Abstract
RNA-binding proteins are customarily regarded as important facilitators of gene expression. In recent years, RNA–protein interactions have also emerged as a pervasive force in the regulation of homeostasis. The compendium of proteins with provable RNA-binding function has swelled from the hundreds to the thousands astride the partnership of mass spectrometry-based proteomics and RNA sequencing. At the foundation of these advances is the adaptation of RNA-centric capture methods that can extract bound protein that has been cross-linked in its native environment. These methods reveal snapshots in time displaying an extensive network of regulation and a wealth of data that can be used for both the discovery of RNA-binding function and the molecular interfaces at which these interactions occur. This review will focus on the impact of these developments on our broader perception of post-transcriptional regulation, and how the technical features of current capture methods, as applied in mammalian systems, create a challenging medium for interpretation by systems biologists and target validation by experimental researchers.
Keywords: biochemical techniques and resources, RNA, RNA-binding proteins
Introduction
RNA-binding proteins (RBPs) are proteins that directly interact with single-stranded or double-stranded RNA. These interactions form complexes, known as ribonucleoproteins (RNPs), which drive the post-transcriptional processes necessary for gene expression. Thus, for the entirety of its lifecycle, RNA is tended to by a host of proteins that govern its synthesis, trafficking, translation, and degradation. This active participation makes the discovery of RBPs a critical first step towards understanding how such post-transcriptional governance impacts homeostasis or disorder. In recent years, some efforts have strayed from traditional strategies such as immunoprecipitation or targeted mutagenesis, towards the global screening of RNA-bound protein using mass spectrometry (MS) based proteomics. These unbiased approaches have begun to reveal an RNA interactome of unexpected complexity that is characterised by widespread participation from the proteome and hints at unknown, yet ubiquitous, molecular processes occurring throughout the cell.
The molecular and physiological landscape of RNA-binding proteins
The attraction of a protein's RNA-binding domains (RBDs) to a RNA's surface of interaction depends on a diverse range of forces that are widely recognised. For instance, the K-homology domains of the fragile X mental retardation 1 (FMR1) and Nova proteins rely on nucleotide recognition via hydrophobic interactions with non-aromatic residues, hydrogen bonding to bases, and shape complementarity with sugar-phosphate backbone contacts [1]. The RNA-recognition motifs (RRM), found in many proteins of the spliceosome, typically rely on a salt bridge between an Arg or Lys residue with the phosphodiester backbone and stacking interactions between aromatic residues and nucleobases [2]. Another well-known group of domains includes the Zinc finger family which allows transcription factor IIIA to discriminate DNA from RNA by leveraging electrostatic interactions with protein side chains that capture RNA loops [3]. Thus, the binding of a single domain can involve a composite event of multiple forces that can guide, or be the product of, complementary structures.
The present canon of conventional RBDs comprises more than a dozen distinct groups in addition to the KH, RRM, and zinc finger families. Among conventional RBPs these domains act as modules which confer different levels of RNA specificity according to their combinatorics. Though affinity can be supported by interaction with a single domain it is often strengthened by the collective effort of many domains that an RNA is targeted by either sequence, structure, or a blend of both. Where distant sites are bound, domain organisation plays a major role, for instance, a linker region that is ordered or disordered will place different constraints on eligible targets based on its topology and length [2]. Conversely, affinity with a single surface of interaction can be expanded by the structural alignment of several domains or even occluded by intervening structures that act as a molecular switch [4].
In recent years, it has come to light that hundreds of new RBPs are intrinsically disordered and frequently lack known RBDs [5,6]. Disordered regions entail the absence of a stable three-dimensional structure and can harbour multiple interaction sites. These domains are composed of low complexity and repetitive amino acid sequences such as the previously reported, non-classical, short linear motifs (SLiMs), RG[G] repeats, and RS/RG-rich sequences [7]. These same features are shared between RBPs that lack known RBDs implying that plasticity could be the centrepiece of RNA interaction and a means of fine-tuning post-transcriptional control. One such domain previously predicted, though lacking experimental evidence at the time, was the RNA-binding domain abundant in Apicomplexans (RAP) common to the fas-activated serine/threonine (FAST) kinase family [5,8]. The FASTK family are regulators of mitochondrial RNA homeostasis and have since been confirmed to possess an RNA targeting region though upstream of RAP's endonucleolytic domain [9]. Another well-known example, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), can exercise different effects through non-canonical interaction with a variety of AU-rich sequences in a number mRNAs including cyclooxygenase-2 (Cox-2) and colony-stimulating factor-1 (CSF-1) [10–12]. Such proteins, found to be engaged in biological processes but without any clear role in the RNA lifecycle, have been called ‘EnigmRBPs’ and they provide further hints that a network of RNA–protein controls play a significant role in homeostasis [13].
The apparent prevalence of RNA-binding activity among metabolic enzymes such as FASTK and GAPDH has put a spotlight on the moonlighting activities of proteins previously thought to perform only one, highly specific function. That disorder sits at the nexus of many unexpected protein–RNA interactions is revealed by growing evidence that unstructured RBPs might depend on multiple, low-affinity binding sites to assemble phase-separated compartments [14–16]. This phenomenon has been linked to the formation of persistent stress granules and the accumulation of fibrillar protein pathologies in the disease-associated RBP hnRPA1 [15]. Such findings reinforce the value of using broad experimental strategies to generate a genome-wide RBP census that could aid in recognising RNA-binding activity among disease-associated proteins or in diagnosing associated pathologies. This list will inevitably grow and so far RBPs which interact with mRNA have been implicated in 221 Mendelian diseases, the preponderance of which include cancers, muscular atrophies, neurological diseases and metabolic disorders [17]. Given that just 1.5% of the human genome comprises protein-coding genes it is likely these numbers will swell as the role of RBPs regulating the behaviour of non-coding RNA is revealed [18].
