The outer membranes of Gram-negative bacteria provide a permeability barrier to antibiotics and other harmful chemicals. The integrity of this barrier relies on the maintenance of the lipid asymmetry of the outer membrane, and studies of suppressors of a decades-old mutant reveal that YejM plays a key regulatory role and provide a model for the maintenance of this asymmetry.
KEYWORDS: FtsH, LpxC, YciM, YejM, lipopolysaccharide, outer membrane
ABSTRACT
The outer membranes of Gram-negative bacteria provide a permeability barrier to antibiotics and other harmful chemicals. The integrity of this barrier relies on the maintenance of the lipid asymmetry of the outer membrane, and studies of suppressors of a decades-old mutant reveal that YejM plays a key regulatory role and provide a model for the maintenance of this asymmetry.
TEXT
One of the hallmarks of Gram-negative bacteria is their intrinsic resistance to antibiotics, especially large and hydrophobic drugs, due to the presence of an outer membrane (OM) (1). This membrane is unique in biology due to the asymmetry of the bilayer and the presence of lipopolysaccharide (LPS). Although the inner leaflet of the OM is rather typical and composed of phospholipids (PL), the outer leaflet contains primarily LPS, which is composed of lipid A, a core oligosaccharide, and an O-antigen polysaccharide (2). Maintaining this asymmetry is one of the keys to the barrier function and requires regulation of the flow of fatty acyl chains from a common precursor pool between two competing biosynthetic pathways that lead to the synthesis of the LPS and PL (Fig. 1A). Recent papers, one of which is in this issue of the journal (3), find a regulatory role for one of the last essential genes (yejM) in Escherichia coli that lacks a clear function (4, 5). The papers provide evidence that YejM senses an intermediate in these biosynthetic pathways to regulate the stability of LpxC, a key enzyme in the biosynthesis of LPS (6, 7), to balance the synthesis of PL with LPS and maintain OM asymmetry.
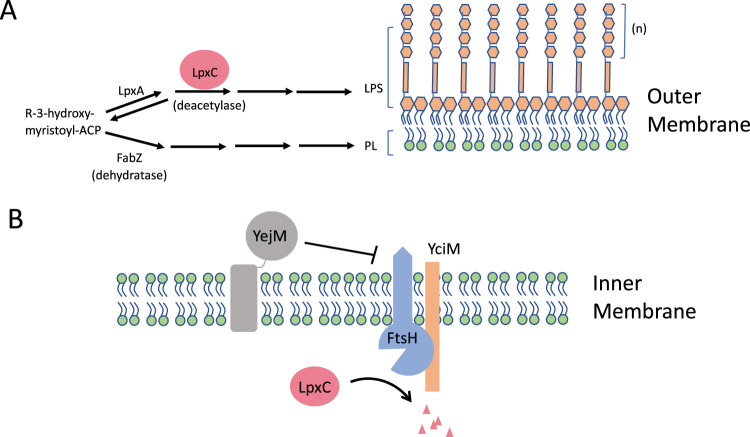
FIG 1.
Regulation of LPS biosynthesis. (A) The biosynthesis of LPS and PL depends upon a common precursor (R-3-hydroxy-myristoyl-acyl carrier protein [ACP]). The first step in LPS biosynthesis is carried out by LpxA and is reversible. The first committed step is carried out by LpxC, a deacetylase. The first committed step in PL biosynthesis is carried out by FabZ, a dehydratase. LPS is in the outer leaflet of the OM, and PL is found in the inner leaflet. (B) The level of LpxC depends upon the activity of the FtsH/YciM protease complex which is regulated by YejM, which may sense the level of an LPS intermediate.
The cell envelopes of Gram-negative bacteria consist of a thin cell wall sandwiched between the OM and the inner or cytoplasmic membrane (IM) in a compartment referred to as the periplasmic space (8). Another key to the barrier function of the OM is that it must be tethered to the cell wall, which occurs through noncovalent interaction between the peptidoglycan (PG) and several OM proteins. One of these is a lipoprotein (lpp product) that is covalently attached to the PG at its carboxy terminus and has its triacylated N terminus embedded in the OM (9). In the absence of this lipoprotein, the outer membrane blebs and the barrier is compromised.
