di- or tri-methylation of lysine residues [5] or mono- and di-methylation of arginine residues [6], can alter the charge of the histone core (especially tri-methylation, which forms a quaternary amine and is a fixed charge). However, the function of both histone acetylation and methylation is primarily directed by modulating specific interactions of chromatin-binding proteins with the nucleosome. These interactions recruit additional functional units that initiate or carry out processes such as gene transcription, replication, and DNA damage response and repair [3]. The synergy between the various histone (PTMs) including phosphorylation, ubiquitination, sumoylation, deacetylation, demethylation, etc. is referred to as the ‘histone code’ [7,8]. Histone PTMs are invariably studied as discrete and isolated signals, however, in the context of a ‘histone code’, histone PTMs almost always operate in concert with one another to elicit a biological purpose. A full set of PTMs concurrent on single histone molecules or more globally within a single nucleosome forms a richer code and maps more directly to functional output [9].
The PTMs that comprise the histone code are laid down by enzymes commonly referred to as ‘writers’, these PTMs/marks are subsequently read by ‘readers’ that catalyze specific biological responses through the recruitment of additional functional units, and the PTMs/marks are finally removed by enzymes referred to as ‘erasers’ [10]. Erasers, similar to writers and readers, are comprised of a large number of enzymes with specificity for particular marks and built-in redundancies. Histone acetyl transferases HATs structurally open chromatin structure and license active transcription [11] and the opposing histone deacetylases (HDACs) remove the transcriptionally permissive acetylation marks laid down by the HATs [12]. In contrast, histone methylation is sometimes transcriptionally silencing or activating. Notably, EZH2 writes the transcriptionally repressive H3K27me3 PTM [13] while MLL (KTM2A) is prominent for writing the transcriptionally active H3K4me3 PTM [14]. With direct relevance to KDM4A, histone H3 lysine 9 is methylated by multiple HMTs [15,16] with SETDB1 [17] and SUV39H1/2 [10,18] capable of methylating H3K9 to all degrees and G9A and its homolog GLP mostly acting as di-methyltransferases [19]. Histone H3 lysine 36 is also methylated by several HMTs, with SETD2 writing H3K36me3 and NSD1/2/3, SMYD2, ASH1L, SETD3, and SETMAR mediating lower methylation degrees [20].
The lysine-specific demethylases (KDMs) (also commonly referred to as histone demethylases) are categorized into two families based on their mechanism of action. The first family of KDMs consist of flavin adenine dinucleotide (FAD)-dependent amine oxidases (KDM1A-B) and the second, much larger family is formed by Fe(II)-dependent dioxygenases [21]. The Fe(II)-dependent dioxygenases comprise the large bulk of the demethylases characterized by the presence of the highly conserved Jumonji C (JmjC)-domain and are further categorized into subfamilies based on sequence homology and methylation state preference [22,23]. This family includes KDM2A-B, KDM3A-B, KDM4A-E, KDM5A-D, KDM6A-B, KDM7A, and KDM8. Importantly, demethylation of tri-methyl lysine (Kme3) is exclusive to only the Fe(II)-dependent dioxygenases, whereas the FAD-dependent KDMs are limited to demethylating mono- and di-methyl lysine (Kme1/2) [24]. KDM4A-E are occasionally referred to as the ‘jhdm3 histone demethylase family’.
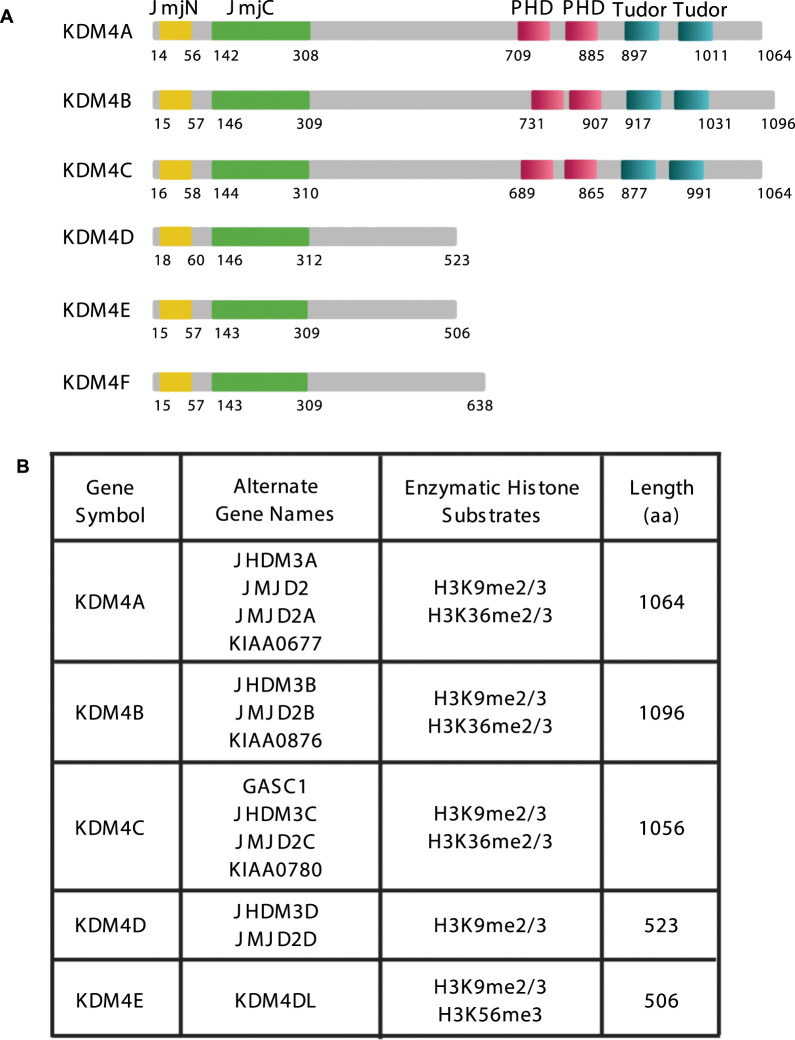
KDM4A
KDM4A is a lysine-specific demethylase that belongs to the Fe(II)-dependent dioxygenase family [23]. KDM4A has been implicated in the demethylation of histone H3 lysine 9 di/tri-methyl marks (H3K9me2/3) and histone H3 lysine 36 di/tri methyl marks (H3K36me2/3). Historically, KDM4A has also been referred to as JMJD2A, JHDM3A, JMJD2, TDRD14B, and KIAA0677. There are five additional KDM4s with close relation to KDM4A itself: KDM4B, KDM4C, KDM4D, KDM4E, and KDM4F (Figure 1). KDM4C is the most homologous to KDM4A and heterodimerizes with KDM4A [25], although homodimerization of KDM4A also modulates enzymatic activity [25]. KDM4D, KDM4E, and KDM4F are approximately half the size of the other KDM4 family members with homology only to JmjC-containing N-terminal demethylase domain (Figure 1A). KDM4E and KDM4F were originally believed to be pseudogenes, although KDM4E has since been discovered to be expressed as a protein with demethylase activity. In addition to sharing structural similarity, KDM4A-C also share enzymatic specificity on chromatin substrates although KDM4A-C display different histone binding preferences [26] (Figure 1B). Kinetic analysis revealed that KDM4A/B/C accept both H3K9 and H3K36 as substrates although demethylation at lysine 9 (K9) occurs preferentially and with a higher efficiency than demethylation at lysine 36 (K36) [27]. KDM4D/E exclusively catalyze demethylation of tri- and di-methylated H3K9 [27] with KDM4E also demethylating the H3K56me3 mark [28]. Moreover, all of these members of the KDM4 family were shown to demethylate lysine 26 on Histone H1.4 (H1.4K26), a linker histone [29]. Kinetic analysis revealed that H1.4K26 is a comparable substrate for KDM4A as H3K9me3 and is a better substrate than H3K36me3 [29]. Side chain residues proximal to the catalytic site are key determinants of specificity [27]. While single-gene knockouts of Kdm4a, Kdm4b, and Kdm4c and double gene knockouts of Kdm4a/b and Kdm4b/c are viable, a triple knockout of Kdm4a/b/c is embryonic lethal [30,31] indicative of functional redundancies within the KDM4 family.
