Abstract
Facilitated by advances in the separation sciences, mass spectrometry and informatics, glycoproteomics, the analysis of intact glycopeptides at scale, has recently matured enabling new insights into the complex glycoproteome. While diverse quantitative glycoproteomics strategies capable of mapping monosaccharide compositions of N- and O-linked glycans to discrete sites of proteins within complex biological mixtures with considerable sensitivity, quantitative accuracy and coverage have become available, developments supporting the advancement of structure-focused glycoproteomics, a recognised frontier in the field, have emerged. Technologies capable of providing site-specific information of the glycan fine structures in a glycoproteome-wide context are indeed necessary to address many pending questions in glycobiology. In this review, we firstly survey the latest glycoproteomics studies published in 2018–2020, their approaches and their findings, and then summarise important technological innovations in structure-focused glycoproteomics. Our review illustrates that while the O-glycoproteome remains comparably under-explored despite the emergence of new O-glycan-selective mucinases and other innovative tools aiding O-glycoproteome profiling, quantitative glycoproteomics is increasingly used to profile the N-glycoproteome to tackle diverse biological questions. Excitingly, new strategies compatible with structure-focused glycoproteomics including novel chemoenzymatic labelling, enrichment, separation, and mass spectrometry-based detection methods are rapidly emerging revealing glycan fine structural details including bisecting GlcNAcylation, core and antenna fucosylation, and sialyl-linkage information with protein site resolution. Glycoproteomics has clearly become a mainstay within the glycosciences that continues to reach a broader community. It transpires that structure-focused glycoproteomics holds a considerable potential to aid our understanding of systems glycobiology and unlock secrets of the glycoproteome in the immediate future.
Keywords: glycopeptide, glycoproteins, glycoproteomics, glycosylation, LC-MS/MS, mass spectrometry
Introduction
The human genome encodes a plethora of enzymes, receptors, and transporters that concertedly facilitate and regulate protein glycosylation, the covalent attachment of complex carbohydrates (glycans) to polypeptides. Glycosylation is a prevalent and structurally diverse type of protein modification central to many biological processes including cellular development [1], lectin-mediated cell–cell communication [2], adhesin-based host-microbial interactions [3,4], and immunity [5–7]. Glycans also aid the folding and impact the solubility and integrity of glycoproteins, as well as regulate their susceptibility to proteolytic cleavage to mention a few of their modulatory functions [8–10].
Dysregulation of the glycosylation machinery often leads to aberrant protein glycosylation. Altered glycophenotypes can arise from and, in other cases, lead to a broad range of pathologies, amongst others, cancer [11–13], infection and inflammation [14–17], and schizophrenia [18]. Furthermore, glycans are functional effectors of genetic and epigenetic disease risk [19]. Consequently, unravelling the dynamics and the glycan remodelling associated with changes to the cellular milieu under normal physiology and during disease conditions have been a long-standing goal of glycobiologists; such advances are required to promote the next generation of glycoprotein-based diagnostics and therapeutics [20,21].
Glycoproteomics, the system-wide study of glycans linked to proteins or peptides, is a powerful analytical discipline capable of identifying and quantifying physiological or disease-driven aberrations in protein glycosylation arising from changes in the glycoprotein level, site occupancy (macro-heterogeneity) and/or glycosylation pattern at individual sites (micro-heterogeneity) [22]. Mass spectrometry (MS)-based analysis of intact glycopeptides as opposed to the study of detached glycans (glycomics) or formerly glycosylated peptides (site mapping) is an attractive analytical strategy since only intact glycopeptide (or glycoprotein) analysis is able to provide direct evidence for the site-specific glycosylation of proteins. Intact glycopeptide profiling, however, remains analytically challenging when applied to crude biological mixtures given the fact that both the chemical structure of the glycan, the peptide modification site, and the identity of the peptide carrier need to be correctly identified and quantified for each glycopeptide.
The field of glycoproteomics has for these reasons received considerable attention over the past years. As previously reviewed [22–34], diverse LC–MS/MS-driven glycoproteomics tools and strategies capable of quantitatively mapping monosaccharide compositions of N- and O-linked glycans to peptides and, in favourable cases, to discrete sites of proteins in complex biological mixtures are now available. This has led to a body of literature exploring the glycoproteome in various biological systems, some with considerable coverage, as demonstrated by studies detecting thousands of intact N- and O-glycopeptides within a single glycoproteomics experiment [35–43]. While the identification accuracy of some of the reported glycopeptides from such studies may be questioned due to the analytical challenges outlined above [44–46], these examples illustrate that the technology is now getting ready to address pending questions in systems glycobiology.
A recognised limitation, however, is that most glycoproteomics strategies, at present, do not provide information of the glycan structure beyond the generic monosaccharide composition i.e. hexose (Hex), N-acetylhexosamine (HexNAc), deoxyhexose (dHex), and N-acetylneuraminic acid (NeuAc) or even, in some cases, simply consider the glycans as a largely undefined protein modification based on their molecular mass. The structural elucidation of peptide-linked glycans beyond their monosaccharide compositions is often inferred from our growing knowledge of the related yet species-specific biosynthetic pathways responsible for the glycoprotein formation that exist across the areas of life [47,48]. This lack of concrete evidence for the glycan fine structure is an important shortcoming given that glycobiological processes are often governed by specific rather than generic glycan structures and substructures (glycoepitopes) occupying distinct sites on their carrier proteins. Detailed site-specific glycan profiling of the glycoproteome, herein referred to as structure-focused glycoproteomics, capable of discriminating isomeric monosaccharide residues e.g. N-acetyl-β-d-glucosamine (β-linked GlcNAc) versus N-acetyl-α-d-galactosamine (α-linked GalNAc), key topological differences e.g. antenna (α1,2/3/4-) versus core (α1,6-) fucosylation and triantennary (β1,4/6-) versus bisecting (β1,4-) GlcNAc-containing glycans, and isomeric glycosidic linkage types e.g. α2,3- vs α2,6-sialylation is consequently considered one of the next important frontiers in glycoproteomics.
This review will first provide an overview of the most recent glycoproteomics studies published in the period 2018–2020, their approaches and their findings, and then summarise important technological developments supporting the emergence of structure-focused glycoproteomics.
Recent N- and O-glycoproteomics studies (2018–2020)
Our survey of the most recent literature identified a total of 28 studies applying LC–MS/MS-based glycoproteomics to biological samples, Figure 1. While our previous review revealed that glycoproteomics studies carried out in the period 2014–2016 typically reported between 100 and 2000 unique (non-redundant) intact N- and O-linked glycopeptides [22], studies published since 2018 are now routinely identifying and quantifying thousands of glycopeptides, some even beyond 10 000 and 20 000 intact glycopeptides [49,50].
Figure 1.
Glycoproteome coverage reported by recent glycoproteomics studies published in the period 2018–2020 arranged by the studied species and publication year. The studies included in this overview all performed large-scale glycoproteomics analysis of complex biological samples from unaltered or only mildly manipulated biological sources and reported a significant number of intact glycopeptides. Studies not matching these criteria and studies reanalysing existing glycoproteomics data were left out. We apologise for any omission of studies published in this period that we may have failed to identify in our survey of the literature. The coverage as measured by the reported unique (non-redundant) intact N-glycopeptides (yellow bars) and O-glycopeptides (blue bars) has been plotted for each study. Details of these 28 glycoproteomics studies including their full references are provided in Table 1.
It is widely recognised that the glycoproteome coverage depends on the complexity and dynamical range of the biological sample(s) being studied, the amount of protein starting material (often high µg/low mg protein levels), and, notably, relates directly to the accuracy and confidence by which the reported glycopeptides are identified [22]. Regardless of these constraints, the recent glycoproteomics studies have clearly reached a greater glycoproteome coverage over a relative short period indicating that the glycoproteomics technologies are rapidly maturing. We therefore decided to carefully mine these recent glycoproteomics papers to explore how the studies were technically performed, how data were collected and how the data provided new insight into the glycoproteome, Table 1.
Table 1. Recent N- and O-glycoproteomics studies published in the period 2018–2020.
| Biological sample (species) | Approach 1) Mass spectrometer 2) MS/MS method (energy level applieda) 3) Data acquisition 4) Data analysis 5) Quantification |
Glycoproteome coverageb 1) Glycopeptides 2) Glycosites 3) Glycoproteins |
Main findings/novelty | Reference |
|---|---|---|---|---|
| Sera from PCa patients and healthy individuals (human) | 1) Q-Exactive 2) SCE-HCD (25 ± 10% NCE) 3) DDA 4) Mascot v2.4 5) Label-free (XICs) |
1) 3447(N) 2) 970(N) 3) 257(N) |
• Site-specific identification and quantification of the sialylated N-glycoproteome in sera from PCa patients and healthy donors | Bollineni et al. [51] |
| Breast cancer PANC1 and PKM2 knockout cell lines (human) | 1) Orbitrap Fusion Lumos Tribrid 2) EThcD (SA 33% NCE) 3) DDA 4) Byonic v2.9.38 in Proteome Discoverer v2.1 5) DiLeu |
1) 1067(N) 2) 311(N) 3) 205(N) |
• Identification of site-specific N-glycosylation differences in PKM2 knockout breast cancer and parental cells | Chen et al. [39] |
| Urine from patients with PCa and BPH (human) | 1) Q-Exactive HF 2) HCD (29% NCE) 3) DDA 4) Byonic v2.6.46 5) TMT |
1) 954(N)/965(O) 2) 1310(N) 3) 729(N)/160(O) |
• Identification of a panel of 56 N-glycopeptides able to discriminate PCa and BPH patients | Kawahara et al. [52] |
| Resting and activated primary T cells (human) | 1) LTQ-Orbitrap Elite 2) HCD (27 eV)-pd-ETD (SA 35 eV) and inclusion list-based HCD (27 eV)/ETD (SA 35 eV)/CID (35 eV) 3) DDA 4) Byonic v2.0 in Proteome Discoverer v1.4 5) Label-free (XIC) |
1) 2219(O) 2) 851(O) 3) 1045(O) |
• Identification and subcellular localisation of O-GlcNAcylated proteins expressed by T cells • Found changes in the site-specific O-GlcNAcylation during T cell activation |
Woo et al.[53] |
| Tumour and adjacent control tissues from patients with kidney cancer, T cells and sera (human) | 1) Q-Exactive HF and Orbitrap Fusion Lumos Tribrid 2) ETD/HCD (36% NCE) 3) DDA 4) Sequestc in Proteome Discoverer v2.2. 5) Label-free (spectral counting) |
1) 4827(O) 2) 3055(O) 3) 1060(O) |
• Specific enrichment of O-glycopeptides in complex clinical samples using OpeRATOR • Identified site-specific O-glycosylation differences between kidney tumour and adjacent tissues |
Yang et al. [40] |
| Serum from patients with (non-) crescentic IgA nephropathy and healthy donors (human) | 1) Q-Exactive HF and Orbitrap Fusion Lumos Tribrid 2) HCD, EThcD 3) DDA 4) Byonic v2.10.21 5) Dimethyl labelling |
1) 499(O) 2) 173(O) 3) 49(O) |
• Identification of site-specific changes in O-glycosylation associated with crescentic IgA nephropathy | Zhang et al. [54] |
| Breast cancer MCF-7 cell line and matching MCF-7 stem cells (human) | 1) Q-Exactive 2) SCE-HCD (30 ± 10% NCE) 3) DDA 4) GPSeekerc 5) Diethyl labelling |
1) 2558(N) 2) 727(N) 3) 640(N) |
• Identified differences in the site-specific N-glycosylation between cancer cells and cancer stem cells | Wang et al. [55] |
| HepG2 liver cancer cell line and LO2 normal liver cell line (human) | 1) Q-Exactive 2) SCE-HCD (30 ± 10% NCE) 3) DDA 4) GPSeekerc 5) Dimethyl labelling |
1) 5405(N) 2) 1218(N) 3) 1077(N) |
• Structure-focused glycoproteomics study revealing site-specific N-glycan differences in HepG2 and LO2 | Xiao and Tian [56] |
| Serum from healthy donors (human) | 1) Orbitrap Fusion Tribrid 2-3) ETD (SA 25% NCE)/SCE-HCD (27 ± 5% NCE)-DDA; SCE-HCD (27 ± 5% NCE)-DIA 4) Sequest TH, ptmRS in Proteome Discoverer v1.4, v2.2 and Spectronaut v11.0 with a large O-glycopeptide spectral library 5) Label-free (XIC) |
1) 269(O) 2) 253(O) 3) 159(O) |
• Amongst the first DIA-based glycoproteomics studies • Deep profiling of the serum O-glycoproteome without glycopeptide enrichment |
Ye et al. [57] |
| Urinary exosomes from male donors (human) | 1) Orbitrap Fusion Lumos Tribrid 2) SCE-HCD (35 ± 5% NCE)-pd-EThcD (SA 15% NCE) 3) DDA 4) Protein Prospector v5.22.1, pGlyco v2.0 5) Label-free (XIC) |
1) 3144(N) 2) 604(N) 3) 378(N) |
• N-glycoproteome and site-specific micro-heterogeneity of human urinary exosomes | Brown et al. [42] |
| DG75 Burkitt's lymphoma cells (human) | 1) Orbitrap Fusion and Lumos Tribrid 2) HCD (25% for MS2, 35% NCE for MS3) 3) DDA 4) pGlyco v2.0 5) TMT |
1) 5367(N) 2) 855(N) 3) 528(N) |
• Optimised strategy for quantitative site-specific glycopeptide analysis • Site-specific glycosylation changes upon inhibition of the fucosylation process |
Fang et al. [58] |
| Tumour and control tissues from high-grade serous ovarian carcinoma patients (human) | 1) Q-Exactive and Orbitrap Fusion Lumos Tribrid 2) HCD (38% NCE) 3) DDA 4) GPQuest v2.1 5) TMT |
1) 3202(N) 2) 1690(N) 3) Not reported |
• Differences in site-specific N-glycosylation and abundance of glycosylation enzymes in tumour and control tissues | Hu et al. [59] |
| PCa and BPH tissues (human) | 1) Q Exactive HF-X 2) HCD (35% NCE) 3) DDA 4) Byonic v2.6.46 5) TMT |
1) 7447(N)/522(O) 2) 1085(N)/308(O) 3) 540(N)/178(O) |
• The use of integrated N- and O-glycome and glycoproteome profiling revealed key glycan changes associated with PCa progression | Kawahara et al. [43] |
| Oncogene-transformed MCF10A breast epithelial cells (human) | 1) Orbitrap Fusion Lumos Tribrid 2) HCD (30% NCE)-pd-AI-ETD 3) DDA 4) Byonicc 5) Label-free (XIC) |
1) 2459(N) 2) 785(N) 3) 480(N) |
• Oncogene-induced remodelling of the surface glycoproteome and the global cellular glycoproteome | Leung et al. [60] |
| Urine from healthy donors (human) | 1) Q-Exactive 2) SCE-HCD (30 ± 10% NCE) 3) DDA 4) GPSeekerc 5) Label-free (spectral counting) |
1) 2986(N) 2) 419(N) 3) 327(N) |
• Site-specific N-glycopeptide analysis of human urinary glycoproteome with putative N-glycan structural information | Shen et al. [61] |
| Serum from healthy donors (human) | 1) Q-Exactive 2) HCD (33% NCE) 3) DDA 4) pMatchGlyco v1.2 (spectral library of de-N-glycopeptides) 5) Label-free (XIC) |
1) 22 677(N) 2) 1036(N) 3) 526(N) |
• Site-specific N-glycosylation of serum glycoproteins with very high glycoproteome coverage | Shu et al.[50] |
| ADR-resistant MCF-7/ADR cell line and matching MCF-7/ADR cancer stem cell line (human) | 1) Q-Exactive 2) SCE-HCD (30 ± 10% NCE) 3) DDA 4) GPSeekerc 5) Diethyl labelling |
1) 4016(N) 2) 1102(N) 3) 1014(N) |
• Differences in site-specific N-glycosylation between cancer cells and cancer stem cells | Xu et al. [62] |
| Plasma from papillary thyroid carcinoma patients and healthy donors (human) | 1) Orbitrap Fusion Lumos Tribrid 2) SCE-HCD (30 ± 10% NCE) 3) DDA 4) pGlyco v2.2.0 5) Qualitative |
1) 1644(N) 2) 862(N) 3) 369(N) |
• Profiling of the medium- and low-abundant plasma glycoproteome (reported circulating glycoproteins down to 0.5 ng/ml levels) | Zhang et al. [63] |
| Liver tumour and para-cancerous tissues from HCC patients (human) | 1) Orbitrap Fusion Lumos Tribrid 2) HCD (37% NCE) 3) DDA 4) GPQuest v2.0 5) TMT |
1) 4741(N) 2) 1184(N) 3) 894(N) |
• Differences in site-specific N-glycosylation in HCC patients with low and high levels of α-fetoprotein | Zhao et al. [64] |
| Urine from healthy male and female donors (human) | 1) Q-Exactive 2) HCD (30% NCE) 3) DDA 4) Byonicc 5) Label-free (XIC) |
1) 1300(O) 2) Not reported 3) 435(O) |
• Gender differences identified in the urine O-glycoproteome | Zhao et al. [65] |
| Milk from healthy donor (human) | 1) Orbitrap Fusion Lumos Tribrid 2) HCD (30% NCE)-pd-EThcD (SA 27% NCE) 3) DDA, SIM, PRM (30% NCE) 4) Byonic v3.2.0 in Proteome Discoverer v2.2 5) Label-free (XIC) |
1) 1697(N) 2) 191(N) 3) 110(N) |
• Dynamics in the site-specific N-glycosylation over the lactation period | Zhu et al. [66] |
| CHO-K1 subclone (mouse) | 1) Q-Exactive 2) HCD (32% NCE) 3) DDA 4) GPQuest v2.0 5) Label-free (spectral counting) |
1) 10 338(N) 2) 1162(N) 3) 530(N) |
• Macro- and micro-heterogeneity of the N-glycoproteome of the lysate and culture media of CHO-K1 cells | Yang et al. [49] |
| Brain tissue (mouse) | 1) Orbitrap Fusion Lumos Tribrid 2) HCD (28% NCE)-pd-AI-ETD 3) DDA 4) Byonicc 5) Qualitative |
1) 5662(N) 2) 1545(N) 3) 771(N) |
• Application of AI-ETD to complex N-glycopeptide mixtures • Site-specific identification of isomeric glycans using oxonium ions • Creative visualisation of glycoproteomics data |
Riley et al. [41] |
| L6 myoblasts cell line and myotubes (rat) | 1) Orbitrap Fusion Tribrid 2) HCD (40% NCE)-pd-EThcD/CID (35% NCE) 3) DDA 4) Byonicc in Proteome Discoverer v2.1.0.801 5) TMT |
1) 2751(N) 2) Not reported 3) Not reported |
• Longitudinal monitoring of site-specific N-glycosylation during myogenesis and muscle development | Blazev et al. [67] |
| Brain tissue from AD APP/PS1 transgenic and wild-type models (mouse) | 1) Triple-TOF 4600 and Orbitrap Fusion Tribrid 2) SCE-HCD (30 ± 10% NCE) 3) DDA 4) pGlyco v2.0 5) Label-free (n/a) |
1) 3524(N) 2) 1493(N) 3) 722(N) |
• Differences in site-specific N-glycosylation in brain tissue from AD and wild type mice | Fang et al. [68] |
| Heart tissue (mouse) | 1) Orbitrap Fusion Lumos Tribrid 2) SCE-HCD (20% and 33% NCE) 3) DDA 4) GPQuestc 5) Label-free (XIC) |
1) 2925(N) 2) 1016(N) 3) 864(N) |
• Site-specific changes in N-glycosylation during the loss of regenerative capacity of neonatal mouse hearts early in life | Li et al. [69] |
| CHO-K1 cells with FUT8 knockout and wild type (mouse) | 1) Q-Exactive and Orbitrap Fusion Lumos Tribrid 2) HCD (32% NCE) 3) DDA 4) GPQuest v2.0 5) Label-free (spectral counting) |
1) 2634(N) 2) 459(N) 3) 243(N) |
• Global alteration in the N-glycosylation including the lack of core fucosylation in CHO-K1 cells upon FUT8 disruption | Yang et al. [70] |
| Arabidopsis thaliana microsomes from wild-type and β-1,2-xylosyltransferase mutant (plant) | 1) Orbitrap Fusion Lumos Tribrid 2) HCD (30% NCE)-pd-ETD or ETD 3) DDA 4) Byonic v2.6 in Proteome Discoverer v2.1 5) Label-free (XIC) |
1) 1110(N) 2) 492(N) 3) 324(N) |
• Mapping of the structural heterogeneity of N-glycosylation in Arabidopsis • Absence of Xyl in xylt mutant plants |
Zeng et al. [71] |
The table provides information of the biological samples, experimental conditions, the glycoproteome coverage and key findings of each study plotted in Figure 1.