Since the OM is a diffusion barrier, it contains channels (porins) that allows nutrients to enter the periplasmic space, where they encounter specific transport systems that mediate transport across the inner membrane and into the cytoplasm (10). Hydrophilic nutrients of less than ∼650 Da are able to diffuse through the well-characterized general porins, whereas larger hydrophilic compounds (various iron chelators or glucose polymers [maltodextrins]) require larger or specialized channels which are gated and energized by TonB (11). Since most antibiotics act in the periplasm or cytoplasm, they must pass through the OM to reach their target. Those that are small enough and hydrophilic can diffuse through the general porins, whereas large and hydrophobic antibiotics have difficulty permeating the OM. However, permeation of the OM is aided by the accumulation of PL in the outer leaflet of the OM. In fact, Gram-negative bacteria have two systems that decrease PL in the outer leaflet, a retrograde transport system (Mla) (12–14) and an enzyme (PldA) that degrades PL in the outer leaflet (12). Nonetheless, a reduction in LPS synthesis compromises the OM barrier through the introduction of PL in the outer leaflet and a reduction in the lateral interactions between LPS molecules. Conditions that compensate for this reduction in LPS synthesis by restoring the LPS/PL ratio reestablish the barrier.
LpxC is the main control point in LPS biosynthesis and, as a consequence, the integrity of the OM. As with other aspects of envelope biogenesis, the study of the OM barrier has been aided by the isolation of suppressors of a defect, an approach also employed in recent papers. This approach has been aided in recent years by the ease of doing whole-genome sequencing (WGS) to identify the affected targets; but first, some history. The envA1 mutation dates to the 1970s and was found as a mutation that confers increased sensitivity to antibiotics (15). Since envA was shown to be an essential gene, it was assumed that envA1 resulted in decreased activity of the gene (16). Indeed, Raetz’s group working on the LPS biosynthetic pathway found that envA1 had decreased deacetylase activity leading to a reduction in the amount of LPS (6). Moreover, this enzyme was found to carry out the first committed step in LPS biosynthesis, and so envA was renamed lpxC (6), in line with other genes in the pathway.
A clue about the role of lpxC came when a suppressor of the antibiotic sensitivity conferred by envA1 was located in fabZ, which encodes a dehydratase that carries out the first committed step in PL biosynthesis (Fig. 1A). This finding genetically linked PL and LPS biosynthesis (17, 18) and suggested that a partial loss of function mutation in fabZ (reduced enzyme activity) rebalances the flux through the pathways. Subsequently, a decrease in LpxC activity was shown to reduce LPS biosynthesis and increase the amount of PL. This decrease in LPS biosynthesis also led to an increase in the level of LpxC, suggesting a feedback loop that senses some intermediate in the LPS pathway to affect the level of LpxC to balance the two pathways (7). This link was further strengthened when it was found that loss of FtsH, a well-known protease (19), was suppressed by a mutation in fabZ; but in this case, the mutation enhanced the enzyme activity, thus compensating for an increase in LpxC to balance flux between the two pathways (20). FtsH was subsequently shown to degrade LpxC with the aid of the essential adaptor YciM (also called LapB) (21, 22). Consequently, deletion of either ftsH or yciM increases the level of the deacetylase (LpxC) and in turn increases LPS causing toxicity and cell death. Thus, ftsH and yciM are both essential genes, because their absence leads to toxic levels of LPS. However, these genes can be bypassed if there is a compensating mutation (such as in fabZ [18] or lpxC [19]) that rebalances the flux between these two pathways. Although FtsH is known to degrade other substrates, it is this role in regulating LPS biosynthesis that makes it essential (19).
Enter YejM. yejM is an essential gene that encodes an IM protein with 5 transmembrane helices and a large C-terminal periplasmic domain (23). The original yejM mutation was found among isolates with increased sensitivity to hydrophobic antibiotics (24). Although yejM is essential, the original mutation truncated the periplasmic domain, indicating this domain was not essential, but in its absence, the OM barrier was not maintained due to decreased LPS (23). Similar mutations were also found in yejM that allowed iron-chelating enterobactin to enter mutant cells defective in its transport across the OM (25). Such mutations also caused the typical array of phenotypes associated with a defective OM barrier, including sensitivity to vancomycin and detergents, temperature sensitivity, and leakage of periplasmic enzymes. This suggested that damage to the OM barrier allowed enterobactin to gain access to the periplasmic space, where it would encounter the IM uptake system. To understand how YejM was affecting the OM barrier, the various groups isolated suppressors that restored the barrier, identified the genes involved using whole-genome sequencing (WGS), and then tried to determine the molecular basis of the suppression. As a result, all three groups propose a similar model to account for YejM’s role in maintaining the OM barrier function (Fig. 1B).
