Abstract
Although transport into the nucleus mediated by the importin (IMP) α/β1-heterodimer is central to viral infection, small molecule inhibitors of IMPα/β1-dependent nuclear import have only been described and shown to have antiviral activity in the last decade. Their robust antiviral activity is due to the strong reliance of many different viruses, including RNA viruses such as human immunodeficiency virus-1 (HIV-1), dengue (DENV), and Zika (ZIKV), on the IMPα/β1-virus interface. High-throughput compound screens have identified many agents that specifically target this interface. Of these, agents targeting IMPα/β1 directly include the FDA-approved macrocyclic lactone ivermectin, which has documented broad-spectrum activity against a whole range of viruses, including HIV-1, DENV1–4, ZIKV, West Nile virus (WNV), Venezuelan equine encephalitis virus, chikungunya, and most recently, SARS-CoV-2 (COVID-19). Ivermectin has thus far been tested in Phase III human clinical trials for DENV, while there are currently close to 80 trials in progress worldwide for SARS-CoV-2; preliminary results for randomised clinical trials (RCTs) as well as observational/retrospective studies are consistent with ivermectin affording clinical benefit. Agents that target the viral component of the IMPα/β1-virus interface include N-(4-hydroxyphenyl) retinamide (4-HPR), which specifically targets DENV/ZIKV/WNV non-structural protein 5 (NS5). 4-HPR has been shown to be a potent inhibitor of infection by DENV1–4, including in an antibody-dependent enhanced animal challenge model, as well as ZIKV, with Phase II clinical challenge trials planned. The results from rigorous RCTs will help determine the therapeutic potential of the IMPα/β1-virus interface as a target for antiviral development.
Keywords: antiviral agents, dengue virus, importin, ivermectin, SARS-CoV-2, Zika virus
Introduction
Compartmentalisation is a key feature of the eukaryotic cell, with transport between the different compartments essential for cellular function [1]. The double membrane of the nuclear envelope encircles and thereby defines the nuclear compartment, which sequesters the cell's genetic material, as well as the processes of RNA transcription and subsequent RNA maturation processes such as mRNA splicing. Protein translation occurs in the cytoplasmic compartment, so that specific signal-dependent transport into and out of the nucleus is necessary for the cell to function; i.e. mRNA needs to be exported to the cytoplasm for translation to occur, while all components of the protein machinery that mediate and/or regulate key nuclear processes such as transcription [2–4] and mRNA splicing/processing need to be imported into the nucleus.
The transport of proteins >45 kDa into and out of the nucleus is mediated by members of the importin (IMP) superfamily, of which there are multiple distinct α and β forms [5,6]. The best-characterised pathway by which host proteins enter the nucleus is that mediated by the IMPα/β1 heterodimeric transport complex [7]. Host proteins dependent on IMPα/β1 for nuclear import include those central to the antiviral response, such as members of the signal transducer and activators of transcription (STATs) and nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) transcription factor families.
Many viruses exploit the host cell nuclear trafficking machinery, and in particular, the pathway mediated by the host IMPα/β1 heterodimer, in order to enhance their ability to replicate or to subvert the host immune response [3]. Nuclear transport of particular viral proteins has been shown to be critically important for infection by flaviviruses (e.g. dengue virus — DENV or Zika virus — ZIKV; non-structural protein 5 — NS5), lentiviruses (e.g. human immunodeficiency virus — HIV-1; integrase — IN), alphaviruses (e.g. Venezuelan equine encephalitis virus — VEEV; capsid protein — CP), alphainfluenza viruses (Influenza A; nucleoprotein — NP), lyssaviruses (e.g. P1 and P3 forms of the Rabies virus — RV; phospho — P — protein), orthopneumoviruses (e.g. respiratory syncytial virus — RSV; matrix — M — protein), and others [3,4,8–13]. Over the last decade or so, studies using high-throughput screening (HTS) approaches targeting the host–virus interface (i.e. IMPα/β1 recognition of viral proteins) have identified small molecular inhibitors [14–17], which target either the host IMPα/β1 heterodimer, or the specific viral protein [18]. Many these are exciting prospects as antiviral drugs.
The host cell nuclear import pathway
To gain access to the nucleus, proteins generally require nuclear localisation signals (NLSs), which are recognised by either the IMPα subunit within the IMPα/β1 heterodimer, or by one of the IMPβs directly [5,6,2,5]. The IMP:cargo complex is then translocated through the nuclear pore complex (NPC). Once within the nucleus, binding of Ran-GTP to IMPβ dissociates the complex, freeing the protein cargo to perform its nuclear function [19,20]. This process is illustrated in Figure 1. As hinted at above, there are multiple distinct IMPαs; these can be grouped into three subfamilies, but all are highly homologous, and function in the same way within the IMPα/β1 heterodimer to mediate nuclear import through a shared domain structure of ten α-helical armadillo (ARM) repeats involved in NLS recognition, and a flexible N-terminal importin-β-binding (IBB) domain (see Figure 2) [21]. X-ray crystallographic evidence indicates that NLS-binding is most commonly mediated by IMPα ARM repeats 2–4 (‘major NLS-binding site’) [22], with IMPα ARM repeats 6–8 (‘minor NLS-binding site’) [23] used to a lesser extent, as well as in combination with the major site in the case of certain NLSs. The IBB domain itself contains an NLS-like sequence, so that when the IMPα IBB domain is not bound to IMPβ1, this sequence is bound in the NLS-binding pocket [24], essentially ‘autoinhibiting’ IMPα to prevent binding to NLS-containing cargoes in the absence of IMPβ1 [20,21]. The distinct IMPαs are believed to have certain differences in NLS-binding specificity, as well as tissue distribution [3,21,25,26], potentially having more specialised roles in transporting particular import cargoes and not others [2]. STAT1, for example, has been reported to interact with IMPα5 and not IMPα1 [27], while the Ran nucleotide exchange factor RCC1 appears to be transported into the nucleus only by IMPα3/α4 [28]. Consistent with this, certain viral proteins have been identified as being recognised only by specific IMPαs; these include Hendra and Nipah virus W protein [25,29] and avian virus Influenza A nucleoprotein (NP) which preferentially bind IMPα3, whereas mammalian virus Influenza A NP binds IMPα7 [30].
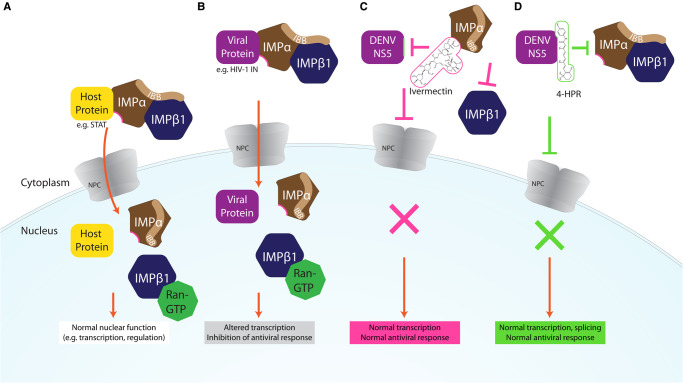
Figure 1. Schematic representation of the host IMPα/β1-dependent nuclear import pathway (A), showing how it is co-opted by viral proteins in viral infection (B), and how small molecule antivirals can impact the pathway (C,D).
(A) Host proteins (e.g. transcription factor STAT-2) contain nuclear localisation signals which are recognised by IMPα (brown), after the autoinhibitory IMPβ-binding (IBB) domain of IMPα binds IMPβ1, forming the IMPα/β heterodimer. This complex is then translocated across the nuclear pore complex (NPC), and the cargo is released after the binding of Ran-GTP to IMPβ1 dissociates the complex. The cargo can then carry out its normal nuclear function, such as transcriptional regulation of the antiviral response. (B) During viral infection, specific NLS-containing viral proteins (e.g. HIV-1 IN, purple) are imported into the nucleus by the same IMPα/β1 dependent mechanism, where they can interfere with normal cellular functions, such as altering transcription to antagonise the antiviral response in order to maximise the rate of virus production. (C) Ivermectin (pink) binds IMPα, dissociating the IMPα/β1 heterodimer and preventing binding to its viral (as well as host) protein target(s), thereby preventing its nuclear import and downstream transcriptional effects. (D) 4-HPR (green) specifically binds viral protein DENV2 NS5, preventing binding of the IMPα/β1 and nuclear import, and associated downstream effects on transcription and splicing.
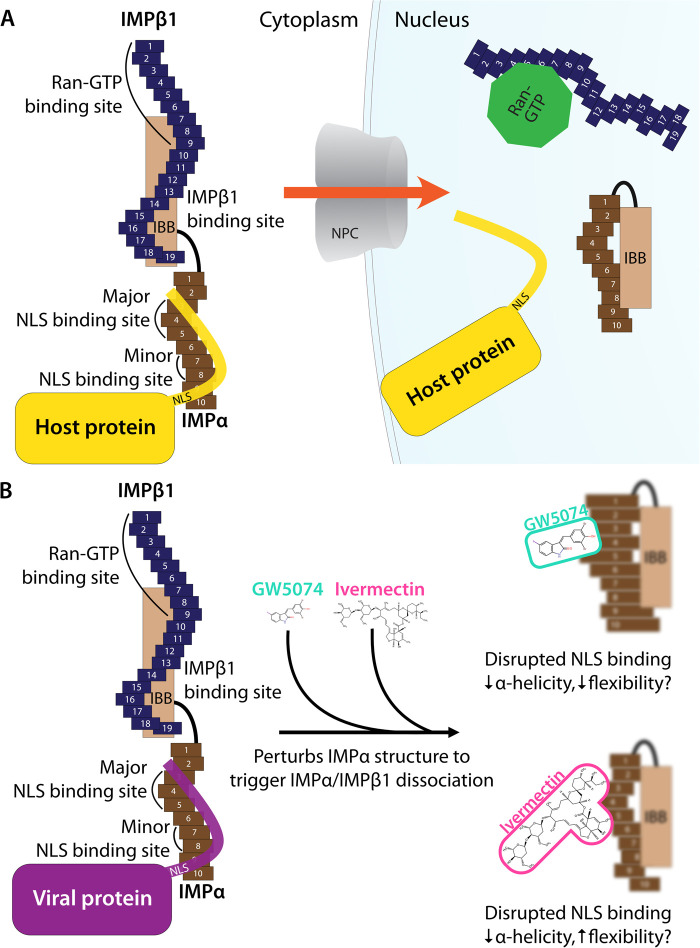
Figure 2. Schematic representation of IMPα and IMPβ1 structural/functional domains; impact of inhibitors.