Experimental strategies for high-throughput discovery
The chemistry underpinning our current gold standards for global RBP discovery rely upon the cross-linking of protein–RNA interactions and RNA capture (Table 1). The observation that a covalent bond can form between an amino acid in contact with an RNA nucleotide following ultraviolet (UV) excitation has been the subject of study for more than 40 years [19,20]. While these interactions can be fixed natively at 254 nm (cCL) it is also possible to discriminate contacts with newly synthesised RNA by first culturing cells with a photo-reactive nucleoside such as 4-thiouridine (4SU) which instead reacts at 365 nm (PAR-CL) [21]. Although numerous other cross-linking strategies exist the popularity of UV methods has endured as a cornerstone of RBP discovery by high-throughput protocols [22]. The earliest of these capture methods, termed RNA interactome capture (RIC), repurposed mRNA capture protocols typically used for RNA-seq library preparation, to isolate bound RBPs that were first covalently cross-linked to RNA by either 254 nm or 365 nm irradiation (Figure 1i) and next analysed by MS to identify RBP en masse. Using quantitative proteomics these studies identified up to 900 proteins as having RNA-binding function based on their relative abundance in non-cross-linked vs cross-linked samples following detergent-based washing [6,23]. Although this depth was dependent on high cellular inputs (>300e6 cells), RIC was immediately recognised for its marriage of proven RNA methods with quantitative MS proteomics. With 900 proteins identified, these surveys demonstrated more than half of all binding sites not only lack similarity with classical RBDs but also map to intrinsically disordered regions not previously considered [5].
Table 1. Technical features of high throughput global and targeted methods for protein–RNA-binding studies using mass spectrometry or RNA sequencing.
| Method | Pub. year | Cross-link | Target /capture | MS label | Capture | Cells used in original study | Ref. |
|---|---|---|---|---|---|---|---|
| RNA interactome capture (RIC) | 2012 | 365 nm UV-4SU 254 nm UV |
RNA; poly-A tail OdT hybridisation |
SILAC Label-free |
OdT hybridisation, magnetic | HEK293T, HeLa | [6,69] |
| Orthogonal organic phase separation (OOPS) | 2019 | 254 nm UV | RNA; global Phase separation |
SILAC TMT |
Phase separation | HEK293, U2Os, MCF10A | [28] |
| Protein-cross-linked RNA extraction (XRNAX) | 2018 | 254 nm UV | RNA; global Phase separation Silica column |
SILAC | Phase separation + silica column | MCF7 | [27] |
| Phenol-toluol extraction (PTEX) | 2019 | 254 nm UV | RNA, global Phase separation |
LFQ | Phase separation | HEK293 | [29] |
| Click chemistry-assisted RNA interactome capture (CARIC) | 2018 | 365 nm UV-4SU + EU label + biotin |
RNA, labelled Click chemistry |
Dimethyl labelling | Click chemistry of metabolically labelled RNA, magnetic | HeLa | [26] |
| RNA interactome capture using click chemistry (RICK) | 2017 | 254 nm UV + EU label + biotin |
RNA, labelled Click chemistry |
Not described | Click chemistry of metabolically labelled RNA, magnetic | HeLa, HEK293T | [70] |
| Capture hybridisation analysis of RNA targets (CHART) | 2011 | Formaldehyde | RNA; sequence Hybridisation |
Western | 25mer biotinylated probe, magnetic | HeLa | [71] |
| Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP) | 2015 | Formaldehyde | RNA; sequence Tiling hybridisation |
Not described | 20mer biotinylated tiling probe, magnetic | E36 | [42] |
| RNA affinity purification MS (RAP-MS) | 2015 | 254 nm UV | RNA; sequence Tiling hybridisation |
SILAC | 90mer biotinylated tiling probe, magnetic | SM33 | [41] |
| CRISPR-based RNA-united interacting system (CRUIS) | 2020 | Formaldehyde | Protein; label Immunoprecipitation | LFQ | sgRNA-Cas13a proximity labelling, magnetic | HEK293T | [43] |
| RNA immunoprecipitation (RIP, RIP-Seq, RIP-chip) | 2006, 2010 | None, native | Protein; epitope Immunoprecipitation | RNA-Seq only | Antibody | mESC | [72,73] |
| Cross-linking and immunoprecipitation (CLIP, CLIP-Seq, HITS-CLIP, iCLIP, eCLIP) | 2003, 2009 | 254 nm UV 365 nm UV-4SU |
Protein; epitope Immunoprecipitation | RNA-Seq only | Antibody | HepG2, K562, HeLa, mouse brain | [50–52] |
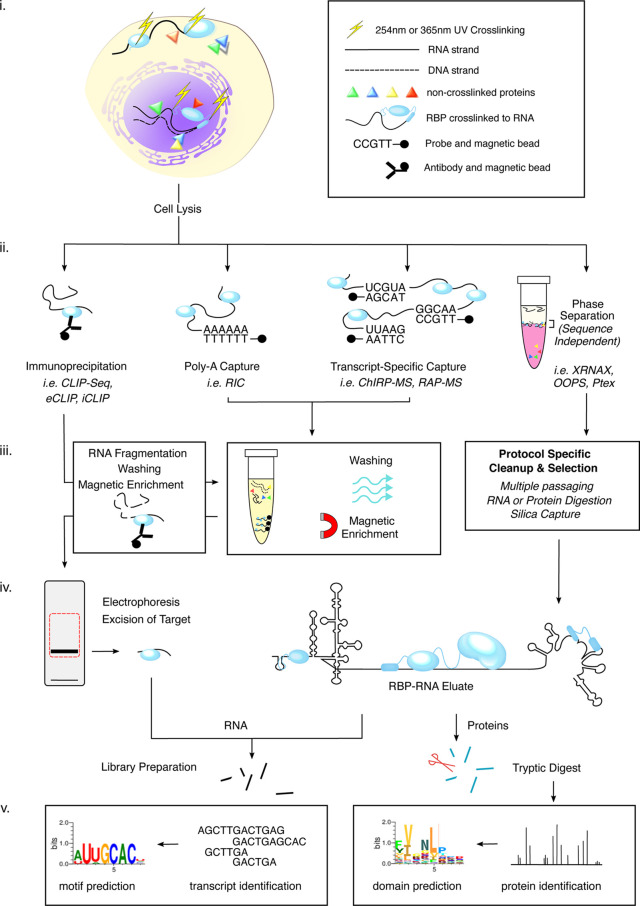
Figure 1. Common features of RBP–RNA capture methods.