In the paper in this issue of the journal, the Misra lab isolated 6 suppressors of YejM truncated for its periplasmic domain that restored the typical resistance of E. coli to large antibiotics, namely, vancomycin and erythromycin (3). Whole-genome sequencing (WGS) revealed that the suppressors mapped to the two components of the protease, FtsH and YciM, involved in LpxC degradation. A straightforward hypothesis was that activity of the protease was reduced and LpxC stabilized. Indeed, LpxC was not detectable in the YejM mutant, but its level was restored to various degrees in the suppressors, which correlated with the strength of the suppressor. How do the suppressors work? The three in YciM result in the instability of YciM, although it is not clear why, but loss of the adaptor explains the stabilization of LpxC. The three mutations in ftsH affect fairly conserved residues and presumably reduce its activity, but the precise mechanism remains to be determined. This leads to a model where YejM senses some LPS precursor to inhibit FtsH/YciM proteolytic activity (Fig. 1B).
YejM showed up in a screen that the Bernhardt lab was doing, and since it was an essential gene of unknown function, they explored it further (4). Depletion of YejM resulted in cell chaining and lysis similar to what is observed when cells are treated with an inhibitor of LPS biosynthesis. Moreover, YejM depletion was rescued by overexpression of LpxC, consistent with YejM being needed to stabilize LpxC. In fact, YejM was able to be deleted in a strain in which LpxC was overexpressed. To better understand the role of YejM, the lab also turned to suppressor analysis. Twelve suppressors of the sensitivity of a YejM depletion to SDS (sensitivity to detergents is another phenotype of mutants with an altered OM) were obtained. WGS revealed quite a variety of mutations, most of which are consistent with enhancing the level of LpxC, such as increased gene dosage of lpxC and missense mutations in yciM. In addition, a novel POLAR-recruitment 2-hybrid assay suggested an interaction between YejM and YciM.
The Silhavy lab obtained suppressors of a truncated YejM mutant by selecting for growth at 42°C in the presence of SDS and EDTA (EDTA weakens the OM barrier by chelating Mg2+, which helps to stabilize lateral interactions between LPS molecules) (5). WGS revealed that the suppressor mutations were in yciM and lpxC. One in lpxC was a frameshift in the stop codon leading to a C-terminal extension known to stabilize LpxC as it is no longer a substrate for FtsH/YciM (26). This suggests that the YejM mutant has lower LPS, which is corrected by the suppressors. Measuring total LPS, in fact, confirmed that this is the case. This left one thing to clarify: how does deletion of lpp (lipoprotein gene) suppress the toxicity of excess LPS caused by loss of the otherwise essential yciM gene (21, 27)? Lipoprotein is perhaps the most abundant protein in E. coli, and it was thought that deletion of lpp preserved acyl chains that could then be used to make PL, helping to restore the PL/LPS ratio. However, a mutant Lpp that is acylated, but cannot be attached to the PG, still suppressed the YejM mutant, ruling out that hypothesis. Instead, the loss of lpp increases vesiculation that sheds excess LPS, reducing its toxicity.
The work in E. coli offers a possible explanation for a finding in Salmonella enterica serovar Typhimurium. In this organism, YejM is called PbgA and was identified as a protein required for the OM barrier function during infection, where the LPS is modified and the phospholipid (cardiolipin) content of the OM is increased (28). PbgA bound cardiolipin, and it was proposed that PbgA shuttled cardiolipin to the OM. The structure of the periplasmic domain revealed striking similarities to lipoteichoic acid synthase (LtaS) but lacked a functional active site, although it had a hydrophobic cleft that could potentially bind LPS precursors (29). More recently, the structure of full-length PbgA was obtained, revealing a cleft within the membrane domain that contained conserved residues suggesting an active site (30). The structure also had two bound cardiolipin molecules consistent with PbgA being involved with cardiolipin. However, cardiolipin is not essential in E. coli, and so it is unlikely that this plays a role in the barrier function (31).
Mice infected with PbgA mutants truncated for the periplasmic domain survive, whereas those infected with the wild type (WT) succumb to the infection (32). Interestingly, survivors were found among organ homogenates of mice infected with the PbgA mutant that restored the LPS defect (based upon a reporter for membrane integrity). In this earlier study, WGS, like in the above-described studies with E. coli, revealed that the survivors contained mutations in yciM, ftsH, and lpxC and suppressed various envelope phenotypes to various degrees. Although various explanations were put forward, the recent studies in E. coli suggest how the suppressor mutations reestablish the balance between PL and LPS biosynthesis in the mutant and thus restore the asymmetry of the OM and reestablish its barrier function (Fig. 1B).