(A) Schematic representation of the IMPα/β1 complex based on the crystal structures of human IMPα and IMPβ1. IMPα comprises 10 armadillo (ARM) repeats and an IMPβ-binding (IBB) domain. IMPα ARM repeats 2–4 form the major cargo NLS binding site, while ARM 7–8 form the minor NLS binding site. IMPβ1 comprises 19 HEAT repeats, where repeats 7–19 bind the IMPα IBB domain, and Ran-GTP binds to the N-terminus/repeat 8. In the cytoplasm, IMPβ1 (blue) binds IMPα (brown) via the IBB domain, exposing the major NLS binding site for cargo protein (yellow) binding. After transport into the nucleus across the NPC, Ran-GTP binds to IMPβ1, altering its conformation and displacing and releasing the IBB domain, which then binds the NLS binding sites in an autoinhibitory manner, preventing NLS binding. (B) Ivermectin and GW5074 bind directly to IMPα to perturb its structural integrity as confirmed by biophysical measurements including circular dichroism, thermostability analysis, and analytical ultracentrifugation [44,47], to reduce IMPα-helicity/impact flexibility, and thereby prevent IMPα binding to NLS-containing proteins or IMPβ1.
Viral proteins subvert the host nuclear import machinery
The host cell nuclear transport can be exploited by viral proteins that target nuclear processes to antagonise the host antiviral response. These include DENV NS5, which accesses the nucleus in order to impact many host nuclear processes (Figure 1); this leads to a range of outcomes such as delay of production of interleukin 8 (IL-8) that favour a host cell environment conducive to viral replication [8,31], effects on host cell spliceosome complexes to reduce the efficiency of pre-mRNA splicing of genes important for the antiviral response [32], and inhibition of recruitment of Paf1/RNA polymerase II complex component PAF1C to suppress interferon (IFN)-stimulated genes [33]. Other nuclear localising viral proteins that antagonise the host antiviral response, include RV P3-protein, in part though binding to STAT-1 and promyelocytic leukaemia tumour suppressor protein to impact IFN signalling [34], and RSV M, which modulates infected host cell transcriptional outcomes [3,35].
In the case of other proteins such as HIV-1 IN and influenza A NP, nuclear access is critical to the viral life cycle [4]. Dependent on IMPα/β1 [36], for example, nuclear import of HIV-1 IN/the HIV-1 preintegration complex (PIC) is essential to enable the DNA form of the HIV-1 genome to be integrated into the host DNA to enable productive infection [4,9,36,37]. VEEV CP, in contrast, appears to bind to the host IMPα/β1 heterodimer, as well as the IMPβ-homologue nuclear export protein EXP1, to form a tetrameric complex that accumulates at the nuclear pore and blocks nuclear import of host proteins, thereby reducing the host IFN-α/β response [38,39]. In the case of SARS-CoV-1, the causative agent of severe acute respiratory syndrome (SARS), open reading frame (ORF) 6 has been shown to target host IMPα, and sequester it at the rough ER/Golgi, to prevent its key role in mediating STAT-1 nuclear import, thus mollifying the host cell antiviral response [40].
The critical importance of the IMPα/β1-virus axis to viral infection has been formally demonstrated for many viruses, including DENV, where mutagenesis of the NS5 NLS nuclear targeting through impaired IMPα/β1 recognition results in an attenuated virus and markedly reduced infectious virus production [8]. In addition, several structurally distinct small molecule inhibitors that inhibit NS5 nuclear import (see below), also reduce infectious virus production, underlining the critical importance of nuclear targeting of NS5 in DENV infection. Similar results have been obtained for HIV, where inhibiting the nuclear import of HIV-1 IN protein dramatically reduces HIV-1 infection [41,42], and for VEEV, where mutations within the CP NLS region result in attenuated nuclear accumulation, and are correlated with a non-pathogenic phenotype [39]. Finally, that nuclear trafficking of RV P-protein is critical to infection is implied by the observation that an attenuated non-lethal chicken embryo (CE) cell-adapted strain (Ni-CE) of the highly pathogenic RV Nishigahara (Ni) strain, harbours mutations that impact nucleocytoplasmic distribution [12].
Inhibitors targeting IMPα/β1: ivermectin
HTS using chemical compound libraries and recombinant proteins has been used to identify many small molecule inhibitors targeting host IMPα/β (see Table 2). Ivermectin (22,23-dihydroavermectin B), the best-studied of these, is a macrocyclic lactone produced by the bacterium Streptomyces avermitilis, which was discovered in 1975, and approved for use in humans as an antiparasitic agent 10 years later. It is US FDA-approved for treating a range of ectoparasitic and endoparasitic infestations, with an excellent safety profile and millions of people treated globally for onchocerciasis (river blindness) and lymphatic filariasis [43]. Ivermectin is one of many agents listed on the WHO List of Essential Medicines, with the work identifying it/establishing its properties recognised by the Nobel Prize for Physiology or Medicine in 2015.
Table 2. In vitro properties of viral protein cargo-targeted inhibitors with antiviral effects.
| Compound | Documented action (IC50) | Antiviral against | Effective concentration (assay, host)/fold reduction in virus production (assay, host cell line) |
|---|---|---|---|
Mifepristone1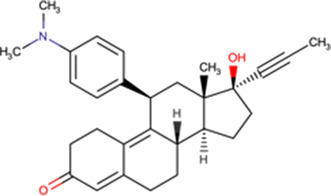
|
Inhibits interaction in vitro of HIV IN with Impα/β (27 μM) [14] Binds to HIV IN core domain but not Impα/β (HSQC NMR) [37] 50 μM inhibits GFP-HIV-1 IN nuclear accumulation in transfected cells [14] 5 μM reduces HIV-1 IN nuclear accumulation > 5-fold in in vitro reconstituted nuclear transport assay [37] 5 μM reduces VEEV CP nuclear accumulation in infected cells [51] Reduces rate/extent of VEEV CP-GFP nuclear import (FRAP) [52] 50 μM reduces HAdV genome nuclear import (cellular fractionation/qPCR) [53] |
HIV-1 | 10 μM > 8-fold (EGFP reporter virus, CEMx174) [50] 10 μM >20-fold (ELISA, PBMC) [50] 200 μM > 3-fold (luciferase, HeLa) [42] |
| VEEV | EC50 = 19.9 μM (PFU, Vero) [52] 10 μM > 10-fold (PFU, U87MG) [51] 10 μM > 5-fold (PFU, Vero) [51] 10 μM > 15-fold (PFU, Vero) [52] |
||
| Human adenovirus HAdV5 |