As stated in the respective papers;
The glycoproteome coverage is here indicated as the reported unique (non-redundant) intact glycopeptides, glycosylation sites and glycoproteins. (N) refers to N-glycosylation and (O) refers to O-glycosylation;
Software version not reported.
With the exception of a single study exploring the plant N-glycoproteome [71], these recent glycoproteomics papers all investigated tissues, bodily fluids (e.g. sera) or cell lines of human origin or from rodent model systems derived under different (patho)physiological conditions. This illustrates that glycoproteomics currently is used to predominantly address glycobiological questions related to human health and disease.
The published studies either used glycoproteomics to qualitatively or quantitatively map a particular glycoproteome of interest, e.g. the serum O-glycoproteome [57] or alternatively performed comparative glycoproteomics to elucidate the site-specific N- or O-glycosylation changes between two or more patient groups e.g. patients with prostate cancer (PCa) and benign prostatic hyperplasia [52] or over a period of interest, e.g. longitudinal profiling during lactation [66] or neonatal heart development [69].
In the surveyed studies, data were most frequently collected using data-dependent acquisition (DDA) by employing higher-energy collision dissociation (HCD-), stepped collision energy (SCE)-HCD-, electron-transfer dissociation (ETD)- and/or electron-transfer/higher-energy collision dissociation (EThcD-) MS/MS acquisition strategies (discussed below). Our survey also illustrated that targeted re-isolation and orthogonal fragmentation of glycopeptide ions are increasingly performed using diagnostic oxonium ions arising from HCD-MS/MS commonly referred to as product-dependent (pd) acquisition [60,66,67,71]. Data-independent acquisition (DIA), albeit less common, has also been used to profile mixtures of glycopeptides [57,72–76]. Various search engines, most commonly the commercially available Byonic [77,78], and academic tools, amongst others, GPQuest [79] and GPSeeker [56], were used to identify glycopeptides from the obtained raw data.
Regarding the sample preparation, most studies used trypsin digestion or, less commonly, combinations of proteases e.g. trypsin and Glu-C, Lys-C or chymotrypsin, and employed some form of glycoprotein or glycopeptide enrichment prior to the LC–MS/MS detection. In fact, glycoproteomics is rarely applied directly to complex biological samples without any type of enrichment [57]. Hydrophilic interaction liquid chromatography (HILIC), lectin affinity chromatography e.g. concanavalin A (ConA), Vicia villosa agglutinin (VVA), Jacalin, wheat germ agglutinin (WGA), Ricinus communis agglutinin (RCA), and less common strategies including mixed anion-exchange chromatography, TiO2, and chemical and chemoenzymatic methods, were used to enrich glycopeptides. HILIC-based solid phase extraction (SPE) represents one of the least biased glycopeptide enrichment methods given the considerable (local) hydrophilicity shared by most N-glycopeptides [80], but may not quantitatively capture the less hydrophilic (shorter) O-glycopeptides and truncated N-glycopeptides [81–83]. To increase the glycoproteome coverage, some studies used multi-step enrichment protocols including sequential glycoprotein enrichment using Jacalin affinity chromatography followed by glycopeptide enrichment using HILIC [54] or employed sequential glycopeptide enrichment using HILIC and multi-lectin affinity (ConA, WGA and RCA 120) [39], while others applied parallel glycopeptide enrichment using ConA and mixed mode strong anion exchange-HILIC [60], or employed glycopeptide prefractionation using high-pH reversed-phase LC after the glycopeptide enrichment step [42].
The enriched glycopeptides were, as expected, most commonly separated and detected using reversed-phase nano-scale LC–MS/MS on various Orbitrap platforms including the Q-Exactive or Tribrid systems. Only few studies used other online separation techniques including HILIC-LC, which reportedly was able to separate sialoglycopeptide isomers in complex mixtures [56]. Although frequently used in proteomics, we did not identify any recent glycoproteomics studies using Q-ToF instrumentation or indeed the more recent timsToF for large-scale glycopeptide detection.
While the pre-2016 glycoproteomics studies were found to be rather qualitative in their approaches by simply reporting lists of identified glycopeptides as reviewed [22], our survey of the post-2018 glycoproteomics papers shows that glycopeptide quantification is now commonly applied. Approaches include label-free quantification by XIC or spectral counting [50,65], isotopic dimethyl [54] or diethyl [55] labelling as well as TMT [52,64] and DiLeu [39] labelling, which collectively enable different types of glycopeptide quantitation strategies all regularly used in the field. While glycoproteome mapping studies typically quantify the relative abundance of all glycoforms observed at each glycosylation site to establish a stoichiometric site-specific glycoprofile, comparative glycoproteomics studies often compare the relative quantity of individual glycopeptides or the glycoprofile at each site between conditions. The glycosylation site occupancy levels are unfortunately still rarely reported in glycoproteomics given the loss of this information in studies using glycopeptide enrichment.
In addition to the above-mentioned glycoproteomics studies performed on glycoprotein samples from unaltered biological sources, the SimpleCell technology, which manipulates cells to homogenously express truncated O-GalNAc glycans (Tn and sialyl-Tn (sTn)) on mucins and mucin-like domains due to the inactivation of the chaperone COSMC, represents an important innovation relevant for glycoproteomics [84–88]. The SimpleCell technology has not only opened exciting avenues to study the challenging mucin glycobiology including how O-glycans impact proteolytic cleavage events, the substrate-specificity of GalNAc-transferases and the role of O-GalNAc glycans in cell–cell interactions and cell differentiation [89–93], but has also improved our ability to profile the critically underexplored O-glycoproteome. Glycoproteomics efforts using the SimpleCell technology to study mucin-type glycosylation in different human and other mammalian cell lines have led to an impressive database (http://www.glycoproteomics.somee.com/), comprising a total of 18 397 unique O-glycopeptides spanning 10 197 O-glycan sites from 2039 O-glycoproteins within the human O-glycoproteome supported by glycopeptide MS/MS evidence.
Innovations in structure-focused glycoproteomics
The following section summarises recent innovations within the emerging field of structure-focused glycoproteomics including improvements in the sample preparation, glycopeptide separation, and LC–MS/MS-based detection. The discussed innovations are summarised in Figure 2. Advances in glycopeptide data analysis and novel search strategies including, amongst several others, MSFragger-Glyco and O-Pair Search [94,95] will not be covered in this review. Readers are instead referred to recent review literature on this topic [96,97].
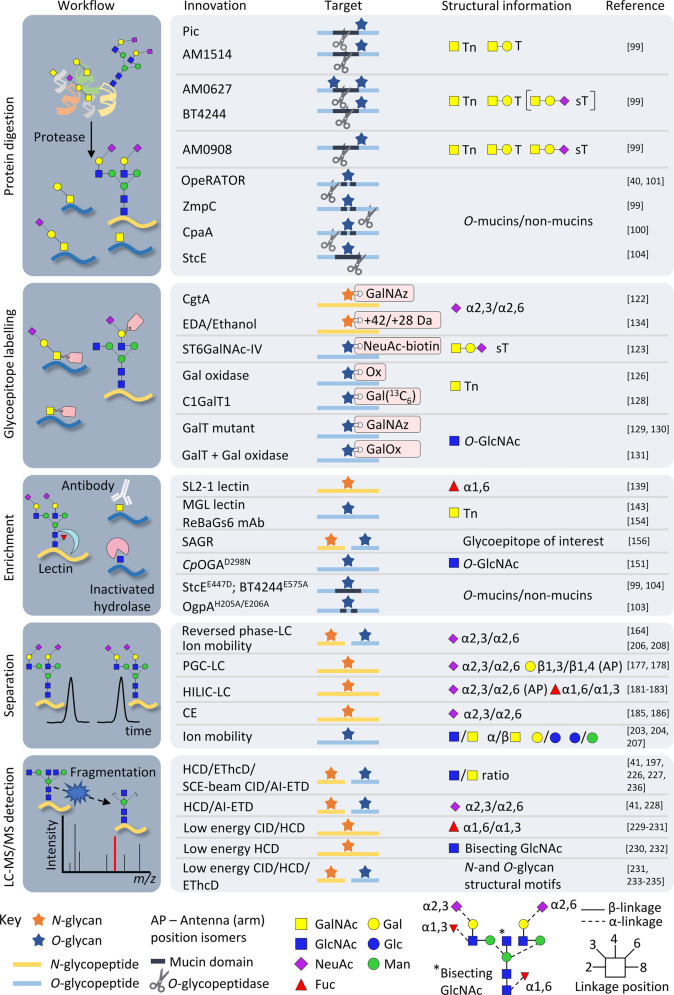
Figure 2. Technical innovations supporting the advancement of structure-focused glycoproteomics.
The advances and the structural information they provide have been separated into sample preparation including protein digestion, glycoepitope labelling and enrichment, online separation and LC–MS/MS detection of intact N-glycopeptides (yellow) and O-glycopeptides (blue). See key for symbol and linkage nomenclature [105].
Innovations in sample preparation
New glycoproteomics-friendly proteases
Bacterial proteases capable of cleaving mucin-type O-glycan motifs and thereby aiding the profiling of densely glycosylated mucin domains and non-mucin O-glycoproteins often left undigested with conventional proteases are rapidly emerging. The commercial OpeRATOR originally derived from the mucin-degrading Akkermansia muciniphila, and the secreted protease of C1 esterase inhibitor (StcE) from Escherichia coli represent prominent examples of recently reported mucinases [98–100].
OpeRATOR cleaves N-terminal to O-glycosylated Ser/Thr in Pro-rich sequences thereby generating peptides carrying N-terminal O-glycosylation for easy site assignment [40,101]. However, site assignment based on the OpeRATOR cleavage pattern alone or with HCD-MS/MS is not ideal given that more than half of all OpeRATOR-generated glycopeptides reportedly contain multiple O-glycans [46] including adjacent O-glycan sites prone to be missed by OpeRATOR [94]. In contrast, EThcD-MS/MS provides, in favourable cases, unambiguous site identification of OpeRATOR-generated O-glycopeptides [46]. Studies exploring the substrate requirements of OpeRATOR indicated that it 1) does not cleave non-glycosylated Ser/Thr [101], 2) preferentially cleaves core 1-type O-glycans and, albeit at a much slower rate, core 3-type O-glycans [40,102,103], and 3) shows poor or no activity against sialylated O-glycans and core 2-type structures [103].
The mucin-selective StcE cleaves the Xxx-Ser/Thr peptide bonds in consensus sequences containing Ser/Thr (P2) — Xxx (P1) — Ser/Thr (P1′) motifs where Xxx is any amino acid residue [104]. Unlike P1 and P1′, glycosylation of Ser/Thr in the P2 position is a strict requirement for StcE activity [104]. StcE displays activity towards motifs carrying various O-glycans including the di-sialyl-T antigens (di-sT) and core 2-type glycopeptide structures and thus complements OpeRATOR as a tool for mucin glycoprofiling.
Other bacterial serine and zinc metalloproteinases have also expanded the toolkit for O-glycoproteome analysis [99]. Pic from E. coli and BT4244 from Bacteroides thetaiotaomicron preferentially cleave N-terminal to glycosylated Ser/Thr (P1′) carrying truncated Tn/T or Tn/T/sT O-glycans, respectively. In contrast, the M60-like protease AM0627 from A. muciniphila preferentially cleaves peptide bonds between two adjacent O-glycosylated Ser/Thr residues [99]. Finally, AM0908 and AM1514 are also M60-like proteases expressed by A. muciniphila that were found to cleave N-terminal to glycosylated Ser/Thr (P1′) with a preference for unoccupied Ser/Thr motifs in the P1 position [99].
While the above mucinases specifically act on glycoproteins containing mucin domains, ZmpC from Streptococcus pneumoniae, which reportedly cleaves peptide bonds in the vicinity of O-glycosylation sites, acts on both mucin and non-mucin proteins [99]. The O-glycopeptidase CpaA expressed by several Acinetobacter strains cleaves N-terminal to glycosylated Ser/Thr in Pro (P1) — Ser/Thr (P1′) motifs irrespective of the sialylation status of the P1′-localised O-glycan substrate on both mucin and non-mucin O-glycoproteins [100].
While the clear substrate preference of most of the newly discovered proteases including OpeRATOR and StcE limits their use for global (unbiased) glycoproteome profiling, they have already proven valuable in studies aiming to map particular subsets of the glycoproteome. Notably, none of the proteases appears to be fully specific for a single glycan structure or glycoepitope emphasizing the importance of downstream glycan characterisation if used for structure-specific glycoproteomics workflows. Collectively, these new proteases have a considerable potential of advancing the field of glycoproteomics; their advent promises to accelerate particularly the analysis of the under-studied O-glycoproteome in the immediate future.
Glycoepitope labelling
Chemoenzymatic and metabolic labelling methods enable visualisation and detection of glycoepitopes of interest in a cellular or tissue setting. These technologies that have been developed over the past two decades are gaining popularity and have a considerable potential for advancing structure-focused glycoproteomics [106]. While metabolic labelling approaches aim to metabolically incorporate functional and easy-to-detect tags into newly synthesised glycoproteins (or other glycoconjugates) in living cellular systems, chemoenzymatic labelling approaches focus on incorporating such probes after glycoprotein formation without making use of the metabolic pathways [106,107].
Recognised limitations of metabolic labelling include the potential of adversely perturbating the living system being explored in particular if artificial metabolic precursors are supplemented at relatively high (micromolar) concentrations [108]. Furthermore, incomplete and unspecific incorporation of the supplemented probes are also commonly experienced [109,110] including the off-target labelling of cysteine residues of some metabolic labelling reagents [111–113]. However, a more specific labelling approach designed to target O-GalNAcylated proteins was recently reported [114]. In that study, precision labelling of O-GalNAcylated proteins was performed by supplementing membrane-permissive caged N-(S)-azidopropionylgalactosamine (GalNAzMe)-1-phosphate probes to cells transfected with a pyrophosphorylase AGX1 mutant in order to biosynthesize uridine diphosphate (UDP)-GalNAzMe donor molecules for downstream GalNAzMe-based labelling of target O-glycoproteins. UDP-GalNAzMe-mediated labelling of O-GalNAc proteins can be further enhanced by transfecting cells with an engineered ‘bump-and-hole' GalNAc-T2 double mutant that contains an enlarged active site and exhibits increased activity for UDP-GalNAzMe as compared with UDP-GalNAc [114–117].
Diverse chemoenzymatic labelling methods that employ various glycosyltransferases and unnatural monosaccharide analogues have been developed over the past decade [107,118–121]. For example, the bacterial β-(1,4)-N-acetylgalactosaminyltransferase CgtA from Campylobacter jejuni that targets NeuAcα2,3Gal-R substrates has been used to label HEK293T cell surface N- and O-glycoproteins with N-azidoacetylgalactosamine (GalNAz), an unnatural analogue of GalNAc [120]. GalNAz incorporation enables convenient click chemistry-driven biotinylation, which, in turn, facilitates the enrichment of labelled targets using streptavidin agarose or detection by western blot, fluorescence microscopy, or LC–MS/MS. Recently, CgtA was used to label NeuAcα2,3Gal-containing glycopeptides derived from human serum glycoproteins with GalNAz [122]. The informative GalNAz reporter ions at m/z 245.09 generated upon SCE-HCD-MS/MS enabled the detection and sialyl-linkage annotation of 1236 N-glycopeptides from 161 human serum glycoproteins. In another study, cell surface-located sT O-glycoproteins expressed by the MCF7 breast cancer and HT29 colon cancer cell lines were labelled, captured and detected using cytidine-5′-monophosphate-NeuAc-biotin and recombinant human ST6GalNAc-IV expressed in baculovirus insect cells [123].