Figure 1. KDM4 structural/functional domains.
(A) Schematic showing the functional and structural domains of KDM4A-F proteins. JmjN-domain residues, JmjC-domain residues, PHD domain residues (PHD1 and PHD2), Tudor domain residues (Tudor1 and Tudor2). (B) Table showing the enzymatic histone substrates of each of the KDM4 family members.
Catalytic domains of KDM4A
KDM4A demethylation activity is achieved similar to other members of the JmjC family. The JmjC domain is located near the N-terminus at residues 142–308 and is accompanied by a JmjN domain (residues 14–56) (Figure 1A) that complements and completes the active site. A single point mutation of histidine 188 (His188) within the JmjC domain completely abolishes demethylase activity on chromatin [32]. In the demethylation process Fe(II)-dependent dioxygenases consume 2-oxoglutarate and oxygen to produce carbon dioxide, succinate and formaldehyde. The JmjC-domain chelates an Fe(II) ion to form a catalytic center. Three amino acid side chains maintain contact with the iron (His188, Glu190, and His276). The iron center co-ordinates oxygen and 2-oxoglutarate and serves as an electron donor and acceptor to achieve its dioxygenase function. The resulting single oxygen atom is then used to oxidize the methyl carbon, ultimately liberating the methyl carbon from the lysine ε-Nitrogen as formaldehyde. In the process 2-oxoglutarate is also cleaved into succinate and carbon dioxide. A new 2-oxoglutarate molecule is used to complete the redox cycle and regenerate Fe(II) in the active site.
An often overlooked but important byproduct of this reaction is the production of highly reactive formaldehyde. Formaldehyde readily forms a variety of intermolecular cross-links, including cross-links between DNA and proteins, and thus has the potential to directly perturb genomic stability [33]. However, dedicated cellular metabolic pathways further oxidize and eliminate this endogenous formaldehyde. Aldehyde dehydrogenase 2 (ALDH2) and alcohol dehydrogenase 3 (ADH3) initiate oxidation of either formaldehyde or formaldehyde-glutathione to formate. Complete oxidation and elimination of formate is a tetrahydrofolate dependent process and connects into the one carbon metabolic pathway, including purine synthesis and indirectly SAM (S-adenosine methionine), which serves as the methyl donor during histone methylation by chromatin methyltransferases [34].
KDM4A demethylates both H3K9me2/3 and H3K36me2/3, although the meaning of H3K36 methylation is less clear than H3K9 methylation. The generally recognized function of H3K9me3, and less clearly H3K9me2, is transcriptional repression. This function is realized in part through recognition by HP1 (heterochromatin protein 1) and downstream processes. H3K36me2/3 is associated with active transcription, although not necessarily as a promoter of transcription, but as a repressor of spurious initiation of transcription [35]. Thus, demethylation of these two distinct marks by KDM4A reflects functions that are somewhat divergent. The coupling of these demethylase functions in KDM4A likely arises from a synergy or antagonism of these signals as part of the histone code with additional nuances than are currently unclear. Despite their nominally opposite meaning, the co-occurrence of H3K9me2/3 and H3K36me2/3 on the same molecule of histone H3 is not uncommon [36]. Although the meaning of this combination remains a mystery, one rational speculation is that the repressive function of H3K36me2/3 is dominant in this configuration. Thus, the dual demethylase activity of KDM4A would be wholly activating. Alternatively, these may function antagonistically to form a bivalent or poised state, wherein KDM4A may reset this poised state and allow transition into a new epigenetic state. Although H3K9me2/3 and H3K36me2/3 marks are directly demethylated by KDM4A, K14ac [37], and K23ac [36] are often commonly found on the same histone [8,36,38,39], and although the precise meaning of these marks remains an enigma, these co-occurring PTMs could ultimately modulate KDM4A activity. For example, H3 threonine 11 phosphorylation (H3T11ph) on the same molecule as H3K9me3 blocks demethylase activity [40]. These combinatorial effects are an essential but often ignored aspect of in vivo chromatin function.
Additional functional domains of KDM4A
In addition to the catalytic JmjC domain, KDM4A contains ancillary structural domains (Figure 1A). It harbors two Tudor domains near the C-terminus at residues 897–954 and residues 955–1011. This arrangement of tandem Tudor domains is a common theme in chromatin-binding proteins and these domains generally function in concert. The Tudor domains are essential in the recognition and binding of methylated histones, including H3K4me2/3 [41] and H4K20me2/3 [26,42–44]. They have also been reported to bind H3K23me3 in vitro [26]. Although H4K20me2 binding is undoubtedly more common [38,45] than binding to H3K4me3 [36,46], the relative abundance of these marks underplays functional importance [36,38,45], and as such the binding of H3K4me3, while infrequent may direct genome localization more strongly to elicit a more consequential effect. For example, the recruitment of KDM4A to H3K4me3 loci would effectively remove the repressive H3K9 methylation and enable the writing of the strongly active mark H3K9ac to establish or maintain persistent transcriptional activity [19].
KDM4A also contains two PHD (plant homeodomain)-type zinc fingers at residues 709–767 and residues 828–885 and one C2HC pre-PHD-type zinc finger (residues 772–805) (Figure 1A). The function of these domains remains unclear; however, PHD domains frequently function to bind methylated lysine residues in a sequence-specific manner. For instance, the homologous domains in KDM7A have been reported to read H3K4me3 [47]. These regions are thus likely involved in chromatin targeting of KDM4A. Thus far, efforts have exclusively focused on the functional characterization of these domains in the context of direct chromatin-associated enzymatic activity; however, these double PHD domains in KDM4A could effectively bind methyl marks on non-histone proteins or promote protein–protein interactions. For instance, KDM4A has been shown to interact with the N-terminal region of N-CoR (nuclear receptor corepressor) through a small NID (N-CoR interaction domain) at residues 597–638 [48]. However, KDM4A is not generally considered a catalytic member of this complex [49,50]. Thus, efforts need to be directed at understanding the precise role of these PHD domains in regulating chromatin and non-chromatin functions of KDM4A.
Post-translational modifications on KDM4A
Large scale proteomic screens identified a variety of putative KDM4A PTMs [51]; however, few have been carefully validated or functionally characterized. KDM4A appears to be N-terminally acetylated [52], which is typically a constitutive co-translational modification of fundamental structural and functional importance rather than a variable modification with regulatory potential [53]. In contrast with acetylation, phosphorylation [54] on tyrosine 547 (Y547), first identified in phospho-proteomic analysis [55], modulates KDM4A activity serving as a critical switch to promote autophagy in mammalian cells [54]. Ubiquitination, a PTM most often correlated to protein degradation, is regulated by multiple ubiquitin ligases on KDM4A including the F-box ubiquitin ligases FBX022 [56] and FBXL4 [57] as well as the RING-finger ubiquitin ligases RNF8 and RNF168 [42]. Although the exact function of ubiquitination by FBX022 is not completely understood, ubiquitination by FBXL4 mediates degradation of KDM4A which is directly associated with its regulation of cell cycle [57] whereas RNF8 and RNF168 regulate KDM4A ubiquitination and degradation in response to DNA damage controlling the recruitment of 53BP1 to sites of damage [42]. In addition to ubiquitination, the deubiquitinase USP1 stabilized KDM4A and promoted prostate cancer proliferation [58]. Another PTM on KDM4A is sumoylation on lysine residue 471 (K471) by a viral Sumo2/3-specific ligase, which stabilizes chromatin association and gene transactivation by KDM4A [59]. Thus, there is growing evidence, that this demethylase is likely tightly regulated at the protein level, with KDM4A undoubtedly having additional PTMs that regulate its diverse biological activity.