EC50 = 2 μM (PFU) [53] | ||
Mifepristone analogue 502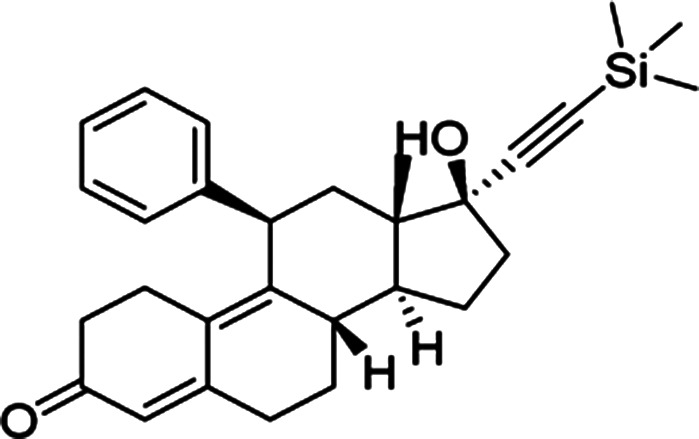
|
50 μM inhibits nuclear import of transfected GFP-CP (FRAP) Reduces recovery of VEEV CP-GFP nuclear fluorescence after FRAP [52] 50 μM inhibits nuclear accumulation of CP in infected cells [52] |
VEEV | EC50 = 7.2 μM (luciferase, Vero) [52] 10 μM > 7-fold (PFU, Vero) [52] |
Budesonide3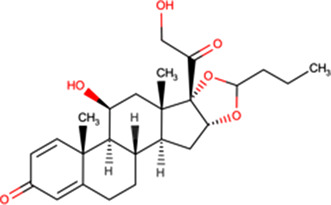
|
Inhibits interaction in vitro of HIV IN with Impα/β (1.2 μM) [37] Binds to HIV IN core domain but not Impα/β (HSQC NMR) [37] 5 μM reduces HIV-1 IN nuclear accumulation > 2-fold in in vitro reconstituted nuclear transport assay [37] 100 μM inhibits HIV PIC nuclear import [37] |
HIV-1 | EC50 = 79.4 μM (luciferase, MT-2, single cycle) [37] EC50 = 49.4 μM (luciferase, TZMbl) [37] |
Flunisolide4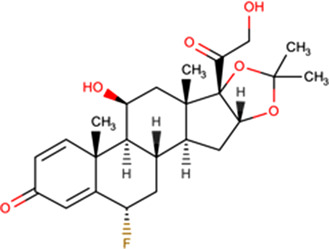
|
Inhibits interaction in vitro of HIV IN with Impα/β (1.4 μM) [37] Binds to HIV IN core domain but not Impα/β (HSQC NMR) [37] 250 μM inhibits HIV PIC nuclear import [37] |
HIV-1 | EC50 = 77.5 μM (luciferase, MT-2, single cycle) [37] EC50 = 105 μM (luciferase, TZMbl) [37] |
G281-1564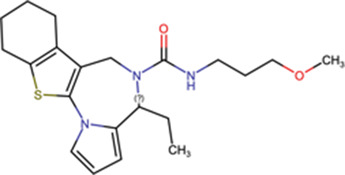
|
Inhibits interaction in vitro of VEEV CP and Impα/β (12 μM) [16] 50 μM inhibits extent and rate of GFP-CP nuclear accumulation in transfected cells (1.8-fold, 1.4-fold, respectively) [16] |
VEEV | EC50 = 10.8 μM (luciferase, Vero)5 [16] 50 μM > 10-fold (PFU, Vero) [16] |
N-(4-hydroxyphenyl) retinamide/fenretinide/4-HPR6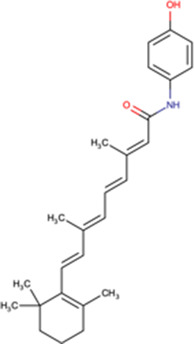
|
Inhibits interaction in vitro of DENV1 NS5:Impα/β (2.3 μM) [82]; DENV2 NS5:Impα/β (0.8 μM) [15]; DENV2 NS5:ImpαΔIBB (1.1 μM) [15]; ZIKV NS5:Impα/β (1.1 μM) [87] Inhibition of DENV2 NS5 nuclear accumulation in infected cells [15,47,77] Destabilises DENV 3 NS5 RdRp (thermostability assay) [44,47] 15 μM inhibits nuclear accumulation of influenza vRNPs (IF) [11] |
Flaviviruses DENV1 (EDEN1) |
EC50 = 2.6 μM (PFU, Huh-7) [15] EC50 (ADE7) = 0.8 μM (PFU, THP-1) [15] EC50 (ADE7) = 0.8 μM (PFU, PBMC) [15] |
| DENV2 (EDEN2) | EC50 = 2.1 μM (PFU, Huh-7) [15] | ||
| DENV2 (NGC) | EC90 = 2.0 μM (PFU, Vero) [77] | ||
| DENV3 (EDEN3) | EC50 = 1.4 μM (PFU, Huh-7) [15] | ||
| DENV4 (EDEN4) | EC50 = 2.1 μM (PFU, Huh-7) [15] | ||
| ZIKV (Asian/Cook Islands/2014) | EC50 = 2.6 μM (PFU, Vero) [87] | ||
| ZIKV (Asian/PF13-251013-1) | EC90 = 1 μM (PFU, multiple hosts) [78] | ||
| ZIKV (African/ MR-766) | 10 μM > 1000-fold (PFU, Vero) [78] | ||
| WNV (Kunjin) | 10 μM > 7-fold (PFU, Vero) [15] 10 μM > 100-fold (PFU, Vero) [77] 10 μM > 10-fold (PFU, Vero) [49] |
||
| YFV | 3 μM > 100-fold (BHK-21, PFU) [46] | ||
| Alphavirus CHIKV |
EC50 = 0.5/3.1 μM (PFU, multiple hosts) [46] | ||
| Modoc | 10 μM >100-fold (PFU, Vero) [77] | ||
| Influenza VLPs8 (avian influenza A/MxA escape mutants) | 10 μM > 4-fold inhibition (luciferase) [11] |
Approved for human use in medical termination of pregnancy, hyperglycaemia treatment in Cushing's syndrome patients;
Analogue of mifepristone ((8S,13S,14S,17S)-17-hydroxy-13-methyl-11-phenyl-17-((trimethylsilyl) ethynyl)-1,2,6,7,8,11,12,13, 14,15,16,17-dodecahydro-3H-cyclopenta[a] phenanthren-3-one)) lacking progesterone receptor antagonism [52];
Approved for asthma, chronic obstructive pulmonary disease, allergic rhinitis and nasal polyps, inflammatory bowel diseases;
Approved for the treatment of asthma, allergic rhinitis;
Selectivity index > 9.3;
Progressed to clinical trials (incl. Phase III) for indications including breast, bladder and paediatric cancers, macular degeneration; other actions include increased phosphorylation of eIF2α, promoting an antiviral state not related to long-chain ceramide biosynthesis, PERK pathway or ATF-4 [15,55,77];
ADE model uses subneutralising concentrations of anti-E protein antibody prior to infection;
Mechanism of action against influenza to be determined.
Abbreviations (see Table 1): ADE, antibody-dependent enhancement of infection; DENV, Dengue virus; CFI, cell-based flavivirus infection assay (immunostaining for virus); CoIP, co-immunoprecipitation; CP, capsid protein; CPE, cytopathogenic effect (host cell); CHIKV, Chikungunya virus; EC50, half-maximal effective concentration; Est., estimated; FRAP, fluorescence recovery after photobleaching; FRET, fluorescence resonance energy transfer; GFP, green fluorescent protein; HAdV, human adenovirus; hCMV, human cytomegalovirus; HCV, hepatitis C virus; HIV, human immunodeficiency virus; HSQ NMR, heteronuclear single quantum coherence nuclear magnetic resonance; IBB, importin β-binding domain; IC50, half-maximal inhibitory concentration; IF, immunofluorescence; IN, integrase; lum, luminescence assay; NS5, non-structural protein 5; PBMC, peripheral blood mononuclear cell; PCR, polymerase chain reaction; PIC, preintegration complex; PFU, plaque forming units (infectious virus); PRV, pseudorabies virus; RdRp, RNA-dependent RNA polymerase; SV40, Simian virus 40; T-ag, SV40 large T-antigen; TCID, tissue culture infectious dose (estimation of viral load based on CPE); VEEV, Venezuelan equine encephalitis virus; VLP, virus-like particle; vRNP, viral ribonucleoprotein complex; VSV-G, vesicular stomatitis virus glycoprotein; WNV, West Nile virus; YFV, Yellow Fever virus; ZIKV, Zika virus.
The first indication that ivermectin might have a role impacting viral protein nuclear import, and thereby potential as an antiviral, was afforded in 2011 in a chemical compound library HTS to identify inhibitors of binding of IMPα/β1 to HIV-1 IN [14]. A nested counterscreening strategy confirmed ivermectin to be an agent likely directly interacting with IMPα/β1 rather than IN to inhibit IMPα/β1:IN binding (IC50 of ∼ 5 μM) [14]. Consistent with this, ivermectin was found to inhibit nuclear accumulation of IN and the well-characterised IMPα/β1-recognised nuclear import cargo SV40 large tumour antigen (T-Ag), but not of cargoes that are transported through IMP-dependent pathways that do not rely on IMPα, such as the IMPβ1-recognised telomere repeat factor-1 [14]. Ivermectin has since been shown to inhibit the binding of IMPα/β to a range of viral proteins, including DENV NS5 (see Figure 1), as well as host cell proteins (see Table 1).
Table 1. In vitro properties of IMPα inhibitors with antiviral effects.