Other approaches have specifically targeted the disease-relevant O-GalNAc glycoepitope (Tn antigen) [124,125]. For example, a study showed that α-linked GalNAc (Tn) and β-linked GlcNAc O-glycopeptides can be discriminated by selective oxidation of the O-GalNAc residues by galactose oxidase from Dactylium dendroides facilitating Tn glycopeptide enrichment using hydrazide solid-phase capture and methoxyamine (CH3ONH2) mediated release [126]. This selective labelling strategy adds a 27 Da mass tag (CH3-O-N=R with an associated water loss (-H2O) upon derivatisation) to all Tn glycopeptides, which, in turn, enables their detection using diagnostic oxonium ions at m/z 171.0765 (144.0656 + 27.0109) and m/z 195.0764 (168.0655 + 27.0109) upon HCD-MS/MS. A similar oxidation-based labelling approach was recently used to enrich O-glycopeptides carrying core 1-type O-glycans including Tn, sTn, T, sT, and di-sT antigens [127]. In that enrichment study, which also employed hydrazide capture and methoxyamine release, tryptic glycopeptides derived from human serum glycoproteins were treated with PNGase F and chemically desialylated, before the exposed Gal and GalNAc residues were oxidised by D. dendroides galactose oxidase.
OpeRATOR was also recently used for the digestion and release of resin-conjugated labelled Tn O-glycoproteins expressed by human Jurkat cells and in pancreatic cancer sera using a method called EXoO-Tn [128]. In short, the EXoO-Tn method involves: (i) trypsinization and desialylation of the extracted proteins, (ii) Tn glycopeptide enrichment using VVA lectin, (iii) isotopic labelling of the Tn glycopeptides with heavy Gal(13C6) by the recombinant human C1GalT1 glycosyltransferase in complex with its chaperone C1GalT1C1 using an artificial UDP-Gal(13C6) nucleotide sugar donor, (iv) digestion by OpeRATOR, and finally v) LC–MS/MS-based detection of the Gal(13C6)-capped Tn glycopeptides using diagnostic oxonium ions at m/z 372.2 (13C6 variant of HexHexNAc) that can discriminate labelled Tn glycopeptides from endogenous T glycopeptides. This approach enabled the identification of 1011 Tn glycopeptides from 480 glycoproteins expressed by Jurkat cells while considerably fewer Tn glycoproteins were identified from pancreatic cancer sera.
Furthermore, selective isotopic labelling and quantification of O-GlcNAcylated glycoproteins was recently achieved by employing UDP-GalNAz, a recombinant mutant of bovine β1,4-galactosyltransferase 1 (GalT1Y289L) and the isotope-tagged cleavable linker (isoTCL) [129] or an isotope-coded photocleavable probe [130]. Both strategies facilitate selective tagging, enrichment, and isotopic labelling of O-GlcNAcylated glycoproteins. Others have used wild-type bovine GalT1 and natural UDP-Gal to label β-linked GlcNAc O-glycopeptides derived from glycoproteins expressed by MCF-7 cells, which were then selectively oxidised by D. dendroides galactose oxidase ahead of enrichment by hydrazide capture [131]. The resulting O-GlcNAcGal glycopeptides could be distinguished from the endogenous O-GalNAcGal glycopeptides by their different oxonium ion ratios (discussed below).
Finally, sialyl linkage information, important to understand many processes related to sialoglycobiology [132,133], can be obtained at the glycoproteome-wide level by applying linkage-selective labelling approaches. For example, α2,6-linked sialic acid residues can be selectively labelled using ethanol creating a 28 Da mass tag by the esterification with C2H5OH (and an associated water loss) while α2,3-sialic acid residues can be selectively labelled using ethylenediamine generating a 42 Da mass tag by the amidation with C2H4(NH2)2 (and an associated water loss) of immobilised glycoproteins [134]. In that study, the derivatised sialoglycoproteins were characterised using on-bead digestion and HILIC-SPE glycopeptide enrichment before the labelled sialoglycopeptide isomers were detected by the chemically unique oxonium ions arising from the derivatised α2,3-(m/z 334.1/316.1) and α2,6-linked (m/z 302.1) sialoglycopeptides upon HCD-MS/MS.
Enrichment strategies
The enrichment methods used in glycoproteomics have been comprehensively reviewed [135]. While some enrichment strategies including HILIC-SPE aim to capture the entire complement of glycopeptides to provide a quantitative view of the glycoproteome [80], structure-specific enrichment approaches aim to capture only a subset of glycopeptides carrying specific glycan structures. In the latter cases, information of the glycan structure can either be inferred from the capture, or, more appropriately if used for structure-focused glycoproteomics, be supported by direct mass spectral evidence of the captured glycopeptides/glycoproteins using downstream detection.
Natural and genetically engineered lectins form a diverse class of glycan-binding proteins that can be used to enrich subsets of glycoepitopes carried by glycoproteins, glycopeptides or other glycoconjugates. For example, BTL from Bryothamnion triquetrum [136], PhoSL from Pholiota squarrosa [137,138] and SL2-1 [139] from Streptomyces rapamycinicus are known to recognise α1,6- (core) fucosylated N-glycans. Thus, binding to any of these lectins indicates that the captured glycoproteins carry core fucosylation, an observation that ideally should be supported by MS or other types of orthogonal evidence. Lectins that recognise other N-glycan structural features include Phaseolus vulgaris erythroagglutinin (PHA-E) and Calystegia sepium lectin (Calsepa) that bind bisecting β1,4-GlcNAc structures, Maackia amurensis lectin (MAL) that recognises α2,3-sialic acid residues and Sambucus nigra agglutinin (SNA) and Sambucus sieboldiana agglutinin (SSA) that both bind α2,6-sialic acid residues [140–142]. Furthermore, the recombinant human macrophage galactose-like C-type lectin (MGL)-Fc chimeric protein was recently used to enrich Tn glycopeptides from ovarian cancer cells and tissues [143]. Finally, engineered lectins displaying enhanced or tailored recognition of particular glycan epitopes including, amongst others, core fucose recognised by the recombinant N224Q mutant of the Aleuria aurantia lectin (AAL), and O-GlcNAc recognised by the Agrocybe aegerita lectin (AANL) mutant AANL6 have the potential of expanding the enrichment tool box for structure-focused glycoproteomics [144–149].
Catalytically inactive hydrolases have also proven valuable for structure-specific glycopeptide enrichment. Recently, a mutant form of the Clostridium perfringens O-GlcNAcase (CpOGAD298N), which retained the binding but not the hydrolytic properties towards O-GlcNAc epitopes [150], was used to enrich β-linked GlcNAcylated O-glycoproteins from HeLa cell lysates and Drosophila embryo lysates [151]. Furthermore, the commercially available catalytically inactive mutant form of the OpeRATOR OgpAH205A/E206A (GlycOCATCH, Genovis AB., Sweden) was shown to bind O-glycopeptides suggesting that this pseudo-protease may serve as another enrichment tool for mucin-type glycosylation [103]. The catalytically-inactive StcEE447D conjugated to beads was also able to enrich MUC16 from an OVCAR-3 cell line [104]. Both the StcEE447D mutant and a BT4244E575A mutant form of the B. thetaiotaomicron BT4244 metalloproteinase (discussed above) were found to be valuable for the detection of mucins using western blotting and for their direct visualisation in tissues [99].
Furthermore, recombinant antibodies displaying affinity towards glycoepitopes are relevant tools for structure-focused glycoproteomics [152–157]. For example, the recombinant mouse anti-Tn IgM ReBaGs6 was used to immunoprecipitate Tn-containing glycoproteins from Tn-positive Colo205 SimpleCells lysates for downstream mass spectrometry identification [154]. Smart anti-glycan reagents (SAGRs) represent another exciting type of emerging affinity tools [156,157]. SAGRs are chimeric proteins comprising murine IgG Fc and the single-chain variable lymphocyte receptors (VLRs) secreted by Petromyzon marinus VLRB+ lymphocytes (B-like cell) upon immunisation with different glycoconjugates, cells and tissues of interest. This innovative technology enables SAGRs to recognise glycan determinants with considerable affinity. Finally, the emergence of artificial glycan-directed antibodies generated by glycopeptide imprinting represents a new interesting avenue that may contribute to structure-focused glycoproteomics in the future [158,159].
Improvements in glycopeptide separation
Glycopeptide separation prior to mass spectrometry detection, commonly performed using reversed-phase nano-scale LC, is an important component of most, if not all, glycoproteomics experiments. Current strategies for the isomeric separation of glycopeptides were recently reviewed [160].
The glycopeptide retention behaviour on reversed-phase LC columns, which is governed primarily by the peptide hydrophobicity, and, secondly, by the properties of the conjugated glycan(s) i.e. size, shape and monosaccharide composition particularly the sialic acid content, was recently studied to generate better prediction tools utilising retention time as orthogonal information for glycopeptide identification and characterisation [161–163]. While typically not allowing for separation of glycopeptide isomers, a recent study found that reversed-phase LC may separate isomeric N- and O-glycopeptides at elevated temperatures [164].
Whilst commonly used for glycomics [165,166], porous graphitised carbon (PGC) LC is still under-utilised in glycoproteomics despite its potential for isomeric separation of relatively small N- and O-glycopeptides generated by nonspecific proteolysis [167–170] or by trypsin [171]. Larger glycopeptides bind irreversibly to PGC LC columns [172,173]. Similar to the PGC-based separation of sialoglycans [174–176], PGC LC reportedly retains α2,3-sialoglycopeptides better than isomeric α2,6-sialoglycopeptides [177], a separation feature that appears to be temperature-dependent [178]. Alternatively, HILIC LC can be used for the separation of intact glycopeptide isomers [56,179] and glycoproteins [180]. HILIC LC coupled with multiple reaction monitoring was used to profile isomeric α2,3- and α2,6-sialoglycopeptides derived from human prostate-specific antigen [181], the latter isomer displaying a greater LC retention [182]. Others have used HILIC LC to separate topological N-glycopeptide isomers of human hemopexin in order to assign the position of the fucose residues on N-glycans (core versus outer arm) and to elucidate the glycan arm position of α2,6-sialic acid residues [183]. A study comparing different HILIC stationary phases recently reported that HALO penta-HILIC provides better isomeric separation of N-glycopeptides than the more widely used ZIC-HILIC phase [184].
Furthermore, isomeric α2,3- and α2,6-sialoglycopeptides possess different electrophoretic mobilities facilitating their separation using capillary electrophoresis (CE)-ESI-MS [185,186]. Recent improvements in the CE hyphenation methods have increased the sensitivity and reproducibility of glycopeptide analysis by CE-MS [187,188] and opened interesting avenues for CE-based glycoproteomics [189]. In fact, CE-MS even showed the potential to separate intact glycoproteoforms [190–193]. LC and CE methods for intact glycoprotein separation were recently reviewed [194].
Aiming to benefit from their orthogonal separation properties, studies employing multi-dimensional separation of glycopeptides are emerging. For example, an online two-dimensional PGC and reversed-phase LC setup was used to separate glycopeptides from simple mixtures [195–198]. Another study used offline reversed-phase LC prefractionation coupled with CE to profile N-glycopeptides generated from human α-1-acid glycoprotein [199]. The potential of multi-dimensional approaches to separate isomeric N-glycopeptides was demonstrated using offline HILIC LC coupled with PGC LC to fractionate a simple mixture of bovine RNase B glycopeptides [200] as well as offline reversed-phase LC prefractionation coupled with online HALO penta-HILIC LC to separate glycopeptides from HepG2 and LO2 cell lysates [56].
Finally, ion mobility spectrometry (IMS), a gas-phase alternative to the chromatographic and electrophoretic separation of glycopeptides, is an emerging separation technique in structure-focused glycoproteomics. When coupled with mass spectrometry, IMS-MS is evidently capable of separating isomeric glycopeptide ions based on their differential conformational size and shape resulting in different drift times [201,202]. Direct IMS-MS analysis showed the potential to separate α-GlcNAc and α-GalNAc-containing glycopeptide epimers [203,204], isomeric O- and N-glycopeptides differing only in the position of the glycosylation site [204–206], α- and β-GalNAc anomeric glycopeptides [204], and epimeric O-glycopeptides carrying either Gal or Glc residues or alternative Glc or Man residues on the same site [207]. The detection of CID-MS/MS-generated B3-type ions (m/z 657.24, corresponding to NeuAcα2,3Galβ1,4GlcNAc or NeuAcα2,6Galβ1,4GlcNAc fragments) by IMS-MS enabled the discrimination of isomeric N- and O-sialoglycopeptides; these two types of B3 ions display NeuAc linkage-specific drift times [206,208]. IMS-MS has also demonstrated a potential to separate intact glycoproteins [209], an observation that deserves to be explored further as the field moves towards top-down glycoprotein analysis.
Improvements in LC–MS/MS detection
Glycoproteomics has recently witnessed many key advances in mass spectrometry-based analysis of glycopeptides [25,32,34]. As discussed below, most advances relevant to structure-focused glycoproteomics involve improvements in the dissociation methods and intelligent glycopeptide-centric LC–MS/MS data acquisition methods, both of which have led to a greater volume and quality of the spectral data generated from glycoproteome profiling experiments.
Hybrid-type fragmentation methods yield high content glycopeptide spectra
Hybrid-type fragmentation methods that combine multiple dissociation methods include EThcD-MS/MS [210–213] widely used in glycoproteomics (see Table 1 for examples) and the less common AI-ETD-MS/MS method restricted to few specialist labs [41,60]. Fragmentation methods that employ multiple energy schemes to target more complete analyte dissociation most prominently the SCE-HCD-MS/MS method are also frequently used in large-scale glycopeptide profiling studies [37,51,56,214,215]. The benefits of hybrid-type fragmentation methods for glycoproteomics were recently summarised [216].
Confident and detailed glycopeptide characterisation from MS/MS spectral evidence rely on the presence of oxonium ions (internal glycan fragments), B-type ions (non-reducing end glycan fragments), Y-type ions (intact glycopeptide fragments), and b-/y- and c/z-type ions (peptide fragments) [217,218]. The peptide fragments may be present with and without the conjugated glycan moieties. These different ion types inform on different parts of the glycopeptide molecule. The SCE-HCD- and EThcD-MS/MS methods have been purpose-build and tailored to intact glycopeptide analysis and are now widely recognised to be well-suited for N- and O-glycopeptide analysis at scale [37,54,219]. Relative to the conventional dissociation methods including resonance activated (ion trap) or beam-type (Q-ToF) CID-, HCD- and ETD-MS/MS, the SCE-HCD- and EThcD-MS/MS fragmentation methods yield more informative and content-rich fragment mass spectra containing both peptide and glycan fragments required for unambiguous or more complete glycopeptide spectral assignment. Glycopeptide-friendly variants of the more common fragmentation methods available on most instruments have also been developed and include the low-energy beam-type CID- and HCD-MS/MS methods that generate spectra rich in oxonium and B-type ions useful in structure-focused glycopeptide analysis (discussed below).
Finally, ultraviolet photodissociation (UVPD)-MS/MS, which utilises high-energy laser photons for the activation and fragmentation of ions represents a less conventional fragmentation method of glycopeptides [220–222]. HCD-triggered-UVPD-MS/MS and EThcD-MS/MS were recently tested for their ability to locate O-GlcNAc sites on peptides from recombinant human and Drosophila melanogaster glycoproteins [223]. In that study, 193 nm UVPD-MS/MS showed a considerable potential for accurate site allocation by generating a range of O-GlcNAc-containing peptide backbone fragments (a-/x- and c-/z-type ions) and in fact outperformed EThcD-MS/MS with respect to the spectral quality and the sequence coverage of the identified glycopeptides [223]. In another study, high-energy UV (213 nm) combined with low-energy infrared photodissociation (10.6 µm) (HiLoPD) was applied to an O-GalNAc-peptide from human erythropoietin [224]. The authors demonstrated that HiLoPD showed a higher backbone ion retention efficiency of GalNAc than UVPD-MS/MS. Despite these promising reports, future experiments are required to evaluate the potential of UVPD-MS/MS for large-scale glycopeptide analysis.
Diagnostic ions for glycoform-specific assignment
Similar to the use of diagnostic ions to identify glycan fine structures in LC–MS/MS-based glycomics [175,176,225], diagnostic ions are important for the glycoform-specific assignment of glycopeptide spectra. Diagnostic ions have been used to discriminate isomeric glycopeptide features such as O-GlcNAc versus O-GalNAc glycopeptides [226,227], sialyl linkage of both N- and O-sialoglycopeptides [228], core versus antenna fucosylated N-glycopeptides [229–231], antenna versus bisecting GlcNAcylated N-glycopeptides [230,232], and glycopeptides containing various other N- and O-glycan motifs [231,233–235]. Some examples of diagnostic ions for the glycoform-specific assignment of glycopeptides are briefly discussed below.
The ratio between the signal intensities of two groups of two HexNAc-specific oxonium ions i.e. (m/z 138 + m/z 168) and (m/z 126 + m/z 144), referred as the GlcNAc/GalNAc ratio, generated using ion trap CID- or HCD-MS/MS, is reportedly able to discriminate between GlcNAc- and GalNAc-containing N- and O-glycopeptides [226,227]. In support, multiple glycoproteomics studies have applied this approach to distinguish extended α-linked GalNAc and β-linked GlcNAc O-glycopeptides [36,131,236,237], glycopeptides carrying either a single O-GlcNAc or O-GalNAc residue [238] or, more crudely, to simply classify O- and N-glycan types on peptides [41,197]. Furthermore, α2,3- and α2,6-sialyl linkage information was obtained by determining the signal intensity ratio of the HCD-MS/MS-derived oxonium ions specific for LacNAc (m/z 204 + m/z 366) and NeuAc (m/z 274 + m/z 292) following an elaborate signal normalisation process that considers both the glycan composition and the LacNAc structure (Galβ1,3GalNAc or Galβ1,4GlcNAc) [228].