KDM4A inhibitors
Several small molecules have been developed that inhibit KDM4A demethylase activity while typically also inhibiting multiple related demethylases (reviewed in-depth [60]). Briefly, 8-Hydroxyquinoline derivatives and pyridine-based inhibitors showed promise as clinically relevant KDM inhibitors but failed to discriminate between KDMs [60,61]. Currently, QC6352 a cell-permeable KDM4 family-specific inhibitor exhibited antiproliferative effects but was unable to discriminate within the KDM4s [62]. Despite the limitations, these inhibitors remain useful as basic research probes and are clinically promising even if the mechanisms of action are obfuscated by the lack of selectivity. One such example is a widely available pan-KDM inhibitor n-Octyl-IOX1 which is 30-fold more effective than its earlier version, with IC50s in the low micromolar range [63]. Jib-04 is yet another useful, commercially available inhibitor, which inhibits multiple KDMs; however, it does so with IC50s in the hundreds of nanomolar range [64].
Role of KDM4A in genomic instability
KDM4A associated copy-number variations in cancer
Genomic instability, a hallmark of cancer, is often linked to copy-number alterations which include amplification or deletion of genomic regions as large as the chromosome arms or small, focused areas such as genomic fragments [65–67]. Methylation state alterations can dictate the predilection of a region to amplify. Analysis of 1770 primary tumor samples from the TCGA database revealed amplification and overexpression of KDM4A in 20% of cancers including uterine, ovarian, bladder, lung, and breast cancers [67]. In addition to genomic amplification, microRNAs such as hsa-mir-23a/b-3p and hsa-mir-137 also regulate KDM4A expression with loss/deletion of these microRNAs resulting in elevated KDM4A [68]. KDM4A overexpression directly correlates to re-replication and copy gains of specific chromosomal regions in multiple cancers, with ovarian cancers showing the most significant correlation between elevated KDM4A levels and copy-number gains [69].
The copy-number variations in cancers with overexpressed KDM4A are attributed to methylation changes (H3K9me3/H3K36me3) on chromatin which lead to the displacement of HP1 and recruitment of the replication machinery at these specific genomic regions resulting in re-replication [69]. Depletion of an alternate member of the same subfamily of demethylases, KDM5A (lysine demethylase for the H3K4me3 mark on histones), resulted in elevated H3K4 methylation and increased KDM4A occupancy at specific chromosomal regions [70], which was further speculated to result in unlicensed replication via bypassing the need for an origin of replication. Intriguingly, the KDM4A-dependent copy-number gains were extra-chromosomal and transient consisting of regions re-replicated during S-phase but lost prior to the end of G2. These copy gains were not integrated or inherited, although the cells retained their ability to re-replicate the same regions during subsequent cell divisions [69,71]. In the case of ovarian cancers, increased KDM4A levels associated directly with co-amplification of chromosomal region 1q12-21. Similarly, the interplay between elevated KDM4A and chromatin methylation status regulated oncogene EGFR copy gains in lung cancer cell lines and in cultured cells overexpressing KDM4A [72]. Similar observations were reported in breast cancer stem-like cells [73]. Stabilization of KDM4A by hypoxia contributed to transient site-specific gains in zebrafish and human cells [74]. Thus, amplification of KDM4A in cancers contributes to genomic instability arising via re-replication and copy-number alterations of genomic loci and is consistent with poor prognosis [75]. Pharmacologic inhibition of demethylase activity in vitro was efficacious in rescuing copy-number gains [74], and currently, concerted efforts are being made in identifying demethylase inhibitors with high specificity for KDM4A.
KDM4A as a regulator of the cell cycle in cancer
KDM4A increases chromatin accessibility and thereby regulates cell cycle progression. KDM4A demethylates H3K9me3 and displaces HP1, which normally binds this methyl mark to maintain a closed conformation, to inhibit firing of replication origins [76]. Overexpression of KDM4A alters replication timing in cells, promotes faster S-phase progression [77] and ultimately results in increased cellular proliferation as observed in bladder [78], prostate [79], colon [80] and human gastric cancers [81] to name a few. Corroborating these data, KDM4A protein levels peak at G1/S, although KDM4A mRNA transcript levels remained steady over the cell cycle [77]. KDM4A directly associates with pRb (phospho-retinoblastoma) and HDACs to mediate transcriptional repression of E2F-responsive promoters, directly linking KDM4A to cellular proliferation [82]. E2F1 in a complex with multiple HDACs negatively regulates ARHI (Aplasia Ras homolog member I) promoter activity [83]. ARHI is a Ras-related small G-protein repressed in breast and ovarian cancers wherein it functions as a tumor suppressor [84–86] to regulate cellular proliferation. In addition to binding pRb, KDM4A was found to interact with the E2F-HDAC complex and transcriptionally repress the ARHI tumor suppressor [87]. In this case, KDM4A acts to recruit co-regulators to the ARHI promoter as opposed to directly modulating promoter methylation status. More recently, the demethylase activity of KDM4A was found to be essential for KDM4A's role as a co-activator of E2F1. In contrast with its role as a recruiter of proteins to the pRb-HDAC complex, the enzymatic activity of KDM4A promoted E2F1 transcriptional activity thereby directly modulating the transcriptional profile of prostate cancer cell proliferation and survival [88].
KDM4A as a modulator of gene transcription
KDM4A in a context-specific fashion results in either transcriptional repression or activation of genes promoted by either KDM4A's role as a demethylase or its role as a recruiter of chromatin factors. In the case of human lung carcinoma, wherein KDM4A expression is elevated, KDM4A repressed the tumor suppressor CHD5 (chromodomain helicase DNA-binding domain 5) which led to reduced activity of p53 [89] and its binding CTCF and recruitment to the first intron of CHD5 reduced CHD5 gene transcription in breast cancer [90]. As part of the N-CoR (nuclear receptor corepressor) complex KDM4A represses the ASCL2 (human achaete scute-like homolog) gene which functions as a transcription factor although this occurs independent of KDM4A's demethylase function and can be attributed to its role as a recruiter of chromatin factors [48]. In addition to its histone substrates, KDM4A demethylated lysine containing peptides from WIZ (Wiz protein), CDYL1 (Chromodomain Y-like protein), CSB (DNA excision repair protein ERCC-6), and G9a (EHMT2, histone lysine methyltransferase) proteins, all constituents of transcription repression complexes [91].
In contrast with repressing gene expression, there are several reports of KDM4A stimulating gene transcription. The first report of KDM4A's role in activating gene expression was identified in prostate cancer where it was established to be a cofactor of androgen receptor (AR) and stimulated AR-dependent transcription of proliferative and survival genes [92] including prostate-specific antigen (PSA) [93] and c-myc [58] via its demethylase activity on H3K9me3. Similarly, KDM4A can form a complex with estrogen receptor alpha (ERα) and coactivate ERα-mediated transcription in breast cancer [94]. In the case of squamous cell carcinoma (SCC) KDM4A contributed to the activation of AP-1 (activator protein-1) by promoting the binding of AP-1 to the promoters of JUN and FOSL1 to maintain a positive feedback loop that activates AP-1 [95]. More recently, KDM4A overexpressed in nasopharyngeal carcinoma (NPC) positively correlated to lactate dehydrogenase (LDHA) levels, which it regulated at the transcriptional level to regulate aerobic glycolysis thereby contributing to NPC progression [96]. In summary, changes in gene expression (repression or activation) arising from epigenetic modulation of chromatin or recruitment of chromatin factors by KDM4A affect key cellular processes tipping the balance in favor of genomic instability and tumor progression.