(i) Intact cells are subjected to UV cross-linking of RBP to RNA either natively at 254 nm or, with assistance from 4SU, at 365 nm. (ii) The subsequent capture protocol will differ according to the study being conducted. From left to right. RBP-centric capture methods utilise immunoprecipitation to extract a population of target proteins in order to analyse their bound transcripts. RNA-centric methods such as poly-A capture or transcript specific capture use complementary probes to target an RNA compartment or RNA species, respectively. The former is frequently followed by global analysis of either RBP or RNA, where the latter is commonly used to investigate specific RBP interactors only. Phase separation methods capture interactions across the total transcriptome by first dissociating DNA, RNA and protein molecules. The cocktail's interphase is relied upon to enrich for protein–RNA complexes based on their compound physico-chemical character. (iii) Each capture method is subject to a clean-up step to improve sample purity. From left to right. Immunoprecipitants are subjected to RNA fragmentation and washing via magnetic separation. RNA-centric captures are also washed by magnetic separation although, because hybridisation anchors the transcript, these methods can tolerate higher stringencies than their immunoprecipitant counterparts. For phase separation methods clean-up steps can vary, even within a single protocol, depending on downstream application. Common aids are multiple passaging through the biphasic cocktail, RNA or protein digestion, and silica capture. In the case of Ptex, cocktail components can also vary. (iv) Immunoprecipitant methods often feature another round of separation by gel electrophoresis and products of a molecular mass close to the target recovered. The RBP–RNA complexes derived from RNA-centric and phase separation methods can be prepared for either RNA Seq or MS proteomics. (v) Sequencing of RNA yields can provide transcript identity or be used to identify RNA motifs based on the read densities flanking the cross-link sites. Similarly, MS proteomics can be used to identify and quantify the RBPs or a domain identification strategy used to find cross-link sites on the protein.
The most obvious criticism of RIC has been its dependence on the use of oligo-dT hybridisation probes intended to target only those transcripts with poly-adenylated tails. A prerequisite of nuclear export, poly-adenylation is common to nearly all cytoplasmic mRNAs and some non-coding RNAs [24]. The tail length has an important role in translation and its gradual shortening over time marks a transcript for enzymatic degradation [25]. These hallmarks of eukaryotic RNAs, and near absence from prokaryotic cells, can respectively bias RIC surveys or render them uninformative. More recently, sequence-agnostic capture approaches have emerged, using either metabolic labelling of cells with nucleosides featuring click chemistry handles or by leveraging the capacity for phenol-chloroform cocktails to separate nucleic acids and proteins. The click chemistry dependent method, termed CARIC, was able to find 597 RBP proteins from HeLa cells while three recently reported phenol-chloroform protocols, XRNAX, OOPS, and PTEX reported 1357, 1838, and 3037 RBPs respectively from extractions of HEK293T cells [26–29]. Where CARIC differs from its phenol-chloroform based counterparts is that the method's RBP yields are restricted to cross-linking events with newly synthesised RNA that have incorporated the alkynyl uridine analogue necessary for capture. Phenol-chloroform methods are distinct in that RBP-isolation relies on the long held observation that protein–RNA complexes tend to collect at the cocktail interphase after centrifugation [30]. These cocktails are a potent mixture of chaotropic salts, anionic detergents, organic, polar, and non-polar components. When combined, their dissociative strength and density gradients can effectively drive protein flocculation and the separation of proteins, RNA, and DNA into different compartments [31]. Concentration of protein–RNA aggregates is thus enriched at the interphase due to the unique biochemical properties of the composite [32]. It is these basic principles that form the basis for the XRNAX, OOPS, and Ptex methods although each protocol does differ in their use of additional processing steps such as partial or complete RNA/protein digestion, silica capture, multiple passaging, or pH manipulation in order to achieve a cleaner product [33]. Overall, these whole-transcriptome approaches to RBP capture represent an important step towards finding a census of protein–RNA interactions (Figure 1ii).
At present, the development of RBP capture methods has mostly been pioneered on immortalised cell lines to provide enough biological material and offset the relative inefficiency of UV cross-linking. Nevertheless, it should be noted that, where UV penetrance is achievable, these methods now also enjoy broader application among organisms as diverse as plants, bacteria, viruses, and single-cell eukaryotes [13,28,34–37]. A common problem for all systems, however, is that bulk RBP capture tends to be greatly enriched for RNPs engaged in transcription, translation, and degradation. For researchers focused on the discovery of RBP interactions outside these processes the sheer abundance and diversity of RNPs can threaten to overwhelm the limited dynamic range of reliable MS quantitation. Nevertheless, studies of translational arrest have gained some insight into unexpected patterns of interaction. For instance, in a model of arsenite-induced stress in the MCF7 cell line, XRNAX was able to identify exosome component 2 (EXOSC2) as rotating its binding activity from cytoplasmic to nuclear RNA transcripts [27]. Moving forward a major challenge for MS-dependent RBP discovery will be the development of methods that can dissect the interactome with increasing granularity especially for materially limited or ex vivo cells, fractionated intracellular compartments and complex experimental models of cellular perturbation [38–40].