| Compound | Documented action in nuclear import (IC50) | Antiviral against | Effective concentration (assay)/fold reduction (assay) |
|---|---|---|---|
Ivermectin1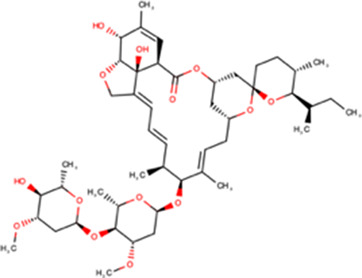
|
Inhibits interaction in vitro of IMPα with HIV IN [14], DENV2 NS5 (1 μM) [42,44], T-ag [15], Hendra V (15 μM) [48], IMPβ1 (7 μM) [44] Inhibits interaction of IMPα with T-ag and NS5 in a cell context as visualised by quantitative BiFc [44] Inhibits CoIP from cell lysates of IMPα with T-ag, Adenovirus EIA [81] Inhibits nuclear accumulation in transfected cells of IMPα/β1- but not β1-recognised viral proteins such as T-ag [16,42], DENV2 NS5 [42], VEEV CP [16], PRV UL42 [60], HIV-1 IN [14,42], hCMV UL44 [42], Influenza A vRNPs [11] as well as host cargoes (e.g. [8,42,46]) Reduces nuclear localisation in infected cells of DENV1-4 NS5 [82], VEEV CP [51] and adenovirus E1A [81] |
Betacoronavirus SARS-CoV-2 |
EC50 = 2.2/2.8 μM (qPCR/ released/cell-associated virus) [59] 5 μM > 5000-fold [59] |
| HIV-1 (VSV-G-pseudotyped NL4-3.Luc.R-E-HIV) | 50 μM > 2-fold (luciferase) [42] | ||
| Influenza VLPs (avian influenza A/MxA escape mutants) | 10 μM total inhibition (luciferase) [11] | ||
| Flaviviruses YFV (17D) |
EC50 = 5/0.5 nM (CPE/qPCR) [83] 3 μM > 50000-fold (PFU) [46] |
||
| DENV1 (EDEN1) | EC50 = 2.3/3.0 μM (CFI, 2 hosts) [82] | ||
| DENV2 (NGC) | EC50 = 0.7 μM (qPCR) [83] EC50 = 3.3/0.4 μM (PFU, 2 hosts2) [44] |
||
| DENV2 (EDEN2) | EC50 = 0.4/0.6 μM (pfu/qPCR) [44] EC50 = 2.1/1.7 μM (CFI, 2 hosts) [82] 50 μM total inhibition (PFU) [42] |
||
| DENV3 (EDEN3) | EC50 = 1.7 μM (CFI) [82] | ||
| DENV4 (EDEN4) | EC50 = 1.9 μM (CFI) [82] | ||
| WNV (NY99) | EC50 = 4 μM (qPCR) [83] | ||
| WNV (MRM61C) | EC50 = 1/0.5 μM (PFU/qPCR) [44] | ||
| ZIKV (Asian/Cook Islands/ 2014) | EC50 = 1.6/1.3 μM (PFU, 2 hosts2) [44] | ||
| Alphaviruses Chikungunya virus (CHIKV-Rluc) Sindbis (HR) Semliki forest virus VEEV (TC83) |
EC50 = 1.9/0.6 μM (luciferase, 2 hosts) [46] 3 μM > 5000-fold (PFU) [46] 3 μM > 200-fold (PFU) [46] 1 μM c. 20-fold (PFU) [51] |
||
| Hendra (Australia/ Horse/1994) |
est. EC50 = 2 μM (TCID/luciferase) [48] | ||
| Adenovirus HAdV-C5 HAdV-B3 |
EC50 = c. 2.5 μM; 10 μM 20-fold (qPCR) [81] 10 μM c. 8-fold (qPCR) [81] |
||
| BK polyomavirus (BKPyV) | Est. EC50 1.5 μM (PFU/CPE/qPCR) [84] | ||
| Pseudorabies | Est. EC50 c. 0.8 μM 1000-fold [60] | ||
Gossypol3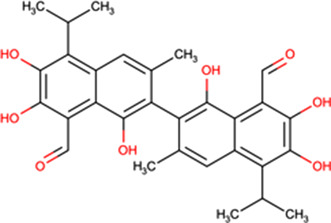
|
Inhibits interaction in vitro of IMPα with Hendra Virus V (10 μM) [48] Inhibits nuclear accumulation in WNV infected cells of NS5 [49] |
WNV (MRM61C) | 10 μM 100-fold (PFU) [49] |
| Hendra (Australia/ Horse/1994) |
10 μM 6-fold (TCID/luciferase) [48] | ||
GW5074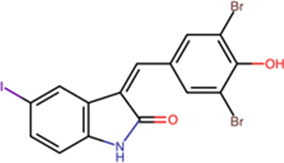
|
Inhibits interaction in vitro of IMPα with DENV2 NS5 (5 μM) [47], Hendra (V 15 uM) [48], IMPβ1 (10 μM) [47] Inhibits nuclear accumulation in DENV2 infected cells of NS5 [47] |
Flaviviruses DENV2 NGC |
EC50 = 0.8/1.4 (PFU/PCR) [47] |
| ZIKV (Asian/Cook Islands/ 2014) | EC50 = 0.3/0.5 (PFU/PCR) [47] | ||
| WNV (MRM61C) | EC50 = 5.2/4.8 (PFU/PCR) [47] | ||
Otava 1111684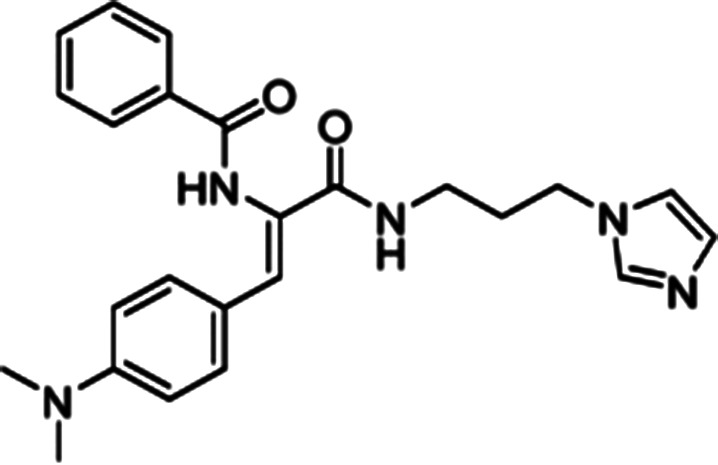
|
Inhibits interaction in vitro of IMPα with VEEV CP [17] Inhibits nuclear accumulation of VEEV CP-GFP in transfected cells [17] Reduces nuclear localisation in infected cells of VEEV CP [17] Reduces rate/extent of VEEV CP-GFP nuclear import (FRAP) [17] |
Alphavirus VEEV (TC83) |
EC50 = 9.9 μM (lum)4 [17] 10 μM > 100-fold [17] |
US Food and Drug Administration (FDA)-approved broad-spectrum antiparasitic agent, including against endoparasitic infestations (strongyloidiasis, onchocerciasis) and ectoparasites causing scabies, pediculosis and rosacea [66]; reported to inhibit helicase activity of DENV2/YFV/WNV NS3 in a FRET-based assay [83];
Includes ex vivo model of human infection in PBMCs;
Abandoned as human male contraceptive due to side effects (hypokalaemic paralysis, testicular damage) [85,86];
Selectivity index of 3.7.
Abbreviations: ADE, antibody-dependent enhancement of infection; BiFc, bimolecular fluorescence complementation; DENV, Dengue virus; CFI, cell-based flavivirus infection assay (immunostaining for virus); CoIP, co-immunoprecipitation; CP, capsid protein; CPE, cytopathogenic effect (host cell); CHIKV, Chikungunya virus; EC50, half-maximal effective concentration; Est., estimated; FRAP, fluorescence recovery after photobleaching; FRET, fluorescence resonance energy transfer; GFP, green fluorescent protein; HAdV, human adenovirus; hCMV, human cytomegalovirus; HCV, hepatitis C virus; HIV, human immunodeficiency virus; IBB, importin β-binding domain; IC50, half-maximal inhibitory concentration; IN, integrase; lum, luminescence assay; NS5, non-structural protein 5; PCR, polymerase chain reaction; PFU, plaque forming units (infectious virus); PRV, pseudorabies virus; RdRp, RNA-dependent RNA polymerase; SV40, simian virus 40; T-ag, SV40 large T-antigen; TCID, tissue culture infectious dose (estimation of viral load based on CPE); VEEV, Venezuelan equine encephalitis virus; VLP, virus-like particle; vRNP, viral ribonucleoprotein complex; VSV-G, vesicular stomatitis virus glycoprotein; WNV, West Nile virus; YFV, Yellow Fever virus; ZIKV, Zika virus.
Recent work has confirmed that the mechanism of action of ivermectin in this context is through direct binding to IMPα, resulting in structural changes that decrease α-helicity and likely impact flexibility, as implied by circular dichroism (CD)/thermostability (TSA) measurements (see Figure 2) [44]. These changes prevent interaction with NLS-containing viral/host protein cargoes, as well as IMPβ1; ivermectin can also dissociate preformed IMPα/β heterodimer, which is the form IMPα needs to be in to mediate nuclear import [44]. Thus, ivermectin prevents recognition of nuclear import cargoes by IMPα and the IMPα/β1 (see Figure 1C), significantly reducing their nuclear accumulation and thereby diminishing suppression of the host antiviral response in infection. Inhibition of nuclear accumulation by ivermectin has been demonstrated for many different viral proteins [14,18] as well as host proteins such as NF-kB p65 [45] in transfected and infected cell systems (see Table 1). Ivermectin's ability to inhibit the binding of IMPα to the viral proteins has also been shown in a cellular context using the technique of biomolecular fluorescence complementation [32].
The IMPα-dependent host-targeted mode of action of ivermectin explains the wide range of viruses for which ivermectin has demonstrated antiviral effects both in vitro and in vivo [18]. In vitro antiviral properties have been demonstrated for a wide range of viruses, including Betacoronavirus SARS-CoV-2 (COVID19), the lentivirus HIV-1, alphainfluenzavirus (influenza A), flaviviruses (DENV, WNV, ZIKV, yellow fever virus — YFV), alphaviruses (CHIKV, Sindbis, Semliki forest virus, VEEV), the Hendra henipavirus, mastadenoviruses (human adenovirus — HAdV — B and C), the BK betapolyomavirus and PSV varicellovirus (Table 1). In the case of DENV and ZIKV, ivermectin antiviral activity has been confirmed in an ex vivo human disease-relevant model of peripheral mononuclear blood monocytes (PBMCs) [15,44]. It is noteworthy that ivermectin was also independently identified by HTS for antiviral agents able to inhibit CHIKV replication using a luciferase reporter-carrying virus [46].
Other inhibitors targeting IMPα/β1
Compound library HTS has enabled the identification of small molecule IMPα-targeting inhibitors other than ivermectin (see Table 1). GW5074 was identified as an inhibitor of IMPα/β1-DENV NS5 binding [15,47]. Recent efforts to characterise the properties of GW5074 have revealed that, in analogous fashion to ivermectin, it binds to IMPα, eliciting changes in α-helicity/flexibility implicated by CD/TSA measurements (see Figure 2), that lead to dissociation of the IMPα/β1 heterodimer/inhibition of NS5 nuclear import [47]. This is likely the basis of GW5074's ability to limit infectious virus production of DENV, ZIKV, and WNV [47]. Another host-targeted inhibitor, gossypol (GSP) discovered in the same HTS that identified ivermectin (see above) [14] has been shown to inhibit interaction of IMPα/β1:Hendra virus (HENV) V protein [48]. GSP has also been shown to have antiviral effects against WNV [49].
In silico structure-based-drug-design HTS has also been successfully applied to identify nuclear transport inhibitors [17]. In silico screening of 1.5 million compounds for the ability to dock into the NLS-binding site of IMPα and thereby mimic binding of the VEEV CP NLS to IMPα identified a range of different small molecules that were subsequently confirmed experimentally as being able to inhibit binding of IMPα/β1 to VEEV CP. Several of the lead compounds, including 1111864 (see Table 1) were confirmed to inhibit CP nuclear accumulation, as well as VEEV replication in infected cells [17].