KDM4A as a regulator of apoptosis
Apoptosis, a mechanism of programmed cell death, is vital to maintaining cellular homeostasis and exerts its protective function by promoting the clearance of defective cells. In cancer, vulnerabilities in either extrinsic or intrinsic apoptotic pathways help tumor cells evade induction of apoptosis, in many cases contributing to the resistance to therapeutic interventions [97,98]. Histone modifications serve a well-established role in modulating apoptosis. KDM4A overexpressed in multiple cancers including human gastric cancer [81], non-small lung cell carcinoma (NSCLC) [99,100] and in glioma cells [101], is associated with poor prognosis in all cases. In the case of gastric cancer, KDM4A knockdown lead to increased cellular apoptosis, arising from up-regulation of pro-apoptotic proteins such as Bax and cleaved caspase 3, and a down-regulation of anti-apoptotic protein Bcl-2 [81]. In addition, KDM4A modulated expression of the pro-apoptotic microRNA miR-34a [81]. Conversely, overexpression of KDM4A in NSCLC cell lines resulted in inhibition of apoptosis [99], highlighting the potential of KDM4A inhibitors to promote apoptosis in a clinical setting. In the case of colon cancer, where KDM4A is amplified, KDM4A directly interacts with p53 [80] to protect cells from apoptosis upon DNA damage [80]. The original report establishing the reversal of histone lysine tri-methylation by the KDM4 family of demethylases was also the first to determine changes in meiotic germ cell apoptosis in Caenorhabditis elegans arising from defects in p53 binding to double-strand breaks (DSBs) dictated by demethylation of H3K9/36me3 by KDM4A [102]. Depletion of KDM4A in C. elegans resulted in slowed DNA replication and increased ATR/p53-dependent apoptosis [77]. In addition to these intrinsic pathways, KDM4A silenced the TNF-related apoptosis-inducing ligand (TRAIL) pathway further highlighting how inhibition of this demethylase may have therapeutic potential in TRAIL-resistant cancers [103]. Importantly, KDM4A amplification and the correlated evasion of apoptosis could lead to persistent replication and/or the induction of replication errors in cells that fail to repair damage resulting in genomic and microsatellite instability.
Role of KDM4A in DNA damage response and repair
DNA damage, especially DSBs, poses unique challenges in terms of accessibility and recruitment of repair factors to sites of damage, with failure to repair giving rise to lethal lesions and mis-repair resulting in genomic instability [104]. Two mechanisms have evolved to protect cells against DSBs —homologous recombination (HR) and non-homologous end-joining (NHEJ). Recently, methylation by SETD2 was implicated in error-free double-strand break repair (DSBR) with H3K36me3 decorating chromatin to recruit key proteins involved in HR [105]. These data predicted that overexpression of KDM4A could drive genomic instability through loss of H3K36me3-mediated HR repair. Importantly, chromatin-associated KDM4A levels dramatically decrease after DNA damage, due in part to RNF8-RNF168-mediated ubiquitination and degradation of KDM4A [42]. Degradation of KDM4A was required to expose the H4K20me2 mark for recruitment of 53BP1, a DNA damage repair protein to sites of DNA damage [42]. Unwarranted binding of KDM4A to H4K20me2 in cells overexpressing this demethylase would presumably preclude DNA-damage repair by impairing binding of p53BP1 and result in genomic instability. Drosophila, dKDM4A (most analogous to human KDM4D) localized to heterochromatin and regulated DNA repair by altering the mobility of DSBs to the heterochromatin periphery and modulating H3K56me3 levels to facilitate repair [106,107]. In addition to DSB repair, H3K36me3 is also involved in DNA mismatch repair (MMR) as it provides a binding site for the MMR protein MSH6 and enables MSH6 foci formation during S-phase [108]. KDM4A overexpression impaired the integrity of MMR and lead to microsatellite instability (MSI) and increased the incidence of spontaneous mutations [109].
Beyond KDM4A nuclear functions
An interesting direction of KDM4A research lies outside the nucleus. Recently, lysine methylation has emerged on several non-histone targets [110] with localization of epigenetic regulators outside the nucleus. For example, KDM4A localizes to the cytoplasm, and interacts with translation initiation factors of polysomes to directly impact protein synthesis [111]. More recently, methyltransferase SETD2 was reported to methylate STAT1 [112], α-tubulin [113], and EZH2 [114]. In addition, the SMYD2 methyltransferase also methylated α-tubulin [115]. Importantly, the identification of these novel non-nuclear targets of chromatin methyltransferases offer exciting possibilities directed at identifying demethylases of these substrates, especially intriguing for KDM4A, the cognate demethylase for the histone mark laid down by SETD2.
Perspective
The interplay between genetics and epigenetics in the onset and progression of human tumors has clearly emerged over the last decade. Whole-genome sequencing efforts have established alterations in several key epigenetic modulators, including methyltransferases and demethylases across multiple cancers establishing the unequivocal role of these epigenetic regulators in tumor pathogenesis. The chromatin demethylase KDM4A is overexpressed in 20% of cancers and is correlated with poor prognosis [67].
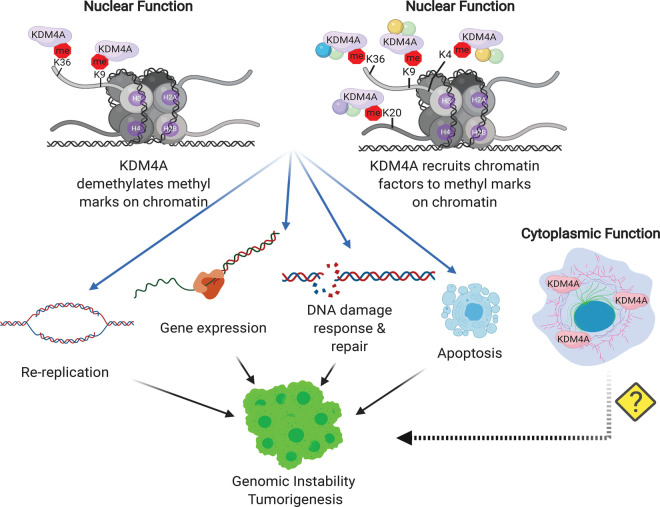
KDM4A enzymatic activity-dependent changes in H3K9 and H3K36 methylation or the recruitment or eviction of epigenetic regulators to marks bound by KDM4A (enzymatic activity independent), co-ordinate cellular processes that maintain genomic stability. Mechanistically, KDM4A driven alterations in the expression of genes regulating cell cycle, recruitment of the replication machinery to permit aberrant initiation of replication, evasion of apoptosis and a failure to correct DNA damage all contribute to genomic instability (Figure 2).
Coordination of signals affecting cytoskeletal and other cytosolic processes with transcription and gene expression seems evolutionarily advantageous. This hypothesis also suggests potential wider cellular coordination during the cell cycle and genome stability processes. It would be fascinating to establish mechanisms by which the non-nuclear functions of KDM4A contribute to genomic instability. Currently, accumulating evidence of the pre-clinical efficacy of several KDM inhibitors suggest that targeting this particular demethylase has potential as co-therapy to suppress genomic alterations in cancer.
Figure 2. Model depicting cellular processes regulated by KDM4A that contribute to genomic instability.
