Lipid homeostasis is critical for proper envelope functions. The level of LpxC, which catalyzes the first committed step of lipopolysaccharide (LPS) synthesis, is controlled by an essential protease complex comprised of FtsH and YciM. Work carried out here suggests YejM, an essential envelope protein, plays a central role in sensing the state of LPS synthesis and controls LpxC levels by regulating the activity of FtsH/YciM. All four essential proteins are attractive targets of therapeutic development.
KEYWORDS: Escherichia coli, FtsH protease, lipid homeostasis, LpxC, YciM, YejM, lipopolysaccharide
ABSTRACT
LpxC is a deacetylase that catalyzes the first committed step of lipid A biosynthesis in Escherichia coli. LpxC competes for a common precursor, R-3-hydroxymyristoyl-UDP-GlcNAc, with FabZ, whose dehydratase activity catalyzes the first committed step of phospholipid biosynthesis. To maintain the optimum flow of the common precursor to these two competing pathways, the LpxC level is controlled by FtsH/YciM-mediated proteolysis. It is not known whether this complex or another protein senses the status of lipid A synthesis to control LpxC proteolysis. The work carried out in this study began with a novel mutation, yejM1163, which causes hypersensitivity to large antibiotics such as vancomycin and erythromycin. Isolates resistant to these antibiotics carried suppressor mutations in the ftsH and yciM genes. Western blot analysis showed a dramatically reduced LpxC level in the yejM1163 background, while the presence of ftsH or yciM suppressor mutations restored LpxC levels to different degrees. Based on these observations, it is proposed that YejM is a sensor of lipid A synthesis and controls LpxC levels by modulating the activity of the FtsH/YciM complex. The truncation of the periplasmic domain in the YejM1163 protein causes unregulated proteolysis of LpxC, thus diverting a greater pool of R-3-hydroxymyristoyl-UDP-GlcNAc toward phospholipid synthesis. This imbalance in lipid synthesis perturbs the outer membrane permeability barrier, causing hypersensitivity toward vancomycin and erythromycin. yejM1163 suppressor mutations in ftsH and yciM lower the proteolytic activity toward LpxC, thus restoring lipid homeostasis and the outer membrane permeability barrier.
IMPORTANCE Lipid homeostasis is critical for proper envelope functions. The level of LpxC, which catalyzes the first committed step of lipopolysaccharide (LPS) synthesis, is controlled by an essential protease complex comprised of FtsH and YciM. Work carried out here suggests YejM, an essential envelope protein, plays a central role in sensing the state of LPS synthesis and controls LpxC levels by regulating the activity of FtsH/YciM. All four essential proteins are attractive targets of therapeutic development.
INTRODUCTION
Integrity of the outer membrane (OM) is vital for Gram-negative enteric bacteria, since this structure is directly exposed to hostile environments—such as in the mammalian gut—where enteric bacteria such as Escherichia coli live. The bacterial OM is an asymmetric lipid bilayer, because its two major lipid constituents, lipopolysaccharide (LPS) and phospholipids (PLs), are distributed asymmetrically, with LPS and PLs occupying the outer and inner leaflets, respectively (1, 2). β-Barrels and lipoproteins are the two major proteinaceous constituents of the OM, with the former class frequently making channels to allow diffusion of nutrients from the external milieu or transport of proteins/LPS into or across the OM (1). Unlike β-barrels, lipoproteins are involved in providing mechanical strength to the envelope structure (3) or in the transport of OM constituents (4, 5). A thin peptidoglycan layer beneath the OM is connected to it through a specific lipoprotein (Lpp) (6) and a β-barrel (OmpA) (7).
The OM function of maintaining the selective permeability barrier is compromised when its lipid asymmetry is perturbed (1, 2). Many factors contribute to this perturbation, including altered LPS packing due to interrupted bridging between LPS’s phosphate groups and cations (8) and processes that are defective in removing PL from the outer leaflet of the OM (9). LPS and PL synthesis is intimately tied to a common acyl-donating precursor, R-3-hydroxymyristoyl-acyl carrier protein (ACP), whose distribution into these two competing pathways is critically controlled (10, 11). LpxA catalyzes the first reversible step of UDP-GlcNAc acylation by R-3-hydroxymyristoyl-ACP (12). The second nonreversible step, involving LpxC-mediated deacetylation of acylated UDP-GlcNAc, commits the pathway to subsequent additions of acyl chains and the completion of the lipid A molecule (13). FabZ is a dehydratase that competes for R-3-hydroxymyristoyl-ACP for PL synthesis (10).
Mutations in lpxC, called envA, were first isolated among smooth ampicillin-sensitive derivatives of a rough ampicillin-resistant parent strain (14). The original envA mutation conferred sensitivity to several antibiotics (15, 16) and carried a missense mutation, resulting in an H19 to Y substitution in the 305-residue LpxC (EnvA) protein (17). Additional missense mutations in lpxC, called asmB, were later obtained among isolates that reversed the assembly defect of a mutant OmpF protein (18). To reveal the genetic interactive network of lpxC, revertants overcoming sensitivity to antibiotics or bile salts were obtained, and the extragenic suppressor alleles called sefA1 (15, 19) and sabA1 (20) were subsequently mapped to fabZ. Independently, mutations in fabZ were also identified as suppressors of a temperature sensitive allele of lpxA (10). These studies provided the genetic evidence of interactions between the two competing pathways involving LPS and PL biosynthesis. Indeed, when mutations in lpxC lowered LPS synthesis, a greater pool of acyl-ACP was driven toward PL synthesis (20). Inhibition of lipid A synthesis either by an lpxA mutation or an inhibitor of LpxC’s deacetylase activity elevated LpxC levels (21). This led to the hypothesis of the existence of a novel feedback loop that senses lipid A precursors to regulate the rate of lipid A biosynthesis (21). To coordinate this regulation, the authors also postulated the existence of a lipid A sensor in the OM or the periplasmic side of the inner membrane (IM). Consistent with the prediction of posttranscription regulation of LpxC, Ogura et al. (11) showed that an IM-bound essential protease, FtsH, is responsible for degrading LpxC and that FtsH’s essentiality may stem from elevated LPS synthesis.