More precise approaches to discriminate the range of RBP activities include extraction methods that depend upon transcript-targeted capture. In these variants, the oligo-dT sequence used by RIC probes are replaced with a library of complementary sequences which tile the target RNA transcript. Coincident studies termed comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) and RNA antisense purification coupled with mass spectrometry (RAP-MS) use this strategy to show hundreds of proteins involved in X-inactive specific transcript (XIST) mediated silencing of gene expression in secondary copies of the X chromosome [41,42]. Key findings associated with these studies included mapping the co-ordinate assembly of XIST RNP from pluripotency to differentiation, and the identification of SHARP, SAF-A, and LBR as RBPs required for transcriptional repression.
RAP-MS and ChIRP-MS hybridisation methods depend on the chance encounter between probe and its complementary target in a complex lysate. An alternative strategy, CRISPR-based RNA-united interacting system (CRUIS), relies on a guide RNA to anchor dCas13a to its RNA target whereupon the proximity labelling enzyme PafA mediates PupE modification of the nearby protein [43]. Subsequently, the PupE substrate is used to enrich the RBP for MS analysis and its utilisation for tracking stress granules or identifying interactors with non-coding RNA activated by DNA damage (NORAD) and p21 mRNA hold promise for the targeting of specific RNAs in a given locale [43]. Indeed, given that each RNA spends the entirety of its lifecycle in association with RBPs, CRUIS, in combination with genetic control of PafA, can offer the opportunity to target interactions at a specific stage or location. Another use for probes includes their capacity to be modified with epitranscriptomic signatures. These synthetic sequences can be used as baits to interrogate total lysates in a search for RBPs whose interaction depend on an RNA modification. When combined with UV cross-linking such chemoproteomic probes have been used to define show that the N1-methyladenosine dependent interaction between YTH N6-methyladenosine RNA-binding protein 2 (YTHDF2) and its target can control transcript destabilisation [44]. Cross-linking events between wholly endogenous interactors risks loss of such information once the nucleotide contact becomes buried or irretrievable from its protein binding partner.
The precision of UV cross-linking and RBP affinity capture
The assignment of RNA-binding function depends on the quantitative comparison between non-cross-linked and cross-linked samples. Thus, confidence in identification relies upon two primary factors; the specificity of cross-linking and the stringency of washing (Figure 1iii). Fortunately, UV cross-linking occurs only at extremely short distances and, coupled with application to intact cells, has been a transformative advance for fixing bona-fide RBP–RNA contacts in their native environment [45,46]. UV cross-linking can be conducted at 254 nm although because efficiency is poor (<1–5%), and has a slight uracil bias, the cross-linked RBP–RNA complexes recovered cannot wholly represent the full complement of complexes engaged in biological processes [22]. Where material limitations are problematic, however, higher yields of RBP–RNA complexes can be achieved by culturing cells with photo-reactive 4SU [47]. The field has, however, begun to move away from 4SU due to toxicity concerns and its cross-linking restriction to synthetic uracil [48]. Despite its low efficiency, the selectivity of a cross-link that is generated only between protein and RNA provides an extraordinary opportunity for RBPs to be identified qualitatively, by purification, rather than inferred via relative quantitation. Traditionally, affinity proteomics brings with it the need for a user to expertly assess both the likelihood that an enriched protein is indirectly associated with their target, and thence co-purifying with it, and the statistical impact that common background proteins, shared between experiment and control samples, might have on finding significance. Because the cross-link is covalent, however, there is every chance that high stringency protocols can be developed in the future to eliminate unwanted passengers altogether (Figure 1iv).
The precision of UV cross-linking can be assessed by RNA sequencing. The covalent bond between RBP and RNA is irreversible and the bound amino acids frequently block reverse transcription on the template strand of enriched transcripts [49]. This results in truncated cDNAs presenting with low sequence diversity or a loss of primer binding sites for qPCR. This inherent incompatibility between reverse transcription enzymes and cross-linked RNA adds value to both the native 254 nm and 4SU-dependent 365 nm approaches. If 80% of reads halt at the site of cross-linking then the sequencing of truncated cDNAs can reliably reveal the proximal site of RBP binding; moreover, where 4-thiouridine has facilitated cross-linking, successful reverse transcription leads to a T > C substitution, essentially presenting as a point mutation that flags binding sites [21]. The motifs at these sites are of particular interest to the study of RNA biology and are the primary focus of RBP–RNA extraction methods that focus on cross-linking and immunoprecipitation (CLIP) analysis and its sequencing-enabled successors CLIP-Seq, iCLIP, and eCLIP [50–54].
MS strategies can also sequence RNA-binding domains that are cross-linked to the site of interaction. The covalent bond between RBD and RNA is irreversible and RNA destruction cannot fully divest a RBD of all attached nucleotides. Thus, when a cross-linked peptide enters the mass spectrometer a mass shift which corresponds with the size of its RNA cargo can be observed. Typical database matching algorithms for popular MS software will not recognise these spectra because they are not calibrated to recognise such modifications. A bioinformatic pipeline that allows for such identifications is RNPXL, is a bioinformatic pipeline which has identified 257 cross-linking sites [55]. Alternative strategies have located up to 1000 sites by instead identifying the native mass of sequences flanking the cross-linking site; methods such as RBDmap (or its variants), pCLAP, and RBR-ID limit MS acquisition to the bound region by conducting a limited digest of the larger protein before discarding these regions and next using tryptic digest to identify peptides proximal, but not tethered, to the RNA moiety [5,28,56,57].