While these recent studies have connected YejM to LPS synthesis by regulating LpxC levels, it remains to be determined how YejM senses the LPS assembly status and communicates with the FtsH/YciM proteolytic machinery to regulate LpxC levels. Also, the more severe phenotype caused by complete loss of YejM compared to the truncated version remains to be explained. Moreover, it is possible that this regulation can be exploited to find compounds that disrupt the OM barrier to overcome the intrinsic resistance to antibiotics. With several labs converging on YejM, the answers may come soon.
The views expressed in this article do not necessarily reflect the views of the journal or of ASM.
Footnotes
For the article discussed, see https://doi.org/10.1128/JB.00303-20.
REFERENCES
- 1.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitfield C, Trent MS. 2014. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem 83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen D, Kelly K, Qiu N, Misra R. 2020. YejM controls LpxC levels by regulating protease activity of the FtsH/YciM complex of Escherichia coli. J Bacteriol 202:e00303-20. doi: 10.1128/JB.00303-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fivenson EM, Bernhardt TG. 2020. An essential membrane protein modulates the proteolysis of LpxC to control lipopolysaccharide synthesis in Escherichia coli. mBio 11:e00939-20. doi: 10.1128/mBio.00939-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guest RL, Same Guerra D, Wissler M, Grimm J, Silhavy TJ. 2020. YejM modulates activity of the YciM/FtsH protease complex to prevent lethal accumulation of lipopolysaccharide. mBio 11:e00598-20. doi: 10.1128/mBio.00598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young K, Silver LL, Bramhill D, Cameron P, Eveland SS, Raetz CR, Hyland SA, Anderson MS. 1995. The envA permeability/cell division gene of Escherichia coli encodes the second enzyme of lipid A biosynthesis. UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase. J Biol Chem 270:30384–30391. doi: 10.1074/jbc.270.51.30384. [DOI] [PubMed] [Google Scholar]
- 7.Sorensen PG, Lutkenhaus J, Young K, Eveland SS, Anderson MS, Raetz CR. 1996. Regulation of UDP-3-O-[R-3-hydroxymyristoyl]-N-acetylglucosamine deacetylase in Escherichia coli. The second enzymatic step of lipid a biosynthesis. J Biol Chem 271:25898–25905. doi: 10.1074/jbc.271.42.25898. [DOI] [PubMed] [Google Scholar]
- 8.Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asmar AT, Collet JF. 2018. Lpp, the Braun lipoprotein, turns 50-major achievements and remaining issues. FEMS Microbiol Lett 365:fny199. doi: 10.1093/femsle/fny199. [DOI] [PubMed] [Google Scholar]
- 10.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/mmbr.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Postle K, Kadner RJ. 2003. Touch and go: tying TonB to transport. Mol Microbiol 49:869–882. doi: 10.1046/j.1365-2958.2003.03629.x. [DOI] [PubMed] [Google Scholar]
- 12.Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc Natl Acad Sci U S A 106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abellón-Ruiz J, Kaptan SS, Baslé A, Claudi B, Bumann D, Kleinekathöfer U, van den Berg B. 2017. Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat Microbiol 2:1616–1623. doi: 10.1038/s41564-017-0046-x. [DOI] [PubMed] [Google Scholar]
- 14.Thong S, Ercan B, Torta F, Fong ZY, Wong HY, Wenk MR, Chng SS. 2016. Defining key roles for auxiliary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry. eLife 5:e19042. doi: 10.7554/eLife.19042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Normark S. 1970. Genetics of a chain-forming mutant of Escherichia coli. Transduction and dominance of the envA gene mediating increased penetration to some antibacterial agents. Genet Res 16:63–78. doi: 10.1017/s0016672300002287. [DOI] [PubMed] [Google Scholar]