A mutation in the putative lipid A sensor, yejM, was first obtained among antibiotic-sensitive isolates with reduced lipid A levels (22). However, the identity of the affected gene in the original isolate LH530 remained unknown until a decade later when De Lay and Cronan (23) identified it to be yejM. The original yejM mutation caused a truncation of the protein segment thought to reside in the periplasm (23). Moreover, it was determined that the IM-anchored region of YejM, predicted to fold into five transmembrane helices, was essential for growth (23). Recently, additional mutations in yejM were isolated through a genetic strategy that sought bypass suppressors to allow iron-chelating enterobactin to enter cells defective in its transport across the OM (24). These mutations also caused truncations of the putative periplasmic domain and conferred hypersensitivity to antibiotics, such as vancomycin, that are normally precluded from crossing the intact OM barrier. Based on the observations of increased OM permeability, elevated levels of lyso-PLs, and a synthetic growth phenotype without PldA, which is responsible for degrading PL from the outer leaflet of the OM, it was concluded that the essentiality of YejM stems from its role in lipid homeostasis (24). YejM is also implicated in cardiolipin transport in Salmonella (25, 26) and Shigella (27); however, YejM’s essentiality is unlikely to be due to this proposed function given that cardiolipin is not essential in these enteric bacteria.
In 2014, an IM-bound modulator of FtsH, YciM, was identified (28–30). yciM is essential, but the gene deletion is possible when cells are grown on a minimal medium at 30°C (29) or on a rich medium in the presence of a suppressor mutation mapping in lpxC (29, 30). These studies implicated that YciM regulates LPS synthesis by modulating FtsH-mediated degradation of LpxC and concluded that ΔyciM’s lethality, like that of ΔftsH, may be linked to LPS overproduction due to unregulated LpxC levels. However, it remained unclear as to how cells sensed excess LPS to activate FtsH/YciM-mediated degradation of LpxC levels and reestablish LPS/PL homeostasis. As concluded in this and two recent studies (31, 32), YejM likely plays a sensory role for LPS synthesis by monitoring the accumulation of LPS intermediates in the IM and coordinates with the FtsH/YciM complex to regulate LpxC levels.
The study presented here took advantage of the antibiotic hypersensitivity phenotype of the mutant yejM1163 allele (24) and sought antibiotic-resistant revertants. Suppressor mutations in these revertants were found in the ftsH and yciM genes. It was shown that yejM1163 caused a dramatic reduction in the LpxC level, while suppressor mutations reelevated LpxC levels to various degrees to normalize lipid homeostasis and repair a breach in the OM permeability barrier.
RESULTS AND DISCUSSION
Isolation of antibiotic-resistant revertants of the ΔtonB yejM1163 strain.
The antibiotic hypersensitivity phenotype of the ΔtonB yejM1163 strain allowed the opportunity to isolate and characterize suppressor mutations that confer antibiotic resistance. Vancomycin and erythromycin were chosen as selective agents, since both are large antibiotics (>700 Da) and thus are useful in probing outer membrane permeability defects. Erythromycin- and vancomycin-resistant mutants arose at a frequency of 10−7 to 10−8, indicating the prevalence of possible missense mutations in genes indispensable under the applied selective condition. Six independently obtained resistant isolates were characterized in some detail for this study.
Whole-genome sequence (WGS) analysis was carried out to locate the mutation responsible for the antibiotic-resistant phenotype. The initial WGS analysis involved four revertants with distinct growth phenotypes on selective and nonselective media. Once mutations were located, they were confirmed by PCR amplification and Sanger sequencing of the affected gene. Genetic location of suppressor mutations in the remaining two revertants were first determined by P1 transductional analysis using markers linked to the targeted genes, followed by direct Sanger sequencing of the amplified DNA fragment. All six suppression mutations were unique and mapped within the two essential genes, ftsH and yciM (lapB) (Table 1). The location of all six suppressors in two essential genes and their missense nature were consistent with the low frequency of occurrence of the antibiotic-resistant revertants.
TABLE 1.
MICs of vancomycin and erythromycin in various genetic backgrounds
| Strain name | Relevant genotype | Codon (no.) change | MIC (μg/ml) of: |
|
|---|---|---|---|---|
| Erythromycin | Vancomycin | |||
| RAM3199 | ΔtonB | NAa | 96 | 128 |
| RAM3200 | ΔtonB yejM1163 | NA | 3 | 4 |
| RAM3201 | ΔtonB yejM1163 ftsH-E367D | GAA (367)→GAC | 48 | 64 |
| RAM3202 | ΔtonB yejM1163 yciM-W377G | UGG (377)→GGG | 48 | 12 |
| RAM3203 | ΔtonB yejM1163 yciM-A126V | GCG (126)→GUG | 96–128 | ≥128 |
| RAM3204 | ΔtonB yejM1163 yciM-L95R | CUA (95)→CGA | 96–128 | ≥128 |
| RAM3205 | ΔtonB yejM1163 ftsH-L78R | CUG (78)→CGG | 64–96 | 96-128 |
| RAM3206 | ΔtonB yejM1163 ftsH-D74A | GAU (74)→GCU | 32–48 | 64-96 |
NA, not applicable.
Phenotypic characterization of revertants.
We first determined MICs against the two antibiotics used for selection, erythromycin and vancomycin, using Etest presoaked antibiotic strips. As expected, all six revertants had significantly higher MICs against erythromycin and vancomycin than the parental strain carrying the yejM1163 allele (Table 1). In two revertants, carrying YciM-L95R and YciM-A126V substitutions, the resistance against these two OM permeability-probing antibiotics reached similar levels as the strain carrying the wild-type yejM allele.
Strains used in this study contained a ΔtonB mutation, which significantly impaired cell growth on a medium not supplemented with 40 μM ferric chloride (24). yejM1163 was isolated among revertants that grew more rapidly than the parent ΔtonB strain on a medium not supplemented with ferric chloride (24). yejM1163 was subsequently shown to increase OM permeability, allowing iron to enter the cell and relieving growth defects on an unsupplemented medium (24). Based on these observations, it stands to reason that suppressor mutations that overcome the antibiotic sensitivity phenotype of the ΔtonB yejM1163 strain would simultaneously reduce the OM permeability defect of the strain. If so, we expect to see a reduced leaky phenotype and somewhat impaired growth on a medium not supplemented with ferric chloride.