Demethylation of H3K9me2/3 or H3K36me3 and enzymatic activity independent recruitment of chromatin factors to methylated genomic loci to which KDM4A are bound can affect processes that maintain genomic stability. Alterations to these functions (usually by over amplification of KDM4A) results in loss of chromatin integrity and contributes to tumor progression. Additional studies focused on non-nuclear targets of KDM4A could reveal additional as yet unknown modes to generate genomic instability.
Acknowledgements
The authors would like to thank Dr. Matthew V. Holt for his insights and critical review of the manuscript.
Abbreviations
- AR
androgen receptor
- ARHI
Aplasia Ras homolog member I
- DSBs
double-strand breaks
- FAD
flavin adenine dinucleotide
- HDACs
histone deacetylases
- HP1
heterochromatin protein 1
- HR
homologous recombination
- NPC
nasopharyngeal carcinoma
- NSCLC
non-small lung cell carcinoma
- PHD
plant homeodomain
- PTMs
post-translational modifications
- TRAIL
TNF-related apoptosis-inducing ligand
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contribution
N.L.Y. and R.D. conceived and wrote the review.
Funding
This work was supported by a NIH R01GM139295-01 to N.L.Y. and a VHL Alliance grant to R.D.
References
- 1.Rando, O.J. and Chang, H.Y. (2009) Genome-wide views of chromatin structure. Annu. Rev. Biochem. 78, 245–271 10.1146/annurev.biochem.78.071107.134639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kouzarides, T. (2007) Chromatin modifications and their function. Cell 128, 693–705 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 3.Zentner, G.E. and Henikoff, S. (2013) Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 20, 259–266 10.1038/nsmb.2470 [DOI] [PubMed] [Google Scholar]
- 4.Mishra, L.N., Pepenella, S., Rogge, R., Hansen, J.C. and Hayes, J.J. (2016) Acetylation mimics within a single nucleosome alter local dna accessibility in compacted nucleosome arrays. Sci. Rep. 6, 34808 10.1038/srep34808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGrath, J. and Trojer, P. (2015) Targeting histone lysine methylation in cancer. Pharmacol. Ther. 150, 1–22 10.1016/j.pharmthera.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 6.Bedford, M.T. and Clarke, S.G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 10.1016/j.molcel.2008.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenuwein, T. and Allis, C.D. (2001) Translating the histone code. Science (New York, NY) 293, 1074–1080 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- 8.Rothbart, S.B. and Strahl, B.D. (2014) Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta 1839, 627–643 10.1016/j.bbagrm.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mishra, L.N. and Hayes, J.J. (2018) A nucleosome-free region locally abrogates histone H1-dependent restriction of linker DNA accessibility in chromatin. J. Biol. Chem. 293, 19191–19200 10.1074/jbc.RA118.005721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyun, K., Jeon, J., Park, K. and Kim, J. (2017) Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 49, e324 10.1038/emm.2017.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howe, L., Brown, C.E., Lechner, T. and Workman, J.L. (1999) Histone acetyltransferase complexes and their link to transcription. Crit. Rev. Eukaryot. Gene Expr. 9, 231–243 10.1615/CritRevEukarGeneExpr.v9.i3-4.80 [DOI] [PubMed] [Google Scholar]
- 12.Seto, E. and Yoshida, M. (2014) Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 6, a018713 10.1101/cshperspect.a018713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chammas, P., Mocavini, I. and Di Croce, L. (2020) Engaging chromatin: PRC2 structure meets function. Br. J. Cancer 122, 315–328 10.1038/s41416-019-0615-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, Y., Han, J., Zhang, Y., Cao, F., Liu, Z., Li, S.et al. (2016) Structural basis for activity regulation of MLL family methyltransferases. Nature 530, 447–452 10.1038/nature16952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brower-Toland, B., Riddle, N.C., Jiang, H., Huisinga, K.L. and Elgin, S.C. (2009) Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster. Genetics 181, 1303–1319 10.1534/genetics.108.100271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rice, J.C., Briggs, S.D., Ueberheide, B., Barber, C.M., Shabanowitz, J., Hunt, D.F.et al. (2003) Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12, 1591–1598 10.1016/S1097-2765(03)00479-9 [DOI] [PubMed] [Google Scholar]
- 17.Wang, H., An, W., Cao, R., Xia, L., Erdjument-Bromage, H., Chatton, B.et al. (2003) mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell 12, 475–487 10.1016/j.molcel.2003.08.007 [DOI] [PubMed] [Google Scholar]
- 18.Shi, Y. and Whetstine, J.R. (2007) Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell 25, 1–14 10.1016/j.molcel.2006.12.010 [DOI] [PubMed] [Google Scholar]
- 19.Plazas-Mayorca, M.D., Bloom, J.S., Zeissler, U., Leroy, G., Young, N.L., DiMaggio, P.A.et al. (2010) Quantitative proteomics reveals direct and indirect alterations in the histone code following methyltransferase knockdown. Mol. Biosyst. 6, 1719–1729 10.1039/c003307c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner, E.J. and Carpenter, P.B. (2012) Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13, 115–126 10.1038/nrm3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maes, T., Carceller, E., Salas, J., Ortega, A. and Buesa, C. (2015) Advances in the development of histone lysine demethylase inhibitors. Curr. Opin. Pharmacol. 23, 52–60 10.1016/j.coph.2015.05.009 [DOI] [PubMed] [Google Scholar]
- 22.Labbe, R.M., Holowatyj, A. and Yang, Z.Q. (2013) Histone lysine demethylase (KDM) subfamily 4: structures, functions and therapeutic potential. Am. J. Transl. Res. 6, 1–15 PMID: [PMC free article] [PubMed] [Google Scholar]
- 23.Tsukada, Y., Fang, J., Erdjument-Bromage, H., Warren, M.E., Borchers, C.H., Tempst, P.et al. (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 10.1038/nature04433 [DOI] [PubMed] [Google Scholar]
- 24.Couture, J.F., Collazo, E., Ortiz-Tello, P.A., Brunzelle, J.S. and Trievel, R.C. (2007) Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 14, 689–695 10.1038/nsmb1273 [DOI] [PubMed] [Google Scholar]