Different diagnostic ion signal intensity ratios were also reported to inform on structural features of the glycan antenna. For example, core (α1,6-) and antenna (α1,2/3/4-) fucosylation can be differentiated based on the intensity ratios of B- or Y-type ions generated upon low-energy beam-type CID-MS/MS as demonstrated on glycopeptides derived from human prostate specific antigen and α-1-acid glycoprotein [229]. Moreover, diagnostic ions for HexNAc1Hex1Fuc1 (m/z 512) and HexNAc1Hex1Fuc1NeuAc1 (m/z 803) were reported to evidence antenna-localised fucosylation [230]. The authors noted that these diagnostic ions were also present, albeit at low levels, in spectra arising from core fucosylated glycopeptides [229,230]. Conversely, core fucosylation can reportedly be identified based on characteristic Y1-type ions (peptide + GlcNAc1Fuc1) generated at relatively high dissociation energies [229].
Furthermore, characteristic Y-type ions (i.e. peptide + HexNAc3Hex1 and peptide + HexNAc3Hex1Fuc1) generated via low-energy HCD-MS/MS can be used to indicate bisecting GlcNAc-containing glycopeptides as demonstrated for different mammalian model glycoproteins and glycoproteins from rat renal tissues [230,232]. B- and Y-type ions generated upon EThcD-MS/MS carried out with low supplemental energy were also found to be useful to establish the glycan topology as was shown for extended O-sialoglycopeptides obtained from human urine [234,235]. Diagnostic B-type ions generated by low-energy HCD-MS/MS were also found to reveal various structural motifs of the glycan antenna of glycopeptides derived from recombinant human glycoproteins and HEK293 secretomes including outer arm N,N-diacetyllactosamine (LacdiNAc), sialylated LacdiNAc, fucosylated LacdiNAc, sialylated GalNAc, and polyLacNAc amongst other motifs [231]. Furthermore, a Hex2HexNAc1 (m/z 528.19) to Hex1HexNAc1 (m/z 366.14) intensity ratio above 0.3 upon SCE-HCD-MS/MS was recently shown to indicate the presence of α-galactosyl epitopes (i.e. Galα1,3Galβ1,4GlcNAc-R) carried by complex-type N-glycopeptides as demonstrated for mice spleen and lung tissues and plasma [233]. The absence of a particular Y-type ion (peptide + HexNAc2Hex4) characteristic for hybrid- or oligomannosidic-type N-glycans represents complementary evidence of α-galactosylation. Finally, negative-mode detection in LC–MS/MS-based glycopeptide analysis may enhance the information of the fine structure of peptide-linked glycans by generating diverse glycan fragment ions upon ion trap CID-, HCD-MS/MS and CID-MS3 [239].
Lastly, albeit not the focus of this review, it is worthwhile to highlight just a few exciting advances of glycopeptide-search algorithms and machine learning tools that have opened interesting avenues for more efficient classification of N- and O-glycopeptides and their isomeric structural features [96,97]. Developments in such informatics areas have for example enabled the crude classification of N- and O-glycopeptides from human serum based on the intensities of various oxonium ions generated by HCD-MS/MS [240]. Machine learning-based classification was also applied to discriminate core- versus antenna-fucosylation using B- and Y-type ions generated by ion trap CID-MS/MS as demonstrated for N-glycopeptides from human plasma proteins [241,242]. Finally, a new computational algorithm named O-GlycoProteome Analyzer was shown to be able to identify mucin-type O-glycosylation (e.g. core 1 versus core 2) in human urine and plasma using the GlcNAc/GalNAc ratio (see above) and other B- and Y-type diagnostic ions generated by HCD-MS/MS [243].
Collectively, the above studies reporting on diverse improvements in the LC–MS/MS-based detection of glycopeptides illustrate the immense potential of using advanced fragmentation methods and diagnostic ions in innovative ways to elucidate glycan fine structural features at the glycopeptide level. It is clear that further optimisation of the dissociation methods including the applied energy setups, activation times and triggering mode(s) as well as comprehensive benchmarking and standardisation across instruments and informatics improvements are still required to enhance the utility of diagnostic ions for large-scale glycopeptide characterisation in future glycoproteomics experiments. Efforts to reduce the co-isolation of glycopeptide precursors and attempts to limit the in-source fragmentation of labile glycopeptide features, both recognised as significant analytical challenges in glycoproteomics [58,227,244,245], are examples of other important areas that should receive attention as the glycoproteomics methods develop.
Conclusion
This review has compiled and surveyed a considerable body of recent literature, some papers literally ‘hot-off-the-press', employing quantitative glycoproteomics to study the complexity and dynamics of the human and other mammalian glycoproteomes with unprecedented sensitivity and coverage. The increasing publication activity and quality in this area of science from an expanding group of research laboratories around the world is highly encouraging and indicates that the glycoproteomics technologies are reaching (and are actively being applied by) the community. As abundantly exemplified in Table 1, quantitative glycoproteomics is now sufficiently mature to tackle a range of complex biological questions from new angles. Human health and disease appear to be principle application areas of the recent glycoproteomics studies, but given the ubiquitous nature of glycoproteins, the technologies discussed herein can readily or with some adjustments be applied more widely to other biological areas of life science.
As the sample handling, instrumentation and bioinformatics solutions for glycopeptide analysis continue to evolve, it is expected that studies achieving an even greater glycoproteome coverage will soon emerge allowing us to access the low-to-medium abundance glycoproteins in complex biological specimens. The innovative use of new mucin-friendly proteases to generate O-glycopeptides with favourable MS properties is particularly exciting since such novel approaches will dramatically benefit studies seeking to improve the coverage of the hitherto ‘dark' mucin O-glycoproteome.
While such advances promise to reveal many of the secrets hidden in the low abundance glycoproteome, it is crucial that the technology improvements also eventually will open avenues for glycopeptide profiling directly from biological samples without the need for glycopeptide enrichment or other types of sample perturbation. Methods compatible with minimal sample handling and processing are required to generate workflows with higher sample throughput and reproducibility, and, importantly, to establish a fully quantitative view of the glycoproteome including information of the site occupancy currently not captured with most existing methods.
Moreover, ensuring high identification accuracy of the reported glycopeptides is imperative. Automated identification of intact glycopeptides is a comparably complex and highly error-prone process relative to the analyte identification in most other areas of proteomics [25,45,97]. Robust probability-based false discovery rate-driven search engines and algorithms for intact glycopeptides are still required to propel the field forward. Adding to the informatics advances needed to achieve such important improvements, ‘glycomics-assisted glycoproteomics' represents a complementary strategy that can guide the glycoproteomics data analysis to lower the glycopeptide false discovery rates while simultaneously addressing the complexity of the glycome [35,43,67,246,247].
Another significant limitation, indeed the focus of this review, is that current glycoproteomics methods still only allow assignment of generic monosaccharide compositions of glycans rather than the glycan fine structural features to discrete protein sites. It is widely recognised that knowledge of the glycan fine structures is a prerequisite to understand the effect of glycosylation on protein function. Technologies that can uncover the intricate details of the heterogenous glycan structures decorating each site of proteins in mixtures, herein referred to as ‘structure-focused glycoproteomics' have remained a holy grail of glycobiology [248].
As discussed in this review, the field has recently witnessed many exciting innovations supporting the advancement of structure-focused glycoproteomics. Importantly, these developments target different parts of the glycoproteomics workflow spanning sample preparation including new glycoform-selective proteases and chemoenzymatic labelling and enrichment methods, over innovations within the separation sciences involving both improved LC, CE and IMS approaches to improvements in diverse areas of mass spectrometry-based detection. The emerging O-glycan-specific mucinases and chemoenzymatic labelling approaches, and advances in the acquisition and fragmentation schemes yielding high content fragment mass spectra appear to dominate the innovation space and are likely to contribute significantly to glycoform-focused O- and N-glycoproteome profiling in the near future. We surprisingly did not identify any large-scale glycopeptide studies using sequential exoglycosidase treatment to obtain information of the terminal glycoepitopes, a strategy often used in glycomics [249], highlighting just one example serving to illustrate that all methodological avenues are still not explored in the context of structure-focused glycoproteomics.
Notwithstanding these exciting developments, structure-focused glycoproteomics remains in its infancy and the field still faces significant challenges ahead. Similar to the existing composition-focused glycopeptide mapping technologies, structure-focused glycoproteomics should ideally move towards unbiased profiling and quantitation of the glycoproteome. This is particularly important when the technology is applied in ‘discovery-mode' as opposed to targeted glycoproteomics studies where a specific glycopeptide form or a class of glycoepitopes are profiled to address a specific research question. Most innovations discussed herein including the developments of new labelling, digestion and enrichment methods target specific subsets of the glycoproteome and are thus incompatible with unbiased quantitative glycoproteome profiling. In contrast, LC–MS/MS methods that can elucidate subtle differences of all introduced glycopeptide forms either by improved separation and/or via mass spectral evidence may in our view have a stronger potential to facilitate fully quantitative yet still structure-centric glycoproteome profiling in the future.
Finally, it is clear that the field of structure-focused glycoproteomics should already now begin to prepare for a downstream integration with the other ‘omics disciplines including glycomics, transcriptomics, and metabolomics, while simultaneously paying close attention to and align with the exciting advances emerging in the area of top-down analysis of intact glycoproteins [250–254].
Glycoscientists have for years recognised that no single technology can provide a holistic and comprehensive understanding of the structural and functional heterogeneity inherently associated with protein glycosylation. While a full implementation of structure-focused glycoproteomics hold a tremendous potential to enhance our insight into pending glycobiological questions, it is clear that such technologies are but few amongst many tools that glycoscientists will need to draw on to explore the fascinating world of glycobiology in years ahead.
Perspectives
Importance: Glycoproteomics is a powerful quantitative technology that enables scientists to study the entire complement of glycoproteins expressed by cells, tissues or organisms at a given time, space and condition. Detailed site-specific characterisation of glycan structures in a glycoproteome-wide context, herein referred to as ‘structure-focused glycoproteomics’ is considered one of the next frontiers to be targeted to advance the field further and to address many pending questions in glycobiology.
Current understanding and challenges: Our survey has revealed a rapidly growing body of literature employing quantitative glycoproteomics to study the dynamics and complexity of the glycoproteome with unprecedented sensitivity and coverage demonstrating that the technology is maturing and ready to be applied to biology. Notably, many novel innovations spanning new chemoenzymatic labelling, digestion, enrichment, separation and mass spectrometry detection strategies compatible with structure-focused glycoproteomics have recently emerged revealing glycan fine structural details with protein site resolution. Challenges in the field include a high false discovery rate of glycopeptides reported in glycoproteomics papers, a lack of standardisation of the data collection, analysis, interpretation, reporting and sharing, and incomplete integration with other ‘omics disciplines (glycomics, transcriptomics..) and top-down MS approaches.
Future directions: Our review illustrates that structure-focused glycoproteomics holds a considerable potential to become a key tool in the emerging area of systems glycobiology and unlock secrets of the glycoproteome in years to come.
Abbreviations
- AANL
Agrocybe aegerita lectin
- AD
Alzheimer's disease
- ADR
Adriamycin
- AI-ETD
activated-ion electron transfer dissociation
- BPH
benign prostatic hyperplasia
- CE
capillary electrophoresis
- CID
collision-induced dissociation
- ConA
concanavalin A
- CSF
cerebrospinal fluid
- Da
Dalton
- DDA
data-dependent acquisition
- DIA
data-independent acquisition
- di-sT
di-sialyl-T antigen
- ETD
electron-transfer dissociation
- EThcD
electron-transfer/higher-energy collision dissociation
- Fuc
L-fucose
- Gal
D-galactose
- GalNAc
N-acetyl-D-galactosamine
- GalNAz
N-azidoacetylgalactosamine
- GlcNAc
N-acetyl-D-glucosamine
- HCC
hepatocellular carcinoma
- HCD
higher-energy collision dissociation
- Hex
hexose
- HexNAc
N-acetylhexosamine
- HILIC
hydrophilic interaction liquid chromatography
- IMS
ion mobility spectrometry
- LacdiNAc
N,N-diacetyllactosamine
- LC
liquid chromatography
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- NCE
normalised collision energy
- NeuAc
N-acetyl-D-neuraminic acid
- NeuGc
N-glycolyl-D-neuraminic acid
- PCa
prostate cancer
- pd
product-dependent
- PGC
porous graphitised carbon
- PRM
parallel reaction monitoring
- Pro
proline
- Q-ToF
quadrupole time-of-flight
- RCA
Ricinus communis agglutinin
- SA
supplemental activation
- SAGR
smart anti-glycan reagent
- SCE
stepped collision energy
- Ser
serine
- SIM
selected ion monitoring
- SPE
solid phase extraction
- sT
sialyl-T antigen
- sTn
sialyl-Tn antigen
- Thr
threonine
- TMT
tandem mass tag
- UDP
uridine diphosphate
- UVPD
ultraviolet photodissociation
- VVA
Vicia villosa agglutinin
- WGA
wheat germ agglutinin
- XIC
extracted ion chromatogram
- ZIC-HILIC
zwitterionic-hydrophilic interaction liquid chromatography
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Open Access
Open access for this article was enabled by the participation of Macquarie University in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with CAUL.
Funding
A.C. was supported by an international Macquarie University Research Excellence Scholarship (iMQRES). R.K. was supported by an Early Career Fellowship from the Cancer Institute NSW. M.T.-A. was supported by a Macquarie University Safety Net Grant.
Author Contributions
A.C., R.K. and M.T.-A. wrote the review.