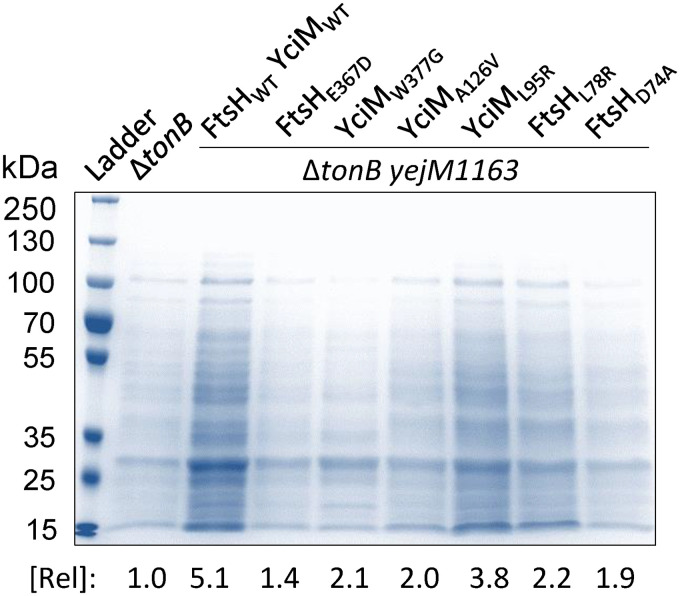
To assess the leaky phenotype, cell-free supernatants were isolated from overnight cultures grown with ferric chloride. Iron was added to ensure normal growth of the ΔtonB control strain and revertants that increase MICs against erythromycin and vancomycin. After separating cells by centrifugation, any remaining cells were removed by filtering supernatants through 0.2-μm-pore size syringe filters. Supernatants were concentrated by centrifugation under vacuum and reconstituted in roughly one-tenth the original volume. Samples of the supernatants, normalized to the culture optical density at 600 nm (OD600), were analyzed by SDS-PAGE, and protein bands were visualized and quantified after Coomassie brilliant blue staining (Fig. 1). As expected, the presence of the yejM1163 mutation in the ΔtonB background caused a dramatic increase the amount of proteins leaked through the outer membrane. It should be noted that we previously showed that leaked proteins in the yejM1163 background are not the result of cell lysis, since, unlike the highly leaky ΔtolA strain (33), there was no trace of the cytoplasmic enzyme in the culture supernatant (24). All six revertants derived from the ΔtonB yejM1163 strain had reduced levels of leaked proteins (Fig. 1), thus supporting the hypothesis that the suppressor mutations mapping in ftsH or yciM lower the OM permeability of the yejM1163 mutant. Based on antibiotic resistance data (Table 1), we were expecting YciM-L95R and YciM-A126V mutants to display the lowest leaky phenotype, but this was not the case (Fig. 1). One possibility for this discrepancy may be that higher protein contents in the culture supernatants of the two YciM mutants in Fig. 1 reflect a combined result of perturbed OM integrity and increased OM vesiculation. In contrast, the antibiotic resistance phenotype in Table 1 mainly reflects restored OM permeability barrier.
FIG 1.
Examination of cell-free culture supernatants by SDS-PAGE. Cells from overnight-grown cultures were spun down, and the supernatants, after filtration and concentration, were analyzed by SDS-PAGE. Supernatant proteins were visualized after staining with Coomassie brilliant blue. Protein bands were quantified using Bio-Rad Quantity One software (amounts relative to that in the ΔtonB strain are shown). The key genetic characteristics of strains used are shown at the top.
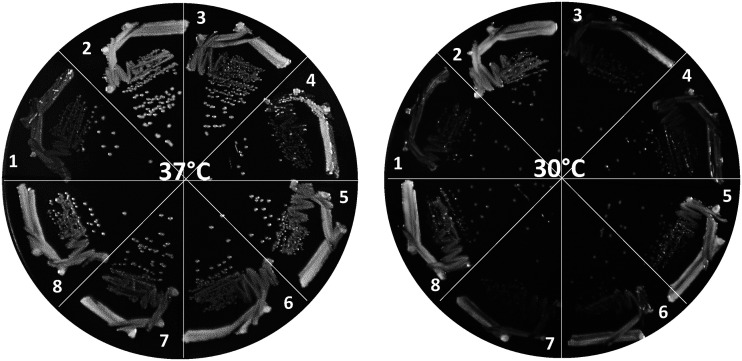
Next, we tested the growth of strains on a medium unsupplemented with ferric chloride (Fig. 2). The ΔtonB strain grew poorly, and faster-growing revertants were occasionally visible after 24 h growth at both 30°C and 37°C. In contrast, the ΔtonB yejM1163 strain grew much faster than the ΔtonB strain, forming larger colonies, and produced no visible revertants. All six revertants had growth patterns in between those of the ΔtonB and ΔtonB yejM1163 strains, with more pronounced growth differences at 30°C than at 37°C (Fig. 2). It is worth noting that we did not determine bacterial growth rates by monitoring growth of liquid cultures due to the concern that faster growing revertants, as seen in the ΔtonB strain and one of the revertants carrying YciM-W377G, may take over the population and thus artificially display better-than-expected growth. Nevertheless, combined with higher MIC (Table 1) and reduced protein leakage data (Fig. 1), the qualitative growth data (Fig. 2) are consistent with the notion that all six suppressor mutations reduce the OM permeability defect of the yejM1163 mutant.
FIG 2.
Bacterial growth on LBA was recorded after incubating petri plates at 37°C and 30°C for 24 h. Bacterial strains used are as follows: 1, RAM3199 (ΔtonB); 2, RAM3200 (ΔtonB yejM1163); 3, RAM3201 (RAM3200 ftsH-E367D); 4, RAM3202 (RAM3200 yciM-W377G); 5, RAM3203 (RAM3200 yciM-A129V); 6, RAM3204 (RAM3200 yciM-L95R); 7, RAM3205 (RAM3200 ftsH-L78R); 8, RAM3206 (RAM3200 ftsH-D74A).
Effect of suppressor mutations on LpxC levels.
We previously concluded that YejM plays an essential role in lipid homeostasis independent of its putative nonessential role in transporting cardiolipin from the inner to the outer membrane (24). This conclusion was further corroborated by the identification of yejM1163 suppressor mutations in two genes—ftsH and yciM—whose products coordinate proteolysis of LpxC (11, 29, 30, 34), an essential enzyme that operates at a critical juncture in the biogenesis of LPS and PL (10). Based on the central role LpxC plays in lipid biogenesis and the fact that yejM1163 suppressors map to genes whose products control LpxC levels, we hypothesized that YejM positively regulates LpxC levels. Accordingly, LpxC would be destabilized in the yejM1163 background but restabilized by suppressor mutations in the ftsH and yciM genes. This hypothesis is also consistent with the membrane permeability data, since destabilization in LpxC is expected to drive the common pool of lipid intermediate toward PL synthesis and interfere with the OM asymmetry and permeability (Table 1 and Fig. 1).