Analytical challenges confronting RNA interactome surveys
An experimental protocol that can identify and measure interactions and simultaneously describe the molecular interface of these binding events has extraordinary potential for both systems biology and target discovery (Figure 1v). Such data, however, also presents enormous experimental and multi-disciplinary challenges. Given the discovery of unconventional RBPs and moonlighting enzymes is a nascent field, it will be some time before many are validated. For researchers investigating a defined molecular event, cellular fate, or disease phenotype, finding that smoking gun is deeply dependent on the stringency of capture and sensitivity of MS acquisition. Additional hard limits can be set by high material demands, which preclude the study of many primary cell types, or use of metabolic labelling or chemical tagging which can be expensive and unwieldy where many biological replicates are desired per condition in both non-cross-linked and cross-linked states [58]. Future RBP capture protocols that solve these problems can provide non-specialist laboratories the opportunity to adopt RBP discovery as a routine tool.
The temptation is to forge ahead with systems-level prediction of interaction networks. Recall, however, that constant interaction with a panoply of proteins is a feature of the RNA lifecycle so, in many cases, the lion's share of quality peptide IDs will be consumed by highly conserved proteins and the proteins that co-purify with them. This bias may lead to key processes being obscured by high levels of physiological redundancy. Other technical influences exist, for instance, the uracil bias of UV cross-linking will have a systemic effect on the relative quantitation of RBP engagement notwithstanding dynamic shifts in RNA secondary structure that will favour the formation of covalent bonds being captured by more exposed strand geometries [59]. Finally, the sheer combinatoric variety of RBPs that can decorate a single transcript presents a formidable set theory problem for networks seeking to disentangle independent contributions to the larger regulatory network.
Common elements of sequence and structure shared between RBD and RNA have been used, with varying degrees of success, as training models for bioinformatic predictions of RBPs [60–64]. Although focused on molecular discovery, rather than network interpretation, such algorithms can offer a valuable sanity-check to interpret the performance and development of interactome capture protocols. The cumulative detail offered by molecular analysis at the binding interface can also support broader hypotheses. Indeed, pattern analyses of binding events on a whole transcript level supports the notion that complex RNA secondary structures exercise evolutionary pressure by using exposed nucleotides to select for an accommodative protein topology [63]. That RNA and protein actively exert reciprocating forces is, in turn, reinforced by complex molecular machines such as the ribosome and RNAse P whose unity each yields a unique function greater than the sum their parts [65,66]. Thus, we should expect that future attempts to increase the stringency of RBP–RNA captures must simultaneously focus on denaturing and unravelling the RNA structures that scaffold so many of these interactions.
Conclusion
For decades, the most compelling scientific narratives behind RNA–protein binding have illuminated events associated with gene expression. High-throughput RBP capture methods have revealed that these interactions are not limited to the manufacture of molecular products by specialised machinery but might, in fact, also serve as a common mechanism for governing metabolic processes [67,68]. As protocols mature, an even greater depth is likely to be revealed and, when combined with lower input requirements, snapshots of homeostatic control will allow these approaches to be extended to more diverse cellular models. The capacity for RBP capture protocols to generate sequence-level evidence of local interactions is an exciting development for molecular biologists and provides an enormous opportunity to catalogue common patterns that might aid bioinformatic prediction. Beyond such consensus, however, the apparent conservation of disorder shows that complementarity is likely to be negotiated by topological flexibility; an observation that will broaden scientific narratives that may confine regulation to being a product of molecular specialisation and fated structure. Moving forwards from these early drafts will demand considerable technical rigor and computational inference from both protein chemists and RNA biologists. But as an experimental method that can both discover networks of intracellular interaction and reveal the molecular interface at which these occur, the future is bright.
Perspective
In recent years, screening for RNA-bound protein using MS based proteomics has revealed that protein–RNA interactions are ubiquitous and widespread. These activities extend beyond common mechanisms of gene expression to include participation from metabolic enzymes and complex, co-evolved molecular machines.
These screening methods, in concert with RNA sequencing, offer an unbiased discovery method that can measure dynamic changes in the interactome and pinpoint the bound sequences of both binding partners. These technologies can be applied to the whole transcriptome, specific transcripts of interest, or networks peculiar to cellular compartments.
Future technical advances will need to increase the stringency of RBP identification, lower the necessary material input, reduce technical complexity and broaden experimental utility. Fully mining data-rich experiments from global RBP–RNA capture protocols will require significant multi-disciplinary knowledge and may benefit from specialised statistical approaches that can adjust conventional search algorithms to further trim false positives. As repositories continue to expand with parameter-rich MS and RNA-Seq data, strategies for RBP classification may also benefit by integrating structural prediction, hypothesis-free machine learning models or network modelling.
Abbreviations
- CLIP
cross-linking and immunoprecipitation
- CRUIS
CRISPR-based RNA-united interacting system
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- MS
mass spectrometry
- RBDs
RNA-binding domains
- RBPs
RNA-binding proteins
- RIC
RNA interactome capture
- RNPs
ribonucleoproteins
- RRM
RNA-recognition motifs
- XIST
X-inactive specific transcript
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contributions
J.S. wrote the manuscript. J.S. and J.J.S. conceived and edited the manuscript. A.I.W. assisted with critical input and advice.
Open Access
Open access for this article was enabled by the participation of the Walter and Eliza Hall Institute in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with CAUL.
Funding
This work was provided support by the Victorian State Government Operational Infrastructure Scheme grant and funding from an Australian Government National Health and Medical Research Council Dora Lush Postgraduate Scholarship.