References
- 1.Tran, D.T. and Ten Hagen, K.G. (2013) Mucin-type O-glycosylation during development. J. Biol. Chem. 288, 6921–6920 10.1074/jbc.R112.418558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaltner, H., Abad-Rodriguez, J., Corfield, A.P., Kopitz, J. and Gabius, H.J. (2019) The sugar code: letters and vocabulary, writers, editors and readers and biosignificance of functional glycan-lectin pairing. Biochem. J. 476, 2623–2655 10.1042/BCJ20170853 [DOI] [PubMed] [Google Scholar]
- 3.Lin, B., Qing, X., Liao, J. and Zhuo, K. (2020) Role of protein glycosylation in host-pathogen interaction. Cells 9, 1022 10.3390/cells9041022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson, A.J., de Vries, R.P. and Paulson, J.C. (2019) Virus recognition of glycan receptors. Curr. Opin. Virol. 34, 117–129 10.1016/j.coviro.2019.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereira, M.S., Alves, I., Vicente, M., Campar, A., Silva, M.C., Padrao, N.A.et al. (2018) Glycans as key checkpoints of T cell activity and function. Front. Immunol. 9, 2754 10.3389/fimmu.2018.02754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ugonotti, J., Chatterjee, S. and Thaysen-Andersen, M. (2020) Structural and functional diversity of neutrophil glycosylation in innate immunity and related disorders. Mol. Aspects Med. 100882 10.1016/j.mam.2020.100882 [DOI] [PubMed] [Google Scholar]
- 7.Reily, C., Stewart, T.J., Renfrow, M.B. and Novak, J. (2019) Glycosylation in health and disease. Nat. Rev. Nephrol. 15, 346–366 10.1038/s41581-019-0129-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goth, C.K., Vakhrushev, S.Y., Joshi, H.J., Clausen, H. and Schjoldager, K.T. (2018) Fine-tuning limited proteolysis: a major role for regulated site-specific O-glycosylation. Trends Biochem. Sci. 43, 269–284 10.1016/j.tibs.2018.02.005 [DOI] [PubMed] [Google Scholar]
- 9.Varki, A. (2017) Biological roles of glycans. Glycobiology 27, 3–49 10.1093/glycob/cww086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sumer-Bayraktar, Z., Grant, O.C., Venkatakrishnan, V., Woods, R.J., Packer, N.H. and Thaysen-Andersen, M. (2016) Asn347 glycosylation of corticosteroid-binding globulin fine-tunes the host immune response by modulating proteolysis by Pseudomonas aeruginosa and neutrophil elastase. J. Biol. Chem. 291, 17727–17742 10.1074/jbc.M116.735258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodrigues, J.G., Balmana, M., Macedo, J.A., Pocas, J., Fernandes, A., de-Freitas-Junior, J.C.M.et al. (2018) Glycosylation in cancer: selected roles in tumour progression, immune modulation and metastasis. Cell. Immunol. 333, 46–57 10.1016/j.cellimm.2018.03.007 [DOI] [PubMed] [Google Scholar]
- 12.Zhang, L. and Ten Hagen, K.G. (2018) Pleiotropic effects of O-glycosylation in colon cancer. J. Biol. Chem. 293, 1315–1316 10.1074/jbc.H117.812826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee, S., Lee, L.Y., Kawahara, R., Abrahams, J.L., Adamczyk, B., Anugraham, M.et al. (2019) Protein paucimannosylation is an enriched N-glycosylation signature of human cancers. Proteomics 19, e1900010 10.1002/pmic.201900010 [DOI] [PubMed] [Google Scholar]
- 14.Loke, I., Kolarich, D., Packer, N.H. and Thaysen-Andersen, M. (2016) Emerging roles of protein mannosylation in inflammation and infection. Mol. Aspects Med. 51, 31–55 10.1016/j.mam.2016.04.004 [DOI] [PubMed] [Google Scholar]
- 15.Bhat, A.H., Maity, S., Giri, K. and Ambatipudi, K. (2019) Protein glycosylation: sweet or bitter for bacterial pathogens? Crit. Rev. Microbiol. 45, 82–102 10.1080/1040841X.2018.1547681 [DOI] [PubMed] [Google Scholar]
- 16.Hare, N.J., Lee, L.Y., Loke, I., Britton, W.J., Saunders, B.M. and Thaysen-Andersen, M. (2017) Mycobacterium tuberculosis infection manipulates the glycosylation machinery and the N-glycoproteome of human macrophages and their microparticles. J. Proteome Res. 16, 247–263 10.1021/acs.jproteome.6b00685 [DOI] [PubMed] [Google Scholar]
- 17.Delannoy, C., Huang, C., Coddeville, B., Chen, J.-Y., Mouajjah, D., Groux-Degroote, S.et al. (2020) Mycobacterium bovis BCG infection alters the macrophage N-glycome. Mol. Omics 16, 345–354 10.1039/C9MO00173E [DOI] [PubMed] [Google Scholar]
- 18.Mealer, R.G., Williams, S.E., Daly, M.J., Scolnick, E.M., Cummings, R.D. and Smoller, J.W. (2020) Glycobiology and schizophrenia: a biological hypothesis emerging from genomic research. Mol. Psychiatry 25, 3129–3139 10.1038/s41380-020-0753-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stambuk, T., Klasic, M., Zoldos, V. and Lauc, G. (2020) N-glycans as functional effectors of genetic and epigenetic disease risk. Mol. Aspects Med. 100891 10.1016/j.mam.2020.100891 [DOI] [PubMed] [Google Scholar]
- 20.Axford, J., Alavi, A., Cummings, R., Lauc, G., Opdenakker, G., Reis, C.et al. (2019) Translational glycobiology: from bench to bedside. J. R. Soc. Med. 112, 424–427 10.1177/0141076819865863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang, D. and Zaia, J. (2019) Why glycosylation matters in building a better Flu vaccine. Mol. Cell. Proteomics 18, 2348–2358 10.1074/mcp.R119.001491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thaysen-Andersen, M., Packer, N.H. and Schulz, B.L. (2016) Maturing glycoproteomics technologies provide unique structural insights into the N-glycoproteome and Its regulation in health and disease. Mol. Cell. Proteomics 15, 1773–1790 10.1074/mcp.O115.057638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thaysen-Andersen, M. and Packer, N.H. (2014) Advances in LC-MS/MS-based glycoproteomics: getting closer to system-wide site-specific mapping of the N- and O-glycoproteome. Biochim. Biophys. Acta 1844, 1437–1452 10.1016/j.bbapap.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 24.Levery, S.B., Steentoft, C., Halim, A., Narimatsu, Y., Clausen, H. and Vakhrushev, S.Y. (2015) Advances in mass spectrometry driven O-glycoproteomics. Biochim. Biophys. Acta 1850, 33–42 10.1016/j.bbagen.2014.09.026 [DOI] [PubMed] [Google Scholar]
- 25.Darula, Z. and Medzihradszky, K.F. (2018) Analysis of mammalian O-glycopeptides-we have made a good start, but there is a long way to go. Mol. Cell. Proteomics 17, 2–17 10.1074/mcp.MR117.000126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khoo, K.H. (2019) Advances toward mapping the full extent of protein site-specific O-GalNAc glycosylation that better reflects underlying glycomic complexity. Curr. Opin. Struct. Biol. 56, 146–154 10.1016/j.sbi.2019.02.007 [DOI] [PubMed] [Google Scholar]
- 27.Yang, Y., Franc, V. and Heck, A.J.R. (2017) Glycoproteomics: a balance between high-throughput and in-depth analysis. Trends Biotechnol. 35, 598–609 10.1016/j.tibtech.2017.04.010 [DOI] [PubMed] [Google Scholar]
- 28.Ruhaak, L.R., Xu, G., Li, Q., Goonatilleke, E. and Lebrilla, C.B. (2018) Mass spectrometry approaches to glycomic and glycoproteomic analyses. Chem. Rev. 118, 7886–7930 10.1021/acs.chemrev.7b00732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao, H., Suttapitugsakul, S., Sun, F. and Wu, R. (2018) Mass spectrometry-based chemical and enzymatic methods for global analysis of protein glycosylation. Acc. Chem. Res. 51, 1796–1806 10.1021/acs.accounts.8b00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narimatsu, H., Kaji, H., Vakhrushev, S.Y., Clausen, H., Zhang, H., Noro, E.et al. (2018) Current technologies for complex glycoproteomics and their applications to biology/disease-driven glycoproteomics. J. Proteome Res. 17, 4097–4112 10.1021/acs.jproteome.8b00515 [DOI] [PubMed] [Google Scholar]
- 31.Yu, A., Zhao, J., Peng, W., Banazadeh, A., Williamson, S.D., Goli, M.et al. (2018) Advances in mass spectrometry-based glycoproteomics. Electrophoresis 39, 3104–3122 10.1002/elps.201800272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen, Z., Huang, J. and Li, L. (2019) Recent advances in mass spectrometry (MS)-based glycoproteomics in complex biological samples. Trends Analyt. Chem. 118, 880–892 10.1016/j.trac.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cipollo, J.F. and Parsons, L.M. (2020) Glycomics and glycoproteomics of viruses: mass spectrometry applications and insights toward structure-function relationships. Mass Spectrom. Rev. 39, 371–409 10.1002/mas.21629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suttapitugsakul, S., Sun, F. and Wu, R. (2020) Recent advances in glycoproteomic analysis by mass spectrometry. Anal. Chem. 92, 267–291 10.1021/acs.analchem.9b04651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker, B.L., Thaysen-Andersen, M., Fazakerley, D.J., Holliday, M., Packer, N.H. and James, D.E. (2016) Terminal galactosylation and sialylation switching on membrane glycoproteins upon TNF-alpha-induced insulin resistance in adipocytes. Mol. Cell. Proteomics 15, 141–153 10.1074/mcp.M115.054221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King, S.L., Joshi, H.J., Schjoldager, K.T., Halim, A., Madsen, T.D., Dziegiel, M.H.et al. (2017) Characterizing the O-glycosylation landscape of human plasma, platelets, and endothelial cells. Blood Adv. 1, 429–442 10.1182/bloodadvances.2016002121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, M.Q., Zeng, W.F., Fang, P., Cao, W.Q., Liu, C., Yan, G.Q.et al. (2017) pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat. Commun. 8, 438 10.1038/s41467-017-00535-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alves, M.J., Kawahara, R., Viner, R., Colli, W., Mattos, E.C., Thaysen-Andersen, M.et al. (2017) Comprehensive glycoprofiling of the epimastigote and trypomastigote stages of Trypanosoma cruzi. J. Proteomics 151, 182–192 10.1016/j.jprot.2016.05.034 [DOI] [PubMed] [Google Scholar]
- 39.Chen, Z.W., Yu, Q., Hao, L., Liu, F.B., Johnson, J., Tian, Z.C.et al. (2018) Site-specific characterization and quantitation of N-glycopeptides in PKM2 knockout breast cancer cells using DiLeu isobaric tags enabled by electron-transfer/higher-energy collision dissociation (EThcD). Analyst 143, 2508–2519 10.1039/C8AN00216A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, W., Ao, M., Hu, Y., Li, Q.K. and Zhang, H. (2018) Mapping the O-glycoproteome using site-specific extraction of O-linked glycopeptides (EXoO). Mol. Syst. Biol. 14, e8486 10.15252/msb.20188486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riley, N.M., Hebert, A.S., Westphall, M.S. and Coon, J.J. (2019) Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis. Nat. Commun. 10, 1311 10.1038/s41467-019-09222-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown, C.J., Gaunitz, S., Wang, Z., Strindelius, L., Jacobson, S.C., Clemmer, D.E.et al. (2020) Glycoproteomic analysis of human urinary exosomes. Anal. Chem. 92, 14357–14365 10.1021/acs.analchem.0c01952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawahara, R., Recuero, S., Srougi, M., Leite, K.R.M., Thaysen-Andersen, M. and Palmisano, G. (2020) The complexity and dynamics of the tissue glycoproteome associated with prostate cancer progression. Mol. Cell. Proteomics 10.1074/mcp.RA120.002320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darula, Z. and Medzihradszky, K.F. (2015) Carbamidomethylation side reactions may lead to glycan misassignments in glycopeptide analysis. Anal. Chem. 87, 6297–6302 10.1021/acs.analchem.5b01121 [DOI] [PubMed] [Google Scholar]
- 45.Lee, L.Y., Moh, E.S.X., Parker, B.L., Bern, M., Packer, N.H. and Thaysen-Andersen, M. (2016) Toward automated N-glycopeptide identification in glycoproteomics. J. Proteome Res. 15, 3904–3915 10.1021/acs.jproteome.6b00438 [DOI] [PubMed] [Google Scholar]
- 46.Riley, N.M., Malaker, S.A. and Bertozzi, C.R. (2020) Electron-based dissociation is needed for O-glycopeptides derived from OpeRATOR proteolysis. Anal. Chem. 92, 14878–14884 10.1021/acs.analchem.0c02950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moremen, K.W., Tiemeyer, M. and Nairn, A.V. (2012) Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462 10.1038/nrm3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tjondro, H.C., Loke, I., Chatterjee, S. and Thaysen-Andersen, M. (2019) Human protein paucimannosylation: cues from the eukaryotic kingdoms. Biol. Rev. Camb. Philos. Soc. 94, 2068–2100 10.1111/brv.12548 [DOI] [PubMed] [Google Scholar]
- 49.Yang, G., Hu, Y., Sun, S., Ouyang, C., Yang, W., Wang, Q.et al. (2018) Comprehensive glycoproteomic analysis of Chinese hamster ovary cells. Anal. Chem. 90, 14294–14302 10.1021/acs.analchem.8b03520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shu, Q., Li, M., Shu, L., An, Z., Wang, J., Lv, H.et al. (2020) Large-scale identification of N-linked intact glycopeptides in human serum using HILIC enrichment and spectral library search. Mol. Cell. Proteomics 19, 672–689 10.1074/mcp.RA119.001791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bollineni, R.C., Koehler, C.J., Gislefoss, R.E., Anonsen, J.H. and Thiede, B. (2018) Large-scale intact glycopeptide identification by mascot database search. Sci. Rep. 8, 2117 10.1038/s41598-018-20331-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawahara, R., Ortega, F., Rosa-Fernandes, L., Guimaraes, V., Quina, D., Nahas, W.et al. (2018) Distinct urinary glycoprotein signatures in prostate cancer patients. Oncotarget 9, 33077–33097 10.18632/oncotarget.26005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woo, C.M., Lund, P.J., Huang, A.C., Davis, M.M., Bertozzi, C.R. and Pitteri, S.J. (2018) Mapping and quantification of over 2000 O-linked glycopeptides in activated human T cells with isotope-targeted glycoproteomics (Isotag). Mol. Cell. Proteomics 17, 764–775 10.1074/mcp.RA117.000261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, Y., Xie, X., Zhao, X., Tian, F., Lv, J., Ying, W.et al. (2018) Systems analysis of singly and multiply O-glycosylated peptides in the human serum glycoproteome via EThcD and HCD mass spectrometry. J. Proteomics 170, 14–27 10.1016/j.jprot.2017.09.014 [DOI] [PubMed] [Google Scholar]
- 55.Wang, Y., Xu, F., Xiao, K., Chen, Y. and Tian, Z. (2019) Site- and structure-specific characterization of N-glycoprotein markers of MCF-7 cancer stem cells using isotopic-labelling quantitative N-glycoproteomics. Chem. Commun. 55, 7934–7937 10.1039/C9CC04114A [DOI] [PubMed] [Google Scholar]
- 56.Xiao, K.J. and Tian, Z.X. (2019) GPSeeker enables quantitative structural N-glycoproteomics for site- and structure-specific characterization of differentially expressed N-glycosylation in hepatocellular carcinoma. J. Proteome Res. 18, 2885–2895 10.1021/acs.jproteome.9b00191 [DOI] [PubMed] [Google Scholar]
- 57.Ye, Z., Mao, Y., Clausen, H. and Vakhrushev, S.Y. (2019) Glyco-DIA: a method for quantitative O-glycoproteomics with in silico-boosted glycopeptide libraries. Nat. Methods 16, 902–910 10.1038/s41592-019-0504-x [DOI] [PubMed] [Google Scholar]
- 58.Fang, P., Ji, Y., Silbern, I., Doebele, C., Ninov, M., Lenz, C.et al. (2020) A streamlined pipeline for multiplexed quantitative site-specific N-glycoproteomics. Nat. Commun. 11, 5268 10.1038/s41467-020-19052-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu, Y., Pan, J., Shah, P., Ao, M., Thomas, S.N., Liu, Y.et al. (2020) Integrated proteomic and glycoproteomic characterization of human high-grade serous ovarian carcinoma. Cell Rep. 33, 108276 10.1016/j.celrep.2020.108276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leung, K.K., Wilson, G.M., Kirkemo, L.L., Riley, N.M., Coon, J.J. and Wells, J.A. (2020) Broad and thematic remodeling of the surfaceome and glycoproteome on isogenic cells transformed with driving proliferative oncogenes. Proc. Natl Acad. Sci. U.S.A. 117, 7764–7775 10.1073/pnas.1917947117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen, Y., Xiao, K. and Tian, Z. (2020) Site- and structure-specific characterization of human urinary N-glycoproteome with site-determining and structure-diagnostic product ions. Rapid Commun. Mass Spectrom. 35, e8952 10.1002/rcm.8952 [DOI] [PubMed] [Google Scholar]
- 62.Xu, F., Wang, Y., Xiao, K., Hu, Y., Tian, Z. and Chen, Y. (2020) Quantitative site- and structure-specific N-glycoproteomics characterization of differential N-glycosylation in MCF-7/ADR cancer stem cells. Clin. Proteomics 17, 3 10.1186/s12014-020-9268-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Y., Mao, Y., Zhao, W., Su, T., Zhong, Y., Fu, L.et al. (2020) Glyco-CPLL: an integrated method for in-depth and comprehensive N-glycoproteome profiling of human plasma. J. Proteome Res. 19, 655–666 10.1021/acs.jproteome.9b00557 [DOI] [PubMed] [Google Scholar]
- 64.Zhao, T., Jia, L., Li, J., Ma, C., Wu, J., Shen, J.et al. (2020) Heterogeneities of site-specific N-glycosylation in HCC tumors with low and high AFP concentrations. Front. Oncol. 10, 496 10.3389/fonc.2020.00496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao, X., Zheng, S., Li, Y., Huang, J., Zhang, W., Xie, Y.et al. (2020) An integrated mass spectroscopy data processing strategy for fast identification, in-depth, and reproducible quantification of protein O-glycosylation in a large cohort of human urine samples. Anal. Chem. 92, 690–698 10.1021/acs.analchem.9b02228 [DOI] [PubMed] [Google Scholar]