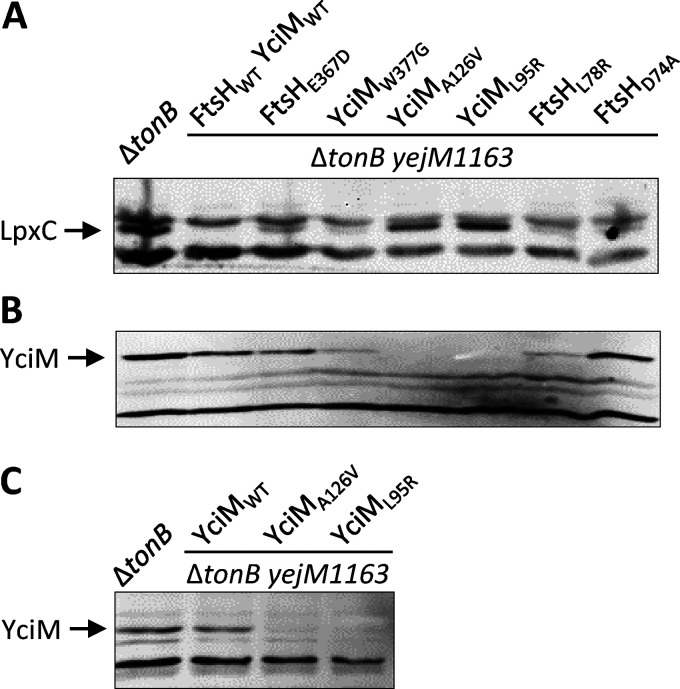
To test our hypothesis, we examined LpxC levels by Western blot analysis using the commercially available LpxC polyclonal antibodies (Fig. 3A). In the wild-type yejM background, the LpxC band, migrating at the 34-kDa position, was present just below a cross-reacting band running above it. Remarkably, no LpxC was detected from cell extracts of the yejM1163 strain. Different levels of LpxC were detected in strains carrying suppressor mutations in the ftsH or yciM gene. Although LpxC was barely visible in four of the suppressor-carrying strains, its levels rose dramatically in two strains carrying the A126V and L95R alterations in the YciM protein (Fig. 3A). The absence of LpxC in the yejM1163 strain supported the hypothesis that wild-type YejM is required for LpxC’s stability. On the other hand, suppressor mutations appear to stabilize LpxC and reestablish lipid homeostasis by downregulating the activities of two negative modulators of LpxC, FtsH and YciM.
FIG 3.
Effects of ftsH and yciM suppressor mutations on LpxC and YciM levels. Western blot analyses were carried out to detect LpxC (A) and YciM (B and C). LpxC was examined from whole-cell extracts, while YciM was examined from either purified envelopes (B) or whole-cell extracts (C).
Effect of suppressor mutations on YciM levels.
The strong suppressive effects of YciM-A126V and YciM-L95R prompted the examination of YciM levels to determine whether suppressors broadly affect YciM’s folding and stability or its specific function. YciM levels were examined from Western blots of envelopes purified from overnight cultures (Fig. 3B). Remarkably, all three yciM suppressor mutants showed significantly reduced levels of YciM, and no YciM was detected in strains carrying the YciM-A126V or YciM-L95R substitution. It is important to note that LpxC levels were the highest in the same two suppressor strains with no detectable YciM (Fig. 3A), thus showing a strong negative correlation between the levels of the two proteins. Curiously, the level of YciM was also significantly reduced in one of the suppressors carrying the FtsH-L78R substitution. Although the reason for this is not clear, it may reflect altered substrate specificity of FtsH (see below).
Because YciM levels in strains carrying the A126V and L95R suppressor alterations were dramatically low, we considered the possibility that mutant YciM was lost during the lengthy envelope isolation procedure. Therefore, YciM levels in these two strains were reexamined from cell extracts obtained by directly boiling pelleted cells in an SDS-based sample buffer (Fig. 3C). Again, unlike the control strains, no YciM was detected in the two suppressor carrying strains, thus confirming the notion that in vivo misfolding/destabilization of the mutant YciM proteins likely reduces FtsH-mediated degradation of LpxC.
Phenotypic verification of the mutant YciM data.
The data presented above suggested that reduced levels of the mutant YciM proteins led to LpxC stabilization and restoration of lipid homeostasis in the yejM1163 background. If so, expression of wild-type YciM in the mutant yciM background should once again stimulate LpxC degradation and destabilize lipid homeostasis. We can phenotypically test this balance/imbalance in lipid biogenesis by determining MICs against erythromycin and vancomycin. The wild-type gene and a mutant yciM allele, yciM-L95R, were cloned into pBAD24 in which an arabinose-inducible promoter controls the expression of the cloned gene (35). Plasmids with cloned yciM genes and an empty vector were transformed into the yejM1163 strain carrying the yciM-L95R mutation, and MICs against erythromycin and vancomycin were measured. The presence of a plasmid carrying the wild-type yciM gene restored the drug sensitivity phenotype in the yejM1163 yciM-L95R strain (Table 2), indicating that (i) yciM-L95R is recessive to the wild-type allele and (ii) the presence of wild-type YciM triggers an imbalance in lipid homeostasis, presumably by inducing FtsH-mediated LpxC proteolysis. As expected, a plasmid carrying the mutant yciM-L95R allele or the empty vector failed to reverse the drug-resistant phenotype of the yejM1163 yciM-L95R strain (Table 2), reflecting the preservation of balanced lipid homeostasis. These data further support the notion that the unstable YciM-L95R and presumably YciM-A126V proteins fail to stimulate FtsH-mediated LpxC proteolysis, thus suppressing the yejM1163 defect. The presence of the pBAD24-yciM plasmid in the ΔtonB background did not significantly change MICs against erythromycin or vancomycin, while the yejM1163 strain carrying this plasmid grew sickly and displayed elevated MICs against the two antibiotics, presumably due to overexpression of YciM (Table 2). It is worth mentioning that during MIC tests, leaky yciM expression from the plasmid was sufficient for complementation. The presence of 0.02% arabinose caused a severe growth defect in the strain expressing wild-type yciM but not the mutant yciM gene (data not shown). We assume that this growth defect likely stems from enhanced proteolysis of LpxC, whose levels are already extremely low due to yejM1163, thus further curtailing the flow of the common lipid precursor pool toward LPS synthesis.