Nuclear import inhibitors targeting specific viral proteins
In addition to the host-targeted inhibitors discussed above, several inhibitors have been identified that specifically target the virus side of the IMPα/β1-virus axis to prevent viral protein nuclear import (see Table 2). The steroid progesterone/glucocorticoid receptor antagonist mifepristone was the first such inhibitor to be identified in HTS, through its ability to inhibit the interaction of recombinant IMPα/β1 with HIV-1 IN [14]. Nested counterscreening was used to establish that its action was specific to IMPα/β1: IN and not due to direct effects on IMPα/β1 [14]), with more recent NMR studies confirming its direct binding to the IN core domain, with effects within the vicinity of key residues within the IN NLS [37]. Importantly, mifepristone was able to inhibit nuclear import of IN in an in vitro reconstituted nuclear import assay [37]. In vitro results had previously indicated strong antiviral activity against HIV-1 [50], with activity also documented against VEEV and HAdV [51–53]; since neither of these viruses possess an IN-homologue, the precise mechanism by which mifepristone impacts nuclear accumulation of CP in VEEV-infected cells [51] and of the HAdV genome in HAdV-infected cells [53] is unclear. A limitation on the clinical utility of mifepristone is the fact that it is abortifacient; medicinal chemistry and SAR analysis were able to be used to generate novel mifepristone analogues that lacked detectable progesterone receptor antagonism, but retained the ability both to inhibit VEEV CP nuclear accumulation and to limit VEEV infection [52], underlining how medicinal chemistry/SAR analysis can produce an inhibitor with improved properties.
The glucocorticoid budesonide was identified in the same HTS campaign as mifepristone (IC50 of c. 1 μM), subsequent characterisation using NMR confirming that budesonide also binds to the HIV-1 IN NLS region [37]. SAR/NMR analysis of closely related molecules showed that flunisolide also bound IN, but causes greater structural perturbation than budesonide, consistent with flunisolide being a more potent inhibitor of IN nuclear accumulation in vitro [37]. Importantly, both flunisolide and budesonide could be confirmed to inhibit nuclear import of the HIV PIC, as inferred from quantitative polymerase chain reaction analysis of unintegrated nuclear viral DNA [25]. Both budesonide and flunisolide also showed robust antiviral activity towards HIV-1, including in a single round replication assay (see Table 2).
From HTS of a > 14 000 compound library for inhibitors of IMPα/β1:VEEV CP binding, followed by a nested counterscreen, many chemical scaffolds were identified that retained potent inhibitory activity [16]. Subsequent in silico-informed SAR analysis focussed on several of these, with compound G281–1564 confirmed to be a specific inhibitor of IMPα/β1:CP interaction. Experiments in transfected cells confirmed inhibition of CP but not SV-40T-Ag nuclear accumulation, with fluorescence recovery after photobleaching experiments demonstrating inhibitory effects on both the rate and extent of CP nuclear import. Importantly, G281–1564 was found to limit VEEV infection in cell culture (EC50 of 10.8 μM) [16]; subsequent work indicated that G281–1564 prevents delays in cell cycle progression, which in part may depend on CP nuclear localisation/ IMPα/β1:CP binding [54].
One of the most promising inhibitors targeting the virus side of the IMPα/β1-virus axis is N-(4-hydroxyphenyl) retinamide (4-HPR, a.k.a. fenretinide), which was identified through HTS as a specific inhibitor of IMPα/β1:DENV NS5 interaction (IC50 of 0.8 μM) [15]. 4-HPR was shown to reduce the nuclear accumulation of DENV NS5 in infected cells, as well as strongly inhibiting viral replication of all four circulating DENV strains (DENV1-4) with EC50 values between ∼1–2 μM, including in an ex vivo model of severe antibody-dependent enhanced (ADE) human infection using PBMCs [15]. 4-HPR also limits infection by other flaviviruses, including ZIKV (EC50 of 1–2 μM), WNV, and YFV (see Table 2). SAR analysis together with the use of other inhibitors confirmed 4-HPR's specificity, and that its mechanism of antiviral action is though inhibiting NS5 nuclear import, rather than other effects (e.g. on ceramide metabolism) [15]. TSA assays confirmed that 4-HPR destabilises DENV2 NS5 RNA-dependent RNA polymerase (RdRp) domain, and not IMPα [44], highlighting its specificity for the viral component of the host–virus interface.
It is of interest that 4-HPR has also been identified in HTS (see above) as an inhibitor of CHIKV [46], and reported to limit infection by the arbovirus Modoc and influenza A viruses. Like CHIKV, Modoc, and influenza virus do not have a correlate of flavivirus NS5 (well-conserved in DENV1-4, ZIKV, YFV, WNV, Japanese Encephalitis Virus) that localises in the nucleus of the infected host cell to impact transcription/splicing, so that 4-HPR's antiviral action in this case may be through other cellular pathways (ceramide biosynthesis/unfolded protein response) [47,55].
Pre-clinical/clinical studies for IMPα targeting inhibitors
Host-directed agents that impact essential cellular activities like nuclear transport must be used with caution in a clinical setting. Of the small molecule inhibitor drugs targeted against the host IMPα/β1 side of the IMPα/β1:virus interface, ivermectin is the only one that has progressed from to human clinical trials for viral indications. Although it has an established safety profile in humans [43,56], and is FDA-approved for many parasitic infections [56–58], ivermectin targets host nuclear import function that is unquestionably important in the antiviral response, with titration of a large proportion of the IMPα repertoire of a cell/tissue/organ is likely to lead to toxicity. Where a host-directed agent like ivermectin can optimally be used to treat viral infection may well be in the initial stages of infection or potentially prophylactically by keeping the viral load low so that the body's immune system has an opportunity to mount a full antiviral response [44,59].
Pre-clinical studies in a lethal mouse PRV challenge study showed that ivermectin administration (0.2 mg/kg) 12 h post-infection rescued 50% of mice, while the same dose at the time of infection rescued 60% of mice [60]. Results from a human Phase III clinical trial for the treatment of patients presenting with fever and confirmed DENV diagnosis showed that daily administration (0.4 mg/kg for 3 days) was safe and effective at improving virus clearance, but the authors concluded that dosing regimen modification was required to achieve the clinical benefit, potentially due to the timing of administration of the doses [61].
The current SARS-CoV-2 pandemic has already seen >67 million infections and >1.5 million deaths (nearing 300 000 in the US alone [62]) worldwide [63]. SARS-CoV-2 is a respiratory virus, so it is noteworthy that pharmacokinetic modelling, based on both the levels of ivermectin achievable in human serum from standard 0.2 mg/kg dosing and robust measurement in large animal experiments, indicates that concentrations of ivermectin 10 times higher than the c. 2.5 μM EC50 indicated by in vitro experiments (Table 1) are likely achievable in the human lung [64]. Modelling based on different assumptions predicts lower values, but highlights the long-term stability of ivermectin in the lung (>30 days) [65]. Currently, almost 80 clinical trials are in progress globally for the treatment or prevention of SARS-CoV-2 [66–68], including variations on dosing regimens, combination therapies (see also [69–71]) and prophylactic protocols. Preliminary results from prophylactic study NCT04422561 of asymptomatic family close-contacts of confirmed COVID-19 patients indicate that two doses of ivermectin 72 h apart (0.25 mg/kg) are efficacious, resulting in an 87% relative risk reduction in the development of symptoms (58.4% of the untreated group vs 7.4% of the ivermectin-treated group).
Results from retrospective/observational trials for ivermectin [72–75] are also consistent with clinical benefit in the context of SARS-CoV-2 infection. Mymensingh Medical College Hospital (Bangladesh) reported that none of 115 subjects receiving a single 12 mg dose of ivermectin developed pneumonia/cardiovascular complications, compared with 9.8% (pneumonia) and 1.5% (ischemic stroke) in 133 control subjects [72]. Ivermectin-treated patients became SARS-CoV-2 negative faster (median 4 compared with 15 days), had shorter hospital stays (median 9 versus 15 days), and lower mortality (0.9 versus 6.8%). Importantly, significantly fewer ivermectin group patients developed respiratory distress (2.6 versus 15.8%), or required oxygen (9.6 versus 45.9%) or intensive care management (0.09 versus 8.3%). Comparable results were obtained in a 196 patient propensity-matched cohort study at Broward Health Medical Centre (Florida, U.S.A.) [73], with significantly lower mortality (13.3%) in subjects receiving ivermectin (0.2 mg/kg), compared with 24.5% mortality in those not receiving it, with more significant differences for patients with severe pulmonary involvement (mortality rates of 38.8 versus 80.7%). These early results are consistent with efficacy, but it is clear that only the results from large rigorous randomised clinical trials (RCTs) [66,67] will establish ivermectin's utility to treat or prevent SARS-CoV-2 infection.
Pre-clinical/clinical studies for viral protein targeting inhibitors
Of the small molecule inhibitors targeting the virus side of the IMPα/β1:virus interface, only mifepristone has thus far been investigated clinically in any detail. Mifepristone was shown to reduce virus production and viral DNA copy number in HAdV-infected mice, concomitant with inhibition of HAdV genome nuclear import [53]. However, although a Phase I/II clinical trial of mifepristone (75–225 mg/day) in HIV patients indicated it was safe and well-tolerated, there was no significant effect on plasma HIV-1 RNA levels or CD4+ lymphocyte counts [76].