- 16.Beall B, Lutkenhaus J. 1987. Sequence analysis, transcriptional organization, and insertional mutagenesis of the envA gene of Escherichia coli. J Bacteriol 169:5408–5415. doi: 10.1128/jb.169.12.5408-5415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kloser AW, Laird MW, Misra R. 1996. asmB, a suppressor locus for assembly-defective OmpF mutants of Escherichia coli, is allelic to envA (lpxC). J Bacteriol 178:5138–5143. doi: 10.1128/jb.178.17.5138-5143.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloser A, Laird M, Deng M, Misra R. 1998. Modulations in lipid A and phospholipid biosynthesis pathways influence outer membrane protein assembly in Escherichia coli K-12. Mol Microbiol 27:1003–1008. doi: 10.1046/j.1365-2958.1998.00746.x. [DOI] [PubMed] [Google Scholar]
- 19.Bittner LM, Arends J, Narberhaus F. 2017. When, how and why? Regulated proteolysis by the essential FtsH protease in Escherichia coli. Biol Chem 398:625–635. doi: 10.1515/hsz-2016-0302. [DOI] [PubMed] [Google Scholar]
- 20.Ogura T, Inoue K, Tatsuta T, Suzaki T, Karata K, Young K, Su LH, Fierke CA, Jackman JE, Raetz CR, Coleman J, Tomoyasu T, Matsuzawa H. 1999. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol Microbiol 31:833–844. doi: 10.1046/j.1365-2958.1999.01221.x. [DOI] [PubMed] [Google Scholar]
- 21.Mahalakshmi S, Sunayana MR, SaiSree L, Reddy M. 2014. yciM is an essential gene required for regulation of lipopolysaccharide synthesis in Escherichia coli. Mol Microbiol 91:145–157. doi: 10.1111/mmi.12452. [DOI] [PubMed] [Google Scholar]
- 22.Nicolaes V, El Hajjaji H, Davis RM, Van der Henst C, Depuydt M, Leverrier P, Aertsen A, Haufroid V, Ollagnier de Choudens S, De Bolle X, Ruiz N, Collet JF. 2014. Insights into the function of YciM, a heat shock membrane protein required to maintain envelope integrity in Escherichia coli. J Bacteriol 196:300–309. doi: 10.1128/JB.00921-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Lay NR, Cronan JE. 2008. Genetic interaction between the Escherichia coli AcpT phosphopantetheinyl transferase and the YejM inner membrane protein. Genetics 178:1327–1337. doi: 10.1534/genetics.107.081836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirvas L, Nurminen M, Helander IM, Vuorio R, Vaara M. 1997. The lipid A biosynthesis deficiency of the Escherichia coli antibiotic-supersensitive mutant LH530 is suppressed by a novel locus, ORF195. Microbiology 143:73–81. doi: 10.1099/00221287-143-1-73. [DOI] [PubMed] [Google Scholar]
- 25.Qiu N, Misra R. 2019. Overcoming iron deficiency of an Escherichia coli tonB mutant by increasing outer membrane permeability. J Bacteriol 201:e00340-19. doi: 10.1128/JB.00340-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuhrer F, Langklotz S, Narberhaus F. 2006. The C-terminal end of LpxC is required for degradation by the FtsH protease. Mol Microbiol 59:1025–1036. doi: 10.1111/j.1365-2958.2005.04994.x. [DOI] [PubMed] [Google Scholar]
- 27.Klein G, Kobylak N, Lindner B, Stupak A, Raina S. 2014. Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J Biol Chem 289:14829–14853. doi: 10.1074/jbc.M113.539494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalebroux ZD, Edrozo MB, Pfuetzner RA, Ressl S, Kulasekara BR, Blanc MP, Miller SI. 2015. Delivery of cardiolipins to the Salmonella outer membrane is necessary for survival within host tissues and virulence. Cell Host Microbe 17:441–451. doi: 10.1016/j.chom.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong H, Zhang Z, Tang X, Huang S, Li H, Peng B, Dong C. 2016. Structural insights into cardiolipin transfer from the Inner membrane to the outer membrane by PbgA in Gram-negative bacteria. Sci Rep 6:30815. doi: 10.1038/srep30815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan J, Petersen EM, Hinds TR, Zheng N, Miller SI. 2020. Structure of an inner membrane protein required for PhoPQ-regulated increases in outer membrane cardiolipin. mBio 11:e03277-19. doi: 10.1128/mBio.03277-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishijima S, Asami Y, Uetake N, Yamagoe S, Ohta A, Shibuya I. 1988. Disruption of the Escherichia coli cls gene responsible for cardiolipin synthesis. J Bacteriol 170:775–780. doi: 10.1128/jb.170.2.775-780.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cian MB, Giordano NP, Masilamani R, Minor KE, Dalebroux ZD. 2019. Salmonella enterica serovar Typhimurium uses PbgA/YejM to regulate lipopolysaccharide assembly during bacteremia. Infect Immun 88:e00758-19. doi: 10.1128/IAI.00758-19. [DOI] [PMC free article] [PubMed] [Google Scholar]