TABLE 2.
MICs of vancomycin and erythromycin in various genetic backgrounds containing an empty pBAD24 vector or the cloned wild-type or mutant yciM gene
| Strain name | Relevant genotype | MIC (μg/ml) of: |
|
|---|---|---|---|
| Erythromycin | Vancomycin | ||
| RAM3212 | ΔtonB (pBAD24) | 64–96 | 128 |
| RAM3213 | ΔtonB (pBAD-yciM) | 64–96 | 128 |
| RAM3214 | ΔtonB yejM1163 (pBAD24) | 6 | 4 |
| RAM3215 | ΔtonB yejM1163 (pBAD-yciM) | 3 | 1.5 |
| RAM3207 | ΔtonB yejM1163 yciM-L95R (pBAD24) | 64–96 | >128 |
| RAM3208 | ΔtonB yejM1163 yciM-L95R (pBAD-yciM) | 4 | 2 |
| RAM3209 | ΔtonB yejM1163 yciM-L95R (pBAD-yciM-L95R) | 96 | >128 |
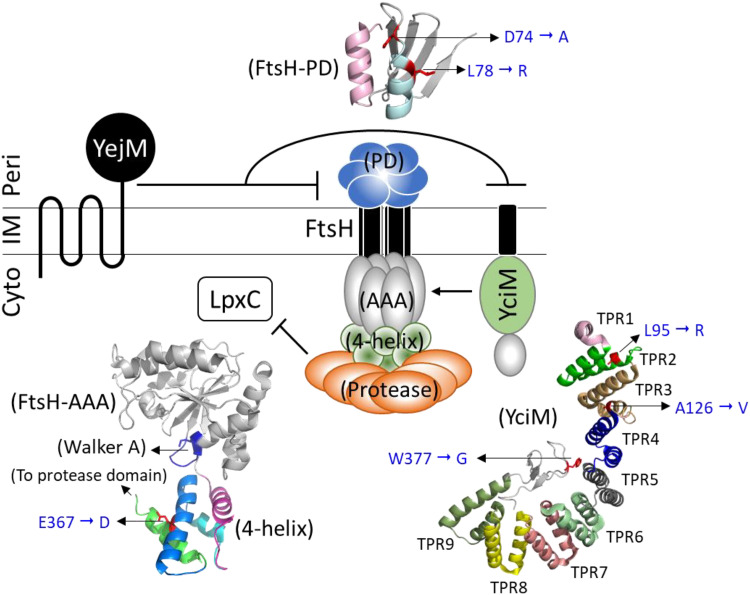
YciM structure and the possible mechanism of suppression.
Modeling (28, 29) and structural (36) analyses revealed that YciM has two distinct domains, with its N terminus anchored to the inner membrane, while the soluble cytoplasmic domain is folded into 9 tetratricopeptide repeat (TPR) motifs, known for their ability to mediate protein-protein interactions (37), and a rubredoxin-like metal-binding motif (38) at its C terminus. The three novel suppressor mutations isolated in this work affect highly conserved residues in TPR-2 (L95R), TPR-3 (A126V), or the metal-binding motif (W377G) (Fig. 4 and 5). The L95R and A126V substitutions drastically lower YciM levels, while the effect of W377G on YciM levels is less severe (Fig. 3). Although the three substitutions could affect specific activities of YciM, e.g., its interaction with FtsH or metal-binding capacity, lower YciM levels in all three cases preclude us from suggesting a specific functional defect. Instead, it is possible that the suppressive effect of these substitutions stems from their lower levels. Since YciM interacts with FtsH, a broad-specificity envelope protease, it is conceivable that structural changes brought upon by the three substitutions make the mutant YciM proteins targets of FtsH. The net result of lower YciM levels would be reduced FtsH-mediated proteolysis of LpxC and, consequently, its stabilization in the yejM1163 background.
FIG 4.
Alignment of selected regions of YciM/LapB and FtsH proteins from different species by ClustalW2 analysis. The YciM sequence alignment included the TPR2, TPR3, and rubredoxin-like domains, while the FtsH sequence alignment included the periplasmic and four-helix bindle domains. YciM/LapB protein sequences used were from E. coli (P0AB58), Alteromonas sp. (A0A5H3A1J7), Haemophilus influenzae (P44130), Aliivibrio fischeri (Q5E3Z5), and Citrobacter werkmanii (A0A090TYQ6). FtsH protein sequences used were from E. coli (P0AAI3), Thermotoga maritima (Q9WZ49), and Arabidopsis thaliana (Q39102).
FIG 5.
A cartoon showing the working model of the involvement of YejM in regulating LpxC levels. It is hypothesized that the periplasmic domain of YejM interacts with the FtsH/YciM complex to lower its proteolytic activity against LpxC. Also shown are the various domains of YciM and FtsH, where six suppressor alterations isolated in this study are located. FtsH-PD, periplasmic domain; FtsH-AAA, ATPase associated with diverse cellular activities; 4-helix, four-helix bundle; TPR, tetratricopeptide repeat; Cyto, cytoplasm; IM, inner membrane; Peri, periplasmic.
It is important to note that YciM is an essential protein, yet at least in two mutants, YciM-L95R and YciM-A126V, YciM levels were undetectable by Western blotting. Previous work has shown that a deletion of yciM is tolerated in a background containing a missense mutation in lpxC (29, 30). Essentiality of YciM in the wild-type background is thought to stem from LPS overproduction due to LpxC stabilization without activated FtsH (29, 30). Consequently, mutations such as those mapping to lpxC, which decrease the flow of the common lipid precursor pool to LPS, are expected to be tolerated in a ΔyciM background. Due to extremely low protein levels, yciM-L95R and yciM-A126V mimic a yciM-null allele. yejM1163 dramatically reduces LpxC levels/stability, thus creating an environment where yciM-L95R and yciM-A126V alleles will be tolerated. Indeed, yciM-L95R and yciM-A126V elevate LpxC levels in the yejM1163 background, but this increase simply normalizes the flow of the common lipid precursor pool between LPS and PL pathways and repairs the OM permeability defect caused by yejM1163.
FtsH structure and the mechanism of suppression.