In the case of 4-HPR as a therapeutic for flavivirus infection, results are thus far restricted to mouse models. In AG129 mice, twice-daily dosage of 4-HPR at 180 mg/kg (resulting in ∼15 μM plasma concentration of 4-HPR), reduced viremia >50-fold in DENV2 infection [77]. In a lethal challenge model of ADE-DENV1 infection in AG129 mice, 20 mg/kg 4-HPR once-daily rescued 20% of mice, whilst twice-daily treatment rescued 70% of mice [15], concomitant with reduced inflammatory cytokine responses (tumour necrosis factor α, IL-6, -10, and -12) [15]. Efficacy was also seen in AG129 mice for ZIKV infection, where 4-HPR at 60 mg/kg twice-daily reduced viremia by ∼10-fold [78]. Previous human Phase II/III trials for a variety of adult and paediatric solid tumour indications have demonstrated 4-HPR plasma trough concentrations exceeding 7 μM (up to a peak of >50 μM) are achievable with a tolerable safety profile [79,80], which compares favourably with EC50 values of 1–2 μM for DENV in human PBMCs [15]. Based on these results as well as 4-HPR's established human safety and pharmacokinetic profiles in adults and children, human clinical trials are planned to investigate 4-HPR's exciting potential as a therapeutic and potentially prophylactic agent against DENV.
Future prospects
It is clear from the last decade that small molecules targeting the host IMPα/β1:virus interface can be found that have potent antiviral activity, and exciting potential to treat human infection [66]. While host-targeted ‘broad spectrum’ inhibitors such as ivermectin can affect a range of viruses that share the need for the same nuclear import pathways as part of their lifecycle, potential toxicity must be considered, due to the essential nature of these import pathways for host cell function [4]. Virus-targeted inhibitors like 4-HPR, on the other hand, do not inhibit host protein nuclear import and hence are not toxic, but inevitably have a more restricted spectrum of antiviral activity as a result [66].
Conventional HTS (e.g. [14–16]) have proven effective at identifying promising lead compounds which are specific and effective at achievable concentrations, with in silico HTS emerging as a complementary approach to identify promising leads, and facilitate focussed library selection (e.g. [17,52]). Drugs such as ivermectin and 4-HPR are clearly exciting prospects, but the possibilities have by no means been yet exhausted in terms of agents that target the host IMPα/β1:virus axis, let alone other host IMP:virus interfaces. It is clear that further HTS and development of novel approaches to target these and other interfaces are likely to prove beneficial in the current landscape, in terms of identifying new possible agents to control existing and emerging viral pathogens.
Perspectives
Nuclear import is essential to the infectious cycles of a range of viruses of medical significance. Inhibitors of nuclear import targeting either the host or virus side of the IMPα/β1:virus interface have been identified in recent years, and shown to possess potent antiviral activity.
Host-targeted inhibitor of nuclear import ivermectin is showing exciting early results as a treatment for COVID-19 infection. Virus-targeted inhibitor of nuclear import of flavivirus NS5, 4-HPR, appears to be an exciting prospect for future clinical trials.
The process of nuclear import, and the IMPα/β1:virus interface in particular, remains a viable target of interest for the future identification and development of antivirals to tackle existing and emerging viral disease.
Acknowledgements
The authors acknowledge the support of an Australian Government Research Training Program (RTP) Scholarship to A.J.M. and National Health and Medical Research Council Australia Senior Principal Research Fellowship (APP1002486/APP1103050) to D.A.J.
Abbreviations
- ADE
antibody-dependent enhanced
- ARM
α-helical armadillo
- CD
circular dichroism
- CE
chicken embryo
- DENV
dengue
- HIV-1
human immunodeficiency virus-1
- HTS
high-throughput screening
- IBB
importin-β-binding
- NLSs
nuclear localisation signals
- NP
nucleoprotein
- PBMCs
peripheral mononuclear blood monocytes
- PIC
preintegration complex
- RCTs
randomised clinical trials
- SARS
severe acute respiratory syndrome
- STATs
signal transducer and activators of transcription
- TSA
thermostability
- WNV
West Nile virus
- ZIKV
Zika
Competing Interests
D.A.J. is an inventor on a patent application for the use of ivermectin for coronavirus infection, and a patent on the use of 4-HPR in flavivirus infection. A.J.M. declares no competing interests.
Author Contributions
A.J.M. drafted the manuscript, D.A.J. provided overall direction, and helped revise the manuscript.
Open Access
Open access for this article was enabled by the participation of Monash University in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with CAUL.
References
- 1.Aguzzi, A. and Altmeyer, M. (2016) Phase separation: linking cellular compartmentalization to disease. Trends Cell Biol. 26, 547–558 10.1016/j.tcb.2016.03.004 [DOI] [PubMed] [Google Scholar]
- 2.Poon, I.K.H. and Jans, D.A. (2005) Regulation of nuclear transport: central role in development and transformation? Traffic (Copenhagen, Denmark) 6, 173–186 10.1111/j.1600-0854.2005.00268.x [DOI] [PubMed] [Google Scholar]
- 3.Fulcher, A.J. and Jans, D.A. (2011) Regulation of nucleocytoplasmic trafficking of viral proteins: an integral role in pathogenesis? Biochim. Biophys. Acta 1813. 2176–2190 10.1016/j.bbamcr.2011.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caly, L., Wagstaff, K.M. and Jans, D.A. (2012) Nuclear trafficking of proteins from RNA viruses: potential target for antivirals? Antivir. Res. 95, 202–206 10.1016/j.antiviral.2012.06.008 [DOI] [PubMed] [Google Scholar]
- 5.Fried, H. and Kutay, U. (2003) Nucleocytoplasmic transport: taking an inventory. Cell Mol. Life Sci. 60, 1659–1688 10.1007/s00018-003-3070-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terry, L.J., Shows, E.B. and Wente, S.R. (2007) Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 318, 1412–1416 10.1126/science.1142204 [DOI] [PubMed] [Google Scholar]
- 7.Smith, K.M., Antonio, V.D., Bellucci, L., Thomas, D.R., Caporuscio, F., Ciccarese, F.et al. (2018) Contribution of the residue at position 4 within classical nuclear localization signals to modulating interaction with importins and nuclear targeting. Biochim. Biophys. Acta Mol. Cell Res. 1865, 1114–1129 10.1016/j.bbamcr.2018.05.006 [DOI] [PubMed] [Google Scholar]
- 8.Pryor, M.J., Rawlinson, S.M., Butcher, R.E., Barton, C.L., Waterhouse, T.A., Vasudevan, S.G.et al. (2007) Nuclear localization of dengue virus nonstructural protein 5 through its importin α/β-recognized nuclear localization sequences is integral to viral infection. Traffic (Copenhagen, Denmark) 8, 795–807 10.1111/j.1600-0854.2007.00579.x [DOI] [PubMed] [Google Scholar]
- 9.Piller, S., Caly, L. and Jans, D. (2003) Nuclear import of the pre-integration complex (PIC): the achilles heel of HIV? Curr. Drug Targets 4, 409–429 10.2174/1389450033490984 [DOI] [PubMed] [Google Scholar]
- 10.Rawlinson, S.M., Pryor, M.J., Wright, P.J. and Jans, D.A. (2006) Dengue virus RNA polymerase NS5: a potential therapeutic target? Curr. Drug Targets 7, 1623–1638 10.2174/138945006779025383 [DOI] [PubMed] [Google Scholar]
- 11.Götz, V., Magar, L., Dornfeld, D., Giese, S., Pohlmann, A., Höper, D.et al. (2016) Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Sci. Rep. 6, 23138–23115 10.1038/srep23138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito, N., Moseley, G.W., Blondel, D., Shimizu, K., Rowe, C.L., Ito, Y.et al. (2010) Role of interferon antagonist activity of rabies virus phosphoprotein in viral pathogenicity. J. Virol. 84, 6699–6710 10.1128/JVI.00011-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiltzer, L., Okada, K., Yamaoka, S., Larrous, F., Kuusisto, H.V., Sugiyama, M.et al. (2014) Interaction of rabies virus P-protein with STAT proteins is critical to lethal rabies disease. J. Infect. Dis. 209, 1744–1753 10.1093/infdis/jit829 [DOI] [PubMed] [Google Scholar]
- 14.Wagstaff, K.M., Rawlinson, S.M., Hearps, A.C. and Jans, D.A. (2011) An AlphaScreen®-based assay for high-throughput screening for specific inhibitors of nuclear import. J. Biomol. Screen. 16, 192–200 10.1177/1087057110390360 [DOI] [PubMed] [Google Scholar]
- 15.Fraser, J.E., Watanabe, S., Wang, C., Chan, W.K.K., Maher, B., Lopez-Denman, A.et al. (2014) A nuclear transport inhibitor that modulates the unfolded protein response and provides in vivo protection against lethal dengue virus infection. J. Infect. Dis. 210, 1780–1791 10.1093/infdis/jiu319 [DOI] [PubMed] [Google Scholar]
- 16.Thomas, D.R., Lundberg, L., Pinkham, C., Shechter, S., DeBono, A., Baell, J.et al. (2018) Identification of novel antivirals inhibiting recognition of Venezuelan equine encephalitis virus capsid protein by the importin α/β1 heterodimer through high-throughput screening. Antivir. Res. 151, 8–19 10.1016/j.antiviral.2018.01.007 [DOI] [PubMed] [Google Scholar]
- 17.Shechter, S., Thomas, D.R., Lundberg, L., Pinkham, C., Lin, S.-C., Wagstaff, K.M.et al. (2017) Novel inhibitors targeting Venezuelan equine encephalitis virus capsid protein identified using in silico structure-based-drug-design. Sci. Rep. 7, 17705 10.1038/s41598-017-17672-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jans, D.A., Martin, A.J. and Wagstaff, K.M. (2019) Inhibitors of nuclear transport. Curr. Opin. Cell Biol. 58, 50–60 10.1016/j.ceb.2019.01.001 [DOI] [PubMed] [Google Scholar]
- 19.Pemberton, L.F. and Paschal, B.M. (2005) Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic (Copenhagen, Denmark) 6, 187–198 10.1111/j.1600-0854.2005.00270.x [DOI] [PubMed] [Google Scholar]
- 20.Stewart, M. (2007) Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195–208 10.1038/nrm2114 [DOI] [PubMed] [Google Scholar]