References
- 1.Lewis, H.A., Musunuru, K., Jensen, K.B., Edo, C., Chen, H., Darnell, R.B.et al. (2000) Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell 100, 323–332 10.1016/S0092-8674(00)80668-6 [DOI] [PubMed] [Google Scholar]
- 2.Lunde, B.M., Moore, C. and Varani, G. (2007) RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8, 479–490 10.1038/nrm2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosphila, R.M., Lu, D., Searles, M.A. and Klug, A. (2003) Crystal structure of a zinc-finger – RNA complex reveals two modes of molecular recognition. Nature 426, 96–100 10.1038/nature02088 [DOI] [PubMed] [Google Scholar]
- 4.Sherwood A, V. and Henkin, T.M. (2016) Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu. Rev. Microbiol. 70, 361–374 10.1146/annurev-micro-091014-104306 [DOI] [PubMed] [Google Scholar]
- 5.Castello, A., Fischer, B., Frese, C.K., Horos, R., Alleaume, A.M., Foehr, S.et al. (2016) Comprehensive identification of RNA-binding domains in human cells. Mol. Cell 63, 696–710 10.1016/j.molcel.2016.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baltz, A.G., Munschauer, M., Schwanhäusser, B., Vasile, A., Murakawa, Y., Schueler, M.et al. (2012) The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 1–34 10.1016/j.molcel.2012.05.021 [DOI] [PubMed] [Google Scholar]
- 7.Balcerak, A., Trebinska-Stryjewska, A., Konopinski, R., Wakula, M. and Grzybowska, E.A. (2019) RNA-protein interactions: Disorder, moonlighting and junk contribute to eukaryotic complexity. Open Biol. 9, 190096 10.1098/rsob.190096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee, I. and Hong, W. (2004) RAP- a putative RNA-binding domain. Trends Biochem. Sci. 29, 565–567 10.1016/j.tibs.2004.09.001 [DOI] [PubMed] [Google Scholar]
- 9.Boehm, E., Zaganelli, S., Maundrell, K., Jourdain, A.A., Thore, S. and Martinou, J.C. (2017) FASTKD1 and FASTKD4 have opposite effects on expression of specific mitochondrial RNAs, depending upon their endonuclease-like RAP domain. Nucleic Acids Res. 45, 6135–6146 10.1093/nar/gkx164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeda, Y., Yamaji, R., Irie, K., Kioka, N. and Murakami, A. (2012) Glyceraldehyde-3-phosphate dehydrogenase regulates cyclooxygenase-2 expression by targeting mRNA stability. Arch. Biochem. Biophys. 528, 141–147 10.1016/j.abb.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 11.Zhou, Y., Yi, X., Stoffer, J.B., Bonafe, N., Gilmore-Hebert, M., McAlpine, J.et al. (2008) The multifunctional protein glyceraldehyde-3-phosphate dehydrogenase is both regulated and controls colony-stimulating factor-1 messenger RNA stability in ovarian cancer. Mol. Cancer Res. 6, 1375–1384 10.1158/1541-7786.MCR-07-2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcin, E.D. (2019) GAPDH as a model non-canonical AU-rich RNA binding protein. Semin. Cell Dev. Biol. 86, 162–173 10.1016/j.semcdb.2018.03.013 [DOI] [PubMed] [Google Scholar]
- 13.Beckmann, B.M., Horos, R., Fischer, B., Castello, A., Eichelbaum, K., Alleaume, A.M.et al. (2015) The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 6, 10127 10.1038/ncomms10127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zagrovic, B., Bartonek, L. and Polyansky, A.A. (2018) RNA-protein interactions in an unstructured context. FEBS Lett. 592, 2901–2916 10.1002/1873-3468.13116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molliex, A., Temirov, J., Lee, J., Coughlin, M., Kanagaraj, A.P., Kim, H.J.et al. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, Y., Currie, S.L. and Rosen, M.K. (2017) Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 292, 19110–19120 10.1074/jbc.M117.800466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castello, A., Fischer, B., Hentze, M.W. and Preiss, T. (2013) RNA-binding proteins in Mendelian disease. Trends Genet. 29, 318–327 10.1016/j.tig.2013.01.004 [DOI] [PubMed] [Google Scholar]
- 18.Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J.et al. (2001) Erratum: Initial sequencing and analysis of the human genome: international human genome sequencing consortium (Nature (2001) 409 (860-921)). Nature 412, 565–566 10.1038/35087627 [DOI] [PubMed] [Google Scholar]
- 19.Meisenheimer, K.M. and Koch, T.H. (1997) Photocross-linking of nucleic acids to associated proteins. Crit. Rev. Biochem. Mol. Biol. 32, 101–140 10.3109/10409239709108550 [DOI] [PubMed] [Google Scholar]
- 20.Wagenmakers, A.J., Reinders, R.J. and van Venrooij, W.J. (1980) Cross-linking of mRNA to proteins by irradiation of intact cells with ultraviolet light. Eur. J. Biochem. 112, 323–330 10.1111/j.1432-1033.1980.tb07207.x [DOI] [PubMed] [Google Scholar]
- 21.Hafner, M., Landthaler, M., Burger, L., Khorshid, M., Hausser, J., Berninger, P.et al. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheeler, E.C., Van Nostrand, E.L. and Yeo, G.W. (2018) Advances and challenges in the detection of transcriptome-wide protein–RNA interactions. Wiley Interdiscip. Rev. RNA 9, 1–11 10.1002/wrna.1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castello, A., Fischer, B., Eichelbaum, K., Horos, R., Beckmann, B.M., Strein, C.et al. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- 24.Coller, J.M., Gray, N.K. and Wickens, M.P. (1998) mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 12, 3226–3235 10.1101/gad.12.20.3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu, X. and Brewer, G. (2012) The regulation of mRNA stability in mammalian cells: 2.0. Gene 500, 10–21 10.1016/j.gene.2012.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang,, R., Han,, M., Meng,, L. and Chen,, X. (2018) Transcriptome-wide discovery of coding and noncoding RNA-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 115, E3879–E3887 10.1073/pnas.1718406115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trendel, J., Schwarzl, T., Horos, R., Prakash, A., Bateman, A., Hentze, M.W.et al. (2019) The human RNA-binding proteome and its dynamics during translational arrest. Cell 176, 391–403.e19 10.1016/j.cell.2018.11.004 [DOI] [PubMed] [Google Scholar]