FtsH is an assemblage of several topologically, structurally, and functionally distinct domains (39–41). The three novel suppressor mutations isolated in this study affect relatively conserved residues (Fig. 5) and map in two distinct domains: D74A and L78R are present in the periplasmic domain (PD), while E367D affects the cytoplasmic 4-helix-bundle domain, which is sandwiched between the nucleotide-binding AAA domain and the protease domain (Fig. 5). Although the primary sequence of the periplasmic domain shows little conservation, the two affected residues, D74 and L78, are relatively conserved within α-helix 2 (Fig. 4). The periplasmic domain of FtsH is shown to be essential neither for oligomerization nor for proteolytic activity; instead, it is proposed to play a role in the recognition of membrane-bound substrates (42). This domain of FtsH interacts with the negative modulators HflKC that are thought to influence proteolysis of membrane-bound substrates from the periplasmic side (42). LpxC is a cytoplasmic protein, and so it is unlikely that the two suppressor alterations in the periplasmic domain of FtsH directly affect LpxC recognition. We consider an alternative that the periplasmic domain of YejM negatively modulates FtsH through interactions with FtsH’s periplasmic domain. If so, a truncation of the periplasmic domain in YejM1163 may activate FtsH and enhance LpxC proteolysis. D74A and L78R may indirectly lower FtsH’s protease activity or LpxC recognition so that stabilized LpxC can direct the common lipid precursor pool toward LPS synthesis.
Function of the 4-helix-bundle domain, where the E367D alteration lies, remains poorly understood. Residues of the E367-containing α-helix 8 are highly conserved (Fig. 5), indicating structural/functional importance. Due to its positioning between the nucleotide-binding and protease domains (Fig. 5), the 4-helix-bundle domain may be crucial for allosteric communications between the two domains during ATP hydrolysis and substrate binding/release. The E367D change may interfere with this communication and thus the proteolytic activity of FtsH. It is also worth considering that E367D may affect FtsH’s interaction with YciM, lessening FtsH activation. Regardless of the precise mechanism, E367D appears to lower FtsH’s protease activity to partially stabilize LpxC (Fig. 3) and mend the OM permeability defect (Table 1).
Comparison of our findings with those from other laboratories.
After our initial conclusion of the essential role of YejM in lipid homeostasis (24), two subsequent publications (31, 43) and one recently submitted manuscript (32) provided independent verifications of this thesis. The work carried out here and that in three other laboratories began with either mutant yejM alleles producing C-terminal truncated proteins (24, 31, 43) or with a strain in which yejM expression could be depleted (32). All four studies sought revertants with improved growth phenotypes, and in all cases, yciM turned out to be a common site of suppressor mutations. Since YciM is a positive modulator of FtsH that degrades LpxC (Fig. 5), it is not surprising that work conducted here (Table 1) and by Cian et al. (43) also found suppressor mutations mapping to ftsH. Moreover, as LpxC is the target of the YciM-FtsH complex, overexpression of LpxC (32) or missense mutations in lpxC (43) also overcame the growth defect caused by YejM truncation or depletion. These genetic findings revealed a crucial role of YejM in stabilizing LpxC, which sits in a critical juncture of LPS and PL synthesis (10, 13, 20, 21).
The question remains as to exactly how YejM stabilizes LpxC. One hypothesis posits direct binding of YejM to the FtsH/YciM complex to lower its activity toward LpxC (Fig. 5). Indeed, the work carried out here (Fig. 3) and that by Fivenson and Bernhardt (32) showed LpxC destabilization in the YejM mutant or YejM-depleted background. Colocalization studies have indicated that YejM interacts with YciM (32). Curiously, however, no studies have suggested any significant part of YciM being exposed to the periplasm. Instead, the available data suggest that YciM is anchored to the inner membrane through its first 20 amino acids, while the rest of the protein resides in the cytoplasm (28). Given this topology, it is difficult to see how YejM’s periplasmic domain, which is partially deleted in YejM1163, would directly interact with YciM to interfere with its function. On the other hand, the periplasmic domain of FtsH, located between its two transmembrane (TM) helices, has been shown to be important for its function and interaction with its negative modulators, HflKC (42). Moreover, two of the three novel ftsH suppressor mutations, which appear to lower FtsH’s activity against LpxC, map to the periplasmic domain of FtsH (Fig. 5). Based on the observations that YciM apparently lacks any significant periplasmically exposed region and that FtsH’s periplasmic domain influences its function, it is worth considering that YejM may interact with FtsH’s periplasmic domain to modulate its activity. While experimental verification for this awaits, a common theme has emerged from the recent studies that YejM plays a crucial role in regulating lipid homeostasis by modulating FtsH/YciM-mediated proteolysis of LpxC. Future work on how an imbalance in lipid homeostasis affects YejM activity will provide a clearer picture on its role as an LpxC regulator.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and antibiotic sensitivity assays.
All Escherichia coli K-12 strains used in this study are derived from RAM1292 (MC4100 Δara714) (44). Lysogenic broth (LB) was prepared from Difco LB EZMix powder. LB agar (LBA) was prepared using LB plus 1.5% agar (Becton, Dickinson and Company). When required, kanamycin (25 μg/ml), ampicillin (50 μg/ml), and FeCl3 (40 μM) were added to LB or LBA. Unless specified, all cultures were incubated at 37°C for 18 h. All chemicals were of analytical grade. Antibiotic sensitivity was assessed by determining MICs of vancomycin and erythromycin. Etest strips (bioMérieux) soaked with a concentration gradient of an antibiotic were placed on LBA plates overlaid with 4 ml of soft agar containing 100 μl of overnight-grown bacterial cultures and ferric chloride. When strains containing a plasmid were used, soft agar was also supplemented with ampicillin. Plates were incubated for 24 h at 37°C, after which the inhibition zones were measured. Two independent cultures were used, and each culture was tested with two duplicates of antibiotics strips.
Genetic and DNA methods.
Bacteriophage P1-mediated transductions, using antibiotic resistance markers, were carried out as described previously (45). The promoterless yciM gene was cloned into pBAD24 (35) by ligating a PCR-amplified yciM open reading frame (ORF). Cloning was enabled by the introduction of NcoI and HindIII restriction sites during PCR amplification of the wild-type or mutant yciM ORF and ligating into pBAD24 that was also restriction digested with NcoI and HindIII. The ligated mixture was transformed into transformation-competent E. coli JM109 cells, and transformed colonies were selected for on LBA containing ampicillin. Plasmids containing the correct-size yciM gene clone were extracted using a QiaPrep Spin MiniPrep kit (Qiagen) and sequenced. pBAD24-yciM was then transformed into appropriate strains for complementation analysis. Sanger sequencing and whole-genome sequencing, as described previously (33), were carried out at the Arizona State University DNA Core Facility. The yciM NcoI forward primer sequence was 5′-ATTCCATGGTGGAGTTGTTGTTTCTGCTTTTGCC-3′; the yciM HindIII rev primer sequence was 5′-ATAAGCTTAGTGGTGGTGGTGGTGGTGCAGGCCATCAAGACCGCG-3′.