- 21.Goldfarb, D.S., Corbett, A.H., Mason, D.A., Harreman, M.T. and Adam, S.A. (2004) Importin α: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14, 505–514 10.1016/j.tcb.2004.07.016 [DOI] [PubMed] [Google Scholar]
- 22.Fontes, M.R.M., Teh, T. and Kobe, B. (2000) Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J. Mol. Biol. 297, 1183–1194 10.1006/jmbi.2000.3642 [DOI] [PubMed] [Google Scholar]
- 23.Chen, M.-H., Ben-Efraim, I., Mitrousis, G., Walker-Kopp, N., Sims, P.J. and Cingolani, G. (2005) Phospholipid scramblase 1 contains a nonclassical nuclear localization signal with unique binding site in importin α. J. Biol. Chem. 280, 10599–10606 10.1074/jbc.M413194200 [DOI] [PubMed] [Google Scholar]
- 24.Kobe, B. (1999) Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat. Struct. Biol. 6, 388–397 10.1038/7625 [DOI] [PubMed] [Google Scholar]
- 25.Smith, K.M., Tsimbalyuk, S., Edwards, M.R., Cross, E.M., Batra, J., Costa, T.et al. (2018) Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses. Nat. Commun. 9, 3703 10.1038/s41467-018-05928-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt, J.E., Ly-Huynh, J.D., Efthymiadis, A., Hime, G.R., Loveland, K.L. and Jans, D.A. (2007) Regulation of nuclear import during differentiation; the IMP α gene family and spermatogenesis. Curr. Genom. 8, 323–334 10.2174/138920207782446151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekimoto, T., Imamoto, N., Nakajima, K., Hirano, T. and Yoneda, Y. (1997) Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 16, 7067–7077 10.1093/emboj/16.23.7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Köhler, M., Speck, C., Christiansen, M., Bischoff, F.R., Prehn, S., Haller, H.et al. (1999) Evidence for distinct substrate specificities of importin α family members in nuclear protein import. Mol. Cell Biol. 19, 7782–7791 10.1128/MCB.19.11.7782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ao, Z., Jayappa, K.D., Wang, B., Zheng, Y., Kung, S., Rassart, E.et al. (2010) Importin α3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J. Virol. 84, 8650–8663 10.1128/JVI.00508-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabriel, G., Klingel, K., Otte, A., Thiele, S., Hudjetz, B., Arman-Kalcek, G.et al. (2011) Differential use of importin-α isoforms governs cell tropism and host adaptation of influenza virus. Nat. Commun. 2, 156 10.1038/ncomms1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rawlinson, S.M., Pryor, M.J., Wright, P.J. and Jans, D.A. (2009) CRM1-mediated nuclear export of dengue virus RNA polymerase NS5 modulates interleukin-8 induction and virus production. J. Biol. Chem. 284, 15589–15597 10.1074/jbc.M808271200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Maio, F.A., Risso, G., Iglesias, N.G., Shah, P., Pozzi, B., Gebhard, L.G.et al. (2016) The dengue virus NS5 protein intrudes in the cellular spliceosome and modulates splicing. PLoS Pathog. 12, e1005841 10.1371/journal.ppat.1005841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah, P.S., Link, N., Jang, G.M., Sharp, P.P., Zhu, T., Swaney, D.L.et al. (2018) Comparative flavivirus-host protein interaction mapping reveals mechanisms of dengue and zika virus pathogenesis. Cell 175, 1931–1945.e18 10.1016/j.cell.2018.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chelbi-Alix, M.K., Vidy, A., Bougrini, J.E. and Blondel, D. (2006) Rabies viral mechanisms to escape the IFN system: the viral protein P interferes with IRF-3, Stat1, and PML nuclear bodies. J. Interf. Cytokine Res. 26, 271–280 10.1089/jir.2006.26.271 [DOI] [PubMed] [Google Scholar]
- 35.Hu, M., Bogoyevitch, M.A. and Jans, D.A. (2020) Impact of respiratory syncytial virus infection on host functions: implications for antiviral strategies. Physiol. Rev. 100, 1527–1594 10.1152/physrev.00030.2019 [DOI] [PubMed] [Google Scholar]
- 36.Anna C, H. and David A, J. (2006) HIV-1 integrase is capable of targeting DNA to the nucleus via an importin α/β-dependent mechanism. Biochem. J. 398, 475–484 10.1042/BJ20060466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagstaff, K.M., Headey, S., Telwatte, S., Tyssen, D., Hearps, A.C., Thomas, D.R.et al. (2018) Molecular dissection of an inhibitor targeting the HIV integrase dependent preintegration complex nuclear import. Cell. Microbiol. 59, e12953 10.1111/cmi.12953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryman, K.D. and Klimstra, W.B. (2008) Host responses to alphavirus infection. Immunol. Rev. 225, 27–45 10.1111/j.1600-065X.2008.00670.x [DOI] [PubMed] [Google Scholar]
- 39.Atasheva, S., Fish, A., Fornerod, M. and Frolova, E.I. (2010) Venezuelan equine encephalitis virus capsid protein forms a tetrameric complex with CRM1 and importin α/β that obstructs nuclear pore complex function. J. Virol. 84, 4158–4171 10.1128/JVI.02554-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frieman, M., Yount, B., Heise, M., Kopecky-Bromberg, S.A., Palese, P. and Baric, R.S. (2007) Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 81, 9812–9824 10.1128/JVI.01012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bukrinsky, M.I., Sharova, N., McDonald, T.L., Pushkarskaya, T., Tarpley, W.G. and Stevenson, M. (1993) Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. U.S.A. 90, 6125–6129 10.1073/pnas.90.13.6125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagstaff, K.M., Sivakumaran, H., Heaton, S.M., Harrich, D. and Jans, D.A. (2012) Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 443, 851–856 10.1042/BJ20120150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laing, R., Gillan, V. and Devaney, E. (2017) Ivermectin – old drug, new tricks? Trends Parasitol. 33, 463–472 10.1016/j.pt.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, S.N.Y., Atkinson, S.C., Wang, C., Lee, A., Bogoyevitch, M.A., Borg, N.A.et al. (2020) The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antivir. Res. 177, 104760 10.1016/j.antiviral.2020.104760 [DOI] [PubMed] [Google Scholar]
- 45.Ci, X., Li, H., Yu, Q., Zhang, X., Yu, L., Chen, N.et al. (2009) Avermectin exerts anti-inflammatory effect by downregulating the nuclear transcription factor kappa-B and mitogen-activated protein kinase activation pathway. Fundam. Clin. Pharm. 23, 449–455 10.1111/j.1472-8206.2009.00684.x [DOI] [PubMed] [Google Scholar]
- 46.Varghese, F.S., Kaukinen, P., Gläsker, S., Bespalov, M., Hanski, L., Wennerberg, K.et al. (2016) Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses. Antivir. Res. 126, 117–124 10.1016/j.antiviral.2015.12.012 [DOI] [PubMed] [Google Scholar]
- 47.Yang, S.N.Y., Atkinson, S.C., Fraser, J.E., Wang, C., Maher, B., Roman, N.et al. (2019) Novel flavivirus antiviral that targets the host nuclear transport importin α/β1 heterodimer. Cells 8, 281 10.3390/cells8030281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atkinson, S.C., Audsley, M.D., Lieu, K.G., Marsh, G.A., Thomas, D.R., Heaton, S.M.et al. (2018) Recognition by host nuclear transport proteins drives disorder-to-order transition in Hendra virus V. Sci. Rep. 8, 23 10.1038/s41598-017-18157-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez-Denman, A.J., Russo, A., Wagstaff, K.M., White, P.A., Jans, D.A. and Mackenzie, J.M. (2018) Nucleocytoplasmic shuttling of the West Nile virus RNA-dependent RNA polymerase NS5 is critical to infection. Cell. Microbiol. 20, e12848 10.1111/cmi.12848 [DOI] [PubMed] [Google Scholar]
- 50.Schafer, E.A., Venkatachari, N.J. and Ayyavoo, V. (2006) Antiviral effects of mifepristone on human immunodeficiency virus type-1 (HIV-1): targeting Vpr and its cellular partner, the glucocorticoid receptor (GR). Antivir. Res. 72, 224–232 10.1016/j.antiviral.2006.06.008 [DOI] [PubMed] [Google Scholar]
- 51.Lundberg, L., Pinkham, C., Baer, A., Amaya, M., Narayanan, A., Wagstaff, K.M.et al. (2013) Nuclear import and export inhibitors alter capsid protein distribution in mammalian cells and reduce Venezuelan equine encephalitis virus replication. Antivir. Res. 100, 662–672 10.1016/j.antiviral.2013.10.004 [DOI] [PubMed] [Google Scholar]
- 52.DeBono, A., Thomas, D.R., Lundberg, L., Pinkham, C., Cao, Y., Graham, J.D.et al. (2019) Novel RU486 (mifepristone) analogues with increased activity against Venezuelan equine encephalitis virus but reduced progesterone receptor antagonistic activity. Sci. Rep. 9, 2634 10.1038/s41598-019-38671-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marrugal-Lorenzo, J.A., Serna-Gallego, A., González-González, L., Buñuales, M., Poutou, J., Pachón, J.et al. (2018) Inhibition of adenovirus infection by mifepristone. Antivir. Res. 159, 77–83 10.1016/j.antiviral.2018.09.011 [DOI] [PubMed] [Google Scholar]
- 54.Lundberg, L., Fontenot, J., Lin, S.-C., Pinkham, C., Carey, B.D., Campbell, C.E.et al. (2018) Venezuelan equine encephalitis virus capsid implicated in infection-induced cell cycle delay in vitro. Fronti. Microbiol. 9, 3126 10.3389/fmicb.2018.03126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fraser, J.E., Wang, C., Chan, W.K.K., Vasudevan, S.G. and Jans, D.A. (2016) Novel dengue virus inhibitor 4-HPR activates ATF4 independent of protein kinase R-like endoplasmic reticulum kinase and elevates levels of eIF2α phosphorylation in virus infected cells. Antivir. Res. 130, 1–6 10.1016/j.antiviral.2016.03.006 [DOI] [PubMed] [Google Scholar]
- 56.Canga, A.G., Prieto, A.M.S., Liébana, M.J.D., Martínez, N.F., Vega, M.S. and Vieitez, J.J.G. (2008) The pharmacokinetics and interactions of ivermectin in humans–a mini-review. AAPS J. 10, 42–46 10.1208/s12248-007-9000-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crump, A. and Ōmura, S. (2011) Ivermectin, ‘Wonder drug’ from Japan: the human use perspective. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 87, 13–28 10.2183/pjab.87.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.World Health Organization. World Health Organization Model List of Essential Medicines 2019. Available from: https://apps.who.int/iris/bitstream/handle/10665/325771/WHO-MVP-EMP-IAU-2019.06-eng.pdf.