- 25.Levin, M., Stark, M. and Assaraf, Y.G. (2018) The JmjN domain as a dimerization interface and a targeted inhibitor of KDM4 demethylase activity. Oncotarget 9, 16861–16882 10.18632/oncotarget.24717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su, Z., Wang, F., Lee, J.H., Stephens, K.E., Papazyan, R., Voronina, E.et al. (2016) Reader domain specificity and lysine demethylase-4 family function. Nat. Commun. 7, 13387 10.1038/ncomms13387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hillringhaus, L., Yue, W.W., Rose, N.R., Ng, S.S., Gileadi, C., Loenarz, C.et al. (2011) Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family. J. Biol. Chem. 286, 41616–41625 10.1074/jbc.M111.283689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jack, A.P., Bussemer, S., Hahn, M., Punzeler, S., Snyder, M., Wells, M.et al. (2013) H3k56me3 is a novel, conserved heterochromatic mark that largely but not completely overlaps with H3K9me3 in both regulation and localization. PLoS One 8, e51765 10.1371/journal.pone.0051765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walport, L.J., Hopkinson, R.J., Chowdhury, R., Zhang, Y., Bonnici, J., Schiller, R.et al. (2018) Mechanistic and structural studies of KDM-catalysed demethylation of histone 1 isotype 4 at lysine 26. FEBS Lett. 592, 3264–3273 10.1002/1873-3468.13231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen, M.T., Kooistra, S.M., Radzisheuskaya, A., Laugesen, A., Johansen, J.V., Hayward, D.G.et al. (2016) Continual removal of H3K9 promoter methylation by Jmjd2 demethylases is vital for ESC self-renewal and early development. EMBO J. 35, 1550–1564 10.15252/embj.201593317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedersen, M.T., Agger, K., Laugesen, A., Johansen, J.V., Cloos, P.A., Christensen, J.et al. (2014) The demethylase JMJD2C localizes to H3K4me3-positive transcription start sites and is dispensable for embryonic development. Mol. Cell. Biol. 34, 1031–1045 10.1128/MCB.00864-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klose, R.J., Yamane, K., Bae, Y., Zhang, D., Erdjument-Bromage, H., Tempst, P.et al. (2006) The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442, 312–316 10.1038/nature04853 [DOI] [PubMed] [Google Scholar]
- 33.Lu, K., Ye, W., Zhou, L., Collins, L.B., Chen, X., Gold, A.et al. (2010) Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J. Am. Chem. Soc. 132, 3388–3399 10.1021/ja908282f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pietzke, M., Meiser, J. and Vazquez, A. (2020) Formate metabolism in health and disease. Mol. Metab. 33, 23–37 10.1016/j.molmet.2019.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venkatesh, S. and Workman, J.L. (2013) Set2 mediated H3 lysine 36 methylation: regulation of transcription elongation and implications in organismal development. Wiley Interdiscip. Rev. Dev. Biol. 2, 685–700 10.1002/wdev.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young, N.L., DiMaggio, P.A., Plazas-Mayorca, M.D., Baliban, R.C., Floudas, C.A. and Garcia, B.A. (2009) High throughput characterization of combinatorial histone codes. Mol. Cell. Proteom. 8, 2266–2284 10.1074/mcp.M900238-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jurkowska, R.Z., Qin, S., Kungulovski, G., Tempel, W., Liu, Y., Bashtrykov, P.et al. (2017) H3k14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1. Nat. Commun. 8, 2057 10.1038/s41467-017-02259-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holt, M.V., Wang, T. and Young, N.L. (2019) High-throughput quantitative top-down proteomics: histone H4. J. Am. Soc. Mass Spectrom. 30, 2548–2560 10.1007/s13361-019-02350-z [DOI] [PubMed] [Google Scholar]
- 39.Young, N.L., Plazas-Mayorca, M.D. and Garcia, B.A. (2010) Systems-wide proteomic characterization of combinatorial post-translational modification patterns. Expert Rev. Proteom. 7, 79–92 10.1586/epr.09.100 [DOI] [PubMed] [Google Scholar]
- 40.Krishnan, S. and Trievel, R.C. (2013) Structural and functional analysis of JMJD2D reveals molecular basis for site-specific demethylation among JMJD2 demethylases. Structure 21, 98–108 10.1016/j.str.2012.10.018 [DOI] [PubMed] [Google Scholar]
- 41.Huang, Y., Fang, J., Bedford, M.T., Zhang, Y. and Xu, R.M. (2006) Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science (New York, NY) 312, 748–751 10.1126/science.1125162 [DOI] [PubMed] [Google Scholar]
- 42.Mallette, F.A., Mattiroli, F., Cui, G., Young, L.C., Hendzel, M.J., Mer, G.et al. (2012) RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 31, 1865–1878 10.1038/emboj.2012.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim, J., Daniel, J., Espejo, A., Lake, A., Krishna, M., Xia, L.et al. (2006) MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 7, 397–403 10.1038/sj.embor.7400625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, J., Thompson, J.R., Botuyan, M.V. and Mer, G. (2008) Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 15, 109–111 10.1038/nsmb1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, T., Holt, M.V. and Young, N.L. (2018) The histone H4 proteoform dynamics in response to SUV4-20 inhibition reveals single molecule mechanisms of inhibitor resistance. Epigenetics Chromatin 11, 29 10.1186/s13072-018-0198-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah, R.N., Grzybowski, A.T., Cornett, E.M., Johnstone, A.L., Dickson, B.M., Boone, B.A.et al. (2018) Examining the roles of H3K4 methylation states with systematically characterized antibodies. Mol. Cell 72, 162–177.e7 10.1016/j.molcel.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horton, J.R., Upadhyay, A.K., Qi, H.H., Zhang, X., Shi, Y. and Cheng, X. (2010) Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat. Struct. Mol. Biol. 17, 38–43 10.1038/nsmb.1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, D., Yoon, H.G. and Wong, J. (2005) JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2). Mol. Cell. Biol. 25, 6404–6414 10.1128/MCB.25.15.6404-6414.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li, J., Wang, J., Wang, J., Nawaz, Z., Liu, J.M., Qin, J.et al. (2000) Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19, 4342–4350 10.1093/emboj/19.16.4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoon, H.G., Chan, D.W., Huang, Z.Q., Li, J., Fondell, J.D., Qin, J.et al. (2003) Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 22, 1336–1346 10.1093/emboj/cdg120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hornbeck, P.V., Zhang, B., Murray, B., Kornhauser, J.M., Latham, V. and Skrzypek, E. (2015) Phosphositeplus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43(Database issue), D512–D520 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gauci, S., Helbig, A.O., Slijper, M., Krijgsveld, J., Heck, A.J. and Mohammed, S. (2009) Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal. Chem. 81, 4493–4501 10.1021/ac9004309 [DOI] [PubMed] [Google Scholar]
- 53.Ree, R., Varland, S. and Arnesen, T. (2018) Spotlight on protein N-terminal acetylation. Exp. Mol. Med. 50, 1–13 10.1038/s12276-018-0116-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernard, A., Jin, M., Gonzalez-Rodriguez, P., Fullgrabe, J., Delorme-Axford, E., Backues, S.K.et al. (2015) Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Curr. Biol. 25, 546–555 10.1016/j.cub.2014.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tao, W.A., Wollscheid, B., O'Brien, R., Eng, J.K., Li, X.J., Bodenmiller, B.et al. (2005) Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat. Methods 2, 591–598 10.1038/nmeth776 [DOI] [PubMed] [Google Scholar]