Protein methods.
To isolate whole-cell envelopes, cells were lysed by the French press method (46). Envelopes were pelleted by centrifuging cell-free lysates for 1 h at 105,000 × g and resuspended in an SDS-based sample buffer. For detection of specific proteins, Western blot analysis was carried out as described previously (46). LpxC and YciM primary rabbit antibodies (purchased from LifeSpan Biosciences, Inc.) were diluted 1:5,000 prior to use. To isolate cell-free supernatant, cells from 1-ml overnight cultures were pelleted by centrifugation at 16,000 × g for 5 min. Eight hundred microliters of the supernatant was collected and passed through a 0.22-μm filter. Filtrate, roughly 600 μl, was dried in a SpeedVac and reconstituted in 50 μl of sterile water. Concentrated supernatants (9 μl) were analyzed by SDS-polyacrylamide (11%) gels, and proteins were visualized by staining the gel with Coomassie brilliant blue.
ACKNOWLEDGMENTS
We thank the Research and Training Initiatives office of the School of Life Sciences and NIH grant R21 AI117150 for partly funding this work.
Footnotes
For a commentary on this article, see https://doi.org/10.1128/JB.00370-20.
REFERENCES
- 1.Nikaido H. 2003. Molecular basis of bacterial outer membrane revisited. Microbiol Mol Biol Rev 67:593–656. 2003. doi: 10.1128/mmbr.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazzaroni JC, Germon P, Ray MC, Vianney A. 1999. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol Lett 177:191–197. doi: 10.1111/j.1574-6968.1999.tb13731.x. [DOI] [PubMed] [Google Scholar]
- 4.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol 61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 5.Chng S-S, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. 2010. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc Natl Acad Sci U S A 107:5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun V, Wolff H. 1970. The murein-lipipoprotein linkage in the cell wall of Escherichia coli. Eur J Biochem 14:387–391. doi: 10.1111/j.1432-1033.1970.tb00301.x. [DOI] [PubMed] [Google Scholar]
- 7.Park JS, Lee WC, Yeo KJ, Ryu K-S, Kumarasiri M, Hesek D, Lee M, Mobashery S, Song JH, Kim SI, Lee JC, Cheong C, Jeon YH, Kim H-Y. 2012. Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J 26:219–228. doi: 10.1096/fj.11-188425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snyder S, Kim D, McIntosh TJ. 1999. Lipopolysaccharide bilayer structure: effect of chemotype, core mutations, divalent cation, and temperature. Biochemistry 38:10758–10767. doi: 10.1021/bi990867d. [DOI] [PubMed] [Google Scholar]
- 9.May KL, Silhavy TJ. 2018. The Escherichia coli phospholipase PldA regulates outer membrane homeostasis via lipid signaling. mBio 9:e00379-18. doi: 10.1128/mBio.00379-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohan S, Kelly TM, Eveland SS, Raetz CRH, Anderson MS. 1994. An Escherichia coli gene (fabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase. J Biol Chem 269:32896–32903. [PubMed] [Google Scholar]
- 11.Ogura T, Inoue K, Tatsuta T, Suzaki T, Karata K, Young K, Su L-H, Fierk CA, Jackman JE, Raetz CRH, Coleman J, Tomoyasu T, Matsuzawa H. 1999. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol Microbiol 31:833–844. doi: 10.1046/j.1365-2958.1999.01221.x. [DOI] [PubMed] [Google Scholar]
- 12.Golloway SM, Raetz C. 1990. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J Biol Chem 265:6394–6402. [PubMed] [Google Scholar]
- 13.Young K, Silver LL, Bramhill D, Cameron P, Eveland SS, Raetz CRH, Hyland SA, Anderson MS. 1995. The envA permeability/cell divison gene of Escherichia coli encodes the second enzyme of lipid A biosynthesis. J Biol Chem 270:30384–30391. doi: 10.1074/jbc.270.51.30384. [DOI] [PubMed] [Google Scholar]
- 14.Normark S, Boman HG, Matsson E. 1969. Mutant of Escherichia coli with anomalous cell division and ability to decrease episomally and chromosomally mediated resistance to ampicillin and several other antibiotics. J Bacteriol 97:1334–1342. doi: 10.1128/JB.97.3.1334-1342.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Normark S. 1970. Genetics of a chain-forming mutant of Escherichia coli: transduction and dominance of the envA gene mediating increased penetration to some antibacterial agents. Genet Res 16:63–78. doi: 10.1017/s0016672300002287. [DOI] [PubMed] [Google Scholar]
- 16.Young K, Silver LL. 1991. Leakage of periplasmic enzymes from envA1 strains of Escherichia coli. J Bacteriol 173:3609–3614. doi: 10.1128/jb.173.12.3609-3614.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beall B, Lutkenhaus J. 1987. Sequence analysis, transcriptional organization, and insertional mutagenesis of the envA gene of Escherichia coli. J Bacteriol 169:5408–5415. doi: 10.1128/jb.169.12.5408-5415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloser AW, Laird MW, Misra R. 1996. asmB, a suppressor locus for assembly-defective OmpF mutants of Escherichia coli, is allelic to envA (lpxC). J Bacteriol 178:5138–5143. doi: 10.1128/jb.178.17.5138-5143.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grundström T, Normark S, Magnusson KE. 1980. Overproduction of outer membrane protein suppresses envA-induced hyperpermeability. J Bacteriol 144:884–890. doi: 10.1128/JB.144.3.884-890.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kloser A, Laird M, Deng M, Misra R. 1998. Modulation in lipid A and phospholipid biosynthesis pathways influence outer membrane protein assembly in Escherichia coli K-12. Mol Microbiol 27:1003–1008. doi: 10.1046/j.1365-2958.1998.00746.x. [DOI] [PubMed] [Google Scholar]
- 21.Sorensen PG, Lutkenhaus J, Young K, Eveland SS, Anderson MS, Raetz C. 1996. Regulation of UDP-3-O-[R-3-hydroxymyristoyl]-N-acetylglucosamine deacetylase in Escherichia coli. J Biol Chem 271:25898–25905. doi: 10.1074/jbc.271.42.25898. [DOI] [PubMed] [Google Scholar]