- 66.Zhu, J., Lin, Y.H., Dingess, K.A., Mank, M., Stahl, B. and Heck, A.J.R. (2020) Quantitative longitudinal inventory of the N-glycoproteome of human milk from a single donor reveals the highly variable repertoire and dynamic site-specific changes. J. Proteome Res. 19, 1941–1952 10.1021/acs.jproteome.9b00753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blazev, R., Ashwood, C., Abrahams, J.L., Chung, L.H., Francis, D., Yang, P.et al. (2020) Integrated glycoproteomics identifies a role of N-glycosylation and galectin-1 on myogenesis and muscle development. Mol. Cell. Proteomics 10.1074/mcp.RA120.002166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fang, P., Xie, J., Sang, S., Zhang, L., Liu, M., Yang, L.et al. (2020) Multilayered N-glycoproteome profiling reveals highly heterogeneous and dysregulated protein N-glycosylation related to Alzheimer's disease. Anal. Chem. 92, 867–874 10.1021/acs.analchem.9b03555 [DOI] [PubMed] [Google Scholar]
- 69.Li, J., Jia, L., Hao, Z., Xu, Y., Shen, J., Ma, C.et al. (2020) Site-specific N-glycoproteomic analysis reveals upregulated sialylation and core fucosylation during transient regeneration loss in neonatal mouse hearts. J. Proteome Res. 19, 3191–3200 10.1021/acs.jproteome.0c00172 [DOI] [PubMed] [Google Scholar]
- 70.Yang, G., Hoti, N., Chen, S.Y., Zhou, Y., Wang, Q., Betenbaugh, M.et al. (2020) One-step enrichment of intact glycopeptides from glycoengineered Chinese hamster ovary cells. Front. Chem. 8, 240 10.3389/fchem.2020.00240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zeng, W., Ford, K.L., Bacic, A. and Heazlewood, J.L. (2018) N-linked glycan micro-heterogeneity in glycoproteins of arabidopsis. Mol. Cell. Proteomics 17, 413–421 10.1074/mcp.RA117.000165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zacchi, L.F. and Schulz, B.L. (2016) SWATH-MS glycoproteomics reveals consequences of defects in the glycosylation machinery. Mol. Cell. Proteomics 15, 2435–2447 10.1074/mcp.M115.056366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin, C.H., Krisp, C., Packer, N.H. and Molloy, M.P. (2018) Development of a data independent acquisition mass spectrometry workflow to enable glycopeptide analysis without predefined glycan compositional knowledge. J. Proteomics 172, 68–75 10.1016/j.jprot.2017.10.011 [DOI] [PubMed] [Google Scholar]
- 74.Pegg, C.L., Phung, T.K., Caboche, C.H., Niamsuphap, S., Bern, M., Howell, K.et al. (2020) Quantitative data independent acquisition glycoproteomics of sparkling wine. Mol. Cell. Proteomics 10.1074/mcp.RA120.002181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou, C. and Schulz, B.L. (2020) Glycopeptide variable window SWATH for improved data independent acquisition glycoprotein analysis. Anal. Biochem. 597, 113667 10.1016/j.ab.2020.113667 [DOI] [PubMed] [Google Scholar]
- 76.Mun, D.G., Renuse, S., Saraswat, M., Madugundu, A., Udainiya, S., Kim, H.et al. (2020) PASS-DIA: a data-Independent acquisition approach for discovery studies. Anal. Chem. 92, 14466–14475 10.1021/acs.analchem.0c02513 [DOI] [PubMed] [Google Scholar]
- 77.Bern, M., Kil, Y.J. and Becker, C. (2012) Byonic: advanced peptide and protein identification software. Curr. Protoc. Bioinformatics 10.1002/0471250953.bi1320s40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roushan, A., Wilson, G.M., Kletter, D., Sen, K.I., Tang, W.H., Kil, Y.J.et al. (2020) Peak filtering, peak annotation, and wildcard search for glycoproteomics. Mol. Cell. Proteomics 10.1074/mcp.RA120.002260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Toghi Eshghi, S., Shah, P., Yang, W., Li, X. and Zhang, H. (2015) GPQuest: a spectral library matching algorithm for site-specific assignment of tandem mass spectra to intact N-glycopeptides. Anal. Chem. 87, 5181–5188 10.1021/acs.analchem.5b00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mysling, S., Palmisano, G., Højrup, P. and Thaysen-andersen, M. (2010) Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics. Anal. Chem. 82, 5598–5609 10.1021/ac100530w [DOI] [PubMed] [Google Scholar]
- 81.Neue, K., Mormann, M., Peter-Katalinić, J. and Pohlentz, G. (2011) Elucidation of glycoprotein structures by unspecific proteolysis and direct nanoESI mass spectrometric analysis of ZIC-HILIC-enriched glycopeptides. J. Proteome Res. 10, 2248–2260 10.1021/pr101082c [DOI] [PubMed] [Google Scholar]
- 82.Ahmad Izaham, A.R., Ang, C.S., Nie, S., Bird, L.E., Williamson, N.A. and Scott, N.E. (2020) What are we missing by using hydrophilic enrichment? improving bacterial glycoproteome coverage using total proteome and FAIMS analyses. J. Proteome Res. 10.1021/acs.jproteome.0c00565 [DOI] [PubMed] [Google Scholar]
- 83.Loke, I., Packer, N. and Thaysen-Andersen, M. (2015) Complementary LC-MS/MS-based N-glycan, N-glycopeptide, and intact N-glycoprotein profiling reveals unconventional Asn71-glycosylation of human neutrophil cathepsin G. Biomolecules 5, 1832–1854 10.3390/biom5031832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Steentoft, C., Vakhrushev, S.Y., Vester-Christensen, M.B., Schjoldager, K.T., Kong, Y., Bennett, E.P.et al. (2011) Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat. Methods 8, 977–982 10.1038/nmeth.1731 [DOI] [PubMed] [Google Scholar]
- 85.Steentoft, C., Vakhrushev, S.Y., Joshi, H.J., Kong, Y., Vester-Christensen, M.B., Schjoldager, K.T.B.G.et al. (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 10.1038/emboj.2013.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vakhrushev, S.Y., Steentoft, C., Vester-Christensen, M.B., Bennett, E.P., Clausen, H. and Levery, S.B. (2013) Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells. Mol. Cell. Proteomics 12, 932–944 10.1074/mcp.O112.021972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang, Z., Halim, A., Narimatsu, Y., Jitendra Joshi, H., Steentoft, C., Schjoldager, K.T.et al. (2014) The GalNAc-type O-glycoproteome of CHO cells characterized by the SimpleCell strategy. Mol. Cell. Proteomics 13, 3224–3235 10.1074/mcp.M114.041541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Campos, D., Freitas, D., Gomes, J., Magalhaes, A., Steentoft, C., Gomes, C.et al. (2015) Probing the O-glycoproteome of gastric cancer cell lines for biomarker discovery. Mol. Cell. Proteomics 14, 1616–1629 10.1074/mcp.M114.046862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.King, S.L., Goth, C.K., Eckhard, U., Joshi, H.J., Haue, A.D., Vakhrushev, S.Y.et al. (2018) TAILS N-terminomics and proteomics reveal complex regulation of proteolytic cleavage by O-glycosylation. J. Biol. Chem. 293, 7629–7644 10.1074/jbc.RA118.001978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lavrsen, K., Dabelsteen, S., Vakhrushev, S.Y., Levann, A.M.R., Haue, A.D., Dylander, A.et al. (2018) De novo expression of human polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) in colon adenocarcinoma inhibits the differentiation of colonic epithelium. J. Biol. Chem. 293, 1298–1314 10.1074/jbc.M117.812826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Narimatsu, Y., Joshi, H.J., Schjoldager, K.T., Hintze, J., Halim, A., Steentoft, C.et al. (2019) Exploring regulation of protein O-glycosylation in isogenic human HEK293 cells by differential O-glycoproteomics. Mol. Cell. Proteomics 18, 1396–1409 10.1074/mcp.RA118.001121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bagdonaite, I., Pallesen, E.M., Ye, Z., Vakhrushev, S.Y., Marinova, I.N., Nielsen, M.I.et al. (2020) O-glycan initiation directs distinct biological pathways and controls epithelial differentiation. EMBO Rep. 21, e48885 10.15252/embr.201948885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dabelsteen, S., Pallesen, E.M.H., Marinova, I.N., Nielsen, M.I., Adamopoulou, M., Rømer, T.B.et al. (2020) Essential functions of glycans in human epithelia dissected by a CRISPR-Cas9-engineered human organotypic skin model. Dev. Cell 54, 669–84.e7 10.1016/j.devcel.2020.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Polasky, D.A., Yu, F., Teo, G.C. and Nesvizhskii, A.I. (2020) Fast and comprehensive N- and O-glycoproteomics analysis with MSFragger-Glyco. Nat. Methods 17, 1125–1132 10.1038/s41592-020-0967-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu, L., Riley, N.M., Shortreed, M.R., Bertozzi, C.R. and Smith, L.M. (2020) O-Pair search with metaMorpheus for O-glycopeptide characterization. Nat. Methods 17, 1133–1138 10.1038/s41592-020-00985-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abrahams, J.L., Taherzadeh, G., Jarvas, G., Guttman, A., Zhou, Y. and Campbell, M.P. (2020) Recent advances in glycoinformatic platforms for glycomics and glycoproteomics. Curr. Opin. Struct. Biol. 62, 56–69 10.1016/j.sbi.2019.11.009 [DOI] [PubMed] [Google Scholar]
- 97.Cao, W., Liu, M., Kong, S., Wu, M., Zhang, Y. and Yang, P. (2020) Recent advances in software tools for more generic and precise intact glycopeptide analysis. Mol. Cell. Proteomics 10.1074/mcp.R120.002090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Noach, I., Ficko-Blean, E., Pluvinage, B., Stuart, C., Jenkins, M.L., Brochu, D.et al. (2017) Recognition of protein-linked glycans as a determinant of peptidase activity. Proc. Natl Acad. Sci. U.S.A. 114, E679–EE88 10.1073/pnas.1615141114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shon, D.J., Malaker, S.A., Pedram, K., Yang, E., Krishnan, V., Dorigo, O.et al. (2020) An enzymatic toolkit for selective proteolysis, detection, and visualization of mucin-domain glycoproteins. Proc. Natl Acad. Sci. U.S.A. 117, 21299–21307 10.1073/pnas.2012196117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Haurat, M.F., Scott, N.E., Di Venanzio, G., Lopez, J., Pluvinage, B., Boraston, A.B.et al. (2020) The glycoprotease CpaA secreted by medically relevant acinetobacter species targets multiple O-Linked host glycoproteins. mBio 11, e02033-20 10.1128/mBio.02033-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang, S., Onigman, P., Wu, W.W., Sjogren, J., Nyhlen, H., Shen, R.F.et al. (2018) Deciphering protein O-Glycosylation: solid-phase chemoenzymatic cleavage and enrichment. Anal. Chem. 90, 8261–8269 10.1021/acs.analchem.8b01834 [DOI] [PubMed] [Google Scholar]
- 102.Yang, W., Song, A., Ao, M., Xu, Y. and Zhang, H. (2020) Large-scale site-specific mapping of the O-GalNAc glycoproteome. Nat. Protoc. 15, 2589–2610 10.1038/s41596-020-0345-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Trastoy, B., Naegeli, A., Anso, I., Sjogren, J. and Guerin, M.E. (2020) Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila. Nat. Commun. 11, 4844 10.1038/s41467-020-18696-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Malaker, S.A., Pedram, K., Ferracane, M.J., Bensing, B.A., Krishnan, V., Pett, C.et al. (2019) The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins. Proc. Natl Acad. Sci. U.S.A. 116, 7278–7287 10.1073/pnas.1813020116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Neelamegham, S., Aoki-Kinoshita, K., Bolton, E., Frank, M., Lisacek, F., Lütteke, T.et al. (2019) Updates to the symbol nomenclature for glycans guidelines. Glycobiology 29, 620–624 10.1093/glycob/cwz045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Palaniappan, K.K. and Bertozzi, C.R. (2016) Chemical glycoproteomics. Chem. Rev. 116, 14277–14306 10.1021/acs.chemrev.6b00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aguilar A, L., Briard, J.G., Yang, L., Ovryn, B., Macauley, M.S. and Wu, P. (2017) Tools for studying glycans: recent advances in chemoenzymatic glycan labeling. ACS Chem. Biol. 12, 611–621 10.1021/acschembio.6b01089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Han, S.S., Shim, H.E., Park, S.J., Kim, B.C., Lee, D.E., Chung, H.M.et al. (2018) Safety and optimization of metabolic labeling of endothelial progenitor cells for tracking. Sci. Rep. 8, 13212 10.1038/s41598-018-31594-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Boyce, M., Carrico, I.S., Ganguli, A.S., Yu, S.H., Hangauer, M.J., Hubbard, S.C.et al. (2011) Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc. Natl Acad. Sci. U.S.A. 108, 3141–3146 10.1073/pnas.1010045108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Woo, C.M., Iavarone, A.T., Spiciarich, D.R., Palaniappan, K.K. and Bertozzi, C.R. (2015) Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat. Methods 12, 561–567 10.1038/nmeth.3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qin, W., Qin, K., Fan, X., Peng, L., Hong, W., Zhu, Y.et al. (2018) Artificial cysteine S-glycosylation induced by per-O-acetylated unnatural monosaccharides during metabolic glycan labeling. Angew. Chem. Int. Ed. Engl. 57, 1817–1820 10.1002/anie.201711710 [DOI] [PubMed] [Google Scholar]
- 112.Darabedian, N., Yang, B., Ding, R., Cutolo, G., Zaro, B.W., Woo, C.M.et al. (2020) O-acetylated chemical reporters of glycosylation can display metabolism-dependent background labeling of proteins but are generally reliable tools for the identification of glycoproteins. Front. Chem. 8, 318 10.3389/fchem.2020.00318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Qin, K., Zhang, H., Zhao, Z. and Chen, X. (2020) Protein S-glyco-modification through an elimination–addition mechanism. J. Am. Chem. Soc. 142, 9382–9388 10.1021/jacs.0c02110 [DOI] [PubMed] [Google Scholar]
- 114.Debets, M.F., Tastan, O.Y., Wisnovsky, S.P., Malaker, S.A., Angelis, N., Moeckl, L.K.R.et al. (2020) Metabolic precision labeling enables selective probing of O-linked N-acetylgalactosamine glycosylation. Proc. Natl. Acad. Sci. U. S. A. 117, 25293–25301 10.1073/pnas.2007297117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Choi, J., Wagner, L.J.S., Timmermans, S., Malaker, S.A., Schumann, B., Gray, M.A.et al. (2019) Engineering orthogonal polypeptide GalNAc-transferase and UDP-sugar pairs. J. Am. Chem. Soc. 141, 13442–13453 10.1021/jacs.9b04695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schumann, B., Malaker, S.A., Wisnovsky, S.P., Debets, M.F., Agbay, A.J., Fernandez, D.et al. (2020) Bump-and-Hole engineering identifies specific substrates of glycosyltransferases in living cells. Mol. Cell 78, 824–34.e15 10.1016/j.molcel.2020.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cioce, A., Malaker, S.A. and Schumann, B. (2020) Generating orthogonal glycosyltransferase and nucleotide sugar pairs as next-generation glycobiology tools. Curr. Opin. Chem. Biol. 60, 66–78 10.1016/j.cbpa.2020.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li, M., Shen, J., Liu, X., Shao, J., Yi, W., Chow, C.S.et al. (2008) Identification of a new alpha1,2-fucosyltransferase involved in O-antigen biosynthesis of escherichia coli O86:B7 and formation of H-type 3 blood group antigen. Biochemistry 47, 11590–7 10.1021/bi801067s [DOI] [PubMed] [Google Scholar]
- 119.Yi, W., Shen, J., Zhou, G., Li, J. and Wang, P.G. (2008) Bacterial homologue of human blood group A transferase. J. Am. Chem. Soc. 130, 14420–1 10.1021/ja805844y [DOI] [PubMed] [Google Scholar]
- 120.Wen, L., Zheng, Y., Jiang, K., Zhang, M., Kondengaden, S.M., Li, S.et al. (2016) Two-step chemoenzymatic detection of N-acetylneuraminic acid-alpha(2-3)-Galactose glycans. J. Am. Chem. Soc. 138, 11473–6 10.1021/jacs.6b07132 [DOI] [PubMed] [Google Scholar]
- 121.Li, Q., Li, Z., Duan, X. and Yi, W. (2014) A tandem enzymatic approach for detecting and imaging tumor-associated thomsen-Friedenreich antigen disaccharide. J. Am. Chem. Soc. 136, 12536–9 10.1021/ja5054225 [DOI] [PubMed] [Google Scholar]
- 122.Zhu, H., Wang, S., Liu, D., Ding, L., Chen, C., Liu, Y.et al. (2020) Identifying sialylation linkages at the glycopeptide level by glycosyltransferase labeling assisted mass spectrometry (GLAMS). Anal. Chem. 92, 6297–6303 10.1021/acs.analchem.9b05068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wen, L., Liu, D., Zheng, Y., Huang, K., Cao, X., Song, J.et al. (2018) A One-step chemoenzymatic labeling strategy for probing sialylated thomsen–Friedenreich antigen. ACS Cent. Sci. 4, 451–457 10.1021/acscentsci.7b00573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fu, C., Zhao, H., Wang, Y., Cai, H., Xiao, Y., Zeng, Y.et al. (2016) Tumor-associated antigens: Tn antigen, sTn antigen, and T antigen. HLA 88, 275–286 10.1111/tan.12900 [DOI] [PubMed] [Google Scholar]
- 125.Rodríguez, E., Schetters, S.T.T. and Van Kooyk, Y. (2018) The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 18, 204–211 10.1038/nri.2018.3 [DOI] [PubMed] [Google Scholar]
- 126.Zheng, J., Xiao, H. and Wu, R. (2017) Specific identification of glycoproteins bearing the Tn antigen in human cells. Angew. Chem. Int. Ed. Engl. 56, 7107–7111 10.1002/anie.201702191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.You, X., Yao, Y., Mao, J., Qin, H., Liang, X., Wang, L.et al. (2018) Chemoenzymatic approach for the proteomics analysis of mucin-type core-1 O-glycosylation in human serum. Anal. Chem. 90, 12714–12722 10.1021/acs.analchem.8b02993 [DOI] [PubMed] [Google Scholar]
- 128.Yang, W., Ao, M., Song, A., Xu, Y., Sokoll, L. and Zhang, H. (2020) Mass spectrometric mapping of glycoproteins modified by Tn-Antigen using solid-Phase capture and enzymatic release. Anal. Chem. 92, 9230–9238 10.1021/acs.analchem.0c01564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Qin, K., Zhu, Y., Qin, W., Gao, J., Shao, X., Wang, Y.-L.et al. (2018) Quantitative profiling of protein O-GlcNAcylation sites by an isotope-tagged cleavable linker. ACS Chem. Biol. 13, 1983–1989 10.1021/acschembio.8b00414 [DOI] [PubMed] [Google Scholar]
- 130.Li, J., Li, Z., Duan, X., Qin, K., Dang, L., Sun, S.et al. (2019) An isotope-coded photocleavable probe for quantitative profiling of protein O-GlcNAcylation. ACS Chem. Biol. 14, 4–10 10.1021/acschembio.8b01052 [DOI] [PubMed] [Google Scholar]