- 56.Tan, M.K., Lim, H.J. and Harper, J.W. (2011) SCF(FBXO22) regulates histone H3 lysine 9 and 36 methylation levels by targeting histone demethylase KDM4A for ubiquitin-mediated proteasomal degradation. Mol. Cell. Biol. 31, 3687–3699 10.1128/MCB.05746-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Rechem, C., Black, J.C., Abbas, T., Allen, A., Rinehart, C.A., Yuan, G.C.et al. (2011) The SKP1-Cul1-F-box and leucine-rich repeat protein 4 (SCF-FbxL4) ubiquitin ligase regulates lysine demethylase 4A (KDM4A)/Jumonji domain-containing 2A (JMJD2A) protein. J. Biol. Chem. 286, 30462–30470 10.1074/jbc.M111.273508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui, S.Z., Lei, Z.Y., Guan, T.P., Fan, L.L., Li, Y.Q., Geng, X.Y.et al. (2020) Targeting USP1-dependent KDM4A protein stability as a potential prostate cancer therapy. Cancer Sci. 111, 1567–1581 10.1111/cas.14375 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Yang, W.S., Campbell, M. and Chang, P.C. (2017) SUMO modification of a heterochromatin histone demethylase JMJD2A enables viral gene transactivation and viral replication. PLoS Pathog. 13, e1006216 10.1371/journal.ppat.1006216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin, H., Li, Q., Li, Q., Zhu, J., Gu, K., Jiang, X.et al. (2018) Small molecule KDM4s inhibitors as anti-cancer agents. J. Enzyme Inhib. Med. Chem. 33, 777–793 10.1080/14756366.2018.1455676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hopkinson, R.J., Tumber, A., Yapp, C., Chowdhury, R., Aik, W., Che, K.H.et al. (2013) 5-Carboxy-8-hydroxyquinoline is a broad spectrum 2-oxoglutarate oxygenase inhibitor which causes iron translocation. Chem. Sci. 4, 3110–3117 10.1039/c3sc51122g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen, Y.K., Bonaldi, T., Cuomo, A., Del Rosario, J.R., Hosfield, D.J., Kanouni, T.et al. (2017) Design of KDM4 inhibitors with antiproliferative effects in cancer models. ACS Med. Chem. Lett. 8, 869–874 10.1021/acsmedchemlett.7b00220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schiller, R., Scozzafava, G., Tumber, A., Wickens, J.R., Bush, J.T., Rai, G.et al. (2014) A cell-permeable ester derivative of the JmjC histone demethylase inhibitor IOX1. ChemMedChem 9, 566–571 10.1002/cmdc.201300428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, L., Chang, J., Varghese, D., Dellinger, M., Kumar, S., Best, A.M.et al. (2013) A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat. Commun. 4, 2035 10.1038/ncomms3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hook, S.S., Lin, J.J. and Dutta, A. (2007) Mechanisms to control rereplication and implications for cancer. Curr. Opin. Cell Biol. 19, 663–671 10.1016/j.ceb.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stratton, M.R., Campbell, P.J. and Futreal, P.A. (2009) The cancer genome. Nature 458, 719–724 10.1038/nature07943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beroukhim, R., Mermel, C.H., Porter, D., Wei, G., Raychaudhuri, S., Donovan, J.et al. (2010) The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 10.1038/nature08822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Black, J.C., Zhang, H., Kim, J., Getz, G. and Whetstine, J.R. (2016) Regulation of transient site-specific copy gain by microRNA. J. Biol. Chem. 291, 4862–4871 10.1074/jbc.M115.711648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Black, J.C., Manning, A.L., Van Rechem, C., Kim, J., Ladd, B., Cho, J.et al. (2013) KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell 154, 541–555 10.1016/j.cell.2013.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mishra, S., Van Rechem, C., Pal, S., Clarke, T.L., Chakraborty, D., Mahan, S.D.et al. (2018) Cross-talk between lysine-modifying enzymes controls site-specific DNA amplifications. Cell 174, 803–817.e16 10.1016/j.cell.2018.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Black, J.C. and Whetstine, J.R. (2015) Too little O2 too much gain. Cell Cycle 14, 2869–2870 10.1080/15384101.2015.1076659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clarke, T.L., Tang, R., Chakraborty, D., Van Rechem, C., Ji, F., Mishra, S.et al. (2020) Histone lysine methylation dynamics control EGFR DNA copy-number amplification. Cancer Discov. 10, 306–325 10.1158/2159-8290.CD-19-0463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Metzger, E., Stepputtis, S.S., Strietz, J., Preca, B.T., Urban, S., Willmann, D.et al. (2017) KDM4 inhibition targets breast cancer stem-like cells. Cancer Res. 77, 5900–5912 10.1158/0008-5472.CAN-17-1754 [DOI] [PubMed] [Google Scholar]
- 74.Black, J.C., Atabakhsh, E., Kim, J., Biette, K.M., Van Rechem, C., Ladd, B.et al. (2015) Hypoxia drives transient site-specific copy gain and drug-resistant gene expression. Genes Dev. 29, 1018–1031 10.1101/gad.259796.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee, D.H., Kim, G.W., Jeon, Y.H., Yoo, J., Lee, S.W. and Kwon, S.H. (2020) Advances in histone demethylase KDM4 as cancer therapeutic targets. FASEB J. 34, 3461–3484 10.1096/fj.201902584R [DOI] [PubMed] [Google Scholar]
- 76.Gerace, E.L. and Moazed, D. (2010) Histone demethylation and timely DNA replication. Mol. Cell 40, 683–684 10.1016/j.molcel.2010.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Black, J.C., Allen, A., Van Rechem, C., Forbes, E., Longworth, M., Tschop, K.et al. (2010) Conserved antagonism between JMJD2A/KDM4A and HP1gamma during cell cycle progression. Mol. Cell 40, 736–748 10.1016/j.molcel.2010.11.008 [DOI] [PubMed] [Google Scholar]
- 78.Wang, F., Li, Y., Shan, F., Zhang, Q., Wang, L., Sheng, B.et al. (2019) Upregulation of JMJD2A promotes migration and invasion in bladder cancer through regulation of SLUG. Oncol. Rep. 42, 1431–1440 10.3892/or.2019.7246 [DOI] [PubMed] [Google Scholar]
- 79.Mu, H., Xiang, L., Li, S., Rao, D., Wang, S. and Yu, K. (2019) MiR-10a functions as a tumor suppressor in prostate cancer via targeting KDM4A. J. Cell. Biochem. 120, 4987–4997 10.1002/jcb.27774 [DOI] [PubMed] [Google Scholar]
- 80.Kim, T.D., Shin, S., Berry, W.L., Oh, S. and Janknecht, R. (2012) The JMJD2A demethylase regulates apoptosis and proliferation in colon cancer cells. J. Cell. Biochem. 113, 1368–1376 10.1002/jcb.24009 [DOI] [PubMed] [Google Scholar]
- 81.Hu, C.E., Liu, Y.C., Zhang, H.D. and Huang, G.J. (2014) JMJD2A predicts prognosis and regulates cell growth in human gastric cancer. Biochem. Biophys. Res. Commun. 449, 1–7 10.1016/j.bbrc.2014.04.126 [DOI] [PubMed] [Google Scholar]
- 82.Gray, S.G., Iglesias, A.H., Lizcano, F., Villanueva, R., Camelo, S., Jingu, H.et al. (2005) Functional characterization of JMJD2A, a histone deacetylase- and retinoblastoma-binding protein. J. Biol. Chem. 280, 28507–28518 10.1074/jbc.M413687200 [DOI] [PubMed] [Google Scholar]
- 83.Lu, Z., Luo, R.Z., Peng, H., Huang, M., Nishmoto, A., Hunt, K.K.et al. (2006) E2F-HDAC complexes negatively regulate the tumor suppressor gene ARHI in breast cancer. Oncogene 25, 230–239 10.1038/sj.onc.1209025 [DOI] [PubMed] [Google Scholar]
- 84.Luo, R.Z., Fang, X., Marquez, R., Liu, S.Y., Mills, G.B., Liao, W.S.et al. (2003) ARHI is a Ras-related small G-protein with a novel N-terminal extension that inhibits growth of ovarian and breast cancers. Oncogene 22, 2897–2909 10.1038/sj.onc.1206380 [DOI] [PubMed] [Google Scholar]
- 85.Yu, Y., Xu, F., Peng, H., Fang, X., Zhao, S., Li, Y.et al. (1999) NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc. Natl. Acad. Sci. U.S.A. 96, 214–219 10.1073/pnas.96.1.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang, L., Hoque, A., Luo, R.Z., Yuan, J., Lu, Z., Nishimoto, A.et al. (2003) Loss of the expression of the tumor suppressor gene ARHI is associated with progression of breast cancer. Clin.Cancer Res. 9(10 Pt 1), 3660–3666 PMID: [PubMed] [Google Scholar]