- 59.Caly, L., Druce, J.D., Catton, M.G., Jans, D.A. and Wagstaff, K.M. (2020) The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 178, 104787 10.1016/j.antiviral.2020.104787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lv, C., Liu, W., Wang, B., Dang, R., Qiu, L., Ren, J.et al. (2018) Ivermectin inhibits DNA polymerase UL42 of pseudorabies virus entrance into the nucleus and proliferation of the virus in vitro and vivo. Antivir. Res. 159, 55–62 10.1016/j.antiviral.2018.09.010 [DOI] [PubMed] [Google Scholar]
- 61.Yamasmith E., Saleh-arong F.A., Avirutnan P., Angkasekwinai N., Mairiang, D., Wongsawat, E., et al. Efficacy and Safety of Ivermectin against Dengue Infection: A Phase III, Randomized, Double-blind, Placebo-controlled Trial. The 34th Annual Meeting The Royal College of Physicians of Thailand. 2018.
- 62. https://covid.cdc.gov/covid-data-tracker/#cases_totalcases Centres for Disease Control and Prevention. CDC COVID Data Tracker 2020. Available from:
- 63. https://coronavirus.jhu.edu/map.html Johns Hopkins University Coronavirus Resource Centre. COVID-19 Dashboard 2020. Available from:
- 64.Arshad, U., Pertinez, H., Box, H., Tatham, L., Rajoli, R.K.R., Curley, P.et al. (2020) Prioritization of anti-SARS-Cov-2 drug repurposing opportunities based on plasma and target site concentrations derived from their established human pharmacokinetics. Clin. Pharmacol. Ther. 108, 775–790 10.1002/cpt.1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmith, V.D., Zhou, J. and Lohmer, L.R.L. (2020) The approved dose of ivermectin alone is not the ideal dose for the treatment of COVID-19. Clin. Pharmacol. Ther. 108, 762–765 10.1002/cpt.1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jans, D.A. and Wagstaff, K.M. (2020) Ivermectin as a broad-spectrum host-directed antiviral: the real deal? Cells 9, 2100 10.3390/cells9092100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jans, D.A. and Wagstaff, K.M. (2020) The broad spectrum host-directed agent ivermectin as an antiviral for SARS-CoV-2? Biochem. Biophys. Res. Commun. In press 10.1016/j.bbrc.2020.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jans, D.A, Wagstaff KM. Ivermectin as Broad-Spectrum Host-Directed Antiviral 2020. Available from: https://encyclopedia.pub/2895 [DOI] [PMC free article] [PubMed]
- 69.Wang, Y., Zhang, D., Du, G., Du, R., Zhao, J., Jin, Y.et al. (2020) Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 395, 1569–1578 10.1016/S0140-6736(20)31022-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cao, B., Wang, Y., Wen, D., Liu, W., Wang, J., Fan, G.et al. (2020) A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 382, 1787–1799 10.1056/NEJMoa2001282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Héctor, C., Roberto, H., Psaltis, A. and Veronica, C. (2020) Study of the efficacy and safety of topical ivermectin + iota-carrageenan in the prophylaxis against COVID-19 in health personnel. J. Biomed. Res. Clin. Investig. 2 10.31546/2633-8653.1007 [DOI] [Google Scholar]
- 72.Khan, M.S.I., Khan, M.S.I., Debnath, C.R., Nath, P.N., Mahtab, M.A., Nabeka, H.et al. (2020) Ivermectin treatment may improve the prognosis of patients with COVID-19. Archivos De Bronconeumología 56, 828–830 10.1016/j.arbres.2020.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rajter, J.C., Sherman, M.S., Fatteh, N., Vogel, F., Sacks, J. and Rajter, J.-J. (2020) Use of ivermectin is associated with lower mortality in hospitalized patients with COVID-19 (ICON study). Chest In press. 10.1016/j.chest.2020.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alam, M.T., Murshed, R., Bhiuyan, E., Saber, S., Alam, R.F. and Robin, R.C. (2020) A case series of 100 COVID-19 positive patients treated with combination of ivermectin and doxycycline. J. Bangladesh Coll. Physicians Surg. 13, 10–15 10.3329/jbcps.v38i0.47512 [DOI] [Google Scholar]
- 75.Rahman, M.A., Iqbal, S.A., Islam, M.A., Niaz, M.K., Hussain, T. and Siddiquee, T.H. (2020) Comparison of viral clearance between ivermectin with doxycycline and hydroxychloroquine with azithromycin in COVID-19 patients. J. Bangladesh Coll. Physicians Surg. 13, 5–9 10.3329/jbcps.v38i0.47514 [DOI] [Google Scholar]
- 76.Para, M.F., Schouten, J., Rosenkranz, S.L., Yu, S., Weiner, D., Tebas, P.et al. (2010) Phase I/II trial of the anti-HIV activity of mifepristone in HIV-infected subjects ACTG 5200. J. Acquir. Immune Defic. Syndr. 53, 491–495 10.1097/QAI.0b013e3181d142cb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carocci, M., Hinshaw, S.M., Rodgers, M.A., Villareal, V.A., Burri, D.J., Pilankatta, R.et al. (2015) The bioactive lipid 4-hydroxyphenyl retinamide inhibits flavivirus replication. Antimicrob. Agents Chemother. 59, 85–95 10.1128/AAC.04177-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pitts, J.D., Li, P.-C., de Wispelaere, M. and Yang, P.L. (2017) Antiviral activity of N-(4-hydroxyphenyl) retinamide (4-HPR) against Zika virus. Antivir. Res. 147, 124–130 10.1016/j.antiviral.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garaventa, A., Luksch, R., Lo Piccolo, M.S., Cavadini, E., Montaldo, P.G., Pizzitola, M.R.et al. (2003) Phase I trial and pharmacokinetics of fenretinide in children with neuroblastoma. Clin. Cancer Res. 9, 2032–2039 PMID: [PubMed] [Google Scholar]
- 80.Cooper, J.P., Reynolds, C.P., Cho, H. and Kang, M.H. (2017) Clinical development of fenretinide as an antineoplastic drug: pharmacology perspectives. Exp. Biol. Med. (Maywood, NJ) 242, 1178–1184 10.1177/1535370217706952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.King, C.R., Tessier, T.M., Dodge, M.J., Weinberg, J.B. and Mymryk, J.S. (2020) Inhibition of human adenovirus replication by the importin α/β1 nuclear import inhibitor ivermectin. J. Virol 94, e00710–20 10.1128/jvi.00710-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tay, M.Y.F., Fraser, J.E., Chan, W.K.K., Moreland, N.J., Rathore, A.P., Wang, C.et al. (2013) Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antivir. Res. 99, 301–306 10.1016/j.antiviral.2013.06.002 [DOI] [PubMed] [Google Scholar]
- 83.Mastrangelo, E., Pezzullo, M., Burghgraeve, T.D., Kaptein, S., Pastorino, B., Dallmeier, K.et al. (2012) Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug. J Antimicrob. Chemoth. 67, 1884–1894 10.1093/jac/dks147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bennett, S.M., Zhao, L., Bosard, C. and Imperiale, M.J. (2015) Role of a nuclear localization signal on the minor capsid proteins VP2 and VP3 in BKPyV nuclear entry. Virology 474, 110–116 10.1016/j.virol.2014.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Porat, O. (1990) Effects of gossypol on the motility of mammalian sperm. Mol. Reprod. Dev. 25, 400–408 10.1002/mrd.1080250414 [DOI] [PubMed] [Google Scholar]
- 86.Yu, Z.H. and Chan, H.C. (1998) Gossypol as a male antifertility agent – why studies should have been continued. Int. J. Androl. 21, 2–7 10.1046/j.1365-2605.1998.00091.x [DOI] [PubMed] [Google Scholar]
- 87.Wang, C., Yang, S.N.Y., Smith, K., Forwood, J.K. and Jans, D.A. (2017) Nuclear import inhibitor N-(4-hydroxyphenyl) retinamide targets Zika virus (ZIKV) nonstructural protein 5 to inhibit ZIKV infection. Biochem. Biophys. Res. Commun. 493, 1555–1559 10.1016/j.bbrc.2017.10.016 [DOI] [PubMed] [Google Scholar]