- 28.Queiroz, R.M.L., Smith, T., Villanueva, E., Marti-Solano, M., Monti, M., Pizzinga, M.et al. (2019) Comprehensive identification of RNA–protein interactions in any organism using orthogonal organic phase separation (OOPS). Nat. Biotechnol. 37, 169–178 10.1038/s41587-018-0001-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urdaneta, E.C., Vieira-Vieira, C.H., Hick, T., Wessels, H.H., Figini, D., Moschall, R.et al. (2019) Purification of cross-linked RNA-protein complexes by phenol-toluol extraction. Nat. Commun. 10, 990 10.1038/s41467-019-08942-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Favre, A., Moreno, G., Blondel, M.O., Kliber, J., Vinzens, F. and Salet, C. (1986) 4-Thiouridine photosensitized RNA-protein crosslinking in mammalian cells. Biochem. Biophys. Res. Commun. 141, 847–854 10.1016/S0006-291X(86)80250-9 [DOI] [PubMed] [Google Scholar]
- 31.Chomczynski, P. and Sacchi, N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 10.1016/0003-2697(87)90021-2 [DOI] [PubMed] [Google Scholar]
- 32.Avison, M. (2007) Measuring Gene Expression, 1st edn, Taylor and Francis, New York [Google Scholar]
- 33.Smith, T., Villanueva, E., Queiroz, R.M.L., Dawson, C.S., Elzek, M., Urdaneta, E.C.et al. (2020) Organic phase separation opens up new opportunities to interrogate the RNA-binding proteome. Curr. Opin. Chem. Biol. 54, 70–75 10.1016/j.cbpa.2020.01.009 [DOI] [PubMed] [Google Scholar]
- 34.Sysoev, V.O., Fischer, B., Frese, C.K., Gupta, I., Krijgsveld, J., Hentze, M.W.et al. (2016) Global changes of the RNA-bound proteome during the maternal-to-zygotic transition in Drosophila. Nat. Commun. 7, 990 10.1038/ncomms12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-moreno, M., Noerenberg, M., Ni, S., Fischer, B., Mohammed, S. and Castello, A. (2019) System-wide profiling of RNA-binding proteins uncovers key regulators of virus infection resource system-wide profiling of RNA-binding proteins uncovers key regulators of virus infection. Mol. Cell 74, 196–211.e11 10.1016/j.molcel.2019.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marondedze, C., Thomas, L., Serrano, N.L., Lilley, K.S. and Gehring, C. (2016) The RNA-binding protein repertoire of Arabidopsis thaliana. Sci. Rep. 6, 1–13 10.1038/srep29766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shchepachev, V., Bresson, S., Spanos, C., Petfalski, E., Fischer, L., Rappsilber, J.et al. (2019) Defining the RNA interactome by total RNA- associated protein purification. Mol. Syst. Biol. 15, e8689 10.15252/msb.20188689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Backlund, M., Stein, F., Rettel, M., Schwarzl, T., Perez-Perri, J.I., Brosig, A.et al. (2020) Plasticity of nuclear and cytoplasmic stress responses of RNA-binding proteins. Nucleic Acids Res. 48, 4725–4740 10.1093/nar/gkaa256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conrad, T., Albrecht, A.S., De Melo Costa, V.R., Sauer, S., Meierhofer, D. and Ørom, U.A. (2016) Serial interactome capture of the human cell nucleus. Nat. Commun. 7, 1–11 10.1038/ncomms11212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Licatalosi, D.D., Ye, X. and Jankowsky, E. (2020) Approaches for measuring the dynamics of RNA–protein interactions. Wiley Interdiscip. Rev. RNA 11, 1–23 10.1002/wrna.1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McHugh, C.A., Chen, C.K., Chow, A., Surka, C.F., Tran, C., McDonel, P.et al. (2015) The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu, C., Zhang, Q.C., Da Rocha, S.T., Flynn, R.A., Bharadwaj, M., Calabrese, J.M.et al. (2015) Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, Z., Sun, W., Shi, T., Lu, P., Zhuang, M. and Liu, J.L. (2020) Capturing RNA-protein interaction via CRUIS. Nucleic Acids Res. 48, e52 10.1093/nar/gkaa143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seo, K.W. and Kleiner, R.E. (2020) YTHDF2 recognition of N 1 - methyladenosine (m 1 A)-modified RNA is associated with transcript destabilization. ACS Chem. Biol. 15, 132–139 10.1021/acschembio.9b00655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ascano, M., Hafner, M., Cekan, P., Gerstberger, S. and Tuschl, T. (2012) Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip. Rev. RNA 3, 159–177 10.1002/wrna.1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ule, J., Jensen, K.B., Ruggiu, M., Mele, A., Ule, A. and Darnell, R.B. (2003) CLIP identifies nova-regulated RNA networks in the brain. Science 302, 1212–1215 10.1126/science.1090095 [DOI] [PubMed] [Google Scholar]
- 47.Spitzer, J., Hafner, M., Landthaler, M., Ascano, M., Farazi, T., Wardle, G.et al. (2014) PAR-CLIP (photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation): a step-by-step protocol to the transcriptome-wide identification of binding sites of RNA-binding proteins. Methods Enzymol. 539, 113–161 10.1016/B978-0-12-420120-0.00008-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burger, K., Mühl, B., Kellner, M., Rohrmoser, M., Gruber-Eber, A., Windhager, L.et al. (2013) 4-thiouridine inhibits rRNA synthesis and causes a nucleolar stress response. RNA Biol. 10, 1623–1630 10.4161/rna.26214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urlaub, H., Hartmuth, K. and Lührmann, R. (2002) A two-tracked approach to analyze RNA-protein crosslinking sites in native, nonlabeled small nuclear ribonucleoprotein particles. Methods 26, 170–181 10.1016/S1046-2023(02)00020-8 [DOI] [PubMed] [Google Scholar]