- 87.Li, L.L., Xue, A.M., Li, B.X., Shen, Y.W., Li, Y.H., Luo, C.L.et al. (2014) JMJD2A contributes to breast cancer progression through transcriptional repression of the tumor suppressor ARHI. Breast Cancer Res. 16, R56 10.1186/bcr3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang, L.Y., Hung, C.L., Chen, Y.R., Yang, J.C., Wang, J., Campbell, M.et al. (2016) KDM4A coactivates E2F1 to regulate the PDK-dependent metabolic switch between mitochondrial oxidation and glycolysis. Cell Rep. 16, 3016–3027 10.1016/j.celrep.2016.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mallette, F.A. and Richard, S. (2012) JMJD2A promotes cellular transformation by blocking cellular senescence through transcriptional repression of the tumor suppressor CHD5. Cell Rep. 2, 1233–1243 10.1016/j.celrep.2012.09.033 [DOI] [PubMed] [Google Scholar]
- 90.Guerra-Calderas, L., Gonzalez-Barrios, R., Patino, C.C., Alcaraz, N., Salgado-Albarran, M., de Leon, D.C.et al. (2018) CTCF-KDM4A complex correlates with histone modifications that negatively regulate CHD5 gene expression in cancer cell lines. Oncotarget 9, 17028–17042 10.18632/oncotarget.24798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ponnaluri, V.K., Vavilala, D.T., Putty, S., Gutheil, W.G. and Mukherji, M. (2009) Identification of non-histone substrates for JMJD2A-C histone demethylases. Biochem. Biophys. Res. Commun. 390, 280–284 10.1016/j.bbrc.2009.09.107 [DOI] [PubMed] [Google Scholar]
- 92.Shin, S. and Janknecht, R. (2007) Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochem. Biophys. Res. Commun. 359, 742–746 10.1016/j.bbrc.2007.05.179 [DOI] [PubMed] [Google Scholar]
- 93.Zhang, J., Li, Q., Zhang, S., Xu, Q. and Wang, T. (2016) Lgr4 promotes prostate tumorigenesis through the Jmjd2a/AR signaling pathway. Exp. Cell Res. 349, 77–84 10.1016/j.yexcr.2016.09.023 [DOI] [PubMed] [Google Scholar]
- 94.Berry, W.L., Shin, S., Lightfoot, S.A. and Janknecht, R. (2012) Oncogenic features of the JMJD2A histone demethylase in breast cancer. Int. J. Oncol. 41, 1701–1706 10.3892/ijo.2012.1618 [DOI] [PubMed] [Google Scholar]
- 95.Ding, X., Pan, H., Li, J., Zhong, Q., Chen, X., Dry, S.M.et al. (2013) Epigenetic activation of AP1 promotes squamous cell carcinoma metastasis. Sci. Signal. 6, ra28.1–13., S0-5 10.1126/scisignal.2003884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Su, Y., Yu, Q.H., Wang, X.Y., Yu, L.P., Wang, Z.F., Cao, Y.C.et al. (2017) JMJD2A promotes the Warburg effect and nasopharyngeal carcinoma progression by transactivating LDHA expression. BMC Cancer 17, 477 10.1186/s12885-017-3473-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wong, R.S. (2011) Apoptosis in cancer: from pathogenesis to treatment. J. Exp. Clin. Cancer Res. 30, 87 10.1186/1756-9966-30-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Emdad, L., Bhoopathi, P., Talukdar, S., Pradhan, A.K., Sarkar, D., Wang, X.Y.et al. (2020) Recent insights into apoptosis and toxic autophagy: the roles of MDA-7/IL-24, a multidimensional anti-cancer therapeutic. Semin. Cancer Biol. 66, 140–154 10.1016/j.semcancer.2019.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu, W., Jiang, K., Shen, M., Chen, Y. and Huang, H.Y. (2016) Jumonji domain containing 2A predicts prognosis and regulates cell growth in lung cancer depending on miR-150. Oncol Rep. 35, 352–358 10.3892/or.2015.4349 [DOI] [PubMed] [Google Scholar]
- 100.Soini, Y., Kosma, V.M. and Pirinen, R. (2015) KDM4A, KDM4B and KDM4C in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 8, 12922–12928 PMID: [PMC free article] [PubMed] [Google Scholar]
- 101.Wang, B., Fan, X., Ma, C., Lei, H., Long, Q. and Chai, Y. (2016) Downregulation of KDM4A suppresses the survival of glioma cells by promoting autophagy. J. Mol. Neurosci. 60, 137–144 10.1007/s12031-016-0796-6 [DOI] [PubMed] [Google Scholar]
- 102.Whetstine, J.R., Nottke, A., Lan, F., Huarte, M., Smolikov, S., Chen, Z.et al. (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467–481 10.1016/j.cell.2006.03.028 [DOI] [PubMed] [Google Scholar]
- 103.Wang, J., Wang, H., Wang, L.Y., Cai, D., Duan, Z., Zhang, Y.et al. (2016) Silencing the epigenetic silencer KDM4A for TRAIL and DR5 simultaneous induction and antitumor therapy. Cell Death Differ. 23, 1886–1896 10.1038/cdd.2016.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jeggo, P. and Lavin, M.F. (2009) Cellular radiosensitivity: how much better do we understand it? Int. J. Radiat. Biol. 85, 1061–1081 10.3109/09553000903261263 [DOI] [PubMed] [Google Scholar]
- 105.Pfister, S.X., Ahrabi, S., Zalmas, L.P., Sarkar, S., Aymard, F., Bachrati, C.Z.et al. (2014) SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 7, 2006–2018 10.1016/j.celrep.2014.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Colmenares, S.U., Swenson, J.M., Langley, S.A., Kennedy, C., Costes, S.V. and Karpen, G.H. (2017) Drosophila histone demethylase KDM4A has enzymatic and non-enzymatic roles in controlling heterochromatin integrity. Dev. Cell 42, 156–169.e5 10.1016/j.devcel.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Janssen, A., Colmenares, S.U., Lee, T. and Karpen, G.H. (2019) Timely double-strand break repair and pathway choice in pericentromeric heterochromatin depend on the histone demethylase dKDM4A. Genes Dev. 33, 103–115 10.1101/gad.317537.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li, F., Mao, G., Tong, D., Huang, J., Gu, L., Yang, W.et al. (2013) The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 153, 590–600 10.1016/j.cell.2013.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Awwad, S.W. and Ayoub, N. (2015) Overexpression of KDM4 lysine demethylases disrupts the integrity of the DNA mismatch repair pathway. Biol. Open 4, 498–504 10.1242/bio.201410991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang, X., Huang, Y. and Shi, X. (2015) Emerging roles of lysine methylation on non-histone proteins. Cell Mol. Life Sci. 72, 4257–4272 10.1007/s00018-015-2001-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Van Rechem, C., Black, J.C., Boukhali, M., Aryee, M.J., Graslund, S., Haas, W.et al. (2015) Lysine demethylase KDM4A associates with translation machinery and regulates protein synthesis. Cancer Discov. 5, 255–263 10.1158/2159-8290.CD-14-1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen, K., Liu, J., Liu, S., Xia, M., Zhang, X., Han, D.et al. (2017) Methyltransferase SETD2-mediated methylation of STAT1 is critical for interferon antiviral activity. Cell 170, 492–506.e14 10.1016/j.cell.2017.06.042 [DOI] [PubMed] [Google Scholar]
- 113.Park, I.Y., Powell, R.T., Tripathi, D.N., Dere, R., Ho, T.H., Blasius, T.L.et al. (2016) Dual chromatin and cytoskeletal remodeling by SETD2. Cell 166, 950–962 10.1016/j.cell.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yuan, H., Han, Y., Wang, X., Li, N., Liu, Q., Yin, Y.et al. (2020) SETD2 restricts prostate cancer metastasis by integrating EZH2 and AMPK signaling pathways. Cancer Cell 350–365.e7 10.1016/j.ccell.2020.05.022 [DOI] [PubMed] [Google Scholar]
- 115.Li, L.X., Zhou, J.X., Wang, X., Zhang, H., Harris, P.C., Calvet, J.P.et al. (2020) Cross-talk between CDK4/6 and SMYD2 regulates gene transcription, tubulin methylation, and ciliogenesis. Sci. Adv. 6, eabb3154 10.1126/sciadv.abb3154 [DOI] [PMC free article] [PubMed] [Google Scholar]