- 131.Xu, S., Sun, F. and Wu, R. (2020) A chemoenzymatic method based on easily accessible enzymes for profiling protein O-GlcNAcylation. Anal. Chem. 92, 9807–9814 10.1021/acs.analchem.0c01284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Varki, A. and Gagneux, P. (2012) Multifarious roles of sialic acids in immunity. Ann. N. Y. Acad. Sci. 1253, 16–36 10.1111/j.1749-6632.2012.06517.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhou, X., Yang, G. and Guan, F. (2020) Biological functions and analytical strategies of sialic acids in tumor. Cells 9, 273 10.3390/cells9020273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang, S., Wu, W.W., Shen, R.F., Bern, M. and Cipollo, J. (2018) Identification of sialic acid linkages on intact glycopeptides via differential chemical modification using IntactGIG-HILIC. J. Am. Soc. Mass Spectrom. 29, 1273–1283 10.1007/s13361-018-1931-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Riley, N.M., Bertozzi, C.R. and Pitteri, S.J. (2020) A pragmatic guide to enrichment strategies for mass spectrometry-based glycoproteomics. Mol. Cell. Proteomics 10.1074/mcp.R120.002277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nascimento ASF, D., Serna, S., Beloqui, A., Arda, A., Sampaio, A.H., Walcher, J.et al. (2015) Algal lectin binding to core (α1–6) fucosylated N-glycans: structural basis for specificity and production of recombinant protein. Glycobiology 25, 607–616 10.1093/glycob/cwv002 [DOI] [PubMed] [Google Scholar]
- 137.Kobayashi, Y., Tateno, H., Dohra, H., Moriwaki, K., Miyoshi, E., Hirabayashi, J.et al. (2012) A novel core fucose-specific lectin from the mushroom Pholiota squarrosa. J. Biol. Chem. 287, 33973–33982 10.1074/jbc.M111.327692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cabanettes, A., Perkams, L., Spies, C., Unverzagt, C. and Varrot, A. (2018) Recognition of complex core-fucosylated N-glycans by a mini lectin. Angew. Chem. Int. Ed. Engl. 57, 10178–10181 10.1002/anie.201805165 [DOI] [PubMed] [Google Scholar]
- 139.Vainauskas, S., Duke, R.M., McFarland, J., McClung, C., Ruse, C. and Taron, C.H. (2016) Profiling of core fucosylated N-glycans using a novel bacterial lectin that specifically recognizes alpha1,6 fucosylated chitobiose. Sci. Rep. 6, 34195 10.1038/srep34195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Allam, H., Aoki, K., Benigno, B.B., McDonald, J.F., Mackintosh, S.G., Tiemeyer, M.et al. (2015) Glycomic analysis of membrane glycoproteins with bisecting glycosylation from ovarian cancer tissues reveals novel structures and functions. J. Proteome Res. 14, 434–446 10.1021/pr501174p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nagae, M., Kanagawa, M., Morita-Matsumoto, K., Hanashima, S., Kizuka, Y., Taniguchi, N.et al. (2016) Atomic visualization of a flipped-back conformation of bisected glycans bound to specific lectins. Sci. Rep. 6, 22973 10.1038/srep22973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lastovickova, M., Strouhalova, D. and Bobalova, J. (2020) Use of lectin-based affinity techniques in breast cancer glycoproteomics: a review. J. Proteome Res. 19, 1885–1899 10.1021/acs.jproteome.9b00818 [DOI] [PubMed] [Google Scholar]
- 143.Napoletano, C., Steentoff, C., Battisti, F., Ye, Z., Rahimi, H., Zizzari, I.G.et al. (2020) Investigating patterns of immune interaction in ovarian cancer: probing the O-glycoproteome by the macrophage galactose-like C-type lectin (MGL). Cancers (Basel) 12, 2841 10.3390/cancers12102841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Norton, P., Comunale, M.A., Herrera, H., Wang, M., Houser, J., Wimmerova, M.et al. (2016) Development and application of a novel recombinant aleuria aurantia lectin with enhanced core fucose binding for identification of glycoprotein biomarkers of hepatocellular carcinoma. Proteomics 16, 3126–3136 10.1002/pmic.201600064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Su, Y., Ye, X., Xu, B., Li, Y., Yang, Q., Yu, W.et al. (2020) CBS homogenization mutation strategy narrows the glycan binding profile of a GlcNAc-specific lectin AANL. Glycobiology 30, 159–173 10.1093/glycob/cwz089 [DOI] [PubMed] [Google Scholar]
- 146.Yabe, R., Suzuki, R., Kuno, A., Fujimoto, Z., Jigami, Y. and Hirabayashi, J. (2006) Tailoring a novel sialic acid-binding lectin from a ricin-B chain-like galactose-binding protein by natural evolution-mimicry. J. Biochem. 141, 389–399 10.1093/jb/mvm043 [DOI] [PubMed] [Google Scholar]
- 147.Yabe, R., Itakura, Y., Nakamura-Tsuruta, S., Iwaki, J., Kuno, A. and Hirabayashi, J. (2009) Engineering a versatile tandem repeat-type alpha2-6sialic acid-binding lectin. Biochem. Biophys. Res. Commun. 384, 204–209 10.1016/j.bbrc.2009.04.090 [DOI] [PubMed] [Google Scholar]
- 148.Hu, D., Tateno, H., Kuno, A., Yabe, R. and Hirabayashi, J. (2012) Directed evolution of lectins with sugar-binding specificity for 6-sulfo-galactose. J. Biol. Chem. 287, 20313–20320 10.1074/jbc.M112.351965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hirabayashi, J. and Arai, R. (2019) Lectin engineering: the possible and the actual. Interface Focus 9, 20180068 10.1098/rsfs.2018.0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mariappa, D., Selvan, N., Borodkin, V., Alonso, J., Ferenbach, A.T., Shepherd, C.et al. (2015) A mutant O-GlcNAcase as a probe to reveal global dynamics of protein O-GlcNAcylation during drosophila embryonic development. Biochem. J. 470, 255–262 10.1042/BJ20150610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Selvan, N., Williamson, R., Mariappa, D., Campbell, D.G., Gourlay, R., Ferenbach, A.T.et al. (2017) A mutant O-GlcNAcase enriches Drosophila developmental regulators. Nat. Chem. Biol. 13, 882–887 10.1038/nchembio.2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Loureiro, L.R., Sousa, D.P., Ferreira, D., Chai, W., Lima, L., Pereira, C.et al. (2018) Novel monoclonal antibody L2A5 specifically targeting sialyl-Tn and short glycans terminated by alpha-2-6 sialic acids. Sci. Rep. 8, 12196 10.1038/s41598-018-30421-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Steentoft, C., Migliorini, D., King, T.R., Mandel, U., June, C.H. and Posey, A.D. (2018) Glycan-directed CAR-T cells. Glycobiology 28, 656–669 10.1093/glycob/cwy008 [DOI] [PubMed] [Google Scholar]
- 154.Matsumoto, Y., Kudelka, M.R., Hanes, M.S., Lehoux, S., Dutta, S., Jones, M.B.et al. (2020) Identification of Tn antigen O-GalNAc-expressing glycoproteins in human carcinomas using novel anti-Tn recombinant antibodies. Glycobiology 30, 282–300 10.1093/glycob/cwz095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Steentoft, C., Fuhrmann, M., Battisti, F., Van Coillie, J., Madsen, T.D., Campos, D.et al. (2019) A strategy for generating cancer-specific monoclonal antibodies to aberrant O-glycoproteins: identification of a novel dysadherin-Tn antibody. Glycobiology 29, 307–319 10.1093/glycob/cwz004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.McKitrick, T.R., Goth, C.K., Rosenberg, C.S., Nakahara, H., Heimburg-Molinaro, J., McQuillan, A.M.et al. (2020) Development of smart anti-glycan reagents using immunized lampreys. Commun. Biol. 3, 91 10.1038/s42003-020-0819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.McKitrick, T.R., Eris, D., Mondal, N., Aryal, R.P., McCurley, N., Heimburg-Molinaro, J.et al. (2020) Antibodies from lampreys as smart anti-glycan reagents (SAGRs): perspectives on their specificity, structure, and glyco-genomics. Biochemistry 59, 3111–3122 10.1021/acs.biochem.9b01015 [DOI] [PubMed] [Google Scholar]
- 158.Bie, Z., Chen, Y., Ye, J., Wang, S. and Liu, Z. (2015) Boronate-affinity glycan-oriented surface imprinting: a new strategy to mimic lectins for the recognition of an intact glycoprotein and Its characteristic fragments. Angew. Chem. Int. Ed. Engl. 54, 10211–5 10.1002/anie.201503066 [DOI] [PubMed] [Google Scholar]
- 159.Bie, Z., Xing, R., He, X., Ma, Y., Chen, Y. and Liu, Z. (2018) Precision imprinting of glycopeptides for facile preparation of glycan-specific artificial antibodies. Anal. Chem. 90, 9845–9852 10.1021/acs.analchem.8b01903 [DOI] [PubMed] [Google Scholar]
- 160.Reyes, C.D.G., Jiang, P., Donohoo, K., Atashi, M. and Mechref, Y. (2020) Glycomics and glycoproteomics: approaches to address isomeric separation of glycans and glycopeptides. J. Sep. Sci. 1–25 10.1002/jssc.202000878 [DOI] [PubMed] [Google Scholar]
- 161.Choo, M.S., Wan, C., Rudd, P.M. and Nguyen-Khuong, T. (2019) GlycopeptideGraphMS: improved glycopeptide detection and identification by exploiting graph theoretical patterns in mass and retention time. Anal. Chem. 91, 7236–7244 10.1021/acs.analchem.9b00594 [DOI] [PubMed] [Google Scholar]
- 162.Ang, E., Neustaeter, H., Spicer, V., Perreault, H. and Krokhin, O. (2019) Retention time prediction for glycopeptides in reversed-phase chromatography for glycoproteomic applications. Anal. Chem. 91, 13360–6 10.1021/acs.analchem.9b02584 [DOI] [PubMed] [Google Scholar]
- 163.Klein, J. and Zaia, J. (2020) Relative retention time estimation improves N-glycopeptide identifications by LC-MS/MS. J. Proteome Res. 19, 2113–2121 10.1021/acs.jproteome.0c00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ji, E.S., Lee, H.K., Park, G.W., Kim, K.H., Kim, J.Y. and Yoo, J.S. (2019) Isomer separation of sialylated O- and N-linked glycopeptides using reversed-phase LC-MS/MS at high temperature. J. Chromatogr. B 1110–1111, 101–107 10.1016/j.jchromb.2019.02.015 [DOI] [PubMed] [Google Scholar]
- 165.Hinneburg, H., Chatterjee, S., Schirmeister, F., Nguyen-Khuong, T., Packer, N.H., Rapp, E.et al. (2019) Post-column make-up flow (PCMF) enhances the performance of capillary-flow PGC-LC-MS/MS-based glycomics. Anal. Chem. 91, 4559–4567 10.1021/acs.analchem.8b05720 [DOI] [PubMed] [Google Scholar]
- 166.Sethi, M.K., Kim, H., Park, C.K., Baker, M.S., Paik, Y.K., Packer, N.H.et al. (2015) In-depth N-glycome profiling of paired colorectal cancer and non-tumorigenic tissues reveals cancer-, stage- and EGFR-specific protein N-glycosylation. Glycobiology 25, 1064–1078 10.1093/glycob/cwv042 [DOI] [PubMed] [Google Scholar]
- 167.Froehlich, J.W., Barboza, M., Chu, C., Lerno, L.A., Clowers, B.H., Zivkovic, A.M.et al. (2011) Nano-LC–MS/MS of glycopeptides produced by nonspecific proteolysis enables rapid and extensive site-specific glycosylation determination. Anal. Chem. 83, 5541–5547 10.1021/ac2003888 [DOI] [PubMed] [Google Scholar]
- 168.Nwosu, C.C., Huang, J., Aldredge, D.L., Strum, J.S., Hua, S., Seipert, R.R.et al. (2013) In-gel nonspecific proteolysis for elucidating glycoproteins: a method for targeted protein-specific glycosylation analysis in complex protein mixtures. Anal. Chem. 85, 956–963 10.1021/ac302574f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Huang, J., Guerrero, A., Parker, E., Strum, J.S., Smilowitz, J.T., German, J.B.et al. (2015) Site-specific glycosylation of secretory immunoglobulin A from human colostrum. J. Proteome Res. 14, 1335–1349 10.1021/pr500826q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lee, J., Hua, S., Lee, S.H., Oh, M.J., Yun, J., Kim, J.Y.et al. (2018) Designation of fingerprint glycopeptides for targeted glycoproteomic analysis of serum haptoglobin: insights into gastric cancer biomarker discovery. Anal. Bioanal. Chem. 410, 1617–1629 10.1007/s00216-017-0811-y [DOI] [PubMed] [Google Scholar]
- 171.Cao, L., Tolic, N., Qu, Y., Meng, D., Zhao, R., Zhang, Q.et al. (2014) Characterization of intact N- and O-linked glycopeptides using higher energy collisional dissociation. Anal. Biochem. 452, 96–102 10.1016/j.ab.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Xue, Y., Xie, J., Fang, P., Yao, J., Yan, G., Shen, H.et al. (2018) Study on behaviors and performances of universal N-glycopeptide enrichment methods. Analyst 143, 1870–1880 10.1039/C7AN02062G [DOI] [PubMed] [Google Scholar]
- 173.Thaysen-Andersen, M., Wilkinson, B.L., Payne, R.J. and Packer, N.H. (2011) Site-specific characterisation of densely O-glycosylated mucin-type peptides using electron transfer dissociation ESI-MS/MS. Electrophoresis 32, 3536–3545 10.1002/elps.201100294 [DOI] [PubMed] [Google Scholar]
- 174.Nakano, M., Saldanha, R., Göbel, A., Kavallaris, M. and Packer, N.H. (2011) Identification of glycan structure alterations on cell membrane proteins in desoxyepothilone B resistant leukemia cells. Mol. Cell. Proteomics 10, M111.009001 10.1074/mcp.M111.009001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Everest-Dass, A.V., Abrahams, J.L., Kolarich, D., Packer, N.H. and Campbell, M.P. (2013) Structural feature ions for distinguishing N- and O-linked glycan isomers by LC-ESI-IT MS/MS. J. Am. Soc. Mass Spectrom. 24, 895–906 10.1007/s13361-013-0610-4 [DOI] [PubMed] [Google Scholar]
- 176.Ashwood, C., Lin, C.-H., Thaysen-Andersen, M. and Packer, N.H. (2018) Discrimination of isomers of released N- and O-glycans using diagnostic product ions in negative Ion PGC-LC-ESI-MS/MS. J. Am. Soc. Mass Spectrom. 29, 1194–1209 10.1007/s13361-018-1932-z [DOI] [PubMed] [Google Scholar]
- 177.Chen, R., Stupak, J., Williamson, S., Twine, S.M. and Li, J. (2019) Online porous graphic carbon chromatography coupled with tandem mass spectrometry for post-translational modification analysis. Rapid Commun. Mass Spectrom. 33, 1240–1247 10.1002/rcm.8459 [DOI] [PubMed] [Google Scholar]
- 178.Zhu, R., Huang, Y., Zhao, J., Zhong, J. and Mechref, Y. (2020) Isomeric separation of N-glycopeptides derived from glycoproteins by porous graphitic carbon (PGC) LC-MS/MS. Anal. Chem. 92, 9556–9565 10.1021/acs.analchem.0c00668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zauner, G., Koeleman, C.A.M., Deelder, A.M. and Wuhrer, M. (2010) Protein glycosylation analysis by HILIC-LC-MS of proteinase K-generated N- and O-glycopeptides. J. Sep. Sci. 33, 903–910 10.1002/jssc.200900850 [DOI] [PubMed] [Google Scholar]
- 180.Dominguez-Vega, E., Tengattini, S., Peintner, C., van Angeren, J., Temporini, C., Haselberg, R.et al. (2018) High-resolution glycoform profiling of intact therapeutic proteins by hydrophilic interaction chromatography-mass spectrometry. Talanta 184, 375–381 10.1016/j.talanta.2018.03.015 [DOI] [PubMed] [Google Scholar]
- 181.van der Burgt, Y.E.M., Siliakus, K.M., Cobbaert, C.M. and Ruhaak, L.R. (2020) HILIC-MRM-MS for linkage-specific separation of sialylated glycopeptides to quantify prostate-specific antigen proteoforms. J. Proteome Res. 19, 2708–2716 10.1021/acs.jproteome.0c00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Huang, Y., Nie, Y., Boyes, B. and Orlando, R. (2016) Resolving isomeric glycopeptide glycoforms with hydrophilic interaction chromatography (HILIC). J. Biomol. Tech. 27, 98–104 10.7171/jbt.16-2703-003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kozlik, P., Goldman, R. and Sanda, M. (2018) Hydrophilic interaction liquid chromatography in the separation of glycopeptides and their isomers. Anal. Bioanal. Chem. 410, 5001–5008 10.1007/s00216-018-1150-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Molnarova, K. and Kozlik, P. (2020) Comparison of different HILIC stationary phases in the separation of hemopexin and immunoglobulin G glycopeptides and their isomers. Molecules 25, 4655 10.3390/molecules25204655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kammeijer, G.S.M., Jansen, B.C., Kohler, I., Heemskerk, A.A.M., Mayboroda, O.A., Hensbergen, P.J.et al. (2017) Sialic acid linkage differentiation of glycopeptides using capillary electrophoresis - electrospray ionization - mass spectrometry. Sci. Rep. 7, 3733 10.1038/s41598-017-03838-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kammeijer, G.S.M., Nouta, J., De La Rosette, J.J.M.C.H., De Reijke, T.M. and Wuhrer, M. (2018) An in-depth glycosylation assay for urinary prostate-specific antigen. Anal. Chem. 90, 4414–4421 10.1021/acs.analchem.7b04281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kammeijer, G.S.M., Kohler, I., Jansen, B.C., Hensbergen, P.J., Mayboroda, O.A., Falck, D.et al. (2016) Dopant enriched nitrogen gas combined with sheathless capillary electrophoresis–electrospray ionization-mass spectrometry for improved sensitivity and repeatability in glycopeptide analysis. Anal. Chem. 88, 5849–5856 10.1021/acs.analchem.6b00479 [DOI] [PubMed] [Google Scholar]
- 188.Qu, Y., Sun, L., Zhu, G., Zhang, Z., Peuchen, E.H. and Dovichi, N.J. (2018) Sensitive and fast characterization of site-specific protein glycosylation with capillary electrophoresis coupled to mass spectrometry. Talanta 179, 22–27 10.1016/j.talanta.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Khatri, K., Klein, J.A., Haserick, J.R., Leon, D.R., Costello, C.E., McComb, M.E.et al. (2017) Microfluidic capillary electrophoresis–mass spectrometry for analysis of monosaccharides, oligosaccharides, and glycopeptides. Anal. Chem. 89, 6645–6655 10.1021/acs.analchem.7b00875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Haselberg, R., De Jong, G.J. and Somsen, G.W. (2013) Low-flow sheathless capillary electrophoresis–mass spectrometry for sensitive glycoform profiling of intact pharmaceutical proteins. Anal. Chem. 85, 2289–2296 10.1021/ac303158f [DOI] [PubMed] [Google Scholar]
- 191.Redman, E.A., Batz, N.G., Mellors, J.S. and Ramsey, J.M. (2015) Integrated microfluidic capillary electrophoresis-electrospray ionization devices with online MS detection for the separation and characterization of intact monoclonal antibody variants. Anal. Chem. 87, 2264–2272 10.1021/ac503964j [DOI] [PubMed] [Google Scholar]
- 192.Le-Minh, V., Tran, N.T., Makky, A., Rosilio, V., Taverna, M. and Smadja, C. (2019) Capillary zone electrophoresis-native mass spectrometry for the quality control of intact therapeutic monoclonal antibodies. J. Chromatogr. A. 1601, 375–384 10.1016/j.chroma.2019.05.050 [DOI] [PubMed] [Google Scholar]