- 22.Hirvas L, Nurminen M, Helander IM, Vuorio R, Vaara M. 1997. The lipid A biosynthesis deficiency of the Escherichia coli antibiotic-supersensitive mutant LH530 is suppressed by a novel locus, ORF195. Microbiology 143:73–81. doi: 10.1099/00221287-143-1-73. [DOI] [PubMed] [Google Scholar]
- 23.De Lay NR, Cronan JE. 2008. Genetic interaction between the Escherichia coli AcpT phosphopantetheinyl transferase and the YejM inner membrane protein. Genetics 178:1327–1337. doi: 10.1534/genetics.107.081836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu N, Misra R. 2019. Overcoming iron deficiency of an Escherichiai coli tonB mutant by increasing outer membrane permeability. J Bacteriol 201:e00340-19. doi: 10.1128/JB.00340-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dalebroux ZD, Edrozo MB, Pfuetzner RA, Ress S, Kulasekara BR, Blanc M-P, Miller SI. 2015. Delivery of cardiolipins to the Salmonella outer membrane is necessary for survival within host tissues and virulence. Cell Host Microbe 17:441–451. doi: 10.1016/j.chom.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong H, Zhang Z, Tang X, Huang S, Li H, Peng B, Dong C. 2016. Structural insights into cardiolipin transfer from the inner membrane to the outer membrane by PbgA in Gram-negative bacteria. Sci Rep 6:30815. doi: 10.1038/srep30815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossi RM, Yum L, Agaisse H, Payne SM. 2017. Cardiolipin synthesis and outer membrane localization are required for Shigella flexneri virulence. mBio 8:e01199-17. doi: 10.1128/mBio.01199-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolaes V, El Hajjaji H, Davis RM, Van der Henst C, Depuydt M, Leverrier P, Aertsen A, Haufroid V, de Choudens SO, De Bolle X, Ruiz N, Collet J-F. 2014. Insights into the function of YciM, a heat shock membrane protein required to maintain envelope integrity in Escherichia coli. J Bacteriol 196:300–309. doi: 10.1128/JB.00921-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein G, Kobylak N, Lindner B, Stupak A, Raina S. 2014. Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J Biol Chem 289:14829–18453. doi: 10.1074/jbc.M113.539494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahalakshmi S, Sunayana MR, SaiSree L, Reddy M. 2014. yciM is an essential gene required for regulation of lipopolysaccharide synthesis in Escherichia coli. Mol Microbiol 91:145–157. doi: 10.1111/mmi.12452. [DOI] [PubMed] [Google Scholar]
- 31.Guest RL, Guerra DS, Wissler M, Grimm J, Silhavy TJ. 2020. YejM modulates activity of YciM/FtsH protease complex to prevent lethal accumulation of lipopolysaccharide. mBio 11:e00598-20. doi: 10.1128/mBio.00598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fivenson EM, Bernhardt TG. 2020. An essential membrane protein modulates the proteolysis of LpxC to control lipopolysaccharide synthesis in Escherichia coli. mBio 11:e00939-20. doi: 10.1128/mBio.00939-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kern B, Leiser OP, Misra R. 2019. Suppressor mutations in degS overcome the acute temperature-sensitive phenotype of ΔdegP and ΔdegP Δtol-pal mutants of Escherichia coli. J Bacteriol 201:e00742-18. doi: 10.1128/JB.00742-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomanek N, Arends J, Lindemann C, Barkovits K, Meyer HE, Marcus K, Narberhaus F. 2019. Intricate crosstalk between lipopolysaccharide, phospholipid, and fatty acid metabolism in Escherichia coli modulates proteolysis of LpxC. Front Microbiol 9:3285. doi: 10.3389/fmicb.2018.03285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prince C, Jia Z. 2015. An unexpected due: rubredoxin binds nine TPR motifs to form LapB, an essential regulator of lipopolysaccharide synthesis. Structure 23:1500–1506. doi: 10.1016/j.str.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 37.D’Andrea LD, Regan L. 2003. TPR proteins: the versatile helix. Trends Biochem Sci 28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Lovenberg W, Sobel BE. 1965. Rubredoxin: a new electron transfer protein from Clostridium pasteurianum. Proc Natl Acad Sci U S A 54:193–199. doi: 10.1073/pnas.54.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krzywda S, Brzozowski AM, Verma C, Karata K, Ogura T, Wilkinson AJ. 2002. The crystal structure of the AAA domain of the ATP-dependent protease FtsH of Escherichia coli at 1.5Å resolution. Structure 10:1073–1083. doi: 10.1016/S0969-2126(02)00806-7. [DOI] [PubMed] [Google Scholar]
- 40.Bieniossek C, Schalch T, Bumann M, Meister M, Meier R, Baumann U. 2006. The molecular architecture of the metalloprotease FtsH. Proc Natl Acad Sci U S A 103:3066–3071. doi: 10.1073/pnas.0600031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scharfenberg F, Serek-Heuberger J, Coles M, Hartmann MD, Habeck M, Martin J, Lupas AN, Alva V. 2015. Structure and evolution of N-domains in AAA metalloproteases. J Mol Biol 427:910–927. doi: 10.1016/j.jmb.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 42.Akiyama Y, Kihara A, Mori H, Ogura T, Ito K. 1998. Roles of the periplasmic domain of Escherichia coli FtsH (HflB) in protein interactions and activity modulation. J Biol Chem 273:22326–22333. doi: 10.1074/jbc.273.35.22326. [DOI] [PubMed] [Google Scholar]
- 43.Cian MB, Giordano NP, Masilamani R, Minor KE, Dalebroux ZD. 2019. Salmonella enterica serovar Typhimurium uses PbgA/YejM to regulate lipopolysaccharide assembly during bacteremia. Infect Immun 88:e00758-19. doi: 10.1128/IAI.00758-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Werner J, Misra R. 2005. YaeT affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol Microbiol 57:1450–1459. doi: 10.1111/j.1365-2958.2005.04775.x. [DOI] [PubMed] [Google Scholar]
- 45.Silhavy TJ, Berman ML, Enquist LW. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 46.Bennion D, Charlson ES, Coon E, Misra R. 2010. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol Microbiol 77:1153–1171. doi: 10.1111/j.1365-2958.2010.07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]