- 50.Van Nostrand, E.L., Pratt, G.A., Shishkin, A.A., Gelboin-Burkhart, C., Fang, M.Y., Sundararaman, B.et al. (2016) Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 10.1038/nmeth.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.König, J., Zarnack, K., Rot, G., Curk, T., Kayikci, M., Zupan, B.et al. (2010) iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 17, 909–915 10.1038/nsmb.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chi, S.W., Zang, J.B., Mele, A. and Darnell, R.B. (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486 10.1038/nature08170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darnell, R.B. (2010) HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip. Rev. RNA 1, 266–286 10.1002/wrna.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin, C. and Miles, W.O. (2019) Survey and summary beyond ClIp: advances and opportunities to measure RBP-RNA and RNA-RNA interactions. Nucleic Acids Res. 47, 5490–5501 10.1093/nar/gkz295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kramer, K., Sachsenberg, T., Beckmann, B.M., Qamar, S., Boon, K.-L., Hentze, M.W.et al. (2014) Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat. Methods. 11, 1064–1070 10.1038/nmeth.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He, C., Sidoli, S., Warneford-Thomson, R., Tatomer, D.C., Wilusz, J.E., Garcia, B.A.et al. (2016) High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol. Cell 64, 416–430 10.1016/j.molcel.2016.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mullari, M., Lyon, D., Jensen, L.J. and Nielsen, M.L. (2017) Specifying RNA-binding regions in proteins by peptide cross-linking and affinity purification. J. Proteome Res. 16, 2762–2772 10.1021/acs.jproteome.7b00042 [DOI] [PubMed] [Google Scholar]
- 58.Larochelle, S. (2019) RNA-binding proteome redux. Nat. Methods 16, 219 10.1038/s41592-019-0349-3 [DOI] [PubMed] [Google Scholar]
- 59.Marchese, D., de Groot, N.S., Lorenzo Gotor, N., Livi, C.M. and Tartaglia, G.G. (2016) Advances in the characterization of RNA-binding proteins. Wiley Interdiscip. Rev. RNA 7, 793–810 10.1002/wrna.1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han, L.Y., Cai, C.Z., Lo, S.L., Chung, M.C.M. and Chen, Y.Z. (2004) Prediction of RNA-binding proteins from primary sequence by a support vector machine approach. RNA 10, 355–368 10.1261/rna.5890304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li, Y.E., Xiao, M., Shi, B., Yang, Y.-C.T., Wang, D., Wang, F.et al. (2017) Identification of high-confidence RNA regulatory elements by combinatorial classification of RNA–protein binding sites. Genome Biol. 18, 169 10.1186/s13059-017-1298-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Nostrand, E.L., Freese, P., Pratt, G.A., Wang, X., Wei, X., Xiao, R.et al. (2020) A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 10.1038/s41586-020-2077-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Groot N, S., Armaos, A., Graña-Montes, R., Alriquet, M., Calloni, G., Vabulas, R.M.et al. (2019) RNA structure drives interaction with proteins. Nat. Commun. 10, 1–13 10.1038/s41467-018-07882-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Corley, M., Burns, M.C. and Yeo, G.W. (2020) How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol. Cell 78, 9–29 10.1016/j.molcel.2020.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klinge, S. and Woolford, J.L. (2019) Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 20, 116–131 10.1038/s41580-018-0078-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jarrous, N. (2017) Roles of RNase P and its subunits. Trends Genet. 33, 594–603 10.1016/j.tig.2017.06.006 [DOI] [PubMed] [Google Scholar]
- 67.Castello, A., Hentze, M.W. and Preiss, T. (2015) Metabolic enzymes enjoying new partnerships as RNA-binding proteins. Trends Endocrinol. Metab. 26, 746–757 10.1016/j.tem.2015.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guiducci, G., Paone, A., Tramonti, A., Giardina, G., Rinaldo, S., Bouzidi, A.et al. (2019) The moonlighting RNA-binding activity of cytosolic serine hydroxymethyltransferase contributes to control compartmentalization of serine metabolism. Nucleic Acids Res. 47, 4240–4254 10.1093/nar/gkz129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Castello, A., Horos, R., Strein, C., Fischer, B., Eichelbaum, K., Steinmetz, L.M.et al. (2013) System-wide identification of RNA-binding proteins by interactome capture. Nat. Protoc. 8, 491–500 10.1038/nprot.2013.020 [DOI] [PubMed] [Google Scholar]
- 70.Bao, X., Guo, X., Yin, M., Tariq, M., Lai, Y., Kanwal, S.et al. (2018) Capturing the interactome of newly transcribed RNA. Nat. Methods 15, 213–220 10.1038/nmeth.4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simon, M.D, Wang, C.I, Kharchenko P, V., West, J.A, Chapman, B.A, Alekseyenko, A.Aet al. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20497–20502 10.1073/pnas.1113536108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao, J., Ohsumi, T.K., Kung, J.T., Ogawa, Y., Grau, D.J., Sarma, K.et al. (2010) Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 40, 939–953 10.1016/j.molcel.2010.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keene, J.D., Komisarow, J.M. and Friedersdorf, M.B. (2006) RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 1, 302–307 10.1038/nprot.2006.47 [DOI] [PubMed] [Google Scholar]