- 193.Giorgetti, J., Lechner, A., Del Nero, E., Beck, A., François, Y.-N. and Leize-Wagner, E. (2019) Intact monoclonal antibodies separation and analysis by sheathless capillary electrophoresis-mass spectrometry. Eur. J. Mass Spectrom. 25, 324–332 10.1177/1469066718807798 [DOI] [PubMed] [Google Scholar]
- 194.Camperi, J., Pichon, V. and Delaunay, N. (2020) Separation methods hyphenated to mass spectrometry for the characterization of the protein glycosylation at the intact level. J. Pharm. Biomed. Anal. 178, 112921 10.1016/j.jpba.2019.112921 [DOI] [PubMed] [Google Scholar]
- 195.Zhao, Y., Szeto, S.S.W., Kong, R.P.W., Law, C.H., Li, G., Quan, Q.et al. (2014) Online two-dimensional porous graphitic carbon/reversed phase liquid chromatography platform applied to shotgun proteomics and glycoproteomics. Anal. Chem. 86, 12172–9 10.1021/ac503254t [DOI] [PubMed] [Google Scholar]
- 196.Stavenhagen, K., Plomp, R. and Wuhrer, M. (2015) Site-specific protein N- and O-glycosylation analysis by a C18-porous graphitized carbon-liquid chromatography-electrospray ionization mass spectrometry approach using pronase treated glycopeptides. Anal. Chem. 87, 11691–9 10.1021/acs.analchem.5b02366 [DOI] [PubMed] [Google Scholar]
- 197.Stavenhagen, K., Kayili, H.M., Holst, S., Koeleman, C.A.M., Engel, R., Wouters, D.et al. (2018) N- and O-glycosylation analysis of human C1-inhibitor reveals extensive mucin-type O-glycosylation. Mol. Cell. Proteomics 17, 1225–1238 10.1074/mcp.RA117.000240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Stavenhagen, K., Gahoual, R., Dominguez Vega, E., Palmese, A., Ederveen, A.L.H., Cutillo, F.et al. (2019) Site-specific N- and O-glycosylation analysis of atacicept. mAbs 11, 1053–1063 10.1080/19420862.2019.1630218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Qu, Y., Sun, L., Zhang, Z. and Dovichi, N.J. (2018) Site-specific glycan heterogeneity characterization by hydrophilic interaction liquid chromatography solid-phase extraction, reversed-phase liquid chromatography fractionation, and capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry. Anal. Chem. 90, 1223–1233 10.1021/acs.analchem.7b03912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lu, J., Fu, D., Yu, L., Cao, C., Zou, L. and Liang, X. (2017) Determination of N-glycopeptides by hydrophilic interaction liquid chromatography and porous graphitized carbon chromatography with mass spectrometry detection. Anal. Lett. 50, 315–324 10.1080/00032719.2016.1181644 [DOI] [Google Scholar]
- 201.Glaskin, R.S., Khatri, K., Wang, Q., Zaia, J. and Costello, C.E. (2017) Construction of a database of collision cross section values for glycopeptides, glycans, and peptides determined by IM-MS. Anal. Chem. 89, 4452–4460 10.1021/acs.analchem.6b04146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gelb, A.S., Lai, R., Li, H. and Dodds, E.D. (2019) Composition and charge state influence on the ion-neutral collision cross sections of protonated N-linked glycopeptides: an experimental and theoretical deconstruction of coulombic repulsion vs. charge solvation effects. Analyst 144, 5738–5747 10.1039/C9AN00875F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Both, P., Green, A.P., Gray, C.J., Šardzík, R., Voglmeir, J., Fontana, C.et al. (2014) Discrimination of epimeric glycans and glycopeptides using IM-MS and its potential for carbohydrate sequencing. Nat. Chem. 6, 65–74 10.1038/nchem.1817 [DOI] [PubMed] [Google Scholar]
- 204.Pathak, P., Baird, M.A. and Shvartsburg, A.A. (2020) High-resolution Ion mobility separations of isomeric glycoforms with variations on the peptide and glycan levels. J. Am. Soc. Mass Spectrom. 31, 1603–1609 10.1021/jasms.0c00183 [DOI] [PubMed] [Google Scholar]
- 205.Campbell, J.L., Baba, T., Liu, C., Lane, C.S., Le Blanc, J.C.Y. and Hager, J.W. (2017) Analyzing glycopeptide isomers by combining differential mobility spectrometry with electron- and collision-based tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 28, 1374–1381 10.1007/s13361-017-1663-6 [DOI] [PubMed] [Google Scholar]
- 206.Hinneburg, H., Hofmann, J., Struwe, W.B., Thader, A., Altmann, F., Varon Silva, D.et al. (2016) Distinguishing N-acetylneuraminic acid linkage isomers on glycopeptides by ion mobility-mass spectrometry. Chem. Commun. (Camb) 52, 4381–4384 10.1039/C6CC01114D [DOI] [PubMed] [Google Scholar]
- 207.Wu, R., Chen, X., Wu, W.J., Wang, Z., Hung, Y.L.W., Wong, H.T.et al. (2020) Fine adjustment of gas modifier loadings for separation of epimeric glycopeptides using differential ion mobility spectrometry mass spectrometry. Rapid Commun. Mass Spectrom. 34, e8751 10.1002/rcm.8751 [DOI] [PubMed] [Google Scholar]
- 208.Guttman, M. and Lee, K.K. (2016) Site-specific mapping of sialic acid linkage isomers by Ion mobility spectrometry. Anal. Chem. 88, 5212–5217 10.1021/acs.analchem.6b00265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Barroso, A., Gimenez, E., Konijnenberg, A., Sancho, J., Sanz-Nebot, V. and Sobott, F. (2018) Evaluation of ion mobility for the separation of glycoconjugate isomers due to different types of sialic acid linkage, at the intact glycoprotein, glycopeptide and glycan level. J. Proteomics. 173, 22–31 10.1016/j.jprot.2017.11.020 [DOI] [PubMed] [Google Scholar]
- 210.Frese, C.K., Altelaar, A.F.M., Van Den Toorn, H., Nolting, D., Griep-Raming, J., Heck, A.J.R.et al. (2012) Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal. Chem. 84, 9668–9673 10.1021/ac3025366 [DOI] [PubMed] [Google Scholar]
- 211.Yu, Q., Wang, B., Chen, Z., Urabe, G., Glover, M.S., Shi, X.et al. (2017) Electron-transfer/higher-energy collision dissociation (EThcD)-enabled intact glycopeptide/glycoproteome characterization. J. Am. Soc. Mass Spectrom. 28, 1751–1764 10.1007/s13361-017-1701-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Čaval, T., Zhu, J., Tian, W., Remmelzwaal, S., Yang, Z., Clausen, H.et al. (2019) Targeted analysis of lysosomal directed proteins and their sites of mannose-6-phosphate modification. Mol. Cell. Proteomics. 18, 16–27 10.1074/mcp.RA118.000967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Cao, Q., Yu, Q., Liu, Y., Chen, Z. and Li, L. (2020) Signature-ion-triggered mass spectrometry approach enabled discovery of N- and O-linked glycosylated neuropeptides in the crustacean nervous system. J. Proteome Res. 19, 634–643 10.1021/acs.jproteome.9b00525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Yang, H., Yang, C. and Sun, T. (2018) Characterization of glycopeptides using a stepped higher-energy C-trap dissociation approach on a hybrid quadrupole orbitrap. Rapid Commun. Mass Spectrom. 32, 1353–1362 10.1002/rcm.8191 [DOI] [PubMed] [Google Scholar]
- 215.Wang, Y. and Tian, Z. (2020) New energy setup strategy for intact N-glycopeptides characterization using higher-energy collisional dissociation. J. Am. Soc. Mass Spectrom. 31, 651–657 10.1021/jasms.9b00089 [DOI] [PubMed] [Google Scholar]
- 216.Reiding, K.R., Bondt, A., Franc, V. and Heck, A.J.R. (2018) The benefits of hybrid fragmentation methods for glycoproteomics. TrAC. Trends Anal. Chem. 108, 260–268 10.1016/j.trac.2018.09.007 [DOI] [Google Scholar]
- 217.Domon, B. and Costello, C.E. (1988) A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate J. 5, 397–409 10.1007/BF01049915 [DOI] [Google Scholar]
- 218.Roepstorff, P. and Fohlman, J. (1984) Letter to the editors. Biol. Mass Spectrom. 11, 601 10.1002/bms.1200111109 [DOI] [PubMed] [Google Scholar]
- 219.Riley, N.M., Malaker, S.A., Driessen, M.D. and Bertozzi, C.R. (2020) Optimal dissociation methods differ for N- and O-glycopeptides. J. Proteome Res. 19, 3286–3301 10.1021/acs.jproteome.0c00218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhang, L. and Reilly, J.P. (2009) Extracting both peptide sequence and glycan structural information by 157 nm photodissociation of N-linked glycopeptides. J. Proteome Res. 8, 734–742 10.1021/pr800766f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Madsen, J.A., Ko, B.J., Xu, H., Iwashkiw, J.A., Robotham, S.A., Shaw, J.B.et al. (2013) Concurrent automated sequencing of the glycan and peptide portions of O-linked glycopeptide anions by ultraviolet photodissociation mass spectrometry. Anal. Chem. 85, 9253–9261 10.1021/ac4021177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Brodbelt, J.S. (2014) Photodissociation mass spectrometry: new tools for characterization of biological molecules. Chem. Soc. Rev. 43, 2757–2783 10.1039/C3CS60444F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Escobar, E.E., King, D.T., Serrano-Negrón, J.E., Alteen, M.G., Vocadlo, D.J. and Brodbelt, J.S. (2020) Precision mapping of O-linked N-acetylglucosamine sites in proteins using ultraviolet photodissociation mass spectrometry. J. Am. Chem. Soc. 142, 11569–11577 10.1021/jacs.0c04710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Halim, M.A., MacAleese, L., Lemoine, J., Antoine, R., Dugourd, P. and Girod, M. (2018) Ultraviolet, infrared, and high-low energy photodissociation of post-translationally modified peptides. J. Am. Soc. Mass Spectrom. 29, 270–283 10.1007/s13361-017-1794-9 [DOI] [PubMed] [Google Scholar]
- 225.Harvey, D.J. (2020) Negative ion mass spectrometry for the analysis of N-linked glycans. Mass Spectrom. Rev. 39, 586–679 10.1002/mas.21622 [DOI] [PubMed] [Google Scholar]
- 226.Halim, A., Westerlind, U., Pett, C., Schorlemer, M., Rüetschi, U., Brinkmalm, G.et al. (2014) Assignment of saccharide identities through analysis of oxonium Ion fragmentation profiles in LC–MS/MS of glycopeptides. J. Proteome Res. 13, 6024–6032 10.1021/pr500898r [DOI] [PubMed] [Google Scholar]
- 227.Yu, J., Schorlemer, M., Gomez Toledo, A., Pett, C., Sihlbom, C., Larson, G.et al. (2016) Distinctive MS/MS fragmentation pathways of glycopeptide-generated oxonium ions provide evidence of the glycan structure. Chemistry 22, 1114–1124 10.1002/chem.201503659 [DOI] [PubMed] [Google Scholar]
- 228.Pett, C., Nasir, W., Sihlbom, C., Olsson, B.M., Caixeta, V., Schorlemer, M.et al. (2018) Effective assignment of alpha2,3/alpha2,6-sialic acid isomers by LC-MS/MS-based glycoproteomics. Angew. Chem. Int. Ed. Engl. 57, 9320–9324 10.1002/anie.201803540 [DOI] [PubMed] [Google Scholar]
- 229.Acs, A., Ozohanics, O., Vekey, K., Drahos, L. and Turiak, L. (2018) Distinguishing core and antenna fucosylated glycopeptides based on low-energy tandem mass spectra. Anal. Chem. 90, 12776–12782 10.1021/acs.analchem.8b03140 [DOI] [PubMed] [Google Scholar]
- 230.Hoffmann, M., Pioch, M., Pralow, A., Hennig, R., Kottler, R., Reichl, U.et al. (2018) The fine art of destruction: a guide to in-depth glycoproteomic analyses-exploiting the diagnostic potential of fragment ions. Proteomics 18, e1800282 10.1002/pmic.201800282 [DOI] [PubMed] [Google Scholar]
- 231.Sanda, M., Benicky, J. and Goldman, R. (2020) Low collision energy fragmentation in structure-specific glycoproteomics analysis. Anal. Chem. 92, 8262–8267 10.1021/acs.analchem.0c00519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Dang, L., Shen, J., Zhao, T., Zhao, F., Jia, L., Zhu, B.et al. (2019) Recognition of bisecting N-glycans on intact glycopeptides by two characteristic ions in tandem mass spectra. Anal. Chem. 91, 5478–5482 10.1021/acs.analchem.8b05639 [DOI] [PubMed] [Google Scholar]
- 233.Lee, H.K., Ha, D.I., Kang, J.G., Park, G.W., Lee, J.Y., Cho, K.et al. (2020) Selective identification of alpha-galactosyl epitopes in N-glycoproteins using characteristic fragment ions from higher-energy collisional dissociation. Anal. Chem. 92, 13144–13154 10.1021/acs.analchem.0c02276 [DOI] [PubMed] [Google Scholar]
- 234.Darula, Z., Pap, A. and Medzihradszky, K.F. (2019) Extended sialylated O-glycan repertoire of human urinary glycoproteins discovered and characterized using electron-transfer/higher-energy collision dissociation. J. Proteome Res. 18, 280–291 10.1021/acs.jproteome.8b00587 [DOI] [PubMed] [Google Scholar]
- 235.Pap, A., Tasnadi, E., Medzihradszky, K.F. and Darula, Z. (2020) Novel O-linked sialoglycan structures in human urinary glycoproteins. Mol. Omics 16, 156–164 10.1039/C9MO00160C [DOI] [PubMed] [Google Scholar]
- 236.Marino, F., Bern, M., Mommen, G.P.M., Leney, A.C., van Gaans-van den Brink, J.A.M., Bonvin, A.et al. (2015) Extended O-GlcNAc on HLA class-I-bound peptides. J. Am. Chem. Soc. 137, 10922–5 10.1021/jacs.5b06586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nilsson, J., Brinkmalm, G., Ramadan, S., Gilborne, L., Noborn, F., Blennow, K.et al. (2019) Synthetic standard aided quantification and structural characterization of amyloid-beta glycopeptides enriched from cerebrospinal fluid of Alzheimer's disease patients. Sci. Rep. 9, 5522 10.1038/s41598-019-41897-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.White, III, C.W., Fan, X., Maynard, J.C., Wheatley, E.G.. Bieri, G., Couthouis, J.et al. (2020) Age-related loss of neural stem cell O-GlcNAc promotes a glial fate switch through STAT3 activation. Proc. Natl Acad. Sci. U.S.A. 117, 22214–22224 10.1073/pnas.2007439117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kuo, C.W. and Khoo, K.H. (2020) Strategic applications of negative-mode LC-MS/MS analyses to expedite confident mass spectrometry-based identification of multiple glycosylated peptides. Anal. Chem. 92, 7612–7620 10.1021/acs.analchem.0c00236 [DOI] [PubMed] [Google Scholar]
- 240.Eshghi S, T., Yang, W., Hu, Y., Shah, P., Sun, S., Li, X.et al. (2016) Classification of tandem mass spectra for identification of N- and O-linked glycopeptides. Sci. Rep. 6, 37189 10.1038/srep37189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Jeong, H.K., Hwang, H., Kang, Y.M., Lee, H.K., Park, G.W., Lee, J.Y.et al. (2020) Computational classification of core and outer fucosylation of N-glycoproteins in human plasma using collision induced dissociation in mass spectrometry. Rapid Commun. Mass Spectrom. 34, e8917 10.1002/rcm.8917 [DOI] [PubMed] [Google Scholar]
- 242.Hwang, H., Jeong, H.K., Lee, H.K., Park, G.W., Lee, J.Y., Lee, S.Y.et al. (2020) Machine learning classifies core and outer fucosylation of N-glycoproteins using mass spectrometry. Sci. Rep. 10, 318 10.1038/s41598-019-57274-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Park, G.W., Lee, J.W., Lee, H.K., Shin, J.H., Kim, J.Y. and Yoo, J.S. (2020) Classification of mucin-type O-glycopeptides using higher-energy collisional dissociation in mass spectrometry. Anal. Chem. 92, 9772–9781 10.1021/acs.analchem.0c01218 [DOI] [PubMed] [Google Scholar]
- 244.Reusch, D., Haberger, M., Falck, D., Peter, B., Maier, B., Gassner, J.et al. (2015) Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles—Part 2: mass spectrometric methods. mAbs 7, 732–742 10.1080/19420862.2015.1045173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Pap, A., Klement, E., Hunyadi-Gulyas, E., Darula, Z. and Medzihradszky, K.F. (2018) Status report on the high-throughput characterization of complex intact O-glycopeptide mixtures. J. Am. Soc. Mass Spectrom. 29, 1210–1220 10.1007/s13361-018-1945-7 [DOI] [PubMed] [Google Scholar]
- 246.Parker, B.L., Thaysen-Andersen, M., Solis, N., Scott, N.E., Larsen, M.R., Graham, M.E.et al. (2013) Site-specific glycan-peptide analysis for determination of N-glycoproteome heterogeneity. J. Proteome Res. 12, 5791–5800 10.1021/pr400783j [DOI] [PubMed] [Google Scholar]
- 247.Zhao, P., Praissman, J.L., Grant, O.C., Cai, Y., Xiao, T., Rosenbalm, K.E.et al. (2020) Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe 28, 586–601.e6 10.1016/j.chom.2020.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Thaysen-Andersen, M., Kolarich, D. and Packer, N.H. (2020) Glycomics & glycoproteomics: from analytics to function. Mol. Omics 10.1039/d0mo90019b [DOI] [PubMed] [Google Scholar]
- 249.Abrahams, J.L., Campbell, M.P. and Packer, N.H. (2018) Building a PGC-LC-MS N-glycan retention library and elution mapping resource. Glycoconj. J. 35, 15–29 10.1007/s10719-017-9793-4 [DOI] [PubMed] [Google Scholar]
- 250.Yang, Y., Wang, G., Song, T., Lebrilla, C.B. and Heck, A.J.R. (2017) Resolving the micro-heterogeneity and structural integrity of monoclonal antibodies by hybrid mass spectrometric approaches. mAbs 9, 638–645 10.1080/19420862.2017.1290033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lin, Y.H., Franc, V. and Heck, A.J.R. (2018) Similar albeit not the same: in-depth analysis of proteoforms of human serum, bovine serum, and recombinant human fetuin. J. Proteome Res. 17, 2861–2869 10.1021/acs.jproteome.8b00318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wohlschlager, T., Scheffler, K., Forstenlehner, I.C., Skala, W., Senn, S., Damoc, E.et al. (2018) Native mass spectrometry combined with enzymatic dissection unravels glycoform heterogeneity of biopharmaceuticals. Nat. Commun. 9, 1713 10.1038/s41467-018-04061-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Reiding, K.R., Franc, V., Huitema, M.G., Brouwer, E., Heeringa, P. and Heck, A.J.R. (2019) Neutrophil myeloperoxidase harbors distinct site-specific peculiarities in its glycosylation. J. Biol. Chem. 294, 20233–20245 10.1074/jbc.RA119.011098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Tjondro, H.C., Ugonotti, J., Kawahara, R., Chatterjee, S., Loke, I., Chen, S.et al. (2020) Hyper-truncated Asn355- and Asn391-glycans modulate the activity of neutrophil granule myeloperoxidase. J. Biol. Chem. 10.1074/jbc.RA120.016342 [DOI] [PMC free article] [PubMed] [Google Scholar]